Abstract
Background
Octamer-binding transcription factor 4 (Oct-4) has been identified to participate in the tumorigenicity and malignancy of non-small cell lung cancer (NSCLC). However, its definite prognostic roles in NSCLC still remain a debate. Therefore, we conducted this meta-analysis to evaluate the prognostic value of Oct-4 expression in NSCLC and its relationship to some major clinicopathological characteristics.
Methods
A comprehensive literature retrieval was performed in PubMed, EMBASE and the Web of Science to identify the full-text articles that met our eligibility criteria. Odds ratio (OR) with 95% confidence interval (CI) severed as the summarized statistics for clinicopathological assessments, and hazard ratio (HR) with 95% CI served as the summarized statistics for prognostic assessments. Q-test and I2-statistic were used to evaluate the level of heterogeneity. Potential publication bias was detected by both Begg’s test and Egger’s test.
Results
There were 16 retried articles with 1,363 NSCLC cases included into this meta-analysis. Oct-4 expression was found to be significantly associated with the unfavorable outcomes for differentiation degree (OR: 3.065; 95% CI: 1.568–5.957; P=0.001), TNM stage (OR: 3.695; 95% CI: 2.252–6.063; P<0.001) and lymphatic metastasis (OR: 2.372; 95% CI: 1.504–3.742; P<0.001), but not associated with the histological subtypes, gender, age and smoking status. Oct-4 expression was also significantly associated with the poor prognosis of NSCLC (HR: 3.030; 95% CI: 2.283–4.021; P<0.001). The prognostic roles of Oct-4 expression in NSCLC still remained statistically reliable in the subgroups stratified by statistical analysis, patients’ origins, positively-stained sites and histological subtypes.
Conclusions
Our meta-analysis indicates that Oct-4 can serve as a strong biomarker predicting the poor clinicopathological and prognostic characteristics of NSCLC. More high-quality studies based on a large sample size will be very helpful to further validate and modify our findings in the future.
Keywords: Octamer-binding transcription factor 4 (Oct-4), non-small cell lung cancer (NSCLC), prognosis, meta-analysis
Introduction
Lung cancer is one of the most lethal malignances. It remains the leading cause of cancer-related deaths and a worldwide challenge to human health, particularly to the heavy smoking peoples in both developed and developing countries (1,2). According to authoritative estimations, non-small cell lung cancer (NSCLC) accounts for more than 85% of all lung cancer cases and its five-year overall survival (OS) rate approximates 15%, suggesting its poor prognosis (1,3). Currently, advanced surgical techniques, anesthetic techniques and perioperative managements have significantly improved the safety of lung cancer surgery but have hardly benefited the prognosis of NSCLC (4). It has been widely accepted that early relapse, metastasis and poor response to treatments are the primary causes of poor survival rate of NSCLC (5). To improve treatment plans and patients’ outcomes, a novel biomarker is urgently required to efficiently predict the clinicopathological and prognostic features of NSCLC based on the current diagnostic and therapeutic regimens.
In recent years, a growing number of evidences have highlighted the biological functions of cancer stem cells (CSCs) during the malignancy progression (6,7). Octamer-binding transcription factor 4 (Oct-4) belongs to the POU-domain transcription factor family which is generally expressed in both embryonic stem cells and CSCs (8,9). The latest evidences have indicated that Oct-4 may play a critical role in maintaining the self-renewal ability and pluripotency of embryonic stem cells (10). Oct-4 is also found to participate in the tumorigenicity and malignancy of NSCLC (11). A recent systematic review has demonstrated that Oct-4 expression is strongly correlated with some common malignant characteristics that result in the poor prognoses of different solid tumors (12). However, the integrated details of NSCLC are not separately reported in this study. Some controversies still exist on the relationship between Oct-4 expression and major clinicopathological characteristics of NSCLC, such as differentiation degree, TNM stage and lymphatic metastasis (13-17). Moreover, the previously published survival data correlated with Oct-4 expression in NSCLC have not been systematically reviewed until now. The definite prognostic roles of Oct-4 remain a debate (13,15-19).
Limited sample availability in individual studies may cause negative effects on clarifying this pending issue accurately. Meta-analysis is generally considered as a well-established method integrating the appropriate evidences from a number of homogeneous studies to formulate a global conclusion on both clinical practice and biomarker functions (20,21). Therefore, we conducted the present systematic review and meta-analysis to determine the clinicopathological and prognostic significance of Oct-4 expression in patients with NSCLC.
Methods
Protocol
No protocol had been previously published for this review. A systematic review and meta-analysis does not require necessary patients’ consent or ethical approval. We declared that this meta-analysis was performed in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) statement (22). The additional PRISMA 2009 checklist is given in the Table S1.
Search strategy
Literature retrieval for this meta-analysis was performed by two of our researchers (J Huang and XD Zhou) between January 21, 2016 and January 24, 2016. No language restriction was considered in this meta-analysis.
Three universal English electronic databases, including PubMed, EMBASE (via Ovid interface) and the Web of Science (via campus network of Sichuan University), and one Chinese native electronic database, China National Knowledge Infrastructure (CNKI), were selected for identifications of eligible articles published up to January 21, 2016.
A comprehensive literature retrieval was based on two search strings combined with six key words and two Boolean Operators (“AND” and “OR”). The key words we used to formulate the search details are listed as follows: (I) “Oct-4” and “octamer-binding transcription factor 4”; (II) “lung cancer”, “lung carcinoma”, “lung neoplasm” and “lung tumor”. The complete records of the search strings in each database are summarized in the Table S2.
In addition, we also manually screened the reference list of each article to identify any possibly included study with no duplication.
Inclusion and exclusion criteria
We established the following inclusion and exclusion criteria to confirm the studies included into our meta-analysis.
Inclusion criteria: (I) the target disease is NSCLC; (II) the expression of Oct-4 is independently studied instead of collaborating with other biomarkers; (III) immunohistochemistry (IHC) is applied for Oct-4 staining in NSCLC specimens; (IV) the demographics or statistics revealing the relationship between Oct-4 expression and clinicopathological characteristics of NSCLC are validly reported; (V) the statistical data correlated with the prognostic value of Oct-4 expression in NSCLC, including hazard ratio (HR), relative risk (RR) and odds ratio (OR) with corresponding 95% confidence interval (CI), are published in the results; (VI) the survival events with log-rank P value or Kaplan-Meier survival curves (K-M curves) are available from the original articles; (VII) OS serves as the key endpoint.
Exclusion criteria: (I) the following articles are immediately excluded: reviews, case reports, animal experiments and conference abstracts; (II) a comparison of Oct-4 expression between cancerous tissues and normal tissues is not included; (III) the continuous variables are not considered; (IV) positive Oct-4 expression is not stained by IHC.
Quality assessment
Newcastle-Ottawa Scale (NOS) was an appropriate assessment tool to estimate the quality of original non-randomized studies (23). Three perspectives including selection, comparability and exposure were considered for a semi-quantitative estimation. The “star system” with a maximum of 9 stars was used to grade all of the included studies. We regarded 8–9 stars as good quality, 6–7 stars as fair quality, and lower than 6 stars as poor quality.
Data collection
A Microsoft Excel sheet (primary document is not given) was designed to collect the following information: (I) publication data including researchers, publication years and languages; (II) experimental data including study design, study period, patients’ origins, investigating categories, experimental materials, detecting methods and sites, cut-off definitions, endpoints and follow-ups; (III) demographic data including sample sizes, the number of patients with positive and negative expression of Oct-4, and the number of adenocarcinoma (AC) and squamous cell carcinoma (SCC) cases; (IV) statistical data including summarized statistics with their sources, and statistical analysis methods (including univariate analysis and multivariate analysis).
Statistical analysis
To assess the relationship between Oct-4 expression and clinicopathological characteristics of NSCLC, we determined OR with 95% CI as the appropriate summarized statistics. In general, OR could be extracted from the demographics or statistics which were reported in the original articles. If HR or RR was published, we could immediately incorporate it into the meta-analysis (24).
To assess the prognostic value of Oct-4 expression in NSCLC, HR with 95% CI served as the summarized estimates because HR was the only appropriate statistic compatible for both censoring and time-to-events (25). It was our first priority to integrate the HR outcomes derived from multivariate analysis because that multivariate analysis using logistic regression or Cox proportional hazards model was generally applied to eliminate the bias risks from other confounding factors in observational studies. In addition, if RR or OR was conducted from multivariate analysis, we could also incorporate it into the meta-analysis (24). If no multivariate statistic was reported, we extrapolated the HR with 95% CI from the survival data according to a practical method described by Tierney et al. (26). The formulas used for HR extractions are given as follows.
| [1] |
| [2] |
| [3] |
Where O-E is the log-rank Observed minus Expected events and V is the log-rank Variance (26). If necessary, we could also extract the survival details by Engauge Digitizer 4.1 (http://sourceforge.net) from the K-M curves to measure the accuracy of estimated HR.
We used Q-test and I2-statistic to determine the level of heterogeneity within this meta-analysis. On the one hand, fine heterogeneity was defined by I2<50% and P>0.1, indicating a standard fixed-effect model test (Mantel-Haenszel method) for the integrations of OR and HR. On the other hand, a random-effect model test (DerSimonian and Laird method) would be determined if a significant heterogeneity was suggested by I2≥50% or P≤0.1 (27).
We performed an additional sensitivity analysis to further evaluate the stability of our summarized outcomes, in which the impact of each study on the overall estimates could be detected by omitting the individual study sequentially. The robustness of our meta-analysis would be verified if there was no substantial variation between the adjusted summarized outcomes and primary summarized outcomes (28).
Both the Begg’s test and Egger’s test were collaborated to detect the potential publication bias in this meta-analysis. On the one hand, the presence of bias was suggested by the visual symmetry of funnel plot conducted from Begg’s test, in which log ORs or log HRs were plotted against their corresponding standard errors (SEs) (29). On the other hand, its significance was also suggested by Egger’s P value. A significant publication bias would be revealed by either visual asymmetry of Begg’s funnel plot or Egger’s P value <0.05. However, if less than ten studies were included into the meta-analysis, publication bias tests were no longer required (24).
Finally, we declared that all of the above statistical analyses were accomplished by STATA 12.0 (STATA Corporation, College Station, TX).
Results
The selection of included studies
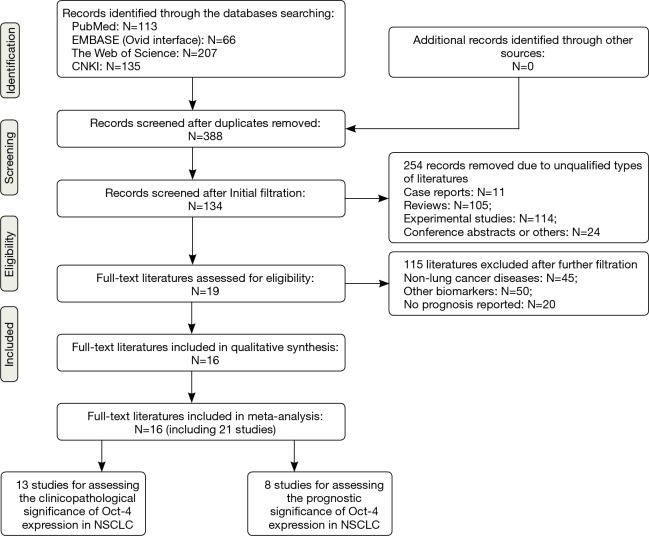
The complete procedures for literature retrieval were shown as a PRISMA flow diagram (Figure 1). A primary retrieval identified a total of 521 items of publications, including 113 citations in PubMed, 66 citations in EMBASE, 207 citations in the Web of Science and 135 citations in CNKI. After excluding 133 duplicates, the titles and abstracts of the remaining 388 items were screened for initial filtration. Then, 254 of them were directly excluded because of their irrelevant styles, including 11 case reports, 105 reviews, 114 experimental studies and 24 conference abstracts. The further filtration was performed by reading through the full-text of the remaining 134 articles. Among them, 19 articles were identified for possible eligibility of qualitative synthesis, after excluding 45 studies about other cancers, 50 studies about other biomarkers and 20 studies irrelevant with the prognostic roles of Oct-4. Then, we further excluded three of them from the qualitative synthesis because that IHC was not used for Oct-4 staining in these studies (30-32). Therefore, the remaining 16 articles met all of the eligibility criteria and were finally included into our meta-analysis (13-19,33-41).
Figure 1.
PRISMA flow diagram of literature retrieval. NSCLC, non-small cell lung cancer; Oct-4, octamer-binding transcription factor 4; PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-analysis.
The quality level of included studies
Two researchers (SJ Li and WB Zhang) were assigned to grade all of the included studies. Their mean NOS score was 7.4 (ranged from 6 to 9), suggesting a generally good quality level. The complete results of quality estimations are tabulated as Table S3.
The basic characteristics of included studies
Baseline characteristics of the 16 eligible articles are summarized in Table S4. A total of 21 retrospective observational studies were reported from these articles, including 13 studies investigating the association between Oct-4 expression and clinicopathological characteristics of NSCLC (13-17,33-36,38-41) and 8 studies evaluating the prognostic value of Oct-4 expression for OS in patients with NSCLC (13,15-19,37,40). They enrolled a total of 1,363 NSCLC patients from China mainland (n=1,092, ratio =80.1%) (13-16,33-41) and Taiwan (n=271, ratio =19.9%) (17-19). Because the histological subtypes of NSCLC were not clearly reported in some studies (14,37,40), the number of AC cases and SCC cases was unable to be calculated. IHC was commonly used for Oct-4 staining in paraffin-embedded specimens but cut-off values and positive-staining sites varied a lot across the included studies (Table S4). Positive Oct-4 expression was detected in 698 patients, with the positive ratio of 51.2%. Eight studies determined OS as the endpoint to assess the prognostic roles of Oct-4 expression in NSCLC, with a maximum follow-up period ranged from 12 to 120 months (13,15-19,37,40). In addition, the other demographics of NSCLC, including ages and clinical stages, were also outlined in Table S4.
The statistical characteristics of included studies
To evaluate the association between Oct-4 expression and clinicopathological characteristics of NSCLC, all of the relevant 13 included studies reported the demographics but none of them published any statistic from multivariate analysis (13-17,33-36,38-41). Therefore, the incorporative ORs were commonly extrapolated from the reported demographic details, which were derived from the univariate analysis (Table S4).
To determine the prognostic significance of Oct-4 expression in NSCLC, two of the eight relevant included studies reported the statistics from multivariate analysis which adequately eliminated the other confounding factors, including HR with 95% CI in one study (17) and OR with 95% CI in another one study (40). The remaining six studies (13,15,16,18,19,37) just published the survival data with log-rank P value and K-M curves, which were used to extrapolate the HR outcomes based on univariate analysis (Table S4).
Association between octamer-binding transcription factor 4 (Oct-4) expression and clinicopathological features of non-small cell lung cancer (NSCLC)
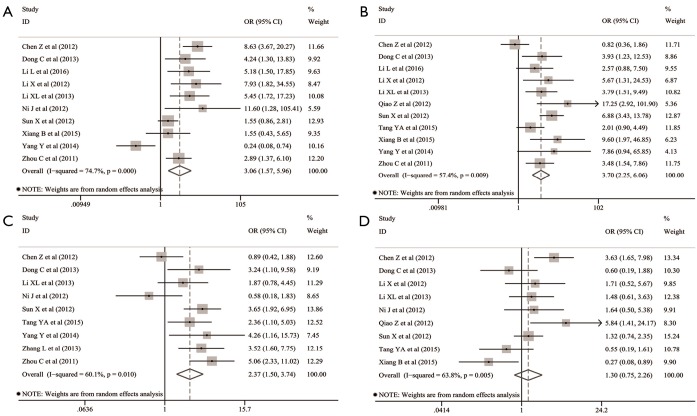
The common clinicopathological parameters of NSCLC involved the gender, age, smoking status, differentiation degree, TNM stage, lymphatic metastasis and histological subtypes in this meta-analysis. The pooled analyses indicated that Oct-4 expression was significantly associated with the increased risk of unfavorable outcomes for differentiation degree (OR: 3.065; 95% CI: 1.568–5.957; P=0.001; I2=74.7%, P<0.001) (Table 1 and Figure 2A), TNM stage (OR: 3.695; 95% CI: 2.252–6.063; P<0.001; I2=57.4%, P=0.009) (Table 1 and Figure 2B) and lymphatic metastasis (OR: 2.372; 95% CI: 1.504–3.742; P<0.001; I2=60.1%, P=0.010) (Table 1 and Figure 2C). However, no significant relationship was observed between Oct-4 expression and histological subtypes of NSCLC (OR: 1.301; 95% CI: 0.749–2.261; P=0.351; I2=63.8%, P=0.005) (Table 1 and Figure 2D), gender (OR: 0.868; 95%CI: 0.650–1.159; P=0.337; I2=0.0%, P=0.999) (Table 1 and Figure 3A), age (OR: 1.033; 95% CI: 0.772–1.383; P=0.828; I2=0.0%, P=0.631) (Table 1 and Figure 3B) and smoking status (OR: 1.099; 95% CI: 0.711–1.700; P=0.671; I2=0.0%, P=0.907) (Table 1 and Figure 3C).
Table 1. Meta-analysis of the association between Oct-4 expression and clinicopathological characteristics of NSCLC.
| Characteristics | N | No. samples | Heterogeneity (I2, P) | Model | OR (95% CI) | P value | Publication bias | Conclusion | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | PE | NE | Begg (P) | Egger (P) | |||||||
| Gender (male vs. female) | 10 | 943 | 470 | 473 | 0.0%, 0.999 | Fixed | 0.868 (0.650–1.159) | 0.337 | 0.371 | 0.273 | Not significant |
| Age (≥60 vs. <60 years) | 9 | 830 | 413 | 417 | 0.0%, 0.631 | Fixed | 1.033 (0.772–1.383) | 0.828 | – | – | Not significant |
| Smoking status (yes vs. no) | 4 | 408 | 214 | 194 | 0.0%, 0.907 | Fixed | 1.099 (0.711–1.700) | 0.671 | – | – | Not significant |
| Differentiation (G3 vs. G1 and G2) | 10 | 875 | 416 | 459 | 74.7%, <0.001 | Random | 3.065 (1.568–5.957) | 0.001 | 0.283 | 0.456 | Significant |
| TNM stage (III/IV vs. I/II) | 11 | 998 | 502 | 496 | 57.4%, 0.009 | Random | 3.695 (2.252–6.063) | <0.001 | 0.087 | 0.216 | Significant |
| Lymphatic metastasis (yes vs. no) | 9 | 965 | 455 | 510 | 60.1%, 0.010 | Random | 2.372 (1.504–3.742) | <0.001 | – | – | Significant |
| Histological subtypes (AC vs. SCC) | 9 | 788 | 405 | 383 | 63.8%, 0.005 | Random | 1.301 (0.749–2.261) | 0.351 | – | – | Not significant |
AC, adenocarcinoma; CI, confidence interval; NE, negative expression; NI, no information; NSCLC, non-small cell lung cancer; Oct-4, octamer-binding transcription factor 4; OR, odds ratio; PE, positive expression; SCC, squamous cell carcinoma.
Figure 2.
Association between Oct-4 expression and the pathological characteristics including (A) differentiation degree, (B) TNM stage, (C) lymphatic metastasis and (D) histological subtypes of NSCLC. CI, confidence interval; NSCLC, non-small cell lung cancer; Oct-4, octamer-binding transcription factor 4; OR, odds ratio.
Figure 3.
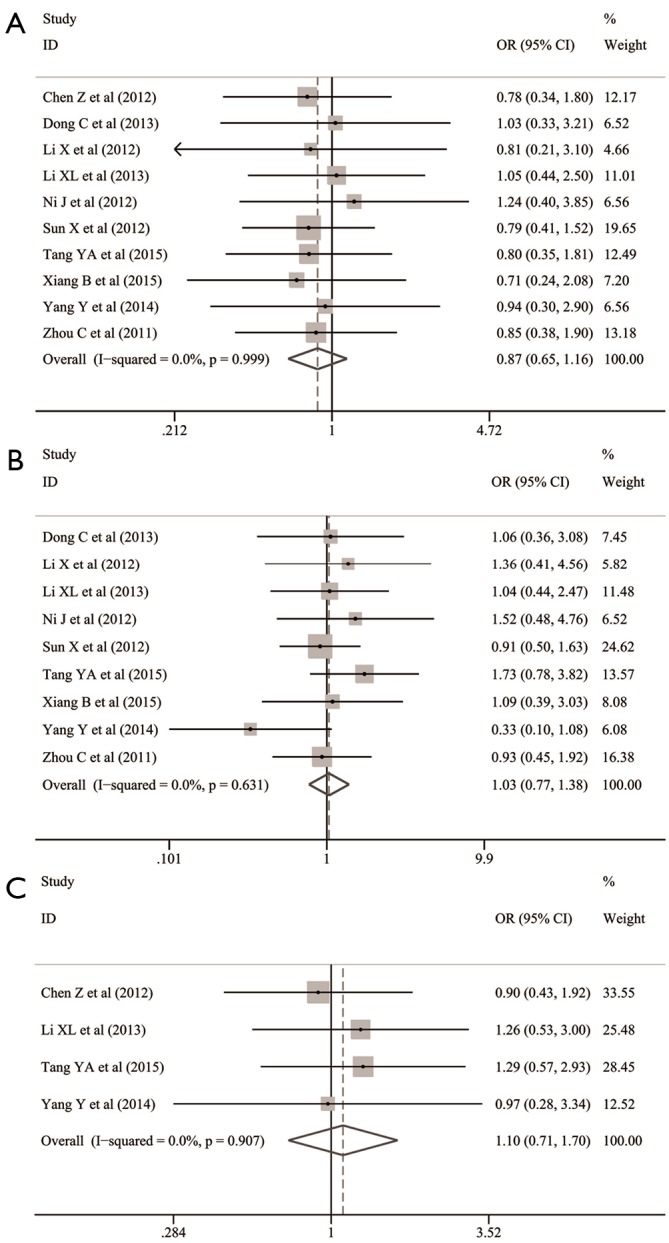
Association between Oct-4 expression and the clinical characteristics including (A) gender, (B) age and (C) smoking status in patients with NSCLC. CI, confidence interval; NSCLC, non-small cell lung cancer; Oct-4, octamer-binding transcription factor 4; OR, odds ratio.
Association between octamer-binding transcription factor 4 (Oct-4) expression and prognosis of non-small cell lung cancer (NSCLC)
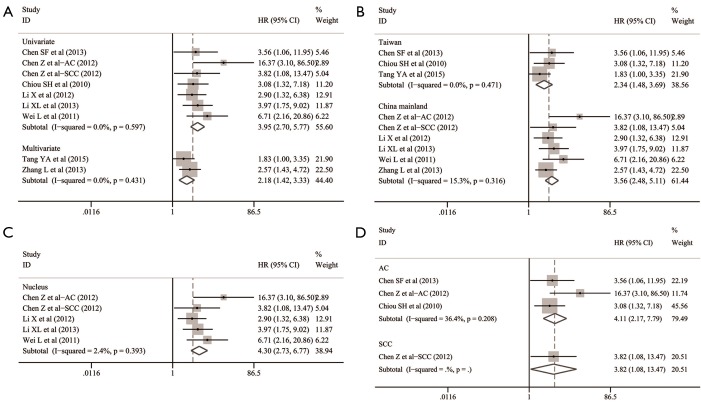
The overall analysis indicated that Oct-4 expression was significantly associated with the lower OS in patients with NSCLC (HR: 3.030; 95% CI: 2.283–4.021; P<0.001; I2=15.1%, P=0.308) (Table 2 and Figure 4).
Table 2. Prognostic significance of Oct-4 expression for OS in patients with NSCLC.
| Groups of outcomes | N | No. samples | Heterogeneity (I2, P) | Model | HR (95% CI) | P value | Conclusion | ||
|---|---|---|---|---|---|---|---|---|---|
| Total | PE | NE | |||||||
| Overall | 8 | 707 | 377 | 330 | 15.1%, 0.308 | Fixed | 3.030 (2.283–4.021) | <0.001 | Significant |
| Statistical analysis | |||||||||
| Multivariate analysis | 2 | 259 | 144 | 115 | 0.0%, 0.431 | Fixed | 2.175 (1.423–3.326) | <0.001 | Significant |
| Univariate analysis | 6 | 448 | 233 | 215 | 0.0%, 0.597 | Fixed | 3.948 (2.701–5.770) | <0.001 | Significant |
| Origins of patients | |||||||||
| China mainland | 5 | 436 | 189 | 247 | 15.3%, 0.316 | Fixed | 3.564 (2.484–5.113) | <0.001 | Significant |
| Taiwan | 3 | 271 | 188 | 83 | 0.0%, 0.471 | Fixed | 2.340 (1.484–3.690) | <0.001 | Significant |
| Positive-staining sites | |||||||||
| Nucleus | 4 | 310 | 137 | 173 | 2.4%, 0.393 | Fixed | 4.301 (2.733–6.767) | <0.001 | Significant |
| Non-nucleus | Given up because of the scarcity of available data | ||||||||
| Histological subtypes1 | |||||||||
| AC | 3 | 196 | 133 | 63 | 36.4%, 0.208 | Fixed | 4.108 (2.166–7.790) | <0.001 | Significant |
| SCC | 1 | 52 | 17 | 35 | – | – | 3.822 (1.084–13.470) | 0.037 | Significant |
1, the clinical data of each histological subtypes of NSCLC was only reported in (13,18,19). AC, adenocarcinoma; CI, confidence interval; HR, hazard ratio; NE, negative expression; NSCLC, non-small cell lung cancer; Oct-4, octamer-binding transcription factor 4; OS, overall survival; PE, positive expression; SCC, squamous cell carcinoma.
Figure 4.
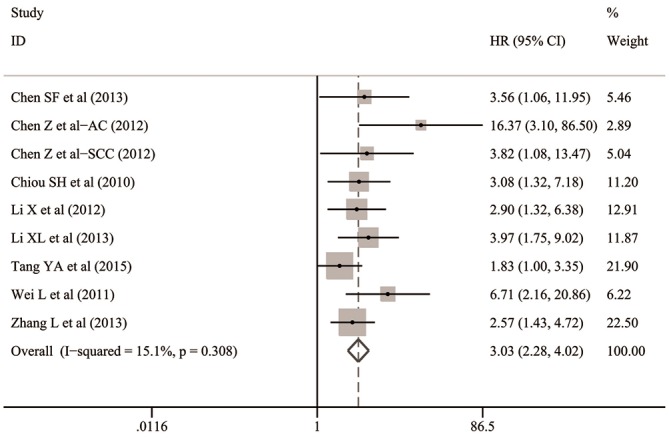
Overall analysis for the prognostic value of Oct-4 expression for OS in patients with NSCLC. AC, adenocarcinoma; CI, confidence interval; HR, hazard ratio; NSCLC, non-small cell lung cancer; Oct-4, octamer-binding transcription factor 4; OS, overall survival; SCC, squamous cell carcinoma.
To further investigate the prognostic roles of Oct-4 in detail, we classified the eight relevant studies into several subgroups according to the statistical analysis, patients’ origins, positive-staining sites and histological subtypes of NSCLC (Table 2).
In the subgroups stratified by statistical analysis, both the summarized HR with 95% CI pooling two studies for multivariate analysis (HR: 2.175; 95% CI: 1.423–3.326; P<0.001; I2=0.0%, P=0.431) (17,40) and six studies for univariate analysis (HR: 3.948; 95% CI: 2.701–5.770; P<0.001; I2=0.0%, P=0.597) (13,15,16,18,19,37) indicated that Oct-4 expression could independently predict the worse OS in patients with NSCLC (Table 2 and Figure 5A).
Figure 5.
Subgroup analysis for the prognostic value of Oct-4 expression in patients with NSCLC stratified by (A) statistical analysis, (B) the origins of patients, (C) positive-staining sites and (D) histological subtypes. AC, adenocarcinoma; CI, confidence interval; HR, hazard ratio; NSCLC, non-small cell lung cancer; Oct-4, octamer-binding transcription factor 4; SCC, squamous cell carcinoma.
In the subgroups stratified by the origins of patients, the pooled HR was 3.564 (95% CI: 2.484–5.113; P<0.001; I2=15.3%, P=0.316) for 1,092 NSCLC cases from China mainland in five studies (13,15,16,37,40). And the pooled HR was 2.340 (95% CI: 1.484–3.690; P<0.001; I2=0.0%, P=0.471) for the remaining 271 NSCLC cases from Taiwan in three studies (17-19). A significant correlation between Oct-4 expression and poor OS of NSCLC was thus revealed in both China mainland group and Taiwan group (Table 2 and Figure 5B).
In the subgroups stratified by positive-staining sites, only the survival data for nuclear expression of Oct-4 was available from four included studies (13,15,16,37). Their pooled HR suggested that nuclear Oct-4 expression could significantly predict the worse OS of NSCLC (HR: 4.301; 95% CI: 2.733–6.767; P<0.001; I2=2.4%, P=0.393) (Table 2 and Figure 5C). A subgroup analysis for non-nuclear Oct-4 expression was given up because of the scarcity of extractable data about Oct-4 staining sites from the other four studies (17-19,40).
In the subgroups stratified by histological subtypes, three studies reported available survival data of 196 AC cases (ratio =14.4%) (13,18,19) and one of them also provided the survival data of 52 SCC cases (ratio =4.0%) (13). The remaining studies enrolled a total of 1,112 NSCLC cases (ratio =81.6%) but analyzed them as a whole. The survival outcomes of either AC or SCC was not available from these studies (15-17,37,40). Finally, both the pooled estimates of AC group (HR: 4.108; 95% CI: 2.166–7.790; P<0.001; I2=36.4%, P=0.208) and SCC group (HR: 3.822; 95% CI: 1.084–13.470; P=0.037) indicated the predictive value of Oct-4 expression for poor OS in patients with NSCLC (Table 2 and Figure 5D).
Sensitivity analysis
Sensitivity analysis was conducted to evaluate the stability of the pooled HR and the derived plot was shown as Figure 6. By visual inspection, none of the individual HRs was out of the estimated ranges, suggesting that no substantial variation would be revealed between the adjusted summarized HR and primary summarized HR if omitting each study sequentially. Therefore, the strong robustness of the prognostic roles of Oct-4 was confirmed.
Figure 6.
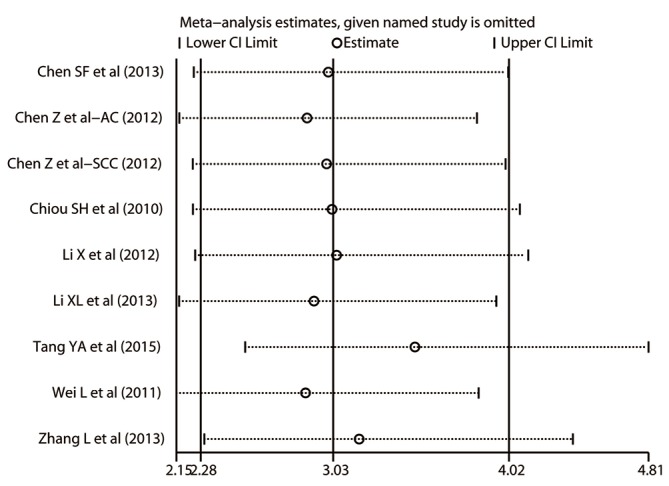
Sensitivity analysis for the correlation between Oct-4 expression and the prognosis in patients with NSCLC. CI, confidence interval; NSCLC, non-small cell lung cancer; Oct-4, octamer-binding transcription factor 4.
Publication bias
For the relationship between Oct-4 expression and clinicopathological characteristics of NSCLC, no less than ten studies were included in the pooled analysis of gender (n=10), differentiation degree (n=10) and TNM stage (n=11). No evidence for significant publication bias was detected by either Egger’s test or Begg’s test across these included studies (Table 1). Nine or fewer studies were included into the pooled analyses for other clinicopathological parameters. Publication bias tests were thus no longer necessary.
Only eight included studies were available within the overall analysis for the prognostic roles of Oct-4 expression in NSCLC (13,15-19,37,40). Therefore, we gave up the further publication bias tests because of the scarcity of enough evidences.
Discussion
Oct-4 is encoded by POU5F1 (POU domain, class 5, transcription factor-1) gene in human beings (42). In 1989, Oct-4 was firstly identified in embryonic stem cells and germ cells during mouse embryogenesis (43). Later, Boyer et al. (44) discovered that Oct-4, Sex determine region Y-box 2 (SOX2) and Nanog co-occupy a substantial portion of their target genes which encode some important transcription factors. Many researches have also indicated that Oct-4, SOX2 and Nanog can collaborate to maintain the pluripotency and self-renewal ability of embryonic stem cells (45-47). Therefore, Oct-4 can act as a master switch by interacting with other transcription factors to activate or repress the expression of specific genes in stem cells (13,48).
In recent years, a growing number of evidences have put the CSC theory in the spotlight for cancer prevention and therapy because of its potential to serve as an ideal model for early diagnosis and treatment of various cancers (7). On the basis of current experimental evidences, tumors may be originated from the malignant transformation of somatic stem cells (6). Oct-4 has been found as a CSC marker and extensively studied in a number of cancer cells. Monk et al. (49) even discovered that the Oct-4 gene is only expressed in CSCs but not in normal somatic cells, indicating a crucial role of Oct-4 in oncogenesis. Down-regulation of Oct-4 results in the loss of CSCs and the reduction of possibility of cancer development (8). Therefore, Oct-4 may be a pivotal regulator for the malignancy potential and growth of cancers (50,51).
Most of the recent studies indicated that Oct-4 can significantly affect the tumorigenicity and malignancy of NSCLC (11). Li et al. (15) collected the clinical data of 44 advanced NSCLC cases and compared Oct-4 expression between cancerous tissues and adjacent benign tissues. Finally, positive staining of Oct-4 was only identified in the cancer cells of NSCLC, with the positive ratio of 54.5%. Oct-4 was thus considered as a valuable biomarker to distinguish cancer from noncancerous lesions. However, Oct-4 is only expressed in CSCs. And CSCs belong to a very small subpopulation of NSCLC cells which are highly enriched with the properties of self-renewal, extensive proliferation and malignancy development. The amount of CSCs identified from the enrolled specimens of NSCLC may vary hugely across different investigations. Therefore, the roles of Oct-4 expression in NSCLC have not been well-defined, especially on the perspective of prognosis.
To our knowledge, our study is the first meta-analysis to systematically summarize the prognostic value of Oct-4 expression in NSCLC and its relationship to some common clinicopathological characteristics. By applying the evidence-based method to a large number of enrolled samples, we found that positive Oct-4 expression was significantly associated with the unfavorable conditions on differentiation degree, TNM stage and lymphatic metastasis. In addition, no significant relationship was revealed between Oct-4 expression and the gender, age, smoking status and histological subtypes of NSCLC. Moreover, Oct-4 expression could independently predict the poorer OS in NSCLC patients. Further analyses indicated that the prognostic value of Oct-4 still remained prominent in the subgroups stratified by statistical analysis methods, origins of patients and histological subtypes, and in nuclear staining cases.
When pooling relevant studies together to address the prognostic roles of Oct-4, we got an initial impression indicating that the current evidences commonly indicated that Oct-4 expression could significantly predict the poor prognosis of NSCLC (13,15-19,37,40). In addition, the correlation between Oct-4 expression and the negative prognosis of NSCLC was not substantially altered by any other endpoint event and Oct-4 staining method (30-32). A prognostic indicator is considered to be of high predictive value when its HR is larger than 2 (52). The overall pooled HR in our meta-analysis was 3.030 (95% CI: 2.283–4.021; P<0.001), suggesting that Oct-4 could serve as a strong biomarker to predict the poor prognosis of NSCLC.
However, the validity of our summarized outcomes might be more or less attenuated by the following two bias risks from the statistical sources.
On the one hand, the majority of incorporative data from included studies was analyzed by univariate analysis (13,15,16,18,19,37). Only two included studies reported the multivariate statistics to evaluate the prognostic significance of Oct-4 expression in NSCLC (17,40). Multivariate analysis using logistic regression or Cox proportional hazards model is an effective method to eliminate the bias risks from other confounding factors in observational studies. Given such concerns, the accuracy of pooled HR with 95% CI might be slightly attenuated because some insufficiently eliminated confounding factors, such as differentiation degree, TNM stage, lymphatic metastasis and other biomarkers, could also affect the prognosis of NSCLC and interfered the identification of the actual roles of Oct-4. Despite a strong linkage between Oct-4 expression and the poor OS of NSCLC was identified from our meta-analysis, this finding need further affirmations and modifications in the future studies without any bias risk originated from other confounding factors.
On the other hand, the studies reporting beneficial intervention effects or a larger effect size are generally considered to have more opportunities to be published, while an equal amount of data leading to the other direction may remain unpublished (24). Therefore, potential publication bias will be an unavoidable issue to be addressed in a meta-analysis. However, fewer than ten included studies may cause some adverse effects on the efficacy of current publication bias tests (20). Therefore, publication bias tests are not really encouraged when integrating few eligible evidences due to the loss of practical significance. In our meta-analysis, all of the eight available studies indicated that Oct-4 expression was significantly associated with the lower OS in patients with NSCLC (13,15-19,37,40). We suspected that potential publication bias might exist between these studies, although no evidence could be validly detected for the moment.
For further assessments, the correlation between Oct-4 expression and the negative OS of NSCLC remained prominent in the subgroups stratified by statistical analysis, patients’ origins and histological subtypes, and in nuclear staining cases. However, we recognized that the following three issues from our meta-analysis should be seriously considered in clinical practice.
Firstly, we gave up a further subgroup analysis for cytoplasmic or membranous staining cases because their details were not reported in the included studies (17-19,40). Oct-4 gene can generate three isoforms, including Oct-4A, Oct-4B, and Oct-4B1, by alternative splicing and alternative translation initiation (53). In general, Oct-4A is primarily located in the nucleus, while Oct-4B and Oct-4B1 are both expressed in the cytoplasm. However, only Oct-4A has been proved to regulate the pluripotency of stem cells (30,53). These findings may explain the scarcity of clinical data for non-nuclear staining cases from previous reports.
Secondly, all the included studies were performed in Chinese populations. During the literature retrieval, we only found one study reported by Cortes-Dericks et al. (30) evaluating the prognostic roles of Oct-4 in 64 Italian patients with AC. Their multivariate data indicated that Oct-4 could be used as an independent predictor for the poor disease-free survival of AC (P=0.047) (30). However, we finally excluded this survival data because quantitative reverse transcriptase-polymerase chain reaction was applied to detect the Oct-4 expression in AC cases. We have imposed a strict eligibility criterion to ensure the great homogeneity of included studies, including the IHC application and key endpoint. Therefore, the validity of our findings should be judiciously evaluated in the clinical settings of other nations.
Finally, it should be noted that only three studies for patients with AC (13,18,19) and one study for patients with SCC (13) were available for the subgroup analysis stratified by histological subtypes of NSCLC. Most of the included studies just analyzed all the enrolled NSCLC cases together instead of classifying them according to different histological subtypes and performing a survival analysis, respectively (15-17,37,40). Considering that only 192 AC cases and 52 SCC cases were enrolled in the subgroups, insufficient sample size might result in some potential biases to the integrated results, although the prognostic roles of Oct-4 were significantly revealed in both AC and SCC according to the current evidences. Therefore, more large-scale studies analyzing the Oct-4 expression in each subtype of NSCLC are urgently required to resolve this issue.
In addition, we also clarified the relationship between Oct-4 expression and some major clinicopathological characteristics of NSCLC by synthesizing the 13 included studies quantitatively (13-17,33-36,38-41). The pooled analyses indicated that positive Oct-4 expression was significantly associated with the unfavorable outcomes for differentiation degree, TNM stage and lymphatic metastasis, which were widely accepted as important factors leading to the poor prognosis of NSCLC. However, no significant difference in Oct-4 expression was observed between AC cases and SCC cases. Both AC and SCC are the most frequent and aggressive histological subtypes of NSCLC (1). Previous laboratorial evidences have also suggested that CSCs are involved in the oncogenesis and maintenance of both AC and SCC (15). According to the pooled estimates of our meta-analysis, Oct-4 expression showed no significant tendency towards either AC or SCC. However, as Chen et al. (13) suggested, there are some substantial differences in the pathogenesis of AC and SCC. Further studies evaluating the expression of Oct-4 in the endothelial precursors and original sites of AC and SCC are needed to confirm this finding in the future.
Limitations
Several limitations in this meta-analysis should be acknowledged. First, the statistics incorporated into quantitative synthesis were mainly derived from univariate analysis instead of multivariate analysis. The insufficiently eliminated confounding factors might decrease the accuracy of integrated results in the clinical practices. Second, the cut-off definitions for positive staining of Oct-4 varied across different studies and might interfere the actual roles of Oct-4 expression in NSCLC. Third, the validity of publication bias tests was largely attenuated because of the scarcity of enough studies included into the present meta-analysis. Four, all of the included studies were performed in Chinese populations. Oncologists should evaluated our discoveries judiciously in the clinical settings of their own countries. Five, although no language restriction was imposed, many additional reports might be identified from more native electronic databases in other languages and further enrich our meta-analysis.
Conclusions
In conclusion, our meta-analysis suggests that Oct-4 expression is significantly associated with the unfavorable outcomes for differentiation degree, TNM stage and lymphatic metastasis in patients with NSCLC. In addition, Oct-4 can also serve as a strong biomarker predicting the poor prognosis of NSCLC. Some controversies are still not well-resolved in this meta-analysis. More high-quality studies based on a large sample size will be very helpful to further validate and modify our discoveries in the future.
Acknowledgements
We thank the assistance of the staff in Department of Thoracic Surgery and Lung Cancer Center, West China Hospital, Sichuan University.
Funding: This study was supported by the Foundation of Science and Technology support plan Department of Sichuan Province (2014SZ0148 and 2015SZ0158).
Table S1. PRISMA 2009 checklist.
| Section/topic | # | Checklist item | Reported on page # |
|---|---|---|---|
| Title | |||
| Title | 1 | Identify the report as a systematic review, meta-analysis, or both | 1 |
| Abstract | |||
| Structured summary | 2 | Provide a structured summary including, as applicable: background; objectives; data sources; study eligibility criteria, participants, and interventions; study appraisal and synthesis methods; results; limitations; conclusions and implications of key findings; systematic review registration number | 2 |
| Introduction | |||
| Rationale | 3 | Describe the rationale for the review in the context of what is already known | 2 |
| Objectives | 4 | Provide an explicit statement of questions being addressed with reference to participants, interventions, comparisons, outcomes, and study design (PICOS) | 2 |
| Methods | |||
| Protocol and registration | 5 | Indicate if a review protocol exists, if and where it can be accessed (e.g., Web address), and, if available, provide registration information including registration number. | 2, 3 |
| Eligibility criteria | 6 | Specify study characteristics (e.g., PICOS, length of follow-up) and report characteristics (e.g., years considered, language, publication status) used as criteria for eligibility, giving rationale | 6, 7 |
| Information sources | 7 | Describe all information sources (e.g., databases with dates of coverage, contact with study authors to identify additional studies) in the search and date last searched | 2 |
| Search | 8 | Present full electronic search strategy for at least one database, including any limits used, such that it could be repeated | 2, Table S2 |
| Study selection | 9 | State the process for selecting studies (i.e., screening, eligibility, included in systematic review, and, if applicable, included in the meta-analysis) | 2–4 (Figure 1) |
| Data collection process | 10 | Describe method of data extraction from reports (e.g., piloted forms, independently, in duplicate) and any processes for obtaining and confirming data from investigators | 3 |
| Data items | 11 | List and define all variables for which data were sought (e.g., PICOS, funding sources) and any assumptions and simplifications made | 3 |
| Risk of bias in individual studies | 12 | Describe methods used for assessing risk of bias of individual studies (including specification of whether this was done at the study or outcome level), and how this information is to be used in any data synthesis | 3 |
| Summary measures | 13 | State the principal summary measures (e.g., risk ratio, difference in means). | 8, 9 |
| Synthesis of results | 14 | Describe the methods of handling data and combining results of studies, if done, including measures of consistency (e.g., I2) for each meta-analysis | 3, 4 |
| Risk of bias across studies | 15 | Specify any assessment of risk of bias that may affect the cumulative evidence (e.g., publication bias, selective reporting within studies) | 4 |
| Additional analyses | 16 | Describe methods of additional analyses (e.g., sensitivity or subgroup analyses, meta-regression), if done, indicating which were pre-specified | 4 |
| Results | |||
| Study selection | 17 | Give numbers of studies screened, assessed for eligibility, and included in the review, with reasons for exclusions at each stage, ideally with a flow diagram | 4, 5 (Figure 1) |
| Study characteristics | 18 | For each study, present characteristics for which data were extracted (e.g., study size, PICOS, follow-up period) and provide the citations | 5, Table S4 |
| Risk of bias within studies | 19 | Present data on risk of bias of each study and, if available, any outcome level assessment (see item 12) | 5, Table S3 |
| Results of individual studies | 20 | For all outcomes considered (benefits or harms), present, for each study: (I) simple summary data for each intervention group (II) effect estimates and confidence intervals, ideally with a forest plot | 5–8 (Figures 2,3,4,5) |
| Synthesis of results | 21 | Present results of each meta-analysis done, including confidence intervals and measures of consistency | 5–8 |
| Risk of bias across studies | 22 | Present results of any assessment of risk of bias across studies (see Item 15) | 6, 8 |
| Additional analysis | 23 | Give results of additional analyses, if done [e.g., sensitivity or subgroup analyses, meta-regression (see item 16)] | 7–9 (Figures 5,6) |
| Discussion | |||
| Summary of evidence | 24 | Summarize the main findings including the strength of evidence for each main outcome; consider their relevance to key groups (e.g., healthcare providers, users, and policy makers) | 9–11 |
| Limitations | 25 | Discuss limitations at study and outcome level (e.g., risk of bias), and at review-level (e.g., incomplete retrieval of identified research, reporting bias) | 11 |
| Conclusions | 26 | Provide a general interpretation of the results in the context of other evidence, and implications for future research | 11, 12 |
| Funding | |||
| Funding | 27 | Describe sources of funding for the systematic review and other support (e.g., supply of data); role of funders for the systematic review | 12 |
From: Moher D et al. (22). For more information, visit: www.prisma-statement.org.
Table S2. Summary of the electronic literature retrieval*.
| Searches | Search details | Items found |
|---|---|---|
| PubMed search strategy | ||
| #1 | Search ((((“lung neoplasms”[MeSH Terms] OR (“lung”[All Fields] AND “neoplasms”[All Fields]) OR “lung neoplasms”[All Fields] OR (“lung”[All Fields] AND “cancer”[All Fields]) OR “lung cancer”[All Fields]) OR ((“lung”[MeSH Terms] OR “lung”[All Fields]) AND (“carcinoma”[MeSH Terms] OR “carcinoma”[All Fields]))) OR (“lung neoplasms”[MeSH Terms] OR (“lung”[All Fields] AND “neoplasms”[All Fields]) OR “lung neoplasms”[All Fields] OR (“lung”[All Fields] AND “neoplasm”[All Fields]) OR “lung neoplasm”[All Fields])) OR (“lung neoplasms”[MeSH Terms] OR (“lung”[All Fields] AND “neoplasms”[All Fields]) OR “lung neoplasms”[All Fields] OR (“lung”[All Fields] AND “tumor”[All Fields]) OR “lung tumor”[All Fields])) AND (“octamer transcription factor-3”[MeSH Terms] OR (“octamer”[All Fields] AND “transcription”[All Fields] AND “factor-3”[All Fields]) OR “octamer transcription factor-3”[All Fields] OR “oct 4”[All Fields]) | 101 |
| #2 | Search ((((“lung neoplasms”[MeSH Terms] OR (“lung”[All Fields] AND “neoplasms”[All Fields]) OR “lung neoplasms”[All Fields] OR (“lung”[All Fields] AND “cancer”[All Fields]) OR “lung cancer”[All Fields]) OR ((“lung”[MeSH Terms] OR “lung”[All Fields]) AND (“carcinoma”[MeSH Terms] OR “carcinoma”[All Fields]))) OR (“lung neoplasms”[MeSH Terms] OR (“lung”[All Fields] AND “neoplasms”[All Fields]) OR “lung neoplasms”[All Fields] OR (“lung”[All Fields] AND “neoplasm”[All Fields]) OR “lung neoplasm”[All Fields])) OR (“lung neoplasms”[MeSH Terms] OR (“lung”[All Fields] AND “neoplasms”[All Fields]) OR “lung neoplasms”[All Fields] OR (“lung”[All Fields] AND “tumor”[All Fields]) OR “lung tumor”[All Fields])) AND (octamer-binding[All Fields] AND (“transcription factors”[MeSH Terms] OR (“transcription”[All Fields] AND “factors”[All Fields]) OR “transcription factors”[All Fields] OR (“transcription”[All Fields] AND “factor”[All Fields]) OR “transcription factor”[All Fields]) AND 4[All Fields]) | 12 |
| EMBASE (via Ovid interface) search strategy | ||
| #1 | Search ((lung cancer or lung carcinoma or lung neoplasm or lung tumor) and Oct-4).af. | 55 |
| #2 | Search ((lung cancer or lung carcinoma or lung neoplasm or lung tumor) and octamer-binding transcription factor 4).af. | 11 |
| The Web of Science (via campus network of Sichuan University) search strategy | ||
| #1 | Search TS=((lung cancer OR lung carcinoma OR lung neoplasm OR lung tumor) AND Oct-4) | 180 |
| #2 | Search TS=((lung cancer OR lung carcinoma OR lung neoplasm OR lung tumor) AND octamer-binding transcription factor 4) | 27 |
*, the complete details of search strings in CNKI database are not given because of the Chinese words used during the literature retrieval.
Table S3. NOS scoring records of the included studies.
| Authors [year] (Ref.) | Selection | Comparability | Exposure | Total score | ||
|---|---|---|---|---|---|---|
| Assessment of outcome | follow-up long enough for outcomes | Adequacy of follow-up of cohorts | ||||
| Chen et al. [2013] (18) | 4 | 1 | 1 | 1 | 1 | 8 |
| Chen et al. [2012] (13) | 4 | 1 | 1 | 1 | 1 | 8 |
| Chiou et al. [2010] (19) | 4 | 1 | 0 | 1 | 1 | 7 |
| Dong et al. [2013] (33) | 4 | 1 | 1 | 0 | 1 | 7 |
| Li et al. [2016] (14) | 4 | 1 | 1 | 0 | 1 | 7 |
| Li et al. [2012] (15) | 4 | 1 | 1 | 0 | 1 | 7 |
| Li et al. [2013] (16) | 4 | 1 | 1 | 1 | 1 | 8 |
| Ni et al. [2012] (34) | 4 | 1 | 1 | 0 | 1 | 7 |
| Qiao et al. [2012] (35) | 4 | 1 | 1 | 0 | 1 | 7 |
| Sun et al. [2012] (36) | 4 | 1 | 0 | 0 | 1 | 6 |
| Tang et al. [2015] (17) | 4 | 2 | 1 | 1 | 1 | 9 |
| Wei et al. [2011] (37) | 4 | 1 | 1 | 1 | 1 | 8 |
| Xiang et al. [2015] (38) | 4 | 1 | 1 | 0 | 1 | 7 |
| Yang et al. [2014] (39) | 4 | 1 | 1 | 0 | 1 | 7 |
| Zhang et al. [2013] (40) | 4 | 2 | 1 | 1 | 1 | 9 |
| Zhou et al. [2011] (41) | 4 | 1 | 0 | 0 | 1 | 6 |
NOS, Newcastle-Ottawa Scale.
Table S4. Baseline characteristics of included studies.
| Authors [year] (Ref.) | Language | Origin | Study design | Study period | NOS | Categories | No. samples | Mean age (years) | Histological subtypes | Stages | Material | Detection | Positive site | Cut-off value | Outcomes | Extractions | Analysis | Endpoint | Follow-up (months) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CP features | Prognosis | Total | PE | NE | AC | SCC | Others | ||||||||||||||||||
| Chen et al. [2013] (18) | English | Taiwan | ROS | 2004–2009 | 8 | × | √ | 20 | 18 | 2 | 70.4 | 20 | – | – | III–IV | Paraffin embedded tissue | IHC | NI | 10% staining | HR | Reported | U | OS | 60 | |
| Chen et al. [2012] (13) | English | China mainland | ROS | 2003–2004 | 8 | √ | √ | 113 | 57 | 56 | 57.2 | 58 | 52 | 3 | I–IV | Paraffin embedded tissue | IHC | Nucleus | Median histoscore | OR, HR | DDE | U | OS | 60 | |
| Chiou et al. [2010] (19) | English | Taiwan | ROS | NI | 7 | × | √ | 118 | 78 | 40 | 58.8 | 118 | – | – | I–IV | Paraffin embedded tissue | IHC | NI | NI | HR | DDE | U | OS | 80 | |
| Dong et al. [2013] (33) | Chinese | China mainland | ROS | 2009–2012 | 7 | √ | × | 60 | 37 | 23 | 68.0 | 17 | 43 | – | I–IV | Paraffin embedded tissue | IHC | Nucleus | 25% staining | OR | DDE | U | – | – | |
| Li et al. [2016] (14] | English | China mainland | ROS | 2013–2014 | 7 | √ | × | 65 | 38 | 27 | 53.0 | NI | NI | NI | I–IV | Paraffin embedded tissue | IHC | Membrane | 25% staining | OR | DDE | U | – | – | |
| Li et al. [2012] (15) | English | China mainland | ROS | 2008–2011 | 7 | √ | √ | 44 | 24 | 20 | 62.6 | 23 | 21 | – | I–IV | Paraffin embedded tissue | IHC | Nucleus | 10% staining | OR, HR | DDE | U | OS | 12 | |
| Li et al. [2013] (16) | English | China mainland | ROS | 2004–2007 | 8 | √ | √ | 102 | 29 | 73 | NI | 60 | 42 | – | I–IV | Paraffin embedded tissue | IHC | Nucleus | 25% staining | OR, HR | DDE | U | OS | 60 | |
| Ni et al. [2012] (34) | Chinese | China mainland | ROS | 2007–2009 | 7 | √ | × | 50 | 21 | 29 | 53.0 | 18 | 29 | 3 | I–IV | Paraffin embedded tissue | IHC | Nucleus | 20% staining | OR | DDE | U | – | – | |
| Qiao et al. [2012] (35) | Chinese | China mainland | ROS | 2003–2009 | 7 | √ | × | 40 | 15 | 25 | NI | 19 | 21 | – | I–III | Paraffin embedded tissue | IHC | Nucleus | 5% staining | OR | DDE | U | – | – | |
| Sun et al. [2012] (36) | Chinese | China mainland | ROS | 2008–2010 | 6 | √ | × | 186 | 87 | 99 | 55.0 | 81 | 105 | – | I–IV | Paraffin embedded tissue | IHC | Cytoplasm | NI | OR | DDE | U | – | – | |
| Tang et al. [2015] (17) | English | Taiwan | ROS | NI | 9 | √ | √ | 133 | 92 | 41 | NI | 97 | 22 | 14 | I–IV | Paraffin embedded tissue | IHC | NI | 50% staining | OR, HR | DDE, reported | U & M | OS | 120 | |
| Wei et al. [2011] (37) | Chinese | China mainland | ROS | 2002–2005 | 8 | × | √ | 51 | 27 | 24 | 55.0 | NI | NI | NI | I–IV | Paraffin embedded tissue | IHC | Nucleus | 0% staining | HR | DDE | U | OS | 98 | |
| Xiang et al. [2015] (38) | Chinese | China mainland | ROS | 2010–2013 | 7 | √ | × | 60 | 43 | 17 | 53.9 | 20 | 30 | 10 | I–IV | Paraffin embedded tissue | IHC | Nucleus | 50% staining | OR | DDE | U | – | – | |
| Yang et al. [2014] (39) | Chinese | China mainland | ROS | 2008–2013 | 7 | √ | × | 60 | 36 | 24 | 61.0 | 23 | 37 | – | I–IV | Paraffin embedded tissue | IHC | NI | 10% staining | OR | DDE | U | – | – | |
| Zhang et al. [2013] (40) | Chinese | China mainland | ROS | 2005–2006 | 9 | √ | √ | 126 | 52 | 74 | NI | NI | NI | NI | I–IV | Paraffin embedded tissue | IHC | NI | 1% staining | OR, HR | DDE, reported | U & M | OS | 90 | |
| Zhou et al. [2011] (41) | Chinese | China mainland | ROS | 2000–2010 | 6 | √ | × | 135 | 44 | 91 | 60.0 | 70 | 49 | 16 | I–IV | Paraffin embedded tissue | IHC | Nucleus | NI | OR | DDE | U | – | – | |
AC, adenocarcinoma; CP, clinicopathological; DDE, demographic data extrapolated; HR, hazard ratio; IHC, immunohistochemistry; M, multivariate; NE, negative expression; NI, no information; OR, odds ratio; OS, overall survival; PE, positive expression; ROS, retrospective observational study; U, univariate.
Ethical Statement: A systematic review and meta-analysis does not require necessary patients’ consent or ethical approval.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet 2000;355:479-85. 10.1016/S0140-6736(00)82038-3 [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917. 10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 4.Baltayiannis N, Chandrinos M, Anagnostopoulos D, et al. Lung cancer surgery: an up to date. J Thorac Dis 2013;5 Suppl 4:S425-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai JR, Liu PL, Chen YH, et al. Ginkgo biloba extract decreases non-small cell lung cancer cell migration by downregulating metastasis-associated factor heat-shock protein 27. PLoS One 2014;9:e91331. 10.1371/journal.pone.0091331 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature 2001;414:105-11. 10.1038/35102167 [DOI] [PubMed] [Google Scholar]
- 7.Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol 2007;18:460-6. 10.1016/j.copbio.2007.10.007 [DOI] [PubMed] [Google Scholar]
- 8.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 2000;24:372-6. 10.1038/74199 [DOI] [PubMed] [Google Scholar]
- 9.Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol 2002;12:432-8. 10.1016/S0962-8924(02)02352-8 [DOI] [PubMed] [Google Scholar]
- 10.Hay DC, Sutherland L, Clark J, et al. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells 2004;22:225-35. 10.1634/stemcells.22-2-225 [DOI] [PubMed] [Google Scholar]
- 11.Chen YC, Hsu HS, Chen YW, et al. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One 2008;3:e2637. 10.1371/journal.pone.0002637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong B, Lin Y, Lai Y, et al. Relationship of Oct-4 to malignant stage: a meta-analysis based on 502 positive/high Oct-4 cases and 522 negative/low case-free controls. Oncotarget 2016;7:2143-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Wang T, Cai L, et al. Clinicopathological significance of non-small cell lung cancer with high prevalence of Oct-4 tumor cells. J Exp Clin Cancer Res 2012;31:10. 10.1186/1756-9966-31-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li LI, Lv Y, Zhang Y, et al. Expression and clinical significance of Oct-4 and E-cad in non-small-cell lung cancer. Oncol Lett 2016;11:234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Wang J, Xu Z, et al. Expression of Sox2 and Oct4 and their clinical significance in human non-small-cell lung cancer. Int J Mol Sci 2012;13:7663-75. 10.3390/ijms13067663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li XL, Jia LL, Shi MM, et al. Downregulation of KPNA2 in non-small-cell lung cancer is associated with Oct4 expression. J Transl Med 2013;11:232. 10.1186/1479-5876-11-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang YA, Chen CH, Sun HS, et al. Global Oct4 target gene analysis reveals novel downstream PTEN and TNC genes required for drug-resistance and metastasis in lung cancer. Nucleic Acids Res 2015;43:1593-608. 10.1093/nar/gkv024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen SF, Lin YS, Jao SW, et al. Pulmonary Adenocarcinoma in Malignant Pleural Effusion Enriches Cancer Stem Cell Properties during Metastatic Cascade. PLoS One 2013;8:e54659. 10.1371/journal.pone.0054659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiou SH, Wang ML, Chou YT, et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res 2010;70:10433-44. 10.1158/0008-5472.CAN-10-2638 [DOI] [PubMed] [Google Scholar]
- 20.Li SJ, Chen DL, Zhang WB, et al. Prognostic value of stromal decorin expression in patients with breast cancer: a meta-analysis. J Thorac Dis 2015;7:1939-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li SJ, Fan J, Zhou J, et al. Diabetes Mellitus and Risk of Bronchopleural Fistula After Pulmonary Resections: A Meta-Analysis. Ann Thorac Surg 2016;102:328-39. 10.1016/j.athoracsur.2016.01.013 [DOI] [PubMed] [Google Scholar]
- 22.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 2009;339:b2535. 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603-5. 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 24.Higgins JP, Green S. editors. Cochrane Handbook for Systematic Reviews of Interventions. Version 5.1.0 [updated March 2011]. The Cochrane Collaboration, 2011. Available online: www.cochrane-handbook.org
- 25.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998;17:2815-34. [DOI] [PubMed] [Google Scholar]
- 26.Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into metaanalysis. Trials 2007;8:16. 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539-58. 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 28.Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557-60. 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50:1088-101. 10.2307/2533446 [DOI] [PubMed] [Google Scholar]
- 30.Cortes-Dericks L, Galetta D, Spaggiari L, et al. High expression of octamer-binding transcription factor 4A, prominin-1 and aldehyde dehydrogenase strongly indicates involvement in the initiation of lung adenocarcinoma resulting in shorter disease-free intervals. Eur J Cardiothorac Surg 2012;41:e173-81. 10.1093/ejcts/ezs170 [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Han B, Huang J, et al. Prognostic significance of OCT4 expression in adenocarcinoma of the lung. Jpn J Clin Oncol 2010;40:961-6. 10.1093/jjco/hyq066 [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Wang H, Jin B, et al. Correlations between OCT4 expression and clinicopathological factors and prognosis of patients with lung adenocarcinoma. Zhongguo Fei Ai Za Zhi 2013;16:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong C, Tu J, Tao L, et al. OCT4 and miRNA-155 expression pattern in non-small cell lung cancer tissues: relationship with clinicopathological characteristics. Cancer Res Prev Treat 2013;40:776-80. [Google Scholar]
- 34.Ni J, Yang L, Zhu X, et al. Expression and significance of OCT4, CD133 in non-small cell lung cancer. J Clin Med Pract 2012;16:19-22. [Google Scholar]
- 35.Qiao Z, Ma Y, Dang C, et al. Expression of OCT-4 in lung cancer and its clinical significance. Mod Oncol 2012;20:299-302. [Google Scholar]
- 36.Sun X, Zong X, Wang D. Study on significance of expression of Nanog and Oct4 by using tissue chip technique in non-small cell lung cancer. J Clin Exp Med 2012;16:1258-60. [Google Scholar]
- 37.Wei L, Liu X, Hu C. Correlation between expression of HIF-2α and OCT-4 and prognosis of NSCLC. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2011;36:854-8. [DOI] [PubMed] [Google Scholar]
- 38.Xiang B, Li L, Cao L, et al. Expression and clinical significance of stem cell transcription factors Oct4 and Sox2 in lung carcinoma. China Med Herald 2015;12:74-7. [Google Scholar]
- 39.Yang Y, Yao H, Bai J, et al. Expression and clinical significance of Octamer 4 in non-small cell lung cancer. Chinese Remed Clin 2014;14:1332-5. [Google Scholar]
- 40.Zhang L, Lin Y. Clinical significance of the expression of Oct-4 gene in lung cancer stem cells. China Med Herald 2013;10:22-5. [Google Scholar]
- 41.Zhou C, Liao D. Expression and significance of Oct-4 in non-small cell lung cancer. J Qiqihar Univ Med 2011;32:1204-5. [Google Scholar]
- 42.Cauffman G, Liebaers I, Van Steirteghem A, et al. POU5F1 isoforms show different expression patterns in human embryonic stem cells and preimplantation embryos. Stem Cells 2006;24:2685-91. 10.1634/stemcells.2005-0611 [DOI] [PubMed] [Google Scholar]
- 43.Schöler HR, Hatzopoulos AK, Balling R, et al. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J 1989;8:2543-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005;122:947-56. 10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chambers I, Colby D, Robertson M, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003;113:643-55. 10.1016/S0092-8674(03)00392-1 [DOI] [PubMed] [Google Scholar]
- 46.Avilion AA, Nicolis SK, Pevny LH, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 2003;17:126-40. 10.1101/gad.224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matin MM, Walsh JR, Gokhale PJ, et al. Specific knockdown of Oct4 and beta2-microglobulin expression by RNA interference in human embryonic stem cells and embryonic carcinoma cells. Stem Cells 2004;22:659-68. 10.1634/stemcells.22-5-659 [DOI] [PubMed] [Google Scholar]
- 48.Matoba R, Niwa H, Masui S, et al. Dissecting Oct3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS One 2006;1:e26. 10.1371/journal.pone.0000026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monk M, Holding C. Human embryonic genes re-expressed in cancer cells. Oncogene 2001;20:8085-91. 10.1038/sj.onc.1205088 [DOI] [PubMed] [Google Scholar]
- 50.Hu T, Liu S, Breiter DR, et al. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer Res 2008;68:6533-40. 10.1158/0008-5472.CAN-07-6642 [DOI] [PubMed] [Google Scholar]
- 51.Hu J, Qin K, Zhang Y, et al. Downregulation of transcription factor Oct4 induces an epithelial-to-mesenchymal transition via enhancement of Ca2+ influx in breast cancer cells. Biochem Biophys Res Commun 2011;411:786-91. 10.1016/j.bbrc.2011.07.025 [DOI] [PubMed] [Google Scholar]
- 52.Pan F, Mao H, Deng L, et al. Prognostic and clinicopathological significance of microRNA-21 overexpression in breast cancer: a meta-analysis. Int J Clin Exp Pathol 2014;7:5622-33. [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Dai J. Concise review: isoforms of OCT4 contribute to the confusing diversity in stem cell biology. Stem Cells 2010;28:885-93. [DOI] [PMC free article] [PubMed] [Google Scholar]