Abstract
Environmental factors clearly influence the pathogenesis of Type 1 diabetes, an autoimmune disease. We have studied gut microbiota as important environmental agents that could affect the initiation or progression of type 1 diabetes especially in the prenatal period. We used neomycin, targeting mainly Gram negative or vancomycin, targeting mainly Gram positive bacteria, to treat pregnant NOD mothers and to study autoimmune diabetes development in their offspring. Neomycin-treated offspring were protected from diabetes, while vancomycin-treated offspring had accelerated diabetes development, and both antibiotics caused distinctly different shifts in gut microbiota composition compared with the offspring from untreated control mice. Our study demonstrated that neomycin treatment of pregnant mothers leads to generation of immune-tolerogenic antigen-presenting cells (APCs) in the offspring and these APCs had reduced specific autoantigen-presenting function both in vitro and in vivo. Moreover, the protection from diabetes mediated by tolerogenic APCs was vertically transmissible to the second generation. In contrast, more diabetogenic inflammatory T cells were found in the lymphoid organs of the offspring from the vancomycin-treated pregnant mothers. This change however was not transmitted to the second generation. Our results suggested that prenatal exposure to antibiotic influenced gut bacterial composition at the earliest time point in life and is critical for consequent education of the immune system. As different bacteria can induce different immune responses, understanding these differences and how to generate self-tolerogenic APCs could be important for developing new therapy for type 1 diabetes.
Keywords: Type 1 diabetes, gut microbiota, antibiotic treatment
1. Introduction
Type 1 diabetes (T1D) is a T cell-mediated autoimmune disease which commonly presents in childeren and young adults [1]. The insulin-producing beta cells of the pancreatic islets are damaged and destroyed by activated autoreactive T cells resulting in the poor regulation of blood sugar and hyperglycemia [2]. Many genes determine susceptibility to the disease although the most important genetic susceptibility gene region, IDDM1 is encoded within the HLA [3-5]. However, a sharp rise in T1D incidence has been seen in recent years in genetically susceptible young children and individuals who do not carry high risk HLA alleles [6] and this has also taken place over a relatively short period of time. This strongly suggests that non-genetic factors, especially environmental factors influence disease development.
The gut microbiota maintain homeostasis of gut epithelia and permeability and the interaction between gut epithelia and the bacteria promotes the development of a normal immune system [7]. Altered composition of gut microbiota has been found to be associated with the development of various diseases including obesity and type 2 diabetes [8], metabolic and liver disease [9], intestinal inflammatory syndrome [10], allergic disorders [11], Alzheimer’s disease [12] and autoimmune diseases [13-15].
Increasing evidence suggests that changes in commensal microbiota correlate with the development of T1D in both mouse models and humans [16, 17]. We and others have demonstrated that T1D development is modulated by gut microbiota [18-21]. A recent longititudinal childhood study suggested a change of the composition of gut microbiota prior to the clinical onset of T1D in the children who were at high risk of developing T1D [22].
Early life exposure to the gut microbiota is critical for the maturation of the host immune system as well as maintaining health later on in life [23] and colonization with specific gut microbiota is strongly influenced by microbial exposures at birth [24]. Thus, the mode of infant delivery and the methods of feeding have profound impact on the composition of gut microbiota of the infants, which in turn affects health later in life [25, 26]. Millions of people have been saved from life-threatening infections since the discovery of antibiotics; however, recent studies have shown that exposure to antibiotics early in life can have undesirable consequences including the development of obesity and asthma [27-29]. Using a short exposure to a combination of Neomycin/Polymyxin B/Streptomycin, reducing Gram negative (G−) gut bacteria in the non-obese diabetic (NOD) mouse model of human T1D, we demonstrated that the mice were significantly protected from T1D development [20]. This protection was most striking when mice were treated in the prenatal period [20]. Thus, the time of life at which antibiotics are administered is crucial in studying the effect of gut microbiota on T1D development. In addition to the time of antibiotic exposure, we hypothesized that the type of antibiotic is also important. To test our hypothesis we investigated the effect of early exposure to two antibiotics, vancomycin and neomycin, that have very different functional features, on T1D development and studied the underlying mechanism.
2. Materials and Methods
2.1 Mice
Female NOD/Caj mice were originally obtained from the Jackson Laboratory and have been maintained at Yale University for many years. BDC2.5NOD and NY8.3NOD T cell receptor (TCR) transgenic mice were purchased from the Jackson Laboratory. The mice used in this study were kept in specific pathogen–free conditions with a 12-hour dark/light cycle, in individually-ventilated filter cages with autoclaved food, at the Yale University animal facility. The use of the animals in this study was approved by the Yale University Institutional Animal Care and Use Committee.
2.2 Antibiotic treatment
We treated pregnant (plugged) NOD mice with neomycin or vancomycin (all from Sigma) via drinking water at a final concentration of 1mg/ml for neomycin, 0.5mg/ml for vancomycin. The treatment was withdrawn within 24 hours after birth and diabetes development was observed in the offspring. The offspring from neomycin- or vancomycin-treated mothers were designated as Neo- or Van-mice.
2.3 Gut permeability assay
Gut permeability assay was performed as previously described [30]. Briefly, the mice were gavaged with FITC-dextran (600mg/kg) (Sigma) after fasting overnight. Blood samples were collected at 4 hours after gavage. After serial dilution, the serum samples were plated in a 96-well plate and read on a fluorescence spectrophotometer. Serum samples from non-FITC-dextran treated NOD mice were used as baseline and different concentrations of FITC-dextran were used to generate the standard curve. The test results were calculated based on the standard curve and baseline levels.
2.4 Intracellular cytokine (ICC) staining
For ICC staining, 106 cells were cultured for 4 hours with 50ng/ml PMA (Sigma), 500ng/ml of ionomycin (Sigma) and 1μl/ml of Golgiplug (BD Bioscience), before staining with mononoclonal antibodies (mAbs) against surface markers. After fixation, cells were stained with different intracellular anti-cytokine mAbs (Biolegend) and their expression was analyzed by flow cytometry.
2.5 Real time quantitative PCR (qPCR)
SFB-specific forward (SFB-F 5′-AGGAGGAGTCTGCGGCACATTAGC-3′) and reverse primers (SFB-R 5′-TCCCCACTGCTGCCTCCCGTAG-3′) were used to determine the relative abundance of SFB in the total bacterial DNA. Total 16S rRNA was used as the control and the results were analyzed by the delta-delta CT method after normalization with 16S rRNA. Each sample was analyzed in triplicate and the experiments were repeated twice.
2.6 16S rRNA sequencing analysis
Fecal samples were collected from both pregnant mothers and their offspring and DNA extraction was performed as previously described [28]. The V4 region of the bacterial 16S ribosomal gene was amplified from each DNA sample using a barcoded, broadly conserved, bacterial forward primer (5′-GTGCCAGCMGCCGCGGTAA-3′) and reverse primer (5′-GGACTACHVGGGTWTCTAAT-3′). The PCR products were purified with a Qiagen gel extraction kit. After quantification of DNA concentration using the Qubit dsDNA HS assay kit, equimolar amounts of each sample were pooled and used for pyrosequencing. Sequencing was performed on the Ion Torrent Personal Genome Machine (PGM) sequencing system (Life Technologies) using 200 bp read chemistry. The sequencing data were analyzed with QIIME software package (http://qiime.org) and UPARSE pipeline to pick operational taxonomic units (OTUs). Taxonomy assignment was performed at various levels using representative sequences of each OTU. G+/G− ratio was calculated using the most abundant Gram positive (G+) Firmicutes plus Actinobacteria contents divided by the most abundant Gram negative (G−) Bacteroidetes plus Proteobacteria contents.
2.7 Cell purification
Total APCs were purified by removing CD4+ (clone GK1.5) and CD8+ T cells (clone T1B105) using mAb hybridoma supernatants, followed by magnetic bead (conjugated with goat anti-Rat IgG) separation. T cells were purified by removing MHC class II+ cells (clone 10.2.16) and B cells (anti-mouse IgM and IgG (Qiagen)) using mAb hybridoma supernatants and magnetic bead separation. The purity of the cells was routinely ≥90%, analyzed by flow cytometry.
2.8 T cell proliferation assay
Purified CD4+ BDC2.5 or CD8+ NY8.3 T cells (105/well) were co-cultured with irradiated APCs (5 × 104/well) from experimental mice, in triplicate, in the presence or absence of specific antigens (Ag) - BDC2.5 mimotope peptide (RTRPLWVRME) or IGRP206-214 peptide (VYLKTNVFL), respectively. Cell proliferation was determined by stimulation index (SI), which was calculated by [3H]-thymidine incorporation (cpm) with Ag/[3H]-thymidine incorporation (cpm) without Ag.
2.9 Lymphocyte Adoptive Transfer
Total splenocytes or purified T cells from diabetic NOD mice were injected into irradiated recipient mice (107/mouse, i.v.). The recipient mice were monitored for diabetes every week. Purified T cells from diabetic NOD mice (3×106/mouse) with or without total APCs from neomycin-treated or untreated NOD mice (3×106/mouse) were injected into irradiated NOD mice. The recipient mice were monitored for diabetes every week.
2.10 Statistics
Statistical analysis was performed using GraphPad Prism software. P values of < 0.05 were considered significant.
3. Results
3.1 Prenatal neomycin or vancomycin treatment has different impacts on diabetes development in NOD mice
To investigate the effect of single antibiotic treatment, targeting different bacterial species, on diabetes development, we treated two NOD mating groups (n=4-6 breeding pairs/group) with neomycin or vancomycin, individually, in the drinking water after the females were plugged (+/− 24 hours). As controls, we included a group of NOD mice without treatment. The antibiotics were withdrawn immediately after delivery of the pups and diabetes development of the offspring was monitored on a weekly basis. In sharp contrast to diabetes incidence in the progeny from neomycin-treated breeders, vancomycin treatment strongly accelerated T1D development and 100% of mice were diabetic by the age of 21 weeks old (Fig.1), while prenatal neomycin treatment significantly protected NOD mice from diabetes development (Fig.1). Our results indicate that exposure to different antibiotics early in life can affect T1D development very differently.
Fig. 1.
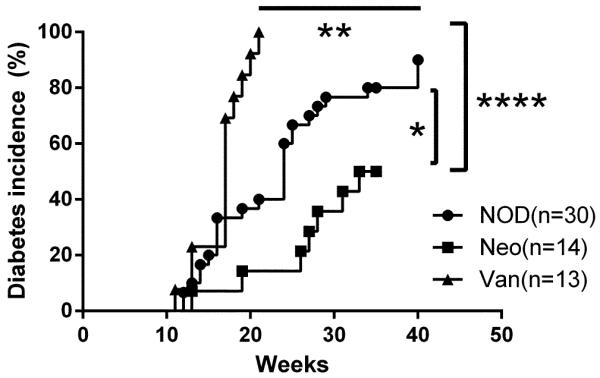
Diabetes incidence in NOD mice prenatally-treated with Neomycin or Vancomycin. NOD: untreated control. Neo: Neomycin-treated. Van: Vancomycin-treated. *: P<0.05, **: P<0.01, ****: P<0.0001 (Log-rank (Mantel-Cox) test).
3.2 Prenatal antibiotic treatment altered the cecum weight and gut permeability of the progeny
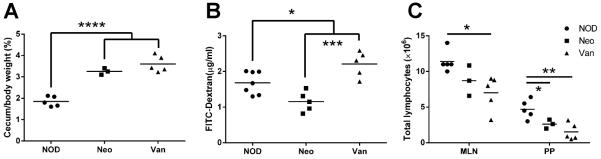
To investigate whether prenatal antibiotic treatment affected the cecal mass, where a large number of gut bacteria reside, we measured the cecal weight and found a significant increase of cecal mass and the ratio of cecal weight to body weight in the progeny of antibiotic-treated breeders compared to the progeny from untreated breeders (Fig.2A). The enlargement of the cecum was greater when the progeny were younger (4-5 weeks old, Fig.2A) and the increased cecal mass was less striking at 16 weeks of age (data not shown). It is interesting that the increased cecal mass was independent of the type of antibiotic used (Fig.2A).
Fig. 2.
Impact of prenatal antibiotic treatment on 4-week-old NOD mice. (A) Cecum weight. (B) Gut permeability assay. (C) Total lymphocytes in MLN and PP. *: P<0.05, **: P<0.01, ***: P<0.001, ****: P<0.0001 (Student’s t-test).
Next, we examined the effect of prenatal antibiotic exposure on gut permeability. It is interesting that prenatal exposure of neomycin reduced gut permeability whereas prenatal exposure to vancomycin increased gut permeability compared to the mice without prenatal antibiotic exposure (Fig.2B). This suggests that prenatal exposure of vancomycin leads to a “leaky” gut.
3.3 Prenatal antibiotic treatment reduced the cellularity of gut associated lymphoid tissue (GALT) of the progeny
To test if prenatal antibiotic exposure affected the development of GALT, we examined mesenteric lymph nodes (MLN) and Peyer’s patches (PP) of the progeny that were exposed to neomycin or vancomycin. As expected, the cellularity of MLN and PP from prenatally antibiotic-exposed mice was significantly reduced compared to the progeny without antibiotic exposure (Fig.2C); however, the reduction was more marked in the mice that were prenatally exposed to vancomycin compared with the neomycin-exposed progeny (Fig.2C). We did not find an obvious difference in the composition of T or B lymphocytes (data not shown). Our results also suggest that diabetes protection or acceleration associated with prenatal antibiotic exposure is most likely mediated by the change in gut bacteria, which affects the immune system.
3.4 Prenatal antibiotic treatment altered the composition of gut microbiota in the breeding mothers and their progeny
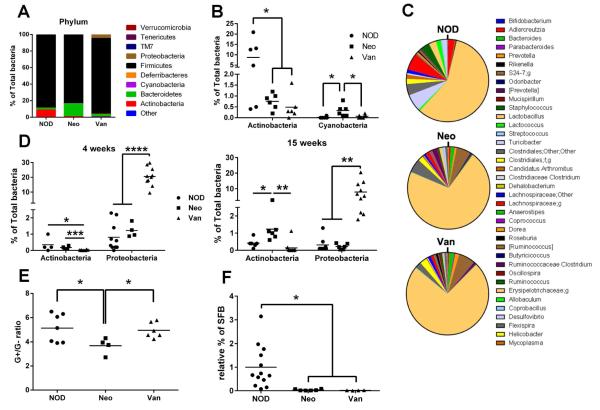
To examine the composition of gut microbiota, we performed 16S rRNA sequencing. We first studied the breeding mothers before and after administration of antibiotic water. There were no obvious differences between any of the breeding mothers before antibiotic administration (data not shown). However, after the 3-week antibiotic treatment, we have observed a clear change in these females. At the phylum level, Actinobacteria, Bacteroidetes, Cyanobacteria from antibiotic-treated breeding females were significantly different from non-treated breeding female mice (Fig.3A). It is interesting that more Bacteroidetes were found in neomycin-treated mice and more Firmicutes were found in vancomycin-treated mice (Fig.3A). Actinobacteria were significantly reduced in both neomycin- and vancomycin-treated breeding mothers compared to untreated mothers (Fig.3B); however, we found an increase in Cyanobacteria in neomycin-treated breeding mothers compared to either vancomycin-treated mothers or untreated controls (Fig.3B). We found more alterations in gut microbial taxa at the genus level (Fig. 3C) as well as other levels (data not shown) in treated mice compared with controls.
Fig. 3.
Bacterial changes after antibiotic treatment in NOD mice. (A) Bacterial composition at phylum level in mothers after 3 weeks of treatment with antibiotics. (B) Actinobacteria and Cyanobacteria percentage in feces from mothers. (C) Bacterial composition at genus level in feces from mothers. (D) Actinobacteria and Cyanobacteria percentage in feces from offspring at 4 weeks and 15 weeks of age. (E) Gram+ to Gram-ratio in feces from offspring. (F) SFB percentage in feces from offspring by qPCR. *: P<0.05, **: P<0.01, ****: P<0.0001 (Student’s t-test).
We further examined the alteration of gut microbiota by 16S rRNA sequencing to identify which specific bacterial taxa were changed after prenatal antibiotic exposure. We found that the composition of gut bacteria in these mice was significantly altered, even though the mice were not directly exposed to the antibiotics. The mothers treated with vancomycin and neomycin had significantly reduced Actinobacteria compared to the control mice. Similar to their mothers, the mice prenatally exposed to vancomycin showed significantly reduced Actinobacteria, but an increase in Proteobacteria compared to the control mice or those treated with neomycin at 4 weeks of age (Fig.3D). However, unlike their mothers, neither Actinobacteria (Fig.3D), nor Cyanobacteria (data not shown) were altered in the mice prenatally exposed to neomycin. As the composition of the gut microbiota changes with age, we also investigated the gut microbiota in the progeny at 15 weeks. It is interesting that the differences in the abundance of Proteobacteria seen in the mice prenatally exposed to vancomycin remained high even at 15 weeks old (Fig.3D), while Actinobacteria remained low (Fig.3D). In contrast, the abundance of Actinobacteria in the mice prenatally exposed to neomycin was elevated with age (Fig.3D). We also examined the overall G+/G− ratio in both Neo- and Van-treated mice. Although the Van group did not show differences compared with the untreated NOD group, a significant reduction was presented in the Neo group (Fig. 3E).
Segmented filamentous bacteria (SFB) have been reported to be associated with T1D protection in NOD mice [31]. To test whether SFB play a role in diabetes protection in the mice prenatally exposed to neomycin, we examined the abundance of SFB in the fecal samples from these mice by qPCR with SFB-specific primers. As controls, we included the fecal samples from the mice prenatally exposed to vancomycin, which accelerated diabetes development and the samples from the untreated progeny. We found that SFB were depleted in both groups of mice prenatally exposed to antibiotics, either neomycin or vancomycin (Fig.3F). Our results suggest that SFB do not play a role in diabetes protection in the mice prenatally exposed to neomycin.
3.5 The effect of antibiotics on T cell phenotype
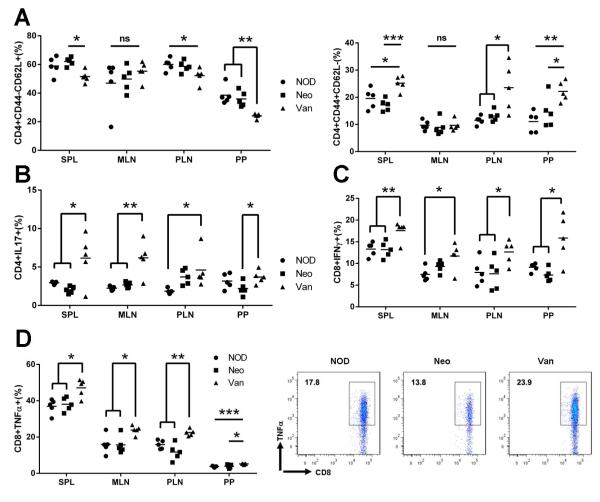
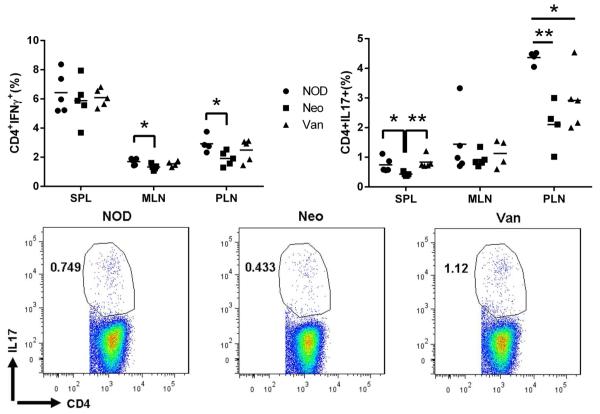
Since T1D is a T cell-mediated disease, we investigated the impact of prenatal exposure to antibiotics on T cell phenotype. We examined T cells from spleen (SPL), mesenteric lymph node (MLN), pancreatic lymph nodes (PLN) and Peyer’s patches (PP) of the mice from the three groups. The mice prenatally exposed to vancomycin showed a significant decrease in naïve T cell markers (CD44-CD62L+) and a highly increased number of CD4+ T cells expressing memory/effector markers in most of the lymphoid tissues examined (Fig.4A). This was even more marked in PP, the gut associated lymphoid tissue (GALT). The same profile was also found for CD8+ T cells (data not shown). In line with this phenotype, CD4+ T cells from the mice prenatally exposed to vancomycin also expressed significantly more inflammatory cytokine IL-17 (Fig.4B) and CD8+ T cells expressed significantly more IFN-γ and TNF-α in the four lymphoid tissues examined (Fig.4C, D). Our results indicated that accelerated diabetes in the mice prenatally exposed to vancomycin was mediated by the alteration of gut microbiota which enhanced inflammatory T cells. In contrast, it is interesting that neomycin treatment did not have an obvious impact on these inflammatory T cells compared to the control mice. We did not observe enhanced anti-inflammatory T cells including FoxP3+ Tregs and IL-10-producing T cells in diabetes-protected mice that were prenatally exposed to neomycin (data not shown).
Fig. 4.
T cell profiling of SPL, MLN, PLN and PP from NOD prenatally-treated with neomycin or vancomycin at 4-5 weeks of age. Lymphocytes were gated on CD4+ or CD8+ and the expression of surface or ICC markers was examined. (A) Naïve and Memory CD4 T cells. (B) IL17-expressing CD4 T cells. (C) IFNγ-expressing CD8 T cells. (D): TNFα-expressing CD8 T cells. Representative FACS plots from PLN. ns: not significant, *: P<0.05, **: P<0.01, ***: P<0.001 (Student’s t-test).
3.6 The effect of prenatal antibiotic exposure on the phenotype of antigen presenting cells (APCs)
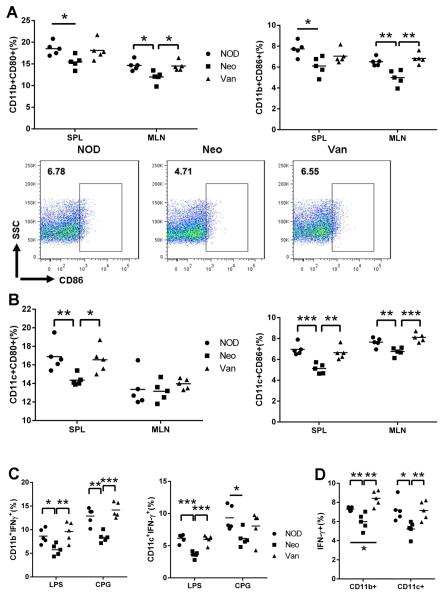
To investigate whether prenatal antibiotic exposure affects the phenotype and function of the APCs, we first examined the phenotype of the APCs in spleen (non-GALT) and MLN (GALT) from the progeny of the three groups. Whereas vancomycin clearly altered the T cell phenotype, neomycin appears to have stronger impact on the APCs (Fig.5A, B). The expression of the co-stimulatory molecules, CD80 and CD86, on CD11b+ (pan macrophage marker) and CD11c+ (pan dendritic cell, (DC) marker) cells were significantly down-regulated in both spleen and MLN from mice prenatally exposed to neomycin. Furthermore, we examined the phenotype of the APCs in response to Toll-like receptor (TLR) agonist stimulation including LPS (TLR4) and CpG (TLR9), both of which are enriched in bacteria. IFN-γ expression was significantly lower in macrophages and DCs from mice prenatally treated with neomycin, but not in APCs from mice prenatally treated with vancomycin (Fig. 5C). Similar to the phenotype found in the in vitro-stimulated APCs, significantly fewer IFN-γ-expressing macrophages and DCs were found in the mice prenatally exposed to neomycin, when studied immediately ex-vivo (Fig.5D).
Fig. 5.
APC profiling of SPL, MLN after prenatal antibiotic exposure. Cells were gated on CD11b+ or CD11c+ cells and CD80 and CD86 expression was examined. (A) CD80 and CD86 expressing CD11b+ cells. Representative FACS plots of CD11b+CD86+ cells from MLN. (B) CD80 and CD86 expressing CD11c+ cells. (C) Total lymphocytes were isolated and stimulated with LPS or CPG and tested for IFNγ-expressing CD11b+ and CD11c+ cells. (D) IFNγ expressing CD11b+ and CD11c+ cells from prenatally antibiotic-treated NOD. *: P<0.05, **: P<0.01, ***: P<0.001 (Student’s t-test).
3.7 The function of APCs from mice born to mothers treated with antibiotics in gestation
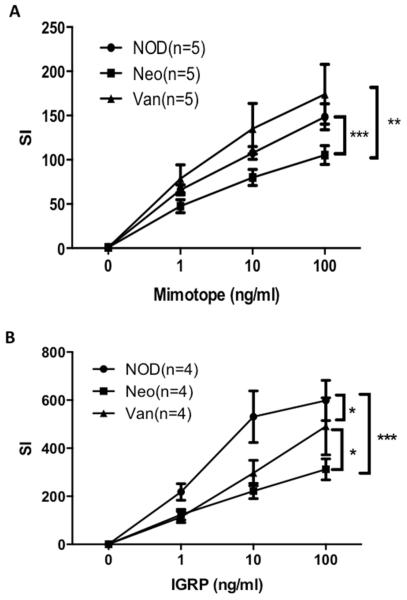
We tested the antigen-presenting function of total APCs from the mice prenatally exposed to neomycin and vancomycin using BDC2.5 CD4+ and NY8.3 CD8+ T cell proliferation as readouts. Here, the proliferative responses of the antigen-specific T cells reflect the antigen-presenting ability of the APCs to the T cells. Our results showed that APCs from the mice prenatally exposed to neomycin had a weaker ability to stimulate both BDC2.5 CD4+ (Fig.6A) and NY8.3 CD8+ (Fig.6.B) T cells compared to the APCs from the control mice or the APC from the mice prenatally exposed to vancomycin (Fig.6). It is interesting that APC from prenatally vancomycin-treated mice appear to stimulate BDC2.5 CD4+ T cells most strongly, correlated with accelerated diabetes development in the vancomycin-treated mice.
Fig. 6.
In vitro function of APC from prenatal antibiotic-treated NOD mice. (A) BDC2.5 CD4 T cell proliferation following stimulation with mimotope peptide presented by APC from the different groups of mice. (B): NY8.3 CD8 T cells proliferation stimulated with specific IGRP peptide presented by APC from the different groups of mice. *: P<0.05, **: P<0.01, ***: p<0.001 (Two way ANOVA).
3.8 Diabetes protection is transferable
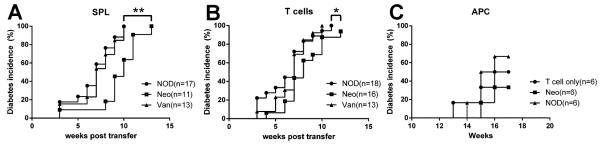
Our results indicated the protection from diabetes in the offspring of neomycin-treated mice was correlated with APC less able to stimulate pathogenic cells. We therefore tested whether the diabetes phenotype generated by prenatal exposure to antibiotics could be transferred by T cells or APCs. To test our hypothesis, we performed three sets of adoptive transfer experiments. In the first set, we used the mice prenatally exposed to antibiotics or controls as the recipients. These mice were irradiated so that their endogenous T cell proliferative function was impaired but not the ability of the APCs to present antigens. Total splenocytes from unmanipulated diabetic NOD mice were transferred to the irradiated recipients. As shown in Fig.7A, the recipients that were prenatally exposed to neomycin had significantly delayed diabetes onset compared to control non-antibiotic-treated or vancomycin-exposed mice, although all the mice in the study ultimately developed diabetes at a later time. In the second set of adoptive transfer experiments, we transferred purified splenic T cells from unmanipulated diabetic NOD mice to the irradiated mice that were prenatally exposed to antibiotics or control non-antibiotic-treated mice. When the APCs were depleted from transferred splenocytes, diabetes development in the recipients that were prenatally exposed to neomycin was also delayed compared to non-antibiotic treated NOD control group (Fig.7B), but the effect was less strong compared to transferring total splenocytes (Fig.7A) and the difference with prenatally vancomycin-exposed mice was diminished. Lastly, we tested the APC function directly. We used irradiated NOD mice as the recipients and purified splenic APCs from the mice prenatally exposed to neomycin or controls as donor cells together with purified T cells from unmanipulated diabetic NOD mice. Although this was not statistically significant, there was a trend to a delay and reduction in diabetes development in the recipients that were transferred with APCs from mice prenatally exposed to neomycin, compared to that from the control mice (Fig.7C). Together with the results shown in Fig. 6, we suggest that diabetes protection induced by prenatal exposure to neomycin could be mediated, in part, by APCs.
Fig. 7.
Adoptive transfer assay. (A) Recipients: prenatal antibiotic-treated or untreated NOD mice at 6 weeks of age. Donor cells: total splenocytes from diabetic NOD mice. (B) Recipients: prenatal antibiotic-treated or untreated NOD mice at 6 weeks of age. Donor cells: purified T cells from diabetic NOD mice. (C) Recipients: irradiated 6-week-old NOD mice. Donor cells: purified T cells from diabetic NOD mice with or without total APC from prenatally neomycin-treated or untreated NOD mice. *: P<0.05, **: P<0.01 (Log-rank (Mantel-Cox) test).
3.9 Cytokine profile of T cells in the recipient mice
In a similar but separate set of adoptive transfer experiments to those shown in Fig.7B, some recipient mice were sacrificed 4 weeks after the adoptive transfer of purified T cells from diabetic NOD mice. We examined the profile of intracellular cytokine expression in the T cells from the spleen, MLN and PLN. Although the recipient mice were adoptively transferred with the same diabetogenic T cells, fewer IFN-γ and IL-17 expressing CD4+ T cells were found in the recipient mice that were prenatally exposed to neomycin, compared to vancomycin and control groups (Fig.8). These results further support the notion that APCs are the link between the suppressed inflammatory phenotype of T cells and diabetes protection in the progeny of neomycin-treated mice.
Fig. 8.
Th1 and Th17 profiling of T cells from mice adoptively transferred with diabetogenic T cells. NOD: NOD mice as recipients; Neo: Prenatal neomycin-treated mice as recipients; Van: Prenatal vancomycin-treated mice as recipients. IFNγ-expressing CD4 T cells (left) and IL17-expressing CD4 T cells (right). Representative FACS plots: IL17-expressing CD4 T cells from SPL. *: P<0.05, **: P<0.01 (Student’s t-test).
3.10 Diabetes protection can be vertically transmitted to the second generation
To investigate whether the protected or accelerated diabetes phenotype, in the mice prenatally exposed to neomycin and vancomycin, respectively, can be inherited by the next generation, we bred the mice that were prenatally exposed to neomycin or vancomycin. As controls, we also bred the progeny of untreated mice. Diabetes development was observed in the offspring of this breeding. It is interesting that diabetes protection due to prenatal neomycin exposure can be passed on to the second generation whereas the phenotype of accelerated diabetes seen in the progeny of vancomycin-treated mice was lost in the second generation (Fig.9). This suggests that not all the impact by antibiotic treatment on immune system can be transferred through reproduction.
Fig. 9.
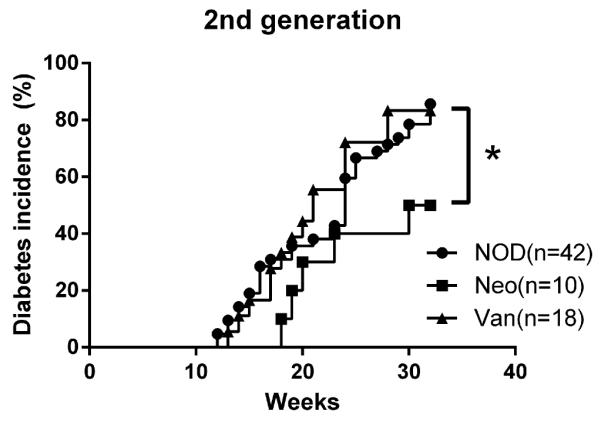
Diabetes incidence in the offspring of prenatal antibiotic-treated NOD mice. NOD: untreated NOD control mice; Neo: 2nd generation of prenatally neomycin-treated NOD mice; Van: 2nd generation of prenatal vancomycin-treated NOD mice. *: P<0.05 (Log-rank (Mantel-Cox) test).
4. Discussion
In this study, we investigated the effects of type and timing of antibiotic treatment, particularly at the earliest stages of life. We showed that neomycin treatment of pregnant NOD mice altered the gut bacterial composition and protected the offspring from diabetes and moreover, this protection could be transmitted to the second generation. This protection with neomycin, which was associated with a reduction in immunogenic phenotype in the APC of the spleen and gut-draining lymph nodes and reduced capacity to stimulate pathogenic autoantigen-specific CD4 and CD8 T cells, is most likely mediated by altered gut microbiota. In sharp contrast, prenatal exposure to vancomycin, by treatment of the mothers, induced changes in the microbiota that accelerated diabetes in the offspring, but this phenotype was not transmitted to the next generation. The vancomycin-treated offspring had more activated/memory inflammatory T cells in the gut and pancreatic-draining lymph nodes. Our results provide evidence that alteration of gut bacteria in the mother has a significant effect on immune development and manifestation of autoimmune diabetes in the offspring.
The use of antibiotics will alter the composition of gut microbiota [32, 33]. This short period of continuous prenatal exposure to both neomycin and vancomycin induced distinct changes in the gut microbiome in the offspring compared with the progeny from the untreated NOD mothers. These differences were maintained far beyond the postnatal period, even after the antibiotics had been withdrawn for a very long time. Our approach allowed us to assess the effect of setting the bacterial composition early in life and how this determined the immune responses later on. However, the individual effect of any given antibiotic on the bacterial composition is subject to other environmental influences, and the composition of the gut microbiota is not static. In our current study, the length of treatment and age at which treatment was given was particularly important. With this treatment protocol, we showed that the clearest effect on diabetes development occurred when the treatment was administered in the prenatal period. The mice were exposed to the specific bacteria from the mother, shaped by the treatment before birth and the bacterial composition in the very early stages of their life was critical for diabetes susceptibility. It is known that early life influences, starting from the flora set at the time of birth, shape the developing immune system both in mice [34, 35] and humans [36, 37]. This indicates that the initial flora, which not only “educate” the immune system but also affect the response to other influences in the environment, are critical. Thus, it is not surprising that some of the studies by different investigators, in different environments have shown divergent effects. Our studies using vancomycin accord with the investigations where the same antibiotic used continuously from birth accelerated diabetes onset, although only in male mice [38], or when given through gestation and continued through life [39]. These continuous treatments maintained the composition of the microbiota, and thus, prolonged the effects, suggesting that the microbiota that is set very early have a substantial effect. However, it should be noted that long-term antibiotic treatment raises a concern about propagation of resistant bacterial strains in the gut [40]. Even the 3-week treatment of the pregnant females, using neomycin or vancomycin, could cause increase of some G− or G+ bacteria, respectively, which are possibly neomycin or vancomycin resistant (data not shown). These studies contrast with the study by Hansen and colleagues who showed when vancomycin was given after birth for 4 weeks or from the age of 8 weeks onwards, there was protection from diabetes [41], and indeed this accords with our data that no acceleration of diabetes occurred when vancomycin was given after birth for 3 weeks (data not shown). Overall, the studies suggest that early timing has a particular effect, but if started later, the length of antibiotic usage might make an important contribution to the immunological phenotype and development of autoimmunity.
How does the composition of bacteria alter the development of diabetes? The bacteria could have a direct or indirect effect. Here, NOD mice exposed to vancomycin at the prenatal stage developed diabetes more rapidly and the mice also had more inflammatory T cells. Several factors could contribute to this. Firstly, DC/macrophages may mature quicker as a result of increased gut permeability, as we demonstrated with vancomycin treatment, and this could lead to increased stimulation with microbial antigens. It is known that DC can interact directly with the gut luminal contents and the bacteria in the layers of the gut wall via pseudopodia. Secondly, in a separate study, we have evidence that some bacteria can stimulate diabetogenic CD8 T cells through the mechanism of molecular mimicry (manuscript submitted). These stimulated CD8 T cells in the gut could then migrate to islets and accelerate diabetes.
However, not all the antibiotics promote diabetes development. Neomycin treatment encouraged predominance of different bacterial genera and this was associated with protection from diabetes. Our approach suggested that prenatal neomycin treatment may induce protective APCs. This result is consistent with our previous study showing that maternal treatment with Neomycin/Polymyxin B/Streptomycin, all targeting Gram-negative bacteria, protected offspring from diabetes by generation of tolerogenic APCs. Importantly, the protection could also be transmitted to the next generation. The mice had reduced gut permeability and the APC had a less inflammatory phenotype. These changes in the immune cells were not related to a direct effect of antibiotic treatment, as firstly, the mice (offspring) did not have direct contact with antibiotics; secondly, both purified T cells and APCs treated in culture with neomycin or vancomycin in vitro had similar profiles of cytokines, co-stimulation and activation markers (data not shown). However, our results showed that the offspring of vancomycin-treated breeders developed accelerated diabetes and they also had increased gut permeability, whereas the offspring of neomycin-treated breeders were protected from diabetes and these mice showed reduced gut permeability. Thus the “leaky gut” is associated with the disease onset but further work would be required to demonstrate causation. However, what has been shown here is that the “leaky gut” was unlikely to be a direct consequence of antibiotic exposure as the mice did not have direct gut exposure to the antibiotics. Furthermore, the gut permeability test was examined in adulthood. It has been reported that antibiotic treatment could affect beneficial bacteria that control the growth of fungi in the gut [42]. Toxins produced by the fungi could lead to the increased gut permeability [42]. It is not clear whether this is the case in the offspring of vancomycin-treated breeders; however, these mice have altered gut microbiota even they have not had direct contact with antibiotics. It is also not clear that whether indirect antibiotic exposure during fetal development could result in epigenetic changes. What is clear, however, is that the composition of gut microbiota in the offspring of antibiotic-treated breeders is very different from the offspring of untreated breeders. It is also noteworthy that the altered gut microbiota in the offspring are very stable. Thus, the changes in the immune cells are most likely due to the changes in gut microbiota, which are known to affect the host immune system, to polarize more towards an inflammatory or tolerant phenotype.
It had been suggested that APC dysfunction leads to diabetes development [43, 44], but this had not previously been correlated with gut bacterial composition. Supporting our previous finding with the combined treatment using three antibiotics [20], we found that prenatal exposure to neomycin alone may induce similar immune tolerance, and the disease protection was transferrable to the second generation or other hosts.
Our study has a number of important implications. Firstly, effects of antibiotics in pregnancy could have important implications for determining gut bacteria at the time of birth and subsequently influence the immunopathogenesis of autoimmune diabetes. Secondly, alteration of gut bacteria could potentially lead to the generation of tolerogenic APCs, which might be used as an indicator to monitor immuno-therapy. Thirdly, our study reinforces the possibility that using probiotics (a combination of beneficial gut microbiota) early in life could have a major immuno-modulatory effect. This approach would provide an easy to administer preventative treatment. It will require, however, that we more precisely identify a “beneficial” collection of bacteria and a controlled clinical trial.
Highlights.
◆ Antibiotic treatment during pregnancy influences T1D development in offspring
◆ Neomycin and Vancomycin treatment significantly change the gut microbiota
◆ Neomycin treatment induces immunotolerogenic APCs and protects from diabetes
◆ Vancomycin treatment induces inflammatory T cell increase, accelerating diabetes
◆ Neomycin protection from diabetes is vertically transmissible
Acknowledgements
We thank Karl Hager (Lab Medicine, Yale) for assistance with 16S rRNA sequencing and all the lab members for their kind technical help and critical scientific comments during the study. This work was supported by NIH grants (DK092882, DK100500, P30 DK945735), ADA (14-BS-222) and JDRF 2015-136.
Abbreviations
- T1D
type 1 diabetes
- HLA
human leukocyte antigen
- IDDM1
insulin-dependent diabetes mellitus 1
- NOD
non-obese diabetic
- TCR
T cell receptor
- Neo
neomycin
- Van
vancomycin
- ICC
intracellular cytokine
- PMA
Phorbol 12-myristate 13-acetate
- SI
stimulation index
- SPL
spleen
- MLN
mesenteric lymph nodes
- PLN
pancreatic lymph nodes
- PP
Peyer’s patches
- APC
antigen presenting cell
- DC
dendritic cell
- IGRP
islet-specific glucose-6-phosphatase catalytic subunit-related protein
- G+
Gram positive
- G−
Gram negative
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Babaya N, Nakayama M, Eisenbarth GS. The stages of type 1A diabetes. Annals of the New York Academy of Sciences. 2005;1051:194–204. doi: 10.1196/annals.1361.061. [DOI] [PubMed] [Google Scholar]
- 2.Bluestone JA, Herold K, Eisenbarth G. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 2010;464(7293):1293–300. doi: 10.1038/nature08933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Achenbach P, et al. Natural history of type 1 diabetes. Diabetes. 2005;54(Suppl 2):S25–31. doi: 10.2337/diabetes.54.suppl_2.s25. [DOI] [PubMed] [Google Scholar]
- 4.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nature genetics. 2009;41(6):703–7. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Concannon P, et al. Genome-wide scan for linkage to type 1 diabetes in 2,496 multiplex families from the Type 1 Diabetes Genetics Consortium. Diabetes. 2009;58(4):1018–22. doi: 10.2337/db08-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patterson CC, et al. Trends in childhood type 1 diabetes incidence in Europe during 1989-2008: evidence of non-uniformity over time in rates of increase. Diabetologia. 2012;55(8):2142–7. doi: 10.1007/s00125-012-2571-8. [DOI] [PubMed] [Google Scholar]
- 7.Natividad JM, et al. Commensal and probiotic bacteria influence intestinal barrier function and susceptibility to colitis in Nod1−/−; Nod2−/− mice. Inflammatory bowel diseases. 2012;18(8):1434–46. doi: 10.1002/ibd.22848. [DOI] [PubMed] [Google Scholar]
- 8.Tai N, Wong FS, Wen L. The role of gut microbiota in the development of type 1, type 2 diabetes mellitus and obesity. Reviews in endocrine & metabolic disorders. 2015;16(1):55–65. doi: 10.1007/s11154-015-9309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llorente C, Schnabl B. The gut microbiota and liver disease. Cellular and molecular gastroenterology and hepatology. 2015;1(3):275–284. doi: 10.1016/j.jcmgh.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walujkar SA, et al. Characterization of bacterial community shift in human Ulcerative Colitis patients revealed by Illumina based 16S rRNA gene amplicon sequencing. Gut pathogens. 2014;6:22. doi: 10.1186/1757-4749-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vital M, et al. Alterations of the Murine Gut Microbiome with Age and Allergic Airway Disease. Journal of immunology research. 2015;2015:892568. doi: 10.1155/2015/892568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Lukiw WJ. Microbiome-generated amyloid and potential impact on amyloidogenesis in Alzheimer's disease (AD) Journal of nature and science. 2015;1(7):e138. [PMC free article] [PubMed] [Google Scholar]
- 13.Goto Y, Kurashima Y, Kiyono H. The gut microbiota and inflammatory bowel disease. Current opinion in rheumatology. 2015;27(4):388–96. doi: 10.1097/BOR.0000000000000192. [DOI] [PubMed] [Google Scholar]
- 14.Johnson BM, et al. Impact of dietary deviation on disease progression and gut microbiome composition in lupus-prone SNF1 mice. Clinical and experimental immunology. 2015;181(2):323–37. doi: 10.1111/cei.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–27. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murri M, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC medicine. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455(7216):1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alkanani AK, et al. Alterations in Intestinal Microbiota Correlate With Susceptibility to Type 1 Diabetes. Diabetes. 2015;64(10):3510–20. doi: 10.2337/db14-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burrows MP, et al. Microbiota regulates type 1 diabetes through Toll-like receptors. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(32):9973–7. doi: 10.1073/pnas.1508740112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y, et al. Maternal Antibiotic Treatment Protects Offspring from Diabetes Development in Nonobese Diabetic Mice by Generation of Tolerogenic APCs. Journal of immunology. 2015;195(9):4176–84. doi: 10.4049/jimmunol.1500884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng J, et al. Long term effect of gut microbiota transfer on diabetes development. Journal of autoimmunity. 2014;53:85–94. doi: 10.1016/j.jaut.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kostic AD, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell host & microbe. 2015;17(2):260–73. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arrieta MC, et al. The intestinal microbiome in early life: health and disease. Frontiers in immunology. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(26):11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dogra S, et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio. 2015;6(1):e02419–14. doi: 10.1128/mBio.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matamoros S, et al. Development of intestinal microbiota in infants and its impact on health. Trends in microbiology. 2013;21(4):167–73. doi: 10.1016/j.tim.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Saari A, et al. Antibiotic exposure in infancy and risk of being overweight in the first 24 months of life. Pediatrics. 2015;135(4):617–26. doi: 10.1542/peds.2014-3407. [DOI] [PubMed] [Google Scholar]
- 28.Munyaka PM, Khafipour E, Ghia JE. External influence of early childhood establishment of gut microbiota and subsequent health implications. Frontiers in pediatrics. 2014;2:109. doi: 10.3389/fped.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cox LM, Blaser MJ. Antibiotics in early life and obesity. Nature reviews. Endocrinology. 2015;11(3):182–90. doi: 10.1038/nrendo.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, et al. Role of IRAK-M in alcohol induced liver injury. PloS one. 2013;8(2):e57085. doi: 10.1371/journal.pone.0057085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kriegel MA, et al. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(28):11548–53. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaarala O. Gut microbiota and type 1 diabetes. The review of diabetic studies : RDS. 2012;9(4):251–9. doi: 10.1900/RDS.2012.9.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikkelsen KH, et al. Effect of Antibiotics on Gut Microbiota, Gut Hormones and Glucose Metabolism. PloS one. 2015;10(11):e0142352. doi: 10.1371/journal.pone.0142352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336(6080):489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell SL, et al. Perinatal antibiotic-induced shifts in gut microbiota have differential effects on inflammatory lung diseases. The Journal of allergy and clinical immunology. 2015;135(1):100–9. doi: 10.1016/j.jaci.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 36.Amarri S, et al. Changes of gut microbiota and immune markers during the complementary feeding period in healthy breast-fed infants. Journal of pediatric gastroenterology and nutrition. 2006;42(5):488–95. doi: 10.1097/01.mpg.0000221907.14523.6d. [DOI] [PubMed] [Google Scholar]
- 37.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell host & microbe. 2014;15(3):382–92. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Candon S, et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PloS one. 2015;10(5):e0125448. doi: 10.1371/journal.pone.0125448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown K, et al. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. The ISME journal. 2016;10(2):321–32. doi: 10.1038/ismej.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Card RM, et al. Impact of Ciprofloxacin and Clindamycin Administration on Gram-Negative Bacteria Isolated from Healthy Volunteers and Characterization of the Resistance Genes They Harbor. Antimicrobial agents and chemotherapy. 2015;59(8):4410–6. doi: 10.1128/AAC.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen CH, et al. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. 2012;55(8):2285–94. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z, Li N, Neu J. Tight junctions, leaky intestines, and pediatric diseases. Acta paediatrica. 2005;94(4):386–93. doi: 10.1111/j.1651-2227.2005.tb01904.x. [DOI] [PubMed] [Google Scholar]
- 43.Jin Y, et al. APC dysfunction is correlated with defective suppression of T cell proliferation in human type 1 diabetes. Clinical immunology. 2009;130(3):272–9. doi: 10.1016/j.clim.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manirarora JN, et al. APC activation restores functional CD4(+)CD25(+) regulatory T cells in NOD mice that can prevent diabetes development. PloS one. 2008;3(11):e3739. doi: 10.1371/journal.pone.0003739. [DOI] [PMC free article] [PubMed] [Google Scholar]