Abstract
Serrated polyposis is a rare disorder characterised by the presence of multiple serrated polyps in the large intestine, and an increased personal and familial risk of colorectal cancer. Knowledge of the molecular characteristics of colonic lesions which develop in this syndrome is fragmented, making it difficult to understand the underlying genetic basis of this condition. We conducted a systematic review and meta‐analysis of all studies which evaluated the molecular characteristics of colorectal neoplasms found in individuals with serrated polyposis. We identified 4561 potentially relevant studies, but due to a lack of consensus in the reporting of findings, only fourteen studies were able to be included in the meta‐analysis. BRAF mutation was found in 73% (95% CI 65–80%) of serrated polyps, 0% (95% CI 0–3%) of conventional adenomas and 49% (95%CI 33–64%) of colorectal cancers. In contrast, KRAS mutation was present in 8% (95% CI 5–11%) of serrated polyps, 3% (95% CI 0–13%) of conventional adenomas and 6% (95% CI 0–13%) of colorectal cancers. Absence of MLH1 immunostaining was found in 3% (95% CI 0–10%) of serrated polyps and 53% (95% CI 36–71%) of colorectal cancers. Overall, microsatellite instability was found in 40% (95% CI 18–64%) of colorectal cancers arising in the setting of serrated polyposis. Our results indicate that diverse molecular pathways are likely to contribute to the increased predisposition for colorectal cancer in individuals with serrated polyposis. We also propose a set of minimum standards for the reporting of future research in serrated polyposis as this is a rare syndrome and collation of research findings from different centres will be essential to identify the molecular mechanisms involved in the pathogenesis of this condition.
Keywords: serrated polyposis, molecular pathway, colorectal cancer, conventional adenoma, serrated polyp
Introduction
Significant advances in understanding the molecular pathways involved in the pathogenesis of colorectal cancer (CRC) have been achieved in the past three decades. The recognition of a ‘serrated’ pathway to CRC has challenged the previously held notion that CRCs developed exclusively from conventional adenomas 1. Serrated polyps, named because of their characteristic ‘saw‐toothed’ histological appearance, are thought to progress to CRC via a sequence of molecular events which frequently include activation of the mitogen‐activated protein kinase (MAPK) pathway through mutations of BRAF or KRAS oncogenes, as well as epigenetic silencing of genes through promoter hypermethylation (termed CpG island methylator phenotype, CIMP) 2, 3, 4, 5.
Serrated polyposis, previously known as hyperplastic polyposis syndrome, is a rare condition characterised by the presence of numerous serrated polyps in the large intestine. The current definition of serrated polyposis is shown in Table 1 6. The syndrome is associated with an increased risk of CRC 7, 8, 9, 10, 11 and has many hallmarks of a disease caused by a pathogenic germline genetic mutation. These include restricted ethnicity to individuals with Northern European ancestry 10, 11, familial clustering, and increased risk of CRC in the relatives of serrated polyposis individuals 12, 13. To date however, the molecular basis of serrated polyposis remains unknown.
Table 1.
World Health Organization clinical criteria for the identification of serrated polyposis 16
| Criterion 1 | At least five serrated polyps located proximal to the sigmoid colon, two of which are larger than 10 mm in diameter |
| Criterion 2 | Any number of serrated polyps located proximal to the sigmoid colon in an individual who has a first degree relative with serrated polyposis |
| Criterion 3 | More than 20 serrated polyps of any size distributed throughout the colon |
For this reason, serrated polyposis is currently defined by an arbitrary set of clinical parameters which describe a heterogeneous group of patients. Studies evaluating the molecular characteristics of colonic lesions in individuals with serrated polyposis have been limited by relatively small sample size, and by heterogeneity in the patients recruited. The aim of this systematic review and meta‐analysis is to summarise the literature on the molecular features of CRCs and their precursor lesions in patients with serrated polyposis, in order to identify patterns which could be used as the basis for future research into this condition.
Methods
Our meta‐analysis adhered to the PRISMA statements for reporting on systematic reviews and the STREGA recommendations for reporting of genetic association studies 14, 15. Given that the diagnostic criteria and nomenclature for serrated polyps were only formalised in 2010 6 and that most sessile serrated adenomas polyps (SSA/Ps) were previously identified as hyperplastic polyps (HPs), the term serrated polyp was used to describe all polyps with a serrated histological architecture, including HPs, SSA/Ps, traditional serrated adenomas and mixed serrated polyps (MP). Lesions proximal to the splenic flexure, including those from the caecum, ascending and transverse colon, were classified as proximal; lesions from the splenic flexure, descending colon, sigmoid colon and rectum were classified as distal.
Search strategy
Three investigators (LW, MS, EH) independently searched Medline and EMBASE for articles published before Jan 1st 2015 relating to serrated polyposis. The search was conducted using the following key words as both MeSH terms and text words: (‘hyperplastic polyposis’ OR ‘intestinal polyposis’ OR ‘serrated polyposis’) AND (‘colon OR colonic OR colorectal’) AND (‘polyp OR neoplasm OR neoplasia OR carcinoma OR cancer’). In addition, reference lists of identified articles were searched for additional relevant references.
Study selection criteria
Each manuscript was reviewed for inclusion according to the following criteria: (1) full article was published in peer reviewed journals, (2) the study participants fulfilled the WHO criteria for serrated polyposis 16 (see Table 1) and (3) the study reported the histological and molecular characteristics of colonic polyps and/or cancers collected from individuals with serrated polyposis. Studies which did not describe study participants according to the WHO criteria were included if the investigators were able to use primary data to retrospectively apply the WHO criteria. Investigators LW, MS and EH independently conducted the search, reviewed the manuscripts, and extracted data from the included studies, with disagreements and queries resolved through re‐evaluation of study and discussion.
Data extraction
Using a structured template, the following data were extracted by each investigator: first author's name, year of publication, recruitment and ascertainment method of individuals, histological and molecular characteristics of colonic neoplasms studied. Molecular characteristics were chosen to reflect key steps in the serrated pathway, including (1) mutation of the BRAF or KRAS oncogenes, (2) CIMP, categorised as CIMP‐high, CIMP‐low or CIMP‐negative, (3) immunohistochemical (IHC) expression of mismatch repair proteins MLH1, MSH2, MSH6 and PMS2, and (4) microsatellite instability (MSI) status categorised as MSI‐high (MSI‐H), MSI‐low (MSI‐L) or MS stable (MSS).
Statistical analysis
We calculated the prevalence of key molecular characteristics by dividing the total number of colonic neoplasms with the molecular alteration of interest by the total number of neoplasms in each histological category (serrated polyp, conventional adenoma and CRC). Serrated polyps contiguous with CRC were excluded from the meta‐analysis as they were likely to have molecular features found in both serrated polyps and CRCs, making interpretation of results difficult 17.
As the majority of studies did not report on the proportion of polyps sampled for molecular analysis from each patient, we therefore calculated the mean number of polyps per patient as a means of assessing heterogeneity in the study populations, and hence the validity of pooling data from the studies. Meta‐analysis was performed if mean polyp counts per patient were comparable across studies. Since the proportion of lesions carrying the molecular abnormality of interest was 0 or 1 in some of the included studies, we enabled the Freeman‐Tukey double arcsine transformation so that these studies would be included in the analysis. We used random effects meta‐analysis to calculate the pooled estimates of the proportion with 95% confidence interval (CI) 18. Heterogeneity in the included studies was assessed using the I2 statistic, where I2 > 50% indicates high heterogeneity, implying that the pooled results should be interpreted with caution. All data analyses were performed with STATA version 13.0 (StataCorp, TX, USA).
Results
We reviewed a total of 4561 reports, 40 of which were potentially eligible studies according to the title/abstract (Figure 1). After full review, 20 studies met the inclusion criteria. A further 6 studies were excluded from the meta‐analysis because data pertaining to patients with serrated polyposis were not extractable from the studies or the results of studies were not relevant to the meta‐analysis. There was no inter‐observer disagreement in the final list of included studies. The study designs, patient characteristics and colonic lesions analysed in each study are summarised in supplementary material, Table S1. The characteristics of studies excluded from the meta‐analysis and the reasons for their exclusion are summarised in supplementary material, Table S2. With the exception of one study, the median ages of study cohorts were in the range 50–66 years 19. There was substantial variability in the rate of CRC in the included primary study groups, which was lowest in individuals recruited from polyposis databases and highest in surgical series. As our calculations showed that the mean number of polyps per patient was comparable across the studies, we were able to proceed with the meta‐analysis.
Figure 1.
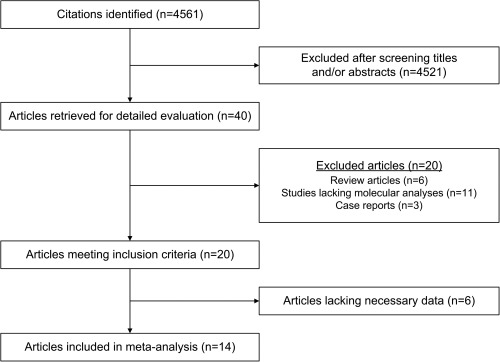
Flow chart showing the results of the search strategy and reasons for exclusion.
Summary estimates
The proportion of serrated polyps, conventional adenomas and CRCs which harboured key molecular characteristics of interest, as analysed using random effects model, are shown in Table 2, and described further below.
Table 2.
Summary estimates of the prevalence (95% CIs) and testing for heterogeneity (I2) for molecular characteristics analysed in serrated polyposis, using a random effects model
| Molecular characteristics analysed | No. of studies | Summary estimate of prevalence % (95% CI) | Heterogeneity I2 statistic, p value |
|---|---|---|---|
| BRAF mutation in serrated polyps | 8 | 73 (65–80) | 75%; p = 0.00 |
| BRAF mutation in conventional adenomas | 6 | 0 (0–3) | 27%; p = 0.23 |
| BRAF mutation in CRCs | 4 | 49 (33–64) | 0%; p = 0.58 |
| KRAS mutation in serrated polyps | 9 | 8 (5–11) | 67%; p = 0.00 |
| KRAS mutation in conventional adenomas | 7 | 3 (0–13) | 45%; p = 0.09 |
| KRAS mutation in CRCs | 7 | 3 (0–16) | 28%, p = 0.23 |
| Loss of MLH1 expression in serrated polyps | 6 | 3 (0–10) | 85%, p = 0.0 |
| Loss of MLH1 expression in CRCs | 8 | 53 (36–71) | 57%, p = 0.02 |
| MSI in CRC | 5 | 40 (18–64) | 45%, p = 0.12 |
Mutations of BRAF and KRAS oncogenes
In the setting of serrated polyposis, the overall estimated rate of BRAF mutation was 73% (95% CI 65–80%) in serrated polyps, 0% (95% CI 0–3%) in conventional adenomas and 49% (95% CI 33–64%) in CRCs (Figure 2A–C). The rate of BRAF mutation was comparable in serrated polyps from the proximal colon (70%, 95% CI 54–85%) with those from the distal colon (68%, 95% CI 59–76%). Nearly all of the BRAF‐mutant cancers (94%, 95% CI 83–100%) were found in the proximal colon.
Figure 2.
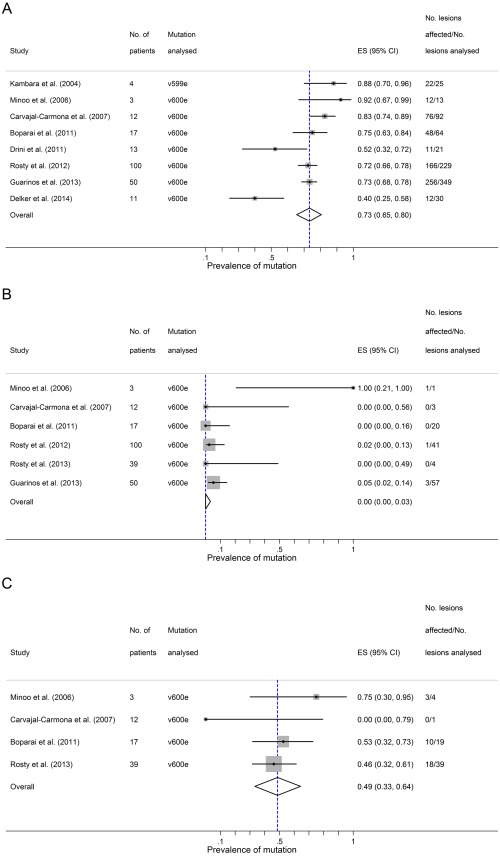
BRAF mutations in the setting of Serrated Polyposis. Three panels showing summary estimates for the proportion of different lesional types carrying BRAF mutations in the setting of Serrated Polyposis. Panel A – serrated polyps; Panel B – conventional adenomas; Panel C – colorectal cancers arising in the setting of Serrated Polyposis. ES = estimated proportion.
The overall estimated rate of KRAS mutation was 8% (95% CI 5–11%) in serrated polyps, 3% (95% CI 0–13%) in conventional adenomas and 3% (95% CI 0–16%) in CRCs from patients with serrated polyposis (Figure 3A–C). KRAS mutation occurred in approximately 4% (95% CI 1–10%) of proximal serrated polyps and 11% (95% CI 3–20%) of distal serrated polyps.
Figure 3.
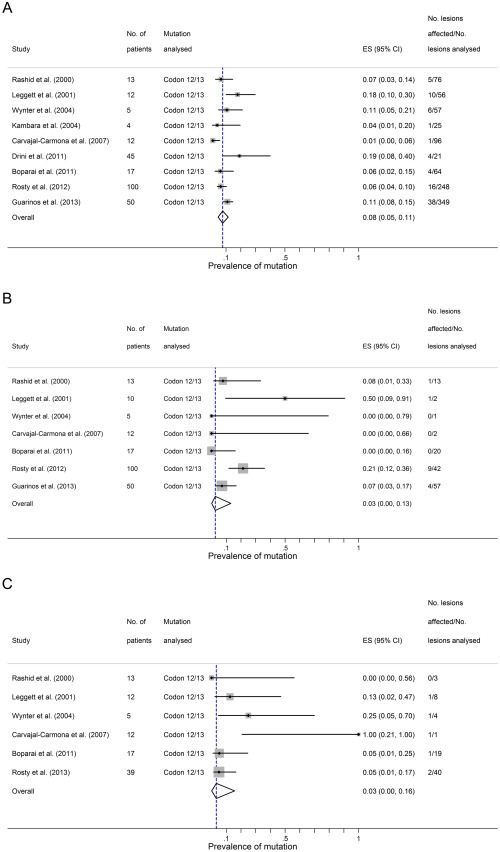
KRAS mutations in the setting of Serrated Polyposis. Three panels showing summary estimate for the proportion different lesional types carrying KRAS mutations in the setting of Serrated Polyposis. Panel A – serrated polyps; Panel B – conventional adenomas; Panel C – colorectal cancers arising in the setting of Serrated Polyposis. ES = estimated proportion.
In the studies which followed the formal diagnostic nomenclature for serrated polyps, the results of their molecular analyses could be stratified according to the histological subtype of serrated polyps 19, 20, 21, 22, 23. All of these studies sampled SSA/Ps and HPs for their molecular analyses, and the overall ratio of SSA/Ps to HPs sampled was approximately 1:1. BRAF mutation was found in 78% (95% CI 66–87%) of SSA/Ps and 54% (95% CI 33–73%) of HPs, whereas KRAS mutation was found in 2% of SSA/Ps (95% CI 0–6%) and 10% of HPs (95% CI 3–21%).
CpG island methylator phenotype
Although several studies reported on the CIMP status of serrated polyps and CRCs in the setting of serrated polyposis 21, 22, 24, 25, a meta‐analysis could not be conducted as studies varied in their definition of CIMP with the use of different gene panels and marker thresholds. Wynter et al used MINT loci (1, 2, 3, 12) to determine the CIMP status of serrated polyps. Of a total of 58 polyps, 35 (60%) were CIMP‐high, 16 (28%) were CIMP‐low, and 7 (12%) were CIMP‐negative 25. On the other hand, Guarinos et al used a commercial CIMP kit to investigate aberrant CpG island methylation in the promoters of CACNA1G, CDKN2A, CRABP1, IGF‐2, MLH1, NEUROG1, RUNX3, and SOCS1. It was reported that CIMP‐high status was found in 71.1% of SSA/Ps, 37.2% of microvesicular HPs and 17.6% of adenomas 22.
IHC analysis of mismatch repair genes and MSI
In the setting of serrated polyposis, absent IHC staining of MLH1 protein was estimated to occur in 3% (95% CI 0–10%) of serrated polyps and 53% (95% CI 36–71%) of CRCs (Figure 4A‐B). There was a paucity of data to permit meta‐analyses of the prevalence of MSH2, MSH6 and PMS2 protein expression. In the only study to have investigated IHC expression of MSH2 and MSH6 in this setting, it was reported that all of the serrated polyps showed normal expression of these proteins 26. Analysis of PMS2 expression was also restricted to a single study, where it was reported that loss of either MLH1 or PMS2 occurred in 2.9% (4 of 138) SSA/Ps 23. None of the studies investigated expression of MSH2, MSH6 and PMS2 protein expression in CRCs from patients with serrated polyposis.
Figure 4.
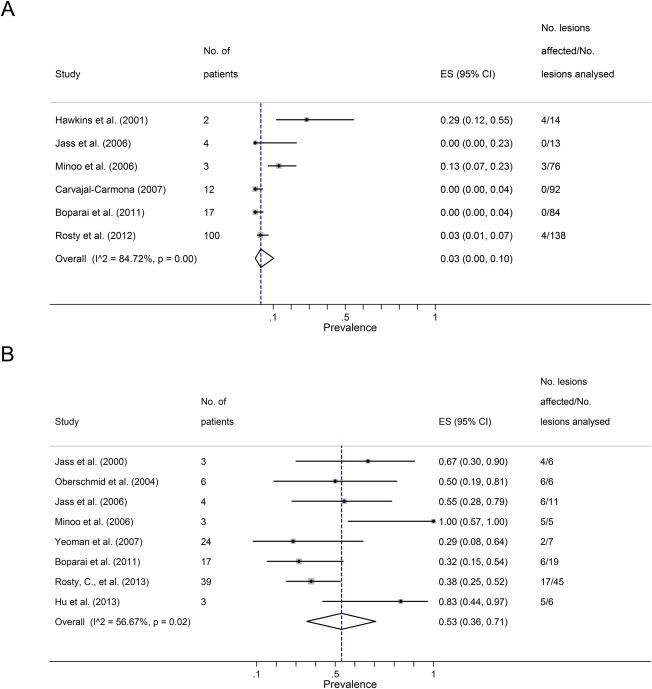
MLH1 loss in the setting of Serrated Polyposis. Panel A shows a summary estimate for the proportion of serrated polyps with MLH1 loss in the setting of Serrated Polyposis. Panel B shows the same data in relation to colorectal cancers arising in the setting of Serrated Polyposis. ES = estimated proportion; I2 = I squared heterogeneity statistic.
MSI‐H was found in approximately 40% (95% CI 18–64%) of all CRCs in serrated polyposis (Figure 5). There were insufficient data to analyse for any overall association between loss of MLH1, MSI‐H and degree of dysplasia. However, a single study found that loss of MLH1 was significantly associated with MSI‐H lesions showing dysplasia or carcinoma compared with those which were MSI‐L or MSS 27.
Figure 5.
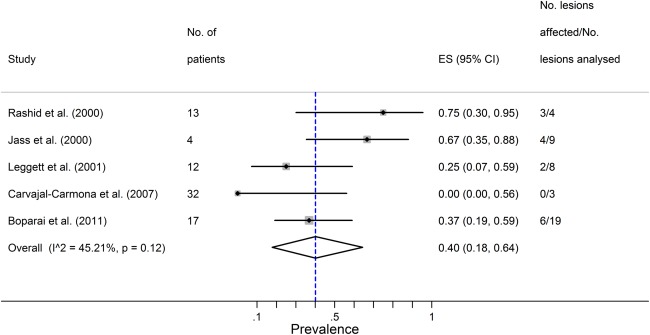
Summary estimate for the proportion of colorectal cancers that are MSI in the setting of Serrated Polyposis. ES = estimated proportion; I2 = I squared heterogeneity statistic.
Sensitivity analysis
We performed sensitivity analysis to assess the impact on our estimates of excluding pre‐2010 studies in which the current definition of serrated polyposis was not used. For the estimates of BRAF and KRAS mutations in serrated polyps, conventional adenomas and CRCs, the results did not change substantially when these studies were excluded. Similarly, the estimated rate of MSI in CRCs did not change significantly with exclusion of pre‐2010 studies. The sensitivity analysis was remarkably robust compared to all of the main study findings. The only qualitative change was a lower rate in the loss of MLH1 expression in CRC, with a prevalence of 44% (95% CI 23–66%) compared to a value of 53% (95% CI 36–71%) in the main analysis.
Discussion
This is the first meta‐analysis focusing on the molecular characteristics of CRCs and their precursor lesions from individuals with serrated polyposis. Individuals with serrated polyposis have a significantly increased risk of developing CRC 7. Earlier studies have shown that serrated polyps in such individuals have higher rates of BRAF mutation compared with their sporadic counterparts 26, 28, 29, 30, 31. In our meta‐analysis, we have shown that CRCs from these individuals have higher rates of BRAF mutation compared to the general population; 49% (95% CI 35–60%) compared to 10–15% 32, 33. This is likely to be accounted for by the multitude of SSA/Ps found in serrated polyposis and is consistent with the hypothesis that CRCs in this setting develop from precursor SSA/Ps through the ‘serrated’ pathway 1.
As mutations in BRAF or KRAS oncogenes are frequently found as mutually exclusive events in serrated carcinomas, we added the rates of BRAF (49%) and KRAS (3%) mutations in CRCs from individuals with serrated polyposis to show that nearly half of all CRCs arising in this setting do not harbour mutations in either BRAF or KRAS. This suggests that mechanisms other than oncogenic mutations of BRAF or KRAS may be involved in the pathogenesis of serrated polyposis. Recent studies suggest that gene mutation status alone is not enough to define the complexity of the underlying biology of CRCs 34. Tian et al developed gene‐specific expression patterns to characterise an activating oncogenic signature for KRAS, BRAF and phosphatidyl inositol 3‐kinase (PIK3CA) in stage II/III CRCs 35. They found that 79 of the 206 tumours with no oncogenic mutations in BRAF, KRAS and PI3KCA could be classified as oncogenic based on their gene expression signatures. Multiple mechanisms were proposed for the oncogenic phenotype in wild‐type patients, including overexpression of key molecules within the MAPK pathway, overexpression of activators of the pathway as well as downregulation of inhibitors of the pathway 35.
Conventional adenomas are found in up to 90% of individuals with serrated polyposis and their presence is associated with an increased risk of colorectal cancer in these individuals 7, 8, 13, 23. However, only a few studies have investigated the molecular characteristics of conventional adenomas from individuals with serrated polyposis. Recently, Pai et al described a new polyp entity, atypical conventional adenoma, found in individuals who also have at least one SSA/P 36. Although these polyps were all wild‐type for BRAF or KRAS mutation, they share some morphological characteristics with serrated polyps including eosinophilic cytoplasm, focal serration and crypt dilatation. These atypical polyps demonstrated low levels of methylation. It is possible that a proportion of CRCs arising in individuals with serrated polyposis develop from conventional adenomas through a process mediated by DNA hypermethylation 32. Furthermore, although it has been proposed that DNA hypermethylation is mediated by activating mutation of the BRAF oncoprotein 5, it is also possible that activation of the BRAF oncoprotein may be mediated by DNA hypermethylation. However, while others have reported higher levels of CpG island methylation in both normal and lesional tissues from individuals with serrated polyposis 25, 37, this systematic review lacked sufficient data to allow meta‐analysis of the CIMP status of tumours. In addition, although epigenetic silencing of MLH1 was shown to occur in approximately half of all CRCs in serrated polyposis, the role of other mismatch repair genes, DNA repair genes or tumour suppressor genes in colorectal carcinogenesis in serrated polyposis is yet unknown due to the limited number of studies which have investigated this.
Our analysis has several limitations, the most important of which is the heterogeneity across studies which have reported on the molecular characteristics of serrated polyposis. There may be multiple reasons for this. The studies were not population based, but drawn from a variety of different patient sources, including colorectal cancer databases, hospital records, polyposis databases, family cancer clinics and genetic clinics. This raises the possibility of ascertainment bias, particularly in those cohorts drawn from individuals presenting with symptomatic bowel cancer. The studies included in this meta‐analysis also had relatively small sample sizes, and showed considerable variability in both the molecular characteristics assessed and the techniques used to analyse them. Another important limitation is that we could not account for potential sampling bias in the original studies as some of the studies did not report on the proportion of total number of polyps sampled for molecular analysis. Further, the criteria used in the selection of polyps sampled for molecular analyses were not specified in any of the studies.
The number of studies which could be included in our meta‐analysis was considerably limited by a lack of conformity and consistency for reporting this rare syndrome. Despite finding 4561 potentially relevant studies, our meta‐analysis was limited to only 14 studies, mostly as a result of reporting discrepancies. These include: participant inclusion criteria not adhering to the WHO definition for serrated polyposis, lack of quantitative description of the total number of serrated polyps identified and the number sampled, and failure to distinguish results pertaining to subjects with serrated polyposis from those without. Furthermore, research in serrated polyposis thus far has focused predominantly on serrated polyps and the ‘serrated’ pathway to CRC. As serrated polyposis is a relatively rare condition, future molecular characterisation of this syndrome will always be hindered by small sample sizes and study population heterogeneity. Pooling of studies from different centres will be crucial in improving the understanding of the molecular pathways involved in the condition. Therefore, we propose a set of minimum standards for the reporting of studies in serrated polyposis (Table 3) to guide future research into the molecular underpinnings of this rare syndrome. In addition to satisfying the WHO criteria for serrated polyposis, future investigators should ensure that they provide basic demographic information about the patients enrolled into the study and, ideally, report the total numbers of serrated polyps, conventional adenomas and CRCs in study participants. In order to minimise sampling bias, it would also be important to report on the proportion of polyps that is sampled for molecular analyses and any specific criteria used in the selection of polyps. Many older studies have restricted their attention to one or two molecular hallmarks of the ‘serrated’ pathway, thereby limiting the interpretation of these findings in the context of the molecular heterogeneity of CRC. It is therefore suggested that a core panel of molecular markers, including MSI or mismatch repair deficiency, CIMP, mutations in BRAF and KRAS be incorporated into any novel molecular markers under investigation. Consideration should also be given as to whether germline sequencing was undertaken and if so by what method. The resultant standardisation of molecular research reporting will facilitate consistent and accurate data collection, comparison of pathological and molecular findings between different studies, and enable research into identifying the molecular mechanisms underlying this enigmatic condition.
Table 3.
New proposed minimum standards for reporting of molecular characteristics in serrated polyposis
| Required data |
|---|
| Subjects must satisfy the WHO criteria for serrated polyposis |
| Age and gender |
| Serrated polyps: histological classification, total number and the number sampled |
| Conventional adenomas (if present): total number and the number sampled |
| CRC (if present): location and proximity to serrated polyp and/or conventional adenoma |
| Recommended data |
| Comment on whether germline sequencing has been undertaken and if so by what method |
| Panel of core molecular markers |
| 1. MSI |
| 2. Mutations in BRAF and KRAS |
| 3. Mismatch repair deficiency (MLH1, MSH2, MSH6, and PMS2) |
| 4. CIMP |
Author contributions
EH participated in data collection, analysis, and writing the article. MS and LW participated in data collection. KC participated in analysing the data and writing the article. RLW participated in designing the research, analysing the data, and writing the article.
Supporting information
SUPPLEMENTARY MATERIAL ONLINE
This supplementary file contains Tables S1 and S2:
Table S1. Characteristics of studies included in the meta‐analysis
Table S2. Characteristics of studies excluded from the meta‐analysis and reasons for exclusion
Acknowledgements
Emily Y. He is supported by a National Health and Medical Research Council (NHMRC) Postgraduate Scholarship (GNT1093776).
The authors declare no conflicts of interest.
References
- 1. Hawkins NJ, Bariol C, Ward RL. The serrated neoplasia pathway. Pathology 2002; 34: 548–555. [PubMed] [Google Scholar]
- 2. Bettington M, Walker N, Clouston A, et al The serrated pathway to colorectal carcinoma: Current concepts and challenges. Histopathology 2013; 62: 367–386. [DOI] [PubMed] [Google Scholar]
- 3. Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum Pathol 2011; 42: 1–10. [DOI] [PubMed] [Google Scholar]
- 4. Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology 2010; 138: 2088–2100. [DOI] [PubMed] [Google Scholar]
- 5. Fang M, Ou J, Hutchinson L, et al The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG island methylator phenotype. Molecular Cell 2014; 55: 904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Snover D, Ahnen D, Burt R, et al Serrated Polyps of the Colon and Rectum and Serrated Polyposis (4th edn). IARC: Lyon, France, 2010. [Google Scholar]
- 7. Boparai KS, Mathus‐Vliegen EM, Koornstra JJ, et al Increased colorectal cancer risk during follow‐up in patients with hyperplastic polyposis syndrome: A multicentre cohort study. Gut 2010; 59: 1094–1100. [DOI] [PubMed] [Google Scholar]
- 8. Buchanan DD, Sweet K, Drini M, et al Risk factors for colorectal cancer in patients with multiple serrated polyps: A cross‐sectional case series from genetics clinics. PLoS One 2010; 5: e11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edelstein DL, Axilbund JE, Hylind LM, et al Serrated polyposis: Rapid and relentless development of colorectal neoplasia. Gut 2013; 62: 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kalady MF, Jarrar A, Leach B, et al Defining phenotypes and cancer risk in hyperplastic polyposis syndrome. Dis Colon Rectum 2011; 54: 164–170. [DOI] [PubMed] [Google Scholar]
- 11. Yeoman A, Young J, Arnold J, et al Hyperplastic polyposis in the New Zealand population: A condition associated with increased colorectal cancer risk and European ancestry. N Z Med J 2007; 120: U2827. [PubMed] [Google Scholar]
- 12. Boparai KS, Reitsma JB, Lemmens V, et al Increased colorectal cancer risk in first‐degree relatives of patients with hyperplastic polyposis syndrome. Gut 2010; 59: 1222–1225. [DOI] [PubMed] [Google Scholar]
- 13. Win AK, Walters RJ, Buchanan DD, et al Cancer risks for relatives of patients with serrated polyposis. Am J Gastroenterol 2012; 107: 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moher D, Liberati A, Tetzlaff J, et al Preferred reporting items for systematic reviews and meta‐analyses: The PRISMA statement. Ann Intern Med 2009; 151: 264–269, w264. [DOI] [PubMed] [Google Scholar]
- 15. von Elm E, Moher D, Little J. Reporting genetic association studies: the STREGA statement. The Lancet; 374: 98–100. [DOI] [PubMed] [Google Scholar]
- 16. Snover D. Serrated Polyps of the Colon and Rectum and Serrated Polyposis WHO Classification of Tumours of the Digestive System (4th edn). IARC: Lyon, France, 2010. [Google Scholar]
- 17. O'Brien MJ, Yang S, Mack C, et al. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol 2006; 30: 1491–1501. [DOI] [PubMed] [Google Scholar]
- 18. Nyaga VN, Arbyn M, Aerts M. Metaprop: A Stata command to perform meta‐analysis of binomial data. Archives of Public Health 2014; 72: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Delker DA, McGettigan BM, Kanth P, et al RNA sequencing of sessile serrated colon polyps identifies differentially expressed genes and immunohistochemical markers. PLoS One 2014; 9: e88367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boparai KS, Dekker E, Polak MM, et al A serrated colorectal cancer pathway predominates over the classic WNT pathway in patients with hyperplastic polyposis syndrome. Am J Pathol 2011; 178: 2700–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drini M, Wong NC, Scott HS, et al Investigating the potential role of genetic and epigenetic variation of DNA methyltransferase genes in hyperplastic polyposis syndrome. PLoS One 2011; 6: e16831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guarinos C, Sanchez‐Fortun C, Rodriguez‐Soler M, et al Clinical subtypes and molecular characteristics of serrated polyposis syndrome. Clin Gastroenterol Hepatol 2013; 11: 705–711; quiz e746. [DOI] [PubMed] [Google Scholar]
- 23. Rosty C, Buchanan DD, Walsh MD, et al Phenotype and polyp landscape in serrated polyposis syndrome: A series of 100 patients from genetics clinics. Am J Surg Pathol 2012; 36: 876–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oberschmid BI, Dietmaier W, Hartmann A, et al Distinct secreted Frizzled receptor protein 1 staining pattern in patients with hyperplastic polyposis coli syndrome. Arch Pathol Lab Med 2004; 128: 967–973. [DOI] [PubMed] [Google Scholar]
- 25. Wynter CV, Walsh MD, Higuchi T, et al Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut 2004; 53: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carvajal‐Carmona LG, Howarth KM, Lockett M, et al Molecular classification and genetic pathways in hyperplastic polyposis syndrome. J Pathol 2007; 212: 378–385. [DOI] [PubMed] [Google Scholar]
- 27. Jass JR, Iino H, Ruszkiewicz A, et al Neoplastic progression occurs through mutator pathways in hyperplastic polyposis of the colorectum. Gut 2000; 47: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beach R, Chan AO, Wu TT, et al BRAF mutations in aberrant crypt foci and hyperplastic polyposis. Am J Pathol 2005; 166: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chan TL, Zhao W, Leung SY, et al BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res 2003; 63: 4878–4881. [PubMed] [Google Scholar]
- 30. Kambara T, Simms LA, Whitehall VL, et al BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 2004; 53: 1137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang S, Farraye FA, Mack C, et al BRAF and KRAS Mutations in hyperplastic polyps and serrated adenomas of the colorectum: Relationship to histology and CpG island methylation status. Am J Surg Pathol 2004; 28: 1452–1459. [DOI] [PubMed] [Google Scholar]
- 32. Rosty C, Walsh MD, Walters RJ, et al Multiplicity and molecular heterogeneity of colorectal carcinomas in individuals with serrated polyposis. Am J Surg Pathol 2013; 37: 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clancy C, Burke JP, Kalady MF, et al BRAF mutation is associated with distinct clinicopathological characteristics in colorectal cancer: A systematic review and meta‐analysis. Colorectal Disease 2013; 15: e711–e718. [DOI] [PubMed] [Google Scholar]
- 34. van Geel RMJM, Beijnen JH, Bernards R, et al Treatment individualization in colorectal cancer. Current Colorectal Cancer Reports 2015; 11: 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tian S, Simon I, Moreno V, et al A combined oncogenic pathway signature of BRAF, KRAS and PI3KCA mutation improves colorectal cancer classification and cetuximab treatment prediction. Gut 2013; 62: 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pai RK, Mackinnon AC, Joseph L, et al Identification of histologically distinct conventional adenomas that arise predominately in patients with sessile serrated adenomas. Am J Surg Pathol 2010; 34: 355–363. [DOI] [PubMed] [Google Scholar]
- 37. Minoo P, Baker K, Goswami R, et al Extensive DNA methylation in normal colorectal mucosa in hyperplastic polyposis. Gut 2006; 55: 1467–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIAL ONLINE
This supplementary file contains Tables S1 and S2:
Table S1. Characteristics of studies included in the meta‐analysis
Table S2. Characteristics of studies excluded from the meta‐analysis and reasons for exclusion


