Abstract
Sunlight’s ultraviolet wavelengths induce cyclobutane pyrimidine dimers (CPDs), which then cause mutations that lead to melanoma or to cancers of skin keratinocytes. In pigmented melanocytes, we found that CPDs arise both instantaneously and for hours after UV exposure ends. Remarkably, the CPDs arising in the dark originate by a novel pathway that resembles bioluminescence but does not end in light: First, UV activates the enzymes nitric oxide synthase (NOS) and NADPH oxidase (NOX), which generate the radicals nitric oxide (NO•) and superoxide (O2•−); these combine to form the powerful oxidant peroxynitrite (ONOO−). A fragment of the skin pigment melanin is then oxidized, exciting an electron to an energy level so high that it is rarely seen in biology. This process of chemically exciting electrons, termed “chemiexcitation”, is used by fireflies to generate light but it had never been seen in mammalian cells. In melanocytes, the energy transfers radiationlessly to DNA, inducing CPDs. Chemiexcitation is a new source of genome instability, and it calls attention to endogenous mechanisms of genome maintenance that prevent electronic excitation or dissipate the energy of excited states. Chemiexcitation may also trigger pathogenesis in internal tissues because the same chemistry should arise wherever superoxide and nitric oxide arise near cells that contain melanin.
Keywords: Ultraviolet, cyclobutane pyrimidine dimer, melanoma, melanin, chemiexcitation, nitric oxide, superoxide, peroxynitrite, triplet state
1. Introduction
The origin of skin cancer – melanoma and basal or squamous cell carcinoma – can be traced to the quantum mechanical level. Electron excitation creates non-DNA structures (“damaged” DNA) that, when copied by a polymerase, write altered information into chemically-normal DNA (mutation). At this quantum mechanical level, our laboratory recently encountered a surprise: Chemical excitation of electrons, termed “chemiexcitation”, is a new source of genome instability in melanocytes and in the keratinocytes that receive melanin from them. This discovery raises questions about how chemiexcitation operates in mammals and whether it underlies additional diseases. It also draws attention to little-noticed mechanisms of genome maintenance that protect us even prior to DNA repair.
The cyclobutane pyrimidine dimer (CPD) is the classic DNA photoproduct created by ultraviolet radiation present in sunlight [1]. It joins two adjacent pyrimidine bases, thymine or cytosine (T or C), by two single bonds that create a 4-carbon ring between the bases (the “cyclobutane” ring) (Fig. 1). This structure disrupts base pairing and distorts the DNA helix from its normal B form. The CPD is lethal and mutagenic, and the “UV signature” of C→T mutations at sites of adjacent pyrimidines is seen in melanomas, non-melanoma skin cancers, precancerous lesions, and sun-exposed skin [2–11]. Poor CPD repair in xeroderma pigmentosum increases childhood melanoma 10,000-fold whereas removing CPDs prevents UV-induced skin cancer in mice. The CPD has long been known to be generated by UV photons; we now find that it can be generated by chemiexcitation when melanin is present [12].
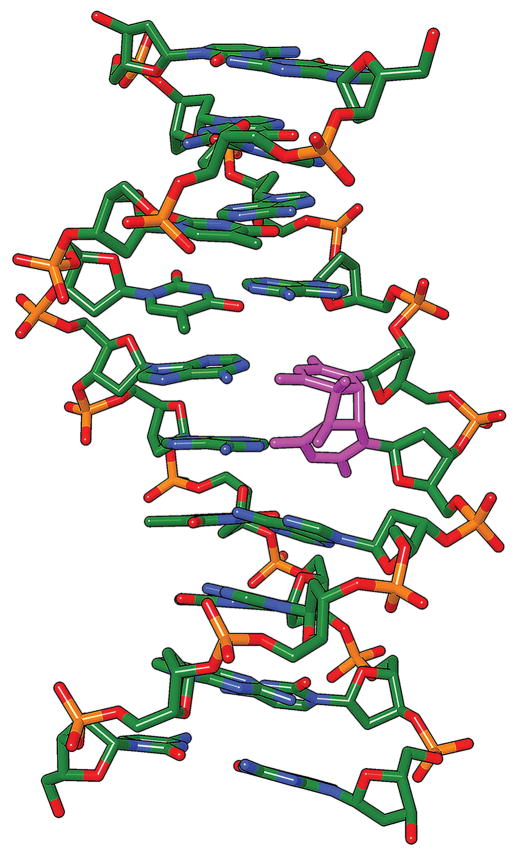
Fig. 1. Thymine-thymine cyclobutane pyrimidine dimer in DNA.
The violet color indicates a four-carbon cyclobutane ring (in the vertical plane) joining two adjacent thymine bases at the site of the former 5–6 double bonds. Orientation is 5′-3′ from bottom to top. The bases are no longer parallel, the 5′ thymine has lost one of its hydrogen bonds with adenine, and the helix is distorted, rendering DNA polymerases error-prone at this site. Modified from [59].
2. CPDs in the dark
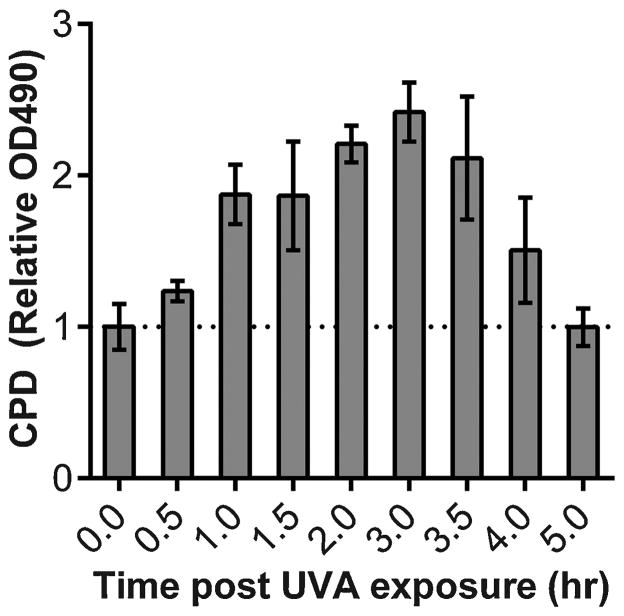
Induction of CPDs in DNA by direct absorption of UVC photons requires < 1 psec [13], so the usual result of irradiation experiments is an immediate peak in CPD formation followed by slow excision repair. We exposed murine melanocytes to UVA radiation and measured CPDs over time using an anti-CPD antibody [14] for ELISA. Surprisingly, melanin-containing murine melanocytes continued to generate CPDs for at least three hours after UVA exposure, after which generation was offset by DNA repair (Fig. 2) [12]. In contrast, melanocytes derived from albino mice reached the peak of CPD induction immediately upon exposure, as expected; so did murine fibroblasts. The delayed CPDs therefore depended on melanin. Similar results were observed using a comet assay after treating lysed cells with a nucleotide excision repair enzyme to induce DNA breaks at CPD sites, also indicating that the delayed CPDs resided in the nucleus.
Fig 2. Cyclobutane pyrimidine dimers (CPDs) generated long after the end of UV exposure.
In melanin-containing melanocytes, CPDs continue to increase for 3 hours after UVA exposure ends. In albino murine melanocytes or in fibroblasts, the peak of CPD induction is at time 0, followed by repair. CPDs were assayed by DNA ELISA and similar results were obtained by an endonuclease-sensitive sites assay using comet and by mass spectrometry. Figure from [12].
These results were intriguing because delayed CPD production had occasionally been reported in other cell types after UVC or UVB exposure [15–19], often with wide variability. It seemed possible that the melanin-dependent mechanism would be more susceptible to experimental study. The important implications for skin cancer prevention and sunbed use made this mechanism worthy of elucidation.
The gold standard assay, mass spectrometry, revealed delayed production of TT, TC, and CT CPDs, only in melanocytes that contained melanin. Cytosine-containing CPDs are the species that generate UV-signature C→T mutations. The effect was larger when concurrent repair was knocked down using siRNA against Xpa or Xpc transcripts. Delayed CPDs constituted half of the total CPDs. This two-fold increase in DNA photoproducts is substantial because xeroderma pigmentosum complementation group D individuals are only 60% defective in CPD repair [20]. With UVB, most CPDs in melanocytes were created in the dark even without repair knockdown.
Human melanocytes also generated delayed CPDs after UVA or UVB, but there was inter-individual variation. For human melanocytes having modest repair of CPD after exposure, but no obvious delayed CPDs, the delayed CPDs were revealed upon siRNA knockdown of XPA or XPC. Individual variation may stem from variable repair or melanin type. The phenomenon could be studied in vivo using transgenic mice that contained epidermal melanocytes due to Kit ligand expression in keratinocytes, thereby mimicking human skin. Delayed CPDs were seen throughout the epidermis. Most cells were keratinocytes, which receive pigment from the melanocytes, implying that the crucial requirement is melanin content rather than melanin synthesis. Both initial and delayed CPDs were more frequent in mice whose melanocytes synthesized red-yellow pheomelanin than in black mice. This suggests that pheomelanin is both a poorer shield against normal CPD formation and a more potent generator of delayed CPDs. This difference could underlie the increased sunburn and skin cancer sensitivity of individuals who have red or blonde hair.
Yet, creating CPDs requires extraordinarily high energies usually found only in ultraviolet light. Where would this energy come from in a cell? To fully appreciate the dilemma and the solution, it is useful to first highlight the photophysics of CPD production by photons.
3. CPDs from photons
Some reactions cannot proceed when the reactants’ electrons are in the ground state, the state in which electrons fill orbitals from the lowest energy upward. The Woodward-Hoffman rules, based on orbital shapes, forbid ground state [2+2] cycloadditions – rings created when 2 atoms connected by a double bond undergo a concerted reaction with 2 atoms of another molecule. Cyclobutane pyrimidine dimer formation requires a [2+2] cycloaddition, so an electron in a DNA base must first be raised to an excited state.
Electron excitation in a well-understood molecule, formaldehyde, is shown in Fig. 3. Once excited, the electron and its former partner are now unpaired; the orbitals are no longer filled from the bottom up and the molecule is in an “electronically excited state”. In many ways, the excited molecule acts as if it contained two radicals (a diradical). Excited states come in two varieties. In the singlet state, the two electrons retain their opposite spins, as the Pauli Exclusion Principle required when they were in the same orbital. In the triplet state, the electrons have opposite spins. Triplet states have lower energies and, because they have longer lifetimes, they are more likely to enter into a reaction. In either case, exciting the electron overcomes the Woodward-Hoffman stricture by achieving two ends: very high energies and very different orbital geometries.
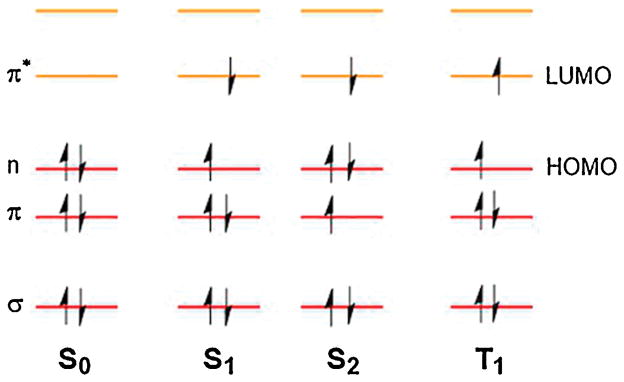
Fig. 3. Exciting electrons in a simple molecule containing a carbonyl.
Electron distribution in the frontier molecular orbitals of the C=O of formaldehyde (CH2O) in the ground state (S0) and excited states (S1, S2, and T1). S, singlet state; T, triplet state. σ and π are bonding orbitals, n is nonbonding, and π* is antibonding. HOMO is the highest occupied molecular orbital; LUMO is the lowest unoccupied molecular orbital, available for entry of an electron or electron pair. Modified from [60].
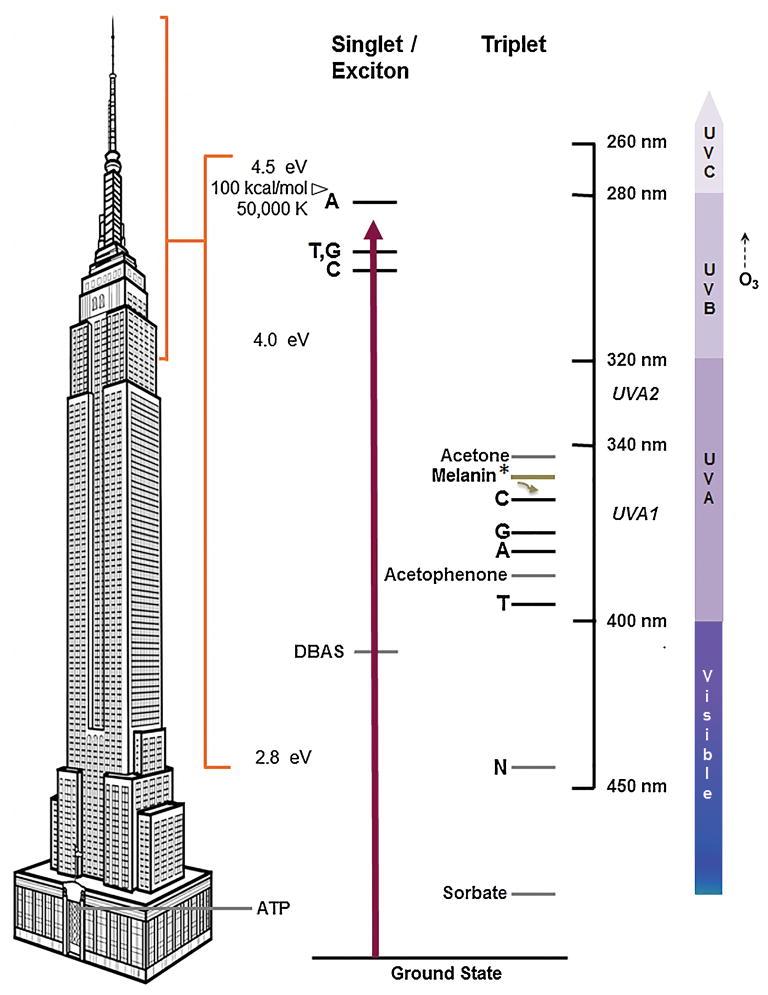
The energies of excited states in DNA are high, on the order of 100 kcal/mol (4.5 eV, corresponding to an ultraviolet wavelength of 280 nm or a temperature of 50,000 K) (Fig. 4). Comparing these energies to storeys of the Empire State Building provides a sense of scale. The amount of energy stored in the high-energy phosphate bond of ATP (a ground-state reaction) is 7 kcal/mol or 0.3 eV, a few storeys above the lobby. The UVC- induced singlet states have energies above the building’s 102nd floor. For this reason, biochemistry has long been thought incapable of making CPDs. How does UV do it?
Fig. 4. Excited states of the DNA bases.
Electronic excitation of the DNA bases involves energies much higher than a typical biochemical reaction, exemplified by the 7 kcal/mol or 0.3 eV released from the high-energy bond of ATP (left). The Empire State Building provides a sense of scale. UVC excites a single base or a delocalized quantum state of 2–3 bases to the singlet state at ES = 4.2–4.4 eV (center) [22, 24, 61]. UVB does this weakly. Stratospheric ozone (O3) prevents excitation by UVC and short-wavelength UVB. Alternatively, UVB excites an intermediary photosensitizer molecule such as acetone or acetophenone to a singlet state (not shown), which then undergoes intersystem crossing to its triplet state; the latter can transfer energy to the lower triplet energy levels of the free bases, above ET = 3.0 eV. In DNA, the triplet energies may be lower (see text). UVA is absorbed by a triplet-like state that may be a mixture between the triplet state and a charge-transfer state, in which partial charge is transferred to another base [30]. The chemically-excited triplet-state melanin fragment (Melanin*) acts as a sensitizer whose energy is well above that of acetophenone. The melanin fragment’s energy can be diverted to the triplet-specific energy acceptor DBAS, which crosses to the singlet state and emits the energy as fluorescence. Or it can be diverted to the triplet-specific quencher sorbate, which isomerizes and dissipates it as heat. Both diversions prevent chemiexcitation from creating CPDs in DNA.
3.1 UVC makes CPDs in DNA via the singlet state
A UVC photon (100–280 nm) has sufficient energy to excite an electron from a ground state π orbital of free thymine or cytosine (T or C) to the singlet state π* orbital (Fig. 4). If another T or C is nearby, the now-unpaired electrons in the excited base form two single bonds with the adjacent base to make a CPD – but only if the two 5–6 double bonds happened to be aligned with each other when the photon was absorbed [13, 21, 22]. The process takes less than a picosecond. In DNA, the photon is more likely to be effective because the bases are already in parallel planes at an angle of only 36°. The singlet described in Fig. 4 is technically an exciton because the photon is initially absorbed by the combined quantum state of two to three adjacent bases.
In contrast, dilute free bases are randomly oriented and this orientation changes quickly. The subpicosecond lifetime of the singlet state is too short for two bases to diffuse near each other or for correct alignments to form at room temperature, so CPDs are formed after the electron undergoes intersystem crossing to the triplet state (Fig. 4), where the lifetime is approximately a microsecond [23]. Triplet state energies are much lower than the singlets and their relative energies are different, so T is now lower than C [24]. In DNA, triplet energies (measured as T- and C-containing CPDs plus apurinic sites [25], hence labeled “N”) appear to be even lower than in free bases or nucleotides [24].
3.2 UVB makes CPDs via the singlet state or by triplet photosensitization
A photon at the short-wavelength end of the UVB region (280–320 nm) can also excite a DNA base to its singlet state and make CPDs [23]. DNA absorbs UVB poorly, so biological effects require 100 fold higher doses. UVB is more effective if it acts through an intermediary molecule that absorbs UVB strongly. Acetone and acetophenone are excited to their singlet state by UVB, then efficiently cross over to their triplet state; this long-lived state allows the excited molecule to collide with DNA bases and transfer the triplet energy radiationlessly to the triplet state of the DNA bases. This type of photosensitization becomes important for chemiexcitation.
3.3 UVA makes CPDs via a triplet-like state
UVA (320–400 nm) is absorbed by DNA even more poorly. Doses 104-fold larger than UVC doses are needed to cause cell death or murine skin cancers [26]. For years, it was believed that UVA was not a significant source of CPDs and that it contributed to pathogenesis through oxidative DNA damage. However, it was recently discovered that UVA-induced CPDs exceed oxidative lesions such as 8-oxo-guanine [27, 28]. UVA-induced CPDs result from direct photon absorption, rather than from photosensitization by an intermediary, because UVA creates CPDs in purified DNA [29]. The excited state must be in the energy realm of the triplet state because UVA energies are too low to reach the DNA singlets. In support of this idea, the CPD species after UVA are biased toward TT CPDs, as they are for triplet photosensitization. The precise situation is more complex. UVA absorption and its CPD production require base pairing, hinting at a mixture between the triplet state and a charge-transfer state in which partial charge is transferred to another base [30].
Melanocytes can evidently reach these high energy levels in the dark, many hours after UV exposure ends. The gulf between ATP-like energies and CPD energies is lessened if DNA triplet states are involved, but the feat that biochemistry must accomplish is still substantial.
4. CPDs from chemiexcitation
To identify the photochemical pathway that produces delayed CPDs after UVA, we first tested kojic acid, an inhibitor of tyrosinase, the rate-limiting enzyme in melanin synthesis. It suppressed production of delayed CPDs, confirming the necessity for melanin. Because cells produce reactive oxygen species for long periods after a UVA exposure [31], radical scavengers were tested. N-acetylcysteine suppressed production of delayed CPDs and vitamin E abolished it. Because melanin generates superoxide radical ion (O2•−) while it is being irradiated [32], we scavenged O2•− with TEMPOL; this also abolished delayed CPDs. A long-lasting source of O2•− in cells is the enzyme NADPH oxidase (NOX), which is rapidly induced by UVA [31]. A NOX inhibitor abolished delayed CPDs. In this experiment, CPDs were even inhibited to a level below that at 0 h, indicating that “delayed” CPDs were also being created during the irradiation.
The O2•− requirement recalled a 1971 experiment in which CPDs were generated in the dark by trimethyl dioxetane. A dioxetane is a strained 4-member ring peroxide that, at modest temperatures, undergoes cleavage to two carbonyls, with one in an electronically excited triplet state. For trimethyl dioxetane, the triplet state had the high energy of a UV photon but transferred its energy to the triplet state of DNA bases by a process independent of radiation [33]. The paper recognized the possibility of chemically-induced CPDs in internal organs, but noted that “No reaction capable of yielding electronically excited products of sufficient energy to induce DNA photochemistry is known to occur in a biological system.” Years later, enzymes producing dioxetanes by using ATP were found to underlie bioluminescence [34]. In subsequent decades, several investigators noticed metabolites that seemed to require reaction mechanisms forbidden in the ground state by the Woodward-Hoffman rules, and suggested that electronic excitation might be an important feature of biochemistry [35, 36]. These groups identified enzymes capable of generating dioxetanes and electronic excitation in a test tube, including horseradish peroxidase (in the presence of H2O2), cytochrome c, and lipoxygenase [37].
One of the few biological molecules that can initiate such “photochemistry in the dark” is peroxynitrite (ONOO−) [38]. It is generated by the spontaneous reaction of O2•− with nitric oxide (NO•). NO• and peroxynitrite are stable enough to diffuse significant distances in the cell [39]. NOX and the NO• generator NOS are UV-inducible within minutes and are expressed in melanocytes, including nuclear NOX4 [40]. A prediction was then that NO• would be required for production of delayed CPDs. Inhibiting iNOS with aminoguanidine indeed resulted in complete suppression. Another prediction was that melanin-containing cells exposed to UV would generate peroxynitrite, which can be detected by its nitration of tyrosines. Exposing melanin-containing melanocytes to UVA triggered an ~400 fold spike in the flux of peroxynitrite per hour.
Were excited triplet states being created? A diagnostic for triplet states is that their energy can also discharge via ultraweak blue or green phosphorescence [36]. This signal can be enhanced several orders of magnitude by diverting the energy to a triplet energy acceptor, sodium 9,10-dibromoanthracene-2-sulfonate (DBAS), which undergoes intersystem crossing to the singlet where it fluoresces strongly. UVA-irradiated melanocytes indeed generated ultraweak chemiluminescence for several hours post-irradiation and only in cells containing melanin. The lifetime of triplet carbonyls is ~10 μs [38], so a signal persistent over the course of hours indicates ongoing creation of the reactants by the two enzymes. To test whether this triplet state was actually required for delayed CPDs, we added a specific quencher of triplet states, ethyl sorbate [41], after UVA exposure. This treatment prevented delayed CPDs in intact melanocytes. Because DBAS diverts triplet energy to luminescence, it also blocked delayed CPDs. These results provided direct evidence that UV-induced superoxide and nitric oxide led to a high-energy triplet state molecule, which then created a delayed CPD by energy transfer. CPDs were being created in vivo by chemiexcitation. We will now adopt the abbreviation “chCPD” for chemiexcitation-induced CPDs. This notation also makes it easier to describe the chemistry-dependent CPDs induced during UV exposure.
The final steps in chCPD generation can now be envisioned, although they have not yet been demonstrated directly. Energy transfer from a triplet carbonyl to the triplet state of a DNA base would use the lower set of DNA energy levels in Fig. 4. For these, a strong bias toward TT CPDs is expected. UVA generates primarily TT CPDs, with TC and CT CPD together contributing only 10–30% of these three CPD types [28]. In the mass spectrometry experiments, we observed such a bias but the ratio of (TC + CT)/TT CPDs in the chCPDs was 4-fold higher than in CPDs induced by UVA directly: In melanin-containing cells it increased from the UVA-like 0.37 at 0 h (directly-induced CPDs) to 1.33 in the CPDs arising during the following 2 h, a ratio more typical of cells irradiated with the higher-energy UVB. This ratio is still too low for a singlet state, but it suggests that the triplet energy of the molecule hosting the dioxetane lies above that of cytosine. The chemiexcited molecule then acts in the same way as a photosensitizer, except that the sensitizer was excited chemically. The mechanism of energy transfer to DNA is unknown, but triplet-triplet energy transfer typically involves a collision between donor and acceptor, with the donor exchanging a high energy electron for a lower energy one (Dexter electron exchange) [42].
5. Melanin as a molecular shuttle
Which molecule hosted the excited triplet state? Melanin is located in the cytoplasm, yet chCPDs were generated in the nucleus. One possible conduit, initiated by O2•− from UV-exposed melanin and ending in a dioxetane, was a radical chain reaction in the lipid membrane stretching from melanosomes to the nuclear membrane [41]. But inducing lipid peroxidation in nuclei did not induce CPDs.
Another candidate was the set of melanin monomers from which the melanin polymer is assembled. These are lipophilic and are therefore potentially able to enter the nucleus. In addition, melanin polymer rapidly solubilizes when exposed to hydrogen peroxide [43] and its degradation by peroxidation or UV-photoionization had been proposed to involve dioxetane and triplet carbonyls [44, 45].
We found that a triplet state could be created in melanin by a cell-free system: synthetic melanin oxidized with peroxynitrite and incubated with DBAS generated chemiluminescence. Replacing peroxynitrite with horseradish peroxidase plus H2O2 [38] also gave chemiluminescence, as did the eumelanin monomer 5,6-dihydroxyindole-2-carboxylic acid (DHICA) and the pheomelanin monomer 5-S-cysteinyldopa (5SCD). Crucially, CPDs were generated in the complete absence of UV when plasmid DNA was incubated with peroxynitrite and melanin, DHICA, or 5SCD. DBAS, which redirects triplet energy toward luminescence, reduced CPD production by up to 90%. These cell-free chCPDs included the mutagenic cytosine-containing CPDs. The level of chCPDs induced in the absence of UV was similar to the level generated in pure DNA by 25 kJ/m2 of UVA, an exposure about one-quarter of that required to produce a barely perceptible sunburn. This value is approximately 1 CPD per 24 kb of DNA created solely by oxidized melanin.
How did melanin overcome the barrier posed by the nuclear membrane? Surprisingly, we found that melanin polymer was rapidly solubilized after exposure to UVA or peroxynitrite. In addition, 3D images of melanocytes revealed opaque granules inside the nucleus after UVA exposure. These may be melanin-containing organelles that moved into the nucleus or spontaneous polymers of melanin precursors, as occurs during normal melanin synthesis.
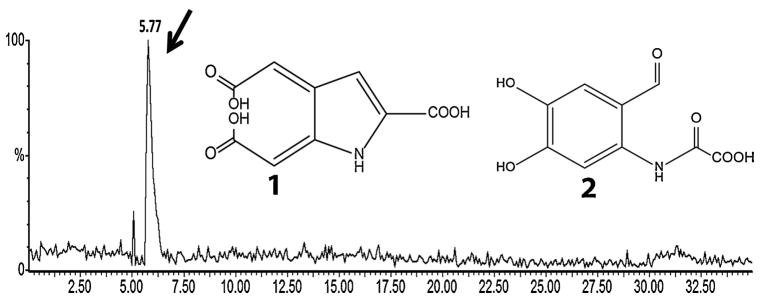
Final confirmation came from identifying carbonyl-containing DHICA reaction products. Two structures had been predicted in the literature. The DHICA analog of the predicted eumelanin degradation product [44] (Fig. 5, structure 1) has a molecular weight of 225 daltons and would be highly polar. An alternative reaction [46] is formation of a dioxetane moiety at the pyrrole ring, also leading to a polar structure of 225 daltons (structure 2). Separating reaction products by HPLC, and scanning the HPLC eluate by mass spectrometry for the corresponding 224 m/z ion, revealed a single peak eluting in the highly polar region. Scanning this fraction of the HPLC eluate by mass spectrometry revealed four m/z peaks, of which the second largest was 224 daltons. Chemical blocking experiments suggested that 2 is the predominant product. These results indicate that melanin or a melanin fragment acts as a molecular shuttle that acquires an electronically excited triplet state, probably at a carbonyl arising from a dioxetane intermediate, and brings this excited state into contact with nuclear DNA. and migration to the nucleus.
Fig. 5. Identification of a putative triplet-carbonyl carrier.
The eumelanin monomer DHICA was oxidized and products were separated by HPLC, monitoring by mass spectrometry. Monitoring for the 224 m/z ion of 225 dalton compounds 1 and 2, expected for triplet-carbonyl derivatives of dioxetane-adducted DHICA, revealed a single, polar peak. Subsequent scanning of the polar HPLC peak revealed four principal m/z peaks, one at 224 daltons. Figure from [12].
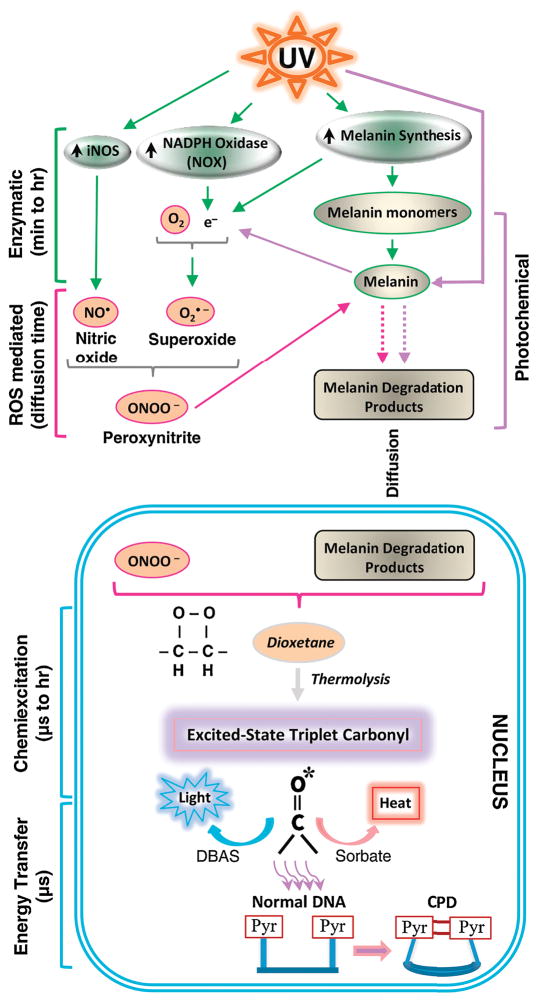
Chemiexcitation in melanocytes, and subsequent generation of chCPDs in the dark, is therefore a multistep process. The various steps are outlined in Fig. 6. The surprisingly sustained time course of chCPD generation can be accounted for by the prolonged steps of UV-induction of NOX and NOS, peroxynitrite-induced solubilization of melanin to fragments or release of pre-melanin monomers from UV-damaged melanosomes, and then migration to the nucleus.
Fig. 6. Generating CPDs in the dark by chemiexcitation.
CPDs are made within picoseconds of UV exposure by direct absorption, but melanocytes generate CPDs by chemiexcitation after UV exposure ends, with melanin being a key participant. Exposing cells to UV radiation up-regulates nitric oxide synthase (NOS), NADPH oxidase (NOX), and enzymes of melanin synthesis, causing sustained generation of nitric oxide (NO•) and superoxide (O2•−). NOS and NOX isoforms are located in both cytoplasm and nucleus. The two radicals produce the powerful oxidant peroxynitrite (ONOO−), which degrades melanin polymer to fragments that appear in the nucleus. Peroxynitrite is also one of the few biological molecules capable of exciting an electron to a triplet state. Peroxynitrite excites an electron in a melanin fragment to a triplet state that has the high energy of a UV photon. The typical triplet-state reaction intermediate, not yet demonstrated (hence indicated in italics), is a cycloaddition of –O–O– to create an unstable dioxetane; dioxetanes undergo spontaneous thermolysis to yield two carbonyls, one of which finishes in a high-energy triplet state (denoted by *). For the melanin-related triplet, the half-life of the reaction intermediate appeared to be minutes, but was generated continuously. A carbonyl consistent with a dioxetane precursor was identified by mass spectrometry. Triplet energy then discharges on a microsecond time scale to generate visible luminescence, or discharges in a radiation-independent manner to an acceptor (“DBAS”) which re-emits it as fluorescence, or to a triplet quencher (“sorbate”) which dissipates the energy via isomerization and heat, or to DNA bases where it makes CPDs. Figure from [12].
6. Consequences of excited states
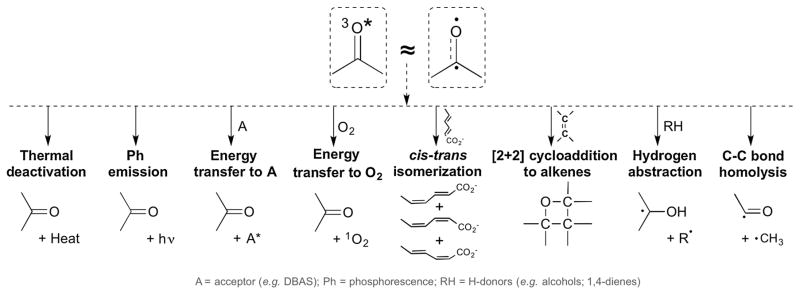
CPDs are not the only consequence of excited states (Fig. 7) [37]. Ideally, the excited molecule would dissipate its energy as heat or light. When the energy is transferred to a molecule having conjugated double bonds, such as sorbate, the diradical character of the excited double bond allows isomerization around the remaining single bond, whereupon the excess energy is dissipated as heat. However, triplet state energy is also readily transferred to ambient O2, which is unusual in being a triplet in the ground state. This transfer creates singlet oxygen – a highly reactive excited state of O2. The excited carbonyl’s diradical character can also initiate [2+2] cycloaddition reactions with molecules other than DNA. It also facilitates hydrogen abstraction from linoleic and arachidonic acid, triggering peroxidation. It facilitates C-C bond homolysis, creating free radicals that undergo further reactions.
Fig. 7. Fates of triplet-state carbonyls.
The energy of a triplet carbonyl is dissipated along both harmless and detrimental pathways. Left to right: Energy is converted to heat (thermal deactivation) or to light emission (phosphorescence, Ph). Transfer of energy to an acceptor A can occur by electron exchange. Fluorescent acceptors such as DBAS are useful for detecting poorly luminescing triplet states. Electron exchange with ambient molecular oxygen, which is a triplet in the ground state, creates singlet oxygen – a highly reactive excited state of O2. When energy is transferred to a molecule having conjugated double bonds, such as sorbate or β-carotene, the diradical character of the excited double bond allows isomerization around the remaining single bond, afterward dissipating the excess energy as heat by vibrational relaxation. This is the function of β-carotene and lycopene in the chloroplast’s reaction center. The excited carbonyl’s diradical character initiates [2+2] cycloaddition reactions (Paterno-Buchi reaction), as well as facilitating hydrogen abstraction from H donors (e.g., alcohols and 1,4-dienes) and C-C bond homolysis. Modified from [37].
In the case of DNA, excited state carbonyls created by dioxetane thermolysis have different results depending on the dioxetane’s substituents. Tri- and tetramethyl dioxetane produce more CPDs than do dioxetanes having larger substituents and higher triplet state energies [47, 48]. Larger substituents instead tend to produce DNA strand breaks, dihydropyrimidines, abasic sites, and 8-oxo-guanine or ring-opened purines, apparently reflecting the dominance of H or e− abstraction over Dexter energy transfer. In all cases, the major product appears to be 8-oxo-guanine or ring-opened purines rather than the CPD. The consequences of a molecule that hosts a dioxetane will therefore depend on that host.
7. Genome maintenance in the presence of excited electrons
The excited states of DNA bases offer overlooked lessons about genome stability. Sunlight’s ability to cause cancer often strikes us as a flaw in the design of DNA, but closer inspection reveals that an occasional tumor at post-reproductive age is a near miss in an otherwise astute design: Nature has installed protections even at the level of chemistry. It is easy to miss the fact that the high energies required to make CPDs mean that evolution has successfully prevented visible light from causing cell death and cancer. Moreover, high energy photons are absorbed by water and ozone, allowing aquatic and terrestrial life [49].
Nature has also arranged to protect DNA after excitation. First, CPDs are formed in low yield. This is because, when the photon is absorbed, the 5–6 double bonds of two adjacent pyrimidines must already be close to each other and transiently parallel rather than at their normal 36° angle. This requirement results from the fact that the singlet lifetime of DNA bases is less than a picosecond, insufficient time for an excited base to align with another [13]. The ultrafast deactivation of the excited state, rather than a more typical lifetime of tens of nanoseconds, in turn results from a remarkable feature of DNA bases – the conical intersection [21]. In some molecules, for particular conformations, the ground state energy is unusually high and the singlet state energy is unusually low, so the excited electron can quickly drop to the ground state instead of instigating a photochemical reaction. The four DNA bases possess conical intersections that appear when the 5–6 double bond is twisted. In contrast, base analogs lack a conical intersection; for example, adenine (6-aminopurine) possesses one but 2-aminopurine does not [50, 51]. This contrast suggests that evolution selected A, T, G, and C for their stability against electronic excitation. The conical intersection also speeds CPD formation, but this is less frequent than deactivation because of the alignment requirement. Second, evolution has arranged backup protection when CPDs do form: DNA’s base stacking facilitates excited-state dimeric complexes of adjacent bases, ensuring that CPDs are created within one strand rather than joining complementary strands. Preventing both strands from being damaged at the same site allows the undamaged complementary strand to be used as a template for DNA excision repair [52]. Third, photoexcited bases repair nearby CPDs [53]. Excitation of an excimer of G and A on the same strand leads to a charge separation, with the excess electron going to A. This electron can transfer to an adjacent CPD, splitting the cyclobutane ring and reverting the CPD to normal in the same way that photolyase does. Because of the relative redox potentials, TA sequences cannot promote this “self-healing” process.
In plants, the function of carotenoids like β-carotene and lycopene is not to act as antioxidants but to protect chlorophyll from excited triplet states created as part of its normal activity. The better known role of β-carotene is in the chloroplast’s Light-Harvesting Complex, which contains the all-trans linear isomer to absorb light at wavelengths not captured by chlorophyll. The carotene is excited to its S2 singlet state and transfers this energy to chlorophyll, exciting the chlorophyll to its singlet state. Energy transfer is efficient because singlet all-trans β-carotene rarely isomerizes, which would waste energy as heat. Problems arise when the excited chlorophyll next transfers the energy to a chlorophyll in the Reaction Center, which ejects an electron to pheophytin at the top of the electron transport chain. If, on a sunny day, too many free electrons are produced, some are recaptured by residual singlet chlorophyll+ molecules and create triplet state chlorophyll. As we’ve seen, triplet states can have undesirable consequences. To quench triplet chlorophylls, nature has cleverly arranged that the Reaction Center contains V-shaped 15-cis β-carotene [54]. The carotene accepts the energy from excited triplet chlorophyll, leaving triplet 15-cis β-carotene. Crucially, excited 15-cis β-carotene isomerizes to all-trans β-carotene, drops to the ground state, and dissipates the excess energy harmlessly as heat – a property that the all-trans isomer does not have. β-carotene and lycopene are just longer versions of sorbate, featuring a lower triplet energy level that cannot then make singlet oxygen. Nature has selected for dissipation of triplet energy, not antioxidant activity.
For human skin, we can imagine intervening with drugs or sunscreens to block photoionized electrons; scavenge superoxide, nitric oxide, or peroxynitrite; or annihilate the melanin radical. These strategies resemble the antioxidant approaches widely advocated for preventing diverse pathologies. However, chemoprevention trials have made it clear that high doses of antioxidants such as β-carotene or selenium can be detrimental, probably due to the role of radical species in normal cell signaling [55]. Quenching the excited triplet states is a novel alternative pharmacologic approach. Should not nature have provided us with endogenous triplet quenchers? Perhaps she has; chemical space has not been searched for triplet quenching activity. Alternatively, nature’s solution may be right in front of us. Most birds derive their feather colors from dietary carotenoids. These feather colors are clearly maintained by selection, presumably for mating efficacy, so natural selection can insist on a species’ diet. In humans, therefore, evolutionary selection for endogenous triplet quenchers is capable of expecting that we eat our vegetables.
8. Beyond melanoma
Is melanin good or bad? It is clearly both, with the tradeoff being less satisfactory in the case of blonde or red melanin. One rationale is that this is the best solution to sunlight protection that evolutionary selection encountered. Because skin tumors arise after reproductive age, when selection pressures are reduced, they would be an example of antagonistic pleiotropy. A second explanation takes note of the fact that, as measured by mass spectrometry in melanocyte DNA, the melanin reduced CPDs by about two-fold compared to albinos – the same as the increase coming from chCPDs. But this result is not a wash: chemiexcitation of melanin has redistributed what would have been a bolus of CPDs made at the instant of sun-exposure, so that instead CPD appearance is spread over hours. The slow trickle of chCPDs would then be easier for nucleotide excision repair to accommodate.
Is chemiexcitation only a concern for skin? Any cell that contains melanin, superoxide, and nitric oxide should embark on the same pathway, with no UV involved. Looking further, dioxetanes are not limited to melanin; they are formed on flavins, tryptophan, and acetoacetate [56–58]. These correlations hint that the remarkable excited-electron events involving melanin in skin are the first glimpse of a wider role for chemiexcitation in mammalian pathogenesis.
Acknowledgments
The writing of this perspective was supported by a research grant to D.E.B. from L’Oreal, Inc. and by NIH grant 5P50CA12197409. We thank Dr. C.H. Kang for Figure 1 and Dr. L.C.P. Goncalves for Figures 3 and 7.
Abbreviations
- CPD
cyclobutane pyrimidine dimer
- chCPD
chemiexcitation-induced CPDs
- NOS
nitric oxide synthase
- NOX
NADPH oxidase
- UVA
ultraviolet A radiation (320–400 nm)
- UVB
280–320 nm
- UVC
100–280 nm
References
- 1.Setlow RB, Carrier WL. Pyrimidine dimers in ultraviolet-irradiated DNA’s. J Mol Biol. 1966;17:237–254. doi: 10.1016/s0022-2836(66)80105-5. [DOI] [PubMed] [Google Scholar]
- 2.Brash DE, Seetharam S, Kraemer KH, Seidman MM, Bredberg A. Photoproduct frequency is not the major determinant of UV base substitution hot spots or cold spots in human cells. Proc Natl Acad Sci U S A. 1987;84:3782–3786. doi: 10.1073/pnas.84.11.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperin AJ, Pontén J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci USA. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ziegler A, Leffell DJ, Kunala S, Sharma HW, Gailani M, Simon JA, Halperin AJ, Baden HP, Shapiro PE, Bale AE, Brash DE. Mutation hotspots due to sunlight in the p53 gene of non-melanoma skin cancers. Proc Natl Acad Sci U S A. 1993;90:4216–4220. doi: 10.1073/pnas.90.9.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ziegler A, Jonason AS, Leffell DJ, Simon JA, Sharma HW, Kimmelman J, Remington L, Jacks T, Brash DE. Sunburn and p53 in the onset of skin cancer. Nature. 1994;372:773–776. doi: 10.1038/372773a0. [DOI] [PubMed] [Google Scholar]
- 6.Jonason AS, Kunala S, Price GJ, Restifo RJ, Spinelli HM, Persing JA, Leffell DJ, Tarone RE, Brash DE. Frequent clones of p53-mutated keratinocytes in normal human skin. Proc Natl Acad Sci USA. 1996;93:14025–14029. doi: 10.1073/pnas.93.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daya-Grosjean L, Sarasin A. The role of UV induced lesions in skin carcinogenesis: an overview of oncogene and tumor suppressor gene modifications in xeroderma pigmentosum skin tumors. Mutat Res. 2005;571:43–56. doi: 10.1016/j.mrfmmm.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, Ariyan S, Narayan D, Dutton-Regester K, Capatana A, Holman EC, Bosenberg M, Sznol M, Kluger HM, Brash DE, Stern DF, Materin MA, Lo RS, Mane S, Ma S, Kidd KK, Hayward NK, Lifton RP, Schlessinger J, Boggon TJ, Halaban R. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge D, Fullam A, Alexandrov L, Tubio J, Stebbings L, Menzies A, Widaa S, Stratton M, Jones P, Campbell P. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015 doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brash DE. UV signature mutations. Photochem Photobiol. 2015:15–26. doi: 10.1111/php.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Premi S, Wallisch S, Mano CM, Weiner AB, Bacchiocchi A, Wakamatsu K, Bechara EJ, Halaban R, Douki T, Brash DE. Chemiexcitation of melanin derivatives induces DNA photoproducts long after UV exposure. Science. 2015;347:842–847. doi: 10.1126/science.1256022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schreier WJ, Schrader TE, Koller FO, Gilch P, Crespo-Hernandez CE, Swaminathan VN, Carell T, Zinth W, Kohler B. Thymine dimerization in DNA is an ultrafast photoreaction. Science. 2007;315:625–629. doi: 10.1126/science.1135428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6–4)photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 15.Carrier W, Snyder R, Regan J. Ultraviolet-induced damage and its repair in human DNA. In: Regan J, Parrish J, editors. The Science of Photomedicine. Plenum; New York: 1982. pp. 91–112. [Google Scholar]
- 16.Beecham EJ, Mushinski JF, Shacter E, Potter M, Bohr VA. DNA repair in the c-myc proto-oncogene locus: possible involvement in susceptibility or resistance to plasmacytoma induction in BALB/c mice. Mol Cell Biol. 1991;11:3095–3104. doi: 10.1128/mcb.11.6.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg RJ, Ruven HJ, Sands AT, de Gruijl FR, Mullenders LH. Defective global genome repair in XPC mice is associated with skin cancer susceptibility but not with sensitivity to UVB induced erythema and edema. J Invest Dermatol. 1998;110:405–409. doi: 10.1111/j.1523-1747.1998.00173.x. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler DL, Martin KE, Ness KJ, Li Y, Dreckschmidt NE, Wartman M, Ananthaswamy HN, Mitchell DL, Verma AK. Protein kinase C epsilon is an endogenous photosensitizer that enhances ultraviolet radiation-induced cutaneous damage and development of squamous cell carcinomas. Cancer Res. 2004;64:7756–7765. doi: 10.1158/0008-5472.CAN-04-1881. [DOI] [PubMed] [Google Scholar]
- 19.Gordon-Thomson C, Gupta R, Tongkao-on W, Ryan A, Halliday GM, Mason RS. 1alpha,25 dihydroxyvitamin D3 enhances cellular defences against UV-induced oxidative and other forms of DNA damage in skin. Photochem Photobiol Sci. 2012;11:1837–1847. doi: 10.1039/c2pp25202c. [DOI] [PubMed] [Google Scholar]
- 20.Kraemer KH, Coon HG, Petinga RA, Barrett SF, Rahe AE, Robbins JH. Genetic heterogeneity in xeroderma pigmentosum: complementation groups and their relationship to DNA repair rates. Proc Natl Acad Sci U S A. 1975;72:59–63. doi: 10.1073/pnas.72.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boggio-Pasqua M, Groenhof G, Schafer LV, Grubmuller H, Robb MA. Ultrafast deactivation channel for thymine dimerization. J Am Chem Soc. 2007;129:10996–10997. doi: 10.1021/ja073628j. [DOI] [PubMed] [Google Scholar]
- 22.Middleton CT, de La Harpe K, Su C, Law YK, Crespo-Hernandez CE, Kohler B. DNA excited-state dynamics: from single bases to the double helix. Annu Rev Phys Chem. 2009;60:217–239. doi: 10.1146/annurev.physchem.59.032607.093719. [DOI] [PubMed] [Google Scholar]
- 23.Ruzsicska BP, Lemaire DGE. DNA photochemistry. In: Horspool WM, Song PS, editors. CRC Handbook of Organic Photochemistry and Photobiology. CRC Press; Boca Raton: 1995. pp. 1289–1317. [Google Scholar]
- 24.Eisinger J, Lamola A. The excited states of nucleic acids. In: Steiner WIRF, editor. Excited States of Proteins and Nucleic Acids. Plenum Press; New York: 1971. pp. 107–185. [Google Scholar]
- 25.Bosca F, Lhiaubet-Vallet V, Cuquerella MC, Castell JV, Miranda MA. The triplet energy of thymine in DNA. J Am Chem Soc. 2006;128:6318–6319. doi: 10.1021/ja060651g. [DOI] [PubMed] [Google Scholar]
- 26.de Laat A, van der Leun JC, de Gruijl FR. Carcinogenesis induced by UVA (365-nm) radiation: the dose-time dependence of tumor formation in hairless mice. Carcinogenesis. 1997;18:1013–1020. doi: 10.1093/carcin/18.5.1013. [DOI] [PubMed] [Google Scholar]
- 27.Rochette PJ, Therrien JP, Drouin R, Perdiz D, Bastien N, Drobetsky EA, Sage E. UVA-induced cyclobutane pyrimidine dimers form predominantly at thymine-thymine dipyrimidines and correlate with the mutation spectrum in rodent cells. Nucleic Acids Res. 2003;31:2786–2794. doi: 10.1093/nar/gkg402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douki T, Reynaud-Angelin A, Cadet J, Sage E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry. 2003;42:9221–9226. doi: 10.1021/bi034593c. [DOI] [PubMed] [Google Scholar]
- 29.Mouret S, Philippe C, Gracia-Chantegrel J, Banyasz A, Karpati S, Markovitsi D, Douki T. UVA-induced cyclobutane pyrimidine dimers in DNA: a direct photochemical mechanism? Org Biomol Chem. 2010;8:1706–1711. doi: 10.1039/b924712b. [DOI] [PubMed] [Google Scholar]
- 30.Markovitsi D. UV-induced DNA damage: the role of electronic excited states. Photochem Photobiol. 2016;92:45–51. doi: 10.1111/php.12533. [DOI] [PubMed] [Google Scholar]
- 31.Valencia A, Kochevar IE. Nox1-based NADPH oxidase is the major source of UVA-induced reactive oxygen species in human keratinocytes. J Invest Dermatol. 2008;128:214–222. doi: 10.1038/sj.jid.5700960. [DOI] [PubMed] [Google Scholar]
- 32.Chedekel MR, Smith SK, Post PW, Pokora A, Vessell DL. Photodestruction of pheomelanin: role of oxygen. Proc Natl Acad Sci USA. 1978;75:5395–5399. doi: 10.1073/pnas.75.11.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamola AA. Production of pyrimidine dimers in DNA in the dark. Biochem Biophys Res Commun. 1971;43:893–898. doi: 10.1016/0006-291x(71)90701-7. [DOI] [PubMed] [Google Scholar]
- 34.Wilson T, Hastings JW. Bioluminescence: Living Lights, Lights for Living. Cambridge, MA: 2013. [Google Scholar]
- 35.White E, Miano J, Watkins C, Breaux E. Chemically produced excited states. Angw Chem Internat Edit. 1974;13:229–243. [Google Scholar]
- 36.Cilento G. Electronic excitation in dark biological processes. In: Adam W, Cilento G, editors. Chemical and Biological Generation of Excited States. Academic Press; New York: 1982. pp. 277–307. [Google Scholar]
- 37.Baader WJ, Stevani CV, Bechara EJH. “Photo” chemistry without light? J Braz Chem Soc. 2015;12:2430–2447. [Google Scholar]
- 38.Knudsen FS, Penatti CA, Royer LO, Bidart KA, Christoff M, Ouchi D, Bechara EJ. Chemiluminescent aldehyde and beta-diketone reactions promoted by peroxynitrite. Chem Res Toxicol. 2000;13:317–326. doi: 10.1021/tx990176i. [DOI] [PubMed] [Google Scholar]
- 39.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romero-Graillet C, Aberdam E, Biagoli N, Massabni W, Ortonne JP, Ballotti R. Ultraviolet B radiation acts through the nitric oxide and cGMP signal transduction pathway to stimulate melanogenesis in human melanocytes. J Biol Chem. 1996;271:28052–28056. doi: 10.1074/jbc.271.45.28052. [DOI] [PubMed] [Google Scholar]
- 41.Velosa AC, Baader WJ, Stevani CV, Mano CM, Bechara EJ. 1,3-diene probes for detection of triplet carbonyls in biological systems. Chem Res Toxicol. 2007;20:1162–1169. doi: 10.1021/tx700074n. [DOI] [PubMed] [Google Scholar]
- 42.Michl J. Photophysics of organic molecules in solution. In: Montalti M, Credi A, Prodi L, editors. Handbook of Photochemistry. 3. CRC / Taylor & Francis; Boca Raton: 2006. p. 650. [Google Scholar]
- 43.Kayatz P, Thumann G, Luther TT, Jordan JF, Bartz-Schmidt KU, Esser PJ, Schraermeyer U. Oxidation causes melanin fluorescence. Investigative ophthalmology & visual science. 2001;42:241–246. [PubMed] [Google Scholar]
- 44.Slawinska D, Slawinski J. Electronically excited molecules in the formation and degradation of melanins. Physiol Chem Phys. 1982;14:363–374. [PubMed] [Google Scholar]
- 45.Wakamatsu K, Nakanishi Y, Miyazaki N, Kolbe L, Ito S. UVA-induced oxidative degradation of melanins: fission of indole moiety in eumelanin and conversion to benzothiazole moiety in pheomelanin. Pigment Cell Melanoma Res. 2012;25:434–445. doi: 10.1111/j.1755-148X.2012.01011.x. [DOI] [PubMed] [Google Scholar]
- 46.Saito I, Matsugo S, Matsuura T. 1,2-Dioxetane formation in an indole system. J Am Chem Soc. 1979;101:4757–4759. [Google Scholar]
- 47.Adam W, Beinhauer A, Mosandl T, Saha-Moller C, Vargas F, Epe B, Muller E, Schiffmann D, Wild D. Photobiological studies with dioxetanes in isolated DNA, bacteria, and mammalian cells. Environmental health perspectives. 1990;88:89–97. doi: 10.1289/ehp.908889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Epe B, Muller E, Adam W, Saha-Moller CR. Photochemical DNA modifications induced by 1,2-dioxetanes. Chem Biol Interact. 1992;85:265–281. doi: 10.1016/0009-2797(92)90067-u. [DOI] [PubMed] [Google Scholar]
- 49.Sagan C. Ultraviolet selection pressue on the earliest organisms. J Theoret Biol. 1973;39:195–200. doi: 10.1016/0022-5193(73)90216-6. [DOI] [PubMed] [Google Scholar]
- 50.Kang H, Lee KT, Jung B, Ko YJ, Kim SK. Intrinsic lifetimes of the excited state of DNA and RNA bases. JACS Commun. 2002;124:12958–12959. doi: 10.1021/ja027627x. [DOI] [PubMed] [Google Scholar]
- 51.Gustavsson T, Improta R, Markovitsi D. DNA/RNA: building blocks of life under UV irradiation. J Phys Chem Lett. 2010;1:2025–2030. [Google Scholar]
- 52.Crespo-Hernandez C, Cohen B, Kohler B. Base stacking controls excited-state dynamics in A.T DNA. Nature. 2005;436:1141–1144. doi: 10.1038/nature03933. [DOI] [PubMed] [Google Scholar]
- 53.Bucher DB, Kufner CL, Schlueter A, Carell T, Zinth W. UV-Induced Charge Transfer States in DNA Promote Sequence Selective Self-Repair. J Am Chem Soc. 2016;138:186–190. doi: 10.1021/jacs.5b09753. [DOI] [PubMed] [Google Scholar]
- 54.Koyama Y. Structures and functions of carotenoids in photosynthetic systems. J Photochern Photobiol B. 1991;9:265–280. [Google Scholar]
- 55.Rhee SG. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 56.Royer LO, Knudsen FS, de Oliveira MA, Tavares MF, Bechara EJ. Peroxynitrite-initiated oxidation of acetoacetate and 2-methylacetoacetate esters by oxygen: potential sources of reactive intermediates in keto acidoses. Chem Res Toxicol. 2004;17:1725–1732. doi: 10.1021/tx049821y. [DOI] [PubMed] [Google Scholar]
- 57.Jung MY, Oh YS, Kim DK, Kim HJ, Min DB. Photoinduced generation of 2,3-butanedione from riboflavin. Journal of agricultural and food chemistry. 2007;55:170–174. doi: 10.1021/jf061999y. [DOI] [PubMed] [Google Scholar]
- 58.Ronsein GE, Oliveira MC, Miyamoto S, Medeiros MH, Di Mascio P. Tryptophan oxidation by singlet molecular oxygen [O2(1Deltag)]: mechanistic studies using 18O-labeled hydroperoxides, mass spectrometry, and light emission measurements. Chem Res Toxicol. 2008;21:1271–1283. doi: 10.1021/tx800026g. [DOI] [PubMed] [Google Scholar]
- 59.Park H, Zhang K, Ren Y, Nadji S, Sinha N, Taylor JS, Kang C. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc Natl Acad Sci U S A. 2002;99:15965–15970. doi: 10.1073/pnas.242422699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lamola AA. Fundamental aspects of the spectroscopy and photochemistry of organic compounds; electronic energy transfer in biologic systems; and photosensitization. In: Pathak MA, Harber LC, Seiji M, Kukita A, editors. Sunlight and Man. University of Tokyo Press; Tokyo: 1972. pp. 17–55. [Google Scholar]
- 61.Schreier WJ, Gilch P, Zinth W. Early events of DNA photodamage. Annu Rev Phys Chem. 2015;66:497–519. doi: 10.1146/annurev-physchem-040214-121821. [DOI] [PubMed] [Google Scholar]