Abstract
MicroRNAs (miRNAs) have been reported to be involved in many neurodegenerative diseases. The present study focused on the role of hsa-miR-144-3p in one of the neurodegenerative diseases, Parkinson’s disease (PD). Our study showed a remarkable down-regulation of miR-144-3p expression in 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine (MPTP)-treated SH-SY5Y cells. MiR-144-3p was then overexpressed and silenced in human SH-SY5Y cells by miRNA-mimics and miRNA-inhibitor transfections, respectively. Furthermore, β-amyloid precursor protein (APP) was identified as a target gene of miR-144-3p via a luciferase reporter assay. We found that miR-144-3p overexpression significantly inhibited the protein expression of APP. Since mitochondrial dysfunction has been shown to be one of the major pathological events in PD, we also focused on the role of miR-144-3p and APP in regulating mitochondrial functions. Our study demonstrated that up-regulation of miR-144-3p increased expression of the key genes involved in maintaining mitochondrial function, including peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α), nuclear respiratory factor 1 (NRF-1) and mitochondrial transcription factor A (TFAM). Moreover, there was also a significant increase in cellular ATP, cell viability and the relative copy number of mtDNA in the presence of miR-144-3p overexpression. In contrast, miR-144-3p silencing showed opposite effects. We also found that APP overexpression significantly decreased ATP level, cell viability, the relative copy number of mtDNA and the expression of these three genes, which reversed the effects of miR-144-3p overexpression. Taken together, these results show that miR-144-3p plays an important role in maintaining mitochondrial function, and its target gene APP is also involved in this process.
Keywords: amyloid precursor protein, hsa-miR-144-3p, mitochondrial functions, Parkinson’s disease
INTRODUCTION
Parkinson’s disease (PD) affects 1–2% of the world’s population and has been regarded as the second most common neurodegenerative disease (de Rijk et al., 1995), first reported by James Parkinson in 1817 (Lees, 2007). Morbidity is reported to be about 1% in people over 65 years old, and significantly increases to 4% for those over 80 years old (de Rijk et al., 1995). The main symptoms of PD are usually characterized by the presence of resting tremor, bradykinesia and rigidity, pathologically mainly due to the progressive loss of dopaminergic neurons in the substantia nigra pars compacta (SN) (Christopher et al., 2015) and the presence of Lewy bodies and neurites (Nussbaum and Ellis, 2003), causing a remarkable reduction of dopamine (DA), which leads to an imbalance between DA and acetyl choline (ACH) in the SN. This imbalance may lead to cell dysfunction and neuron death. The pathogenesis of PD is still not fully clear and there is no cure.
Neurons are energy-consumptive due to their quick response to surroundings and stimuli, and require a mass of mitochondria to produce plenty of adenosine triphosphate (ATP). Hence, mitochondrial homeostasis is extremely important for neurons, and mitochondrial dysfunction could promote neuronal dysfunction and degeneration (Exner et al., 2012). As a result, mitochondrial dysfunction is a main pathological event and manifestation of PD. In particular, there is clear anatomical evidence that PD brains, particularly in the SN, have consistently showed mitochondrial dysfunction (Owen et al., 1996; Schapira, 1995; 2009). MPTP is a kind of positively charged compound that may affect the process of oxidative phosphorylation in mitochondria (Tsou et al., 2015), and ultimately lead to the over-consumption of ATP and cell death (Radad et al., 2015). Hence, MPTP is a contaminator of mitochondrial functions. It is feasible to mimic PD with the treatment of MPTP because the main symptoms of PD are similar to the effects of MPTP (Liu et al., 2015).
MicroRNAs (miRNAs) are a group of small, single-stranded and non-coding RNAs which have been identified as post-transcriptional regulators of gene expression (Graves and Zeng, 2012). Previous studies have demonstrated that miRNAs are pathologically altered in the progress of neurodegenerative diseases (Tan and Yu, 2014). For example, Vallelunga et al. (2014) observed that miR-339-5p was down-regulated, but miR-223, miR-324-3p and miR-24 were up-regulated in PD tissues, which may be regarded as specific diagnosis biomarkers of PD. Burgos et al. (2014) investigated the miRNAs in the cerebrospinal fluid and serum, and found that 13 novel miRNAs could be used to assess disease progression and therapeutic efficacy. In addition, miR-34b/c expression was identified to be decreased in brain samples from PD patients compared with healthy samples (Minones-Moyano et al., 2011). However, to our knowledge the role of miR-144-3p in PD has not been elucidated. In addition, the bioinformatic analysis in our study showed that APP was a target gene of hsa-miR-144-3p, which needs further verification in the present study.
The β-Amyloid precursor protein (APP) is a kind of trans-membrane protein, which is widely expressed in various tissues (Westmark, 2013). In particular, APP can be cleaved by membrane-associated proteases into two segments: sAPPα and sAPPβ. sAPPβ can be further cleaved by γ-secretase into amyloid-β (Aβ) peptides. Aβ was shown to induce oxidative stress and mitochondrial dysfunction in MC-65 and SH-SY5Y cells (Gray et al., 2015). Aggregation of Aβ peptides forms extracellular amyloid plaques, which is a major histopathological hallmark of Alzheimer’s disease (AD); in other words, Alzheimer’s-associated APP facilitates neurodegeneration (Duce et al., 2010; McCarthy et al., 2014). It is worth noting that AD pathology is found in a significant percentage of patients with PD (Alves et al., 2010). Thus, APP is supposed to play a role in the pathogenesis of PD contributing to the death of DA neurons. We indeed learned from some papers on the role of APP in PD (another kind of neurodegenerative disease). Ayton et al. (2015) reported markedly decreased APP expression in dopaminergic neurons of human PD nigra, and that loss of APP promoted neurodegeneration in PD; conversely, APP-overexpressing mice are protected in the PD model. Schulte et al. (2015) also focused on the role of APP in modifying the PD phenotype that contributed to the development of dementia in individuals with PD. In addition, Aβ was also reported to be involved in PD dementia due to its plaques in brain tissues (Irwin et al., 2013). However, the potential role of APP in PD remains poorly elucidated and needs further investigation.
In the present study, we have further confirmed the role of miR-144-3p in the PD model of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-treated SH-SY5Y cells. The results showed that miR-144-3p was also significantly down-regulated in MPTP-treated SH-SY5Y cells. Moreover, miR-144-3p was confirmed to be involved in regulating mitochondrial functions, along with its target gene APP. To be specific, miR-144-3p overexpression could inhibit mitochondrial dysfunction induced by MPTP treatment in SH-SY5Y cells; on the contrary, APP overexpression counteracted the effects of miR-144-3p, both of which may be promising therapeutic targets for PD.
MATERIALS AND METHODS
Cell culture
Human SH-SY5Y cells purchased from the American Type Culture Collection (ATCC, USA) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Hyclone, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone) in an atmosphere of 37°C with 5% CO2. The cell cultures were rinsed with pre-warmed phosphate-buffered saline (PBS) before they were dissociated using Trypsin-EDTA Solution (Hyclone).
Cell treatment protocol
SH-SY5Y cells were treated with 300 μM of MPTP (Sigma-Aldrich, USA) for 12 h to mimic PD. The transfection was performed prior to the treatment with MPTP.
MiR-144-3p target prediction
The putative target gene of miR-144-3p was predicted using TargetScan (http://www.targetscan.org/) and microRNA.org (http://www.microrna.org/microrna/home.do/). The interaction between the mRNA 3′-untranslated region (UTR) of the predicted gene and miR-144-3p was analyzed by RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/).
MiR-144-3p mimics and inhibitor
For the manual alteration of miR-144-3p expression, negative control miRNA (NC miRNA), miRNA-mimics and miRNA-inhibitor (GenePharma, China) were transfected into SH-SY5Y cells using Lipofectamine 2000 Reagent (Invitrogen, USA) according to the manufacturer’s instructions.
Dual-luciferase reporter assay
The possible binding sites of miR-144-3p in the 3′-UTR of APP mRNA were predicted by RNAhybrid. Firstly, complementary DNA (cDNA) fragments containing the predicted miR-144-3p binding sites were amplified and subcloned into pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, USA; named pmirGLO-APP). The control plasmids were also constructed using cDNA fragments containing mutated nucleotides instead of some miR-144-3p binding sites (named pmirGLO-mutAPP). The pre-miR-144-3p and pre-miR-scramble plasmids were constructed and amplified in preparation for use. Subsequently, HEK-293T cells were co-transfected with 100 ng of pmirGLO-APP or pmirGLO-mutAPP vector in the presence of pre-miR-144-3p and pre-miR-scramble using Lipofectamine 2000 Reagent (Invitrogen) and incubated for 48 h. The cells were harvested and the luciferase activities were measured using a Dual-Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s instructions.
APP plasmid construction and transfection
For the overexpression of APP, the sequences encoding APP were amplified through reverse-transcription PCR using a PCR Amplification Kit (TaKaRa Biotechnology, China) and subcloned into pCMV vectors (named pCMV-APP; Beyotime, China). The empty vector served as negative control (NC). Subsequently, SH-SY5Y cells were transfected with pCMV-APP and empty vector using Lipofectamine 2000 Reagent (Invitrogen), whose effects were determined 48 h later using quantitative PCR and Western blot.
RNA extraction and real-time quantitative PCR
Total RNA was collected using the TRIzol® Plus RNA Purification Kit (Ambion, USA) following the manufacturer’s instructions, and quantified using a spectrophotometer at 260 nm. cDNA was synthesized using a RevertAid First Strand cDNA Synthesis Kit (ThermoFisher Scientific, USA). Real-time quantitative PCR was performed using SYBR® Premix Ex TaqTM (TaKaRa Biotechnology) and the Bio-Rad CFX96 touch q-PCR system (Bio-Rad, USA) with the following components: 1 μl of cDNA, 5 μl of SYBR® Premix Ex TaqTM, 1 μl of forward primer, 1 μl of reverse primer and 2 μl of sterile water. β-actin served as an internal reference gene for non-microRNA molecules, and U6 small nuclear RNA (snRNA) for miR-144-3p. Fold induction was calculated using the 2−ΔΔCt method.
Western blot
RIPA buffer (Sigma-Aldrich) was used to collect the protein, and the concentration was measured using Bradford’s method; it was then separated by sodium dodecyl sulfatepolyacrylamide gel electrophoresis (SDS-PAGE). Subsequently, protein bands were electro-blotted onto polyvinylidene difluoride (PVDF) membranes (Invitrogen) and blocked with 10% (w/v) bovine serum albumin (BSA) overnight at 4°C. The membranes were then incubated with primary antibodies for 2 h at room temperature, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies. Specific binding was revealed by a DAB Horseradish Peroxidase Color Development Kit (Beyotime). Densitometry analysis was performed using Image-Pro Plus 6.0 (Media Cybernetics Inc., USA).
Measurement of intracellular ATP
ATP levels were measured using an ATP Assay Kit (Beyotime) according to the manufacturer’s instructions. Briefly, 200 μl of cell lysis solution was centrifuged (12,000 r/min at 4°C for 5 min), and the supernatants were collected as samples. Subsequently, 100 μl of detection reagent was added into a centrifuge tube and kept for 5 min at room temperature, and 10 μl of samples were then added and mixed immediately. The relative light units were measured using a GloMaxTM 96 Microplate Luminometer (Promega, USA). The standard curve was used to calculate the ATP concentration.
MTT assay
Cells were seeded into 96-well dishes with a density of 2 × 104 cells per well, and were then subjected to MPTP treatment and indicated transfections as described above. Next, each well was incubated with 20 μl of MTT solution (5 mg/ml) for 4 h. 150 μl of dimethyl sulfoxide (DMSO; Sigma-Aldrich) was then added to dissolve the formed formazan. The optical density (OD) value of each well at 490 nm was recorded for cell viability.
Mitochondrial DNA (mtDNA) quantification
The relative mtDNA copy number was calculated as ratio of mtDNA/nuclear DNA (nDNA) according to previous studies (Guo et al., 2009; Zhang et al., 2014). Briefly, cells were lysed in RIPA buffer (Sigma-Aldrich) and DNA was extracted with phenol/chloroform followed by ethanol precipitation. Concentrations of extracted DNA were measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA), and then diluted in nuclease-free water to a final concentration of 40 ng/ml. Quantitative PCR was performed as described above for the detection of mtDNA that targets cytochrome c oxidase subunit I (COX1) gene and nDNA that targets hemoglobin subunit beta (β-globin) gene. The primer sequences are as follows: COX1 forward 5′-GCCAACTTCACCATCCAG-3′ and reverse 5′-GCTATCACCTTCACCCTCA-3′; β-globin forward 5′-GCACGTGGATCCTGAGAACT-3′ and reverse 5′-AGCAAGAAAGCGAGCTTAGTG-3′. PCR sample was composed of 5 μl of DNA, 1 μl of forward primer, 1 μl of reverse primer, 3 μl of nuclease-free water and 10 μl of SYBR® Premix Ex TaqTM (a total of 20 μl). PCR reaction was performed following the next procedures: 94°C for 5 min; 94°C for 30 s, 55°C for 30 s, 72°C for 1 min (40 cycles); 72°C for 10 min. Amplification curves and cycle threshold (Ct) were used to determine the relative mtDNA: nDNA ratio in each sample.
Statistical analysis
The data were represented as mean ± standard deviation (SD). One-way Analysis of Variance (ANOVA) or Student’s t-test was used to calculate the statistical significance between groups; p < 0.05 and p < 0.01 represented the statistical significance level.
RESULTS
MiR-144-3p expression is down-regulated by MPTP
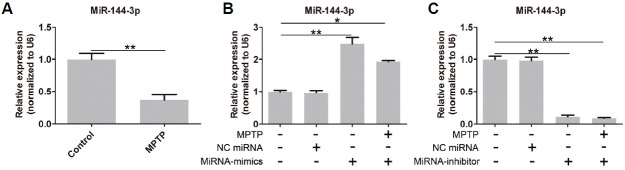
As shown in Fig. 1A, after treatment with MPTP in SH-SY5Y cells, the expression of miR-144-3p was significantly down-regulated. To further identify the specific function of miR-144-3p in PD, we needed to uncover the target gene. As shown in Fig. 1B, miR-144-3p was overexpressed after miRNA-mimics treatment, but its expression was significantly inhibited by miRNA-inhibitor (Fig. 1C).
Fig. 1.
APP is the target gene of miR-144-3p. (A) MiR-144-3p expression was significantly down-regulated by MPTP. (B) MiR-144-3p was overexpressed in SH-SY5Y cells. (C) MiR-144-3p expression was knocked out by miRNA-inhibitor transfection. Each experiment was performed in triplicate. * represents p < 0.05 and ** represents p < 0.01.
MiR-144-3p overexpression restores mitochondrial functions
The level of ATP is a reflection of mitochondrial function. As shown in Fig. 2A, there was a significant decrease in the level of ATP after MPTP and miRNA-inhibitor treatment. However, this inhibitory effect was reversed by miR-144-3p overexpression (Fig. 2A). Cell viability is an indirect index for mitochondrial function. As shown in Fig. 2B, cell viability was significantly inhibited by MPTP and the silencing of miR-144-3p. However, miR-144-3p overexpression notably increased cell viability. In addition, the relative copy number of mtDNA was significantly decreased by MPTP treatment; in contrast, miR-144-3p overexpression markedly recovered the copy number of mtDNA (Fig. 2C). These results implied that miR-144-3p overexpression contributed to improving mitochondrial functions.
Fig. 2.
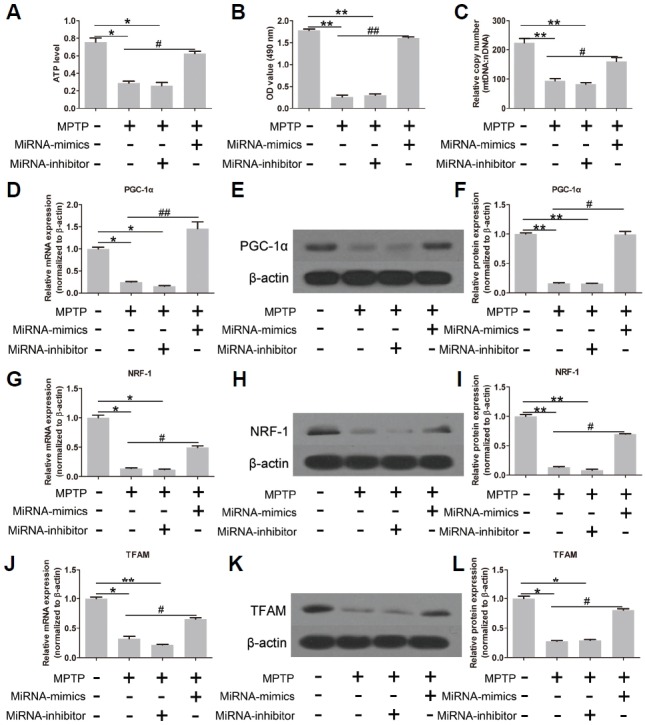
MiR-144-3p contributes to the recovery from MPTP-induced mitochondrial dysfunction. (A) The cellular ATP level was remarkably reduced by MPTP. However, miR-144-3p overexpression restored the ATP level. Similar results in cell viability to ATP level (B) and the relative copy number of mtDNA (C). The expression of PGC-1α (D–F), NRF-1 (G–I) and TFAM (J–L) was significantly decreased by MPTP, but restored by miR-144-3p overexpression. (D, G, and J) Real-time quantitative PCR. (E, H, and K) Western strips of PGC-1α, NRF-1 and TFAM. (F, I and L) Corresponding densitometry analysis of Western strips. Each experiment was performed in triplicate. * and # represent p < 0.05; ** and ## represent p < 0.01.
Subsequently, mitochondrial function-related genes were also investigated. As shown in Figs. 2D, 2G and 2J, MPTP treatment markedly inhibited the mRNA expression of mitochondria-functional genes, including peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α), nuclear respiratory factor 1 (NRF-1) and mitochondrial transcription factor A (TFAM). On the contrary, miR-144-3p overexpression enhanced the mRNA expression of PGC-1α, NRF-1 and TFAM, whereas miRNA-inhibitor showed inhibitory effects. The protein expression of PGC-1α (Fig. 2E), NRF-1 (Fig. 2H) and TFAM (Fig. 2K) was also promoted by miR-144-3p overexpression, which was almost invisible when MPTP and miRNA-inhibitor were present. The corresponding densitometry analysis of Western strips is shown in Figs. 2F, 2I and 2L. The above results showed that miR-144-3p overexpression contributed to the recovery of mitochondrial functions under MPTP conditions. The effects of miR-144-3p-mimics and inhibitor alone (without MPTP treatment) on mitochondrial functions were shown in Supplementary Fig. S1.
APP is the target gene of hsa-miR-144-3p
The predicted binding sites of miR-144-3p in the 3′-UTR of APP are shown in Fig. 3A. Based on this, the relative luciferase activity of pmirGLO-APP in the presence of miR-144-3p overexpression was significantly inhibited, confirming that APP was the target gene of miR-144-3p (Fig. 3B). Since APP was proved to be the target gene of miR-144-3p, we wanted to further investigate the variation of APP expression after treatment with MPTP, miRNA-mimics and miRNA-inhibitor. Interestingly, as shown in Fig. 3C, APP expression was not significantly altered by miR-144-3p via PCR analysis at the mRNA level. However, MPTP markedly promoted mRNA expression of APP. Western blot results showed that, at the protein level, MPTP treatment and miR-144-3p silencing markedly promoted APP expression, whereas miR-144-3p overexpression inhibited APP expression (Fig. 3D), indicating that miR-144-3p was a negative regulator of APP, and APP was one of the target genes of miR-144-3p. The corresponding densitometry analysis of Western strips is shown in Fig. 3E. The effects of miR-144-3p-mimics and inhibitor alone (without MPTP treatment) on mRNA and protein expression of APP were shown in Supplementary Fig. S2.
Fig. 3.
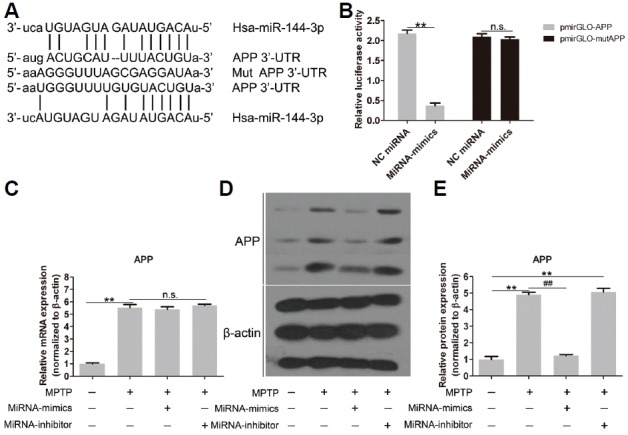
APP expression is regulated by miR-144-3p. (A) The predicted binding sites of miR-144-3p in the 3′-UTR of APP mRNA are shown. (B) MiR-144-3p overexpression significantly inhibited the luciferase activity of APP, but not mutated APP. (C) mRNA expression of APP was significantly promoted by MPTP, but not influenced by miR-144-3p. (D) Western blot strips of APP expression. The protein expression of APP was inhibited by miR-144-3p overexpression, which was reinforced by MPTP and miR-144-3p silencing. (E) Corresponding densitometry analysis of Western strips. Each experiment was performed in triplicate. ** and ## represent p < 0.01; n.s. means no statistical significance.
APP plays a role in the regulation of mitochondrial function
In this experiment, we aimed to up-regulate the expression of APP. SH-SY5Y cells were co-transfected with miRNA-mimics and pCMV-APP harboring no specific binding sites of miR-144-3p. As shown in Fig. 4A, the expression of APP was similar in control and NC groups, but had a sharp increase in the pCMV-APP group at the mRNA level. Expression at the protein level after pCMV-APP transfection also had a remarkable increase (Figs. 4B and 4C), demonstrating that APP expression was factitiously improved. Moreover, pCMV-APP transfection restored the protein expression of APP in the presence of miR-144-3p overexpression (Fig. 4C).
Fig. 4.
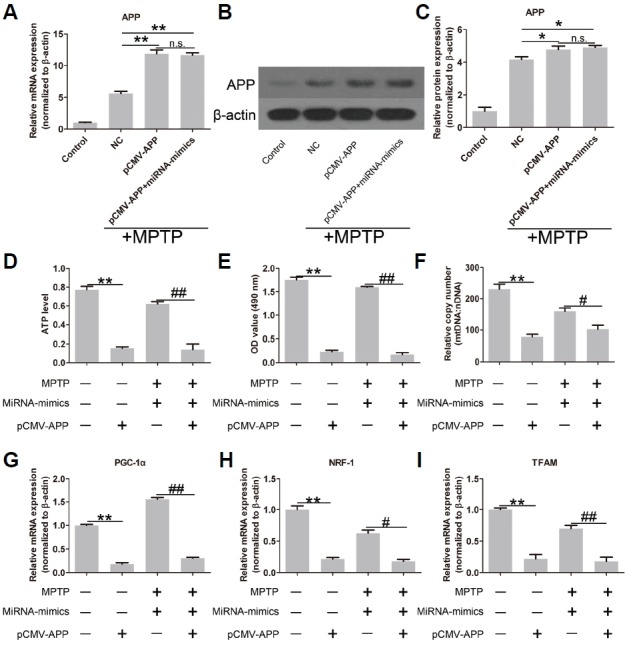
APP overexpression reverses the effects of miR-144-3p overexpression on mitochondrial functions. APP was overexpressed after pCMV-APP transfection at mRNA (A) and protein (B) levels. (C) Quantitative analysis of Western strips confirmed the significantly increased expression of APP. (D) APP overexpression significantly decreased ATP level; APP overexpression also significantly reversed the miR-144-3p overexpression-induced ATP level under MPTP conditions. Similar results in cell viability indicated by MTT (E) and the relative copy number of mtDNA (F). The expression of PGC-1α (G), NRF-1 (H) and TFAM (I) was markedly suppressed by APP overexpression in the presence and absence of miR-144-3p overexpression. Each experiment was performed in triplicate. * and # represent p < 0.05; ** and ## represent p < 0.01; n.s. means no statistical significance.
To further explore the role of APP in mitochondrial function, the effects of APP overexpression on cellular ATP level, cell viability, relative copy number of mtDNA and the expression of PGC-1α, NRF-1 and TFAM were evaluated. As expected, APP overexpression significantly reversed the effects of miR-144-3p overexpression on mitochondrial function. To be specific, APP overexpression remarkably decreased the ATP level (Fig. 4D) and cell viability (Fig. 4E). The relative copy number of mtDNA was also significantly decreased by APP overexpression (Fig. 4F). Moreover, APP overexpression showed inhibitory effects on the expression of PGC-1α (Fig. 4G), NRF-1 (Fig. 4H) and TFAM (Fig. 4I). These results indicated that APP was a negative regulator of mitochondrial function.
DISCUSSION
In the present study, hsa-miR-144-3p was found to be down-regulated in MPTP-treated SH-SY5Y cells. APP was identified as one of the target genes of miR-144-3p. Overexpression of miR-144-3p was accompanied by decreased expression of APP. In addition, miR-144-3p was found to play a role in relieving MPTP-induced mitochondrial dysfunction. We also proved that APP overexpression reversed the effects of miR-144-3p overexpression on mitochondrial function. Taken together, our results provided a promising therapeutic target for the treatment of PD, depending on miR-144-3p and its target gene APP.
MiRNAs play a role in regulating physiological processes by inhibiting the expression of their target genes in post-transcriptional-translation. To explore the underlying mechanisms, the target gene of miR-144-3p had to be discovered. With the help of bioinformatic analysis and dual-luciferase reporter assay, APP was found to have interactions with miR-144-3p; in other words, APP was the target gene of hsa-miR-144-3p. APP has previously been reported to be involved in AD. In fact, AD is usually characterized by Aβ plaques which were discovered by Alois Alzheimer in 1906 (Westmark, 2013). More importantly, Aβ is formed from the cleavage of APP by secretases. Hence, APP production and cleavage determines the level of Aβ in the brain interstitial fluid. Due to the similarities between AD and PD and the fact that over 50% of AD patients showed the pathological features of PD patients, APP has also been reported to play a role in the development of PD (Ayton et al., 2015; Irwin et al., 2013; Schulte et al., 2015). Thus, a clearer understanding of the regulation of APP will provide novel targets for therapeutic intervention of PD. Our results showed that APP was highly expressed in the mimicked PD cell model, and the overexpression of APP contributed greatly to mitochondrial dysfunction. These results were consistent with the results from Schulte et al. (2015).
Pathogenic mtDNA mutations are also associated with PD (Pickrell and Youle, 2015). Excessively accumulated mutations in mtDNA have an increased risk of developing PD (Luoma et al., 2004; Reeve et al., 2013). As a result, mitochondrial dysfunction is a main symptom of PD, indicated by the review article (Burte et al., 2015; Exner et al., 2012; Ruszkiewicz and Albrecht, 2015). In the present study as shown in Fig. 2, there was a significant decrease in the ATP level and cell viability after MPTP treatment, which is similar to the symptoms of PD, indicating that MPTP led to mitochondrial dysfunction. In contrast, miR-144-3p overexpression restored the ATP level and cell viability. It is also worth noting that APP overexpression could reverse the effects of miR-144-3p overexpression, suggesting that APP was an important effector of miR-144-3p.
PGC-1α, NRF-1 and TFAM are three key genes that promote mitochondrial function, because mitochondrial biogenesis that helps maintain mitochondrial homeostasis is regulated by the PGC-1α-NRF-TFAM pathway. To be specific, NRF-1 controls the synthesis of mitochondrial proteins and TFAM drives the transcription and replication of mtDNA. Above all, both NRF-1 and TFAM are regulated by PGC-1α (Wu et al., 1999). Sheng et al. have reported the decreased expression of PGC-1α, NRF-1 and TFAM in AD (Sheng et al., 2012). Exploration of these three genes could help us understand the mechanisms underlying the roles of miR-144-3p and APP in PD-induced mitochondrial dysfunction. In the present study, the decreased expression of PGC-1α, NRF-1 and TFAM by MPTP demonstrated mitochondrial dysfunction. Moreover, miR-144-3p overexpression could help restore mitochondrial functions, indicated by the up-regulated expression of PGC-1α, NRF-1 and TFAM. APP overexpression reversed the miR-144-3p overexpression-induced high expression of these three genes, implying that APP was a negative regulator of mitochondrial function, and APP was the effector of miR-144-3p.
In conclusion, in this present study, with the help of an MPTP-induced cell model, we discovered the interactions between hsa-miR-144-3p and APP. MiR-144-3p was found to be down-regulated in this MPTP-mimicked PD model, along with its target gene APP that participates in regulating MPTP-induced mitochondrial dysfunction.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Alves G., Bronnick K., Aarsland D., Blennow K., Zetterberg H., Ballard C., Kurz M.W., Andreasson U., Tysnes O.B., Larsen J.P., et al. CSF amyloid-beta and tau proteins, and cognitive performance, in early and untreated Parkinson’s disease: the Norwegian ParkWest study. J. Neurol. Neurosurg. Psychiatry. 2010;81:1080–1086. doi: 10.1136/jnnp.2009.199950. [DOI] [PubMed] [Google Scholar]
- Ayton S., Lei P., Hare D.J., Duce J.A., George J.L., Adlard P.A., McLean C., Rogers J.T., Cherny R.A., Finkelstein D.I., et al. Parkinson’s disease iron deposition caused by nitric oxide-induced loss of beta-amyloid precursor protein. J. Neurosci. 2015;35:3591–3597. doi: 10.1523/JNEUROSCI.3439-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos K., Malenica I., Metpally R., Courtright A., Rakela B., Beach T., Shill H., Adler C., Sabbagh M., Villa S., et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One. 2014;9:e94839. doi: 10.1371/journal.pone.0094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burte F., Carelli V., Chinnery P.F., Yu-Wai-Man P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol. 2015;11:11–24. doi: 10.1038/nrneurol.2014.228. [DOI] [PubMed] [Google Scholar]
- Christopher L., Duff-Canning S., Koshimori Y., Segura B., Boileau I., Chen R., Lang A.E., Houle S., Rusjan P., Strafella A.P. Salience network and parahippocampal dopamine dysfunction in memory-impaired Parkinson disease. Ann. Neurol. 2015;77:269–280. doi: 10.1002/ana.24323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rijk M.C., Breteler M.M., Graveland G.A., Ott A., Grobbee D.E., van der Meche F.G., Hofman A. Prevalence of Parkinson’s disease in the elderly: the Rotterdam study. Neurology. 1995;45:2143–2146. doi: 10.1212/wnl.45.12.2143. [DOI] [PubMed] [Google Scholar]
- Duce J.A., Tsatsanis A., Cater M.A., James S.A., Robb E., Wikhe K., Leong S.L., Perez K., Johanssen T., Greenough M.A., et al. Iron-export ferroxidase activity of beta-amyloid precursor protein is inhibited by zinc in Alzheimer’s disease. Cell. 2010;142:857–867. doi: 10.1016/j.cell.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner N., Lutz A.K., Haass C., Winklhofer K.F. Mitochondrial dysfunction in Parkinson’s disease: molecular mechanisms and pathophysiological consequences. EMBO J. 2012;31:3038–3062. doi: 10.1038/emboj.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves P., Zeng Y. Biogenesis of mammalian microRNAs: a global view. Genomics Proteomics Bioinformatics. 2012;10:239–245. doi: 10.1016/j.gpb.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N.E., Sampath H., Zweig J.A., Quinn J.F., Soumyanath A. Centella asiatica attenuates amyloid-beta-induced oxidative stress and mitochondrial dysfunction. J. Alzheimers Dis.. 2015;45:933–946. doi: 10.3233/JAD-142217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Jiang L., Bhasin S., Khan S.M., Swerdlow R.H. DNA extraction procedures meaningfully influence qPCR-based mtDNA copy number determination. Mitochondrion. 2009;9:261–265. doi: 10.1016/j.mito.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin D.J., Lee V.M., Trojanowski J.Q. Parkinson’s disease dementia: convergence of alpha-synuclein, tau and amyloid-beta pathologies. Nat. Rev. Neurosci. 2013;14:626–636. doi: 10.1038/nrn3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees A.J. Unresolved issues relating to the shaking palsy on the celebration of James Parkinson’s 250th birthday. Mov. Disord. 2007;22(Suppl 17):S327–334. doi: 10.1002/mds.21684. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang R.Y., Zhao J., Dong Z., Feng D.Y., Wu R., Shi M., Zhao G. Ginsenoside Rd protects SH-SY5Y cells against 1-Methyl-4-phenylpyridinium induced Injury. Int. J. Mol. Sci. 2015;16:14395–14408. doi: 10.3390/ijms160714395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma P., Melberg A., Rinne J.O., Kaukonen J.A., Nupponen N.N., Chalmers R.M., Oldfors A., Rautakorpi I., Peltonen L., Majamaa K., et al. Parkinsonism, premature menopause, and mitochondrial DNA polymerase gamma mutations: clinical and molecular genetic study. Lancet. 2004;364:875–882. doi: 10.1016/S0140-6736(04)16983-3. [DOI] [PubMed] [Google Scholar]
- McCarthy R.C., Park Y.H., Kosman D.J. sAPP modulates iron efflux from brain microvascular endothelial cells by stabilizing the ferrous iron exporter ferroportin. EMBO Rep. 2014;15:809–815. doi: 10.15252/embr.201338064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minones-Moyano E., Porta S., Escaramis G., Rabionet R., Iraola S., Kagerbauer B., Espinosa-Parrilla Y., Ferrer I., Estivill X., Marti E. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- Nussbaum R.L., Ellis C.E. Alzheimer’s disease and Parkinson’s disease. N. Engl. J. Med. 2003;348:1356–1364. doi: 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- Owen A.D., Schapira A.H.V., Jenner P., Marsden C.D. Oxidative stress and Parkinson’s diseasea. Anna. N Y Acad. Sci. 1996;786:217–223. doi: 10.1111/j.1749-6632.1996.tb39064.x. [DOI] [PubMed] [Google Scholar]
- Pickrell A.M., Youle R.J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radad K.S., Al-Shraim M.M., Moustafa M.F., Rausch W.D. Neuroprotective role of thymoquinone against 1-methyl-4-phenylpyridinium-induced dopaminergic cell death in primary mesencephalic cell culture. Neurosciences. 2015;20:10–16. [PMC free article] [PubMed] [Google Scholar]
- Reeve A., Meagher M., Lax N., Simcox E., Hepplewhite P., Jaros E., Turnbull D. The impact of pathogenic mitochondrial DNA mutations on substantia nigra neurons. J. Neurosci. 2013;33:10790–10801. doi: 10.1523/JNEUROSCI.3525-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruszkiewicz J., Albrecht J. Changes in the mitochondrial antioxidant systems in neurodegenerative diseases and acute brain disorders. Neurochem. Int. 2015;88:66–72. doi: 10.1016/j.neuint.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Schapira A.H.V. Oxidative stress in Parkinson’s disease. Neuropathol. Appl. Neurobiol. 1995;21:3–9. doi: 10.1111/j.1365-2990.1995.tb01022.x. [DOI] [PubMed] [Google Scholar]
- Schapira A.H. Etiology and pathogenesis of Parkinson disease. Neurol. Clin. 2009;27:583–603. v. doi: 10.1016/j.ncl.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Schulte E.C., Fukumori A., Mollenhauer B., Hor H., Arzberger T., Perneczky R., Kurz A., Diehl-Schmid J., Hull M., Lichtner P., et al. Rare variants in beta-amyloid precursor protein (APP) and Parkinson’s disease. Eur. J. Hum. Genet. 2015;23:1328–1333. doi: 10.1038/ejhg.2014.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng B., Wang X., Su B., Lee H.G., Casadesus G., Perry G., Zhu X. Impaired mitochondrial biogenesis contributes to mitochondrial dysfunction in Alzheimer’s disease. J. Neurochem. 2012;120:419–429. doi: 10.1111/j.1471-4159.2011.07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Yu J.T. Causes and consequences of microRNA dysregulation in neurodegenerative diseases. Mol. Neurobiol. 2014;51:1249–1262. doi: 10.1007/s12035-014-8803-9. [DOI] [PubMed] [Google Scholar]
- Tsou Y.H., Shih C.T., Ching C.H., Huang J.Y., Jen C.J., Yu L., Kuo Y.M., Wu F.S., Chuang J.I. Treadmill exercise activates Nrf2 antioxidant system to protect the nigrostriatal dopaminergic neurons from MPP+ toxicity. Exp. Neurol. 2015;263:50–62. doi: 10.1016/j.expneurol.2014.09.021. [DOI] [PubMed] [Google Scholar]
- Vallelunga A., Ragusa M., Di Mauro S., Iannitti T., Pilleri M., Biundo R., Weis L., Di Pietro C., De Iuliis A., Nicoletti A., et al. Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and multiple system atrophy. Front Cell Neurosci. 2014;8:156. doi: 10.3389/fncel.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmark C.J. What’s hAPPening at synapses? The role of amyloid beta-protein precursor and beta-amyloid in neurological disorders. Mol. Psychiatry. 2013;18:425–434. doi: 10.1038/mp.2012.122. [DOI] [PubMed] [Google Scholar]
- Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zuo X., Yang B., Li Z., Xue Y., Zhou Y., Huang J., Zhao X., Zhou J., Yan Y., et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.