Abstract
Small interfering RNA (siRNA) has gained attention as a potential therapeutic reagent due to its ability to inhibit specific genes in many genetic diseases. For many years, studies of siRNA have progressively advanced toward novel treatment strategies against cancer. Cancer is caused by various mutations in hundreds of genes including both proto-oncogenes and tumor suppressor genes. In order to develop siRNAs as therapeutic agents for cancer treatment, delivery strategies for siRNA must be carefully designed and potential gene targets carefully selected for optimal anti-cancer effects. In this review, various modifications and delivery strategies for siRNA delivery are discussed. In addition, we present current thinking on target gene selection in major tumor types.
Keywords: Small interfering RNA (siRNA), Delivery strategy of siRNAs, Cancer, siRNA therapeutics, Gene target
Graphical Abstract
1. Introduction
The discovery of RNA interference (RNAi) has opened doors that might introduce a novel therapeutic tool to the clinical setting [1]. For many decades, small molecules have been developed and utilized in cancer therapy; however, critical problems, such as undesirable toxicity against normal tissues due to a lack of selectivity, still remain today. Using RNAi as a therapeutic tool will allow targeting previously unreachable targets with its potential to silence the function of any cancer causing gene [2]. This unique advantage is made possible by utilizing the biological functions of double-stranded RNA molecules (dsRNA). Endogenous dsRNA is recognized by a ribonuclease protein, termed dicer, and cleaved into small double stranded fragments of 21 to 23 base pairs in length with 2-nucleotide overhangs at the 3’ ends. The cleaved products are referred to as small interfering RNAs (siRNAs). The siRNAs consist of a passenger strand and a guide strand, and are bound by an active protein complex called the RNA-induced silencing complex (RISC). After binding to RISC, the guide strand is directed to the target mRNA, which is cleaved between bases 10 and 11 relative to the 5’ end of the siRNA guide strand, by the cleavage enzyme argonaute-2. Thus, the process of mRNA translation can be interrupted by siRNA [3-5].
The therapeutic application of siRNA has the potential to treat various diseases including cancer [6, 7]. Cancer is a genetic disease caused by the generation of mutated genes within tumor cells; multiple gene mutations both activate disease driving oncogenes and inactivate tumor suppressor genes in cancer [8-10]. Small interfering RNAs that can inactivate specific cancer driving genes have shown great potential as novel cancer therapeutics. Several anti-cancer siRNA based drugs have entered clinical trials, and many are actively sought after in preclinical research [11-13].
Even though the usage of siRNA as therapy has shown promise in the treatment of cancer, many obstacles that hinder the ultimate functionality of siRNAs in the clinic remain to be solved [14, 15]. In order to make this therapy effective, the first and most crucial step is to ensure the delivery of siRNA to the tumor cells from the injection site. In practice, siRNAs face physiological and biological barriers that prevent their delivery to the active site when administered systemically [16-18]. These barriers include, but are not limited to, intravascular degradation, recognition by the immune system, renal clearance, impediments to tumor tissue penetration and uptake into tumor cells, endosomal escape once in tumor cells, and off-target effects [19-21]. Delivery formulations as well as chemical modification of siRNA are required to overcome these challenges and facilitate siRNAs in reaching their target cells [22]. Furthermore, selection of gene targets in cancer is also crucial in designing siRNA therapeutic strategies. Discoveries of mechanisms in cancer provide innovative targets for siRNA therapy that in many cases cannot be targeted with conventional drugs. However, the particular gene pool that drives cancer varies depending on the origins and types of the tumors. Thus, careful selection of gene targets according to their cancer type is essential in siRNA therapeutic strategies.
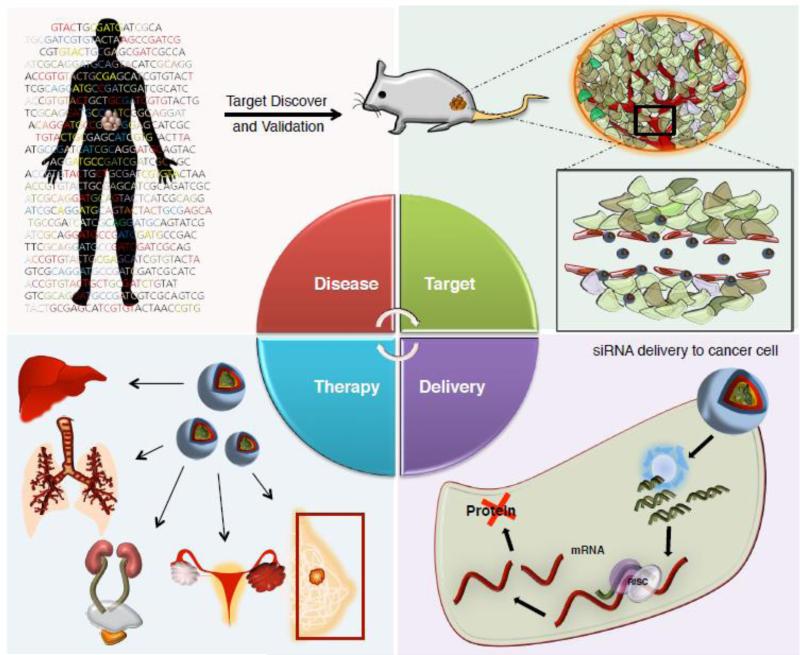
To summarize, target discovery in cancer leads to the selection of siRNA gene targets, followed by their incorporation of the siRNAs into suitable delivery systems that allow access to the desired sites. Once therapeutic effect is observed, further application in varying organs and tissues can be anticipated as shown in Figure 1. This review examines current thoughts on the therapeutic potential of siRNA delivery strategies and the optimal targets for siRNA in major cancer types.
Figure 1.
Development process of siRNA therapeutics for cancer treatment
2. Strategies for siRNA based Therapeutics
In order to activate the RNAi pathway, double stranded siRNA must travel through the bloodstream and gain access to the cytosol of target cells. The hydrophilic nature and large molecular weight of siRNAs prevent the molecules from diffusing across the cellular membrane into the cell; therefore, modifications to the nucleic acid and generation of clever delivery strategies are necessary for the creation of siRNA therapeutics.
2.1. Chemical Modifications
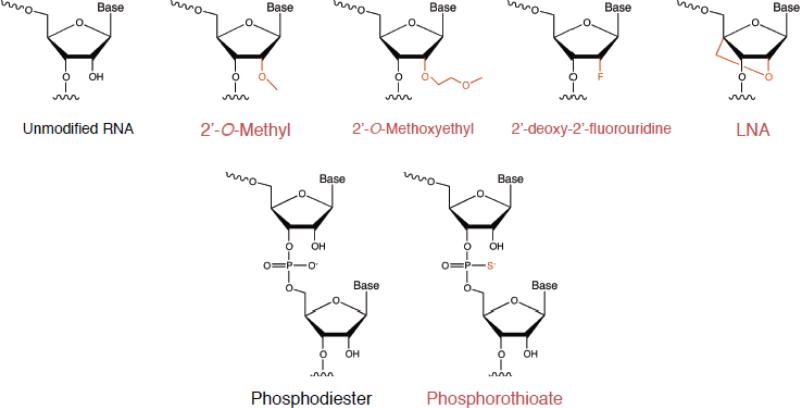
With current bioorganic techniques, oligonucleotides can be synthesized and modified as single strands, then annealed into the desired double stranded material. Customizable oligonucleotide synthesis incorporating artificial modifications enhances the potential of RNA therapeutics by overcoming problems associated with administration of naked siRNA. In particular, unmodified siRNA exposed in the bloodstream stimulates the innate the immune response and is readily degraded by serum nucleases. One of the methods to increase stability in serum and potency of gene silencing efficacy is to employ chemical modifications on the RNA-backbone of siRNA. A wide variety of chemical modifications, listed in Figure 2, have been proposed to overcome existing challenges of siRNA therapeutics.
Figure 2.
Chemical Modifications and siRNA
One of the most common alterations of RNA is modification of the 2’ position on the ribose backbone. These modifications include 2’-O-methyl, 2’-O-methoxyethyl, 2’-deoxy-2’-fluorouridine, locked nucleic acid (LNA), and many more [23-26]. These chemical modifications increase stability against nucleases and improve thermal stability. As a naturally occurring RNA variant, 2’-O-methyl RNA has shown reduced potency or even inactivation in siRNA activity in the RNAi pathway upon heavy modification [27]. The 2’-fluoro modification is compatible with siRNA function and lends stability in presence of nucleases. Combined modification with 2’-fluoro pyrimidines and 2’-O-methyl purines results in highly stable RNA duplexes in serum and improved in vivo activity [28]. The 2’-O-methoxyethyl RNA modification has also shown significant nuclease resistance as well as increased thermal stability (Tm). Nevertheless, this modification is not generally used as frequently as the 2’-O-methyl and 2’-Fluoro RNAs. LNA contains a methylene bridge that connects the 2’-O with the 4’-C positions of the ribose backbone. This causes the siRNA to have “locked” sugar that results in higher stability with increased Tm. Though incorporation of LNA also interferes with the siRNA activity, limited modification retains the functionality [27].
In addition to the sugar modifications, variations in phosphate linkage of siRNA are also accepted as an alternative strategy to overcome functional limitations. The phosphorothioate (PS) linkage, perhaps the most commonly modified linkage in siRNA, often displays cytotoxicity when used extensively; however, PS incorporation does not appear to have a major effect on biodistribution of siRNA. [29]
Apart from modifications made on the backbone, chemical modifications are also made on other parts of siRNA to facilitate delivery to the target site. One of the hurdles in siRNA delivery is that weak negative charge and high molecular weight makes the nucleic acid more prone to serum degradation and capture by the reticuloendothelial system (RES). In order to form more stable delivery complexes, polymerized siRNA can be synthesized, resulting in greater electrostatic interactions and facilitating incorporation into nanoparticles. Lee et al. developed polymerized siRNA using a thiol group to form a stable complex with glycol chitosan via not only electrostatic interaction but also disulfide bond crosslinking. Polymerized siRNA synthesized with thiol groups was also shown to form stable complexes with PEI, albumin, transferrin, hyaluronic acid, and other nanoparticles [30-35]. This delivery reagent was shown to have an anti-tumor effect in xenograft cancer models when systemically injected.
Other chemical alterations of siRNA include base modification, change in overhangs and termini of the RNA duplexes, and varying tertiary structure of the siRNA. In an attempt to develop siRNA for use in clinical trials as drugs, various chemical modifications are being investigated to improve qualities such as serum stability, siRNA potency, low immunostimulation, off-target effects, and target organ/cell delivery [36].
2.2. Polymeric Nanoparticles
Incorporation of siRNAs into nanoparticles is widely used to overcome limitations of nucleic acid formulations. Biodegradable polymeric nanoparticles synthesized via a variety of methods have shown significant therapeutic potential allowing improved stability in serum, better delivery and controlled release. Nanoparticles used in siRNA delivery studies are divided into two categories—natural polymers and synthetic polymers. Cyclodextrin, chitosan, atelocollagen, albumin, gelatin and others are promising natural polymer candidates for siRNA delivery in cancer and other diseases [31, 33, 37]. Synthetic polymers such as polyethylene glycol (PEG), polyethyleneimine (PEI), poly (d,l-lactide-co-glycolic acid) (PLGA), and others have also been extensively investigated as siRNA delivery agents.
Cyclodextrin polymer (CDP) was used as a raw material for first nanoparticles delivery system for siRNA used in clinical trials [38]. CDP is a polycationic oligosaccharide produced during bacterial digestion of cellulose and used by the pharmaceutical industry to deliver small molecules [39]. As a siRNA delivery formulation, self-assembled CPD with PEG and human transferrin (Tf), denoted as CALAA-01, has demonstrated improved targeting ability for cancer cells in preclinical tests. Subsequently, this system was tested in a clinical phase trial 1. Another polymeric nanoparticle class frequently used in nanomedicine is chitosan-based systems. Chitosan, a polysaccharide originally derived from chitin, is comprised of β-(1-4)-linked D-glucosamine (deacetylated unit) and N-acetyl-D-glucosamine (acetylated unit). The cationic nature of the resulting nanoparticle allows electrostatic interactions with negatively charged siRNAs facilitating formulation into the delivery system. Various modifications of chitosan, such as PEGylation and thiolation, have also shown potential in cancer models via a variety of routes of administration [30, 40-43]. Atelocollagen/siRNA complexes have also exhibited anti-cancer effects in several xenograft cancer models. Collagen is a fibrous structural protein in connective tissue. By pepsin treatment, the telopeptide of collagen is removed to reduce immunogenicity generating highly purified type I collagen, also known as atelocollagen. Atelocollagen particles have shown improved cellular uptake, nuclease resistance, and controlled release of nucleic acids [44]. Inhibition in tumor growth of an orthotopic xenograft cancer model has been seen when atelocollagen/siRNA complexes were intratumorally administered [45]. Several factors, such as low toxicity, biodegradability, reduction in immunostimulation, and facile condensation with nucleic acids, contribute to advantages of using natural polymers as delivery agents for siRNA.
Synthetic polymers have been extensively investigated in the drug delivery field because of their well-defined chemistries and the high degree of molecular diversity obtainable via chemical modifications [46]. PEI is considered the most potent in its ability to form stable complexes with nucleic acids due to its highly positive charged nature. Branched PEI (bPEI), which has been found to have lower toxicity than linear PEI, and its derivatives have demonstrated successful siRNA delivery to the tumor sites with resultant anti-cancer effects in preclinical studies. In addition to PEI/siRNA complexes, PEGylation is often incorporated into nanoparticles to increase stability in biological fluids and lend protection against nucleases. Several groups have demonstrated a PEI-PEG platform for effective siRNA delivery in various cancer models [32, 47-49]. Another widely studied synthetic polymer is PLGA. PLGA is a copolymer made from lactic acid and glycolic acid that is approved in therapeutic applications due to its high biodegradability and biocompatibility. With a sustained drug release profile due to slow hydrolytic degradation, PLGA was linked with siRNA through disulfide linkage to form spherical micelles which showed gene silencing effects in breast cancer cells [50].
Though other delivery strategies are gaining attention, particularly including lipid-based siRNA delivery, many investigators continue to use and seek to improve polymeric nanoparticles, based on their stability, flexibility of modification, and high targetability. Nevertheless, deeper understanding into the toxicity and immunogenicity of these delivery methods is necessary to develop polymers as a siRNA delivery strategy for clinical use.
2.3. Lipid-based Delivery
Among various strategies to overcome challenges of siRNA therapeutics, lipid-based nanoparticles have great potential due to their biocompatibility and low toxicity in comparison to inorganic, viral and synthetic polymers. In particular, cationic lipids have emerged as attractive siRNA delivery vehicles owing to their electrostatic interaction with nucleic acids, high transfection efficiency into mammalian cells, and improved pharmacokinetic profiles. 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) is a type of cationic lipid that is commonly used in laboratories and is also commercially available. Kim et al. demonstrated a liposomal siRNA/DOTAP/Cholesterol platform for liver-targeting delivery of siRNAs against HBV [51]. Yagi et al. demonstrated a siRNA delivery complex utilizing cationic DOTAP attached to egg phosphatidylcholine (egg-PC) and PEG lipid in a weight ratio of 24:14.8. This complex has been shown to inhibit tumor growth in a xenograft cancer model via systemic injection [52].
For in vivo delivery studies, stable nucleic acid-lipid particles (SNALPs) have been formulated and tested in multiple disease models. SNALPs consist of a lipid bilayer of fusogenic and cationic lipids entrapping nucleic acids in the core. The surface of the SNALP is coated with PEG to provide enhanced hydrophilicity for improved stability in the serum. The half-life of a siRNA-SNALP complex is much longer compared to unformulated siRNA. An HBV targeted siRNA-SNALP has shown specific reduction in HBV mRNA when intravenously administered in a mouse model of HBV replication at a dosage of 3 mg/kg/day [28]. A siRNA-SNALP delivery complex was also tested against Ebola virus (EBOV) related genes in a guinea pig model [53]. Furthermore, an ApoB specific siRNA encapsulated in a SNALP has shown to have >90% maximal silencing effect of ApoB mRNA in liver upon a single systemic dosage of 2.5 mg/kg in cynomolgus monkeys [54]. Thus, RNAi-mediated gene silencing in non-human primates has clearly demonstrated the therapeutic potential of this new class of drug using SNALP technology.
Although cationic lipid-based siRNA delivery has demonstrated potential in therapy in various disease models, several hurdles remain to enter commercialization of this class of drugs. Toxicity and immediate immune responses elicited by lipid-based delivery designs must be further investigated, and it is likely that further thoughtful modifications will need to be devised.
2.4. Bioconjugated siRNAs
In addition to chemically modifying siRNA or incorporating it into nanoparticles, covalently conjugating biological agents to siRNA cargo is an alternative method to overcome barriers to siRNA efficacy in vivo. Such conjugated delivery systems currently include cholesterol, various peptides, antibodies, aptamers, and biopolymers with various physicochemical profiles, as summarized in Table 1.
Table 1.
Types of Conjugated siRNA
| Conjugation | Structure | Function | Example |
|---|---|---|---|
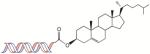
| Cholesterol |
|
Increases hydrophobicity for stability, free nucleases resistance of siRNA in serum, and mRNA suppression by polyplexes | Cholesterol (C27) [55, 57, 163, 164] Cholesteryloxypropan-1-amine (COPA) and Cholesteryl-2-aminoethylcarbamate (CAEC) [165] |
| Peptide |
|
Cell-penetration peptides (CPP) is able to cross the biological membrane for intracellular delivery | TAT [166, 167] MPG8 [168] Pep-3 [169] Penetratin [57] Transportan [170] |
| Antibody |
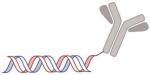
|
Antibody increases the target ability by ligand binding to specific receptors of cancer | Trastuzumab anti-TENB2, anti-NaPi2b [171] F105-P[172] F5-P [173] HIRMAb, TfRMAb [174] Anti-EGFR [59] |
| Aptamer |
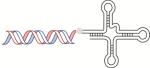
|
Aptamer selectively deliver siRNA to affected tissue via specific binding with reduced side effects | PSMA aptamer [61, 175-177] BAFF-R aptamer [178] HER2 aptamer [179] |
Cholesterol conjugation of siRNA facilitates cellular import and improves intracellular activity of siRNA when injected systemically [22]. Cholesterol in circulation is transported by lipoproteins in serum and taken up by hepatocytes through low density lipoprotein (LDL) receptor-mediated endocytosis. Apolipoprotein B (apoB) targeted siRNA conjugated with cholesterol showed enhanced stability in serum and greater suppression in apoB mRNA levels in the liver [55].
Biofunctional or cell-penetrating peptides can be covalently conjugated to siRNA for improved targeting of cancer cells. In a study by Choi et al., siRNA conjugated with branched PEG was functionalized with a cell penetrating peptide, Hph1, as well as a cationic self-crosslinked fusogenic KALA peptide to form a polyelectrolyte complex micelle for gene silencing in MDA-MB-435 breast cancer cells [56]. In an attempt to reduce innate immune responses, siRNAs directed against the p38 MAP kinase conjugated to the HIV TAT cell penetrating peptide were intratracheally administered to in the lung [57].
For targeted delivery of siRNA to specific tissues or cell types, antibodies or aptamers are being conjugated directly to siRNAs. A considerable number of antibody based drugs including Trastuzumab, Pertuzumab, and Cetuximab are currently given to cancer patients with great success. Antibodies conjugated to chemotherapeutic small molecules have also shown successful therapeutic results, with TDM-1 serving as a prominent example of this class of antibody conjugate [58]. Recently, an anti-EGFR antibody conjugated to a siRNA targeted to KRAS has shown activity in vitro and in vivo in colon cancer resistant to EGFR inhibitors [59].
An alternative target agent that can be conjugated to siRNA is the nucleic acid based aptamer. These aptamers consist of synthetic short single-stranded RNA or DNA ligands that have been selected for target binding with high affinity and specificity. Ever since the generation of aptamers that target the extracellular domains of transmembrane receptors overexpressed in cancer cells, aptamers have gained extensive attention as active targeting moieties for cancer therapeutic agents including siRNA [60]. Meyerholz et al. developed an aptamer conjugated RNA-only approach for prostate cancer therapy. When siRNAs targeting the pro-survival genes, Plk1 and Bcl2, were conjugated with aptamers that specifically binds to prostate-specific membrane antigen (PSMA) and injected intratumorally in a xenograft cancer model, inhibition in tumor growth was observed [61]. Despite the high specificity and binding affinity of aptamers, aptamer-siRNA conjugation faces barriers arising from, among other causes, stability issues due to unprotected negative charge.
In addition to being used as for direct coupling to siRNAs both antibodies and aptamers can be used to target nanoparticles containing siRNAs. As a surface targeting moiety, different types of aptamer facilitate nanoparticle delivery to the specific tumor sites.
3. Current Targets for siRNA in Cancer
Cancer occurs as a result of a series of gene mutations in a cell. Generally, a combination of activating mutations in so-called oncogenes and the loss of tumor suppressor genes lead to uncontrolled cell growth and blockage of natural apoptotic processes [62, 63]. Because many key gene mutations involved in driving cancer, also known as driver genes, have been identified [64, 65], it is easy to see that siRNA therapeutics could be effective in cancer treatment [66, 67]. A major advantage of using siRNA in cancer treatment is its ability to specifically inhibit any of the large set of cancer-associated genes without regard to the druggability of their protein products [67]. This allows us to potentially drug the undruggable. Furthermore, a diverse set of therapeutic siRNA molecules can be developed to target genes associated with the multiple signaling pathways which are aberrantly activated in tumors [68]. Table 1 summarizes the current status of siRNA targets for major cancers, which are discussed in more detail below.
3.1. Lung Cancer
Worldwide, lung cancer is the most frequently occurring tumor type with the highest incidence of cancer-related mortality. Among three main types of lung cancer, non-small cell lung cancer (NSCLC) is the most common, comprising approximately 85% of all lung cancers [69, 70]. Lung cancers often metastasize, leading to treatment failures. Despite the development of novel molecular therapies, aggressive surgery and radio–chemical therapy, prognosis for most of these patients is still very poor. It is possible to deliver siRNA based therapeutics to the lung either by systemically or via intrapulmonary administration [71, 72]. The latter route allows lower doses of siRNAs, reducing undesirable systemic side effects, and also improves the half-life of siRNAs in tumors. Hence, siRNA based therapeutics should be considered for lung cancer treatment [73].
Since cancer is a genetic disease it is always worth asking which genes are most frequently altered in a given tumor type. Epidermal growth factor receptor (EGFR) nucleotide variants are found in various types of cancer including lung cancer [74]. NSCLC in particular displays frequent EGFR mutations, which occurs on exons 18-21, encoding a portion of the EGFR kinase domain. The most common mutation class consists of exon 19 deletions, which activate the tyrosine kinase activity of EGFR, resulting in induction of downstream of pro-growth and survival signaling pathways [75]. Notably, it has been possible to construct allele specific siRNAs against certain oncogenic EGFR mutants, which have displayed significant therapeutic effects in lung tumor models with mutant EGFR alleles. The wet-weight of tumors treated with siRNAs targeting mutant EGFR were observed to be much lower than those of the control siRNA treated group. Furthermore, caspase-3 activity was upregulated in EGFR treated tumor tissue, indicating the induction of apoptosis. The allele specific EGFR siRNA treatment inhibited specific oncogenic EGFR alleles without affecting the normal EGFR allele, leading to a safe and effective treatment [76].
Activating mutations in KRAS frequently occur in lung tumors. Notably, in EGFR mutant lung tumors KRAS mutations can render them resistant to treatment with EGFR directed therapies. A seminal recent publication explored the anti-cancer therapeutic effect of targeting of both KRAS activation and loss of p53 function, another common lesion in many tumor types including lung tumors. The authors used siRNA and miRNA loaded polymer based nanoparticles in a genetically engineering mouse (GEM) model of lung cancer [77]. Nanoparticles loaded with siRNAs targeting KRAS and with a miRNA-34a mimic, which partially restores p53 downstream functions, were intravenously injected in a kras/p53 GEM model (KrasLSL-G12D/wt;p53flox/flox). Combination treatment with miR-34a and KRAS siRNA resulted in average lung tumor regression of 63% of its original volume and increased apoptotic cells. This form of targeted multi-gene combination therapy is perfectly suited for siRNA strategies and allows personalized cancer therapy adaptable to various mutations identified in a particular patient cancer type. The mice which were treated both cisplatin and the nanoparticle formulation consisting of a combination of miR-34a and a KRAS siRNA survived significantly longer compared with mice treated with either single treatment. (Combination of miR-34a/siKRAS and cisplatin; 159.5±19.5d, siLuc ; 93.7±16.1 d, cisplatin; 127.4±9.0d, miR-34a/siKRAS; 129.2±16.2d)
Human-ribophorin II (RPN2) is part of an N- oligosaccharyltransferase complex. Generally, this protein is known to facilitate human cancer resistance against chemotherapy drugs such as docetaxel. In addition, it has been reported RPN2 can serve as an anti-apoptotic protein that regulates tumor survival and cancer stem cell properties through the stabilization of mutant p53 [78]. To deliver naked RPN2 siRNA to the lung through inhalation, specifically astructured siRNAs termed, PnkRNATM and, nkRNATM were used [79]. The RPN2 targeting siRNAs inhibited the growth of a A549 luc-C8 lung xenograft and suppressed RPN2 expression.
The oncoprotein mouse double minute 2 (MDM2) is a negative regulator of the p53 tumor suppressor, which inhibits the transactivation activity of p53 via a direct interaction. A high level of MDM2 gene amplification has been detected in many human tumor types, including lung carcinomas [80, 81]. While inhibition of MDM2 function could obviously be beneficial in tumors with wild type p53, it has also been reported that MDM2 can play a significant role in tumors featuring mutant p53. Thus MDM2 could be a potential therapeutic target for treatment of human NSCLC with either wild type or mutant p53 [82]. In this regard it has been found that MDM2 siRNA modified with a pH-responsive diblock copolymer (termed MDM2 siRNA/PMPC-b-PDPA), showed significant downregulation of MDM2 expression in a xenograft of the p53 mutant H2009 cell line. MDM2 knockdown also resulted in impaired growth of H2009 tumors through cell cycle arrest and apoptosis. The tumor sizes of MDM2 siRNA/PMPC-b-PDPA treated H2009 tumor xenograft mice were significantly smaller than the tumors in control siRNA treated mice [83].
3.2. Liver Cancer
The total set of cancers in each organ type almost invariably constitutes a family of unique diseases. The major form of liver cancer is hepatocellular carcinoma (HCC), which is one of the most frequently occurring cancers worldwide. HCC is a complicated disease due to its broad epidemiology. Its causes include not only mutagenic environmental insults common to many tumors but also viral insults including those arising from hepatitis viruses. Hepatitis B and C infections are major risk factors for the development of HCC. Currently the treatment of HCC is limited. Surgical resection and transplantation are available to a minority of HCC patients who have an organ donor and are free of preexisting liver conditions [84, 85]. In the search for alternative modes of liver cancer treatment, many nucleotide based therapies, including siRNA therapeutics, that target signaling pathways specific to liver tumors, are now undergoing clinical trials [86].
HDACs, histone deacetylases, are crucial enzymes that regulate gene expression by deleting acetyl groups from histone substrates [87]. HDACs interact with key cancer-associated transcription factors, such as ß-catenin, Myc, p53, Stat3, NF-Kb, TFIIE, etc., to regulate the expression of numerous proteins implicated in tumor formation and development. Thus, altered expression and pathological activity of HDACs can lead to tumor onset and progression. HDAC2, which is overexpressed in solid tumors, promotes cell cycle progression and prevents apoptosis by inhibiting action of the tumor suppressor p53 [88]. HDAC2 targeting siRNAs have demonstrated a reduction of liver cancer cell proliferation in vitro. The therapeutic effect of HDAC siRNA/Lipid nanoparticles (LNP) was demonstrated in effectively reduced liver tumor growth in an orthotopic xenograft model [89].
Integrins are a superfamily of cell adhesion receptors that bind to and interact with the extracellular matrix (ECM). They are assembled from small families of α and ß glycoprotein subunits, generating 24 unique noncovalent α/ß heterodimers. The roles of integrins are both important and diverse; they take part in the regulation of cell motility, differentiation, survival, and proliferation. Among the integrin subunits, Itgb1 (ß1-integrin subunit) plays a critical role in liver formation and in the proliferation of liver tumor cells [90]. Overexpression of Itgb1 promotes survival and induces resistance to chemotherapy. It has been reported that activating mutations in Itgb1 affect tumorigenesis [91, 92]. Daniel G. Anderson's group reported that HCC progression can be delayed by targeting Itgb1 with siRNAs. In this study human MET and δN90-ß-catenin were delivered to mice hepatocytes to construct a mouse model of spontaneous HCC [93, 94]. After the introduction of the oncogenes, injection of Itgb1 siRNA/LNP inhibited HCC progression [95]. Liver weights of the Itgb1 siRNA/LNP treated group were significantly reduced compared to those of control groups. HCC tissues also showed a significant reduction in the size and number of tumor foci and enlarged hepatocytes in the residual tumor nodules.
Survivin is a protein inhibitor of apoptosis that is strongly expressed in hepatocellular carcinomas, but is not expressed in differentiated adult tissues [96]. Over-expression of survivin protein can inhibit caspase activation, thereby leading to inhibition of apoptosis and stimulation of HCC cell proliferation [97]. A tumor growth inhibition study using survivin targeting siRNA delivered by RGD-PEG-g-PEI-SPION nanoparticles was reported [98]. Mice bearing tumors arising from human HCC cell line, Bel-7402, were injected with survivin siRNA/RGD-PEG-g-PEI-SPION and exhibited delay in tumor growth. Inhibition of the survivin gene expression by siRNA resulted in an increase of cleaved caspase-3 expression.
Pokemon, a member of the POK family of transcriptional repressors, has been observed to have aberrant overexpression in multiple human cancers including liver tumors [99]. Pokemon plays a critical role in cellular transformation, as a central regulator of the ARF (Alterative reading frame)/p53 pathway and the Rb (retinoblastoma)/ E2F (Early- region −2 transcription factor) pathway [100]. A reconstituted high density lipoprotein (rHDL) based delivery system was used for Pokemon siRNAs in a tumor model with HepG2 cells overexpressing scavenger receptor class B type I (SR-BI). SR-BI, which is expressed in the liver and most malignant cells, interacts with the major component of HDL, apoprotein A-I, to maintain cellular cholesterol homeostasis. The relative expression levels of the Pokemon and Bcl-2 protein were markedly reduced in tumor tissues of the mice treated with cholesterol conjugated Pokemon siRNA/ rHDL as compared to the control group resulting in tumor growth inhibition [101].
Human telomerase reverse transcriptase (hTERT) is an essential component of human telomerase, which is often required to maintain stable telomere length in cancer cells. In liver and breast cancer cells, hTERT is highly expressed, which correlates with telomerase activity. Treatment with a siRNA targeting hTERT conjugated with bioreducible polyethylenimine(SS-PEI) reduced telomerase activity levels and proliferation of HepG2 cells in vitro. Also, when hTERT siRNA/SS-PEI was injected into HepG2 tumor bearing mice by intratumoral (i.t.) injection, tumor sizes were significantly smaller compared to the control siRNA treated tumors [102].
The CSN5 protein is the catalytic center of the mammalian COP9 signalosome (CSN) complex and plays a crucial role in cell proliferation and senescence. In particular, CSN5 binds to transcription factors c-Jun and JunD. It also regulates the stability and controls the function of numerous intracellular regulators of cell proliferation and/or apoptotic signaling such as MYC, c-Jun, JunD, NFkB, and p53 [103]. Generally, high expression of CSN5 has been found in numerous human cancers, suggesting that knockdown of CSN5 could be an effective anti-cancer strategy [104, 105]. Therapeutic efficacy of CSN5 gene knockdown was reported as early as 2011 [106]. In orthotopic mouse model of hepatocarcinoma, Huh-luc cells, were treated with either ß-galactosidase siRNA/SNALP or CSN5 siRNA/SNALP (2mg/kg) by intravenous injection. The CSN5 siRNA/SNALP effectively inhibited the hepatic tumor growth whereas the ß-galactosidase siRNA/SNALP injected group showed partial liver parenchyma. Tumor growth inhibition effect arising from CSN5 knockdown was driven by induction of apoptotic cell death and delay of cell cycle progression.
3.3. Prostate Cancer
Prostate cancer is the most frequent malignant cancer in men. Several therapies are recommended for prostate cancers including prostatectomy, anti-androgenic hormone therapy, chemotherapy and radiotherapy. However, these therapies often permanently lower the quality of life or result in negative impact to healthy organs. A frequent genetic lesion in prostate cancer is the loss of the PTEN tumor suppressor gene, the protein product of which antagonizes the PI3K pathway. However in clinical trials, PI3K inhibitors have showed little effect as monotherapies. Thus the area is wide open for new targets and therapies directed toward them. Notably, oncogenes related to proliferation and metastasis of prostate cancer, have been identified as potential targets for RNAi based prostate cancer therapy [107].
Myc is a transcription factor associated with various biological processes, including replication, transcription, protein synthesis, cell division, and more [108]. Overexpression of Myc is observed frequently in primary and metastatic prostate cancers. A recent study by the Catapano lab provides insight into the efficacy of Myc inhibition by siRNA in prostate cancer-stem like cells using in vitro and in vivo models of human prostate cancer. Treatment with Myc targeting siRNAs resulted in a reduction of stem-like properties such as self-renewal and tumor-initiation, while further inducing cell senescence in monolayer cultures of human prostate cancer cells. Tumor masses of Myc siRNA/jet-PEI treated mice group were almost completely suppressed, whereas the control group had a sharp increase in tumor growth [109].
Casein kinase II (CK2) is a serine/threonine kinase, which exists as tetramer of two α catalytic subunits and two ß regulatory subunits. CK2 is amplified in various cancer types and can activate and regulate the stability of several tumor suppressor proteins, oncogenes, and even key transcription factors like c-Myc, c-Jun, and NFκB [110, 111]. To test whether CK2 might be a suitable target for therapy, mice bearing human prostate cancer cell xenografts were treated with CK2 siRNA using nanocapsules. The treated mice resulted in about 50% less tumor mass and reduced tumor nodules compared to the vehicle treated animals [112].
As noted above prostate cancer is largely promoted by altered signaling in the androgen receptor (AR) and the PI3K/PKB/mTOR pathways. Notably, signaling in both pathways is coordinated by KLK4 (Kallikrein-related peptidase 4). Thus KLK4 has an important role in PCa progression via its role in regulating cell-cycle gene expression. Although KLK4 is expressed in the normal prostate gland, it is significantly more expressed in malignant prostate cancer. [113, 114]. Therefore, it is not surprising that LNCaP or VCaP prostate cancer cells treated with siRNA/liposome targeting KLK4 showed down-regulated AR signaling and KLK4 gene expression. Furthermore, the KLK4 siRNA treated mice showed a dramatic tumor regression of 90% [115].
Notch signaling is associated with multiple cellular processes including differentiation, proliferation, and apoptosis. Dysfunctional regulation of the Notch pathway has been demonstrated in a range of cancers. In prostate cancer cells, abundant Notch1 expression influences tumor invasion. Recent studies indicate that siRNA mediated Notch1 knockdown inhibited invasion and proliferation of prostate cancer cells [116]. Down-regulation of Notch1 expression by a Notch1 targeting siRNA/PSAM-protamine inhibited tumor growth and increased apoptosis in a LNCaP subcutaneous murine xenograft model [117].
3.4. Breast Cancer
Breast cancer is the most frequently diagnosed type of cancer and the leading cause of cancer related death in women [118]. Breast cancer can be divided into distinct subtypes by using either immunohistochemical (IHC) staining patterns for key receptors or, more recently, molecular profiling. The resulting subtypes display different clinical behaviors and responses to treatment [119]. There are many treatments for breast cancer including surgery, chemotherapy, and radiotherapy as well as the use of targeted therapies. Notably, inhibition of various genes that cause breast cancer through siRNA has been tested in animal models.
As one common way of subdividing breast cancers, tumors are classified by their expression of three receptor molecules: the estrogen receptor, the progesterone receptor, and the human epidermal growth factor receptor 2 (HER2). The estrogen receptor alpha (ER-α), a ligand-activated transcription factor, is one of two types of estrogen receptor. It plays a crucial role in regulation of the cell cycle progression of mammary epithelial cells. Approximately 70% of breast cancer cases are observed to be ER-α positive often with overexpression of estrogen receptors [120]. To test siRNA targeting the estrogen receptor, MCF-7 cells, an ER-α positive cell line, were used to create xenografts. Intravenous injection with ER-α targeted siRNA-encapsulated stealth nanocapsules composed of PEG-co-poly(ε-caprolactone-co-dodecyl ß-malate) reduced tumor growth and downregulated the level of ER-α [121].
Approximately 15 % of breast tumors are known as triple-negative breast cancer (TNBC), because they lack IHC expression in HER2, estrogen receptor, and progesterone receptor [122, 123]. Since the growth of TNBC is not dependent on the hormones and/or HER2 receptors, common therapies such as anti-hormonal or HER2 targeting therapies are ineffective. Goga et al. provided a potential target for TNBC treatment by discovering that inhibition of cyclin-dependent kinase 1(CDK1) induced synthetic lethality in TNBC overexpressing c-Myc. As a synthetic lethality-based TNBC treatment, CDK1 targeting siRNA were used with cationic lipid assisted poly(ethylene glycol)-b-poly(D,L-lactide) (PEG-PLA) nanoparticles (lipid NP) as the siRNA carrier . CDK siRNA/lipid NP delivered by systemic injection significantly reduced tumor growth in mice bearing c-Myc overexpressing SUM149 and BT549 xenografts without any systemic toxicity or innate immune response [124]. Obviously, siRNA directly targeting Myc described earlier could also be tried in this setting.
Another kinase target that has been examined in breast cancer is the ataxia-telangiectasia mutated (ATM) protein, a serine/threonine kinase that regulates DNA damage repair and cell cycle checkpoints and activates downstream signals including p53, CHK2, and BRCA following as DNA damage. For breast cancer therapy, an ATM targeting siRNA/porous silicon-based multistage vector (MSV) was administered in an orthotopic MDA-MB 231 mouse model. Biweekly treatment of ATM siRNA/MSV inhibited tumor growth and reduced ATM expression in IHC assay of tumor tissue [125].
Aberrant epigenetic regulation is frequently associated with the tumorigenic process. DNA methylation is a key epigenetic marker that serves as an important regulator of gene transcription. DNA methylation is organized and maintained by DNA methyltransferases (DNMTs). Aberrant patterns of DNA methylation have been identified in many types of human malignancies; for example, hypermethylation of tumor suppressor genes at CpG islands is associated with gene inactivation [126]. Jun Wang et al. reported that a DNMT targeted siRNA coupled with a fusion protein consisting of an anti-HER2 single-chain antibody fragment with a positively charged protamine carrier has successfully suppressed DNMTs in the HER2-expressing BT474 breast tumor model. Down regulation of DNMTs by siRNA induced re-expression of the RASSF1A tumor suppressor gene, which led to inhibition of tumor growth [127].
The osteopontin (OPN) protein binds to multiple cell surface receptors to induce cell adhesion and migration. OPN has been considered as a potential prognostic marker in breast cancer progression because elevated levels of OPN have been found in blood and plasma of patients with metastatic breast cancer. For this reason, suppression of OPN may be utilized as a therapeutic strategy. When mice bearing MDA-MB-231 xenografts were treated with an OPN targeting siRNA encapsulated in glycerol propoxylate triacrylate (GPT) and spermine (SPE) nanoparticles, significant inhibition of breast tumor growth with accompanying knockdown of OPN were observed [128].
3.5. Ovarian cancer
Ovarian cancer is one of the most common types of cancers in women and the leading cause of death in gynecologic cancers. Debulking surgery and chemotherapies with platinum-taxane drugs are generally used in the treatment of ovarian cancer. Despite treatment, the majority of ovarian cancer patients develop recurrant tumors that are resistant to chemotherapy. Consequently, there is an important need for alternative therapeutics for ovarian cancers [129, 130]. However, in contrast to the situation discussed above for TNBC, for ovarian cancers there is a relative dearth of genetically defined targets. One frequent genetic lesion is the loss of the tumor suppressor BRCA1, which has been attacked via PARP inhibitors. Genetic or epigenetic loss of expression of the tumor suppressor PTEN is also frequent, a lesion that can be attacked with PI3K inhibitors. However, there is a clear need for new targets and strategies in this disease.
Recently, Cheung et al. used genome-scale pooled shRNA screens in human cancer cell lines to define 54 overexpressed and essential genes in ovarian cancer that require further validation in vivo [131]. Among them, ID4, a helix-loop-helix (HLH) transcriptional regulator, is highly expressed in most primary ovarian cancers and is, moreover, overexpressed in 32% of high-grade serous ovarian cancers but not in normal tissues including ovary. In a flank xenograft tumor model using OVCAR-8 cells, both intravenous and intraperitoneal injection of ID4 siRNA in a tumor penetrating nanocomplex (TPN) lowered tumor burden compared to control groups. Histological analysis of tumor tissues from ID4 siRNA/TPN treated mice revealed suppression of ID4 expression levels and higher levels of apoptosis in tumor tissues [132].
Ephrin type-A receptor 2 (EphA2) is a tyrosine kinase receptor in the ephrin family that functions as an oncoprotein. EphA2 is highly expressed in many cancer types, but expressed at low levels in normal tissue in adults. This is particularly true for ovarian cancer, where approximately 70% of human ovarian tumors overexpress EphA2. Treatment of several orthotopic ovarian cancer models with paclitaxel and an EphA2 siRNA/DOPC liposome via intraperitoneal injection significantly reduced tumor growth by 81% in the case of a SKOV3ip1 model and by 48% in a HeyA8 model [133].
Enhancer of Zeste Homolog 2 (EZH2) functions as an epigenetic regulator of gene expression that works via histone methylation. Overexpression of EZH2 appears to be involved in cancer progression, functioning by silencing the expression of tumor suppressor genes via specific histone modifications [134]. Recently, EZH2 targeting siRNA were incorporated into chitosan nanoparticles and utilized along with docetaxel conjugated PLGA-PRINT nanoparticles as a combination treatment for ovarian cancer. The combination therapy resulted in 95% reduction in tumor weight and further reduced metastasis in orthotopic mice models of ovarian cancer [135].
CD44 is a cell-surface glycoprotein and a major receptor for hyaluronic acid. Hyaluronic acid is a main component of the peritoneum, where ovarian cancer metastases frequently occur. CD44 is involved in cancer progression and metastases but is not expressed in normal cells [136]. For ovarian cancer treatment, paclitaxel and CD44 targeting siRNA with a tumor specific targeting peptide conjugated dendrimer were intraperitoneally administered in mice bearing human cancer cells directly isolated from malignant ascites of patients with advanced ovarian carcinoma. The combination treatment group showed almost complete tumor inhibition of tumor growth compared to tumors in mice treated with paclitaxel and a control siRNA [137].
C-Jun-NH2-kinases (JNKs) are serine/threonine kinases that bind and phosphorylate c-Jun. They are members of the mitogen-activated protein kinase family that regulates cell proliferation, apoptosis, and differentiation. Continuous activation of JNKs leads to cancer initiation and progression [138]. Of the two major JNK isoforms, JNK1and JNK2, JNK1 specifically plays an important part in cell survival by controlling cell cycle arrest and apoptosis [139]. To determine whether JNK1 knockdown by siRNA is indeed capable of eliciting anti-ovarian tumor effects in vivo, JNK1 siRNA packaged in DOPC-liposomes was used to treat to HeyA8-bearing and SKOV3ip1-bearing nude mice in combination with docetaxel, the current standard of care. Significant decreases in tumor weight and the number of metastatic nodules were observed in the combination treatment group in comparison to the groups treated with either JNK1 siRNA/DOPC or docetaxel alone [140].
X-linked inhibitor of apoptosis protein (XIAP) is an anti-apoptotic protein that prevents apoptotic cell death in tumors. XIAP binds to caspase proteases that are primarily responsible for cell death and stops the proteolytic activation of caspases. Because deregulation of XIAP can contribute to cancer, high level of XIAP expression has been identified as a tumor marker. In SKOV-3 tumor bearing mice, administration of XIAP-siRNA/HER2-PLI (HER2 targeting PEI and PLL based copolymer) via tail vein injection significantly delayed tumor growth and increased survival time compared to the control groups [141].
4. Future prospects
Despite our ever-increasing knowledge of cancer, cancer is still the second leading cause of death, overall, and is predicted to become the leading killer as heart disease therapies improve. Thanks to a multitude of large scale sequencing efforts, numerous genetic alterations have been identified in tumors, opening the way for the generation of siRNA therapeutics targeting both the mutant genes and in lesions in cancer signaling pathways arising from these genetic defects. Small molecule or antibody drugs have proven quite effective for targeting certain cell surface and intracellular protein targets. Recently small molecules and antibodies for cancer immunotherapy have been shown to have strong antitumour effects [142, 143]. However, transcription factors and certain key oncoproteins such as Ras have proven difficult to access and block with conventional drug-based or antibody mediated approaches. In terms of “undruggable” disease targets, siRNA therapeutics have the potential to specifically target and silence almost any gene target [144]. In addition, their process of identifying and optimizing an siRNA for a target is relatively rapid and siRNAs are easy to synthesize [145]. These characteristics strongly support the need to develop siRNA therapeutics for cancer treatment.
Key challenges to siRNA usage in clinic lie both in selecting the best of this vast selection of possible targets and in optimizing delivery of the siRNA agents to individual tumors. Choice of target and delivery route are very important for enhancing therapeutic efficacy in cancer, and should be done in a manner designed to minimize side effects in normal tissue. Since delivery and target selection are perhaps the ultimate limitations for siRNA based therapy we will conclude with a few thoughts on these final challenges.
4.1. Delivery challenge
Problems with pharmacokinetics and delivery are known to limit the usefulness of many of the small molecule cancer therapeutics that have been developed in recent years. However, delivery of siRNA into target cells is even more challenging than delivery of conventional molecular drugs to deliver into target cells. Inherent characteristics of siRNAs, including negative charge, rigid structure, size, and stability, complicate their passive diffusion across the cell membrane; thus, endocytosis is the major mechanism for intracellular delivery of siRNA. Another challenge in siRNA therapy is the off-target silencing of unintended genes. While the design of an siRNA is meant to maximize knockdown of a specific gene target, nevertheless, marginal mismatch of the complementary sequence in mRNA is often tolerated, leading to non-specific knockdown of genes [146].
Exogenous siRNA triggers Toll-like receptor (TLR) mediated innate immune response in both sequence-dependent and independent manners. The TLR-3 pathway is activated by siRNA independent of sequence, whereas TLR-7 in dendritic cells and TLR-8 in monocytes are activated to produce proinflammatory cytokine in a sequence-specific manner [147-149]. Several modification strategies, including 2’-O-methyl chemical modification, have proved to avoid stimulation of the innate immune response; however, the mechanism behind the activation of the immune system still needs further explanation to help avoid side effects in clinical trials.
The pharmacokinetics and pharmacodynamics of siRNA based therapeutics in vivo are just beginning to be studied in the clinic. Optimal dosage levels, timing and duration are expected to vary depending on delivery and targeting strategy, choice of target genes, and disease. Thus our understanding of siRNA based therapies is in its infancy.
4.2. Targeting challenge
As can be seen in the previous sections on specific diseases, cancer is a target rich disease. A major question is which targets deserve the most attention, in the target rich environment that we have described. Here perhaps we can rely on a combination of our knowledge of the genetic basis of cancer and common sense to help make these difficult choices. Cancer is a genetic disease and many successful small molecule therapeutics have directly targeted key genetic lesions. Thus, we should strongly consider potential genetic targets but perhaps concentrate our efforts on targets that are difficult to reach with small molecules. For example, oncoproteins like c-Myc or KRAS are presented in mutated form in about 50% of human cancer types [150]. However, no drugs targeting either c-Myc or KRAS directly have been approved, mainly because of their chemically intractable characteristics. Therefore, RNAi therapeutics against these genes justifiably rank high on target lists.
Further targets are being identified that will allow marshalling of the immune system against tumors and blocking cancer support systems such as the vasculature that facilitate tumor growth. Immunotherapies targeting programmed cell death protein 1 (PD1, Nivolumab), programmed cell death ligand 1 (PDL1) and cytotoxic T lymphocyte antigen 4 (CTLA4, Ipilimumab) have shown exceptional success in melanoma, Hodgkin lymphoma, non-small-cell lung cancer (NSCLC) and bladder cancer, by blocking immune-inhibitory signals, thus enhancing the immune response against cancer [151]. SiRNA therapy is also suited for targeting immune cells because it can be modified to target a specific cell type and used for individual or multiple targets. Anti-tumor immunotherapies with siRNAs against immunosuppressive factors have been reported in dendritic cells [152-155], monocytes[156] and tumor associated macrophage [157] and have exhibited significant results in cancer treatment. In study by Dolina et al., Pdl1 was effectively silenced in Kuffer Cells via in vivo administration of PD-L1 siRNA encapsulated in lipidoid nanoparticles (LNP) into mice. Silencing of Pdl1 effectively enhanced the NK cell and CD8+ T cell intrahepatic accumulation, viral clearance, and CD8+ T cell memory to hepatotropic viral infection. This study demonstrated that transient knockdown of PD-L1 using siRNA directly on the disease-causing cell type might even have benefits when compared to monoclonal antibody usage [158].
Alternatively, a high priority should be placed on investigating multi-targeted gene therapy utilizing a combination of siRNAs to block several pathways. This approach parallels efforts to generate combinations of small molecule therapies, but can be more easily achieved using siRNAs. As mentioned earlier, different types of small RNAs (e.g., siRNA and miRNA) can be partnered in combination therapy [77].
Combination therapy with siRNAs and chemotherapeutic drugs is a credible alternative method to combat tumor heterogeneity and chemo-resistant tumors [159-161]. The combination therapy can overcome multi-drug resistance and improve drug therapeutic response. Amiji group tested combination treatment with two anti-apoptotic genes, bcl-2 and survivin, targeting siRNA and cisplatin to overcome drug resistance in NSCLC [162]. Overexpressed bcl-2 in NSCLC is involved in tumorigenesis and drug resistance as an activator of anti-apoptotic cellular defenses. Using cisplatin-resistant tumor-bearing mice, combination therapy exhibited more effective tumor growth inhibition compared to single therapies as proved by following % inhibition: control siRNA + cisplatin 29%, bcl-2 siRNA + cisplatin 58%, and survivin siRNA + cisplatin 52%. Furthermore, the combination of bcl-2 and survivin siRNA and cisplatin treatment suppressed tumor growth by 62%.
Finally, a combination approach using siRNA with a variety of cancer therapies such as chemotherapy, immunotherapy, radiation therapy, or photodynamic therapy may dramatically improve the efficacy of cancer therapy. In this strategy, each form of therapy can be used on targets particularly suited to the therapy type, such as small molecules inhibitors for kinase targets and siRNAs for targets that are structurally unsuited to small molecule attack. Moreover since different therapeutic modalities may trigger different forms of resistance mechanisms such as P-glycoprotein (P-gp), multidrug resistance-associated proteins (MRP1, MRP2) for small molecules drugs and other yet to be determined modes for siRNAs, such multimodal therapies may be harder for tumors to circumvent. It is our strong hope that by skillful delivery and careful target selection siRNA nanoparticles may take a prominent place in the armamentarium that is being assembled to treat the many diseases that constitute cancer.
Table 2.
siRNA Targets for Treatment of Major Cancers
| Cancer type | siRNA/target gene | Route | Animal model | siRNA modification/Delivery system | Ref. |
|---|---|---|---|---|---|
| Lung | RPN2 | inhalation | A549-luc-C8 cells via I.V. | naked PnkRNA and nKRNAs | [79] |
| C7orf24 | jet injection | EBC-1, S.C. | naked siRNA | [180] | |
| Mcl1 | Intratracheal | B16F10 or LLC, metastatic lung cancer | liposome | [71] | |
| CD31 | i.v. | LLC, metastatic lung cancer | loposome | [181] | |
| Bcl2 | i.v | B16, metastatic lung cancer | Protein, cationic bovine serum albumin (CBSA) | [182] | |
| NPT2b | inhalation | K-ras LA1 model | polymer | [183] | |
| MDM2 | i.v. | H2009, S.C. | Polymer | [83] | |
| Survivin/cyclin B1 | i.v. | TSA-Luc, metastatic lung cancer | stickty siRNA/polymer | [184] | |
| HDM2/C-Myc/VEFG | i.v. | H460 or A546 NSCLC, S.C. | liposome | [185] | |
| Survivin/Bcl-2+Cisplatin | i.v. | A549 resistant, S.C. | Polymer | [162] | |
| Liver | HDAC2 | i.v. | Huh7-luc+, orthotopic liver tumor | 2′-OMe modification / LNPs | [89] |
| Polo-like kinases (PLKs), | i.v. | induction by Diethyl nitrosamine (DEN) | fusogenic liposome | [186] | |
| β1-integrin | i.v. | MET/DN90-β-catenin-induced tumour | 2′-OMe-modification/ LNPs | [95] | |
| RRM2, adriamycin | i.v. | HepG2 Orthotopic HCC | liposome | [187] | |
| various siRNA (KRAS, NHP2L1, BIRC5,CDCA1,PSMA2, Aurora B, etc) | i.v. | HuH7 liver tumors | liposome | [188] | |
| Survivin | i.v. | Bel-7402, S.C. | RGD-Polymer | [98] | |
| Pokemon | i.v. | HepG2, S.C. | Cholesterol conjugation/Liposome | [101] | |
| hTERT | i.t. | HepG2, S.C. | Cationic Polymer | [102] | |
| RhoA | i.v. | SMMC-7721, S.C. | anti-EGFR Fab’-liposomes | [189] | |
| CSN5 | i.v. | Huh7, orthotopic transplantation | 2′-Ome modifications / LNPs | [106] | |
| COP1 | i.v. | HepG2, Huh7, orthotopic | 2′-Ome modifications / LNPs | [190] | |
| Prostate | HSP 27 | i.t. | PC-3, S.C. | amphiphilic dendrimer | [191] |
| survivin | i.t. | PC-3, S.C. | lipid+PEI hybrid nanocarrier (LPN) | [192] | |
| Bcl2 | i.v. | PC-3, Orthotopic | cholesterol conjugation/ lipid nanoplatform, | [193] | |
| Myc | i.p. | PC-3, S.C. | in vivo-jetPEI | [109] | |
| plk-1 | i.t. | PC-3 and LNCaP, S.C. | Aptamer | [175] | |
| YB- 1+ rapamycin | retro orbital | PC3, S.C. | trilayer polymeric micelle | [194] | |
| notch1 | i.v. | LNCaP, S.C. | anti-PSMA scFv fusion proteins | [117] | |
| HIF-1α+ Dox | i.v. | PC3, S.C. | Micellar nanoparticle (MNP) | [195] | |
| VEGF | i.v. | PC3, S.C. | thiolated siRNA/chitosan based polymer | [30] | |
| i.v. | PC3, S.C. | cyclodextrin modified polymer | [196] | ||
| i.v. | TRAMP C1, S.C. | pH-triggered amphiphilic polymer | [197] | ||
| AR | i.v. | LNCaP, S.C. | LNP | [198] | |
| Bcl-Xl+ cisplatin | i.v. | PC-3, S.C. | atelocollagen | [199] | |
| PKN3 | i.v. | DU1-45, S.C. / PC-3, Orthotopic | Atu027 | [200] | |
| AURKB, and EGFR | i.t. | PC3, S.C. | Hiperfect | [201] | |
| cyclin B1 | i.v. | PC3, S.C. | Amphipathic peptide carrier MPG-8 | [168] | |
| EZH2, and p110a plk-1+ paclitaxel Plk1 | i.v. | PC-3M-luc-C6, heart injection/bone metastasis | atelocollagen | [202] | |
| Breast | AURKB, and EGFR | i.v | MDA-MB-435s, S.C. | polymer based Micelleplex | [203] |
| cyclin B1 | i.v. | MDA-MB-435s, S.C. | cationic lipid assisted PEG-PLA nanoparticles | [204] | |
| CDK1 | i.v | SUM149 or BT549, S.C. | cationic lipid assisted PEG-PLA nanoparticles | [124] | |
| Erα | i.v. | MCF-7, S.C. | polymer nanocapsule | [121] | |
| DNMTs | i.v. | BT474, S.C. | anti-Her2 ScFv-protamine (F5-P) | [127] | |
| RhoA | i.t. | MDA-MB-231, S.C. | Cytofectin™ Transfection Reagent | [205] | |
| ataxia-telangiectasia mutated (ATM) | i.v. | MDA-MB-231, orthotopic | porous silicon-based multistage vector (MSV) | [125] | |
| osteopontin (OPN) | i.t. | MDA-MB-231, S.C. | glycerol propoxylate triacrylate-spermine copolymer | [128] | |
| Mcl-1 +RPS6KA5 | i.t. | MDA435_WT and MDA435_resistant (R), S.C. | lipid-substituted polymer | [206] | |
| P-gp +DOX | i.v. | MCF7/A, S.C. | RGD-Liposome | [207] | |
| MnSOD | _ | MCF7-BK-TR cells co-transplanted with siRNA/NPs | PAMAM dendrimer based nanoparticles | [208] | |
| Raf-1 | i.t. | MDA-MB-435, S.C. | histidine–lysine carrier | [209] | |
| VEGF | i.v. | MCF-7 and HT1080, S.C. | cholesterol conjugation/ high density lipoprotein | [210] | |
| Ovary | EZH2+Docetaxel | i.v. | HeyA8 or SKOV3ip1, Orthotopic | PLGA-PRINT nanoparticles | [210] |
| VEGF | i.t. | A2780, S.C. | arginine-grafted polymer-microbubble | [211] | |
| EphA2+pacritaxel | i.p. | HeyA8, orthotopic | neutral liposomes | [133] | |
| Akt+pacritaxel | i.t. | SKOV-3, S.C. | dendrimer | [212] | |
| XIAP | i.v. | SKOV-3, S.C. | ternary copolymer | [141] | |
| Vasohibin2 | i.v. | DISS and SKOV-3, S.C. | atelocollagen | [213] | |
| CD44+paclitaxel | i.p. | human ascitic cells, S.C. | Dendrimer | [137] | |
| POSTN, FAK, and PLXDC1 | i.v | SKOV3ip1, HeyA8, and A2780, Orthotopic | RGD-Labeled Chitosan Nanoparticles | [214] | |
| EphA2 | i.v. | SKOV3ip1 or HeyA8, Orthotopic | Mesoporous Silicon Particles | [215] | |
| c-Jun-NH2-kinases (JNK)+Docetaxel | i.p. | HeyA8 or SKOV3ip1, Orthotopic | liposomes | [140] | |
| KLF6-SV1+Cisplatin | i.p. | SKOV3-Luc, Orthotopic | Accell chemically synthesized siRNAs (Dharmacon) | [216] | |
| FAK+Docetaxel | i.p. | SKOV3ip1, A2780-CP20, and HeyA8MDR, Orthotopic | Neutral liposome | [217] |
Acknowledgements
This study was supported by Global Innovative Research Center (GiRC) project (2012K1A1A2A01055811) of the National Research Foundation of Korea, the Intramural Research Program (Global RNAi Carrier Initiative) of KIST, and a grant from the Women's Cancers Program of Dana Farber Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nature reviews. Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 4.Sashital DG, Doudna JA. Structural insights into RNA interference. Current opinion in structural biology. 2010;20:90–97. doi: 10.1016/j.sbi.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 6.Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nature chemical biology. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iorns E, Lord CJ, Turner N, Ashworth A. Utilizing RNA interference to enhance cancer drug discovery. Nature reviews. Drug discovery. 2007;6:556–568. doi: 10.1038/nrd2355. [DOI] [PubMed] [Google Scholar]
- 8.Van de Veire S, Stalmans I, Heindryckx F, Oura H, Tijeras-Raballand A, Schmidt T, Loges S, Albrecht I, Jonckx B, Vinckier S, Van Steenkiste C, Tugues S, Rolny C, De Mol M, Dettori D, Hainaud P, Coenegrachts L, Contreres JO, Van Bergen T, Cuervo H, Xiao WH, Le Henaff C, Buysschaert I, Kharabi Masouleh B, Geerts A, Schomber T, Bonnin P, Lambert V, Haustraete J, Zacchigna S, Rakic JM, Jimenez W, Noel A, Giacca M, Colle I, Foidart JM, Tobelem G, Morales-Ruiz M, Vilar J, Maxwell P, Vinores SA, Carmeliet G, Dewerchin M, Claesson-Welsh L, Dupuy E, Van Vlierberghe H, Christofori G, Mazzone M, Detmar M, Collen D, Carmeliet P. Further pharmacological and genetic evidence for the efficacy of PlGF inhibition in cancer and eye disease. Cell. 2010;141:178–190. doi: 10.1016/j.cell.2010.02.039. [DOI] [PubMed] [Google Scholar]
- 9.Zhao M, Sun J, Zhao Z. Synergetic regulatory networks mediated by oncogene-driven microRNAs and transcription factors in serous ovarian cancer. Molecular bioSystems. 2013;9:3187–3198. doi: 10.1039/c3mb70172g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doebele RC. Targeted therapies: Time to shift the burden of proof for oncogene-positive cancer? Nature reviews. Clinical oncology. 2013;10:492–493. doi: 10.1038/nrclinonc.2013.135. [DOI] [PubMed] [Google Scholar]
- 11.Collins I, Workman P. New approaches to molecular cancer therapeutics. Nature chemical biology. 2006;2:689–700. doi: 10.1038/nchembio840. [DOI] [PubMed] [Google Scholar]
- 12.Resnier P, Montier T, Mathieu V, Benoit JP, Passirani C. A review of the current status of siRNA nanomedicines in the treatment of cancer. Biomaterials. 2013;34:6429–6443. doi: 10.1016/j.biomaterials.2013.04.060. [DOI] [PubMed] [Google Scholar]
- 13.DeVincenzo J, Cehelsky JE, Alvarez R, Elbashir S, Harborth J, Toudjarska I, Nechev L, Murugaiah V, Van Vliet A, Vaishnaw AK, Meyers R. Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV) Antiviral research. 2008;77:225–231. doi: 10.1016/j.antiviral.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 14.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nature reviews. Drug discovery. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behlke MA. Progress towards in vivo use of siRNAs. Molecular therapy : the journal of the American Society of Gene Therapy. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gewirtz AM. On future's doorstep: RNA interference and the pharmacopeia of tomorrow. The Journal of clinical investigation. 2007;117:3612–3614. doi: 10.1172/JCI34274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nature reviews. Genetics. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 18.Aigner A. Nonviral in vivo delivery of therapeutic small interfering RNAs. Current opinion in molecular therapeutics. 2007;9:345–352. [PubMed] [Google Scholar]
- 19.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, Marshall W, Khvorova A, Linsley PS. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. Rna. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marques JT, Williams BR. Activation of the mammalian immune system by siRNAs. Nature biotechnology. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- 21.Lee SJ, Son S, Yhee JY, Choi K, Kwon IC, Kim SH, Kim K. Structural modification of siRNA for efficient gene silencing. Biotechnology advances. 2013;31:491–503. doi: 10.1016/j.biotechadv.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 23.Blidner RA, Hammer RP, Lopez MJ, Robinson SO, Monroe WT. Fully 2′-deoxy-2′-fluoro substituted nucleic acids induce RNA interference in mammalian cell culture. Chemical biology & drug design. 2007;70:113–122. doi: 10.1111/j.1747-0285.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- 24.Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends in biotechnology. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 25.Dorn G, Patel S, Wotherspoon G, Hemmings-Mieszczak M, Barclay J, Natt FJ, Martin P, Bevan S, Fox A, Ganju P, Wishart W, Hall J. siRNA relieves chronic neuropathic pain. Nucleic acids research. 2004;32:e49. doi: 10.1093/nar/gnh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. Journal of medicinal chemistry. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 27.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 28.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, Vaish N, Zinnen S, Vargeese C, Bowman K, Shaffer CS, Jeffs LB, Judge A, MacLachlan I, Polisky B. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nature biotechnology. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 29.Braasch DA, Paroo Z, Constantinescu A, Ren G, Oz OK, Mason RP, Corey DR. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorganic & medicinal chemistry letters. 2004;14:1139–1143. doi: 10.1016/j.bmcl.2003.12.074. [DOI] [PubMed] [Google Scholar]
- 30.Lee SJ, Huh MS, Lee SY, Min S, Lee S, Koo H, Chu JU, Lee KE, Jeon H, Choi Y, Choi K, Byun Y, Jeong SY, Park K, Kim K, Kwon IC. Tumor-homing poly-siRNA/glycol chitosan self-cross-linked nanoparticles for systemic siRNA delivery in cancer treatment. Angewandte Chemie. 2012;51:7203–7207. doi: 10.1002/anie.201201390. [DOI] [PubMed] [Google Scholar]
- 31.Lee SJ, Yhee JY, Kim SH, Kwon IC, Kim K. Biocompatible gelatin nanoparticles for tumor-targeted delivery of polymerized siRNA in tumor-bearing mice. Journal of controlled release : official journal of the Controlled Release Society. 2013;172:358–366. doi: 10.1016/j.jconrel.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Lee SY, Huh MS, Lee S, Lee SJ, Chung H, Park JH, Oh YK, Choi K, Kim K, Kwon IC. Stability and cellular uptake of polymerized siRNA (poly-siRNA)/polyethylenimine (PEI) complexes for efficient gene silencing. Journal of controlled release : official journal of the Controlled Release Society. 2010;141:339–346. doi: 10.1016/j.jconrel.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Son S, Song S, Lee SJ, Min S, Kim SA, Yhee JY, Huh MS, Chan Kwon I, Jeong SY, Byun Y, Kim SH, Kim K. Self-crosslinked human serum albumin nanocarriers for systemic delivery of polymerized siRNA to tumors. Biomaterials. 2013;34:9475–9485. doi: 10.1016/j.biomaterials.2013.08.085. [DOI] [PubMed] [Google Scholar]
- 34.Yhee JY, Lee SJ, Lee S, Song S, Min HS, Kang SW, Son S, Jeong SY, Kwon IC, Kim SH, Kim K. Tumor-targeting transferrin nanoparticles for systemic polymerized siRNA delivery in tumor-bearing mice. Bioconjugate chemistry. 2013;24:1850–1860. doi: 10.1021/bc400226b. [DOI] [PubMed] [Google Scholar]
- 35.Yoon HY, Kim HR, Saravanakumar G, Heo R, Chae SY, Um W, Kim K, Kwon IC, Lee JY, Lee DS, Park JC, Park JH. Bioreducible hyaluronic acid conjugates as siRNA carrier for tumor targeting. Journal of controlled release : official journal of the Controlled Release Society. 2013;172:653–661. doi: 10.1016/j.jconrel.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug discovery today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Li Z, Han Y, Liang LH, Ji A. Nanoparticle-based delivery system for application of siRNA in vivo. Current drug metabolism. 2010;11:182–196. doi: 10.2174/138920010791110863. [DOI] [PubMed] [Google Scholar]
- 38.Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Molecular pharmaceutics. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 39.Davis ME, Brewster ME. Cyclodextrin-based pharmaceutics: past, present and future. Nature reviews. Drug discovery. 2004;3:1023–1035. doi: 10.1038/nrd1576. [DOI] [PubMed] [Google Scholar]
- 40.Han HD, Mora EM, Roh JW, Nishimura M, Lee SJ, Stone RL, Bar-Eli M, Lopez-Berestein G, Sood AK. Chitosan hydrogel for localized gene silencing. Cancer biology & therapy. 2011;11:839–845. doi: 10.4161/cbt.11.9.15185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Z, Yang C, Song W, Wang Q, Kjems J, Gao S. Chitosan hydrogel as siRNA vector for prolonged gene silencing. Journal of nanobiotechnology. 2014;12:23. doi: 10.1186/1477-3155-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salva E, Kabasakal L, Eren F, Ozkan N, Cakalagaoglu F, Akbuga J. Local delivery of chitosan/VEGF siRNA nanoplexes reduces angiogenesis and growth of breast cancer in vivo. Nucleic acid therapeutics. 2012;22:40–48. doi: 10.1089/nat.2011.0312. [DOI] [PubMed] [Google Scholar]
- 43.de Martimprey H, Bertrand JR, Fusco A, Santoro M, Couvreur P, Vauthier C, Malvy C. siRNA nanoformulation against the ret/PTC1 junction oncogene is efficient in an in vivo model of papillary thyroid carcinoma. Nucleic acids research. 2008;36:e2. doi: 10.1093/nar/gkm1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochiya T, Nagahara S, Sano A, Itoh H, Terada M. Biomaterials for gene delivery: atelocollagen-mediated controlled release of molecular medicines. Current gene therapy. 2001;1:31–52. doi: 10.2174/1566523013348887. [DOI] [PubMed] [Google Scholar]
- 45.Inaba S, Nagahara S, Makita N, Tarumi Y, Ishimoto T, Matsuo S, Kadomatsu K, Takei Y. Atelocollagen-mediated systemic delivery prevents immunostimulatory adverse effects of siRNA in mammals. Molecular therapy : the journal of the American Society of Gene Therapy. 2012;20:356–366. doi: 10.1038/mt.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu ZW, Chien CT, Liu CY, Yan JY, Lin SY. Recent progress in copolymer-mediated siRNA delivery. Journal of drug targeting. 2012;20:551–560. doi: 10.3109/1061186X.2012.699057. [DOI] [PubMed] [Google Scholar]
- 47.DeRouchey J, Schmidt C, Walker GF, Koch C, Plank C, Wagner E, Radler JO. Monomolecular assembly of siRNA and poly(ethylene glycol)-peptide copolymers. Biomacromolecules. 2008;9:724–732. doi: 10.1021/bm7011482. [DOI] [PubMed] [Google Scholar]
- 48.Malek A, Czubayko F, Aigner A. PEG grafting of polyethylenimine (PEI) exerts different effects on DNA transfection and siRNA-induced gene targeting efficacy. Journal of drug targeting. 2008;16:124–139. doi: 10.1080/10611860701849058. [DOI] [PubMed] [Google Scholar]
- 49.Mao S, Neu M, Germershaus O, Merkel O, Sitterberg J, Bakowsky U, Kissel T. Influence of polyethylene glycol chain length on the physicochemical and biological properties of poly(ethylene imine)-graft-poly(ethylene glycol) block copolymer/SiRNA polyplexes. Bioconjugate chemistry. 2006;17:1209–1218. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- 50.Lee SH, Mok H, Lee Y, Park TG. Self-assembled siRNA-PLGA conjugate micelles for gene silencing. Journal of controlled release : official journal of the Controlled Release Society. 2011;152:152–158. doi: 10.1016/j.jconrel.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 51.Kim SI, Shin D, Choi TH, Lee JC, Cheon GJ, Kim KY, Park M, Kim M. Systemic and specific delivery of small interfering RNAs to the liver mediated by apolipoprotein A-I. Molecular therapy : the journal of the American Society of Gene Therapy. 2007;15:1145–1152. doi: 10.1038/sj.mt.6300168. [DOI] [PubMed] [Google Scholar]
- 52.Yagi N, Manabe I, Tottori T, Ishihara A, Ogata F, Kim JH, Nishimura S, Fujiu K, Oishi Y, Itaka K, Kato Y, Yamauchi M, Nagai R. A nanoparticle system specifically designed to deliver short interfering RNA inhibits tumor growth in vivo. Cancer research. 2009;69:6531–6538. doi: 10.1158/0008-5472.CAN-08-3945. [DOI] [PubMed] [Google Scholar]
- 53.Geisbert TW, Hensley LE, Kagan E, Yu EZ, Geisbert JB, Daddario-DiCaprio K, Fritz EA, Jahrling PB, McClintock K, Phelps JR, Lee AC, Judge A, Jeffs LB, MacLachlan I. Postexposure protection of guinea pigs against a lethal ebola virus challenge is conferred by RNA interference. The Journal of infectious diseases. 2006;193:1650–1657. doi: 10.1086/504267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, McClintock K, Nechev LV, Palmer LR, Racie T, Rohl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, MacLachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 55.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, Zimmermann T, Koteliansky V, Manoharan M, Stoffel M. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nature biotechnology. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 56.Choi SW, Lee SH, Mok H, Park TG. Multifunctional siRNA delivery system: polyelectrolyte complex micelles of six-arm PEG conjugate of siRNA and cell penetrating peptide with crosslinked fusogenic peptide. Biotechnology progress. 2010;26:57–63. doi: 10.1002/btpr.310. [DOI] [PubMed] [Google Scholar]
- 57.Moschos SA, Jones SW, Perry MM, Williams AE, Erjefalt JS, Turner JJ, Barnes PJ, Sproat BS, Gait MJ, Lindsay MA. Lung delivery studies using siRNA conjugated to TAT(48-60) and penetratin reveal peptide induced reduction in gene expression and induction of innate immunity. Bioconjugate chemistry. 2007;18:1450–1459. doi: 10.1021/bc070077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong DJ, Hurvitz SA. Recent advances in the development of anti-HER2 antibodies and antibody-drug conjugates. Annals of translational medicine. 2014;2:122. doi: 10.3978/j.issn.2305-5839.2014.08.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baeumer S, Baeumer N, Appel N, Terheyden L, Fremerey J, Schelhaas S, Wardelmann E, Buchholz F, Berdel WE, Muller-Tidow C. Antibody-mediated delivery of anti-KRAS-siRNA in vivo overcomes therapy resistance in colon cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015 doi: 10.1158/1078-0432.CCR-13-2017. [DOI] [PubMed] [Google Scholar]
- 60.Cerchia L, de Franciscis V. Targeting cancer cells with nucleic acid aptamers. Trends in biotechnology. 2010;28:517–525. doi: 10.1016/j.tibtech.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 61.Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, 2nd, Giangrande PH. Systemic administration of optimized aptamersiRNA chimeras promotes regression of PSMA-expressing tumors. Nature biotechnology. 2009;27:839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McLeod HL. Cancer pharmacogenomics: early promise, but concerted effort needed. Science. 2013;339:1563–1566. doi: 10.1126/science.1234139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farooqi AA, Rehman ZU, Muntane J. Antisense therapeutics in oncology: current status. OncoTargets and therapy. 2014;7:2035–2042. doi: 10.2147/OTT.S49652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walther W, Schlag PM. Current status of gene therapy for cancer. Current opinion in oncology. 2013;25:659–664. doi: 10.1097/CCO.0000000000000004. [DOI] [PubMed] [Google Scholar]
- 66.Bora RS, Gupta D, Mukkur TK, Saini KS. RNA interference therapeutics for cancer: challenges and opportunities (review) Molecular medicine reports. 2012;6:9–15. doi: 10.3892/mmr.2012.871. [DOI] [PubMed] [Google Scholar]
- 67.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21- nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 68.Miele E, Spinelli GP, Miele E, Di Fabrizio E, Ferretti E, Tomao S, Gulino A. Nanoparticle-based delivery of small interfering RNA: challenges for cancer therapy. International journal of nanomedicine. 2012;7:3637–3657. doi: 10.2147/IJN.S23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clinic proceedings. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herbst RS, Heymach JV, Lippman SM. Lung cancer. The New England journal of medicine. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shim G, Choi HW, Lee S, Choi J, Yu YH, Park DE, Choi Y, Kim CW, Oh YK. Enhanced intrapulmonary delivery of anticancer siRNA for lung cancer therapy using cationic ethylphosphocholine-based nanolipoplexes. Molecular therapy : the journal of the American Society of Gene Therapy. 2013;21:816–824. doi: 10.1038/mt.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lam JK, Liang W, Chan HK. Pulmonary delivery of therapeutic siRNA. Advanced drug delivery reviews. 2012;64:1–15. doi: 10.1016/j.addr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Merkel OM, Rubinstein I, Kissel T. siRNA delivery to the lung: what's new? Advanced drug delivery reviews. 2014;75:112–128. doi: 10.1016/j.addr.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. The FEBS journal. 2010;277:301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 75.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 76.Takahashi M, Chiyo T, Okada T, Hohjoh H. Specific inhibition of tumor cells by oncogenic EGFR specific silencing by RNA interference. PloS one. 2013;8:e73214. doi: 10.1371/journal.pone.0073214. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Xue W, Dahlman JE, Tammela T, Khan OF, Sood S, Dave A, Cai W, Chirino LM, Yang GR, Bronson R, Crowley DG, Sahay G, Schroeder A, Langer R, Anderson DG, Jacks T. Small RNA combination therapy for lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E3553–3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurashige J, Watanabe M, Iwatsuki M, Kinoshita K, Saito S, Nagai Y, Ishimoto T, Baba Y, Mimori K, Baba H. RPN2 expression predicts response to docetaxel in oesophageal squamous cell carcinoma. British journal of cancer. 2012;107:1233–1238. doi: 10.1038/bjc.2012.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fujita Y, Takeshita F, Mizutani T, Ohgi T, Kuwano K, Ochiya T. A novel platform to enable inhaled naked RNAi medicine for lung cancer. Scientific reports. 2013;3:3325. doi: 10.1038/srep03325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 81.Iwakuma T, Lozano G. MDM2, an introduction. Molecular cancer research : MCR. 2003;1:993–1000. [PubMed] [Google Scholar]
- 82.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS letters. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 83.Yu H, Zou Y, Jiang L, Yin Q, He X, Chen L, Zhang Z, Gu W, Li Y. Induction of apoptosis in non-small cell lung cancer by downregulation of MDM2 using pH-responsive PMPC-b-PDPA/siRNA complex nanoparticles. Biomaterials. 2013;34:2738–2747. doi: 10.1016/j.biomaterials.2012.12.042. [DOI] [PubMed] [Google Scholar]
- 84.Schutte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma--epidemiological trends and risk factors. Digestive diseases. 2009;27:80–92. doi: 10.1159/000218339. [DOI] [PubMed] [Google Scholar]
- 85.Bartosch B, Thimme R, Blum HE, Zoulim F. Hepatitis C virus-induced hepatocarcinogenesis. Journal of hepatology. 2009;51:810–820. doi: 10.1016/j.jhep.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 86.Sehgal A, Vaishnaw A, Fitzgerald K. Liver as a target for oligonucleotide therapeutics. Journal of hepatology. 2013;59:1354–1359. doi: 10.1016/j.jhep.2013.05.045. [DOI] [PubMed] [Google Scholar]
- 87.Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Molecular oncology. 2007;1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kramer OH. HDAC2: a critical factor in health and disease. Trends in pharmacological sciences. 2009;30:647–655. doi: 10.1016/j.tips.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 89.Lee YH, Seo D, Choi KJ, Andersen JB, Won MA, Kitade M, Gomez-Quiroz LE, Judge AD, Marquardt JU, Raggi C, Conner EA, MacLachlan I, Factor VM, Thorgeirsson SS. Antitumor effects in hepatocarcinoma of isoform-selective inhibition of HDAC2. Cancer research. 2014;74:4752–4761. doi: 10.1158/0008-5472.CAN-13-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park CC, Zhang H, Pallavicini M, Gray JW, Baehner F, Park CJ, Bissell MJ. Beta1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer research. 2006;66:1526–1535. doi: 10.1158/0008-5472.CAN-05-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aoudjit F, Vuori K. Integrin signaling inhibits paclitaxel-induced apoptosis in breast cancer cells. Oncogene. 2001;20:4995–5004. doi: 10.1038/sj.onc.1204554. [DOI] [PubMed] [Google Scholar]
- 92.Speicher T, Siegenthaler B, Bogorad RL, Ruppert R, Petzold T, Padrissa-Altes S, Bachofner M, Anderson DG, Koteliansky V, Fassler R, Werner S. Knockdown and knockout of beta1-integrin in hepatocytes impairs liver regeneration through inhibition of growth factor signalling. Nature communications. 2014;5:3862. doi: 10.1038/ncomms4862. [DOI] [PubMed] [Google Scholar]
- 93.Tward AD, Jones KD, Yant S, Cheung ST, Fan ST, Chen X, Kay MA, Wang R, Bishop JM. Distinct pathways of genomic progression to benign and malignant tumors of the liver. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14771–14776. doi: 10.1073/pnas.0706578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stauffer JK, Scarzello AJ, Andersen JB, De Kluyver RL, Back TC, Weiss JM, Thorgeirsson SS, Wiltrout RH. Coactivation of AKT and beta-catenin in mice rapidly induces formation of lipogenic liver tumors. Cancer research. 2011;71:2718–2727. doi: 10.1158/0008-5472.CAN-10-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bogorad RL, Yin H, Zeigerer A, Nonaka H, Ruda VM, Zerial M, Anderson DG, Koteliansky V. Nanoparticle-formulated siRNA targeting integrins inhibits hepatocellular carcinoma progression in mice. Nature communications. 2014;5:3869. doi: 10.1038/ncomms4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nature medicine. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 97.Ito T, Shiraki K, Sugimoto K, Yamanaka T, Fujikawa K, Ito M, Takase K, Moriyama M, Kawano H, Hayashida M, Nakano T, Suzuki A. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology. 2000;31:1080–1085. doi: 10.1053/he.2000.6496. [DOI] [PubMed] [Google Scholar]
- 98.Wu C, Gong F, Pang P, Shen M, Zhu K, Cheng D, Liu Z, Shan H. An RGD-modified MRI-visible polymeric vector for targeted siRNA delivery to hepatocellular carcinoma in nude mice. PloS one. 2013;8:e66416. doi: 10.1371/journal.pone.0066416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maeda T, Hobbs RM, Pandolfi PP. The transcription factor Pokemon: a new key player in cancer pathogenesis. Cancer research. 2005;65:8575–8578. doi: 10.1158/0008-5472.CAN-05-1055. [DOI] [PubMed] [Google Scholar]
- 100.Apostolopoulou K, Pateras IS, Evangelou K, Tsantoulis PK, Liontos M, Kittas C, Tiniakos DG, Kotsinas A, Cordon-Cardo C, Gorgoulis VG. Gene amplification is a relatively frequent event leading to ZBTB7A (Pokemon) overexpression in non-small cell lung cancer. The Journal of pathology. 2007;213:294–302. doi: 10.1002/path.2222. [DOI] [PubMed] [Google Scholar]
- 101.Ding Y, Wang W, Feng M, Wang Y, Zhou J, Ding X, Zhou X, Liu C, Wang R, Zhang Q. A biomimetic nanovector-mediated targeted cholesterol-conjugated siRNA delivery for tumor gene therapy. Biomaterials. 2012;33:8893–8905. doi: 10.1016/j.biomaterials.2012.08.057. [DOI] [PubMed] [Google Scholar]
- 102.Xia W, Wang P, Lin C, Li Z, Gao X, Wang G, Zhao X. Bioreducible polyethylenimine- delivered siRNA targeting human telomerase reverse transcriptase inhibits HepG2 cell growth in vitro and in vivo. Journal of controlled release : official journal of the Controlled Release Society. 2012;157:427–436. doi: 10.1016/j.jconrel.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 103.Yoshida A, Yoneda-Kato N, Kato JY. CSN5 specifically interacts with CDK2 and controls senescence in a cytoplasmic cyclin E-mediated manner. Scientific reports. 2013;3:1054. doi: 10.1038/srep01054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fukumoto A, Ikeda N, Sho M, Tomoda K, Kanehiro H, Hisanaga M, Tsurui Y, Tsutsumi M, Kato JY, Nakajima Y. Prognostic significance of localized p27Kip1 and potential role of Jab1/CSN5 in pancreatic cancer. Oncology reports. 2004;11:277–284. [PubMed] [Google Scholar]
- 105.Ivan D, Diwan AH, Esteva FJ, Prieto VG. Expression of cell cycle inhibitor p27Kip1 and its inactivator Jab1 in melanocytic lesions. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2004;17:811–818. doi: 10.1038/modpathol.3800123. [DOI] [PubMed] [Google Scholar]
- 106.Lee YH, Judge AD, Seo D, Kitade M, Gomez-Quiroz LE, Ishikawa T, Andersen JB, Kim BK, Marquardt JU, Raggi C, Avital I, Conner EA, MacLachlan I, Factor VM, Thorgeirsson SS. Molecular targeting of CSN5 in human hepatocellular carcinoma: a mechanism of therapeutic response. Oncogene. 2011;30:4175–4184. doi: 10.1038/onc.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guo J, Evans JC, O'Driscoll CM. Delivering RNAi therapeutics with non-viral technology: a promising strategy for prostate cancer? Trends in molecular medicine. 2013;19:250–261. doi: 10.1016/j.molmed.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 108.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Civenni G, Malek A, Albino D, Garcia-Escudero R, Napoli S, Di Marco S, Pinton S, Sarti M, Carbone GM, Catapano CV. RNAi-mediated silencing of Myc transcription inhibits stem-like cell maintenance and tumorigenicity in prostate cancer. Cancer research. 2013;73:6816–6827. doi: 10.1158/0008-5472.CAN-13-0615. [DOI] [PubMed] [Google Scholar]
- 110.Litchfield DW. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. The Biochemical journal. 2003;369:1–15. doi: 10.1042/BJ20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guerra B, Issinger OG. Protein kinase CK2 in human diseases. Current medicinal chemistry. 2008;15:1870–1886. doi: 10.2174/092986708785132933. [DOI] [PubMed] [Google Scholar]
- 112.Trembley JH, Unger GM, Korman VL, Abedin MJ, Nacusi LP, Vogel RI, Slaton JW, Kren BT, Ahmed K. Tenfibgen ligand nanoencapsulation delivers bi-functional anti-CK2 RNAi oligomer to key sites for prostate cancer targeting using human xenograft tumors in mice. PloS one. 2014;9:e109970. doi: 10.1371/journal.pone.0109970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xi Z, Klokk TI, Korkmaz K, Kurys P, Elbi C, Risberg B, Danielsen H, Loda M, Saatcioglu F. Kallikrein 4 is a predominantly nuclear protein and is overexpressed in prostate cancer. Cancer research. 2004;64:2365–2370. doi: 10.1158/0008-5472.can-03-2025. [DOI] [PubMed] [Google Scholar]
- 114.Lai J, Lehman ML, Dinger ME, Hendy SC, Mercer TR, Seim I, Lawrence MG, Mattick JS, Clements JA, Nelson CC. A variant of the KLK4 gene is expressed as a cis sense-antisense chimeric transcript in prostate cancer cells. Rna. 2010;16:1156–1166. doi: 10.1261/rna.2019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jin Y, Qu S, Tesikova M, Wang L, Kristian A, Maelandsmo GM, Kong H, Zhang T, Jeronimo C, Teixeira MR, Yuca E, Tekedereli I, Gorgulu K, Alpay N, Sood AK, Lopez-Berestein G, Danielsen HE, Ozpolat B, Saatcioglu F. Molecular circuit involving KLK4 integrates androgen and mTOR signaling in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2572–2581. doi: 10.1073/pnas.1304318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang Z, Li Y, Banerjee S, Kong D, Ahmad A, Nogueira V, Hay N, Sarkar FH. Down-regulation of Notch-1 and Jagged-1 inhibits prostate cancer cell growth, migration and invasion, and induces apoptosis via inactivation of Akt, mTOR, and NF-kappaB signaling pathways. Journal of cellular biochemistry. 2010;109:726–736. doi: 10.1002/jcb.22451. [DOI] [PubMed] [Google Scholar]
- 117.Su Y, Yu L, Liu N, Guo Z, Wang G, Zheng J, Wei M, Wang H, Yang AG, Qin W, Wen W. PSMA specific single chain antibody-mediated targeted knockdown of Notch1 inhibits human prostate cancer cell proliferation and tumor growth. Cancer letters. 2013;338:282–291. doi: 10.1016/j.canlet.2013.05.035. [DOI] [PubMed] [Google Scholar]
- 118.Zhang J, Li X, Huang L. Non-viral nanocarriers for siRNA delivery in breast cancer. Journal of controlled release : official journal of the Controlled Release Society. 2014;190:440–450. doi: 10.1016/j.jconrel.2014.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO, Gelmon K. Metastatic behavior of breast cancer subtypes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:3271–3277. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 120.Howell A, Dowsett M. Endocrinology and hormone therapy in breast cancer: aromatase inhibitors versus antioestrogens. Breast cancer research : BCR. 2004;6:269–274. doi: 10.1186/bcr945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bouclier C, Moine L, Hillaireau H, Marsaud V, Connault E, Opolon P, Couvreur P, Fattal E, Renoir JM. Physicochemical characteristics and preliminary in vivo biological evaluation of nanocapsules loaded with siRNA targeting estrogen receptor alpha. Biomacromolecules. 2008;9:2881–2890. doi: 10.1021/bm800664c. [DOI] [PubMed] [Google Scholar]
- 122.Bosch A, Eroles P, Zaragoza R, Vina JR, Lluch A. Triple-negative breast cancer: molecular features, pathogenesis, treatment and current lines of research. Cancer treatment reviews. 2010;36:206–215. doi: 10.1016/j.ctrv.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 123.Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52:108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- 124.Liu Y, Zhu YH, Mao CQ, Dou S, Shen S, Tan ZB, Wang J. Triple negative breast cancer therapy with CDK1 siRNA delivered by cationic lipid assisted PEG-PLA nanoparticles. Journal of controlled release : official journal of the Controlled Release Society. 2014;192:114–121. doi: 10.1016/j.jconrel.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 125.Xu R, Huang Y, Mai J, Zhang G, Guo X, Xia X, Koay EJ, Qin G, Erm DR, Li Q, Liu X, Ferrari M, Shen H. Multistage vectored siRNA targeting ataxia-telangiectasia mutated for breast cancer therapy. Small. 2013;9:1799–1808. doi: 10.1002/smll.201201510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Baylin SB. DNA methylation and gene silencing in cancer. Nature clinical practice. Oncology. 2005;2(Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 127.Dou S, Yao YD, Yang XZ, Sun TM, Mao CQ, Song EW, Wang J. Anti-Her2 single-chain antibody mediated DNMTs-siRNA delivery for targeted breast cancer therapy. Journal of controlled release : official journal of the Controlled Release Society. 2012;161:875–883. doi: 10.1016/j.jconrel.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 128.Minai-Tehrani A, Jiang HL, Kim YK, Chung YS, Yu KN, Kim JE, Shin JY, Hong SH, Lee JH, Kim HJ, Chang SH, Park S, Kang BN, Cho CS, Cho MH. Suppression of tumor growth in xenograft model mice by small interfering RNA targeting osteopontin delivery using biocompatible poly(amino ester) International journal of pharmaceutics. 2012;431:197–203. doi: 10.1016/j.ijpharm.2012.04.028. [DOI] [PubMed] [Google Scholar]
- 129.Bast RC, Jr., Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nature reviews. Cancer. 2009;9:415–428. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Goff BA, Mandel L, Muntz HG, Melancon CH. Ovarian carcinoma diagnosis. Cancer. 2000;89:2068–2075. doi: 10.1002/1097-0142(20001115)89:10<2068::aid-cncr6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 131.Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, Jiang G, Hsiao J, Mermel CH, Getz G, Barretina J, Gopal S, Tamayo P, Gould J, Tsherniak A, Stransky N, Luo B, Ren Y, Drapkin R, Bhatia SN, Mesirov JP, Garraway LA, Meyerson M, Lander ES, Root DE, Hahn WC. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ren Y, Cheung HW, von Maltzhan G, Agrawal A, Cowley GS, Weir BA, Boehm JS, Tamayo P, Karst AM, Liu JF, Hirsch MS, Mesirov JP, Drapkin R, Root DE, Lo J, Fogal V, Ruoslahti E, Hahn WC, Bhatia SN. Targeted tumor-penetrating siRNA nanocomplexes for credentialing the ovarian cancer oncogene ID4. Science translational medicine. 2012;4:147ra112. doi: 10.1126/scitranslmed.3003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Landen CN, Merritt WM, Mangala LS, Sanguino AM, Bucana C, Lu C, Lin YG, Han LY, Kamat AA, Schmandt R, Coleman RL, Gershenson DM, Lopez-Berestein G, Sood AK. Intraperitoneal delivery of liposomal siRNA for therapy of advanced ovarian cancer. Cancer biology & therapy. 2006;5:1708–1713. doi: 10.4161/cbt.5.12.3468. [DOI] [PubMed] [Google Scholar]
- 134.Suva ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, Baumer K, Le Bitoux MA, Marino D, Cironi L, Marquez VE, Clement V, Stamenkovic I. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer research. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 135.Gharpure KM, Chu KS, Bowerman CJ, Miyake T, Pradeep S, Mangala SL, Han HD, Rupaimoole R, Armaiz-Pena GN, Rahhal TB, Wu SY, Luft JC, Napier ME, Lopez-Berestein G, DeSimone JM, Sood AK. Metronomic docetaxel in PRINT nanoparticles and EZH2 silencing have synergistic antitumor effect in ovarian cancer. Molecular cancer therapeutics. 2014;13:1750–1757. doi: 10.1158/1535-7163.MCT-13-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Naor D, Sionov RV, Ish-Shalom D. CD44: structure, function, and association with the malignant process. Advances in cancer research. 1997;71:241–319. doi: 10.1016/s0065-230x(08)60101-3. [DOI] [PubMed] [Google Scholar]
- 137.Shah V, Taratula O, Garbuzenko OB, Taratula OR, Rodriguez-Rodriguez L, Minko T. Targeted nanomedicine for suppression of CD44 and simultaneous cell death induction in ovarian cancer: an optimal delivery of siRNA and anticancer drug. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:6193–6204. doi: 10.1158/1078-0432.CCR-13-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 139.Sabapathy K, Hochedlinger K, Nam SY, Bauer A, Karin M, Wagner EF. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Molecular cell. 2004;15:713–725. doi: 10.1016/j.molcel.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 140.Vivas-Mejia P, Benito JM, Fernandez A, Han HD, Mangala L, Rodriguez-Aguayo C, Chavez-Reyes A, Lin YG, Carey MS, Nick AM, Stone RL, Kim HS, Claret FX, Bornmann W, Hennessy BT, Sanguino A, Peng Z, Sood AK, Lopez-Berestein G. c-Jun-NH2-kinase-1 inhibition leads to antitumor activity in ovarian cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:184–194. doi: 10.1158/1078-0432.CCR-09-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li J, Cheng D, Yin T, Chen W, Lin Y, Chen J, Li R, Shuai X. Copolymer of poly(ethylene glycol) and poly(L-lysine) grafting polyethylenimine through a reducible disulfide linkage for siRNA delivery. Nanoscale. 2014;6:1732–1740. doi: 10.1039/c3nr05024f. [DOI] [PubMed] [Google Scholar]
- 142.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nature reviews. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber WJ, Mulder GE, Toebes M, Vesely MD, Lam SS, Korman AJ, Allison JP, Freeman GJ, Sharpe AH, Pearce EL, Schumacher TN, Aebersold R, Rammensee HG, Melief CJ, Mardis ER, Gillanders WE, Artyomov MN, Schreiber RD. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN, Frazier J, Chau BN, Loboda A, Linsley PS, Cleary MA, Park JR, Grandori C. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9545–9550. doi: 10.1073/pnas.1121119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Milhavet O, Gary DS, Mattson MP. RNA interference in biology and medicine. Pharmacological reviews. 2003;55:629–648. doi: 10.1124/pr.55.4.1. [DOI] [PubMed] [Google Scholar]
- 146.Snove O, Jr., Holen T. Many commonly used siRNAs risk off-target activity. Biochemical and biophysical research communications. 2004;319:256–263. doi: 10.1016/j.bbrc.2004.04.175. [DOI] [PubMed] [Google Scholar]
- 147.Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, Albuquerque RJ, Yamasaki S, Itaya M, Pan Y, Appukuttan B, Gibbs D, Yang Z, Kariko K, Ambati BK, Wilgus TA, DiPietro LA, Sakurai E, Zhang K, Smith JR, Taylor EW, Ambati J. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nature biotechnology. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 149.Forsbach A, Nemorin JG, Montino C, Muller C, Samulowitz U, Vicari AP, Jurk M, Mutwiri GK, Krieg AM, Lipford GB, Vollmer J. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. Journal of immunology. 2008;180:3729–3738. doi: 10.4049/jimmunol.180.6.3729. [DOI] [PubMed] [Google Scholar]
- 150.Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:7264–7270. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 151.Cubillos-Ruiz JR, Engle X, Scarlett UK, Martinez D, Barber A, Elgueta R, Wang L, Nesbeth Y, Durant Y, Gewirtz AT, Sentman CL, Kedl R, Conejo-Garcia JR. Polyethylenimine-based siRNA nanocomplexes reprogram tumor-associated dendritic cells via TLR5 to elicit therapeutic antitumor immunity. The Journal of clinical investigation. 2009;119:2231–2244. doi: 10.1172/JCI37716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Troegeler A, Lastrucci C, Duval C, Tanne A, Cougoule C, Maridonneau-Parini I, Neyrolles O, Lugo-Villarino G. An efficient siRNA-mediated gene silencing in primary human monocytes, dendritic cells and macrophages. Immunology and cell biology. 2014;92:699–708. doi: 10.1038/icb.2014.39. [DOI] [PubMed] [Google Scholar]
- 153.Alshamsan A, Hamdy S, Haddadi A, Samuel J, El-Kadi AO, Uludag H, Lavasanifar A. STAT3 Knockdown in B16 Melanoma by siRNA Lipopolyplexes Induces Bystander Immune Response In Vitro and In Vivo. Translational oncology. 2011;4:178–188. doi: 10.1593/tlo.11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hobo W, Novobrantseva TI, Fredrix H, Wong J, Milstein S, Epstein-Barash H, Liu J, Schaap N, van der Voort R, Dolstra H. Improving dendritic cell vaccine immunogenicity by silencing PD-1 ligands using siRNA-lipid nanoparticles combined with antigen mRNA electroporation. Cancer immunology, immunotherapy : CII. 2013;62:285–297. doi: 10.1007/s00262-012-1334-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Warashina S, Nakamura T, Harashima H. A20 silencing by lipid envelope-type nanoparticles enhances the efficiency of lipopolysaccharide-activated dendritic cells. Biological & pharmaceutical bulletin. 2011;34:1348–1351. doi: 10.1248/bpb.34.1348. [DOI] [PubMed] [Google Scholar]
- 156.Leuschner F, Dutta P, Gorbatov R, Novobrantseva TI, Donahoe JS, Courties G, Lee KM, Kim JI, Markmann JF, Marinelli B, Panizzi P, Lee WW, Iwamoto Y, Milstein S, Epstein-Barash H, Cantley W, Wong J, Cortez-Retamozo V, Newton A, Love K, Libby P, Pittet MJ, Swirski FK, Koteliansky V, Langer R, Weissleder R, Anderson DG, Nahrendorf M. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nature biotechnology. 2011;29:1005–1010. doi: 10.1038/nbt.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Conde J, Bao CC, Tan YQ, Cui DX, Edelman ER, Azevedo HS, Byrne HJ, Artzi N, Tian FR. Dual Targeted Immunotherapy via In Vivo Delivery of Biohybrid RNAi-Peptide Nanoparticles to Tumor-Associated Macrophages and Cancer Cells. Advanced functional materials. 2015;25:4183–4194. doi: 10.1002/adfm.201501283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dolina JS, Sung SS, Novobrantseva TI, Nguyen TM, Hahn YS. Lipidoid Nanoparticles Containing PD-L1 siRNA Delivered In Vivo Enter Kupffer Cells and Enhance NK and CD8(+) T Cell-mediated Hepatic Antiviral Immunity. Molecular therapy. Nucleic acids. 2013;2:e72. doi: 10.1038/mtna.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Meng H, Mai WX, Zhang H, Xue M, Xia T, Lin S, Wang X, Zhao Y, Ji Z, Zink JI, Nel AE. Codelivery of an optimal drug/siRNA combination using mesoporous silica nanoparticles to overcome drug resistance in breast cancer in vitro and in vivo. ACS nano. 2013;7:994–1005. doi: 10.1021/nn3044066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.MacDiarmid JA, Amaro-Mugridge NB, Madrid-Weiss J, Sedliarou I, Wetzel S, Kochar K, Brahmbhatt VN, Phillips L, Pattison ST, Petti C, Stillman B, Graham RM, Brahmbhatt H. Sequential treatment of drug-resistant tumors with targeted minicells containing siRNA or a cytotoxic drug. Nature biotechnology. 2009;27:643–651. doi: 10.1038/nbt.1547. [DOI] [PubMed] [Google Scholar]
- 161.Abbasi M, Lavasanifar A, Uludag H. Recent attempts at RNAi-mediated P-glycoprotein downregulation for reversal of multidrug resistance in cancer. Medicinal research reviews. 2013;33:33–53. doi: 10.1002/med.20244. [DOI] [PubMed] [Google Scholar]
- 162.Ganesh S, Iyer AK, Weiler J, Morrissey DV, Amiji MM. Combination of siRNA-directed Gene Silencing With Cisplatin Reverses Drug Resistance in Human Non-small Cell Lung Cancer. Molecular therapy. Nucleic acids. 2013;2:e110. doi: 10.1038/mtna.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ambardekar VV, Han HY, Varney ML, Vinogradov SV, Singh RK, Vetro JA. The modification of siRNA with 3' cholesterol to increase nuclease protection and suppression of native mRNA by select siRNA polyplexes. Biomaterials. 2011;32:1404–1411. doi: 10.1016/j.biomaterials.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Oe Y, Christie RJ, Naito M, Low SA, Fukushima S, Toh K, Miura Y, Matsumoto Y, Nishiyama N, Miyata K, Kataoka K. Actively-targeted polyion complex micelles stabilized by cholesterol and disulfide cross-linking for systemic delivery of siRNA to solid tumors. Biomaterials. 2014;35:7887–7895. doi: 10.1016/j.biomaterials.2014.05.041. [DOI] [PubMed] [Google Scholar]
- 165.Han SE, Kang H, Shim GY, Suh MS, Kim SJ, Kim JS, Oh YK. Novel cationic cholesterol derivative-based liposomes for serum-enhanced delivery of siRNA. International journal of pharmaceutics. 2008;353:260–269. doi: 10.1016/j.ijpharm.2007.11.026. [DOI] [PubMed] [Google Scholar]
- 166.Kanazawa T, Morisaki K, Suzuki S, Takashima Y. Prolongation of life in rats with malignant glioma by intranasal siRNA/drug codelivery to the brain with cell-penetrating peptide-modified micelles. Molecular pharmaceutics. 2014;11:1471–1478. doi: 10.1021/mp400644e. [DOI] [PubMed] [Google Scholar]
- 167.Malhotra M, Tomaro-Duchesneau C, Saha S, Kahouli I, Prakash S. Development and characterization of chitosan-PEG-TAT nanoparticles for the intracellular delivery of siRNA. International journal of nanomedicine. 2013;8:2041–2052. doi: 10.2147/IJN.S43683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Crombez L, Morris MC, Dufort S, Aldrian-Herrada G, Nguyen Q, Mc Master G, Coll JL, Heitz F, Divita G. Targeting cyclin B1 through peptide-based delivery of siRNA prevents tumour growth. Nucleic acids research. 2009;37:4559–4569. doi: 10.1093/nar/gkp451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Morris MC, Gros E, Aldrian-Herrada G, Choob M, Archdeacon J, Heitz F, Divita G. A non- covalent peptide-based carrier for in vivo delivery of DNA mimics. Nucleic acids research. 2007;35:e49. doi: 10.1093/nar/gkm053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ishihara T, Goto M, Kodera K, Kanazawa H, Murakami Y, Mizushima Y, Higaki M. Intracellular delivery of siRNA by cell-penetrating peptides modified with cationic oligopeptides. Drug delivery. 2009;16:153–159. doi: 10.1080/10717540902722774. [DOI] [PubMed] [Google Scholar]
- 171.Cuellar TL, Barnes D, Nelson C, Tanguay J, Yu SF, Wen X, Scales SJ, Gesch J, Davis D, van Brabant Smith A, Leake D, Vandlen R, Siebel CW. Systematic evaluation of antibody-mediated siRNA delivery using an industrial platform of THIOMAB-siRNA conjugates. Nucleic acids research. 2015;43:1189–1203. doi: 10.1093/nar/gku1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, Marasco WA, Lieberman J. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nature biotechnology. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 173.Yao YD, Sun TM, Huang SY, Dou S, Lin L, Chen JN, Ruan JB, Mao CQ, Yu FY, Zeng MS, Zang JY, Liu Q, Su FX, Zhang P, Lieberman J, Wang J, Song E. Targeted delivery of PLK1-siRNA by ScFv suppresses Her2+ breast cancer growth and metastasis. Science translational medicine. 2012;4:130ra148. doi: 10.1126/scitranslmed.3003601. [DOI] [PubMed] [Google Scholar]
- 174.Xia CF, Zhang Y, Zhang Y, Boado RJ, Pardridge WM. Intravenous siRNA of brain cancer with receptor targeting and avidin-biotin technology. Pharm Res. 2007;24:2309–2316. doi: 10.1007/s11095-007-9460-8. [DOI] [PubMed] [Google Scholar]
- 175.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nature biotechnology. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 176.Pastor F, Kolonias D, Giangrande PH, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature. 2010;465:227–230. doi: 10.1038/nature08999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wullner U, Neef I, Eller A, Kleines M, Tur MK, Barth S. Cell-specific induction of apoptosis by rationally designed bivalent aptamer-siRNA transcripts silencing eukaryotic elongation factor 2. Current cancer drug targets. 2008;8:554–565. doi: 10.2174/156800908786241078. [DOI] [PubMed] [Google Scholar]
- 178.Zhou J, Tiemann K, Chomchan P, Alluin J, Swiderski P, Burnett J, Zhang X, Forman S, Chen R, Rossi J. Dual functional BAFF receptor aptamers inhibit ligand-induced proliferation and deliver siRNAs to NHL cells. Nucleic acids research. 2013;41:4266–4283. doi: 10.1093/nar/gkt125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, Rothman AM, Hernandez FJ, McNamara JO, 2nd, Giangrande PH. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic acids research. 2012;40:6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hama S, Arata M, Nakamura I, Kasetani T, Itakura S, Tsuchiya H, Yoshiki T, Kogure K. Prevention of tumor growth by needle-free jet injection of anti-C7orf24 siRNA. Cancer gene therapy. 2012;19:553–557. doi: 10.1038/cgt.2012.31. [DOI] [PubMed] [Google Scholar]
- 181.Fehring V, Schaeper U, Ahrens K, Santel A, Keil O, Eisermann M, Giese K, Kaufmann J. Delivery of therapeutic siRNA to the lung endothelium via novel Lipoplex formulation DACC. Molecular therapy : the journal of the American Society of Gene Therapy. 2014;22:811–820. doi: 10.1038/mt.2013.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Han J, Wang Q, Zhang Z, Gong T, Sun X. Cationic bovine serum albumin based self-assembled nanoparticles as siRNA delivery vector for treating lung metastatic cancer. Small. 2014;10:524–535. doi: 10.1002/smll.201301992. [DOI] [PubMed] [Google Scholar]
- 183.Hong SH, Minai-Tehrani A, Chang SH, Jiang HL, Lee S, Lee AY, Seo HW, Chae C, Beck GR, Jr., Cho MH. Knockdown of the sodium-dependent phosphate co-transporter 2b (NPT2b) suppresses lung tumorigenesis. PloS one. 2013;8:e77121. doi: 10.1371/journal.pone.0077121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bonnet ME, Gossart JB, Benoit E, Messmer M, Zounib O, Moreau V, Behr JP, Lenne-Samuel N, Kedinger V, Meulle A, Erbacher P, Bolcato-Bellemin AL. Systemic delivery of sticky siRNAs targeting the cell cycle for lung tumor metastasis inhibition. Journal of controlled release : official journal of the Controlled Release Society. 2013;170:183–190. doi: 10.1016/j.jconrel.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 185.Yang Y, Hu Y, Wang Y, Li J, Liu F, Huang L. Nanoparticle delivery of pooled siRNA for effective treatment of non-small cell lung cancer. Molecular pharmaceutics. 2012;9:2280–2289. doi: 10.1021/mp300152v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chauhan A, Zubair S, Nadeem A, Ansari SA, Ansari MY, Mohammad O. Escheriosome- mediated cytosolic delivery of PLK1-specific siRNA: potential in treatment of liver cancer in BALB/c mice. Nanomedicine (Lond) 2014;9:407–420. doi: 10.2217/NNM.13.21. [DOI] [PubMed] [Google Scholar]
- 187.Gao J, Chen H, Yu Y, Song J, Song H, Su X, Li W, Tong X, Qian W, Wang H, Dai J, Guo Y. Inhibition of hepatocellular carcinoma growth using immunoliposomes for co-delivery of adriamycin and ribonucleotide reductase M2 siRNA. Biomaterials. 2013;34:10084–10098. doi: 10.1016/j.biomaterials.2013.08.088. [DOI] [PubMed] [Google Scholar]
- 188.Li L, Wang R, Wilcox D, Sarthy A, Lin X, Huang X, Tian L, Dande P, Hubbard RD, Hansen TM, Wada C, Zhao X, Kohlbrenner WM, Fesik SW, Shen Y. Developing lipid nanoparticle-based siRNA therapeutics for hepatocellular carcinoma using an integrated approach. Molecular cancer therapeutics. 2013;12:2308–2318. doi: 10.1158/1535-7163.MCT-12-0983-T. [DOI] [PubMed] [Google Scholar]
- 189.Gao J, Yu Y, Zhang Y, Song J, Chen H, Li W, Qian W, Deng L, Kou G, Chen J, Guo Y. EGFR- specific PEGylated immunoliposomes for active siRNA delivery in hepatocellular carcinoma. Biomaterials. 2012;33:270–282. doi: 10.1016/j.biomaterials.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 190.Lee YH, Andersen JB, Song HT, Judge AD, Seo D, Ishikawa T, Marquardt JU, Kitade M, Durkin ME, Raggi C, Woo HG, Conner EA, Avital I, Maclachlan I, Factor VM, Thorgeirsson SS. Definition of ubiquitination modulator COP1 as a novel therapeutic target in human hepatocellular carcinoma. Cancer research. 2010;70:8264–8269. doi: 10.1158/0008-5472.CAN-10-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Liu C, Liu X, Rocchi P, Qu F, Iovanna JL, Peng L. Arginine-terminated generation 4 PAMAM dendrimer as an effective nanovector for functional siRNA delivery in vitro and in vivo. Bioconjugate chemistry. 2014;25:521–532. doi: 10.1021/bc4005156. [DOI] [PubMed] [Google Scholar]
- 192.Xue HY, Wong HL. Solid lipid-PEI hybrid nanocarrier: an integrated approach to provide extended, targeted, and safer siRNA therapy of prostate cancer in an all-in-one manner. ACS nano. 2011;5:7034–7047. doi: 10.1021/nn201659z. [DOI] [PubMed] [Google Scholar]
- 193.Lin Q, Jin CS, Huang H, Ding L, Zhang Z, Chen J, Zheng G. Nanoparticle-enabled, image- guided treatment planning of target specific RNAi therapeutics in an orthotopic prostate cancer model. Small. 2014;10:3072–3082. doi: 10.1002/smll.201303842. [DOI] [PubMed] [Google Scholar]
- 194.Zeng S, Xiong MP. Trilayer micelles for combination delivery of rapamycin and siRNA targeting Y-box binding protein-1 (siYB-1) Biomaterials. 2013;34:6882–6892. doi: 10.1016/j.biomaterials.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Liu XQ, Xiong MH, Shu XT, Tang RZ, Wang J. Therapeutic delivery of siRNA silencing HIF-1 alpha with micellar nanoparticles inhibits hypoxic tumor growth. Molecular pharmaceutics. 2012;9:2863–2874. doi: 10.1021/mp300193f. [DOI] [PubMed] [Google Scholar]
- 196.Guo J, Ogier JR, Desgranges S, Darcy R, O'Driscoll C. Anisamide-targeted cyclodextrin nanoparticles for siRNA delivery to prostate tumours in mice. Biomaterials. 2012;33:7775–7784. doi: 10.1016/j.biomaterials.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 197.Guo J, Cheng WP, Gu J, Ding C, Qu X, Yang Z, O'Driscoll C. Systemic delivery of therapeutic small interfering RNA using a pH-triggered amphiphilic poly-L-lysine nanocarrier to suppress prostate cancer growth in mice. European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences. 2012;45:521–532. doi: 10.1016/j.ejps.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 198.Lee JB, Zhang K, Tam YY, Tam YK, Belliveau NM, Sung VY, Lin PJ, LeBlanc E, Ciufolini MA, Rennie PS, Cullis PR. Lipid nanoparticle siRNA systems for silencing the androgen receptor in human prostate cancer in vivo. International journal of cancer. Journal international du cancer. 2012;131:E781–790. doi: 10.1002/ijc.27361. [DOI] [PubMed] [Google Scholar]
- 199.Mu P, Nagahara S, Makita N, Tarumi Y, Kadomatsu K, Takei Y. Systemic delivery of siRNA specific to tumor mediated by atelocollagen: combined therapy using siRNA targeting Bcl-xL and cisplatin against prostate cancer. International journal of cancer. Journal international du cancer. 2009;125:2978–2990. doi: 10.1002/ijc.24382. [DOI] [PubMed] [Google Scholar]
- 200.Aleku M, Schulz P, Keil O, Santel A, Schaeper U, Dieckhoff B, Janke O, Endruschat J, Durieux B, Roder N, Loffler K, Lange C, Fechtner M, Mopert K, Fisch G, Dames S, Arnold W, Jochims K, Giese K, Wiedenmann B, Scholz A, Kaufmann J. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer research. 2008;68:9788–9798. doi: 10.1158/0008-5472.CAN-08-2428. [DOI] [PubMed] [Google Scholar]
- 201.Addepalli MK, Ray KB, Kumar B, Ramnath RL, Chile S, Rao H. RNAi-mediated knockdown of AURKB and EGFR shows enhanced therapeutic efficacy in prostate tumor regression. Gene therapy. 2010;17:352–359. doi: 10.1038/gt.2009.155. [DOI] [PubMed] [Google Scholar]
- 202.Tang W, Weidner DA, Hu BY, Newton RJ, Hu XH. Efficient delivery of small interfering RNA to plant cells by a nanosecond pulsed laser-induced stress wave for posttranscriptional gene silencing. Plant science : an international journal of experimental plant biology. 2006;171:375–381. doi: 10.1016/j.plantsci.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 203.Sun TM, Du JZ, Yao YD, Mao CQ, Dou S, Huang SY, Zhang PZ, Leong KW, Song EW, Wang J. Simultaneous delivery of siRNA and paclitaxel via a “two-in-one” micelleplex promotes synergistic tumor suppression. ACS nano. 2011;5:1483–1494. doi: 10.1021/nn103349h. [DOI] [PubMed] [Google Scholar]
- 204.Yang XZ, Dou S, Sun TM, Mao CQ, Wang HX, Wang J. Systemic delivery of siRNA with cationic lipid assisted PEG-PLA nanoparticles for cancer therapy. Journal of controlled release : official journal of the Controlled Release Society. 2011;156:203–211. doi: 10.1016/j.jconrel.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 205.Pille JY, Denoyelle C, Varet J, Bertrand JR, Soria J, Opolon P, Lu H, Pritchard LL, Vannier JP, Malvy C, Soria C, Li H. Anti-RhoA and anti-RhoC siRNAs inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer cells in vitro and in vivo. Molecular therapy : the journal of the American Society of Gene Therapy. 2005;11:267–274. doi: 10.1016/j.ymthe.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 206.Aliabadi HM, Maranchuk R, Kucharski C, Mahdipoor P, Hugh J, Uludag H. Effective response of doxorubicin-sensitive and -resistant breast cancer cells to combinational siRNA therapy. Journal of controlled release : official journal of the Controlled Release Society. 2013;172:219–228. doi: 10.1016/j.jconrel.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 207.Jiang J, Yang SJ, Wang JC, Yang LJ, Xu ZZ, Yang T, Liu XY, Zhang Q. Sequential treatment of drug-resistant tumors with RGD-modified liposomes containing siRNA or doxorubicin. European journal of pharmaceutics and biopharmaceutics : official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik e.V. 2010;76:170–178. doi: 10.1016/j.ejpb.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 208.Cho SK, Pedram A, Levin ER, Kwon YJ. Acid-degradable core-shell nanoparticles for reversed tamoxifen-resistance in breast cancer by silencing manganese superoxide dismutase (MnSOD) Biomaterials. 2013;34:10228–10237. doi: 10.1016/j.biomaterials.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Leng Q, Mixson AJ. Small interfering RNA targeting Raf-1 inhibits tumor growth in vitro and in vivo. Cancer gene therapy. 2005;12:682–690. doi: 10.1038/sj.cgt.7700831. [DOI] [PubMed] [Google Scholar]
- 210.Ding Y, Wang Y, Zhou J, Gu X, Wang W, Liu C, Bao X, Wang C, Li Y, Zhang Q. Direct cytosolic siRNA delivery by reconstituted high density lipoprotein for target-specific therapy of tumor angiogenesis. Biomaterials. 2014;35:7214–7227. doi: 10.1016/j.biomaterials.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 211.Florinas S, Kim J, Nam K, Janat-Amsbury MM, Kim SW. Ultrasound-assisted siRNA delivery via arginine-grafted bioreducible polymer and microbubbles targeting VEGF for ovarian cancer treatment. Journal of controlled release : official journal of the Controlled Release Society. 2014;183:1–8. doi: 10.1016/j.jconrel.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kala S, Mak AS, Liu X, Posocco P, Pricl S, Peng L, Wong AS. Combination of dendrimernanovector-mediated small interfering RNA delivery to target Akt with the clinical anticancer drug paclitaxel for effective and potent anticancer activity in treating ovarian cancer. Journal of medicinal chemistry. 2014;57:2634–2642. doi: 10.1021/jm401907z. [DOI] [PubMed] [Google Scholar]
- 213.Koyanagi T, Suzuki Y, Saga Y, Machida S, Takei Y, Fujiwara H, Suzuki M, Sato Y. In vivo delivery of siRNA targeting vasohibin-2 decreases tumor angiogenesis and suppresses tumor growth in ovarian cancer. Cancer science. 2013;104:1705–1710. doi: 10.1111/cas.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Han HD, Mangala LS, Lee JW, Shahzad MM, Kim HS, Shen D, Nam EJ, Mora EM, Stone RL, Lu C, Lee SJ, Roh JW, Nick AM, Lopez-Berestein G, Sood AK. Targeted gene silencing using RGD-labeled chitosan nanoparticles. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:3910–3922. doi: 10.1158/1078-0432.CCR-10-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tanaka T, Mangala LS, Vivas-Mejia PE, Nieves-Alicea R, Mann AP, Mora E, Han HD, Shahzad MM, Liu X, Bhavane R, Gu J, Fakhoury JR, Chiappini C, Lu C, Matsuo K, Godin B, Stone RL, Nick AM, Lopez-Berestein G, Sood AK, Ferrari M. Sustained small interfering RNA delivery by mesoporous silicon particles. Cancer research. 2010;70:3687–3696. doi: 10.1158/0008-5472.CAN-09-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Difeo A, Huang F, Sangodkar J, Terzo EA, Leake D, Narla G, Martignetti JA. KLF6-SV1 is a novel antiapoptotic protein that targets the BH3-only protein NOXA for degradation and whose inhibition extends survival in an ovarian cancer model. Cancer research. 2009;69:4733–4741. doi: 10.1158/0008-5472.CAN-08-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Halder J, Kamat AA, Landen CN, Jr., Han LY, Lutgendorf SK, Lin YG, Merritt WM, Jennings NB, Chavez-Reyes A, Coleman RL, Gershenson DM, Schmandt R, Cole SW, Lopez- Berestein G, Sood AK. Focal adhesion kinase targeting using in vivo short interfering RNA delivery in neutral liposomes for ovarian carcinoma therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:4916–4924. doi: 10.1158/1078-0432.CCR-06-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]