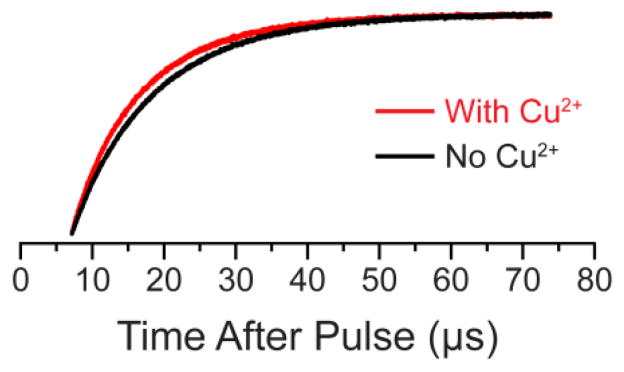
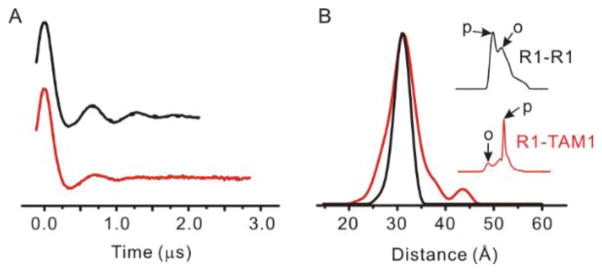
Abstract
Site-directed spin labeling (SDSL) in combination with Electron Paramagnetic Resonance (EPR) spectroscopy has become an important tool for measuring distances in proteins on the order of a few nm. For this purpose pairs of spin labels, most commonly nitroxides, are site-selectively introduced into the protein. Recent efforts to develop new spin labels are focused on tailoring the intrinsic properties of the label to either extend the upper limit of measurable distances at physiological temperature, or to provide a unique spectral lineshape so that selective pairwise distances can be measured in a protein or complex containing multiple spin label species. Triarylmethyl (TAM) radicals are the foundation for a new class of spin labels that promise to provide both capabilities. Here we report a new methanethiosulfonate derivative of a TAM radical that reacts rapidly and selectively with an engineered cysteine residue to generate a TAM containing side chain (TAM1) in high yield. With a TAM1 residue and Cu2+ bound to an engineered Cu2+ binding site, enhanced T1 relaxation of TAM should enable measurement of interspin distances up to 50 Å at physiological temperature. To achieve favorable TAM1-labeled protein concentrations without aggregation, proteins are tethered to a solid support either site-selectively using an unnatural amino acid or via native lysine residues. The methodology is general and readily extendable to complex systems, including membrane proteins.
Keywords: Triarylmethyl radical, spin labeling, saturation recovery, distance measurement at physiological temperature, relaxation enhancement
Graphical abstract
Distance constraints in proteins provide direct information on structure and structural transitions therein. Therefore, methodological development and optimization of distance measurements in proteins is an important endeavor in protein science and structural biology. Typical distances of interest in proteins are on the order of tens of Angstroms (Å). Among the available experimental methods that can measure distances on this scale, site-directed spin labeling (SDSL) combined with electron paramagnetic resonance (EPR) spectroscopy stands out as a unique and powerful approach, applicable to proteins and protein complexes of any degree of complexity or size, and with relatively high sensitivity (μg of proteins required).
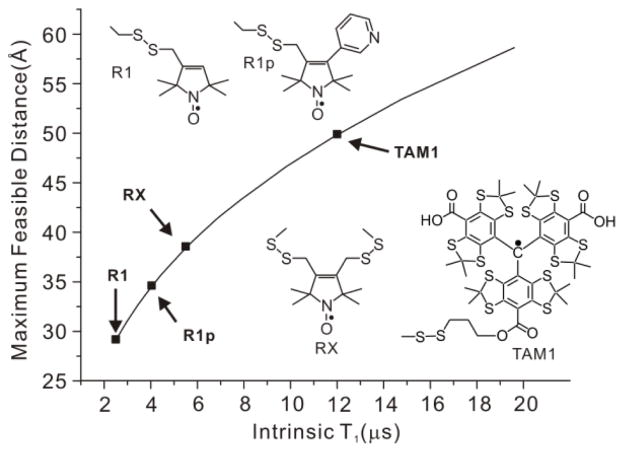
In the SDSL-EPR strategy, spin label pairs are introduced site-selectively [1,2], and the magnetic interactions between the pairs are measured as a metric for the interspin distance. Nitroxides are commonly used as spin labels, and the nitroxide side chain R1 [3,4] is the most popular (Figure 1). Conformationally constrained nitroxide side chains R1p [5] and RX [6] that limit the contribution of side chain flexibility to the determined distances have also been developed (Fig. 1).
Figure 1.
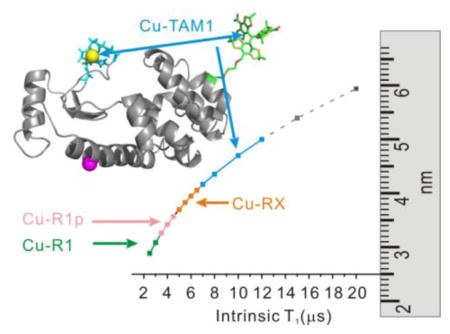
Maximum measurable spin label-to-Cu2+ distance measured by relaxometry as a function of spin label T1. The maximum distances were estimated by taking the minimum measurable drop in T1 due to the presence of Cu2+ to be 0.5 μs with a signal-to-noise ratio >50:1, typical of the data acquired (cf. ref. 16). The arrows indicate the T1 values for the indicated nitroxide side chains (cf. ref. 16) and for TAM1 attached to an immobile protein, reported here to be ≈ 12 μs.
Magnetic dipolar interactions are directly measured by double electron-electron resonance (DEER) [7] and double quantum coherence (DQC) [8]. In either case, the probability distribution of interspin distances in the range of 15–70 Å is obtained. However, in most cases both DEER and DQC are limited to cryogenic temperatures to slow electron spin relaxation rates and rotational tumbling of the entire protein, which would average out the orientation-dependent magnetic dipolar interaction [9]. However, it is desirable to measure distances at physiological temperatures, where potential complications caused by the freezing process and/or shifts in conformational equilibria due to the presence of cryoprotectant are eliminated. Attempts are being made to adapt DEER and DQC for room temperature distance measurements in proteins and DNA using the slowly-relaxing TAM radical as a spin label and immobilizing the macromolecule on a solid support [9–11]. For proteins, the maximum distance so far measured with this strategy is ≈ 30 Å. Room temperature DEER measurements have been made using a spirocyclohexyl nitroxyl spin label on a protein trapped in trehalose glass [10], but this approach does not eliminate a potential structural perturbation in the glassy environment.
Dipolar broadening of continuous wave (CW) lineshapes of interacting nitroxides can be employed to measure distances up to 20 Å at room temperature [12,13]. Hyde and coworkers recently extended this upper limit to 30 Å using non-adiabatic rapid sweep (NARS) EPR at S band [14]. The major barrier preventing the measurement of longer distances by CW methods is the difficulty in detecting very marginal spectral broadening.
An alternative approach to measuring distances at room temperature is based on the T1 relaxation enhancement (RE) of nitroxyl radicals by Cu2+. Using this strategy, Jun et al. reported that RE of an R1 nitroxide side chain in a rapidly tumbling peptide (T1 ≈ 2 μs) could measure Cu2+-nitroxide distances up to ~25 Å [15]. Yang et al. recently verified the theoretical prediction that the maximum range of distance measured by Cu2+-induced RE increases with increasing T1 of the nitroxide (Fig. 1) [16]. In that study, increased T1 was achieved using nitroxides R1p and RX (T1 ≈ 6–8 μs).
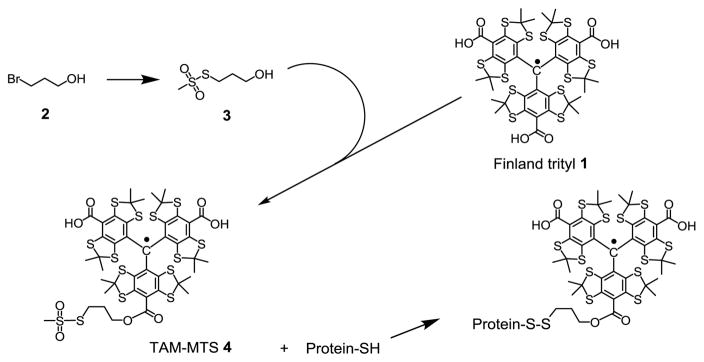
Here we show that a new class of spin labels based on triarylmethyl (TAM) radicals that have T1 ≈ 12 μs when attached to a protein can in principle measure distances up to ≈ 50 Å at room temperature using RE (Fig. 1). For this purpose, a novel TAM-methanethiosulfonate (MTS) reagent that undergoes a selective and high yield reaction with cysteine was prepared. The reaction generates a TAM-labeled side chain designated TAM1 (Scheme 1); details of the TAM-MTS synthesis and reaction conditions with the protein are included in Supporting Information. In an earlier study, a TAM radical for use in DQC was introduced at cysteine via reaction of an activated disulfide TAM reagent [9]. That reagent was limited for general use by a low reactivity, a problem now solved by the highly reactive MTS function.
Scheme 1.
Preparation of TAM-MTS reagent and TAM-labeling of protein
The general application of TAM relaxation enhancement by Cu2+ requires site-selective introduction of both TAM1 and a high affinity Cu2+ binding site in the protein of interest. Recent advances make this possible with the facile introduction of high-affinity Cu2+ binding motifs consisting either of a genetically engineered pentapeptide in loops [16] or histidine pairs introduced into regular secondary structure [17]. A problem encountered upon the introduction of TAM1 is a reduction of the protein solubility [9] at least for some small soluble proteins, a problem that is solved by tethering the protein to a solid support prior to labeling. In this communication, we demonstrate the combined application of the new TAM1 side chain, a high affinity Cu2+ motif and protein tethering to measure the longest range distances yet obtained by EPR methods at ambient temperatures.
In the present study, T4 Lysozyme (T4L) mutants containing the high affinity Cu2+ binding pentapeptide (GGGHG) inserted in loop sequences and a single cysteine residue for introduction of TAM1 are used to illustrate the overall strategy; the site of peptide insertion is indicated with the residue number and the suffix L. Tethering of the mutants to a solid support is most simply carried out by direct covalent attachment to commercially available CNBr-activated Sepharose via protein lysine residues [18]. This procedure has the potential disadvantage for some studies that the protein is attached in random orientation with respect to the matrix. Site-selective attachment can be achieved with the use of genetically incorporated unnatural amino acids as recently described [19]. Here we employ para-acetyl phenylalanine (p-AcPhe) as the unnatural amino acid to which biotin is conjugated via a hydroxylamine derivative [20]. The biotinylated protein is quantitatively adsorbed in high yield to commercially available streptavidin Sepharose, yielding effective concentrations as high as 700 μM without aggregation [20] Tethering by either approach results in compete immobilization of the protein on the CW EPR time scale [18,20]. Details of protein labeling and tethering are provided in the Supporting Information.
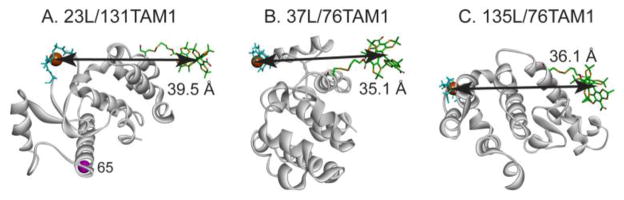
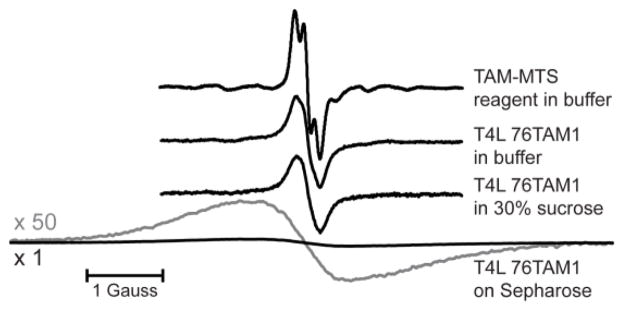
The three Cu2+/TAM1 pairs investigated in this study are shown in Figure 2. Each was tethered using CNBr-activated sepharose, and one (23L/131TAM1) was additionally coupled site-selectively using p-AcPhe at site 65 for comparison (Fig. 2A). The CW spectra of the TAM-MTS reagent alone and attached to T4L at site 76 under the indicated conditions are shown in Figure 3. Spectral line-broadening due to the increased correlation time of TAM1 on the protein is evident, particularly in the tethered sample, where the overall linewidth is similar to those observed for TAM radicals in frozen solution [21], which suggests strong immobilization of the TAM radical with respect to the protein. The major contribution to the linewidth is unresolved anisotropic hyperfine interactions within TAM1 [22]. Similar spectra are obtained for TAM1 at site 131.
Figure 2.
Models of the TAM1-Cu2+ pairs investigated. The 23L/131TAM1 mutant was tethered to Sepharose randomly at native Lys residues by reaction with CNBr-activated Sepharose, or site-selectively using the unnatural amino acid p-AcPhe introduced at site 65, indicated by the magenta sphere in (A) (see text). The TAM1 spin label and Cu2+ in the binding loop were modeled as described in the Supporting Information.
Figure 3.
Normalized CW EPR spectra for TAM-MTS in solution and for the T4 Lysozyme mutant 76TAM1 in various conditions. Spectral line broadening is apparent in the labeled protein samples due to the reduced rotational correlation time of the TAM1 side chain. The spectrum for the Sepharose-immobilized protein via the CNBr method is also shown with the vertical axis scaled up by 50 times (gray). The 13C-satellite lines are clearly evident in the TAM-MTS spectrum (cf. ref. 26).
The spin lattice relaxation times for TAM1 in the tethered T4L mutants in the absence and presence of a stoichiometric amount of Cu2+ were measured using long-pulse Saturation Recovery (SR) EPR following established protocols [23,24]. Figure 4 shows representative SR relaxation curves for T4L 23L/131TAM1 attached site-selectively at residue 65.
Figure 4.
Representative relaxation enhancement data. The saturation recovery traces obtained for the 23L/65p-AcPhe/131TAM1 sample in the absence (black) and presence (red) of Cu2+ at a microwave observe power of 50 μW. A clear “enhancement” in the relaxation rate can be seen upon the addition of Cu2+. SR curves, best fits, and residual traces for the four mutants studied in all conditions are given the SI (Figures S1–S4).
For each mutant, copper-free relaxation curves were well-fit with single exponentials, whereas mono-exponential fits of copper-bound samples were poor. For immobilized proteins containing Cu2+ and TAM1, the apparent T1 of TAM1 depends on the orientation of the interspin vector with respect to the external field. If the interacting spins can be assumed to have isotropic g values, the orientation-dependent T1 is given by Hirsch et al. [25]:
| (Eq. 1) |
where T1s and T1s0 are the spin-lattice relaxation times for TAM in the presence and absence of Cu2+, respectively, ωs and ωf are the resonant frequencies of TAM and Cu2+, T1f and T2f are the spin relaxation times of Cu2+, and βe and h are the Bohr magneton and the Planck constant, respectively. The TAM radical has an essentially isotropic g-tensor, but Cu2+ complexes do not. Nevertheless, it is found that use of gf = 2.1166, the average g for a 3N1O-bound copper state [26] and gs = 2.0058 for TAM (determined by characterization of TAM MTSL; see SI) in Eq. 1 can account well for the experimental data and reasonably corresponds to TAM1 modeled in the protein (Figure 2 and SI), when taking T1f = T2f = 3 ns. The resonant frequency of Cu2+, ωf, was calculated using the isotropic value of gf. Averaging of the copper g-tensor may arise from localized fast motion of the Cu2+ complex in the protein, as suggested by MD simulations as well as DEER distance distributions between nitroxides and Cu2+ in binding loops [16]. The values for Cu2+ relaxation times have been reported to be in the range of 1 to 5 ns [27], and a value of 3 ns has been found to account for copper-induced relaxation of nitroxides in a number of earlier studies [15,16,28]. Calculation of r from relaxation enhancement requires fits of the experimental relaxation curves to a weighted sum of exponentials as a function of the angle θ as described in detail in the SI. The experimentally-determined copper-free intrinsic T1 values and average interspin distances for each mutant studied are given in Table 1.
Table 1.
Intrinsic copper-free T1 values and interspin distances measured for four TAM1-spin labeled mutants of T4 Lysozyme, and modeled interspin distances (See Figure 2).
| T4 Lysozyme Mutanta | Intrinsic T1, (μs)b | Interspin Distance, r (Å) | Modeled Distance (Å) |
|---|---|---|---|
| 23L/131TAM1 | 12.0 | 37.9 | 39.5 |
| 23L/131TAM1 (p-AcPhe at 65) | 12.5 | 37.7 | 39.5 |
| 37L/76TAM1 | 12.1 | 32.6 | 35.1 |
| 135L/76TAM1 | 12.1 | 32.8 | 36.1 |
All proteins immobilized via random attachment to CNBr-activated Sepharose solid support, except for the sample labeled ‘p-AcPhe at 65’, which was site-specifically attached via biotin linkage.
Reported intrinsic T1 values obtained by measuring copper-free T1 values at various observe powers with extrapolation to zero power; complete data in Supporting Information.
The method reported herein is only valid if the occupancy of the Cu2+ binding site is close to 100%. Low affinity sites (Kd in the μM range) can in principle be saturated with high Cu2+ concentrations, but in this case Heisenberg exchange with free Cu2+ will shorten T1; the i and i+4 bi-histidine helical motif employed by Voss et al. [28] has an affinity too low for the present method. Various high affinity Cu2+ peptide binding sites have been reported in the literature; for example, studies described in References [16], [17], and [29] all involve Cu2+ binding sites with nanomolar Kd values.
As mentioned above, the broad linewidth of the TAM1 in solid support-tethered T4L suggests strong immobilization of the spin label. The degree to which this reflects spatial localization of TAM1 was investigated by comparing an R1-R1 distance distribution with the corresponding R1-TAM1 distribution, both being obtained using DEER at cryogenic temperatures. If this comparison is done at sites where the R1-R1 distribution is narrow, a difference in the distribution widths can be tentatively assigned to disorder in the TAM1 side chain. The sample selected for this purpose was the mutant T4L 68C/109C which has a mono-modal distribution of narrow width (<5 Å at half-height) when labeled with R1, as seen in Figure 5 [19].
Figure 5.
DEER data reveal the spatial localization of the TAM1 spin label. (A) The time domain signal of the 68C/109C mutant labeled with R1 (black trace) and with a R1-to-TAM1 ratio of 1:3 (red trace). (B) The distance distributions obtained from analysis of the DEER data in (A) for the R1-R1 (black) and R1-TAM1 (red) labeled mutants. The inset in (B) shows the field-swept electron spin echo spectra (200 G wide) at Q-band for the R1-R1 (black) and R1-TAM1 (red) labeled 68C/109C mutant. The arrows marked “p” and “o” indicate field positions of pump and observe pulses, respectively, for each experiment (see text).
To prepare the T4L R1-TAM hybrid, 25% of cysteines were labeled with R1, followed by reaction with excess TAM-MTS. Statistically, this yields 1/16 R1-R1, 9/16 TAM-TAM spin pair, and 3/8 R1-TAM spin pair. Consequentially, the R1-R1 spin pairs only contribute minimally in the R1-TAM distance distribution. The 4-pulse DEER data are shown in Figure 5. As shown in the figure inset, the pump (inversion) pulse was adjusted to excite primarily TAM spins, and the observe pulse was selected so that primarily R1 spins were observed. The resultant distance distribution of the R1-TAM pair shows essentially the same mean distance and a slightly broader distance distribution compared to that of the R1-R1 pair. The data suggest that the inherent flexibility of the TAM1 side chain is comparable to the R1 side chain commonly employed in DEER distance mapping in proteins.
In summary, the TAM1 side chain has useful advantages over a nitroxide side chain for distance mapping using T1 relaxation enhancement. First, the long T1 for TAM1 attached to a protein enables significantly longer distances to be measured at ambient temperatures, up to ≈ 50 Å (Fig. 1). Although the size of the T4L molecule did not permit sites to be selected with interspin distance measurement beyond ≈ 40 Å, the high signal to noise of the SR data should make the longer distance measurement feasible. Second, the narrow single central resonance line has a ~30 fold higher intensity compared to the same concentration of an immobile nitroxide, providing a comparably higher signal-to-noise in a saturation recovery experiment. Disadvantages of TAM1 compared to a nitroxide include the much larger steric bulk and the instability of the ester linked TAM due to hydrolysis. The half-life of hydrolysis for the ester linkage is ~50 hours and does not severely limit is use [30]. In future studies, other more stable linkages will be explored, and the degree to which the TAM1 side chain destabilizes the protein will be investigated. Regarding the latter point, we note that the measured TAM1-Cu2+ distances agree with those modeled in the native structure, so apparently the presence of TAM1 at surface sites does not lead to significant unfolding. The requirement for tethering the protein to a solid support may be viewed as a disadvantage, but in reality we advocate the general use of tethering in protein spin labeling due to the ability to provide high concentrations of spin with no perturbation to the protein structure and the absence of protein-protein interactions [20].
Supplementary Material
Highlights.
A new cysteine-reactive triarylmethyl spin label for proteins is reported.
Relaxation enhancement of the spin label (TAM1) by Cu2+ is distance dependent.
Interspin distances up to ~50 Å are measurable at physiological temperature.
Acknowledgments
This work was supported by the National Institute of Health (USA) grants NIH/NEI EY005216, NIH/NEI EY00331 and Jules Stein Professorship Endowment (W.L.H.), and Russian Science Foundation (No. 14-14-00922), Russian Foundation for Basic Research (grant 14-03-93180) and the National Institute of Biomedical Imaging and Bioengineering (NIH No. 5P41EB002034) (V.M.T.).
Footnotes
The authors declare no competing financial interests.
Details of synthesis of the TAM-MTS, preparation of protein samples, EPR experiments, discussion of modeling procedure, and additional data analysis are provided in the Supplementary Material.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hubbell WL, López CJ, Altenbach C, Yang Z. Technological advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 2013;23:725–733. doi: 10.1016/j.sbi.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hubbell WL, Cafiso D, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 3.Todd AP, Cong J, Levinthal F, Levinthal C, Hubbell WL. Site-directed mutagenesis of colicin E1 provides specific attachment sites for spin labels whose spectra are sensitive to local conformation. Proteins. 1989;6:294–305. doi: 10.1002/prot.340060312. [DOI] [PubMed] [Google Scholar]
- 4.Fleissner MR, Cascio D, Hubbell WL. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 2009;18:893–908. doi: 10.1002/pro.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fawzi N, Fleissner MR, Anthis N, Kálai T, Hideg K, Hubbell WL, Clore GM. A rigid disulfide-linked nitroxide side chain simplifies the quantitative analysis of PRE data. J Biomol NMR. 2011;51:105–114. doi: 10.1007/s10858-011-9545-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleissner MR, Bridges MD, Brooks EK, Cascio D, Kálai T, Hideg K, Hubbell WL. Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proc Nat Acad Sci USA. 2011;108:16241–16246. doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 8.Borbat P, Freed JH. Pulse Dipolar ESR: Distance Measurements. In: Harmer J, Timmel C, editors. Structural Information from Spin-Labels and Intrinsic Paramagnetic Centers in the Biosciences. Structure and Bonding. Vol. 152. Springer, Heidelberg, Germany; New York, USA: 2014. p. 1. [Google Scholar]
- 9.Yang Z, Liu Y, Borbat P, Zweier JL, Freed JH, Hubbell WL. Pulsed ESR dipolar spectroscopy for distance measurements in immobilized spin labeled proteins in liquid solution. J Am Chem Soc. 2012;134:9950–9952. doi: 10.1021/ja303791p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer V, Swanson MA, Clouston LJ, Boratyński PJ, Stein RA, Mchaourab HS, Rajca A, Eaton SS, Eaton GR. Room-temperature distance measurements of immobilized spin-labeled protein by DEER/PELDOR. Biophys J. 2015;108:1213–1219. doi: 10.1016/j.bpj.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shevelev GY, Krumkacheva OA, Lomzov AA, Kuzhelev AA, Rogozhnikova OY, Trukhin DV, Troitskaya TI, Tormyshev VM, Fedin MV, Pyshnyi DV, Bagryanskaya EG. Physiological-temperature distance measurement in nucleic acid using triarylmethyl-based spin labels and pulsed dipolar EPR spectroscopy. J Am Chem Soc. 2014;136:9874–9877. doi: 10.1021/ja505122n. [DOI] [PubMed] [Google Scholar]
- 12.Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: Experimental strategies and practical limitations. Biochemistry. 2001;40:15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- 13.Rabenstein MD, Shin YK. Determination of the distance between two spin labels attached to a macromolecule. Proc Nat Acad Sci USA. 1995;92:8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kittell AW, Hustedt EJ, Hyde JS. Inter-spin distance determination using L-band (1–2 GHz) non-adiabatic rapid sweep electron paramagnetic resonance (NARS EPR) J Magn Reson. 2012;221:51–56. doi: 10.1016/j.jmr.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jun S, Becker JS, Yonkunas M, Coalson R, Saxena S. Unfolding of alanine-based peptides using electron spin resonance distance measurements. Biochemistry. 2006;45:11666–11673. doi: 10.1021/bi061195b. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, Jiménez-Osés G, López CJ, Bridges MD, Houk KN, Hubbell WL. Long-range distance measurements in proteins at physiological temperatures using saturation recovery EPR spectroscopy. J Am Chem Soc. 2014;136:15356–15365. doi: 10.1021/ja5083206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham TF, Putterman MR, Desai A, Horne WS, Saxena S. The double-histidine Cu2+-binding motif: a highly rigid, site-specific spin probe for electron spin resonance distance measurements. Angew Chem Int Ed. 2015;54:6330–6334. doi: 10.1002/anie.201501968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.López CJ, Fleissner MR, Guo Z, Kusnetzow AK, Hubbell WL. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Sci. 2009;18:1637–1652. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleissner MR, Brustad EM, Kálai T, Altenbach C, Cascio D, Peters FB, Hideg K, Peuker S, Schultz PG, Hubbell WL. Site-directed spin labeling of a genetically encoded unnatural amino acid. Proc Nat Acad Sci USA. 2009;106:21637–21642. doi: 10.1073/pnas.0912009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.López CJ, Fleissner MR, Brooks EK, Hubbell WL. Stationary-phase EPR for exploring protein structure, conformation, and dynamics in spin-labeled proteins. Biochemistry. 2014;53:7067–7075. doi: 10.1021/bi5011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fielding AJ, Carl PJ, Eaton GR, Eaton SS. Multifrequency EPR of four triarylmethyl radicals. Appl Magn Reson. 2005;28:231. [Google Scholar]
- 22.Owenius R, Eaton GR, Eaton SS. Frequency (250 MHz to 9.2 GHz) and viscosity dependence of electron spin relaxation of triarylmethyl radicals at room temperature. J Magn Reson. 2005;172:168–175. doi: 10.1016/j.jmr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Hyde JS. Saturation recovery methodology. In: Kevan L, Schwartz RN, editors. Time domain electron spin resonance. John Wiley & Sons; New York: 1979. pp. 1–30. [Google Scholar]
- 24.Yang Z, Bridges MD, Lerch MT, Altenbach C, Hubbell WL. Saturation Recovery EPR and Nitroxide Spin Labeling for Exploring Structure and Dynamics in Proteins. In Electron Paramagnetic Resonance Investigations of Biological Systems by Using Spin Labels, Spin Probes, and Intrinsic Metal Ions, Part B. In: Qin PZ, Warncke K, editors. Method Enzymol. Vol. 564. Academic Press; UK: 2015. pp. 1–27. [DOI] [PubMed] [Google Scholar]
- 25.Hirsh DJ, Beck WF, Innes JB, Brudvig GW. Using saturation-recovery EPR to measure distances in proteins: Applications to photosystem II. Biochemistry. 1992;31:532–541. doi: 10.1021/bi00117a033. [DOI] [PubMed] [Google Scholar]
- 26.Aronoff-Spencer E, Burns CS, Avdievich NI, Gerfen GJ, Peisach J, Antholine WE, Ball HL, Cohen FE, Prusiner SB, Millhauser GL. Identification of the Cu2+ Binding Sites in the N-Terminal Domain of the Prion Protein by EPR and CD Spectroscopy. Biochemistry. 2000;39:13760–13771. doi: 10.1021/bi001472t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertini I, Luchinat C, Parigi G. Solution NMR of Paramagnetic Molecules: Applications to metallobiomolecules and models. Vol. 2 Elsevier Science; New York: 2001. [Google Scholar]
- 28.Voss J, Salwinski L, Kaback HR, Hubbell WL. A method for distance determination in oproteins using a designed metal ion binding site and site-directed spin labeling: Evaluation with T4 Lysozyme. Proc Nat Acad Sci USA. 1995;92:12295–12299. doi: 10.1073/pnas.92.26.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunningham TF, Shannon MD, Putterman MR, Arachchige RJ, Sengupta I, Gao M, Jaroniec CP, Saxena S. Cysteine-Specific Cu2+ Chelating Tags Used as Paramagnetic Probes in Double Electron Electron Resonance. J Phys Chem B. 2015;119:2839–2843. doi: 10.1021/jp5103143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuzhelev AA, Trukhin DV, Krumkacheva OA, Strizhakov RK, Rogozhnikova OY, Troitskaya TI, Fedin MV, Tormyshev VM, Bagryanskaya EG. Room-temperature electron spin relaxation of triarylmethyl radicals at the X- and Q-bands. J Phys Chem B. 2015;119:13630–13640. doi: 10.1021/acs.jpcb.5b03027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.