Abstract
The relationship between the DNA methylation status of the CpG islands of multiple genes in blood leukocytes in CRC susceptibility and prognosis, as well as possible interactions with dietary factors on CRC risk are unclear. We carried out a case-control study including 421 CRC patients and 506 controls to examine the associations between six genes (AOX-1, RARB2, RERG, ADAMTS9, IRF4, and FOXE-1), multiple CpG site methylation (MCSM) and susceptibility to CRC. High-level MCSM (MCSM-H) was defined as methylation of greater than or equal to 2 of 5 candidate genes (except for RARB2); low-level MCSM (MCSM-L) was when 1 candidate gene was methylated; non-MCSM was when none of the candidate genes were methylated. Blood cell-derived DNA methylation status was detected using methylation-sensitive high-resolution melting analysis. The hypermethylation status of each individual gene was statistically significantly associated with CRC. MCSM status was also associated with CRC (OR = 1.54, 95% CI: 1.15–2.05, P = 0.004). We observed interactions between a high level of dietary intake of cereals, pungent food, and stewed fish with brown sauce, age (older than 60 yrs), smoking and hypermethylation on risk of CRC. MCSM in peripheral blood DNA may be an important biomarker for susceptibility to CRC.
The development of colorectal cancer (CRC) is characterized by genetic and epigenetic dysregulation of signal transduction cascades in a multi-step fashion. The long-term accumulation of epigenetic changes can lead to the occurrence of CRC1. DNA methylation, a major epigenetic modification, induces transcriptional silencing of tumor suppressor genes2 and the subsequent expression of disease phenotypes without any changes to the primary DNA sequences3, which has been identified as a critical step in tumor initiation4, including in CRC. Genes known to be methylated that are detected and studied in tumor-derived DNA in colorectal cancer are CDKN2A5, CDH46, NEUROG17 and MLH18,9 and we tested a gene panel consisting of APC, MGMT, RASSF2A and WIF-110 in our previous study. As a follow-up study, we then searched for genes with aberrant promoter hypermethylation associated with CRC in the PubMeth11, MethDB12 and MethyCancer13 literature databases. It has been reported that gene-specific hypermethylation changes in promoter CpG islands are related to biological processes of tumor progression including metastasis suppressors (AOX-114 and RARB215), angiogenesis inhibitors (RERG16 and ADAMTS917) and signal transcription factors (IRF418 and FOXE-119).
RAS-like estrogen-regulated growth inhibitor (RERG) was first detected in breast cancer using microarray analysis16, after which significantly hypermethylated RERG was observed in colorectal adenocarcinomas but not in adenomas and normal mucosa20. Aldehyde oxidase 1 (AOX-1) was found to be down-regulated and hypermethylated in CRC tissues, suggesting a potential functional role of this gene in cancer development21. ADAM metallopeptidase with thrombospondin type 1 motif, 9 (ADAMTS9) which has been characterized as a novel tumor suppressor gene, has been epigenetically silenced in lymph node metastases in nasopharyngeal carcinoma17. Interferon regulatory factor 4 (IRF4) was found to be highly silenced by DNA hypermethylation in both gastric cancers and gastric mucosa from cancer patients, which suggested that IRF4 hypermethylation may be a useful molecular marker for diagnosing recurring gastric cancer22. Hypermethylation of forkhead box E1 (FOXE1) has also been explored as a noninvasive biomarker for the detection of CRC in stool samples19. Finally, retinoic acid receptor beta 2 (RARB2) was frequently inactivated in cancers of epithelial origin and has been shown to have tumor suppressive activity in lung, breast, and colon cancer cell lines15. In addition, promoter hypermethylation of RARB2 was found in stool samples (13%) of patients with inflammatory bowel disease, but with no aberrant hypermethylation in healthy subjects23.
Recently, researchers have frequently focused on tumor tissues to explore the relationship between DNA methylation status and CRC as potential biomarkers17,19,20,24,25. Furthermore, studies have shown that dietary habits and lifestyle habits, such as drinking and smoking may impact DNA methylation26,27,28. In addition, tumors do not only develop as an isolated phenomenon in their target tissues29 and emerging data suggests that leukocyte DNA methylation might be linked to susceptibility to CRC30. CpG island methylation phenotypes (CIMP) were first introduced by Toyota et al. to define extensive hypermethylation of multiple gene CpG islands from tumor tissues, which is currently recognized as one of the major mechanisms in CRC carcinogenesis31,32. Multiple genetic and epigenetic alterations that contribute to chemoresistance occur during chemotherapy and may eventually impact the disease outcome33. Therefore, we propose that the DNA methylation status of multiple gene CpG islands from blood leukocytes may be associated with the risk and prognosis of CRC. Thus, we carried out this study to explore the interaction between environmental factors and DNA methylation status on the risk of CRC.
Results
Characteristics of the study subjects
This study consisted of 421 cases (163 females and 258 males) with a mean age of 59.46 ± 11.41 and 506 controls (227 females and 279 males) with a mean age of 56.62 ± 10.97 (P = 0.000; Table 1). The proportion of age in groups of 60 yrs to 69 yrs and greater than 70 yrs in cases were both higher than those in controls. The proportion of overweight (BMI ≥ 23.0) in controls (63.4%) was higher than that in cases (52.4%; P = 0.003). The distribution of different occupational categories and family history of other cancers were also different between cases and controls (P = 0.021 and P = 0.004, respectively). The proportion of mental workers in controls (52.9%) was higher than that in cases (43.9%). The proportion of patients with family history of other cancers in cases (78.4%) was lower than that in controls (86.5%).
Table 1. Characteristics of colorectal cancer patients and controls.
| Variable | Cases (%) (n = 421) | Controls (%) (n = 506) | P values |
|---|---|---|---|
| Age (mean ± SD) | 59.46 ± 11.41 | 56.62 ± 10.97 | 0.000 |
| <50 | 79 (18.8) | 133 (26.3) | 0.001 |
| 50–59 | 129 (30.6) | 175 (34.6) | |
| 60–69 | 126 (29.9) | 132 (26.1) | |
| ≥70 | 87 (20.7) | 66 (13.0) | |
| Gender | |||
| Male | 258 (61.3) | 279 (55.1) | 0.059 |
| Female | 163 (38.7) | 227 (44.9) | |
| BMI (kg/m2) | |||
| ≤18.50 | 32 (7.7) | 26 (5.2) | 0.003 |
| 18.5–23.00 | 167 (40.0) | 156 (31.4) | |
| ≥23.00 | 219 (52.4) | 315 (63.4) | |
| Education | |||
| Primary and below | 114 (29.2) | 157 (32.2) | 0.576 |
| Junior high school | 115 (29.4) | 148 (30.4) | |
| Senior middle school | 84 (21.5) | 100 (20.5) | |
| College and above | 78 (19.9) | 82 (16.8) | |
| Occupation | |||
| Mental worker | 170 (43.9) | 246 (52.9) | 0.021 |
| Manual worker | 116 (30.0) | 127 (27.3) | |
| Combined | 101 (26.1) | 92 (19.8) | |
| Family history of cancer | |||
| No | 312 (78.4) | 307 (86.5) | 0.004 |
| Yes | 86 (21.6) | 48 (13.5) | |
| Tumor location | |||
| Colon | 117 (33.5) | — | — |
| Rectum | 232 (66.5) | — | |
Association between the methylation status of six individual genes and CRC
Statistically significant associations between the methylation of AOX-1, RERG, ADAMTS9, IRF4, FOXE-1 and CRC were observed in univariate logistic regression analyses (Table 2). Further multivariate logistic regression analyses with adjustment for age, BMI, occupation and family history of cancer showed that the hypermethylation of AOX-1 (OR = 1.72, 95% CI: 1.30–2.27, P = 0.00), RERG (OR = 2.08, 95% CI: 1.56–2.77, P = 0.00), ADAMTS9 (OR = 1.85, 95% CI: 1.37–2.49, P = 0.00) and IRF4 (OR = 16.96, 95% CI: 5.15–55.84, P = 0.00) were still significantly associated with CRC. However, no statistically significant associations were found between the methylation status of FOXE-1 (P = 0.06) or RARB2 (P = 0.78) and CRC.
Table 2. Associations between methylation of individual genes, MCSM and the risk of CRC.
| Gene | Case (%) | Control (%) | OR | 95% CI | Pvalue | ORa | 95% CI | Pvalue | |
|---|---|---|---|---|---|---|---|---|---|
| IRF4 | Unmethylation | 381 (90.5) | 495 (99.4) | 1.00 | 1.00 | ||||
| Methylation | 40 (9.5) | 3 (0.6) | 17.32 | 5.32–56.42 | 0.000 | 16.96 | 5.15–55.84 | 0.00 | |
| FOXE-1 | Unmethylation | 301 (71.5) | 388 (78.5) | 1.00 | 1.00 | ||||
| Methylation | 120 (28.5) | 106 (21.5) | 1.46 | 1.08–1.97 | 0.014 | 1.35 | 0.99–1.85 | 0.06 | |
| AOX-1 | Unmethylation | 214 (51.2) | 312 (63.5) | 1.00 | 1.00 | ||||
| Methylation | 204 (48.8) | 179 (36.5) | 1.66 | 1.27–2.17 | 0.000 | 1.72 | 1.30–2.27 | 0.00 | |
| ADAMTS9 | Unmethylation | 268 (64.0) | 387 (77.4) | 1.00 | 1.00 | ||||
| Methylation | 151 (36.0) | 113 (22.6) | 1.93 | 1.45–2.58 | 0.000 | 1.85 | 1.37–2.49 | 0.00 | |
| RERG | Unmethylation | 240 (57.3) | 370 (74.1) | 1.00 | 1.00 | ||||
| Methylation | 179 (42.7) | 129 (25.9) | 2.14 | 1.62–2.83 | 0.000 | 2.08 | 1.56–2.77 | 0.00 | |
| RARB2 | Unmethylation | 314 (74.8) | 377 (75.2) | 1.00 | 1.00 | ||||
| Methylation | 106 (25.2) | 124 (24.8) | 1.03 | 0.76–1.39 | 0.865 | 0.96 | 0.70–1.30 | 0.78 | |
| MCSM | Non-MCSM | 106 (25.5) | 167 (34.5) | 1.00 | 1.00 | ||||
| MCSM-L | 125 (30.1) | 161 (33.3) | 1.22 | 0.87–1.71 | 0.242 | 1.23 | 0.87–1.75 | 0.24 | |
| MCSM-H | 184 (44.3) | 156 (32.2) | 1.86 | 1.34–2.57 | 0.000 | 1.79 | 1.28–2.52 | 0.00 | |
| MCSM | 309 (74.5) | 317 (65.5) | 1.54 | 1.15–2.05 | 0.004 | 1.50 | 1.11–2.03 | 0.01 | |
aAdjusted for age, BMI, occupation and family history of cancer.
The results of stratification analysis by age suggested that hypermethylation of FOXE-1 and AOX-1 were associated with risk of CRC only in the older group (aged 60 yrs and older). Hypermethylation of ADAMTS9 and RERG were significantly associated with risk of CRC in both the young and old group, with stronger associations in the old group (Table 3).
Table 3. Association between methylation of genes and risk of CRC by age.
| Gene | <60 |
≥60 |
||||
|---|---|---|---|---|---|---|
| ORa | 95% CI | Pvalue | ORa | 95% CI | Pvalue | |
| IRF4 | 9.618 | 1.187–77.931 | 0.034 | − | − | − |
| FOXE-1 | 0.977 | 0.571–1.670 | 0.932 | 1.700 | 1.023–2.824 | 0.040 |
| AOX-1 | 1.540 | 0.972–2.440 | 0.066 | 3.791 | 2.301–6.246 | 0.000 |
| ADAMTS9 | 1.803 | 1.086–2.992 | 0.023 | 2.025 | 1.244–3.296 | 0.005 |
| RERG | 2.036 | 1.236–3.355 | 0.005 | 4.821 | 2.824–8.231 | 0.000 |
| RARB2 | 0.666 | 0.405–1.096 | 0.110 | 0.921 | 0.556–1.526 | 0.750 |
| MSCM-L | 1.104 | 0.661–1.844 | 0.706 | 1.938 | 1.027–3.655 | 0.041 |
| MSCM-H | 1.527 | 0.886–2.634 | 0.128 | 5.128 | 2.744–9.584 | 0.000 |
| MSCM | 1.276 | 0.814–2.002 | 0.288 | 3.334 | 1.907–5.830 | 0.000 |
aAdjusted for BMI, occupation and family history of cancer.
aAll the ORs were calculated by selecting unmethylation as a reference group.
In another stratification analysis by family history of cancer, AOX-1 hypermethylation was associated with risk of CRC regardless of the presence of a family history of cancer. Hypermethylation of ADAMTS9 and RERG were associated with risk of CRC only without a family history of cancer (Table 4).
Table 4. Association between methylation of genes and risk of CRC by family history of cancer.
| Gene | No |
Yes |
||||
|---|---|---|---|---|---|---|
| ORa | 95% CI | Pvalue | ORa | 95% CI | Pvalue | |
| IRF4 | 25.143 | 3.350–188.705 | 0.002 | — | — | — |
| FOXE-1 | 1.425 | 0.954–2.130 | 0.084 | 0.845 | 0.345–2.066 | 0.712 |
| AOX-1 | 2.340 | 1.622–3.374 | 0.000 | 2.467 | 1.012–6.010 | 0.047 |
| ADAMTS9 | 1.871 | 1.277–2.740 | 0.001 | 1.868 | 0.773–4.517 | 0.165 |
| RERG | 3.269 | 2.195–4.869 | 0.000 | 2.215 | 0.905–5.423 | 0.082 |
| RARB2 | 0.757 | 0.512–1.121 | 0.165 | 0.932 | 0.412–2.107 | 0.866 |
| MSCM-L | 1.311 | 0.846–2.032 | 0.225 | 1.288 | 0.518–3.201 | 0.585 |
| MSCM-H | 2.606 | 1.669–4.069 | 0.000 | 2.661 | 0.984–7.194 | 0.054 |
| MSCM | 1.839 | 1.253–2.701 | 0.002 | 1.793 | 0.803–4.002 | 0.154 |
aAdjusted for age, BMI and occupation.
aAll the ORs were calculated by selecting unmethylation as a reference group.
Association between MCSM methylation status and CRC
These candidate gene biomarkers were integrated into the multiple CpG site methylation (MCSM) panel. High-level MCSM (MCSM-H) was defined when 2 or more of the 5 candidate genes (AOX-1, RERG, ADAMTS9, IRF4 or FOXE-1) were methylated; low-level MCSM (MCSM-L) was defined as 1 of the 5 candidate genes being methylated; and non-MCSM was defined as none of the 5 candidate genes being methylated.
Multivariate logistic regression analysis suggested that there was a statistically significant difference in the distribution of cases and controls in the non-MCSM and MCSM groups (OR = 1.50, 95% CI: 1.11–2.03, P = 0.01). Moreover, after dividing MSCM into two groups, MCSM-L and MCSM-H, a stronger association between MCSM-H (compared with non-MCSM) and CRC (OR = 1.79, 95% CI: 1.28–2.52, P = 0.00) was observed (Table 2).
Compared to the non-MCSM group, MCSM was associated with risk of CRC in the older group (aged older than 60 yrs) and without family history of cancer in a stratification analysis (P = 0.000 and P = 0.002, respectively; Tables 3 and 4). MCSM-L was not associated with risk of CRC without family history of cancer (P = 0.225; Table 4).
The population attributable risk percentage (PAR%) of MCSM on the risk of CRC was 26.1%.
Interaction between the methylation of individual genes, MCSM and environmental factors on the risk of CRC
Antagonistic effects of ADAMTS9 hypermethylation and consumption of stewed fish with brown sauce (≥1 times/week) on the risk of CRC were observed (OR = 0.50, 95% CI: 0.25–0.99, P = 0.046; Supplementary Data Table S10) compared with unmethylated ADAMTS9 and consumption of stewed fish with brown sauce (<1 times/week).
Significant synergistic effects between AOX-1 hypermethylation and intake of cereals (≥100 g/week) and age (older than 60 yrs) on risk of CRC were observed (OR = 1.82, 95% CI: 1.01–3.26, P = 0.045 and OR = 3.21, 95% CI: 1.83–5.64, P = 0.00, respectively; Table 5 and Supplementary Data Table S5).
Table 5. Effects of combination and interaction between age and methylation of genes, MCSM on the risk of CRC.
| Age |
||||
|---|---|---|---|---|
| <60 | ≥60 | Interaction |
||
| ORega (95% CI) | ORia (95% CI) | P | ||
| FOXE-1 | ||||
| Unmethylation | 1.00 | 1.28 (1.12–1.48) | ||
| Methylation | 1.11 (0.91–1.35) | 2.23 (1.85–2.70) | 1.57 (0.84–2.94) | 0.16 |
| IRF4 | ||||
| Unmethylation | 1.00 | 1.40 (1.24–1.59) | ||
| Methylation | 10.66 (5.43–20.95) | 41.76 (17.00–102.59) | 2.80 (0.23–34.61) | 0.42 |
| ADAMTS9 | ||||
| Unmethylation | 1.00 | 1.25 (1.08–1.45) | ||
| Methylation | 1.54 (1.28–1.86) | 2.87 (2.39–3.45) | 1.49 (0.82–2.72) | 0.19 |
| AOX1 | ||||
| Unmethylation | 1.00 | 0.83 (0.70–0.98) | ||
| Methylation | 1.03 (0.87–1.22) | 2.75 (2.32–3.25) | 3.21 (1.83–5.64) | 0.00 |
| RERG | ||||
| Unmethylation | 1.00 | 1.02 (0.88–1.19) | ||
| Methylation | 1.36 (1.14–1.62) | 3.68 (3.06–4.43) | 2.65 (2.07–3.38) | 0.00 |
| RARB2 | ||||
| Unmethylation | 1.00 | 1.51 (1.31–1.74) | ||
| Methylation | 0.99 (0.82–1.19) | 1.46 (1.21–1.78) | 1.02 (0.79–1.32) | 0.87 |
| MCSM | ||||
| Unmethylation | 1.00 | 0.66 (0.52–0.83) | ||
| Methylation | 1.01 (0.85–1.19) | 1.98 (1.66–2.35) | 2.99 (1.60–5.59) | 0.00 |
aAdjusted for BMI, occupation and family history of cancer.
For RARB2 hypermethylation, antagonistic interactions with pungent food (≥4 times/week), or intake of food overnight (≥3 times/week) were observed (OR = 0.52, 95% CI: 0.28–0.99, P = 0.047 and OR = 0.48, 95% CI: 0.25–0.93, P = 0.03, respectively; Supplementary Data Table S13 and S15).
Additionally, a synergistic effect between RERG hypermethylation and age (older than 60 yrs) on risk of CRC was observed (OR = 2.65, 95% CI: 2.07–3.38, P = 0.00; Table 5).
Furthermore, MCSM showed a statistically significant synergistic interaction with age (older than 60 yrs; OR = 2.99, 95% CI: 21.60–5.59, P = 0.00; Table 5).
The models assessing the association between the six genes, MCSM and environmental factors on CRC were presented in Tables 5 and Supplementary Data Table S5–S16.
Association between the methylation of individual genes, MCSM and CRC prognosis
At the end of the follow-up time (109 months), 113/256 (44.3%) of the patients were confirmed to be alive and 113/256 (44.3%) deaths had occurred. The mean overall survival (OS) of CRC patients was 59.2 months. Multiple Cox regression analysis suggested that Dukes staging, preoperative CA19-9 level and intestinal anastomosis were significantly associated with prognosis of CRC (Supplementary Data Table S3). However, no statistically significant association between the methylation status of the six genes, MCSM and the prognosis of CRC were observed either before or after adjusting for Dukes staging, preoperative carbohydrate antigen 19-9 (CA19-9) level and intestinal anastomosis (Supplementary Data Table S4).
Discussion
DNA methylation occurs in genomic regions rich in cytosine and guanosine (CG) dinucleotides, called CpG islands. This is known to be a well-characterized event in tumor biology and has been extensively documented in CRC tissues24,34,35. Aberrant DNA hypermethylation of specific genes may result in abnormal expression in normal cells, especially at tumor suppressor genes, whose hypermethylation is related to the down-regulation of gene expression, leading to the proliferation and differentiation of tumor cells. Therefore, we proposed that DNA hypermethylation in leukocytes in the peripheral blood may be used as a biomarker of CRC, as described previously36. Ally et al. studied the relationship between estrogen receptor gene methylation in leukocytes and normal colonic tissue DNA in subjects with and without colorectal neoplasia, and found that methylation in leukocytes was 60% lower than that in normal colonic tissue (P < 0.001)37. To explore the association between methylation status in leukocyte DNA and CRC, Gao et al. found 31 CpG sites that were significantly related to CRC risk, especially at two sites in the DSP gene in male smokers38. The candidate biomarkers involved in our studies have been shown to be involved in multiple molecular events associated with tumorigenesis, among which IRF4 hypermethylation associated with the highest risk of CRC (OR = 16.96), while the lowest risk was seen for FOXE-1 hypermethylation (OR = 1.35). Next, we defined a gene panel to assess methylation called MCSM, which included 5 candidate genes, and we found that people with MCSM hypermethylation were 1.54 times more susceptible to CRC compared with non-MCSM hypermethylation (P = 0.004). The high PAR% (26.1%) of MCSM hypermethylation suggests that these methylated genes play a key role in CRC risk.
Dietary factors are commonly recognized as modifiable factors that profoundly influence cancer and tumor behavior39, accounting for 70% the risk of CRC. To date, many researchers have reported that dietary factors and lifestyle not only play a crucial role in tumorigenesis, but are also associated with inducing epigenetic changes40. Therefore, we explored the interaction of dietary factors exposure and peripheral blood DNA methylation level on susceptibility to CRC.
Although the antagonistic interactions between increased vegetable intake (≥100 g/day) and individual genes, MCSM showed no statistical significance, the combinations showed a protective tendency on risk of CRC. Folic acid, rich in vegetables, plays an important role in the provision of methyl moieties that are used to synthesize S-adenosyl methionine, which is the universal methyl donor for DNA methylation41. Inadequate folate availability during cell division can result in the compromised production of thymidine, such that uracil may be substituted in the DNA sequence, which may trigger repair attempts that increases the frequency of chromosomal breaks42. Moreover, methionine, as the substrate for SAM and human essential amino acids, is also critical for maintaining the flux of methyl groups for rementhylation41. Therefore, low dietary intake levels with long-term methionine and folate deficiency could result in methionine cycle disorder, which can be confused with DNA methylation, leading normally non-methylated DNA situation to be aberrantly hypermethylated43. In addition, we observed antagonistic effects between ADAMTS9 hypermethylation and consumption of stewed fish with brown sauce, likely due to the abundant methionine found in fish.
Carcinogenic N-nitroso compounds, which could induce mutations by alkylating DNA and thus activating oncogenes, can be found in food left overnight and are endogenously formed after ingesting red meat in the intestines with the help of the colonic flora44,45. In vivo studies in rat livers showed alterations of DNA methylation patterns by N-Nitrosodimethylamine (NDMA) and it was found that 6.6% of O6-methyl-guanine-O6 meG (O6) position conferred a high mutagenic and carcinogenic susceptibility46, which suggested the possibility that the interaction between eating a lot of food left overnight and RARB2 hypermethylation observed in our study was associated with the risk of CRC.
Some of the effects of the qualitative and quantitative aspects of fat intake have imputed to be a modification of the transcription of key genes involved in pathways related to lipid and glucose metabolism47. Abundant amounts of AOX1 have been observed in adipose tissue and were proposed to play a critical role in adipogenesis and lipid metabolism by modulating peroxisome proliferator-activated receptor-α48,49, which may provide a basis for explaining why fat intake showed an antagonistic interaction with AOX-1 hypermethylation on the risk of CRC. However, the molecular mechanisms relating dietary factors and DNA hypermethylation to effects on tumor carcinogenic process are very complex, and further studies are needed.
No significant relationships were observed between the methylation status of individual genes, MCSM and prognosis of CRC. It has been shown that aberrant hypermethylation of RARB2 in tumor tissue was associated with a shortened OS in 73 CRC tissues50. A meta-analysis showed that CIMP was independently associated with significantly worse prognosis in CRC patients51, suggesting that the mechanism by which CIMP from CRC tissues and MCSM from peripheral blood cells influences CRC prognosis may be different.
Although we did not validate the results of methylation-sensitive high-resolution melting analysis (MS-HRM) with another technique in this study, we did test DAPK-1 and MLH-1 methylation status using MS-HRM, validated with pyrosequecing in our previous study. For DAPK-1, the sensitivity was 89.74% and the specificity was 72.34%. For MLH-1, the sensitivity was 100% and the specificity was 78.69%. This suggests that using MS-HRM to test methylation status was stable and the results were credible.
There were several limitations in our study. First, recall bias may be inevitable for the collection of information on environmental factors, although we did our best to minimize this bias. Second, the collection of information about dietary intake without detailed amounts may limit our power to detect gene-dietary interactions more precisely. Until now, it has not been clear how DNA methylation produces effects on gene expression and to what degree specific DNA methylation can lead to changes in gene expression, eventually resulting in changes in individual susceptibility to diseases.
In summary, our study suggests that both individual gene methylation of IRF4, FOXE-1, AOX-1, ADAMTS9 and RERG as well as MCSM in peripheral blood leukocytes are associated with increased risk of CRC but are not associated with the prognosis of CRC. Hypermethylation variability detected in leukocytes may interact with dietary factors and affect susceptibility to CRC. Additional large-scale studies will be required to validate these observations. Screening additional candidate genes using a more systematic method and synthesizing additional experimental results for further study of MCSM biomarkers in peripheral blood from CRC cases are needed.
Materials and Methods
Study population
We carried out this study after obtaining informed written consent from study subjects and approval from the Human Research and Ethics Committee of Harbin Medical University. All experiments including all relevant details were performed in accordance with relevant guidelines and regulations. We identified 421 CRC patients who underwent surgery at the Cancer Hospital (from June 1, 2004 to May 15, 2005, and May 15, 2007 to January 1, 2008) and the second Affiliated Hospital of Harbin Medical University (from October 15, 2010 to June 15, 2011). All patients were diagnosed based on pathology. Patients with neuroendocrine carcinomas, malignant melanomas, non-Hodgkin’s lymphoma, gastrointestinal stromal tumors, metastatic CRC, and Lynch syndrome CRC were excluded. The 506 control subjects were selected from the Departments of Orthopedics and Ophthalmology at the Second Affiliated Hospital of Harbin Medical University during the same period. Patients with gastrointestinal disease or CRC history were excluded. Samples of peripheral blood (5 ml) were collected and stored at −80 °C immediately. Each participant was interviewed in person by a well-trained interviewer using a structured questionnaire according to the dietary habits in north China and a nutrition survey from the Chinese National Survey Data Archive with information of on demographic characteristics (age, gender, height and weight, education and occupation), lifestyle factors (use of tobacco, alcohol and smoking) and family history of cancer as well as dietary status during the past 12 months.
Postoperatively, we carried out a cohort study within the 256 patients from the Cancer Hospital. The patients were followed at 3–6-month intervals for the first year after resection, and annually thereafter. Clinical information about Dukes stage, chemotherapy, histological and pathological types, the level of serum carcinoembryonic antigen (CEA) and CA19-9 before surgery were collected from medical records. OS that was defined as the time from surgery to the patient’s death or the last follow-up visit was used as a measure of prognosis. The date of the last follow-up was March 15, 2014 (the 109th month). The date and cause of death of CRC patients were validated. The rate of loss to follow-up was 11.4% (29/256).
Genomic DNA extraction and sodium bisulfite modification
Genomic DNA was successfully extracted from the peripheral blood samples of 421 CRC patients and 506 controls using the QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany) and was stored at −80 °C. DNA quantity was measured using the Nanodrop 2000 Spectrophotometer (Thermo Scientific).
Bisulfite treatment was used to transform unmethylated cytosine nucleotides into thymidine without changing methylated cytosines. Genomic DNA was chemically modified with a sodium bisulfite modification kit (EpiTect Fast DNA Bisulfite Kit, Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The bisulfate- modified DNA was stored at −20 °C until use.
Analysis of the methylation status of candidate genes
Methylation-sensitive high-resolution melting analysis (MS-HRM) was performed on a LightCycler480 machine (Roche Applied Science, Mannheim, Germany) as previously described36. The primers are listed in Table 6. HRM was performed in a 10 μl volume system consisting of 1X LightCycler480 High Resolution Melting Master Mix (Roche), 200 nmol/l of each primer, and 5 ng of a sodium bisulfite-modified DNA template. The final MgCl2 concentration was adjusted to 3 mmol/l. The PCR amplification protocol consisted of denaturation for 10 min at 95 °C for 1 cycle, denaturation for 10 s at 95 °C, annealing for 30 s with a touchdown of each primer annealing temperature and extension for 30 s at 72 °C for 45 cycles. The HRM melting protocol then consisted of 95 °C for 1 min, 40 °C for 1 min, 74 °C for 5 s and continuous acquisition to 90 °C at 25 acquisitions per 1 °C (LightCycler480, Roche, Mannheim, Germany).
Table 6. Primers for MS-HRM.
| Gene | Primer sequences (5′-3′) | Location | Number of CpG-sites/Length of amplified fragment | Annealing temperature (°C) |
|---|---|---|---|---|
| AOX-1 | F: GAACGTTGGATTTTAATTAAGGTTTTTR: CTAAAAAATAACGAACACCTAAAACC | 2q33 | 8/124 | 64–56 |
| ADAMTS9 | F: ATTTGGTCGAGATGGGGAGTTR: CTACGAAATACCCCTACCCAA | 3p14 | 16/151 | 62–56 |
| FOXE-1 | F: GATTTTTTAGTGAATGGTTAGGGR: CTACTATCCCTAAACCGAAACTT | 9q22 | 6/96 | 68–58 |
| IRF4 | F: CGTTGTAGTTTAGTGATTGATTGR: GCTTCGAAAACTATCACTAAAAC | 6p25.3 | 8/111 | 62–56 |
| RARB2 | F: CGAGTTGTTTGAGGATTGGGATGTR: AATACGTTCCGAATCCTACCCC | 3p24 | 7/89 | 66–56 |
| RERG | F: CGGTTTTGGTCGGGTTTAGTTR: CGCAAAAACAAATACCAATAACC | 12p12 | 11/107 | 64–56 |
Universal methylated (100% methylated) and unmethylated (0% methylated) human whole genomic DNA samples (Zymo Research) were used as a calibrator and positive control, respectively. DNA-free distilled water was used as a negative control. A series of methylated DNA, including 100%, 5%, 1%, 0.5% and 0% on a background of universal unmethylated DNA were constructed as standard curves by serially diluting the methylated control DNA into the unmethylated control according to mass concentration (Figs 1, 2, 3, 4, 5, 6).
Figure 1. Profile of fluorescence obtained at the melting temperature for serial dilutions of methylated DNA (5%, 1%, and 0%) in ADAMTS9 gene.
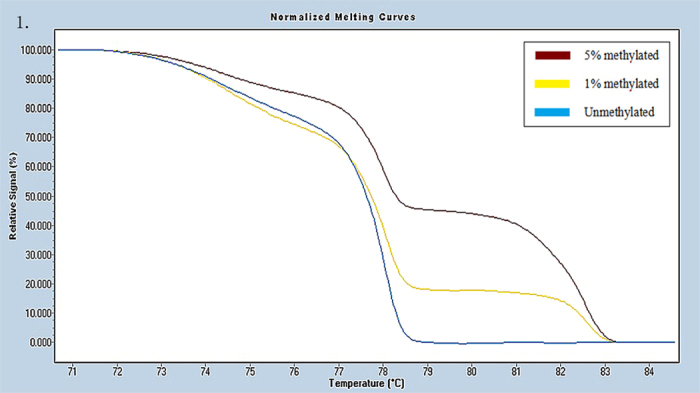
Figure 2. Profile of fluorescence obtained at the melting temperature for serial dilutions of methylated DNA (5%, 1%, 0.5% and 0%) in AOX-1 gene.
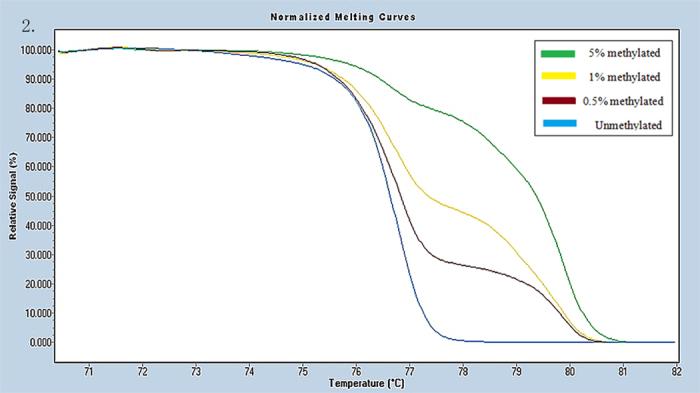
Figure 3. Profile of fluorescence obtained at the melting temperature for serial dilutions of methylated DNA (100%, 5%, 1%, and 0%) in FOXE-1 gene.
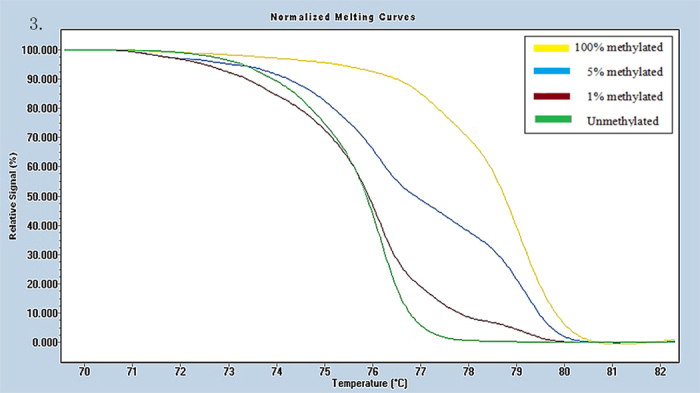
Figure 4. Profile of fluorescence obtained at the melting temperature for serial dilutions of methylated DNA (5%, 1%, 0.5% and 0%) in IRF4 gene.
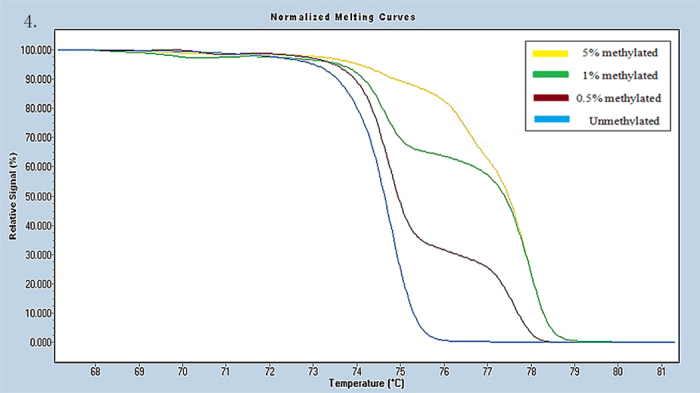
Figure 5. Profile of fluorescence obtained at the melting temperature for serial dilutions of methylated DNA (5%, 1%, 0.5% and 0%) in RARB2 gene.
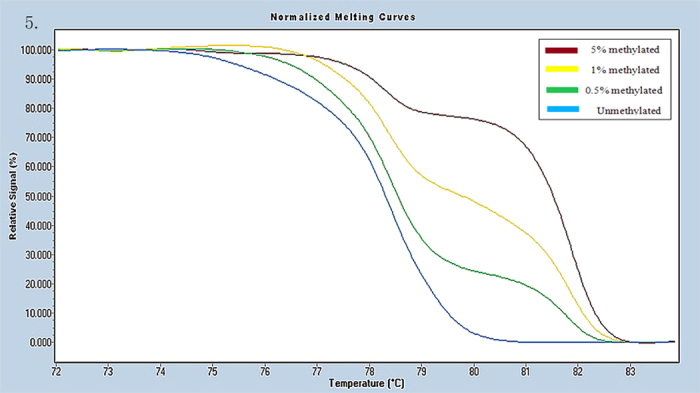
Figure 6. Profile of fluorescence obtained at the melting temperature for serial dilutions of methylated DNA (5%, 1%, 0.5% and 0%) in RERG gene.
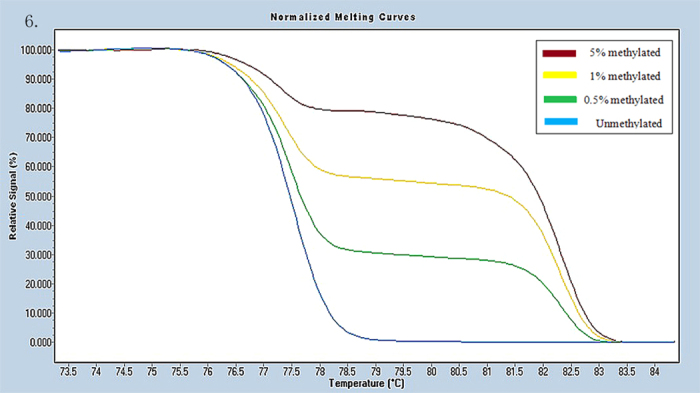
HRM data were analyzed using Gene Scanning Software (version 2.0). Data processing included normalization and temperature shifting using a LightCycler480. The methylation status of the candidate genes was determined by comparing the curves of each sample to the series of standard dilutions in the gene scanning module by two independent observers. Disagreements were settled by consensus or a third opinion.
Statistical analyses
Continuous variables such as age were analyzed by two-sample t-tests. The statistical significance of the association between categorical variables was assessed with chi-square (χ2) tests. We determined 0% as the cutoff value of different methylation status for comparisons between CRC patients and controls. Univariate and multivariate logistic regression analyses were used to assess if relationships existed between the methylation status of AOX-1, ADAMTS9, FOXE-1, IRF4, RARB2 and RERG, as well as MCSM and CRC. Multiple imputation was applied to generate possible values for missing values of dietary factors by creating several “complete” sets of data. We selected the pooled output of each “complete” dataset analysis for subsequent analyses. Multivariate logistic regression analysis and crossover analysis were performed to evaluate the role of candidate genes and MCSM methylation status in interaction with and in combination with environmental factors on the risk of CRC with 4 types of ORs: one comparing exposed and unexposed subjects to the environmental risk factor (ORe); one for the association between gene methylation and risk of CRC (ORg); one for combining the effects of both gene hypermethylation and environmental factos (OReg); and one for the interaction between DNA methylation and environmental factors (ORi = OReg/OReORg). The reference groups consisted of subjects without exposure to the environmental factors or gene methylation. OS was estimated using the life table method. Survival curves were constructed using the Kaplan-Meier method. The Cox regression model was performed to compute the hazard ratio (HR) and 95% CI for the potential prognostic factors. The population attributable risk percentage (PAR %) was calculated by Pexposed (OR − 1)/[1 + Pexposed(OR − 1)]. All statistical analyses were performed using SAS 9.2 software (SAS Institute, Cary, NC, USA). The results were considered significantly different if statistical tests produced a two-tailed P-value of < 0.05.
Additional Information
How to cite this article: Luo, X. et al. Methylation of a panel of genes in peripheral blood leukocytes is associated with colorectal cancer. Sci. Rep. 6, 29922; doi: 10.1038/srep29922 (2016).
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China (NSFC 30972539, 30671801 and 81473055).
Footnotes
Author Contributions Y.Z. and B.C. conceived the hypothesis. R.H. designed the primers of candidate genes. X.L., H.S., J.L. and H.Y. performed the experiments. J.S. and S.L. performed the questionnaire interview. H.B. and X.L. performed subsequent data analysis. Y.L. participated in helpful discussions. X.L. wrote the manuscript with help from R.H. Y.Z. revised the manuscript.
References
- Grady W. M. CIMP and colon cancer gets more complicated. Gut 56, 1498–1500, 10.1136/gut.2007.125732 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa J. P. CpG island methylator phenotype in cancer. Nature reviews. Cancer 4, 988–993, 10.1038/nrc1507 (2004). [DOI] [PubMed] [Google Scholar]
- Berger S. L., Kouzarides T., Shiekhattar R. & Shilatifard A. An operational definition of epigenetics. Genes & development 23, 781–783, 10.1101/gad.1787609 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. et al. Hypermethylation of EDNRB promoter contributes to the risk of colorectal cancer. Diagnostic pathology 8, 199, 10.1186/1746-1596-8-199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H. Z. et al. Detection of aberrant p16 methylation in the serum of colorectal cancer patients. Clinical cancer research: an official journal of the American Association for Cancer Research 8, 188–191 (2002). [PubMed] [Google Scholar]
- Miotto E. et al. Frequent aberrant methylation of the CDH4 gene promoter in human colorectal and gastric cancer. Cancer research 64, 8156–8159, 10.1158/0008-5472.CAN-04-3000 (2004). [DOI] [PubMed] [Google Scholar]
- Herbst A. et al. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. The American journal of gastroenterology 106, 1110–1118, 10.1038/ajg.2011.6 (2011). [DOI] [PubMed] [Google Scholar]
- Grady W. M., Rajput A., Lutterbaugh J. D. & Markowitz S. D. Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer research 61, 900–902 (2001). [PubMed] [Google Scholar]
- Leung W. K. et al. Quantitative detection of promoter hypermethylation in multiple genes in the serum of patients with colorectal cancer. The American journal of gastroenterology 100, 2274–2279, 10.1111/j.1572-0241.2005.50412.x (2005). [DOI] [PubMed] [Google Scholar]
- Lee B. B. et al. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clinical cancer research: an official journal of the American Association for Cancer Research 15, 6185–6191, 10.1158/1078-0432.CCR-09-0111 (2009). [DOI] [PubMed] [Google Scholar]
- Ongenaert M. et al. PubMeth: a cancer methylation database combining text-mining and expert annotation. Nucleic acids research 36, D842–D846, 10.1093/nar/gkm788 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunau C., Renault E., Rosenthal A. & Roizes G. MethDB–a public database for DNA methylation data. Nucleic acids research 29, 270–274 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. et al. MethyCancer: the database of human DNA methylation and cancer. Nucleic acids research 36, D836–D841, 10.1093/nar/gkm730 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand S. H., Orntoft T. F. & Sorensen K. D. Prognostic DNA methylation markers for prostate cancer. International journal of molecular sciences 15, 16544–16576, 10.3390/ijms150916544 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain S. et al. Signalling with retinoids in the human lung: validation of new tools for the expression study of retinoid receptors. BMC cancer 9, 423, 10.1186/1471-2407-9-423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlin B. S. et al. RERG is a novel ras-related, estrogen-regulated and growth-inhibitory gene in breast cancer. The Journal of biological chemistry 276, 42259–42267, 10.1074/jbc.M105888200 (2001). [DOI] [PubMed] [Google Scholar]
- Zhang C. et al. High-resolution melting analysis of ADAMTS9 methylation levels in gastric, colorectal, and pancreatic cancers. Cancer genetics and cytogenetics 196, 38–44, 10.1016/j.cancergencyto.2009.08.016 (2010). [DOI] [PubMed] [Google Scholar]
- Forero A., McCormick K. D., Jenkins F. J. & Sarkar S. N. Downregulation of IRF4 induces lytic reactivation of KSHV in primary effusion lymphoma cells. Virology 458–459, 4–10, 10.1016/j.virol.2014.04.020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadia C. et al. FOXE1 and SYNE1 genes hypermethylation panel as promising biomarker in colitis-associated colorectal neoplasia. Inflammatory bowel diseases 20, 271–277, 10.1097/01.MIB.0000435443.07237.ed (2014). [DOI] [PubMed] [Google Scholar]
- Oster B. et al. Identification and validation of highly frequent CpG island hypermethylation in colorectal adenomas and carcinomas. International journal of cancer. Journal international du cancer 129, 2855–2866, 10.1002/ijc.25951 (2011). [DOI] [PubMed] [Google Scholar]
- Haldrup C. et al. DNA methylation signatures for prediction of biochemical recurrence after radical prostatectomy of clinically localized prostate cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 31, 3250–3258, 10.1200/JCO.2012.47.1847 (2013). [DOI] [PubMed] [Google Scholar]
- Yamashita M. et al. DNA methylation of interferon regulatory factors in gastric cancer and noncancerous gastric mucosae. Cancer science 101, 1708–1716, 10.1111/j.1349-7006.2010.01581.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara D. et al. Novel methylation panel for the early detection of colorectal tumors in stool DNA. Clinical colorectal cancer 9, 168–176, 10.3816/CCC.2010.n.023 (2010). [DOI] [PubMed] [Google Scholar]
- Chan A. O. et al. CpG island methylation in aberrant crypt foci of the colorectum. The American journal of pathology 160, 1823–1830, 10.1016/S0002-9440(10)61128-5 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 28, 605–613, 10.1200/JCO.2009.23.4781 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breda S. G., van Delft J. H., Engels L. G., Kleinjans J. C. & Mathers J. C. Methylation status of CpG islands in the promoter region of genes differentially expressed in colonic mucosa from adenoma patients and controls in response to altered vegetable intake. The British journal of nutrition 101, 1295–1299 (2009). [DOI] [PubMed] [Google Scholar]
- Slattery M. L. et al. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. International journal of cancer. Journal international du cancer 120, 656–663, 10.1002/ijc.22342 (2007). [DOI] [PubMed] [Google Scholar]
- Limsui D. et al. Cigarette smoking and colorectal cancer risk by molecularly defined subtypes. Journal of the National Cancer Institute 102, 1012–1022, 10.1093/jnci/djq201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit C. & Christensen B. Blood-derived DNA methylation markers of cancer risk. Advances in experimental medicine and biology 754, 233–252, 10.1007/978-1-4419-9967-2_12 (2013). [DOI] [PubMed] [Google Scholar]
- Anjum S. et al. A BRCA1-mutation associated DNA methylation signature in blood cells predicts sporadic breast cancer incidence and survival. Genome medicine 6, 47, 10.1186/gm567 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S. & Goel A. Molecular classification and correlates in colorectal cancer. The Journal of molecular diagnostics: JMD 10, 13–27, 10.2353/jmoldx.2008.070082 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady W. M. & Carethers J. M. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 135, 1079–1099, 10.1053/j.gastro.2008.07.076 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho C. et al. Epigenetic inactivation of the BRCA1 interactor SRBC and resistance to oxaliplatin in colorectal cancer. Journal of the National Cancer Institute 106, djt322, 10.1093/jnci/djt322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. S. & Deng G. Epigenetic changes (aberrant DNA methylation) in colorectal neoplasia. Gut and liver 1, 1–11, 10.5009/gnl.2007.1.1.1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer research 60, 4366–4371 (2000). [PubMed] [Google Scholar]
- Wojdacz T. K., Dobrovic A. & Hansen L. L. Methylation-sensitive high-resolution melting. Nature protocols 3, 1903–1908, 10.1038/nprot.2008.191 (2008). [DOI] [PubMed] [Google Scholar]
- Ally M. S., Al-Ghnaniem R. & Pufulete M. The relationship between gene-specific DNA methylation in leukocytes and normal colorectal mucosa in subjects with and without colorectal tumors. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 18, 922–928, 10.1158/1055-9965.EPI-08-0703 (2009). [DOI] [PubMed] [Google Scholar]
- Gao Y. et al. Leukocyte DNA methylation and colorectal cancer among male smokers. World journal of gastrointestinal oncology 4, 193–201, 10.4251/wjgo.v4.i8.193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman M. The second World Cancer Research Fund/American Institute for Cancer Research expert report. Food, nutrition, physical activity, and the prevention of cancer: a global perspective. The Proceedings of the Nutrition Society 67, 253–256, 10.1017/S002966510800712X (2008). [DOI] [PubMed] [Google Scholar]
- Jaenisch R. & Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics 33 Suppl, 245–254, 10.1038/ng1089 (2003). [DOI] [PubMed] [Google Scholar]
- Crider K. S., Yang T. P., Berry R. J. & Bailey L. B. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role. Advances in nutrition 3, 21–38, 10.3945/an.111.000992 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht S. A. & Lipkin M. Chemoprevention of colon cancer by calcium, vitamin D and folate: molecular mechanisms. Nature reviews. Cancer 3, 601–614, 10.1038/nrc1144 (2003). [DOI] [PubMed] [Google Scholar]
- Zhu K. et al. Methyl-group dietary intake and risk of breast cancer among African-American women: a case-control study by methylation status of the estrogen receptor alpha genes. Cancer causes & control: CCC 14, 827–836 (2003). [DOI] [PubMed] [Google Scholar]
- Bingham S. A. et al. Does increased endogenous formation of N-nitroso compounds in the human colon explain the association between red meat and colon cancer? Carcinogenesis 17, 515–523 (1996). [DOI] [PubMed] [Google Scholar]
- Lijinsky W. N-Nitroso compounds in the diet. Mutation research 443, 129–138 (1999). [DOI] [PubMed] [Google Scholar]
- Chikan N. A., Shabir N., Shaff S., Mir M. R. & Patel T. N. N-nitrosodimethylamine in the Kashmiri diet and possible roles in the high incidence of gastrointestinal cancers. Asian Pacific journal of cancer prevention: APJCP 13, 1077–1079 (2012). [DOI] [PubMed] [Google Scholar]
- Kremmyda L. S., Tvrzicka E., Stankova B. & Zak A. Fatty acids as biocompounds: their role in human metabolism, health and disease: a review. part 2: fatty acid physiological roles and applications in human health and disease. Biomedical papers of the Medical Faculty of the University Palacky, Olomouc, Czechoslovakia 155, 195–218, 10.5507/bp.2011.052 (2011). [DOI] [PubMed] [Google Scholar]
- Neumeier M. et al. Aldehyde oxidase 1 is highly abundant in hepatic steatosis and is downregulated by adiponectin and fenofibric acid in hepatocytes in vitro. Biochemical and biophysical research communications 350, 731–735, 10.1016/j.bbrc.2006.09.101 (2006). [DOI] [PubMed] [Google Scholar]
- Weigert J. et al. Small-interference RNA-mediated knock-down of aldehyde oxidase 1 in 3T3-L1 cells impairs adipogenesis and adiponectin release. FEBS letters 582, 2965–2972, 10.1016/j.febslet.2008.07.034 (2008). [DOI] [PubMed] [Google Scholar]
- Miladi-Abdennadher I. et al. Hypermethylation of RARbeta2 correlates with high COX-2 expression and poor prognosis in patients with colorectal carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine 31, 503–511, 10.1007/s13277-010-0063-3 (2010). [DOI] [PubMed] [Google Scholar]
- Juo Y. Y. et al. Prognostic value of CpG island methylator phenotype among colorectal cancer patients: a systematic review and meta-analysis. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO 25, 2314–2327, 10.1093/annonc/mdu149 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


