Abstract
During hippocampal neuron differentiation, the expression of critical inducers of non-neuronal cell lineages must be efficiently silenced. Runx2 transcription factor is the master regulator of mesenchymal cells responsible for intramembranous osteoblast differentiation and formation of the craniofacial bone tissue that surrounds and protects the central nervous system (CNS) in mammalian embryos. The molecular mechanisms that mediate silencing of the Runx2 gene and its downstream target osteogenic-related genes in neuronal cells have not been explored. Here, we assess the epigenetic mechanisms that mediate silencing of osteoblast-specific genes in CNS neurons. In particular, we address the contribution of histone epigenetic marks and histone modifiers on the silencing of the Runx2/p57 bone-related isoform in rat hippocampal tissues at embryonic to adult stages. Our results indicate enrichment of repressive chromatin histone marks and of the Polycomb PRC2 complex at the Runx2/p57 promoter region. Knockdown of PRC2 H3K27-methyltransferases Ezh2 and Ezh1, or forced expression of the Trithorax/COMPASS subunit Wdr5 activates Runx2/p57 mRNA expression in both immature and mature hippocampal cells. Together these results indicate that complementary epigenetic mechanisms progressively and efficiently silence critical osteoblastic genes during hippocampal neuron differentiation.
Keywords: Runx2, Epigenetic regulation of gene expression, Hippocampus
Introduction
Craniofacial bones and the brain ultimately originate from the dorsal neural tube which arises during early stages of fetal development. Formation and maturation of the mammalian head during late gestation involves intricate interactions between neural crest cells that generate facial primordia (e.g., jaw and cheek bones) and the developing brain (1-3). Perturbations in the cellular and molecular pathways that control development of osseous and neural tissues in the developing head are observed in a number of congenital disorders (2). During craniofacial development, neural crest cells are initially indistinguishable from other neuro-epithelial cells, but then they undergo an epithelial-to-mesenchymal transition and migrate to form future craniofacial structures (4). The signaling molecules controlling neural crest formation are the same as those regulating normal osteogenesis and include bone morphogenetic proteins (BMP), fibroblast growth factors (FGF), and Wnt signaling factors. The fundamental gene regulatory mechanisms by which cells maintain a neural cell fate while silencing programs required for generating mesenchymal phenotypes (e.g., in cells with osteogenic lineage potential) remain a major hiatus in our understanding of head development.
Osteoblast lineage commitment is promoted and regulated by a coordinated set of extracellular stimuli and developmentally-regulated signaling pathways (5-8). Following activation in osteoprogenitor cells, these signaling pathways modulate the expression and function of osteoblast master transcription factors. The latter control the expression of downstream bone-phenotypic genes and attenuating microRNAs that together establish the osteoblastic cell component of the mammalian skeleton (9, 10). Commitment of precursor cells to the osteoblast lineage mainly requires the function of the master transcriptional regulator Runx2 (Runt-related transcription factor 2) (11, 12), which in turn controls the expression of numerous target genes (like osteocalcin/Bglap) encoding regulatory proteins that re-enforce the osteoblast phenotype and structural proteins that support formation of a mineralized extracellular matrix (13-19).
During development Runx2/p57 expression is initiated at days 8.5-9.5 post-coitum (p.c.) at the notochord and caudal somite tissues that then lead to formation of the vertebral cord (15, 20, 21). Later, Runx2/p57 is found at the mesenchymal tissues that generate the axial skeleton, ribs and long bones. Osteoblasts in many of the craniofacial bones descend from neural crest cells. The Runx2/p57 gene is primed for transcriptional activation during the epithelial-mesenchymal transition that generates the progenitor cells required for the formation of skull bones (22). Once Runx2 reaches threshold concentrations, it initiates the expression of a plethora of downstream osteogenic target genes that maintain and execute the phenotypic functions of mature osteoblasts (23-25). In contrast, other neural ectoderm progenitors engage differentiation towards multiple neuronal and non-neuronal cell types of the CNS. In these cells, the osteogenic genetic program remains silent throughout post-natal development. The epigenetic mechanisms mediating transcriptional silencing of osteogenic genes in the CNS have not been explored.
Polycomb Group (PcG) proteins are particularly relevant in this context as they are critical epigenetic controllers of gene expression during mammalian development by regulating target genes that are involved in specification of multiple cell lineages (26, 27) and neuronal functions (28). The Polycomb Repressive Complex 2 (PRC2) contains several protein subunits, including Its core components Enhancer of Zeste Homolog (Ezh), Embryonic Ectoderm Development (Eed) and Suppressor of Zeste 12 (Suz12) (29). Mammalian PRC2 complexes can contain either Ezh1 or Ezh2, which function as the catalytic subunits mediating tri-methylation of histone H3 lysine residue 27 (H3K27me3), a modification that is associated with gene repression (30, 31). We have previously reported that during hippocampal development the two PRC2 catalytic subunits Ezh1 and Ezh2 are differentially expressed in post-mitotic mammalian hippocampal neurons (28). Moreover, during maturation of these hippocampal cells both Ezh1 and Ezh2 can have opposing effects on the transcriptional status of specific genes. For example, Ezh1 is recruited to the PSD95/Dlg4 promoter to up-regulate its transcription in mature neurons, but Ezh2 binds to this promoter sequence during early stages of hippocampal development, where it mediates PSD-95 gene repression (28).
While the repressive H3K27me3 epigenetic mark is generated in neuronal tissue by the PRC2 complex, activation of gene expression is associated with tri-methylation of lysine 4 in histone H3 (H3K4me3) (32, 33). The latter modification is mediated by the mammalian Set1 (1A and 1B)- and Mixed Lineage Leukemia (MLL1, 2, 3, 4 and 5)-containing complexes that are collectively referred to as COMPASS (for Complex of Proteins Associated with Set1) and COMPASS-like complexes, respectively (34). COMPASS and COMPASS-like complexes contain the Wdr5 (WD Repeat Domain 5) protein subunit which is required for assembly and stability of these complexes, as well as for full methyltransferase activity (35, 36). In addition, MLL3 and MLL4 complexes are particularly enriched in the H3K27 demethylase UTX (KDM6A), a Jumonji C domain-containing enzyme (37, 38). Because transcriptional repression is associated with reduced H3K4me3 and enhanced H3K27me3 epigenetic marks, we postulate that transcriptional silencing of osteogenic genes in neuronal cells may involve repressive mechanisms that favor recruitment of PRC2 complexes to bone-related genes.
Wdr5 is expressed in immortalized marrow stromal cells, osteoblasts, osteocytes, and chondrocytes both in culture and in vivo. Over-expression of Wdr5 has been shown to accelerate the osteoblast and chondrocyte differentiation programs both in vivo and in cell culture models (39, 40) in part by activating both the canonical Wnt- and the BMP2-signaling pathways during skeletal development (41, 42). Moreover, Wdr5 has been shown to bind to the Runx2 P1 promoter in osteoblastic cells (41, 43) where it can mediate the Runx2 transcriptional up-regulation induced by Wnt signaling. In addition, H3K27me3 can be demethylated by JmjD3 (KDM6B), a Jumonji domain-containing enzyme that interacts with different protein complexes compared to UTX (KDM6A), including transcriptional regulators like p53 (38, 44). The latter protein controls RUNX2 expression by a microRNA-mediated mechanism and is important for bone formation, as well as normal growth and differentiation of osteoblasts (45, 46).
Beyond histone lysine methylation, PRMT5-mediated symmetric di-methylation of arginine 3 in histone H4 (H4R3Me2) also may mediate transcriptional repression (47-49). The ability of PRMT5 to repress transcription has been mainly attributed to its interaction with DNA methyltransferases like Dnmt3A (50) and may perhaps be biologically linked to suppression of transposable elements in pre-implantation embryos (48). Therefore, we hypothesize that the epigenetic interplay between H3K27, H3K4 and H4R3 methyltransferases, as well as H3K27 demethylases may collectively or independently determine repression versus activation of osteogenic lineage-determining factors in neural cells and tissues.
In this work, we examined the mechanisms that repress osteoblast-associated genes during rat hippocampal maturation. We find that the Polycomb PRC2 complex and its associated epigenetic mark H3K27me3 are enriched at the Runx2-P1 promoter region from embryo to adult. We also find presence of H3K9me3 at the Runx2-P1 promoter from post-natal to adult stages, while H4R3me2s is only found enriched at Runx2-P1 in adult animals. Moreover, we demonstrate that silencing of Polycomb H3K27-methyltransferases Ezh2 and Ezh1, as well as forced expression of the Trithorax/COMPASS subunit Wdr5 in hippocampal cells, activates Runx2/p57 mRNA expression. This Wdr5-dependent derepression is accompanied by a significant reduction in PRC2-Ezh2 binding to the P1 promoter and the H3K27me3 mark. Our data are consistent with a model in which osteogenic gene silencing in the hippocampus requires the concerted epigenetic interplay of PRC2 and COMPASS.
Materials and methods
Rat hippocampus isolation and primary hippocampal cell culture
All protocols involving rats were in accordance with NIH guidelines and were approved by the Bioethical and Biosafety Committees of Andres Bello University, Santiago, Chile. Sprague–Dawley rats were deeply anesthetized with CO2 and hippocampi were removed using forceps from 18 days-embryos (E18), 10-days postnatal animals (P10), P30 or adults (P>90). Hippocampal cell cultures were prepared from E18 pups as described previously (28). As shown by several laboratories, including ours (28), these primary hippocampal cultures contain mainly post-mitotic neurons as the proliferation of the astrocytes is inhibited by treatment with 2 μM cytosine arabinoside (AraC). The hippocampus was removed bilaterally, dissected free of meninges and placed into Phosphate buffered saline buffer (PBS) containing 50 μg/mL penicillin/streptomycin (Life Technologies, CA, USA). The tissue was minced, incubated with 0.25% (w/v) trypsin (Life Technologies) for 20 min at 37 °C, and the cells were transferred to a new 15 mL tube containing 8 mL plating medium (Dulbecco's modified Eagle's medium (DMEM, Life Technologies), supplemented with 10% horse serum (Life Technologies), 100 U/mL penicillin, and 100 μg/mL streptomycin). Cells were re-suspended by mechanical agitation through fire-polished glass Pasteur pipettes of decreasing diameters and were seeded in plating medium onto 30-70 kDa poly-L-lysine-coated (Sigma, MO, USA) six-well culture plates at a density of 8 × 105 cells per well. Cultures were maintained at 37°C in 5% CO2 for 4 h before replacing the plating medium with Neurobasal growth medium (Life Technologies) supplemented with B27 (Life Technologies), 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. On day 1, cultured neurons were treated with 2 μM AraC (Sigma) for 24 h. Half of the medium was replaced every 3 days.
Primary rat osteoblasts culture
Rat osteoblasts were isolated from E20 embryos according to the procedure described by Owen et al. (51): calvarias were obtained and suture lines were removed. Successive digestions using 2.5 mg/mL trypsin (Life Technologies) and 2 mg/mL Collagenase P (Roche, Germany) in PBS 1× were performed in a 37°C-water bath as follows: first, 8 min without agitation; second, 10 min at 25 rpm, and finally, 35 min at 60 rpm. Supernatant containing released cells from bone tissue was filtered through a metallic-13 mm and a polypropylene-25 mm Swinnex device (Merck Millipore, Germany). Cells were recovered, centrifuged at 500g for 5 min and re-suspended in αMEM medium (Life Technologies) supplemented with 10% FBS. 6×105 cells were seeded on each 35 mm plate.
ROS17/2.8 and HEK293FT cells culture
Rat osteosarcoma-derived ROS 17/2.8 cells (52) were maintained in F-12 medium (Life Technologies) supplemented with 5% FBS, 1.176 g/liter NaHCO3, 0.118 g/liter CaCl2×2H2O, 6.670 g/liter HEPES (Life Technologies), and Penicillin/Streptomycin. Human embryonic kidney-derived HEK293FT cells were grown in DMEM medium (Life Technologies) supplemented with 10% FBS, 3.7 g/liter NaHCO3, 500 μg/mL G418, and Penicillin/Streptomycin.
Transient transfections
Cells were transfected at 3 days of culture in vitro (DIV3) with a modified CaPO4 transfection protocol to reduce cell toxicity (53, 54). For Wdr5 over expression, protein coding sequence was cloned from mouse-derived total RNA using specific primers carrying restriction sites for XbaI and EcoRI. Primer sequences were Fw: GGC TTC TCT AGA ATG GCC ACA GAG GA and Rev: TGG AGC GAA TTC TTA GCA GTC ACT CT. The amplified sequence was inserted downstream of the citomegalovirus (CMV) promoter in a pCDH vector that also codes for the copGFP under control of the EF1 promoter (cat. no. CD511B, System Biosciences, CA, EEUU).
Lentivirus production
Lentivirus for chromatin modifiers silencing were produced in HEK293FT cells as described before (43). For over expression experiments, pSL3, pSL4, pSL5 and pCDH plasmids were used in a 1:2:2:3 ratio. pLKO.1 and pCDH (empty vectors) were used as controls. After 16–18 h, the culture medium was replaced with fresh Optimem medium (Life Technologies) and cells were maintained at 32 °C for 48 h. Supernatants containing pseudo-typed lentiviral particles were collected, filtered through a 0.45μm-pore size PVDF filter and concentrated using an 100 kDa-MWCO Amicon Ultra-15 filter device (Merck Millipore). Aliquots were immediately stored at −80 °C. Primary hippocampal cells were plated in 6-well culture plates and infected for 96 h with 5-20 μL lentiviral particles containing shRNA-Ezh2 (TRCN0000040075/39040), shRNA-Ezh1 (TRCN0000095697/95), shRNA-PRMT5 (TRCN0000182229), or empty vector plasmids. All short hairpin-containing plasmids were purchased from Open Biosystems (GE Healthcare Dharmacon, UK)
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed using hippocampal tissue as described earlier (55, 56), with the following modifications: isolated hippocampal tissue from E18, P10, P30 or P>90 rats was finely minced with scissors and cross-linked with 1% PFA in PBS (bioWORLD, OH, EEUU) and incubated at room temperature for 30 min with gentle agitation. The reaction was stopped by adding 0.125 M glycine. The tissue was washed three times with cold PBS. For ChIP against chromatin-modifying enzymes, we used double cross-linking with EGS (Ethylene glycol-bis(succinic acid N-hydroxysuccinimide ester), E3257, Sigma) (57): after PFA and glycine, cells were washed three times, incubated with 2mM EGS in PBS1× for 1 h at room temperature with gentle agitation and washed three times with cold PBS. Cross-linked cells were re-suspended in 1 mL of cell lysis buffer (CLB; 5 mM Hepes, pH 8.0, 85 mM KCl, Triton X-100 and proteinase inhibitors), and homogenized with a Dounce homogenizer ∼150 times using a tight pestle. The tissue extract was collected by centrifugation at 3000 × g for 5 min, and was re-suspended in 1 mL of CLB. Remaining cell clumps were disaggregated mechanically by passing 5 times through a syringe with a 25G needle. To get rid of RNA, 10 μg of DNase-free RNase were added and incubated on ice for 1 hour. Extracts were centrifuged at 3000 × g for 5 min at 4 °C, the supernatant was discarded and the pelleted nuclei were re-suspended in 600 μL sonication buffer (50 mM Hepes, pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate acid, 0.1% SDS, and a mixture of proteinase inhibitors). Chromatin was sheared in a water bath sonicator Bioruptor (Diagenode, NJ, USA) to obtain fragments of 200-500 bp. Extracts were sonicated at high power for four to six pulses of 5 min each, 30 sec on, 30 sec off, and centrifuged at 16,000 × g for 15 min at 4 °C. Supernatant was collected, aliquoted, frozen in liquid nitrogen, and stored at − 80 °C; one aliquot was used for A260 measurements to determine concentration and chromatin size was confirmed by electrophoretic analysis. Cross-linked extracts (500 A260 units) were re-suspended in sonication buffer to a final volume of 500 μL. Samples were pre-cleared by incubating with 2–4 μg of normal IgG and 50 μL of protein A/G-agarose beads (Santa Cruz Biotechnology, CA, USA) for 1-2 h at 4 °C with agitation. Chromatin was centrifuged at 4000 × g for 5 min, the supernatant was collected and then immunoprecipitated with specific antibodies (see list below for antibodies used) for 12–16 h at 4 °C. The immunocomplexes were recovered with addition of 50 μL of protein A or G-agarose beads, followed by incubation for 1 h at 4 °C with gentle agitation. Immunoprecipitated complexes were washed once with sonication buffer, twice with LiCl buffer (100 mM Tris–HCl, pH 8.0, 500 mM LiCl, 0.1% Nonidet P40, and 0.1% deoxycholic acid), and once with Tris–EDTA (TE) buffer pH 8.0 (2 mM EDTA and 50 mM Tris–HCl, pH 8.0), each time for 5 min at 4 °C; this was followed by centrifugation at 4000 × g for 5 min. Protein–DNA complexes were eluted by incubation with 100 μL of elution buffer (50 mM NaHCO3 and 1% SDS) for 15 min at 65 °C. Extracts were centrifuged at 16,000 × g for 30 sec, and the supernatant was collected and incubated for 12–16 h at 65 °C, to revert the cross-linking. Proteins were digested with 25 μg of proteinase K (Merck Millipore) for 2 h at 50 °C, and the DNA was recovered by phenol/chloroform extraction and ethanol precipitation using glycogen (20 μg/mL) as a precipitation carrier. The qPCR primers used to quantify the immunoprecipitated DNA were: rat Runx2/p57 promoter region: Fw -118 (GTG GTA GGC AGT CCC ACT TT) and Rev +29 (TGT TTG TGA GGC GAA TGA AG), and rat PSD95 promoter region: Fw −33 (GGA GGG GTG AGA ACC CAC CGA) and Rev +118 (CTC CCC CTC CCC ACT GCT CC). The following antibodies were used for ChIP assays: polyclonal anti histone H3 – total (ab1791, Abcam, MA, USA), polyclonal anti H3K27me3 (07-449, Merck Millipore), polyclonal anti H3K27ac (ab4729, Abcam), polyclonal anti H4R3me2-symmetric (ab5823, Abcam), monoclonal anti H3K4me3 (ab1012, Abcam), polyclonal anti H3K4me1 (ab8895, Abcam), polyclonal anti H3K9me3 (ab8898, Abcam), monoclonal anti HP1α (05-689, Merck Millipore), polyclonal anti Ezh2 (07-689, Merck Millipore), polyclonal anti Ezh1 (ab13665, Abcam), monoclonal anti JBP1/PRMT5 (611539, BD Transduction Laboratories, CA, USA), monoclonal anti Wdr5 (ab56919, Abcam), monoclonal anti Suv39H1 (ab12405, Abcam), polyclonal anti UTX (ab36938, Abcam), polyclonal anti JMJD3 (ab38113, Abcam), normal rabbit IgG (12-370, Merck Millipore), normal mouse IgG (12-371, Merck Millipore).
Western blotting
Nuclear extracts were prepared from hippocampal cells using a buffer that contains 420 mM NaCl, 25% glycerol, 0.2 mM EDTA, 1 mM DTT, 20 mM HEPES pH 7.9, 1.5 mM MgCl2, and protease inhibitors as reported previously (28, 58). Proteins were detected by western blot using specific antibodies: monoclonal Ezh2 (AC22, Cell Signaling), polyclonal Ezh1 (ab13665, Abcam), monoclonal JBP1/PRMT5 (611539, BD Transduction Laboratories), monoclonal Wdr5 (ab56919, Abcam), polyclonal RNA Pol II (sc-899, Santa Cruz Biotechnology), polyclonal TFIIB (sc-225, Santa Cruz Biotechnology).
Reverse transcriptase, PCR and quantitative real-time PCR (qPCR)
Total RNA was extracted with TRIzol (Life Technologies), according to the manufacturer's protocol. Equal amounts of each sample (1-2 μg) were used for reverse transcription. qPCR was performed using Brilliant II SYBR® Green QPCR Master Mix (Agilent Technologies, Santa Clara CA, USA) in a Stratagene Mx3000P thermal cycler Agilent tech. All primer concentrations in qPCR reactions were previously standardized for 100% efficiency. Primer pairs used were: Total Runx2 Fw: CGG GAA TGA TGA GAA CTA CTC, Rev: GTG AAG ACC GTT ATG GTC AA; Runx2/p57 Fw: TCT GGA AAA AAA AGG AGG GAC TAT G, Rev: GGT GCT CGG ATC CCA AAA GAA; Sp7/Osterix: CTC TGG CTA TGC CAA TGA CTA C, Rev: ACA TAT CCA AGG ACG TGT AGA; Bglap/Osteocalcin: CTG AGT CTG ACA AAG CCT TC, Rev: GTG GTC CGC TAG CTC GTC AC; Ezh2 Fw: GCC AGA CTG GGA AGA AAT CTG, Rev: TCA CTG GTG ACT GAA CAC TCC; Ezh1 Fw: AGT GAC AAG AAA GCG GAA ACG CCA, Rev: GCC ATC GAT GTT GGG TGT GCA CTG; PRMT5 Fw: GCT GTG GTG ACG CTA GAG AAC, Rev: AGC CCA GAA GTT CAC TGA CAA; Wdr5 Fw: CCA GTC CAA CCT CAT CGT CT, Rev: CAT CAC GGT TGA AAT GAA CG; GAPDH Fw: CAT GGC CTT CCG TGT TCC TA, Rev: CCT GCT TCA CCA CCT TCT TGA T.
Statistical analyses
For ChIP assays through rat development (E18 to P>90 stage), we used a one-way ANOVA analysis followed by the Dunnett post-test to compare significant changes with respect to E18 samples. For mRNA expression and ChIP analyses where two groups were compared (e.g. silencing vs. control), we used the Student's t-test. In those ChIP analyses where only two columns are shown, a Student's t-test was also performed between the antibody and the control IgG. In all figures, error bars represent the mean ± S.D.; *: P < 0.05, **: P < 0.01, ***: P < 0.001.
Results
Master regulator of the osteogenic lineage gene program is efficiently silenced during hippocampal development
Many studies have assessed epigenetic mechanisms mediating transcriptional activation of genes associated with the osteogenic lineage program during osteoblast differentiation in mesenchymal cell types that are osteogenic or can trans-differentiate into osteoblastic cells (43, 59-71). However, specific mechanisms mediating epigenetic silencing of these genes in non-mesenchymal cells have not been systematically addressed. This study leverages the experimental potential of post-mitotic rat hippocampal brain tissue, a well-characterized and developmentally-regulated differentiation system, to define the critical epigenetic mechanisms that maintain repressed genes of the osteoblastic lineage.
As shown in Figure 1A, using Reverse Transcriptase qPCR analysis we found that mRNA expression of the bone-related Runx2/p57, Sp7 and Bglap genes remains largely repressed in hippocampal tissue obtained at different stages of embryonic and post-natal development. Importantly, these genes are found repressed at stages as early as E18 (Figure 1A), indicating that these immature neuronal and glial cells possess mechanisms in place to efficiently silence these genes. As a control, we show in the same graph (Figure 1A) that mRNAs coding for these three osteogenic genes are readily detected under our experimental conditions in the rat osteoblastic cell line ROS17/2.8 (52, 72, 73). Moreover, in agreement with previous reports (28), the expression of the hippocampal plasticity-related gene PSD-95 progressively increases during hippocampal development and, as expected, is absent in the mRNA samples from the ROS17/2.8 cells (Figure 1A). Together, these results indicate that the hippocampal development sequence represents a valid experimental model to study mechanisms that repress non-neuronal genes and, in particular, to address the contribution of epigenetic control during silencing of the osteogenic lineage program.
Figure 1. Epigenetic post-translational modifications at histones associated with the Runx2/p57 promoter in developing hippocampus.
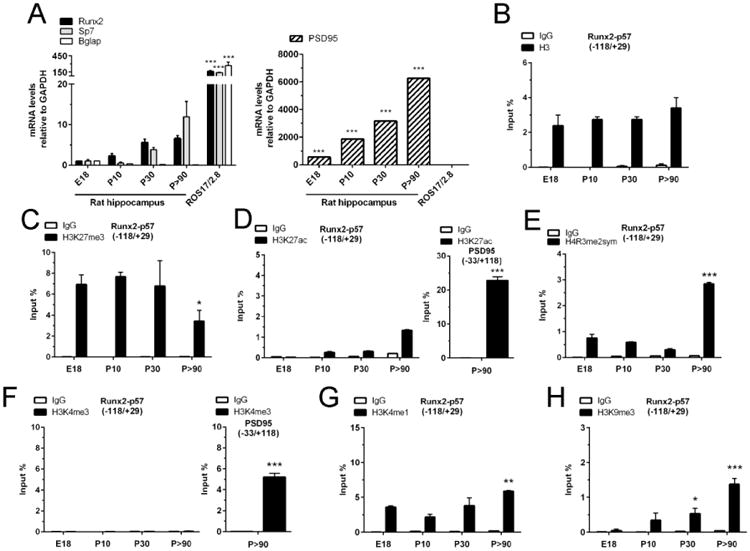
(A): Total RNA was extracted from E18, P10, P30 and P>90 rat hippocampus and mRNA levels were determined by qRT-PCR using specific primers. Results were normalized against GAPDH and expressed as fold change with respect to E18. Samples from rat osteosarcoma ROS17/2.8 cells were used as positive controls. PSD95 mRNA levels are represented with respect to Runx2 E18 expression (B-H): Chromatin immunoprecipitation (ChIP) assays were performed on samples obtained from rat hippocampus at the indicated stages and incubated with antibodies against total histone H3 (B), H3K27me3 (C), H3K27ac (D), H4R3me2-symmetrical (E), H3K4me3 (F), H3K4me1 (G), and H3K9me3 (H). The precipitated DNA was quantified using specific primers against the indicated region of the Runx2/p57promoter. Primers for the PSD95 promoter region were used as positive controls. Results are shown as Input (%) ± SD of at least three independent experiments. Normal IgG was used as specificity control. *: P < 0.05, **: P < 0.01, ***: P < 0.001, with respect to E18 (ANOVA). For the right panel of D and F, ***: P < 0.001, with respect to IgG (Student's t-test).
We next assessed whether a specific landscape of epigenetic histone marks accompanies repression of the osteoblastic master regulator Runx2/p57 during hippocampal development. Using Chromatin Immunoprecipitation (ChIP) analysis, it was determined that the proximal promoter region of this gene (P1 promoter of Runx2) exhibits significant levels of histone H3 (Figure 1B). This enrichment remains relatively constant in samples from subsequent post-natal and adult stages like P30 and P90 (Figure 1B).
Our results also show that there is differential enrichment of specific histone marks at the Runx2/p57 (P1) promoter during hippocampal development. Thus, the Runx2 P1 promoter region exhibits high levels of the H3K27me3 repressive mark at all the developmental stages analyzed, with a significant reduction (50%) in this mark at the adult (P90) stage (Figure 1C). As the H3K27 residue can also be acetylated at promoters that are either transcriptionally active or poised for expression (74, 75), we then examined whether the presence of H3K27ac could be detected in these osteogenic promoters. As shown in Figure 1D, the Runx2-P1 promoter does not exhibit significantly detectable levels of the H3K27ac mark at any stage of the developmental sequence analyzed (Figure 1D, left panel). This reduced enrichment is in contrast to that detected in promoters of transcriptionally active genes like PSD-95 (Figure 1D, right panel).
The symmetric di-methylation of arginine 3 residue in histone H4 (H4R3me2s) is a mark normally associated with the promoter region of transcriptionally repressed genes (47, 48, 76). This mark was found enriched at the Runx2-P1 promoter, particularly in hippocampal cells isolated from adult individuals (P90) (Figure 1E).
In agreement with their transcriptionally inactive status, the Runx2-P1 gene promoter showed reduced enrichment of the H3K4me3 mark (Figure 1F, left panel) a modification that is typically associated with the promoter regions of actively expressed genes (32, 33). As a required control, we demonstrated that in these same hippocampal cells the transcriptionally active PSD-95 gene promoter exhibits high enrichment of this mark (Figure 1F, right panel). Interestingly, we also found that instead the H3K4 residue is modified by mono-methylation (H3K4me1) in the Runx2-P1 promoter (Figure 1G). Similar analyses indicated that the H3K9me3 mark, normally associated with transcriptional silencing at different regions of the genome (30, 31) is poorly enriched in the Runx2-P1 promoter at E18, and then gradually increases as the post-natal development sequence progresses (Figure 1H).
Epigenetic regulators bind to the transcriptionally inactive master osteogenic Runx2 gene promoter during hippocampal development
To gain insight into the enzymatic complexes that can be mediating the deposition of the observed histone mark profiles at the Runx2-P1 gene promoter in hippocampal cells, we carried out ChIP analyses using specific antibodies against well-established regulatory components of candidate complexes.
We find that the PRC2 complex binds to the Runx2 gene, because its catalytic subunit Ezh2 interacts with the P1 promoter region at all the stages analyzed (E18, P10, P30 and p90). This binding of Ezh2 is significantly reduced in hippocampal tissue from adult (P90) animals (Figure 2A). Importantly, the apparent dissociation of Ezh2 from the Runx2-P1 promoter is paralleled by enrichment of the PRC2 catalytic subunit Ezh1 at developmental stages P30 and P90 (Figure 2B). These results indicate that during post-natal development of the hippocampus, binding of PRC2 involves an exchange of the catalytic subunits, which is accompanied by a reduction in the H3K27me3 mark (see Figure 1C).
Figure 2. Chromatin-modifying enzymes associated with the Runx2/p57 promoter in developing hippocampus.
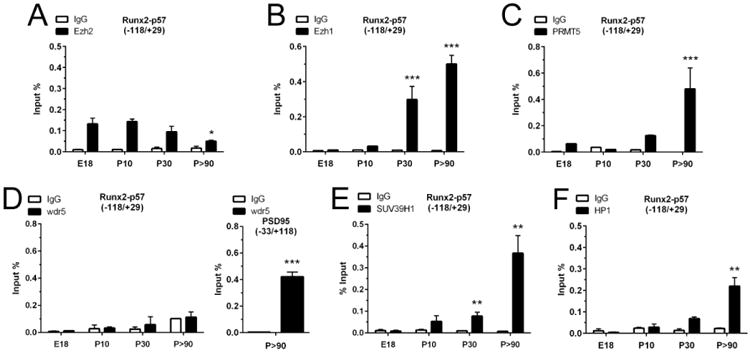
Chromatin immunoprecipitation (ChIP) assays were performed on samples obtained from E18, P10, P30 and P>90 rat hippocampus and incubated with antibodies against Ezh2 (A), Ezh1 (B), PRMT5 (C), Wdr5 (D), SUV39H1 (E), and HP1 (F). The precipitated DNA was quantified using specific primers against the indicated region of the Runx2/p57 promoter. Primers for PSD95 promoter region were used as positive controls. Results are shown as Input (%) ± SD of at least three independent experiments. Normal IgG was used as specificity control. *: P < 0.05, **: P < 0.01, ***: P < 0.001, with respect to E18 (ANOVA). For the right panel of D, ***: P < 0.001, with respect to IgG (Student's t-test).
In agreement with the increased levels of the H4R3me2s mark at the Runx2-P1 promoter in adult hippocampal cells (Figure 1E), we found that the H4R3 methyltransferase PRMT5 is significantly enriched on this promoter in samples from adult (P90) animals (Figure 2C).
In accordance with the absence of H3K4me3 at the Runx2-P1 osteogenic promoter (Figure 1F), we did not find interaction of Wdr5, a critical component shared by all MLL/COMPASS complexes (Figure 2D, left panel). As a necessary control, we demonstrated that Wdr5 is enriched at the PSD95 gene promoter (Figure 2D, right panel), thereby confirming that this protein is present in brain cells and is capable of interacting with transcriptionally active genes (77).
Because the H3K9me3 repressive mark is enriched at the Runx2-P1 promoter in samples of hippocampus from adult individuals (Figure 1H), we next addressed whether this promoter interacts with proteins associated with the deposition of this mark in brain tissue (78). As shown in Figure 2E, binding of Suv39H1 an enzyme that mediates mono-, di- and tri-methylation of the H3K9 residue is detected at the Runx2-P1 gene promoter in samples from adult hippocampus (Figure 2E). Similarly, HP1, a protein that has been shown to interact with chromatin regions containing the H3K9me3 mark (79) was found enriched at the Runx2-P1 promoter in adult hippocampal cells (Figure 2F). Taken together, these results indicate that during post-natal development the Runx2-P1 gene is maintained in a repressed state through a mechanism that involves progressive deposition of the H3K9me3 histone mark, which is well-correlated with heterochromatin formation and transcriptional silencing (80).
Critical contribution of PRC2 during silencing of the Runx2 osteogenic gene in hippocampal cells
Because the H3K27me3 mark is the enzymatic product of the PRC2-Ezh1 and PRC2-Ezh2 complexes (29), we next assessed the contribution of PRC2 binding to this mark as well as to silencing of osteoblast gene expression in hippocampal cells. Similar to the hippocampal tissue studied here, we previously demonstrated that Ezh2 and Ezh1 are differentially expressed during maturation of hippocampal neuron-enriched primary cultures (28). Hence, both complexes PRC2-Ezh1 and PRC2-Ezh2 are abundantly expressed at early stages of neuronal maturation (3 days in culture in vitro, 3DIV) whereas PRC2-Ezh1 is expressed predominantly in mature (17 days, 17DIV) neuronal cells.
Using lentiviruses expressing specific shRNAs against Ezh2 (28), the expression of this enzyme was effectively reduced (at both mRNA and protein levels, see Figure 3A) in immature hippocampal neuron-enriched primary cultures (7DIV). This decreased Ezh2 expression was also reflected by a significant reduction in H3K27me3 levels at the Runx2-P1 promoter (Figure 3C), confirming that a PRC2-Ezh2 containing complex could mediate the deposit of this repressive mark at this transcriptionally-silent P1 promoter. Importantly, this knockdown is not accompanied by changes in histone H3 enrichment (Figure 3B), or by an increased enrichment in the H3K4me3 and H3K27ac marks at this osteogenic master gene promoter (see left panels in Figure 3D and 3E, respectively). These two active marks, however, were found at the transcriptionally active PSD95 gene promoter, detecting a slight increase in their enrichment following Ezh2 knockdown (see right panels in Figures 3D and 3E, respectively).
Figure 3. Knockdown of Ezh2 results in the expression of Runx2/p57 in immature primary hippocampal cells.
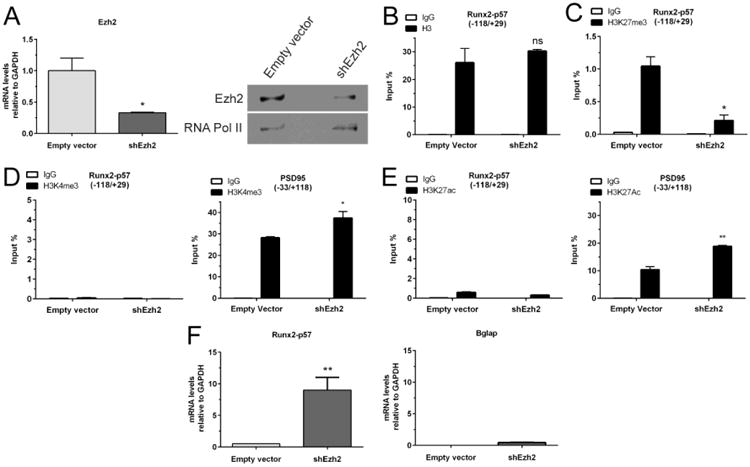
Hippocampal cells were infected at 3DIV with a lentivirus carrying a shRNA against Ezh2 expression. (A): Total RNA samples were obtained 96h later (7DIV) to confirm the Ezh2 silencing by qRT-PCR (left panel). Results were normalized against GAPDH and expressed as fold change with respect to cells infected with a lentivirus packed with an empty vector. Ezh2 knockdown was also confirmed by Western blot using specific antibodies (right panel). Detection of RNA Polymerase II with a specific antibody was used to control for equal protein loading. (B-E): ChIP experiments were performed on samples using specific antibodies against total H3 (B), H3K27me3 (C), H3K4me3 (D) and H3K27ac (E). Results are expressed as Input (%) ± SD of at least three independent experiments. Normal IgG was used as specificity control. Primers for PSD95 promoter region were used as positive controls. (F): Changes in mRNA expression of osteoblastic genes were determined by qRT-PCR. *: P < 0.05, **: P < 0.01, ns: non-significant differences with respect to the corresponding control (empty vector) value (Student's t-test).
Strikingly, the reduced H3K27me3 enrichment found at the Runx2-P1 promoter results in transcriptional activation of the Runx2-p57 gene (Figure 3F, left panel). However, no reversion was detected in the repressed condition of the late bone phenotypic Bglap gene (Figure 3E, right panel). Although the Bglap gene is a downstream target of the Runx2 factor, it is well known to be temporally activated much later than Runx2 both in vivo and in cell culture (81). Together, these results suggest that the PRC2-Ezh2-mediated deposition of the H3K27me3 mark is a principal component of the mechanisms that maintain the expression of the the main osteogenic master regulator Runx2 repressed in immature hippocampal neurons. Additionally, these results indicate that transcriptional activation of this key osteogenic gene can occur in these cells in the absence of the two main histone marks associated with transcription in mammal cells, H3K4me3 and H3K27ac (32, 33, 74, 75).
Our results indicate that in adult hippocampus the osteogenic Runx2 gene promoter here analyzed retains binding of a PRC2 complex containing Ezh1 instead of Ezh2 (Figure 2A and 2B). Therefore, we next determined whether knockdown of Ezh1 expression in mature hippocampal neurons (17DIV through 21DIV) also alters the repressed state of Runx2 gene expression. As shown in Figure 4A, lentiviral-driven shRNA-mediated knockdown of Ezh1 expression (at mRNA and protein levels), results in a significant decrease of the H3K27me3 mark at the Runx2-P1 promoter (Figure 4C) without affecting the overall enrichment of histone H3 (Figure 3B). Lack of enrichment in the H3K4me3 and H3K27ac marks following Ezh1 knockdown was also confirmed (Figures 4D and 4E, respectively). Importantly, the reduction in H3K27me3 was found accompanied by transcriptional activation of the Runx2 gene (Figure 4C, left panel), further confirming that in these hippocampal cells Runx2 gene repression depends on PRC2-mediated H3K27me3 deposition at the P1 promoter. As expected, Bglap gene repression is not modified by Ezh1 knockdown (Figure 4C, right panel).
Figure 4. Knockdown of Ezh1 results in the expression of Runx2/p57 in mature primary hippocampal cells.
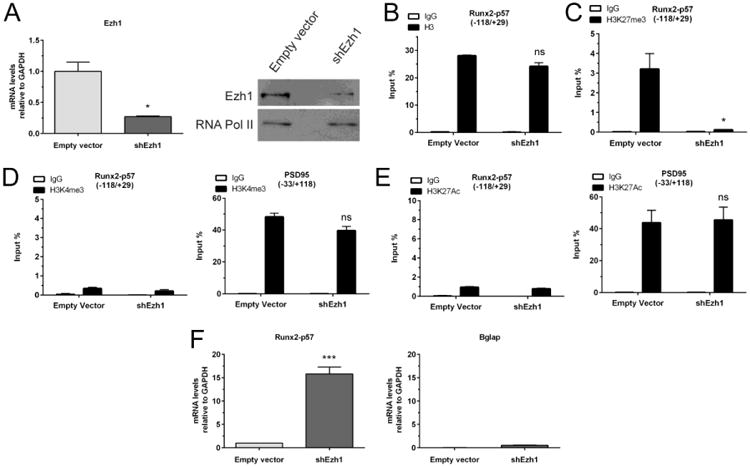
Hippocampal cell cultures were infected at 17DIV with a lentivirus carrying a shRNA against Ezh1 expression. (A): Total RNA samples were obtained 96h later (21DIV) to confirm the Ezh1 silencing by qRT-PCR (left panel). Results were normalized against GAPDH and expressed as fold change with respect to cells infected with a lentivirus packed with an empty vector. Ezh1 knockdown was also confirmed by Western blot using specific antibodies (right panel). Detection of RNA Polymerase II was used to control for equal protein loading. (B-E): ChIP experiments were performed on samples using specific antibodies against total H3 (B), H3K27me3 (C), H3K4me3 (D), and H3K27ac (E). Results are expressed as Input (%) ± SD of at least three independent experiments. Normal IgG was used as specificity control. Primers for PSD95 promoter region were used as positive controls. (F): Changes in mRNA expression of osteoblastic genes were determined by qRT-PCR. *: P < 0.05, ***: P < 0.001, ns: non-significant differences with respect to the corresponding control (empty vector) value (Student's t-test)
We initially observed that one significant change at the Runx2-P1 gene promoter during hippocampal development was the enrichment of the H4R3me2s mark and of its catalyzing enzyme PRMT5 (Figures 1E and 2C, respectively). Therefore, we assessed a putative functional contribution of this mark to the mechanism of epigenetic silencing of Runx2 in mature cells. Using shRNAs against PRMT5, we generated a knockdown of this enzyme that effectively reduces its mRNA and protein levels (Figure 5A) at late stages of hippocampal neuron maturation (17DIV through 21DIV). The resulting loss of PRMT5 causes a drastic decrease of the H4R3me2s mark at the Runx2-P1 promoter (Figure 5B). Interestingly, this strong reduction of the H4R3me2s mark was not reflected in Runx2 gene transcription (Figure 5C, left panel) or Bglap gene expression (Figure 5C, right panel). These results indicate that although binding of PRMT5 and enrichment in H4R3me2s at the Runx2-P1 promoter represent a hallmark of a transcriptionally inert state of this gene (see Discussion), it is not a critical component during maintenance of this repression.
Figure 5. Knockdown of PRMT5 does not alter repression of the Runx2/p57 gene in primary hippocampal cells.
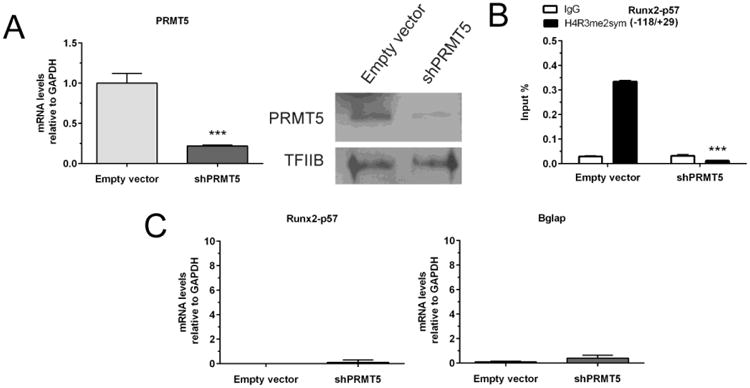
Mature hippocampal cells were infected at 17DIV with a lentivirus carrying a shRNA against PRMT5 expression. (A): Total RNA samples were obtained 96h later to confirm the PRMT5 silencing by qRT-PCR (left panel). Results were normalized and expressed as described in Figure 3. PRMT5 knockdown was also confirmed by Western blot using specific antibodies (right panel). Detection of TFIIB was used to control for equal protein loading. (B): ChIP experiments were performed on samples using specific antibodies against H3K27me3. Results are expressed as Input (%) ± SD of at least three independent experiments. Normal IgG was used as specificity control. (C): Potential changes in mRNA expression of the indicated osteoblastic genes were determined by qRT-PCR. ***: P < 0.001, with respect to the corresponding control (empty vector) value (Student's t-test).
Wdr5-associated H3K4 methylase activity can reactivate transcription of the Runx2/p57 osteogenic master regulator in hippocampal cells
Trithorax/MLL/COMPASS complexes mediate mono-, di-, and tri-methylation of H3K4 at promoter regions that surround the transcriptional start site (34, 82). Wdr5 functions as a critical assembling component that maintains the integrity and functionality of each complex (35). Targeted over expression in vivo of Wdr5 promotes bone formation (39, 40). Corroborating these earlier findings, we find that forced expression of Wdr5 in proliferating rat calvaria-derived primary osteoblasts (Figure 6A, left and central panels) suffices to increase Runx2/p57 mRNA expression (Figure 6A, right panel).
Figure 6. Overexpression of Wdr5 results in the expression of Runx2/p57 in primary hippocampal cells.
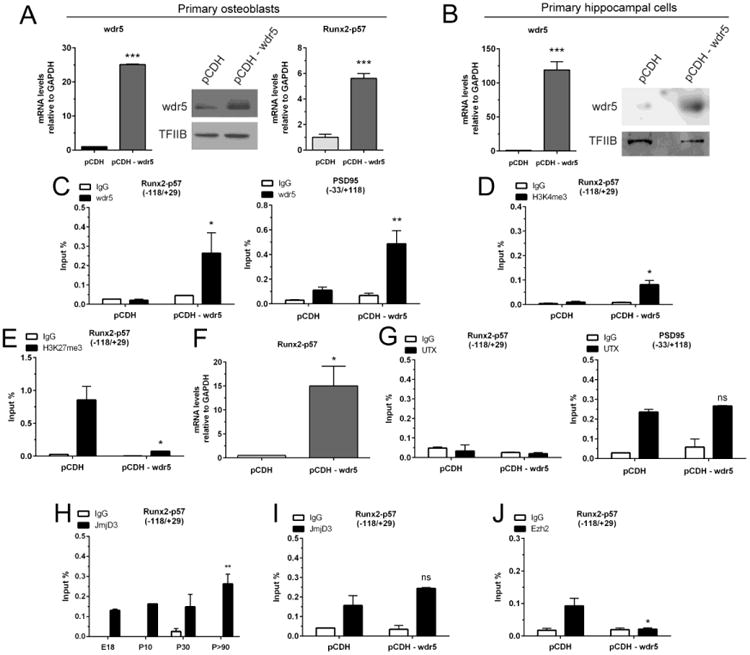
2DIV rat calvaria-derived osteoblasts (A) and 3DIV hippocampal cells were infected or transfected, respectively, with a vector coding for the COMPASS subunit Wdr5 (B-J). (A, B): Total RNA samples were obtained 96h later to confirm Wdr5 mRNA overexpression by qRT-PCR (left panels). Results were normalized against GAPDH and expressed as fold change with respect to cells transfected with an empty vector. Wdr5 overexpression was also confirmed by Western blot using specific antibodies (right panels). TFIIB protein detection was used to control for equal loading. (C-E, G-J): ChIP experiments were performed on samples using specific antibodies against Wdr5 (C), H3K4me3 (D), H3K27me3 (E), UTX (G), JmjD3 (H, I), and Ezh2 (J). Results are expressed as Input (%) ± SD of at least three independent experiments. Normal IgG was used for specificity control. (A [right panel] and F): Changes in mRNA expression of Runx2/p57 were determined by qRT-PCR. *: P < 0.05, **: P <0.01, ***: P<0.001, ns: non-significant differences with respect to the corresponding control (pCDH) value (Student's t-test). For panel H, **: P<0.01, with respect to E18 (ANOVA).
Therefore, we evaluated whether a forced increase in Wdr5-containing Trithorax/MLL/COMPASS-like complexes in hippocampal cells parallels its binding to a silent osteogenic promoter and, through an enrichment of the H3K4me3 mark, produce an activation of the transcription of these genes. As shown in Figure 6B, immature hippocampus neuronal cells (3DIV) were efficiently transfected with a CMV-driven plasmid coding for Wdr5, producing significantly increased levels of Wdr5 mRNA and protein after 96h (Figure 6B, left and right panels, respectively). Forced Wdr5 expression resulted in binding of this protein to the Runx2-P1 promoter. Enrichment of Wdr5 at the Runx2-P1 region is equivalent to that found at the transcriptionally-active neuronal-specific PSD95 promoter (compare Figure 6C, left and right panels). Importantly, binding of Wdr5 at the Runx2-P1 promoter is accompanied by the presence of reduced, but detectable levels of H3K4me3 enrichment (Figure 6D, left panel).
It has been well-established that at least some of the Trithorax/MLL/COMPASS complex members carry UTX-associated H3K27me3 demethylase activity. Hence, once bound to regulatory target regions these multi enzymatic complexes can promote transcription by both increasing the H3K4me3 and reducing the H3K27me3 marks (34). Accordingly, we next addressed whether increased recruitment of Wdr5 at the Runx2-P1 promoter also affected the H3K27me3 enrichment in this region. As shown in Figure 6E, binding of Wdr5 is paralleled by a drastic decrease of the H3K27me3 mark. Most importantly, this H3K27me3 reduction is accompanied by transcriptional activation of the Runx2 gene (Figure 6F). Together, these results further establish that elevated expression of Wdr5 and subsequent association, likely as part of a Trithorax/MLL/COMPASS complex, with the P1 promoter region of the Runx2 gene represents a critical step during activation of transcription of this osteogenic master regulator.
Although it has been demonstrated that the H3K27me3 demethylase UTX is a component of Trithorax/MLL/COMPASS complexes including MLL3/4 as the H3K4 methylase, we did not find UTX bound to the Runx2-P1 promoter in hippocampal cells overexpressing Wdr5 (Figure 6G, left panel). Similarly, UTX was not found associated, in vitro or in vivo, with the Runx2-P1 promoter at any other stage of hippocampal development (data not shown). As a required control, we determined that UTX is bound to the transcriptionally active PSD95 gene promoter in these cells (Figure 6G, right panel). The reduced levels of H3K27me3 found at the Runx2-P1 promoter in hippocampal cells over-expressing Wdr5 could also be the result of an increased interaction of the JMJD3 demethylase (38, 83) and/or reduction in PRC2-Ezh2. It was found that the JMJD3 enzyme interacts continuously with the Runx2-P1 promoter in hippocampal samples isolated at the developmental stages E18, P10, P30, and P90 (Figure 6H). Importantly, we determined that following Wdr5 over expression in hippocampal cells, binding of JMJD3 to the Runx2-P1 promoter remains unchanged (Figure 6I), whereas the interaction of Ezh2 with this promoter is substantially reduced (Figure 6J). Taken together, these results indicate that elevation of Wdr5 levels and subsequent binding of Wdr5 to the Runx2-P1 promoter leads to transcriptional activation through a mechanism that involves PRC2-Ezh2 displacement and H3K27me3 decrease.
Discussion
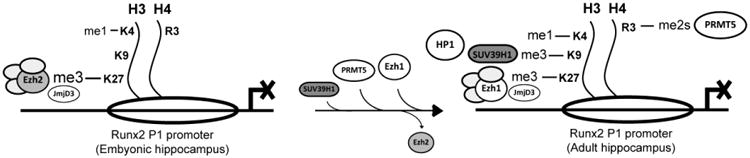
In this work, we have identified epigenetic mechanisms that prevent transcription of the osteogenic gene program in the hippocampus of embryonic, post-natal, and adult rats. We demonstrate that during embryonic development (E18), the Ezh2-containing PRC2 complex principally contributes to suppression of Runx2/p57 gene expression by mediating the deposition of the H3K27me3 mark at the promoter regions near their transcriptional start sites. Moreover, knockdown of Ezh2 in immature embryonic hippocampal cells decreases H3K27me3 enrichment at the Runx2-P1 promoter and activates transcription of this gene. Interestingly, PRC2 continues to principally mediate transcriptional repression of the Runx2/p57 gene in mature hippocampal cells. Moreover, in these cells tri-methylation of the H3K27 is mainly maintained by Ezh1 that replaces Ezh2 in the PRC2 complex as hippocampal maturation progresses (28). Repression of Runx2 as the most critical osteoblast master regulator is also characterized by later enrichment of complementary epigenetic silencing mechanisms in post-natal and adult hippocampus; Runx2/p57 gene silencing may be reinforced at the P1 promoter by deposition of the H3K9me3 and HP1. Together, the results support a model (see Figure 7) in which silencing of this master regulator of osteoblast lineage commitment is maintained by accumulation of repressive epigenetic marks at its osteogenic P1 promoter, as hippocampal neuron maturation proceeds from embryonic development to adulthood. We propose that these biochemical events may represent a fail-safe mechanism that operates in hippocampal tissue to further prevent any potential transcriptional awakening of this gene and of the down-stream osteoblast genetic program in adult CNS cells. We anticipate that functional impairments in vivo of these silencing mechanisms may have physiological consequences with potential pathological implications for human congenital disorders of the head.
Figure 7. Schematic diagram representing the epigenetic mechanisms regulating Runx2/p57 gene silencing during hippocampal maturation.
Late bone phenotypic gene markers like Bglap are not transcriptionally activated following Runx2/p57 expression. This result suggests that additional repressive mechanisms that cannot be overcome by Runx2/p57 must be operating to maintain transcriptional inhibition of Bglap-like genes. Previous reports have established that Bglap silencing in mesenchymal non-osteoblastic or pre-osteoblastic cells involves CpG methylation at the promoter region (66). Unpublished results from our team also indicate that the Bglap gene locus exhibits elevated CpG methylation in hippocampal cells (not shown). Because increased CpG methylation may prevent transcriptional activation of the Bglap gene promoter in mature hippocampal neurons, it is tempting to speculate that DNA methylation at regulatory promoter sequences of genes activated downstream of Runx2 may serve as a fundamental mechanism to further consolidate repression of non-neuronal gene transcription in hippocampal tissue.
A recent study showed that Ezh2 knockdown in undifferentiated mesenchymal C3H10T1/2 cells increases Runx2 expression and potentiates BMP2-dependent osteoblast commitment (84). In contrast, our group recently demonstrated that decreased levels of H3K27me3 at the Runx2 P1 promoter upon loss of Ezh2 in pro-myogenic mesenchymal cells does not suffice to prevent Runx2/p57 repression during myogenic differentiation (43). Rather, repression of the Runx2/p57 master gene during differentiation of mesenchymal cells to the myoblastic lineage is principally mediated by the H3K4me3 demethylase Jarid1B (KDM5B). Importantly, knockdown of Jarid1B prevents repression of the Runx2-P1 promoter during myogenic lineage commitment (43). The latter results in the unscheduled expression of the osteogenic gene program under conditions that induce myogenic differentiation and coincides with enrichment of the H3K4me3 and H3K27ac marks at the Runx2-P1 promoter. Hence, these results suggest that repression of the Runx2/p57 gene during neuronal and muscle lineage commitment may occur through alternative and/or complementary epigenetic mechanisms.
Conditional genetic loss of Ezh2 in uncommitted mouse mesenchymal cells results in multiple defects in skeletal patterning and bone formation (85). Extensive transcriptomic analyses revealed that Ezh2-null cells exhibit elevated levels of Runx2 mRNA expression, consistent with accelerated cessation of cell growth and precocious maturation of osteoblasts. Consistent with these cellular consequences, mice that in which Ezh2 is conditionally deleted in mesenchymal precursor cells have shortened forelimbs, craniosynostosis and clinodactyly. Furthermore, PRC2-Ezh2 has a critical role in epigenetic control during development of craniofacial skeletal structures. Conditional ablation of Ezh2 in neural crest cells (NCCs) during embryonic development significantly affects the formation of neural crest-derived craniofacial structures (86). Interestingly, the absence of Ezh2 and concomitant decrease in chromatin-enrichment of H3K27me3 does not appear to alter differentiation of NCCs during formation of early structures within the peripheral nervous system, even though Ezh2 is clearly critical for embryonic differentiation of cells in the CNS (26). These studies together indicate that PRC2 can only effectively control the expression of non-neuronal genes in particular neuronal tissues and/or at specific developmental stages.
One question that arises from our studies is how tri-methylation of H3K27 by Ezh2 in the embryonic hippocampus dominates over demethylation of H3K27me3, which may be mediated by enzymes like JMJD3/KDM6B and/or UTX/KDM6A (38, 87). We have previously shown that Ezh2 binds together with UTX and JMJD3 to the actively transcribed PSD95/DLG4 gene promoter in hippocampal neurons (28). Continuous recruitment of both demethylases significantly contributes to the removal of the H3K27me3 mark deposited by Ezh2. In this study, we find that JMJD3, but not UTX, binds to the osteogenic Runx2-P1 promoter in hippocampal cells. This result is consistent with the earlier finding that JMJD3 is essential during neuronal lineage differentiation (88) and spinal cord formation (89). Although tight control of H3K27me3 demethylase activity is critical in the CNS, the sole presence of JMJD3 does not suffice to conclude that this enzyme mediates a reduction of this silencing mark from the Runx2-P1 promoter in the hippocampus. Future functional analyses must confirm this possibility as there are several examples of histone demethylases that albeit being enriched at target sequences do not contribute to transcriptional regulation through their intrinsic enzymatic activities (38, 90-92). One alternative attractive possibility that still remains to be experimentally addressed is that the enzymatic activity of JMJD3 may be negatively modulated within the context of the Runx2 P1 promoter in hippocampal neurons.
Wdr5 expression and function is strongly linked to osteoblastic gene expression in mesenchymal cells (39, 41, 42). Its accumulation in at least some non-osteogenic cells may affect the maintenance of the lineage identity. For example, Wdr5 protein levels are actively reduced by rapid ubiquitination and subsequent proteasomal degradation in brain cells to avoid undesired transcriptional activation of non-neuronal genes (77). We find that forced expression of Wdr5 increases its binding to the Runx2-P1 promoter, displaces PRC2/Ezh2 and decreases H3K27me3 enrichment in hippocampal cells. Thus, our results reveal that PRC2/Ezh2-mediated repression of the osteoblast-related master regulator Runx2 is critically responsive to Wdr5 modulation in non-osseous cells.
The methylase PRMT5 contributes to neuronal maturation and prominently expressed in brain. PRMT5 expression gradually increases throughout post-natal brain development (93). Corroborating these results, we find that PRMT5 and its epigenetic product H4R3me2 are significantly enriched at the Runx2-P1 promoter in adult hippocampus. Even though enriched, our results indicate that this epigenetic mark is not critical for maintaining Runx2/p57 gene repression. Rather, PRMT5 may exert its function in the hippocampus, in part, by recruiting the de novo DNA methyltransferase Dnmt3a that mediates transcriptional repression by CpG methylation (50). Because the Runx2-P1 promoter lacks methylated CpG (61), it is plausible that Runx2 silencing may be independent of PRMT5 during hippocampal maturation. Nevertheless, it is also important to keep in mind several reports indicating that PRMT5 may not always function as a transcriptional repressor (43, 94-97).
The ability to de-repress the Runx2/p57 gene in post-mitotic immature hippocampal neurons following Ezh2 knockdown and Wdr5 over expression, is easily understood if activators of this master osteoblastic gene are present in hippocampal cells. Indeed, transcription factor Dlx5 is enriched in the CNS (98). Dlx5 is a critical BMP2 responsive activator of Runx2/p57 gene transcription during osteoblast differentiation (23, 99, 100). Furthermore, Wnt signaling, which is required for brain development and neuronal maintenance (101, 102), also strongly activates Runx2/p57 gene expression in mesenchymal cells (103-105). Wdr5 mediates Wnt signaling pathway in mesenchymal cells and supports induction of Runx2-P1 expression (42). Hence, regulatory cross-talk between both WNT-WDR5 and BMP2-DLX5 dependent pathways may prime activation of Runx2 in non-osseous neural cells.
In conclusion, our results provide novel insights into epigenetic silencing events that are operative in hippocampal cells. The findings from this study may be relevant to mesenchymal gene activation during the epithelial to mesenchymal transition that generates neural crest cells during head development and that is fundamental to formation of craniofacial bone tissues. The work presented here enhances our appreciation for epigenetic pathways that coordinately control bone and brain development in the head and that may be deregulated in the hundreds of currently known congenital craniofacial syndromes.
Highlights.
The osteogenic master gene Runx2 is epigenetically repressed in hippocampus.
Complementary epigenetic mechanisms progressively silence Runx2.
Knockdown of PRC2 activates Runx2 expression in hippocampal cells.
Overexpression of Wdr5-containing Trithorax complexes overrules Runx2 silencing.
Acknowledgments
This work was supported by grants from FONDAP 15090007 (to M.M. and M.L.A.), FONDECYT 1130706 (to M.M.), FONDECYT 3150694 (to MS) and FONDECYT 1140301 (to B.vZ.). Additional funding was provided by NIH (R01 AR049069 to A.J.vW.). R.A. and F.B. were supported by Doctoral Fellowships from CONICYT, Chile. A.R. was partially funded by Doctoral Fellowships from COLCIENCIAS and Pontificia Universidad Javeriana, Colombia.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ishii M, Sun J, Ting MC, Maxson RE. The Development of the Calvarial Bones and Sutures and the Pathophysiology of Craniosynostosis. Current topics in developmental biology. 2015;115:131–56. doi: 10.1016/bs.ctdb.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Marcucio R, Hallgrimsson B, Young NM. Facial Morphogenesis: Physical and Molecular Interactions Between the Brain and the Face. Current topics in developmental biology. 2015;115:299–320. doi: 10.1016/bs.ctdb.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trainor PA, Andrews BT. Facial dysostoses: Etiology, pathogenesis and management. American journal of medical genetics Part C, Seminars in medical genetics. 2013;163C(4):283–94. doi: 10.1002/ajmg.c.31375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhatt S, Diaz R, Trainor PA. Signals and switches in Mammalian neural crest cell differentiation. Cold Spring Harbor perspectives in biology. 2013;5(2) doi: 10.1101/cshperspect.a008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Andre P, Ye L, Yang YZ. The Hedgehog signalling pathway in bone formation. International journal of oral science. 2015;7(2):73–9. doi: 10.1038/ijos.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong Z, Ethen NJ, Williams BO. WNT signaling in bone development and homeostasis. Wiley interdisciplinary reviews Developmental biology. 2014;3(6):489–500. doi: 10.1002/wdev.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G, Deng C, Li YP. TGF-beta and BMP signaling in osteoblast differentiation and bone formation. International journal of biological sciences. 2012;8(2):272–88. doi: 10.7150/ijbs.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin GL, Hankenson KD. Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. Journal of cellular biochemistry. 2011;112(12):3491–501. doi: 10.1002/jcb.23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marie P. Transcription factors controlling osteoblastogenesis. Archives of biochemistry and biophysics. 2008;473(2):98–105. doi: 10.1016/j.abb.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 10.van Wijnen AJ, van de Peppel J, van Leeuwen JP, Lian JB, Stein GS, Westendorf JJ, et al. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Current osteoporosis reports. 2013;11(2):72–82. doi: 10.1007/s11914-013-0143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer MB, Benkusky NA, Pike JW. The RUNX2 cistrome in osteoblasts: characterization, down-regulation following differentiation, and relationship to gene expression. The Journal of biological chemistry. 2014;289(23):16016–31. doi: 10.1074/jbc.M114.552216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H, Whitfield TW, Gordon Ja, Dobson JR, Tai PW, van Wijnen AJ, et al. Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome biology. 2014;15(3):R52. doi: 10.1186/gb-2014-15-3-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee C, Hiebert SW, Stein JL, Lian JB, Stein GS. An AML-1 consensus sequence binds an osteoblast-specific complex and transcriptionally activates the osteocalcin gene. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(10):4968–73. doi: 10.1073/pnas.93.10.4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–64. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 15.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89(5):765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 16.Nishio Y, Dong Y, Paris M, O'Keefe RJ, Schwarz EM, Drissi H. Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene. 2006;372:62–70. doi: 10.1016/j.gene.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 17.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108(1):17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 18.Harada H, Tagashira S, Fujiwara M, Ogawa S, Katsumata T, Yamaguchi A, et al. Cbfa1 isoforms exert functional differences in osteoblast differentiation. The Journal of biological chemistry. 1999;274(11):6972–8. doi: 10.1074/jbc.274.11.6972. [DOI] [PubMed] [Google Scholar]
- 19.Merriman HL, van Wijnen AJ, Hiebert S, Bidwell JP, Fey E, Lian J, et al. The tissue-specific nuclear matrix protein, NMP-2, is a member of the AML/CBF/PEBP2/runt domain transcription factor family: interactions with the osteocalcin gene promoter. Biochemistry. 1995;34(40):13125–32. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 20.Lengner CJ, Drissi H, Choi JY, van Wijnen AJ, Stein JL, Stein GS, et al. Activation of the bone-related Runx2/Cbfa1 promoter in mesenchymal condensations and developing chondrocytes of the axial skeleton. Mech Dev. 2002;114(1-2):167–70. doi: 10.1016/s0925-4773(02)00050-3. [DOI] [PubMed] [Google Scholar]
- 21.Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nature reviews Molecular cell biology. 2012;13(1):27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- 22.Mishina Y, Snider TN. Neural crest cell signaling pathways critical to cranial bone development and pathology. Exp Cell Res. 2014;325(2):138–47. doi: 10.1016/j.yexcr.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holleville N, Mateos S, Bontoux M, Bollerot K, Monsoro-Burq AH. Dlx5 drives Runx2 expression and osteogenic differentiation in developing cranial suture mesenchyme. Developmental biology. 2007;304(2):860–74. doi: 10.1016/j.ydbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Vaes BL, Ducy P, Sijbers AM, Hendriks JM, van Someren EP, de Jong NG, et al. Microarray analysis on Runx2-deficient mouse embryos reveals novel Runx2 functions and target genes during intramembranous and endochondral bone formation. Bone. 2006;39(4):724–38. doi: 10.1016/j.bone.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 25.Choi KY, Kim HJ, Lee MH, Kwon TG, Nah HD, Furuichi T, et al. Runx2 regulates FGF2-induced Bmp2 expression during cranial bone development. Dev Dyn. 2005;233(1):115–21. doi: 10.1002/dvdy.20323. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Ji F, Liu Y, Lei X, Li H, Ji G, et al. Ezh2 regulates adult hippocampal neurogenesis and memory. J Neurosci. 2014;34(15):5184–99. doi: 10.1523/JNEUROSCI.4129-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7(3):288–98. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Henriquez B, Bustos FJ, Aguilar R, Becerra A, Simon F, Montecino M, et al. Ezh1 and Ezh2 differentially regulate PSD-95 gene transcription in developing hippocampal neurons. Molecular and Cellular Neuroscience. 2013;57:130–43. doi: 10.1016/j.mcn.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. The EMBO journal. 2004;23(21):4286–96. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121(6):873–85. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 33.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annual review of biochemistry. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature structural & molecular biology. 2006;13(8):713–9. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 36.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121(6):859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 37.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science (New York, NY) 2007;318(5849):447–50. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 38.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–4. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 39.Gori F, Divieti P, Demay MB. Cloning and characterization of a novel WD-40 repeat protein that dramatically accelerates osteoblastic differentiation. The Journal of biological chemistry. 2001;276(49):46515–22. doi: 10.1074/jbc.M105757200. [DOI] [PubMed] [Google Scholar]
- 40.Gori F, Demay MB. BIG-3, a novel WD-40 repeat protein, is expressed in the developing growth plate and accelerates chondrocyte differentiation in vitro. Endocrinology. 2004;145(3):1050–4. doi: 10.1210/en.2003-1314. [DOI] [PubMed] [Google Scholar]
- 41.Gori F, Friedman LG, Demay MB. Wdr5, a WD-40 protein, regulates osteoblast differentiation during embryonic bone development. Developmental biology. 2006;295(2):498–506. doi: 10.1016/j.ydbio.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 42.Zhu ED, Demay MB, Gori F. Wdr5 is essential for osteoblast differentiation. The Journal of biological chemistry. 2008;283(12):7361–7. doi: 10.1074/jbc.M703304200. [DOI] [PubMed] [Google Scholar]
- 43.Rojas A, Aguilar R, Henriquez B, Lian JB, Stein JL, Stein GS, et al. Epigenetic Control of the Bone-master Runx2 Gene during Osteoblast-lineage Commitment by the Histone Demethylase JARID1B/KDM5B. The Journal of biological chemistry. 2015;290(47):28329–42. doi: 10.1074/jbc.M115.657825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burchfield JS, Li Q, Wang HY, Wang RF. JMJD3 as an epigenetic regulator in development and disease. The international journal of biochemistry & cell biology. 2015;67:148–57. doi: 10.1016/j.biocel.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lengner CJ, Steinman HA, Gagnon J, Smith TW, Henderson JE, Kream BE, et al. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. The Journal of cell biology. 2006;172(6):909–21. doi: 10.1083/jcb.200508130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Deen M, Taipaleenmaki H, Zhang Y, Teplyuk NM, Gupta A, Cinghu S, et al. MicroRNA-34c inversely couples the biological functions of the runt-related transcription factor RUNX2 and the tumor suppressor p53 in osteosarcoma. The Journal of biological chemistry. 2013;288(29):21307–19. doi: 10.1074/jbc.M112.445890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fabbrizio E, El Messaoudi S, Polanowska J, Paul C, Cook JR, Lee JH, et al. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO reports. 2002;3(7):641–5. doi: 10.1093/embo-reports/kvf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S, Gunesdogan U, Zylicz JJ, Hackett JA, Cougot D, Bao S, et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Molecular cell. 2014;56(4):564–79. doi: 10.1016/j.molcel.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Molecular and cellular biology. 2004;24(21):9630–45. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nature structural & molecular biology. 2009;16(3):304–11. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Owen Ta, Aronow M, Shalhoub V, Barone LM, Wilming L, Tassinari MS, et al. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. Journal of cellular physiology. 1990;143(3):420–30. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- 52.Majeska RJ, Rodan SB, Rodan GA. Parathyroid hormone-responsive clonal cell lines from rat osteosarcoma. Endocrinology. 1980;107(5):1494–503. doi: 10.1210/endo-107-5-1494. [DOI] [PubMed] [Google Scholar]
- 53.Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nature protocols. 2006;1(2):695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- 54.Sepulveda FJ, Bustos FJ, Inostroza E, Zuniga FA, Neve RL, Montecino M, et al. Differential roles of NMDA Receptor Subtypes NR2A and NR2B in dendritic branch development and requirement of RasGRF1. J Neurophysiol. 2010;103(4):1758–70. doi: 10.1152/jn.00823.2009. [DOI] [PubMed] [Google Scholar]
- 55.Henriquez B, Hepp M, Merino P, Sepulveda H, van Wijnen AJ, Lian JB, et al. C/EBPbeta binds the P1 promoter of the Runx2 gene and up-regulates Runx2 transcription in osteoblastic cells. Journal of cellular physiology. 2011;226(11):3043–52. doi: 10.1002/jcp.22652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soutoglou E, Talianidis I. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science. 2002;295(5561):1901–4. doi: 10.1126/science.1068356. [DOI] [PubMed] [Google Scholar]
- 57.Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. BioTechniques. 2006;41(6):694, 6, 8. doi: 10.2144/000112297. [DOI] [PubMed] [Google Scholar]
- 58.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic acids research. 1983;11(5):1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee JY, Lee YM, Kim MJ, Choi JY, Park EK, Kim SY, et al. Methylation of the mouse DIx5 and Osx gene promoters regulates cell type-specific gene expression. Mol Cells. 2006;22(2):182–8. [PubMed] [Google Scholar]
- 60.Yang D, Okamura H, Nakashima Y, Haneji T. Histone demethylase Jmjd3 regulates osteoblast differentiation via transcription factors Runx2 and osterix. The Journal of biological chemistry. 2013;288(47):33530–41. doi: 10.1074/jbc.M113.497040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ezura Y, Sekiya I, Koga H, Muneta T, Noda M. Methylation status of CpG islands in the promoter regions of signature genes during chondrogenesis of human synovium-derived mesenchymal stem cells. Arthritis and rheumatism. 2009;60(5):1416–26. doi: 10.1002/art.24472. [DOI] [PubMed] [Google Scholar]
- 62.Montecino M, Lian J, Stein G, Stein J. Changes in chromatin structure support constitutive and developmentally regulated transcription of the bone-specific osteocalcin gene in osteoblastic cells. Biochemistry. 1996;35(15):5093–102. doi: 10.1021/bi952489s. [DOI] [PubMed] [Google Scholar]
- 63.Montecino M, Frenkel B, Lian J, Stein J, Stein G. Requirement of distal and proximal promoter sequences for chromatin organization of the osteocalcin gene in bone-derived cells. Journal of cellular biochemistry. 1996;63(2):221–8. doi: 10.1002/(SICI)1097-4644(19961101)63:2%3C221::AID-JCB9%3E3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 64.Montecino M, Pockwinse S, Lian J, Stein G, Stein J. DNase I hypersensitive sites in promoter elements associated with basal and vitamin D dependent transcription of the bone-specific osteocalcin gene. Biochemistry. 1994;33(1):348–53. doi: 10.1021/bi00167a045. [DOI] [PubMed] [Google Scholar]
- 65.Shen J, Hovhannisyan H, Lian JB, Montecino MA, Stein GS, Stein JL, et al. Transcriptional induction of the osteocalcin gene during osteoblast differentiation involves acetylation of histones h3 and h4. Mol Endocrinol. 2003;17(4):743–56. doi: 10.1210/me.2002-0122. [DOI] [PubMed] [Google Scholar]
- 66.Villagra A, Gutierrez J, Paredes R, Sierra J, Puchi M, Imschenetzky M, et al. Reduced CpG methylation is associated with transcriptional activation of the bone-specific rat osteocalcin gene in osteoblasts. Journal of cellular biochemistry. 2002;85(1):112–22. [PubMed] [Google Scholar]
- 67.Shen J, Montecino M, Lian JB, Stein GS, Van Wijnen AJ, Stein JL. Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D-dependent, bone tissue-specific transcription. The Journal of biological chemistry. 2002;277(23):20284–92. doi: 10.1074/jbc.M112440200. [DOI] [PubMed] [Google Scholar]
- 68.Sierra J, Villagra A, Paredes R, Cruzat F, Gutierrez S, Javed A, et al. Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Molecular and cellular biology. 2003;23(9):3339–51. doi: 10.1128/MCB.23.9.3339-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Villagra A, Cruzat F, Carvallo L, Paredes R, Olate J, van Wijnen AJ, et al. Chromatin remodeling and transcriptional activity of the bone-specific osteocalcin gene require CCAAT/enhancer-binding protein beta-dependent recruitment of SWI/SNF activity. The Journal of biological chemistry. 2006;281(32):22695–706. doi: 10.1074/jbc.M511640200. [DOI] [PubMed] [Google Scholar]
- 70.Tai PW, Wu H, Gordon JA, Whitfield TW, Barutcu AR, van Wijnen AJ, et al. Epigenetic landscape during osteoblastogenesis defines a differentiation-dependent Runx2 promoter region. Gene. 2014;550(1):1–9. doi: 10.1016/j.gene.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hemming S, Cakouros D, Isenmann S, Cooper L, Menicanin D, Zannettino A, et al. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells. 2014;32(3):802–15. doi: 10.1002/stem.1573. [DOI] [PubMed] [Google Scholar]
- 72.Varanasi SS, Datta HK. Characterisation of cytosolic FK506 binding protein 12 and its role in modulating expression of Cbfa1 and osterix in ROS 17/2.8 cells. Bone. 2005;36(2):243–53. doi: 10.1016/j.bone.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 73.Gouveia CH, Schultz JJ, Bianco AC, Brent GA. Thyroid hormone stimulation of osteocalcin gene expression in ROS 17/2.8 cells is mediated by transcriptional and post-transcriptional mechanisms. J Endocrinol. 2001;170(3):667–75. doi: 10.1677/joe.0.1700667. [DOI] [PubMed] [Google Scholar]
- 74.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(50):21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136(18):3131–41. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. The EMBO journal. 2007;26(15):3558–69. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakagawa T, Xiong Y. X-linked mental retardation gene CUL4B targets ubiquitylation of H3K4 methyltransferase component WDR5 and regulates neuronal gene expression. Molecular cell. 2011;43(3):381–91. doi: 10.1016/j.molcel.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu DX, Nath N, Chellappan SP, Greene LA. Regulation of neuron survival and death by p130 and associated chromatin modifiers. Genes & development. 2005;19(6):719–32. doi: 10.1101/gad.1296405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 80.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Molecular cell. 2003;12(6):1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 81.Owen TA, Aronow M, Shalhoub V, Barone LM, Wilming L, Tassinari MS, et al. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. Journal of cellular physiology. 1990;143(3):420–30. doi: 10.1002/jcp.1041430304. [DOI] [PubMed] [Google Scholar]
- 82.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature reviews Genetics. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 83.Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, et al. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. The EMBO journal. 2010;29(8):1401–11. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang YX, Sun HL, Liang H, Li K, Fan QM, Zhao QH. Dynamic and distinct histone modifications of osteogenic genes during osteogenic differentiation. Journal of biochemistry. 2015 doi: 10.1093/jb/mvv059. [DOI] [PubMed] [Google Scholar]
- 85.Dudakovic A, Camilleri ET, Xu F, Riester SM, McGee-Lawrence ME, Bradley EW, et al. Epigenetic Control of Skeletal Development by the Histone Methyltransferase Ezh2. The Journal of biological chemistry. 2015 doi: 10.1074/jbc.M115.672345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schwarz D, Varum S, Zemke M, Scholer A, Baggiolini A, Draganova K, et al. Ezh2 is required for neural crest-derived cartilage and bone formation. Development. 2014;141(4):867–77. doi: 10.1242/dev.094342. [DOI] [PubMed] [Google Scholar]
- 87.Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17(10):850–7. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- 88.Burgold T, Voituron N, Caganova M, Tripathi PP, Menuet C, Tusi BK, et al. The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell reports. 2012;2(5):1244–58. doi: 10.1016/j.celrep.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 89.Akizu N, Estarás C, Guerrero L, Martí E, Martínez-Balbás Ma. H3K27me3 regulates BMP activity in developing spinal cord. Development (Cambridge, England) 2010;137(17):2915–25. doi: 10.1242/dev.049395. [DOI] [PubMed] [Google Scholar]
- 90.Przanowski P, Dabrowski M, Ellert-Miklaszewska A, Kloss M, Mieczkowski J, Kaza B, et al. The signal transducers Stat1 and Stat3 and their novel target Jmjd3 drive the expression of inflammatory genes in microglia. Journal of molecular medicine. 2014;92(3):239–54. doi: 10.1007/s00109-013-1090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pedersen MT, Agger K, Laugesen A, Johansen JV, Cloos PA, Christensen J, et al. The demethylase JMJD2C localizes to H3K4me3-positive transcription start sites and is dispensable for embryonic development. Molecular and cellular biology. 2014;34(6):1031–45. doi: 10.1128/MCB.00864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pereira F, Barbachano A, Silva J, Bonilla F, Campbell MJ, Munoz A, et al. KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells. Human molecular genetics. 2011;20(23):4655–65. doi: 10.1093/hmg/ddr399. [DOI] [PubMed] [Google Scholar]
- 93.Huang J, Vogel G, Yu Z, Almazan G, Richard S. Type II arginine methyltransferase PRMT5 regulates gene expression of inhibitors of differentiation/DNA binding Id2 and Id4 during glial cell differentiation. The Journal of biological chemistry. 2011;286(52):44424–32. doi: 10.1074/jbc.M111.277046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kanno Y, Inajima J, Kato S, Matsumoto M, Tokumoto C, Kure Y, et al. Protein arginine methyltransferase 5 (PRMT5) is a novel coactivator of constitutive androstane receptor (CAR) Biochem Biophys Res Commun. 2015;459(1):143–7. doi: 10.1016/j.bbrc.2015.02.085. [DOI] [PubMed] [Google Scholar]
- 95.LeBlanc SE, Wu Q, Lamba P, Sif S, Imbalzano AN. Promoter-enhancer looping at the PPARgamma2 locus during adipogenic differentiation requires the Prmt5 methyltransferase. Nucleic acids research. 2016 doi: 10.1093/nar/gkw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tarighat SS, Santhanam R, Frankhouser D, Radomska HS, Lai H, Anghelina M, et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia. 2016;30(4):789–99. doi: 10.1038/leu.2015.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tsai WW, Niessen S, Goebel N, Yates JR, 3rd, Guccione E, Montminy M. PRMT5 modulates the metabolic response to fasting signals. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(22):8870–5. doi: 10.1073/pnas.1304602110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kimura MI, Kazuki Y, Kashiwagi A, Kai Y, Abe S, Barbieri O, et al. Dlx5, the mouse homologue of the human-imprinted DLX5 gene, is biallelically expressed in the mouse brain. Journal of human genetics. 2004;49(5):273–7. doi: 10.1007/s10038-004-0139-2. [DOI] [PubMed] [Google Scholar]
- 99.Lee MH, Kim YJ, Yoon WJ, Kim JI, Kim BG, Hwang YS, et al. Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. The Journal of biological chemistry. 2005;280(42):35579–87. doi: 10.1074/jbc.M502267200. [DOI] [PubMed] [Google Scholar]
- 100.Lee MH, Kwon TG, Park HS, Wozney JM, Ryoo HM. BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem Biophys Res Commun. 2003;309(3):689–94. doi: 10.1016/j.bbrc.2003.08.058. [DOI] [PubMed] [Google Scholar]
- 101.Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JL, Grosschedl R. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development (Cambridge, England) 2000;127(3):469–82. doi: 10.1242/dev.127.3.469. [DOI] [PubMed] [Google Scholar]
- 102.Varela-Nallar L, Grabowski CP, Alfaro IE, Alvarez AR, Inestrosa NC. Role of the Wnt receptor Frizzled-1 in presynaptic differentiation and function. Neural Dev. 2009;4:41. doi: 10.1186/1749-8104-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gaur T, Lengner CJ, Hovhannisyan H, Bhat Ra, Bodine PVN, Komm BS, et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. The Journal of biological chemistry. 2005;280(39):33132–40. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- 104.Kang S, Bennett CN, Gerin I, Rapp LA, Hankenson KD, Macdougald OA. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma. The Journal of biological chemistry. 2007;282(19):14515–24. doi: 10.1074/jbc.M700030200. [DOI] [PubMed] [Google Scholar]
- 105.Cho YD, Yoon WJ, Kim WJ, Woo KM, Baek JH, Lee G, et al. Epigenetic modifications and canonical wingless/int-1 class (WNT) signaling enable trans-differentiation of nonosteogenic cells into osteoblasts. The Journal of biological chemistry. 2014;289(29):20120–8. doi: 10.1074/jbc.M114.558064. [DOI] [PMC free article] [PubMed] [Google Scholar]


