Abstract
In their search to understand the evolution of biological complexity, John Maynard Smith and Eörs Szathmáry put forward the notion of major evolutionary transitions as those in which elementary units get together to generate something new, larger and more complex. The origins of chromosomes, eukaryotic cells, multicellular organisms, colonies and, more recently, language and technological societies are examples that clearly illustrate this notion. However, a transition may be considered as anecdotal or as major depending on the specific level of biological organization under study. In this contribution, I will argue that transitions may also be occurring at a much smaller scale of biological organization: the viral world. Not only that, but also that we can observe in real time how these major transitions take place during experimental evolution. I will review the outcome of recent evolution experiments with viruses that illustrate four major evolutionary transitions: (i) the origin of a new virus that infects an otherwise inaccessible host and completely changes the way it interacts with the host regulatory and metabolic networks, (ii) the incorporation and loss of genes, (iii) the origin of segmented genomes from a non-segmented one, and (iv) the evolution of cooperative behaviour and cheating between different viruses or strains during co-infection of the same host.
This article is part of the themed issue ‘The major synthetic evolutionary transitions’.
Keywords: emerging viruses, experimental evolution, genome complexity and evolution, virus evolution, virus sociology
1. Introduction
RNA viruses are the most abundant parasites infecting humans, wildlife, livestock and crops. Despite tremendous efforts, the number of eradicated viruses is quite limited and the perspectives for future eradications are counterbalanced by the emergence of new viruses [1]. The fact that few viruses can be effectively controlled and the pervasive emergence of new ones are probably consequences of viruses' remarkable ability to evolve. As a consequence of their error-prone replication, RNA viruses exist as complex mutant swarms [2]. Indeed, owing to mutational coupling among genotypes, complex mutant swarms and not individual genomes become the target of selection. This high genetic variability combined with very short generation times and large population sizes bestow a tremendous evolutionary potential (e.g. evolvability) on viral populations. Major events in RNA virus, such as their capacity to change host ranges, alter cell tropisms and overcome resistance genes, antiviral responses or drug therapies, have their origin in the repertoire of genetic variants contained in the mutant swarm. Our ability to predict the evolution of viral populations and to design efficient and durable antiviral strategies is quite limited and is being constantly jeopardized by this evolvability.
However, we can take advantage of this tremendous evolutionary potential and use RNA viruses as model systems for experimentally tackling evolutionary questions in a short period of time. Not surprisingly, RNA viruses have become one of the preferred tools in the arsenal of experimental evolutionary biologists [3,4]. RNA viruses have been used to tackle many evolutionary questions (reviewed in [3,4]), most of which will be well described at the scale of microevolutionary processes, i.e. changes in allele frequencies that occur over time within an evolving viral population and that are due to the action of mutation, selection, gene flow and genetic drift. In a few cases, these microevolution processes may result in big genetic and phenotypic changes, as for example the origin of a new virus species or a new genomic organization. Such innovations would be clearly dubbed as evolutionary transitions whether observed for higher organisms. Some other studies have tackled questions that may be better described at the macroevolutionary scale, as for example the evolution of mutualism among viral species that co-infect a common host, although after all, they result from the same fundamental forces than govern microevolution. Again, the rise in complexity in such systems may well be akin to some major evolutionary transition, though not necessarily taken in the strict sense used by Maynard Smith and Szathmáry, which implies the rise of a new level of biological organization by cooperation of simpler units. In this article, I will review some examples of evolution experiments that I believe illustrate evolutionary transitions in the (experimental) viral world. I will start by describing the evolution of a new viral species. Then I will provide an overview of experiments that tackled evolution of novelty in genomic organizations: from the acquisition and loss of genes to the origin of a segmented genome (chromosomes). Finally, I will describe the evolution of cooperation and cheating in viral systems, and a case resulting in a novel, more complex organization.
2. Experimental emergence of a new RNA virus
The definition of a virus species is elusive. According to the International Committee on Taxonomy of Viruses (ICTV), a virus species is ‘a polythetic class of viruses that constitute a replicating lineage and occupy a particular ecological niche’ (www.ictvonline.org). A polythetic class is one whose members share some common properties, although not all of them may share even a single property in common. Putting it in different terms, the members of a virus species are collectively defined by a consensus group of properties. In the case of plant viruses, two ecological factors are key elements of this ‘consensus group of properties': the natural host range that the virus successfully infects in nature and the set of vector species that the virus uses to be efficiently transmitted among hosts. In the example used to illustrate this first evolutionary transition, Tobacco etch virus (TEV), the virus natural host range is limited to plants from the Solanaceae family, including important crops such as tobacco (Nicotiana tabacum), pepper (Capsicum annuum) and tomato (Solanum lycopersicum), and some weed species such as nightshade (Solanum nigrum), soda apple (Solanum aculeatissimum) and jimson weed (Datura stramonium) that can act as reservoirs. The severity of symptoms induced varies widely between species and even between varieties of the same species: from vein clearing, mottling, necrotic etching and plant stunting to less severe symptoms. In a series of evolution experiments [5–11], we have explored the ability of TEV to expand its host range to a new species that is completely outside its natural host range, the brassica Arabidopsis thaliana ecotype Ler-0. Brassicaceae belong to a different order than Solanaceae within the class Magnoliopsida, hence, adaptation of TEV to A. thaliana represents a jump in host species at the taxonomic level of orders. This species jump would be equivalent to the one experienced by the Ebola virus from its reservoir host, bats (order Chiroptera), to humans (order Primates). The Ler-0 ecotype was chosen for this experiment because it allows for systemic movement of TEV, while other ecotypes do not do so [12,13].
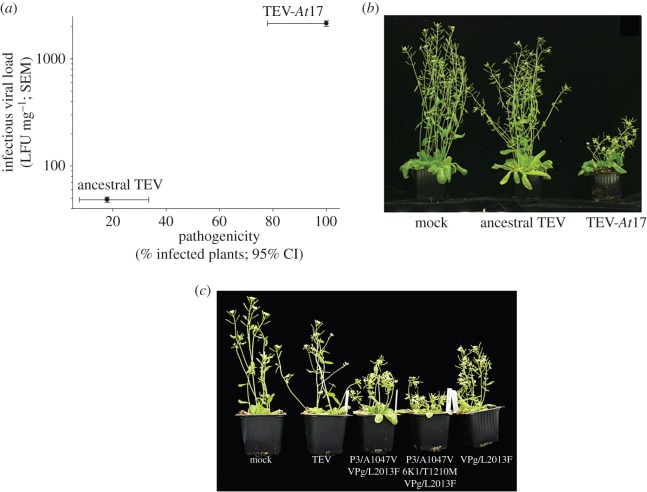
To set up the evolution experiments, a tobacco-adapted clone of TEV was used to manually inoculate a very large number of Arabidopsis Ler-0 plants [5]. Less than 20% of the inoculated plants were systematically infected with no apparent or very mild symptoms. Virus was purified from the infected plants and used to establish a number of independent evolutionary lineages maintained by serial passages. All but one of these lineages went extinct upon a variable number of passages. Extinction events were probably associated with a combination of very low accumulation of viral particles on each plant, the stochastic effect of the transmission bottleneck and the ability of the plant to mount an efficient antiviral response. The only lineage that survived for 17 serial passages, named as TEV-At17, was phenotypically and genetically characterized [5]. Several phenotypic properties were evaluated (figure 1): pathogenicity, infectious viral load and virulence. Pathogenicity is a measure of the efficiency of infecting a plant using a fixed amount of viral particles. The pathogenicity of the Arabidopsis-adapted virus dramatically increased from less than 20% to almost 100% (figure 1a). Likewise, the infectious viral load, a measure of the amount of infectious particles produced per a given amount of plant tissue, increased nearly two logarithms (figure 1a). Finally, symptoms evolved from an asymptomatic infection (central column in figure 1b) to strong, including vein clearing, very severe stunting, silique curling and distortion, and leaf deformation (right column in figure 1b).
Figure 1.
Phenotypic differences between the ancestral tobacco-adapted TEV and the evolved Arabidopsis-adapted virus. (a) Changes in pathogenicity and infectious viral load. (b) Illustration of the symptoms induced by both viruses. (c) Symptoms induced by combinations of the three non-synonymous mutations fixed in TEV-At17 genome. Adapted from [5].
Next, we determined the population consensus sequence for TEV-At17 and found only six point mutations, half of which were non-synonymous: P3/A1047-V, 6K1/T1210M and VPg/L2013F [5]. We focused our attention on these non-synonymous mutations. When recreated by site-directed mutagenesis on the ancestral tobacco-adapted genetic background, only viruses carrying mutation VPg/L2013F-induced symptoms in infected plants, although the symptoms were milder than those generated by TEV-At17 [5] (figure 1c). However, when mutations P3/A1047V and 6K1/T1210M were combined with VPg/L2013F in a pairwise manner, the severity of symptoms increased (figure 1c). Combining the three mutations into a single genome fully phenocopied the symptoms induced by TEV-At17 (figure 1c). These results suggest strong epistatic interactions among them: the additive effects of P3/A1047V and 6K1/T1210M were only expressed in the presence of mutation VPg/L2013F [5,11].
The above experiments demonstrate adaptation of TEV-At17 to its novel host Arabidopsis. Indeed, this adaptation was associated with a negative pleiotropic fitness effect in the ancestral host tobacco, promoting specialization [14]. To evaluate the mechanistic basis of such specialization, we decided to explore at the transcriptomic level the way Arabidopsis plants interacted with the tobacco-adapted and the Arabidopsis-adapted viruses. To do so, the transcriptomes of mock-inoculated control plants, plants infected with the ancestral TEV and plants infected with the evolved TEV-At17 were compared [5,6,8]. After substracting genes whose expression was affected by both evolved and ancestral viruses, which indicated instances of a general response of the plant to infection, 505 genes were upregulated and 1335 downregulated upon infection with TEV-At17 that were not altered by the ancestral virus. Among the downregulated genes, it is worth noting that they were significantly enriched in functional categories involved in the response to biotic stress, signal transduction and, more interestingly, in systemic acquired resistance and RNA-silencing innate immune resistance, the two main antiviral defence responses of plants [5,6,8]. These results suggest that the plant perceived the presence of both viruses in quite different manners: it was able to recognize the presence of the ancestral one and correspondingly mounting an efficient response while it was not capable of mounting such a response when infected with the evolved virus: TEV-At17 replicated, accumulated and spread ahead of the plant response.
Before closing this example, it is interesting to mention that by adapting to the particular ecotype used in this study, Ler-0, TEV-At17 also gained access to other ecotypes that were not susceptible to infection by the ancestral TEV isolate [7,9], thus further broadening its host range.
In conclusion, through experimental evolution of a virus in a novel host, we have been able to generate a new virus with an altered host range and completely different biological properties, perhaps matching the definition of a new viral species, despite very few genetic changes. Renaming TEV-At17 as Arabidopsis etch virus (AEV) or similar might be justified.
3. Flexible RNA genomes: gene gains and losses
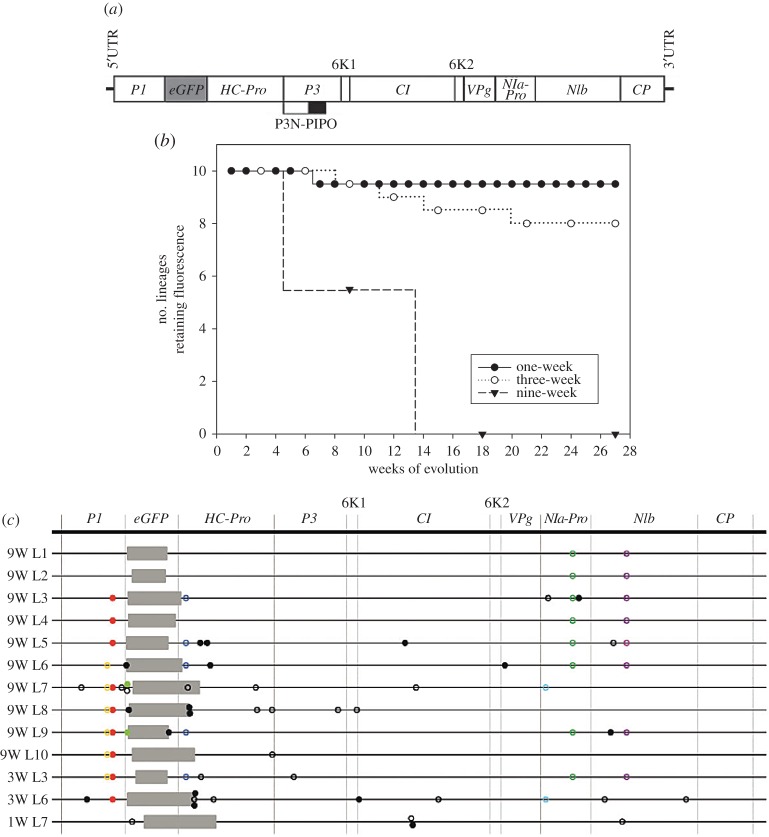
One of the advantages for evolutionary studies of RNA viruses is that if an infectious cDNA clone exists, then it is possible to manipulate the genome to create all sorts of newly engineered genomes. In the first of a series of recent experiments, Zwart et al. [15] explored the dynamics of gene acquisition and loss. In an initial study, a genome carrying an additional gene in-frame with TEV single open reading frame was generated (figure 2a). This additional gene encoded for a functional copy of the green fluorescent protein (eGFP), allowing for visual tracking of the presence of the exogenous gene. To determine the demographic conditions in which this gene was stably maintained in the viral population, three different passage experiments were performed. In the first experiment (figure 2b), passages took place every week for a total of 27 weeks. In this demographic regime, bottlenecks happened often and the effective viral population size was small in comparison with the other two regimes. Therefore, genetic drift was very strong. In the second experiment (figure 2b), passages took place every three weeks for a total of 27 weeks. In this case, bottlenecks were separated in time and the effective viral population size reached was larger than in the one-week passages regime, with genetic drift a less important factor. Finally, in the third evolution experiment (figure 2b), passages took place every nine weeks for a total of 27 weeks. Under this demographic regime, transmission bottlenecks were infrequent and the effective population size reached was larger than in previous regimes. Therefore, we expect genetic drift to be less and less important as transmission bottlenecks separate in time, and natural selection to be more and more efficient as effective population size (and hence genetic diversity) increases. As shown in figure 2b, the number of lineages that lost the exogenous gene during the evolution experiment varied widely among demographic conditions. Under strong-drift, weak-selection conditions, only one lineage showed a partial loss of fluorescence. Under the moderate-drift, moderate-selection regime, two lineages fully lost the exogenous gene. Finally, under the weak-drift, strong-selection demographic conditions, all lineages lost the exogenous eGFP gene. When the populations that lost the eGFP were analysed by Illumina NGS, we found that all had fixed deletions in the eGFP gene (figure 2c). However, variation in the location and size of the deletion was observed. None had a clean removal of the exogenous gene and most had deletion inside the eGFP that retained the reading frame. In some cases, however, deletions included parts of the HC-Pro cistron located downstream of the inserted eGFP. These deletions affected the N-terminal part of the HC-Pro protein, which is involved in aphid-mediated transmission of the virus. Since in our experimental set-up transmission was mechanical, this domain of the HC-Pro protein was dispensable. By contrast, none of the deletions affected part of the C-terminus of the P1 protein, as this protease plays essential roles in the early stages of processing the polyprotein and in interfering with the RNAi antiviral response of the plant.
Figure 2.
Molecular evolution of a viral genome with an exogenous gene inserted. (a) Schematic of the engineered genome TEV-eGFP. Lines represent the viral 5′- and 3′-untranslated regions (UTRs), the grey box represents the inserted eGFP gene, open boxes represent the viral cistrons P1, HC-Pro, P3, 6K1, CI, 6K2, VPg, NIa-Pro, NIb and CP, the lower box indicates the embedded P3N-PIPO cistron. (b) Stability of the eGFP cistron upon passages that represent different demographic conditions: one-week = strong drift, weak selection, three-week = moderate drift, moderate selection, nine-week = weak drift, strong selection. (c) Genome sequence of the evolved lineages that lost the eGFP gene. Grey boxes indicate genomic deletions; full circles and open circles are non-synonymous and synonymous substations, respectively. Black substitutions were observed in only one lineage while coloured substitutions were repeated in two or more lineages. (c) Adapted from [15].
The fitness of all 30 evolved lineages was assessed by means of in planta competition experiments against the wild-type TEV. A negative and significant correlation was observed between fitness and genome length: the longer the genomes, the lower their fitness. This fitness penalty is consistent with the notion of a trade-off between genome length and replication time: longer genomes need longer times to be replicated and therefore are in selective disadvantage compared with shorter genomes. This explains why in demographic regimes where selection was the dominant factor any deletion mutant quickly outcompetes its parental TEV-eGFP.
Non-retroviral integrated RNA viruses (NIRVs) are genes of non-retroviral RNA viruses found in the genomes of many eukaryotic organisms [16,17]. In some cases, NIRVs have become functional genes [18]. It has been suggested that the presence of NIRVs can confer a host with resistance against the cognate virus. Indeed, it has been a common practice to generate commercial transgenic plants resistant to viruses by inserting full or partial viral genes into the genome of plants [19]; these transgenic plants can be considered as synthetic cases of NIRVs. Because the reading frame is conserved in many NIRVs, they can be expressed [20]. This expression may have important implications for the evolution of the cognate virus. To tackle this issue, Tromas et al. [21] evolved TEV in transgenic tobacco plants expressing the NIb gene that encodes for the viral replicase (figure 2a). We hypothesized that during infection, plant-derived NIb expression would make the virus-encoded NIb expression redundant and, therefore, selection on the virus to maintain its own copy would be relaxed. Indeed, the results from the evolution experiments (five lineages, four three-week passages), followed by Illumina NGS characterization of the evolved populations, found that the TEV copy of NIb gene was deleted in several lineages, yet preserving the reading frame of the resulting genomes. Indeed, competition experiments between these shorter-genome viruses and the wild-type virus in the NIb-transgenic tobacco plants showed that the shortened viruses had a 3% per day selective advantage. By contrast, not surprisingly, these NIb-deficient viruses were unviable in wild-type tobacco plants; the evolved viruses become absolutely dependent on the transgenic plants for their survival.
All these experiments illustrate the plasticity of viral genomes. Given the appropriate environmental (host) and demographic conditions, genes can be incorporated and stably maintained in viral genomes or they can be removed because the cell provides their products in a cost-free manner. Thus, it may be to the benefit of a virus to transfer its most relevant genes to the host genome, a phenomenon that would clearly represent a major evolutionary transition from a viral perspective.
4. Evolution of genome architecture: the rise of segmented genomes
The previous section describes a particular form of innovation in the genetic architecture of RNA viruses: the acquisition and loss of genes. Another major transition that viral genomes can undergo is genome segmentation, that is, splitting the gene content into different segments, very much equivalent to chromosomes in eukaryotic organisms. Segmented viruses are more common among plants than among animal RNA viruses and may be packed into the same or into different capsids. The advantages of RNA genome segmentation have been discussed in the past and they range from better resisting the deleterious effect of high mutation rates [22], bolstering evolvability by segment reassortment during mixed infections [22] and selection of shorter and faster replicators, [23,24] to optimizing the co-regulation of the expression of linked genes [25,26].
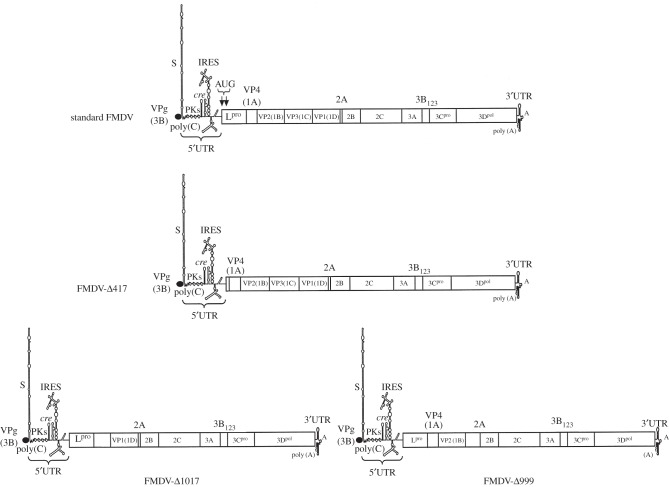
García-Arriaza et al. [27,28] have documented the first step of an evolutionary transition toward genome segmentation of an unsegmented RNA virus, the foot-and-mouth disease virus (FMDV). A clone of FMDV was serially passaged at high multiplicity of infection (MOI) in cell culture for 143 passages. A high MOI means that, on average, each cell can be initially infected by more than one viral particle. A defective genome, still fully dependent on the standard virus, was detected at this evolutionary passage [29]. This viral population was further passaged under the same experimental conditions until reaching passage 260. At this time, the population was analysed and three RNA molecules smaller than the standard genome were detected [27]: Δ417, Δ999 and Δ1017 (figure 3). Deletion of Δ417 affected the L protease gene, whereas the other two deletions affected an overlapping sequence of the VP capsid genes. None of the deletions affected the reading frame of downstream genes, meaning that the rest of gene products were still viable. The three deleted genomes were present at approximately equimolar concentrations per cell and the standard virus was still present, though at least 10 000-fold less than the defective genomes. Interestingly, when the viral population was plaqued out in cell monolayer and single plaques analysed, 98% of plaques contained Δ417 and either Δ999 or Δ1017; but none contained the standard FMDV genome, suggesting that both deleted genomes can complement each other in completing the infection cycle [27]. This result was further confirmed by transfection experiments showing that combinations of RNAs from Δ417 and either Δ999 or Δ1017 were able to produce cytopathology in cells but none of them alone did so [27]. Furthermore, electron microscopy showed that defective RNAs were encapsidated into independent particles, each containing one defective genome [27]. Consistently with this observation, dose-response experiments showed that the shape of the virulence curve of an infection made with the standard virus was as expected for a one-hit model, in which one particle is enough for inducing infection; while the virulence curves observed for the population of defective genomes were compatible with a two-hit model (i.e. more than one particle is necessary to induce infection; [27]).
Figure 3.
Schematic of the standard FMDV and of the three deletion mutants generated after 260 undiluted passages. The standard FMDV shows the names of all 12 genes and a representation of the 5′- and 3′-UTRs, with details of the rich secondary structure of the 5′-UTR, which has activity that plays a role as ribosomal entry sites (IRES) during translation. The Δ417 defective genome lacks practically the entire gene coding for the L protease. The Δ1017 defective genome lacks from the 3′ extreme of the VP4 to half of the VP3 coat proteins. The Δ999 defective genome is missing approximately half of the VP3 and the entire VP1 coat proteins. The remaining genes are all in frame for translation.
Genome segmentation comes with an obvious cost in terms of a reduced probability of infection, because two particles are required to trigger infection. This may not be a problem in the above experimental set-up at high MOI, but in order for a segmented genome to persist for long evolutionary periods, a direct benefit of segmentation should exist that overcomes this penalty. Ojosnegros et al. [30] explored potential mechanisms providing such advantage to the segmented Δ417/Δ999 virus in comparison with the standard FMDV one. After ruling out differences in RNA replication rates and protein expression, the authors concluded that a higher stability of viral particles containing the shorter genomes could provide the required fitness advantage [30]. In this sense, genome segmentation may provide a molecular mechanism to overcome the trade-off between genome size and particle stability, as it allows increases in the amount of genetic information encoded by the complementing genomes as a whole while relaxing packaging constraints for each individual segment.
In conclusion, these studies show that a major evolutionary transition from single-molecule genomes to segmented genomes is possible in RNA viruses under the appropriate environmental conditions. Nonetheless, this still represents a first step in the transition towards a real segmented genome, like those observed nowadays, as the shorter segments Δ417, Δ999 and Δ1017 still share a big chunk of their genes and only differ in the presence or absence of the L protease and the VP capside genes, respectively (figure 3).
5. Conflict and cooperation in the viral world
Co-infection of a single host by several different viruses is common in nature. Co-infection deeply impacts the fitness and virulence of the partner viruses, ranging from antagonism (i.e. viruses negatively impact each other) to synergism (i.e. each virus exerts a positive effect on its partners) [31,32]. In some cases, as for instance the case of plant umbraviruses [33], where the degree of dependence is so tight that the viruses cannot be transmitted in the absence of their assistor luteovirus and of a satellite RNA. In such cases, the viral complex, and not each of its members, has full pathological meaning. Evolutionary transitions to new levels of biological complexity often require cooperation among individuals, but selection among components may favour selfish behaviours that jeopardize the evolution of cooperation.
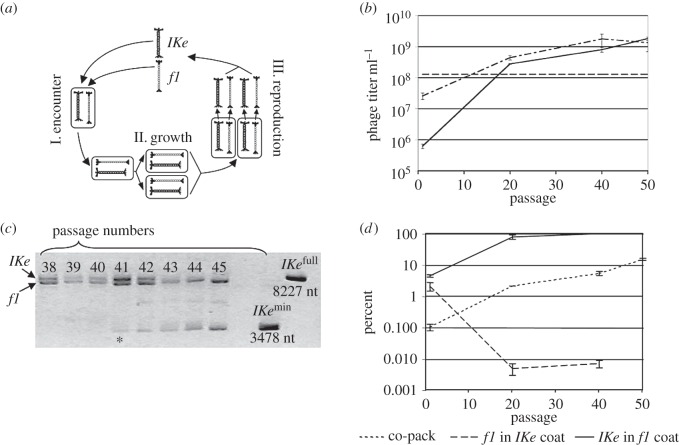
To tackle the rise of cooperation in the viral world, Sachs & Bull [34] investigated the evolution of two very divergent DNA bacteriophages (f1 and IKe) experimentally forced to obey a life cycle with elements of cooperation and conflict during the infection of their natural host Escherichia coli. A priori, the outcome of the experiment could have ranged from extinction of the population (due to selection of selfish elements) to extreme cooperation. The evolution experiment consisted of 50 serial repetitions of the following three-step cycle (figure 4a): (i) encounter of the two viruses within an E. coli cell, (ii) bacterial growth, and (iii) viral replication and competition for cellular resources to generate phage progeny for the next cycle. The trick of the experimental design was that each phage was engineered to encode for a different antibiotic resistance gene (kanamycin and chloramphenicol) and that both antibiotics were present at the same time in the growth medium [34]. By doing so, the researchers ensured that only co-infected cells were able to support viral replication, while singly infected cells would die due to the effect of the alternative antibiotic. Steps (i) and (ii) reinforced cooperation between both viruses; step (iii) favoured selfish reproduction at the expense of the partner. What solution did evolution find to this conundrum? The first observation is that both viruses increased their fitness during the evolution experiment (figure 4b), proving that selection was in operation. The second, essential observation was that the genome size of IKe was decreasing with time (figure 4c). At passage 41, a very short version of IKe, named as IKemin, was first detected. Gradually IKemin displaced the standard full-genome IKe virus. Cloning and sequencing of IKemin showed that it had lost all viral genes but one; likewise, sequencing of the evolved f1 showed the presence of three non-synonymous mutations. Finally, the pivotal observation to understand the solution of this evolutionary conflict was that both f1 and IKemin were packaged together into f1 capsids (figure 4d). The advantage of this solution for both viruses is obvious: co-packaging ensures a high and efficient co-transmission of both viruses, maximizing the presence of both antibiotic resistances in the same cell while minimizing the conflict that was associated with the third step in the life cycle.
Figure 4.
An example of the evolution of cooperation in the viral world. (a) Representation of the three steps in which each experimental evolutionary passage could be subdivided. (b) Increase in viral fitness (measured as titre) for both phages during the course of experimental evolution. The solid line corresponds to IKe and the dotted-dashed line to f1. (c) Emergence and expansion of the genome-reduced IKemin. The asterisk indicates the passage in which IKemin was first detected. (d) Evolution of the frequency of co-packaging (dotted line) and of heterologous packaging (both dashed lines). Adapted from [34] with permission from the authors.
The above example illustrates the evolution of a novel cooperation mechanism between two different viral species co-infecting the same host. A classic example of the opposite outcome can be found in the study done by Tuner & Chao [35] on the interactions between genotypes of bacteriophage ϕ6 during infections at high and low MOI. At high MOI, bacterial cells are infected with more than one viral particle and they compete with each other for cellular resources. Viruses that evolve under such circumstances become selfish, gaining an additional fitness advantage during co-infection with the ancestral virus or with viruses evolved at low MOI, indicating that they evolved a defection strategy for intracellular competition. Not only this, but when the fitness data of the evolved selfish viruses are expressed in the form of a payoff matrix, its structure corresponds to the prisoner's dilemma strategy of game theory. In the prisoner's dilemma game, mutual defection is the dominant strategy and results in the long-term persistence of only one of the contenders.
In conclusion, cooperation and defection are evolvable traits in virus experimental evolution. Cooperation, in the form illustrated by the f1/IKemin system, represents a bona fide major evolutionary transition as it involves the creation of a novel, more complex biological entity by combining simpler units. The properties of this new biological assemble could not be predicted from the information of its constitutive parts.
6. Concluding remarks
I believe that this short review illustrates the usefulness of evolution experiments with viruses to tackle big evolutionary questions and not only microevolutionary processes. The emergence of new species and the appearance of new forms of biological organization and of new forms of genome architecture have been illustrated here with a selection of highly relevant studies.
Before closing my review, I would like to apologize to those colleagues whose viral work may also fit within the umbrella of ‘major evolutionary transitions' but have not been mentioned here. Space constraints, my incomplete knowledge of the field and my personal taste are the reasons.
Acknowledgements
I thank Ricard V. Solé and David Wolpert for organizing the very stimulating Santa Fe Institute Working Group on Major Transitions in Natural, Synthetic and Artificial Evolution and for inviting me to participate. The work presented here would not be possible without the contribution of many past and present members of my laboratory.
Competing interests
I have no competing interests.
Funding
My research is supported by grants BFU2012-30805 from Spain Ministry of Economy and Competitiveness, PROMETEOII/2014/021 from Generalitat Valenciana and EvoEvo (ICT610427) from the European Commission 7th Framework Program.
References
- 1.Jiang S, Hotez PJ. 2015. Combating the emerging viral infectious diseases. Microb. Infect. 17, 83 ( 10.1016/j.micinf.2014.12.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 76, 159–216. ( 10.1128/MMBR.05023-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elena SF, Sanjuán R. 2007. Virus evolution: insights from an experimental approach. Annu. Rev. Ecol. Evol. Syst. 38, 27–52. ( 10.1146/annurev.ecolsys.38.091206.095637) [DOI] [Google Scholar]
- 4.Elena SF, Agudelo-Romero P, Carrasco P, Codoñer FM, Martín S, Torres-Barceló C, Sanjuán R. 2008. Experimental evolution of plant RNA viruses. Heredity 100, 478–483. ( 10.1038/sj.hdy.6801088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agudelo-Romero P, Carbonell P, Pérez-Amador MA, Elena SF. 2008. Virus adaptation by manipulation of host's gene expression. PLoS ONE 3, e2397 ( 10.1371/journal.pone.0002397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agudelo-Romero P, Carbonell P, de la Iglesia F, Carrera J, Rodrigo G, Jaramillo A, Pérez-Amador MA, Elena SF. 2008. Changes in gene expression profile of Arabidopsis thaliana after infection with Tobacco etch virus. Virol. J. 5, 92 ( 10.1186/1743-422X-5-92) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lalić J, Agudelo-Romero P, Carrasco P, Elena SF. 2010. Adaptation of tobacco etch potyvirus to a susceptible ecotype of Arabidopsis thaliana capacitates it for systemic infection of resistant ecotypes. Phil. Trans. R. Soc. B 365, 1997–2007. ( 10.1098/rstb.2010.0044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillung J, Cuevas JM, Elena SF. 2012. Transcript profiling of different Arabidopsis thaliana ecotypes in response to tobacco etch potyvirus infection. Front. Microbiol. 3, 229 ( 10.3389/fmicb.2012.00229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillung J, Cuevas JM, Valverde S, Elena SF. 2014. Experimental evolution of an emerging plant virus in host genotypes that differ in their susceptibility to infection. Evolution 68, 2467–2480. ( 10.1111/evo.12458) [DOI] [PubMed] [Google Scholar]
- 10.Hillung J, Cuevas JM, Elena SF. 2015. Evaluating the within-host fitness effects of mutations fixed during virus adaptation to different ecotypes of a new host. Phil. Trans. R. Soc. B 370, 20140292 ( 10.1098/rstb.2014.0292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lalić J, Elena SF. 2015. The impact of high-order epistasis in the within-host fitness of a positive-sense plant RNA virus. J. Evol. Biol. 28, 2236–2247. ( 10.1111/jeb.12748) [DOI] [PubMed] [Google Scholar]
- 12.Chisholm ST, Parra MA, Anderberg RJ, Carrington JC. 2001. Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long-distance movement of tobacco etch virus. Plant Physiol. 127, 1667–1675. ( 10.1104/pp.010479) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosson P, et al. 2010. RTM3, which controls long-distance movement of potyviruses, is a member of a new plant gene family encoding a MEPRIN and TRAF homology domain-containing protein. Plant Physiol. 154, 222–232. ( 10.1104/pp.110.155754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bedhomme S, Hillung J, Elena SF. 2015. Emerging viruses: why they are not jacks of all trades? Curr. Opin. Virol. 10, 1–6. ( 10.1016/j.coviro.2014.10.006) [DOI] [PubMed] [Google Scholar]
- 15.Zwart MP, Willemsen A, Daròs JA, Elena SF. 2014. Experimental evolution of pseudogenization and gene loss in a plant RNA virus. Mol. Biol. Evol. 31, 121–134. ( 10.1093/molbev/mst175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiba S, Kondo H, Tani A, Saisho D, Sakamoto W, Kanematsu S, Suzuki N. 2011. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog. 7, e1002146 ( 10.1371/journal.ppat.1002146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet. 6, e1001191 ( 10.1371/journal.pgen.1001191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koonin EV. 2010. Taming of the shrewd: novel eukaryotic genes from RNA viruses. BMC Biol. 8, 2 ( 10.1186/1741-7007-8-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertsch C, Beuve M, Dolja VV, Wirth M, Pelsy F, Herrbach E, Lemaire O. 2009. Retention of the virus-derived sequences in the nuclear genome of grapevine as a potential pathway to virus resistance. Biol. Direct 4, 21 ( 10.1186/1745-6150-4-21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor DJ, Bruenn J. 2009. The evolution of novel fungal genes from non-retroviral RNA viruses. BMC Biol. 7, 88 ( 10.1186/1741-7007-7-88) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tromas N, Zwart MP, Forment J, Elena SF. 2014. Shrinkage of genome size in a plant RNA virus upon transfer of an essential viral gene into the host genome. Genome Biol. Evol. 3, 538–550. ( 10.1093/gbe/evu036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao L. 1991. Levels of selection, evolution of sex in RNA viruses, and the origin of life. J. Theor. Biol. 153, 229–246. ( 10.1016/S0022-5193(05)80424-2) [DOI] [PubMed] [Google Scholar]
- 23.Chao L, Tran TT, Matthews C. 1992. Muller's ratchet and the advantage of sex in the RNA virus ϕ6. Evolution 46, 289–299. ( 10.2307/2409851) [DOI] [PubMed] [Google Scholar]
- 24.Nee S. 1987. The evolution of multicompartmental genomes in viruses. J. Mol. Evol. 25, 277–281. ( 10.1007/BF02603110) [DOI] [PubMed] [Google Scholar]
- 25.Pressing J, Reanney DC. 1984. Divided genomes and intrinsic noise. J. Mol. Evol. 20, 135–146. ( 10.1007/BF02257374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sicard A, Yvon M, Timchenko T, Gronenborn B, Michalakis Y, Gutiérrez S, Blanc S. 2013. Gene copy number is differentially regulated in a multipartite virus. Nat. Commun. 4, 2248 ( 10.1038/ncomms3248) [DOI] [PubMed] [Google Scholar]
- 27.García-Arriaza J, Manrubia S, Toja M, Domingo E, Escarmís C. 2004. Evolutionary transition toward defective RNAs that are infectious by complementation. J. Virol. 78, 11 678–11 685. ( 10.1128/JVI.78.21.11678-11685.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García-Arriaza J, Domingo E, Escarmís C. 2005. A segmented form of Foot-and-mouth disease virus interferes with standard virus: a link between interference and competitive fitness. Virology 335, 155–164. ( 10.1016/j.virol.2005.02.012) [DOI] [PubMed] [Google Scholar]
- 29.Charpentier N, Dávila M, Domingo E, Escarmís C. 1996. Long-term, large-population passage of aphtovirus can generate and amplify defective noninterfering particles deleted in the leader protease gene. Virology 223, 10–18. ( 10.1006/viro.1996.0450) [DOI] [PubMed] [Google Scholar]
- 30.Ojosnegros S, García-Arriaza J, Escarmís C, Manrubia SC, Perales C, Arias A, García-Mateu M, Domingo E. 2011. Viral genome segmentation can result from a trade-off between genetic content and particle stability. PLoS Genet. 7, e1001344 ( 10.1371/journal.pgen.1001344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asatryan A, Wodarz D, Komarova NL. 2015. New virus dynamics in the presence of multiple infection. J. Theor. Biol. 377, 98–109. ( 10.1016/j.jtbi.2015.04.014) [DOI] [PubMed] [Google Scholar]
- 32.Elena SF, Bernet GP, Carrasco JL. 2014. The games plant viruses play. Curr. Opin. Virol. 8, 62–67. ( 10.1016/j.coviro.2014.07.003) [DOI] [PubMed] [Google Scholar]
- 33.Robinson DJ, Ryabov EV, Raj SK, Roberts IM, Taliansky ME. 1999. Satellite RNA is essential for encapsidation of groundnut rosette umbravirus RNA by groundnut rosette assistor luteovirus coat protein. Virology 254, 105–114. ( 10.1006/viro.1998.9527) [DOI] [PubMed] [Google Scholar]
- 34.Sachs JL, Bull JJ. 2005. Experimental evolution of conflict mediation between genomes. Proc. Natl Acad. Sci. USA 102, 390–395. ( 10.1073/pnas.0405738102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner PE, Chao L. 1999. Prisoner's dilemma in an RNA virus. Nature 398, 441–443. ( 10.1038/18913) [DOI] [PubMed] [Google Scholar]