Summary
N6-methyladenosine (m6A) is a prevalent, reversible chemical modification of functional RNAs, and is important for central events in biology. The core m6A writers are Mettl3 and Mettl14, which both contain methyltransferase domains. How Mettl3 and Mettl14 cooperate to catalyze methylation of adenosines has remained elusive. We present crystal structures of the complex of Mettl3/Mettl14 methyltransferase domains in apo form as well as with bound S-adenosylmethionine (SAM) or S-adenosylhomocysteine (SAH) in the catalytic site. We determine that the heterodimeric complex of methyltransferase domains, combined with CCCH motifs constitute the minimally required regions for creating m6A modifications in vitro. We also show that Mettl3 is the catalytically active subunit while Mettl14 plays a structural role critical for substrate recognition. Our model provides a molecular explanation for why certain mutations of Mettl3 and Mettl14 lead to impaired function of the methyltransferase complex.
eTOC blurb
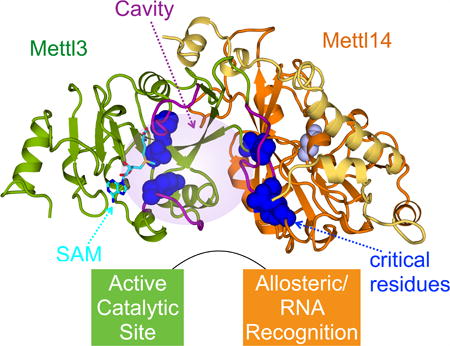
Wang et al. reveal crystal structures of Mettl3/Mettl14 complexes. Extensive intermolecular contact enables Mettl3 and Mettl14 to work cooperatively. Mettl3 is the catalytic subunit, and Mettl14 activates Mettl3 via allostery and recognition of RNA substrates. They explain why certain mutations including those involved in cancer affect RNA methylation.
Introduction
Specific, controlled methylation of nucleic acids is essential for proper gene regulation. One of the most prevalent modifications observed for mRNAs is N6-methyladenosine (m6A). Even though m6A was discovered decades ago, investigation of its role in gene expression lagged behind until it was proved to be reversible, through the discovery of specific m6A demethylases (Jia et al., 2011; Zheng et al., 2013). Recent studies have intensely investigated how m6A-modification of RNA contributes to central events in biology (Fu et al., 2014; Liu and Pan, 2016; Meyer and Jaffrey, 2014). Impaired function of m6A writers, readers, and erasers have been linked to problems in self-renewal of stem cells, circadian clock and developmental defects, obesity, synaptic signaling, and cancers (Ben-Haim et al., 2014; Chen et al., 2015; Fustin et al., 2013; Geula et al., 2015; Peng et al., 2016; Wang et al., 2014a; Zheng et al., 2013).
The molecular role of m6A is still being uncovered. For most mRNAs, m6A modification is most frequently observed in long exons, near stop codons, and in 3′ UTRs (Dominissini et al., 2012; Meyer et al., 2012). Addition of m6A can alter mRNA stability, induce RNA conformational changes, modulate protein-RNA interactions, and even modify microRNA processing (Alarcón et al., 2015; Liu et al., 2015b; Wang et al., 2014a; Wang et al., 2015; Zhou et al., 2016). Several studies investigating the sequence specificity of m6A modification discovered RRACH (R represents A or G, and H represents A, C or U) as the consensus sequence, with the central A as the methyl acceptor (Csepany et al., 1990; Dominissini et al., 2012; Harper et al., 1990; Meyer et al., 2012; Rottman et al., 1994; Schibler et al., 1977). The methyltransferase activity has not been shown to be sensitive to any RNA structural context for creating the m6A mark in vitro (Liu et al., 2014).
Mettl3 was identified as a catalytic component of the methyltransferase complex responsible for creating m6A modifications due to its ability to crosslink with S-adenosylmethionine (SAM) (Bokar et al., 1997). Mettl3 also forms stable complexes with Mettl14, which contains its own methyltransferase domain with a variant catalytic motif (Bujnicki et al., 2002; Liu et al., 2014; Wang et al., 2014a). Although isolated Mettl14 purified from insect cells was reported to have weak methyltransferase activity (Liu et al., 2014), phylogenetic studies of the active site motif suggests that it might have lost catalytic activity (Iyer et al., 2016). For normal m6A modification to occur in cells, Mettl3 and Mettl14 also need to associate with additional factors, such as Wilm's tumor 1-associated protein (WTAP), that may aid with proper localization (Ping et al., 2014; Schwartz et al., 2014). In vitro studies showed that Mettl3 and Mettl14 help each other's protein stability, and that they synergize to produce m6A marks on cognate RNA oligonucleotides (Liu et al., 2014; Wang et al., 2014b). However, the molecular basis for the cooperation between Mettl3 and Mettl14 is unknown.
Here we present crystal structures of the complex of Mettl3 and Mettl14 methyltransferase catalytic domains. We reveal a high-resolution view of the catalytic site of Mettl3 in apo form as well as when occupied by SAM or S-adenosylhomocysteine (SAH). The Mettl14 catalytic site is relatively occluded and shows no sign of binding the necessary cofactor, SAM. In vitro methylation assays with full-length polypeptides containing point mutations in the catalytic motifs of Mettl3 and Mettl14 show that Mettl3 is the only catalytically active subunit. While Mettl14 active site is not required, it has a critical role to structurally support Mettl3, and to interact with substrate RNA. We reveal an extensive set of favorable interactions between Mettl3 and Mettl14, which is important to stabilize the structure of both domains as well as interdomain coordination. We determine that the crystallized methyltransferase domain heterodimer and the two Cys-Cys-Cys-His (CCCH)-type zinc binding motifs of Mettl3 are necessary and sufficient for full in vitro methylation activity. Finally, we identify and provide a molecular explanation for point mutations of Mettl3 and Mettl14 that impair m6A writing, some of which were previously linked to cancer.
Results
Structure of Mettl3/Mettl14 methyltransferase domain complex
To investigate the mechanism of how human Mettl3 and Mettl14 cooperate to methylate RNA, we sought to obtain a three-dimensional structure of the complex. Previous reports state that Mettl3 and Mettl14 must be expressed in insect cells for proper folding (Liu et al., 2014; Liu et al., 2015a). However, when the two proteins were co-expressed in E. coli, we were able to obtain reasonable quantities of homogeneously purified full-length complexes (Figures S1A and S1B). We attempted to crystallize the full-length complex but could not obtain suitable crystals after screening a wide range of conditions. Mettl3 and Mettl14 both contain a predicted methyltransferase catalytic domain (Figure 1A). Through limited proteolysis we determined that the methyltransferase domains of Mettl3 (MTD3, residues 357-580) and Mettl14 (MTD14, residues 111-456) are necessary and sufficient to form stable complexes that can co-elute from a gel-filtration column (Figure S1C). In solution, full-length Mettl3 and Mettl14 polypeptides form a 1:1 complex, as apparent from the molecular weight measured using multi-angle light scattering (Figure 1B). The truncated complex containing MTD3 and MTD14 also maintains the same stoichiometry as the full-length polypeptides (Figure 1C).
Figure 1. Structure of the Mettl3/Mettl14 methyltransferase domain complex.
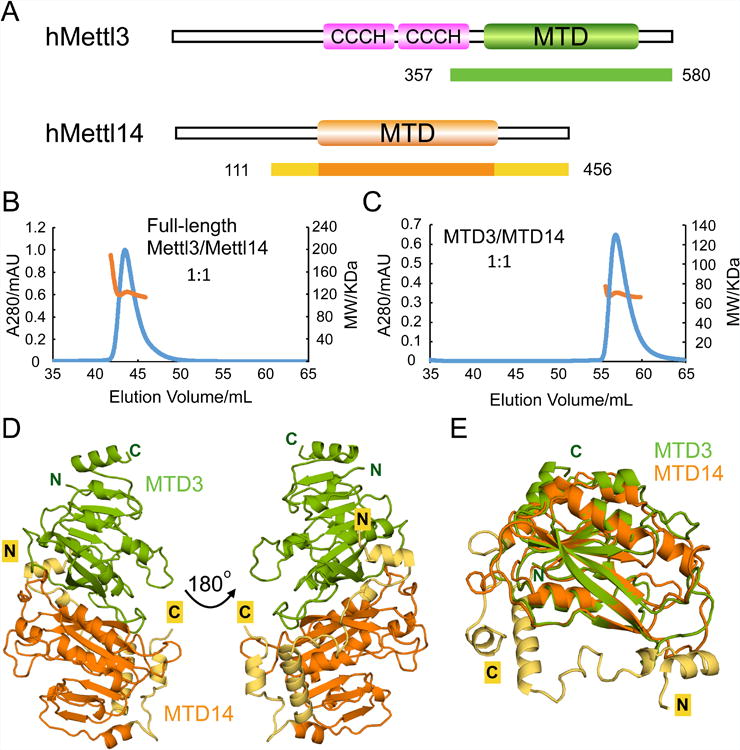
(A) Domain architecture of human Mettl3 and Mettl14. Crystallization constructs are indicated with color-coded bars underneath, with corresponding residue numbers. The terminal extensions of MTD14 are colored in yellow. CCCH indicates a Cys-Cys-Cys-His motif.
(B) Size-exclusion chromatography with Multi-Angle Light Scattering (SEC-MALS) profile for the full-length Mettl3/Mettl14 complex. The left axis represents the absorbance at 280 nm (blue peak) and the right axis represents the measured molecular weight from the scattering (orange line) at each elution volume. The theoretical molecular weight for 1:1 Mettl3/Mettl14 complex is 118 KDa and the average measured molecular weight from MALS is 122 KDa.
(C) SEC-MALS profile for the MTD3/MTD14 complex. Similar representation as (B). The theoretical molecular weight for 1:1 MTD3/MTD14 complex is 65KDa and the average measured molecular weight from MALS is 68KDa.
(D) Cartoon representation of the overall structure of the MTD3/MTD14 complex. MTD3 is shown in green and MTD14 is colored orange. The N-terminal (N) and C-terminal (C) termini are as indicated, and the terminal extensions of MTD14 are colored yellow.
(E) Superimposition of MTD3 and MTD14 in cartoon representation.
See also Figure S1.
When the proteolytically labile portions were removed, the MTD3/MTD14 complex could produce well-ordered crystals that diffracted to 1.65 Å resolution (Table 1). Due to low sequence similarity with known structures of MTDs, molecular replacement failed to solve the phase problem. Thus, we determined the structure of MTD3/MTD14 complex using single anomalous dispersion using selenomethionine-labeled complexes. The overall structure of the complex shows that MTD3 and MTD14 engage in extensive contact with each other to form a stable heterodimer (Figure 1D). The two MTDs share ∼25% sequence identity, and superimpose well with core RMSD of ∼0.9 Å for 118 Cα atoms (Figure 1E). MTD14 has extra terminal extensions (shown in yellow), including an unusual N-terminal extension of about 50aa (Figure S2), which forms a long helix to traverse across one face of MTD14 and makes intimate contact with MTD3 via loops and shorter helical segments (Figure 1D, yellow). The C-terminal helix of MTD14 also lies antiparallel to the N-terminal extension helix to support its position. Therefore, a large interdomain binding interface, combined with additional favorable contacts through the terminal extensions, secures the relative positions of the two domains for coordinated function.
Table 1. Data collection and structure refinement statistics.
| Ligand | Se-Met | Apo | SAM | SAH |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 0.97924 | 0.97934 | 0.97934 | 0.97934 |
| Resolution range (Å) | 50 -1.90 (1.93 - 1.90)a | 50 -1.65 (1.68-1.65) | 50 - 1.70 (1.73 - 1.70) | 50 - 1.65 (1.68 - 1.65) |
| Space group | P 41212 | P 41212 | P 41212 | P 41212 |
| Unit cell (Å, °) | 101.3, 101.3, 117.9 90, 90, 90 | 101.9, 101.9, 117.7 90, 90, 90 | 101.7, 101.7, 117.5 90, 90, 90 | 101.9, 101.9, 118.0 90, 90, 90 |
| Total reflections | 986215 | 1561173 | 1157172 | 1217153 |
| Unique reflections | 48686 | 74698 | 68473 | 75134 |
| Multiplicity | 20.3 (19.5) | 20.9(20.6) | 16.9 (16.6) | 16.2 (16.4) |
| Completeness (%) | 100 (100) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (99.9) |
| Mean I/sigma (I) | 28.0 (4.1) | 47.3 (3.75) | 38.9 (3.4) | 32.8 (3.3) |
| Rmerge | 0.104 (0.889) | 0.063 (0.943) | 0.073 (0.839) | 0.078 (0.767) |
| Structure refinement | ||||
| R-factor/ R-freeb | 0.1602/0.1831 | 0.1634/0.1873 | 0.1598/0.1794 | |
| RMS (bonds) | 0.009 | 0.007 | 0.006 | |
| RMS (angles) | 1.055 | 0.952 | 0.924 | |
| No. of atoms | 4486 | 4584 | 4587 | |
| Macromolecules atoms | 3998 | 4027 | 4007 | |
| Ligands atoms | 0 | 27 | 26 | |
| Waters | 488 | 530 | 554 | |
| Average B-factor | 25.9 | 22.6 | 22.8 | |
| Macromolecules B-factor | 25.1 | 21.3 | 21.4 | |
| Ligand B-factor | 0 | 20.2 | 23.6 | |
| Waters B-factor | 32.8 | 32.6 | 33.1 | |
| Ramachandran plot statistics | ||||
| Most favored regions (%) | 89.2 | 90.0 | 88.6 | |
| Allowed regions (%) | 10.5 | 10.0 | 11.2 | |
| Generously allowed regions (%) | 0.2 | 0 | 0.2 | |
| Disallowed regions (%) | 0 | 0 | 0 | |
The values for the data in the highest resolution shell are shown in parentheses.
Rfree = Σ Test‖Fobs| - |Fcalc‖/Σ Test |Fobs|, where “Test” is a test set of about 5% of the total reflections randomly chosen and set aside prior to refinement for the structure.
Mettl3 catalytic site is critical for methyltransferase activity
Mettl3 and Mettl14 are known to cooperate to produce specific m6A marks on RNA, but the specific role of each component has remained elusive. Sequence analysis suggests that Mettl3 catalytic site contains a more conserved DPPW motif, while Mettl14 has a more divergent EPPL sequence, which is not well conserved even within the Mettl14 family (Figure 2A) (Bujnicki et al., 2002; Iyer et al., 2016). When we compare the catalytic sites of MTD3 and MTD14 after structural superimposition, MTD3 has the more open cavity. MTD14 has an inserted helical segment after its catalytic motif, EPPL (Figures 2A and S2), which occludes the space that is hollow in MTD3 (Figure 2B). Furthermore, the proposed active site cavity in MTD3 is highly conserved, but such conservation is not observed in MTD14 (Figure 2C).
Figure 2. Mettl3 is the active subunit, and Mettl14 catalytic motif is dispensable.
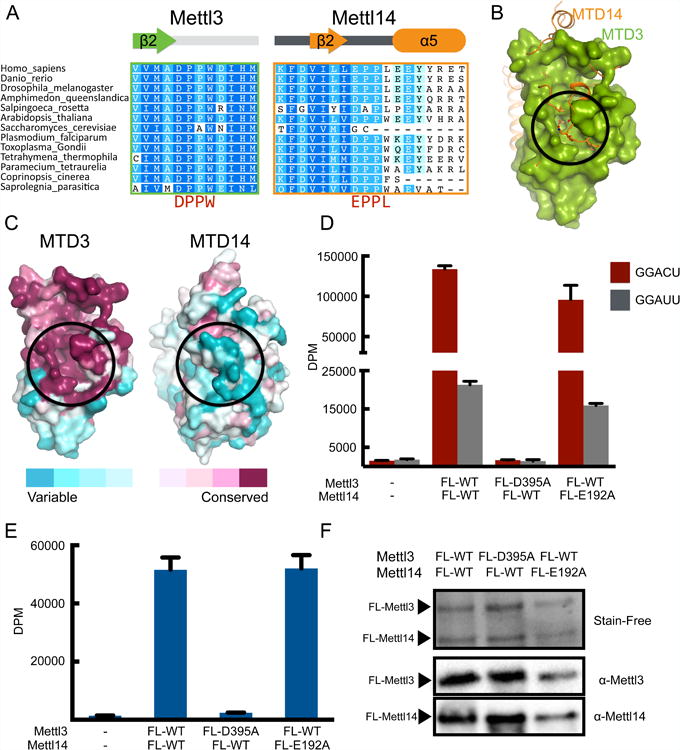
(A) Sequence alignment of the region containing the catalytic motifs DPPW (Mettl3) and EPPL (Mettl14). The secondary structures of the segments are indicated above the sequences.
(B) Superimposition of MTD3 (green surface representation) and MTD14 (orange cartoon and sticks representation). The proposed catalytic cavity (black circle) is more occluded in MTD14.
(C) Sequence conservation shown on surface representation of MTD3 and MTD14. Both MTD3 and MTD14 are colored according to the conservation score. The proposed active site is marked with a black circle.
(D) In vitro methyltransferase activity of the full-length Mettl3/Mettl14 complexes expressed in E. coli, with indicated point mutations for each polypeptide. Bars correspond to amounts of tritium incorporated into methylated RNA substrates shown as disintegrations per minute (DPM) with cognate (GGACU=red) or mutant (GGAUU=gray) sequence. Data shown as mean +/- SD from three replicates.
(E) In vitro methyltransferase activity of the full-length Mettl3/Mettl14 complex with indicated point mutations purified from HEK293 cells (blue bars). Data shown as mean +/- SD from three replicates.
(F) SDS-PAGE analysis (visualized by Stain-Free dye) followed by western blot of the proteins expressed in HEK293 cells used in the in vitro methylation assay in Figure 2E.
See also Figures S2 and S3.
From our structural model we hypothesized that Mettl3 may be the only active methyltransferase in the heterodimeric complex. We tested the ability of each polypeptide to catalyze methylation of RNA in vitro, using a synthetic RNA oligonucleotide containing the consensus sequence GGACU, previously established to be a target for m6A modification (Liu et al., 2014). Mettl3 and Mettl14 proteins are more stable as a complex. We thus generated point mutations of the catalytic motif in full-length Mettl3 Mettl14 complexes, and measured their ability to transfer methyl groups from SAM to RNA. While changing DPPW catalytic motif to APPW (D395A) in Mettl3 abrogates the ability of its complex with wild type Mettl14 to methylate the RNA probe, changing EPPL to APPL in Mettl14 (E192A) has little effect if wild type Mettl3 is present (Figure 2D). A single transition mutation in the consensus sequence for m6A modification, GGACU to GGAUU, is sufficient to significantly reduce methylation levels, in agreement with known RNA specificity for m6A writers. Thus, our in vitro methylation assay is likely to only detect specific methylation events of m6A targets by Mettl3/Mettl14 complexes.
The stark difference in how catalytic mutations of Mettl3 and Mettl14 affect in vitro methylation is not expected from previous results that showed that Mettl14 by itself can have slightly more activity than Mettl3 (Liu et al., 2014). Because it is possible that post-translational modifications can impact methyltransferase activity of Mettl14, we also tested similar constructs purified from HEK293 cells. We consistently observe that mutating the catalytic motif of Mettl3 obliterates methylation activity, while mutant Mettl14 retains full activity (Figures 2E and 2F). Post-translational modifications do not seem to change in vitro methylation activity of Mettl3/Mettl14 complexes significantly, because we observe similar levels of methylated RNA with wild type complexes purified from E. coli, insect cells, and mammalian cells (Figures S3A and S3B). When we purify wild type Mettl3 and Mettl14 polypeptides individually from insect cells, we can reproduce the previously reported activity for Mettl14 (Figure S3C). However, the activity of singly overexpressed Mettl14 is higher than Mettl3, because more endogenous Mettl3 co-purifies with Mettl14 (Figures S3D and S3E).
Structure of MTD3/MTD14 with bound SAM and SAH
Unlike some other bacterially expressed MTDs, our MTD3/MTD14 complex does not co-purify with any cofactor. In order to investigate the non-overlapping roles of Mettl3 and Mettl14, we determined structures of MTD3/MTD14 in complex with a cofactor substrate (SAM) and its product (SAH), to 1.70 Å and 1.65 Å resolution, respectively. For both ligand bound structures, clear electron density is visible in the conserved cavity near the catalytic motif in Mettl3 (Figures 3A and 3B), but no such density is visible for Mettl14. When the catalytic site of MTD14 is superimposed on MTD3 with bound SAM, there are obvious clashes between modeled SAM and the MTD14 pocket (Figures S4A and S4B). As predicted from the more occluded pocket in the apo structure and mutagenesis data, these structures are consistent with Mettl3 being the catalytic subunit, not Mettl14.
Figure 3. The SAM and SAH binding pockets of MTD3.
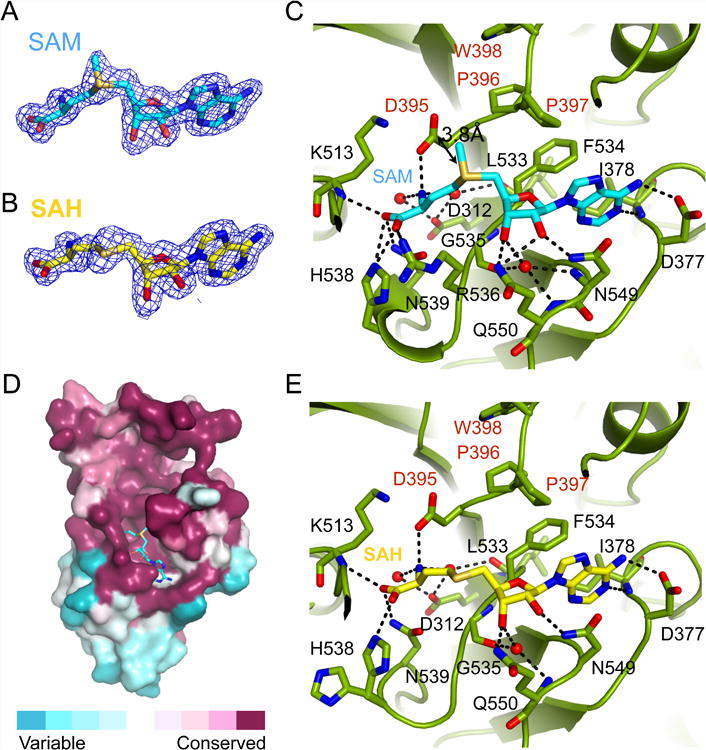
(A) Composite omit map contoured at 1.0 σ shows clear electron density (blue mesh) for SAM, shown in stick representation with backbone colored in cyan.
(B) Composite omit map contoured at 1.0 σ shows clear electron density (blue mesh) for SAH, shown in stick representation with backbone colored in yellow.
(C) Detailed interactions of SAM (cyan sticks) and the active site of MTD3 (green cartoon representation). Specific residues making contacts with SAM are shown in green stick representation and labeled with black text. The catalytic motif DPPW is labeled with red text. Important water molecules involved in SAM binding are shown as red spheres. Hydrogen bonds are indicated with black dashed lines. The distance between the methyl group of SAM and the proposed catalytic residue D395 is marked with a double-headed arrow (3.8Å).
(D) Evolutionary sequence conservation projected on the surface of MTD3 shows high conservation near the SAM binding site. SAM is shown in stick representation, backbone colored in cyan.
(E) Detailed interactions of SAH (yellow sticks) and the active site of MTD3 (green cartoon representation). Specific residues making contacts with SAH are shown in green stick representation and labeled with black text. The active site DPPW motif is labeled with red text. Important water molecules involved in SAM binding are shown as red spheres. Hydrogen bonds are marked as black dashed lines.
See also Figure S4.
SAM is stabilized in the binding pocket by numerous favorable interactions. The adenine ring is sandwiched between Phe 534 and Asn 549, while many polar contacts help to hold the hydroxyl groups on the ribose as well as the amino and carboxyl groups of SAM (Figure 3C). The conserved Asp of the DPPW motif, the proposed catalytic residue to activate the N6 of the modified adenine base, is poised close (3.8 Å) to the methyl group that needs to be transferred. SAM binding site is on one side of a large cavity that is highly conserved at the primary sequence level throughout evolution (Figure 3D). The rest of the conserved cavity is likely for recognizing the RNA substrates. Similar to other MTDs, binding SAM or SAH does not induce a large conformational change of the domain, and most of the interactions are retained after the methyl transfer (Figure 3E) (Guja et al., 2013; Thomas et al., 2003; Yang et al., 2010). Nevertheless, when the apo structure is compared with the SAM or SAH bound states, we observe the largest movement in the loops that “fence” the perimeter of the cavity, especially near the carboxyl group of the bound cofactor (Figure S4C).
Mettl14 stabilizes Mettl3 conformation to increase catalytic activity
The buried surface area between MTD3 and MTD14 is large (2460 Å2) with low temperature factors suggesting a highly stable interface (Figure 4A). Such extensive interaction between MTD3 and MTD14 helps explain why both domains help each other for proper folding and stability. Residues involved in intermolecular contact are spread throughout the surface, and the interacting surfaces tend to be more conserved throughout evolution (Figures 4B-E). Many favorable interactions of various kinds between MTD3 and MTD14 are visualized by our atomic model, including a large hydrophobic patch lined with polar contacts (Figure 4F). When a dramatic mutation in this patch, such as F429A of Mettl3, is introduced, the yield for soluble complex decreases significantly (Figures S5A and S5B).
Figure 4. Mettl14 stabilizes and facilitates Mettl3 catalytic activity.
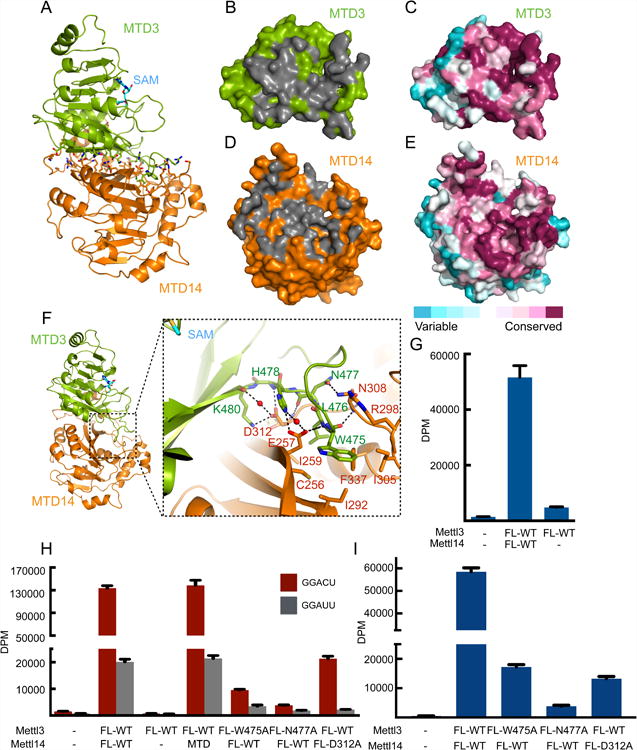
(A) The overall structure of the SAM-bound MTD3/MTD14 complex shown in cartoon representation. SAM is shown in stick representation with backbone colored in cyan. Residues identified by Ligplot to be interdomain contacts are shown with sticks for side chains (green for MTD3 and orange for MTD14).
(B) Green surface representation of MTD3 with residues that contact MTD14 shown in gray.
(C) MTD3 in the same orientation as in (B), but colored by sequence conservation.
(D) Orange surface representation of MTD14 with residues that contact MTD3 shown in gray.
(E) MTD14 in the same orientation as in (D), but colored by sequence conservation.
(F) The overall structure of the SAM-bound MTD3/MTD14 complex shown in cartoon representation. SAM is shown as cyan sticks. The dashed rectangle provides a close-up view of the MTD3/MTD14 binding interface. Critical residues for heterodimer formation are shown as green sticks and labeled with green text (MTD3) or orange sticks and labeled with red text (MTD14). Water molecules mediating hydrogen bonds are shown as small red spheres and hydrogen bonds are shown as black dashed lines.
(G) In vitro methyltransferase activity of the full-length Mettl3/Mettl14 complex or Mettl3 alone expressed in HEK293 cells. Data shown as mean +/- SD from three replicates.
(H) In vitro methyltransferase activity of the full-length Mettl3/Mettl14 complexes expressed in E. coli with indicated point mutations or truncation (MTD). Data shown as mean +/- SD from three replicates.
(I) In vitro methyltransferase activity of the full length Mettl3/Mettl14 complexes expressed in HEK293 cells with indicated point mutations. Data shown as mean +/- SD from three replicates.
See also Figure S5.
Even though Mettl3 is the catalytically active subunit, its activity is negligible without forming stoichiometric complexes with Mettl14, regardless of whether it is expressed in bacteria, insect cells, or mammalian cells (Figures 4G, 4H, S1B, S3C, S3D and S5C). Thus, physically partnering with Mettl14 is necessary for Mettl3 to manifest its methyltransferase activity. In fact, the crystallized construct of Mettl14 (MTD14) is sufficient to rescue Mettl3's activity completely (Figure 4H). Thus, the MTD3/MTD14 interface revealed by our crystal structures includes the features of Mettl14 relevant for potentiating Mettl3's catalytic activity in vitro. We asked how MTD14 might activate Mettl3 by testing intermolecular contacts. In Mettl3, the catalytic cavity is fenced in by three major loops: the two loops near the perimeter of the SAM binding site (Figure S4C), and a third, larger loop which makes extensive contact with Mettl14 (Figure 4F).
We mutated evolutionarily conserved (Figures S5D and S5E), key interacting residues in full-length constructs of Mettl3 (W475A and N477A) and Mettl14 (D312A) to disrupt the intermolecular interactions near this third loop. Although the overall protein solubility does not change noticeably (Figure S1B), the mutant complexes have drastically lowered methylation activity compared to the wild type complexes (Figures 4H,4I, S1B and S5F). Therefore, Mettl14 seems to have a structural role that is critical to support Mettl3's catalytic function.
RNA substrate interactions
MTD3/MTD14 complexes by themselves cannot catalyze m6A modification in vitro (Figure 5A). In Mettl3, there are two CCCH motifs adjacent to MTD3 in Mettl3 (Figure S6A). Since zinc-binding motifs such as CCCHs are known for their abilities to bind nucleic acids (Brown, 2005; Hall, 2005; Lunde et al., 2007), we postulated that they are required for RNA methylatransferase activity. Indeed, when CCCH motifs are attached to the crystallization construct, the complex can be as active as the full-length complexes (Figures 5A and S6B). Consistently, when either of the CCCH motifs is disrupted with point mutations (C294A and C326A), full-length complexes lose the ability to catalyze m6A modification in vitro (Figure 5A). The CCCH motifs are also required for complexes expressed in mammalian cells to show methyltransferase activity (Figures 5B and S6C). As a result, we identify the minimal complex required to reconstitute m6A modification: our crystallized heterodimer of methyltransferase domains (MTD3 and MTD14) and two CCCH motifs of Mettl3.
Figure 5. Proposed RNA substrate interactions with the Mettl3/Mettl14 complex.
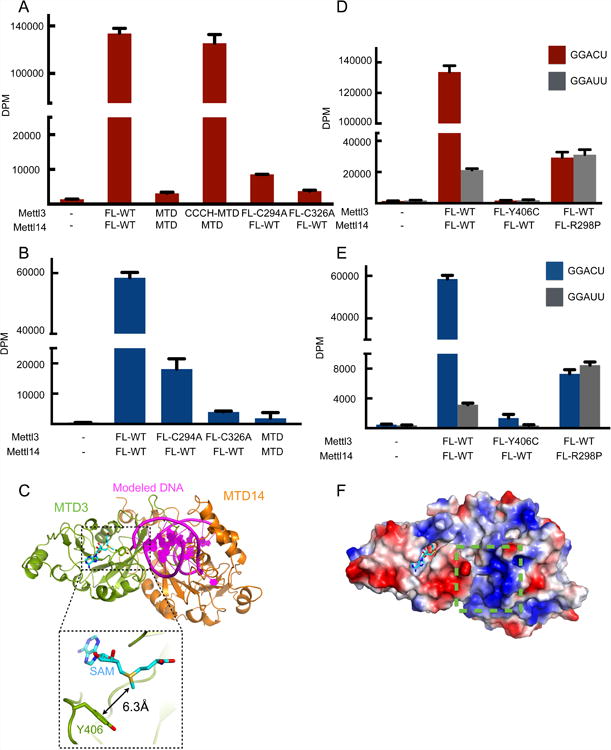
(A) In vitro methyltransferase activity of the full-length Mettl3/Mettl14 complexes expressed in E. coli with indicated point mutations or truncations (MTD).
(B) In vitro methyltransferase activity of the full length Mettl3/Mettl14 complexes expressed in HEK293 cells with indicated point mutations. Data shown as mean +/- SD from three replicates.
(C) Modeling of the RNA binding site by superimposition of the mDNMT1-DNA complex structure (PDB: 4DA4) onto the MTD3/MTD14 complex. The modeled DNA substrate of the complex structure of mDNMT1-DNA is shown in purple, cartoon representation and MTD of mDNMT1 is omitted for simplicity. Close-up view (dashed rectangle) shows the Y406 residue 6.3Å from the methyl group of SAM (cyan sticks).
(D) In vitro methyltransferase activity of the full-length Mettl3/Mettl14 complexes expressed in E. coli with indicated point mutations. Data shown as mean +/- SD from three replicates.
(E) In vitro methyltransferase activity of the full length Mettl3/Mettl14 complexes expressed in HEK293 cells with indicated point mutations. Data shown as mean +/- SD from three replicates.
(F) Surface representation colored by vacuum electrostatic potential of the MTD3/MTD14 complex in the same orientation as in (C). The basic patch close to the modeled DNA is indicated by a green dashed box. SAM is shown in stick representation, colored in cyan.
See also Figure S6.
Despite low sequence identity (∼10%), DNA methyltransferase structures can be superimposed onto our structure of MTD3. The previously determined structure of DNMT1 in complex with substrate DNA was used to provide insight into where RNA may bind in our MTD3/MTD14 model (Figure 5C) (Song et al., 2012). In the Mettl3 active site, an evolutionarily conserved tyrosine residue (Y406) sits on one of the fence loops near the cavity opening, only about 6 Å away from the methyl group that needs to be transferred to the adenine base (Figures 5C and S5D). The phenol ring also has potential to stack against the rings of RNA bases. In an independent study, the Y406C mutation of Mettl3 was identified in large intestine cancer patients (Colon Adenocarcinoma - TCGA, US (reanalyzed by Cancer Genome Project, Sanger Institute); Study ID: COSU376) (Forbes et al., 2015). Thus, we tested how the Y406C mutation affects methyltransferase activity. When full-length Mettl3 with the Y406C mutation is in complex with wild type Mettl14, we can obtain stable protein complexes but cannot observe any significant level of methylation in vitro (Figures 5D, 5E, S1B and S6D). Loss of the tyrosine ring may prevent Mettl3 from interacting properly with the RNA substrate near the adenine to be methylated and catalyzing the methyl transfer.
In our structures, a highly basic patch is found on MTD14, near the MTD3/MTD14 junction close to the catalytic cavity (Figure 5F). One of these basic residues is the highly conserved arginine 298 of Mettl14, which is frequently mutated to proline in endometrial cancer (Uterine Corpus Endometrioid Carcinoma (TCGA, US) import from ICGC; Study ID: COSU419)(Bell, 2014; Forbes et al., 2015) (Figure S5E). Wild type Mettl3 in complex with full-length Mettl14 with the R298P mutation shows significantly decreased methylation activity (Figures 5D, 5E, S1B and S6D). In addition, these complexes can no longer distinguish the cognate RNA target from the mutant RNA substrate. R298 also lies close to the third “fence” loop discussed above, and may have a complex role to affect both rate and RNA specificity. Thus, Mettl14 seems to play a role in substrate recognition as well as structurally supporting the catalytic cavity in Mettl3.
Discussion
Division of labor between Mettl3 and Mettl14
Our structural and biochemical data allow us to present a model for how Mettl3 and Mettl14 collaborate (Figure 6). The complex structure shows extensive interactions between the two methyltransferase domains which keep their conformations coupled. In addition to a large surface-to-surface interaction with various hydrophobic and polar contacts, unusual terminal extensions allow MTD14 to maintain tight interactions with MTD3 and maintain rigid interdomain orientation. Near the intermolecular binding interface, there is a large cavity where we expect the RNA substrate to bind close to the observed SAM binding site. This active site is fenced in by loops (purple) unique to MTD3 (Figure S2). One of the loops depends directly on MTD14 for conformation, which in turn may affect the other two loops and thereby affect catalytic activity or substrate recognition. Thus, although MTD3 and MTD14 have similar overall domain folds, Mettl3 and Mettl14 have two distinct roles in producing m6A marks on RNA.
Figure 6. Annotated model of the MTD3/MTD14 complex structure.
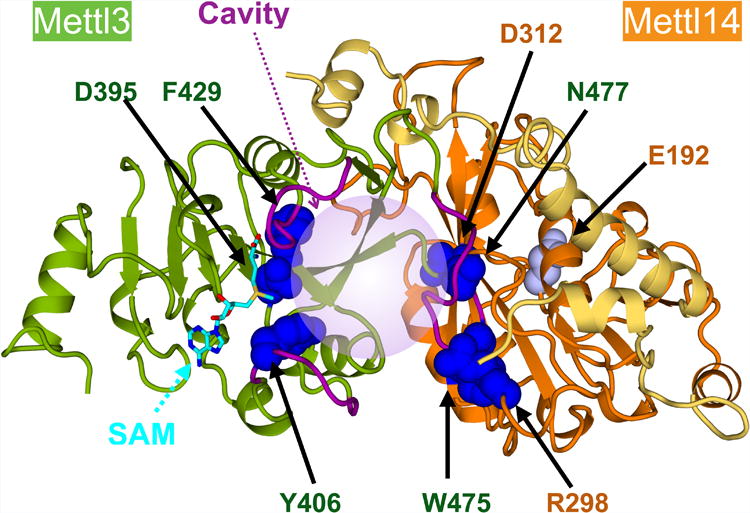
The MTD3 (green cartoon) and MTD14 (orange cartoon, terminal extensions in yellow) complex structure is shown. The terminal extensions of MTD14 reach over to MTD3 (yellow ribbon). SAM (cyan sticks) binds one end of a catalytic cavity, which extends to the interdomain interface (light purple sphere). Concave space between the MTDs is surrounded by three “fence” loops (purple). Specific residues mutated in this study and shown to dramatically reduce enzymatic activity are shown with dark blue sphere representation, and the weak mutation of MTD14 catalytic site is shown with gray spheres. All mutated residues are labeled (MTD3: green, MTD14: orange).
Mettl3 is the catalytic center. Disruption of the Mettl3 catalytic residue with a point mutation (D395A) abrogates any detectable methylation activity of the full-length, otherwise wild type Mettl3/Mettl14 complex, while an analogous mutation of Mettl14 catalytic site has a negligible effect. Moreover, the Mettl3 catalytic domain is the only one with a cavity that can accommodate the necessary methyl donor, SAM, as visualized in our structures. Thus, the catalytic site of Mettl3 is most likely to contain the sole active center for writing m6A modification in Mettl3/Mettl14 complex. In addition to being catalytically active, Mettl3 also contains CCCH-type zinc binding motifs that are critical for in vitro methylation of RNA, probably needed for interacting with RNA substrates. In fact, when the CCCH motifs are added to our crystallized complex, full enzymatic activity can be reconstituted. Therefore, Mettl3 plays a major role in substrate recognition and catalysis. However, isolated Mettl3 without Mettl14 is not active.
Mettl14 is structurally required to activate Mettl3. Mettl14 makes intimate contact with Mettl3 to support a distinct conformation near the active site, which is critical for Mettl3 to carry out catalysis. To determine why Mettl14 is necessary for Mettl3 activity, we tested many structure-guided single point mutations for their effect on heterodimerization and enzymatic function. We found that certain intermolecular contacts are critical for methyltransferase activity while not contributing to protein stability. We report three mutations near the third “fence” loop around the catalytic cavity that have a debilitating effect on the methyltransferase activity. Major changes in the conformation of the third loop may affect the accessibility of the active site to substrates. Furthermore, the three “fence” loops are interconnected and likely affect each other's conformation. Therefore, the shape of the entire cavity and the ability to bind SAM and the RNA substrate may be affected by the Mettl3/Mettl14 junction we observe in the structure. In addition, Mettl14 is also likely to contact RNA substrate through the basic patch that we identified in our structures. Not only is the patch highly basic and proximal to the active site opening, but a mutation (R298P) in this region changes the sequence specificity for the RNA substrate. Therefore, even though Mettl14 does not carry out the catalysis itself, it has an allosteric role to support Mettl3 active site and a critical role in substrate RNA recognition.
Both Mettl3 and Mettl14 are necessary for specific and efficient methyltransferase activity. Previous reports suggested that individually purified recombinant Mettl3 and Mettl14 polypeptides both have some catalytic activity, and that recombinant Mettl14 has higher methylation activity than Mettl3 in vitro (Liu et al., 2014; Wang et al., 2014b). It is likely that some endogenous Mettl homologs co-purify with the recombinant proteins. Mettl14 is prone to aggregation in the absence of Mettl3, regardless of the expression system. During purification, overexpressed Mettl14 in complex with endogenous Mettl3 is enriched in the soluble fraction, rendering it more active overall. On the other hand, eukaryotic expression of Mettl3 yields soluble and stable polypeptide even in the absence of Mettl14. Since Mettl3 can stay soluble even when it is in excess, no such enrichment of the complex with endogenous Mettl14 is observed. Consequently, most of purified Mettl3 after overexpression is free of Mettl14 and overall less active. When catalytic mutations are introduced to one subunit at a time, protein stability does not vary and the functional differences are easier to compare. Using point mutations of the catalytic sites, we conclude that Mettl3, and not Mettl14, is the active subunit. Although unlikely, additional factors or unusual modifications may restore the methyltransferase activity to Mettl14. And since the catalytic site of Mettl14 is more divergent in different species (Figure 2A), other Mettl14 homologs might have retained the catalytic activity.
The ability of Mettl14 to activate Mettl3 is reminiscent of certain DNA methyltransferases, such as Dnmt3. The Dnmt3 family of methyltransferases is responsible for establishing initial CpG methylation de novo, which is important for regulation of transcription of most genes. Dnmt3L is an inactive methyltransferase that binds Dnmt3a or Dnmt3b to enhance their methylation activity. Structure of Dnmt3L in complex with Dnmt3a suggests that Dnmt3L may also stabilize the active site loop conformation (Jia et al., 2007). Therefore, broadly, Dnmt3L and Mettl14 are similar in that they both contain inactive MTDs with structural roles to activate a partner, catalytic MTD. However, the orientation of MTD14 relative to MTD3 is drastically different from that of Dnmt3L to Dnmt3a. And while Dnmt3s form heterotetramers, we only observe heterodimers of Mettl3 and Mettl14.
Conclusion
Our structural and biochemical investigation of Mettl3/Mettl14 complex shows that Mettl3 is a catalytically active methyltransferase while Mettl14 provides structural support to Mettl3 close to its active site to enable catalysis. Using high-resolution three-dimensional information from crystal structures, we identify specific contacts that are important for maintaining the Mettl3/Mettl14 complex intact, as well as the residues that are critical for efficient production of m6A modification in RNA substrates. We also identify the minimal m6A writing complex as MTD3/MTD14 complex with CCCH motifs of Mettl3. Our progress in understanding the molecular mechanism of adenosine methylation of RNAs allows us to provide an explanation for why certain mutations of Mettl3 and Mettl14 are debilitating for creating m6A modifications.
Experimental Procedures
Overexpression of target proteins
For proteins expressed in E coli, specified constructs of human Mettl3 and Mettl14 were subcloned into pETDuet vector and transformed into Rosetta (DE3) pLysS cells (Novagen). Target proteins were expressed in cultures grown in autoinduction media at 20°C overnight (Studier, 2005). For proteins expressed in insect cells, constructs were subcloned into the pFASTBACDual vector and Bac-to-bac (Thermo) system was used to produce baculoviruses to infect Hi5 cells. For proteins expressed in HEK293 cells, constructs were subcloned into the pcDNA3 vector. Transient transfections were performed using Lipofectamine 2000 according to manufacturer's protocol and cells were harvested 48 hours post-transfection.
Protein purification for crystallization
All cultures were harvested and sonicated in lysis buffer (50 mM Bis-Tris (pH7.0), 1 M NaCl, 1 mM DTT and supplemented with protease inhibitors). The supernatant was loaded onto a Ni-NTA affinity column (Qiagen) and the beads were washed with wash buffer (50 mM Bis-Tris (pH7.0), 1 M NaCl, 1 mM DTT and 20mM Imidazole (pH 7.0)) and eluted with elution buffer (20 mM Bis-Tris (pH 7.0), 1 M NaCl, 1 mM DTT and 250 mM Imidazole (pH 7.0)). The eluate was digested with TEV protease at 4 °C overnight to remo ve the N terminal 6x-His affinity tag. Target proteins were further purified by ion-exchange chromatography and finally by gel filtration chromatography. The peak fractions were collected and concentrated to about 15 mg/mL for crystallization screening.
Crystallization and structure determination/refinement
The crystals of apo MTD3/MTD14 complex were obtained using the hanging-drop, vapor-diffusion method by mixing 1 uL protein (15 mg/mL) with 1 uL reservoir solution containing 0.1M Tris (pH8.0) and 20% PEG 3350 and incubating at 18°C. The complex crystals with SAM and SAH were generated by soaking the apo crystals with 2 mM compound for 3 hours at 18°C. Se-SAD and native datasets were collected at APS-19-ID at wavelengths of 0.97924 Å and 0.97934 Å, respectively. Data were indexed, integrated and scaled by the program HKL3000 (Minor et al., 2006). Phases were determined by single-wavelength anomalous dispersion (SAD) using Phenix. Autosol, and density modification and automatic modeling were performed by Phenix. Autobuild (Adams et al., 2010). The model was further manually built with COOT (Emsley et al., 2004) and was refined using Phenix.refine. The PROCHECK program was used to check the quality of the final model, which shows good stereochemistry according to the Ramachandran plot (Laskowski et al., 1993). All structure figures were generated by PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC. Ligplot was used to identify residues involved in interdomain interaction (Figure 4A) (Wallace et al., 1995). Software used in this project was curated by SBGrid (Morin et al., 2013).
In vitro methylation assay
The in vitro methylation assay was carried out in triplicate with a 15 uL reaction mixture containing: 200 nM RNA oligonucleotides (“GGACU”= 5′-UACACUCGAUCUGGACUAAAGCUGCUC; “GGAUU” = 5′-UACACUCGAUCUGGAUUAAAGCUGCUC), 20 mM Tris (pH 7.5), 0.01% Triton-X, 1 mM DTT, 50 uM ZnCl2, 0.2 U/uL RNasin, 1% glycerol and 460 nM [3H]-SAM (Li et al., 2016). All recombinant Mettl3/Mettl14 complexes purified from E. coli were analyzed by SDS-PAGE (Figures S1B and S6B), quantified by UV absorbance at 280nm, and used at 50nM in the final reaction. All other protein complexes were purified by affinity chromatography using Ni-NTA (Qiagen) and normalized by comparing Stain-Free™ intensities (Biorad) after SDS-PAGE. Western blots were performed to positively identify the bands using anti-Mettl3 (Thermo-#PIPA541599) and anti-Mettl14 (Thermo-#PA5-43606) antibodies. Each reaction was incubated at room temperature for 1 hour. Half of the reaction mixture was blotted on Biodyne B nylon membranes and crosslinked with UV (254 nm). The membranes were washed with reaction buffer, deionized water, and 95% ethanol, in that order, and then subjected to liquid-scintillation counting using the TriCarb 2010 TR Scintillation Counter (Perkin Elmer). Levels of RNA with incorporated 3H-methyl group are shown as disintegrations per minute (DPM). All in vitro methylation data are shown as mean +/- SD from three replicates.
Supplementary Material
Highlights.
Structures of Mettl3 and Mettl14 complexes show an extensive interface.
Crystal structures with bound SAM or SAH identify the active site of Mettl3.
Catalytic site of Mettl3, but not Mettl14, is critical for methylation.
Mettl14 is critical to support Mettl3 structurally and recognize RNA substrates.
Cancer mutations of Mettl3/Mettl14 interfere with methylation activity in vitro.
Acknowledgments
We thank support from the Cecil H. and Ida Green Center Training Program in Reproductive Biology Sciences Research and also members of the Structural Biology Laboratory at UT Southwestern for help with data collection. Y.N. is a Southwestern Medical Foundation Scholar in Biomedical Research, a Pew Scholar, and a Packard Fellow (2013-39275). This study was supported by grants from NIH NIGMS NRSA (2T32GM008297 to K.A.D.), the Welch Foundation (I-1851), Cancer Prevention Research Institute of Texas (R1221), and American Cancer Society/ the Harold C. Simmons Comprehensive Cancer Center (IRG-02-196). The use of SBC 19ID beamline at Advanced Photon Source is supported by U.S. Department of Energy contract DE-AC02-06CH11357.
Footnotes
Accession Numbers: Coordinates and structure factors for apo, SAM-bound, and SAH-bound MTD3/MTD14 complexes have been deposited with the Protein Data Bank under accession codes 5K7M, 5K7U, and 5K7W, respectively.
Author Contributions: P.W., K.A.D. and Y.N. designed experiments, analyzed data, and prepared the manuscript. P.W. and K.A.D. made expression constructs, purified proteins, and performed in vitro methylation assays. P.W. determined and refined crystal structures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DW. Novel genetic targets in endometrial cancer. Expert Opinion on Therapeutic Targets. 2014;18:725–730. doi: 10.1517/14728222.2014.909414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haim M, Moshitch-Moshkovitz S, Rechavi G. FTO: linking m6A demethylation to adipogenesis. Cell Research. 2014;25:3–4. doi: 10.1038/cr.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Brown RS. Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Bujnicki JM, Feder M, Radlinska M, Blumenthal RM. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. Journal of molecular evolution. 2002;55:431–444. doi: 10.1007/s00239-002-2339-8. [DOI] [PubMed] [Google Scholar]
- Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, et al. m6A RNA Methylation Is Regulated by MicroRNAs and Promotes Reprogramming to Pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- Csepany T, Lin A, Baldick CJ, Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. The Journal of biological chemistry. 1990;265:20117–20122. [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K, Iucr Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nature reviews Genetics. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Guja KE, Venkataraman K, Yakubovskaya E, Shi H, Mejia E, Hambardjieva E, Karzai WA, Garcia-Diaz M. Structural basis for S-adenosylmethionine binding and methyltransferase activity by mitochondrial transcription factor B1. Nucleic Acids Research. 2013;41:7947–7959. doi: 10.1093/nar/gkt547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TM. Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol. 2005;15:367–373. doi: 10.1016/j.sbi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Harper JE, Miceli SM, Roberts RJ, Manley JL. Sequence specificity of the human mRNA N6-adenosine methylase in vitro. Nucleic acids research. 1990;18:5735–5741. doi: 10.1093/nar/18.19.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Zhang D, Aravind L. Adenine methylation in eukaryotes: Apprehending the complex evolutionary history and functional potential of an epigenetic modification. BioEssays. 2016;38:27–40. doi: 10.1002/bies.201500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YGG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature chemical biology. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Li F, Kennedy S, Hajian T, Gibson E, Seitova A, Xu C, Arrowsmith CH, Vedadi M. A Radioactivity-Based Assay for Screening Human m6A-RNA Methyltransferase, METTL3-METTL14 Complex, and Demethylase ALKBH5. J Biomol Screen. 2016;21:290–297. doi: 10.1177/1087057115623264. [DOI] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature chemical biology. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, He C. Chapter Six Preparation of Human Nuclear RNA m6A Methyltransferases and Demethylases and Biochemical Characterization of Their Catalytic Activity. Methods in Enzymology. 2015a;560:117–130. doi: 10.1016/bs.mie.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015b;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Pan T. N6-methyladenosine-encoded epitranscriptomics. Nature Structural & Molecular Biology. 2016;23:98–102. doi: 10.1038/nsmb.3162. [DOI] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nature Reviews Molecular Cell Biology. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nature reviews Molecular cell biology. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution – from diffraction images to an initial model in minutes. Acta Crystallographica Section D: Biological Crystallography. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P. Collaboration gets the most out of software. eLife Sciences. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Yuan X, Jiang B, Tang Z, Li GC. LncRNAs: key players and novel insights into cervical cancer. Tumour Biol. 2016;37:2779–2788. doi: 10.1007/s13277-015-4663-9. [DOI] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell research. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottman FM, Bokar JA, Narayan P, Shambaugh ME, Ludwiczak R. N6-adenosine methylation in mRNA: substrate specificity and enzyme complexity. Biochimie. 1994;76:1109–1114. doi: 10.1016/0300-9084(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. Journal of molecular biology. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science (New York, NY) 2012;335:709–712. doi: 10.1126/science.1214453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier WF. Protein production by auto-induction in high-density shaking cultures. Protein Expression and Purification. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Thomas CB, Scavetta RD, Gumport RI, Churchill MEA. Structures of Liganded and Unliganded RsrI N6-Adenine DNA Methyltransferase a Distinct Orientation for Active Cofactor Binding. Journal of Biological Chemistry. 2003;278:26094–26101. doi: 10.1074/jbc.M303751200. [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein engineering. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014a;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N(6)-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014b;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang Z, Shen Y, Wang P, Jia X, Zhao L, Zhou P, Gong R, Li Z, Yang Y, et al. Crystal Structure of the Nosiheptide-Resistance Methyltransferase ofStreptomyces actuosus. Biochemistry. 2010;49:6440–6450. doi: 10.1021/bi1005915. [DOI] [PubMed] [Google Scholar]
- Zheng G, Dahl J, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou KI, Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, Pan T. N(6)-Methyladenosine Modification in a Long Noncoding RNA Hairpin Predisposes Its Conformation to Protein Binding. J Mol Biol. 2016;428:822–833. doi: 10.1016/j.jmb.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


