Abstract
Lysosomes are cellular organelles containing diverse classes of catabolic enzymes that are implicated in diverse cellular processes including phagocytosis, autophagy, lipid transport, and aging. Lysosome-associated membrane proteins (LAMP-1 and LAMP-2) are major glycoproteins important for maintaining lysosomal integrity, pH, and catabolism. LAMP-1 and LAMP-2 are constitutively expressed in Salmonella-infected cells and are recruited to Salmonella-containing vacuoles (SCVs) as well as Salmonella-induced filaments (Sifs) that promote the survival and proliferation of the Salmonella. LAMP-3, also known as DC-LAMP/CD208, is a member of the LAMP family of proteins, but its role during Salmonella infection remains unclear. DNA microarray analysis identified LAMP-3 as one of the genes responding to LPS stimulation in THP-1 macrophage cells. Subsequent analyses reveal that LPS and Salmonella induced the expression of LAMP-3 at both the transcriptional and translational levels. Confocal Super resolution N-SIM imaging revealed that LAMP-3, like LAMP-2, shifts its localization from the cell surface to alongside Salmonella. Knockdown of LAMP-3 by specific siRNAs decreased the number of Salmonella recovered from the infected cells. Therefore, we conclude that LAMP-3 is induced by Salmonella infection and recruited to the Salmonella pathogen for intracellular proliferation.
Keywords: LAMP-3, LPS, lysosome, Salmonella typhimurium
INTRODUCTION
Lysosomes are membranous cell organelles that are involved in endocytosis, exocytosis, and intracellular trafficking of vesicles (Appelqvist et al., 2013) and autophagy (Ryter et el., 2013). These organelles play a major metabolic role in the intracellular digestion of macromolecules such as carbohydrates, proteins, lipids, and nucleic acids. A spectrum of 50 hydrolytic enzymes is involved in the lysosomal catabolic process. Abnormal regulation of this pathway appears to be linked to lysosomal storage diseases such as Danon disease, Tay-Sachs disease, and Gaucher disease (Cox et al., 2012; Eskelinen and Saftig, 2009).
In response to bacterial infection, cellular pattern recognition receptors, such as Toll-like receptors (TLRs) and Nod-like receptors (NLRs), are known to be activated to initiate the autophagic (or xenophagic) process (Shroff et al., 2014). Lysosomes play a key role in innate immune defense by fusing with and degrading pathogens contained in autophagic vesicles directly, or fusing with the pathogen-containing vesicles (Knodler and Celli, 2011; Levine and Deretic, 2007). These processes generally result in the destruction of the invasive bacteria, although some bacteria, including Salmonella Typhimurium and Mycobacterium tuberculosis, can block the maturation of phagosomes into degradative organelles called phagolysosomes (Castillo et al., 2012; Jo et al., 2013).
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a gram negative bacteria that can infect and proliferate inside the intestinal epithelium and macrophages, triggering gastrointestinal disease (Coburn et al., 2007; Fabrega and Vila, 2013). The process of Salmonella infection is known to be controlled by Salmonella Pathogenicity Island-1 (SPI-1), which encodes a set of the bacterial proteins, known as the type III secretion system (TTSS or T3SS). SPI-1 is required for the invasion of non-phagocytic cells (Schmidt and Hensel, 2014). Intracellular proliferation proceeds inside the Salmonella containing vacuole (SCV) or the Sif (Salmonella-induced filament) and requires Salmonella Pathogenicity Island-2 (SPI-2) for protection against the functioning of intracellular effectors such as phagocyte oxidase (Vazquez-Torres et al., 2000) and inducible nitric oxide synthase (Chakravortty et al., 2002; Schmidt and Hensel, 2014).
LAMP-1 and LAMP-2 are major lysosomal membrane proteins that account for about half of the total lysosomal membrane proteins. These glycoproteins are responsible for maintaining lysosomal integrity, pH, and catabolism (Andrejewski et al., 1999; Eskelinen 2006). Aberrant localization or expression of LAMP-2 has been implicated in human disease. Expression of LAMP-2 on the cell surface has been implicated in cancer cell survival and metastasis (Agarwal et al., 2015; Saha, 2012). Moreover, LAMP-2 deficiency is associated with Danon disease, a lysosomal storage disease, characterized by vacuolar cardiomyopathy and myopathy (Nishino et al., 2000).
Compared to LAMP-1 and LAMP-2, LAMP-3 is poorly understood. LAMP-3 exhibits 30% and 28% amino acid sequence identity to LAMP-1 and LAMP-2 respectively. LAMP-3 carries numerous potential glycosylation sites in comparison to those of LAMP-1 or LAMP-2 (de Saint-Vis et al., 1998; Wilke et al., 2012). In contrast to the constitutive expression of LAMP-1 and LAMP-2 in many cell types, LAMP-3 expression is regulated temporally and spatially, or is induced by hypoxic conditions. Overexpression of LAMP-3 is known to be associated with cancer metastasis (Liao et al., 2015; Mujcic 2009; Nagelkerke et al., 2013). Recently, LAMP-3 was reported to be associated with modulation of autophagy and autophagy-mediated drug sensitivity (Dominguez-Bautista et al., 2015; Nagelkerke et al., 2014). Compared to LAMP-1 and LAMP-2, LAMP-3 remains poorly understood for its role in the bacterial infection process and we investigated whether LAMP-3 expression plays a role in the Salmonella infection cycle.
MATERIALS AND METHODS
Reagents
Antibodies against phospho-p38 MAPK, phospho-NF-κB p65 were purchased from Cell Signaling (USA). Antibodies against p38MAPK, Anti-NF-κB, LAMP-1, LAMP-2 and LAMP-3 were purchased from Santa Cruz (USA). Anti-GAPDH antibody was purchased from Ab Frontier (Korea). RNA Blood Mini kit (QIAGEN, USA) and cDNA synthesis kit were purchased respectively from QIAGEN (Netherlands) and Invitrogen (USA). LPS and PEI transfection reagents were purchased from Sigma-Aldrich, USA. LAMP-3 shRNA and scrambled shRNA constructs (29mer each) in pRS plasmid were purchased from Origene (USA).
Chemical Inhibitors of NF-κB (PDTC) and JNK II were purchased from Calbiochem (USA). ERK inhibitor (FR180204) and p38 inhibitor (AMG548) were purchased from Tocris Bioscience (UK).
Salmonella typhimurium 1406S(EGFP) was provided by Dr. Choy HE (Chonnam Natl’ Univ., Korea). Overnight culture of the bacteria was inoculated in fresh LB broth containing 100 ug/ml Ampicillin at a ratio of 1/33 dilution and cultured for 4 h before harvest and use for the cell infection, confocal assay and cell proliferation assay.
Cell culture
HeLa cells were cultured in DMEM media supplemented with 10% FBS and penicillin/streptomycin (P/S) antibiotics. THP-1 cells, a pro-monocytic cell line, were culture in RPMI 1640 (Life Technologies, USA) containing 10% FBS (WellGene, Korea), P/S antibiotics (Life Technologies, USA) and 0.05mM mercaptoethanol (Sigma-Aldich, USA). All cell cultures were maintained at 5% CO2 and 37°C.
RNA isolation and RT-PCR
After treatment with 1ug/ml LPS or Salmonella bacteria, total RNAs were isolated from THP-1 cells using the RNA Blood Mini kit according to the manufacturer’s instructions (QIAGEN, USA). cDNA synthesis and RT-PCR analysis were performed simultaneously in a single tube using 10ng/ml of total RNA according to the manufacture’s instruction (AccuPower® RocketScriptTM RT Premix (Bioneer, Korea); cDNA was synthesized using the primers listed below at 42°C for 40 min., followed by pre-denaturation at 95°C for 5min., and 35 cycles (95°C/30 s → 54°C /30 s → 72°C /30 s) of PCR reaction before the final extension (72°C/10 min.) step. The primers used in this study were designed according to “Primer 3 program (Whithead Institute, USA)” and are listed in Table 1.
Stable LAMP-3 knock-down cell line
Hela cells were transfected by PEI transfection reagent with two LAMP-3 shRNA plasmids containing the following oligonucleotides (5′-ACTGGAACCAGTTCATCAACCGTCAGCC-3′ and 5′-GTGGATGAGT GCTCGTCTGACTACACAAT-3′). Two days later, puromycin selection was followed at 1 ug/ml concentration and the emerging colonies were checked for the expression level of LAMP-3 knock-down by western blotting with anti LAMP-3 antibody.
Intracellular survival assay of Salmonella typhimurium
HeLa cells, at about 60% confluency, were infected with salmonella (eGFP) for 30 min. and treated with gentamycin (50 ug/ml) for 30 min. before PBS washing 3 times. Subsequently, the infected HeLa cells were washed 3 times with PBS buffer and lyzed with lysis buffer (0.1% Triton X-100, 0.1% SDS, PBS). The lysates were serially diluted in PBS and plated on LB plate containing ampicillin (100 ug/ml). After overnight, the number of bacteria colonies was counted for CFU.
N-SIM super resolution microscopy
HeLa cells (1 × 106/well) were cultured in DMEM growth media overnight on coverglass in 35 mm dish (NUNC) before infection with Salmonella (eGFP) at about 1:40 MOI. To remove any extracellular bacteria, gentamicin was added 30 min later at the final concentration of 50 ug/ml and after an hour, the media was removed and replaced with fresh DMEM growth media for the indicated time. After washing with PBS 3 times, the HeLa cells were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 10 min. After PBS washing, the cells were blocked with 10% FBS in PBS for 30 min. and reacted with primary LAMP-2 and LAMP-3 antibodies for 3 h and after PBS washing, incubated with secondary antibody (Alexa-594 or Alexa-633) for 2 h and washed with PBS buffer before mounting on slide glass for N-SIM microscope analysis with NIS-Elements Viewer.
RESULTS
LPS and Salmonella infection increases LAMP-3 expression
To identify lipopolysaccharide (LPS) responsive genes, we used the Illumina HumanHT-12 v4 Expression BeadChip Kit. We identified LAMP-3 as a gene that is upregulated in response to LPS. Then, we characterized the regulation of LAMP-3 expression and its effects on the intracellular proliferation of Salmonella Typhimurium.
In order to confirm that LPS induces LAMP-3 gene expression, we treated THP-1 cells with 1 μg/ml of LPS for 1, 4, or 8 h, isolated total RNA, and performed RT-PCR analysis. LAMP-3 was up-regulated 2-fold following 4–8 h of LPS stimulation (Fig. 1A). On the other hand, LAMP-1 and LAMP-2 expression were unchanged despite LPS stimulation. These results were confirmed further by western blot analysis; LAMP-3 expression increased as early as 1 h after LPS stimulation and reached the maximum expression level 8 h after LPS stimulation from 1.2-fold to 1.5-fold repectively. LAMP-1 and LAMP-2 expression were similar to the unstimulated control (Fig. 1B). To determine whether the difference in LAMP gene expression and protein production was LPS-specific, we tested expression levels in response to Salmonella bacteria. Salmonella caused increased expression of LAMP-3 and LAMP-2 in both RT-PCR and Western blot analysis, but LAMP-1 was unaffected (Figs. 1C and 1D). Several bands of LAMP-3, at approximately 70 kDa in size, appeared depending on the time after the stimulation and may be related to differences in the glycosylation process. We obtained similar results when we tested LAMP expression in the Salmonella infected HeLa cells; LAMP-3 was the most upregulated among the three LAMPs in both RT-PCR and western blot analyses (Figs. 1E and 1F).
Fig. 1.
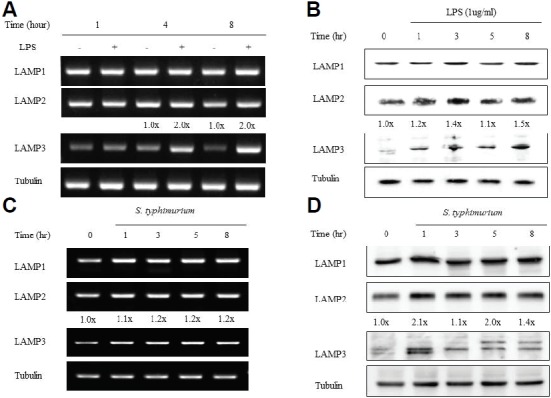
LAMP-3 expression is controlled at the transcription and translation level by LPS and Salmonella in THP-1 and HeLa cells. THP-1 cells, at a density of 1.0 × 106 cells/well, were treated with 1 ug/ml of LPS (A, B) or infected with S. typhimurium (C, D), or HeLa cells were infected with S. typhimurium (E, F). (A, C, and E) At each time point, total cellular RNA was extracted and subjected to RT-PCR with gene specific primers. Tubulin gene was used as an internal control. (B, D, and F) Protein levels of LAMP-1, LAMP-2 and LAMP-3 were examined by Western blotting with specific antibodies. GAPDH was included as a loading control.
Upregulation of LAMP-3 is closely associated with the activation of the NF-κB and p38 MAPK signaling pathways
Given the central role of TLR4 in the LPS-mediated signaling pathway including NF-κB, ERK, JNK and p38 (Guha and Mackman, 2002), we investigated which of these signaling components might control the LAMP-3 expression. We treated THP-1 cells for 30 min with the following chemical inhibitors; ammonium pyrrolidinedithiocarbamate (PDTC: (NF-κB inhibitor, 20 μg/ml)), FR180204 (ERK inhibitor, μM), 420119 (JNK II inhibitor, 40 μM), and AMG548 (p38 inhibitor, 20 μM). Then, cells were stimulated with 1 μg/ml of LPS for 4 hrs and RNA or protein was isolated from the cells. As expected, RT-PCR analysis indicated that LPS stimulation caused a 2-fold increase in LAMP-3 transcript levels (PBS+LPS vs. PBS). However, inhibition of NF-κB and p38 MAPK caused reduced levels of LAMP-3 transcripts. Using phospho-specific antibodies, we examined whether NF-κB and p38 were phosphorylated in response to LPS stimulation (Figs. 2A and 2B). LPS stimulation caused increased phosphorylation of p38, but not NF-κB (Figs. 2C and 2D). In the presence of each inhibitor, phosphorylation was decreased dramatically, but the total amount of protein was unchanged. When THP-1 cells were treated with chemical inhibitors followed by Salmonella infection for 4 h, LAMP-3 protein levels were reduced in the following order: NF-κB (PDTC), p38 MAPK, JNK, and ERK (Figs. 2E and 2F). In the PBS control, LAMP-3 protein levels were increased. Tubulin was used as a protein loading control.
Fig. 2.
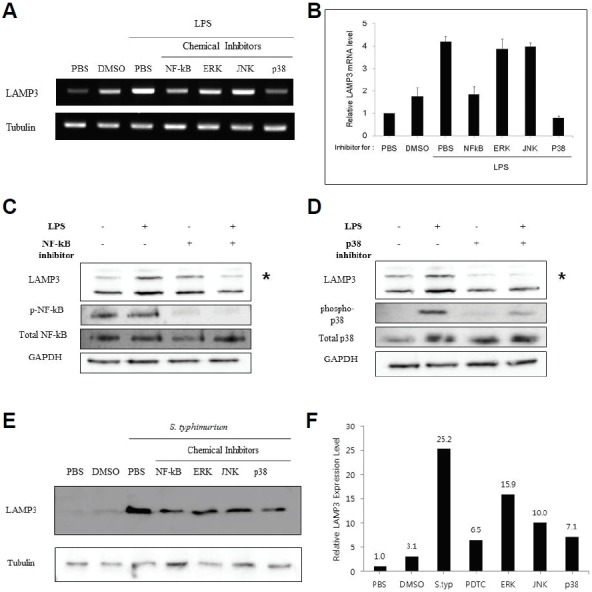
Inhibition of NF-κB and p38 MAPK down-regulates the expression of LAMP-3. THP-1 cells were treated with each inhibitor at the indicated concentration (PDTC / NF-κB inhibitor, 20 ug/ml; FR180204 / ERK inhibitor, 50 uM; JNK II inhibitor / 420119 ; 40 uM, and AMG548 / p38 inhibitor, 20 uM) 30 min. prior to cell stimulation with LPS (1 ug/ml), PBS or DMSO for 4 h. (A, B) Extraction and analysis of mRNA were performed as described in “Materials and Methods”. Levels of LAMP-3 transcripts were quantified by RT-PCR using gene specific primers and normalized against tubulin (an internal control). Results from a representative experiment are shown as mean ± S.E. (n = 3 of a single experiment). (C, D) Analysis of LAMP-3, NF-κB p65, and p38 MAPK protein expression was performed by western blotting with normal or phospho/activation-specific antibodies as indicated. (E) Experiments were done by adding inhibitors at indicated concentrations 30 min. prior to Salmonella infection for 4 h. LAMP-3 protein expression was examined by Western blotting with anti LAMP-3 antibody.
LAMP-3 colocalizes with Salmonella Typhimurium
The N-SIM super-resolution microscope was used to investigate the colocalization of LAMP-3 with LAMP-2 and Salmonella Typhimurium (eGFP) in HeLa cells. In uninfected control cells, LAMP-3 (blue) was apparent as dots that were evenly distributed at the cell periphery, while LAMP-2 was observed at the cell periphery as larger aggregates (Fig. 3, top row). Five hours after infection, a portion of the LAMP-3 colocalized with Salmonella bacteria (green) in the cytosol, which was observed as light blue, whereas the remainder of the LAMP-3 signal appeared as dots in the cytosol. On the other hand, some LAMP-2 protein surrounded the Salmonella and also LAMP-3, while the remainder of the LAMP-2 signal appeared as aggregates. It is interesting to note that LAMP-3 was almost absent in the plasma membrane area following Salmonella infection (Fig 3, middle row). At 8 h post-infection, LAMP-3 colocalized with Salmonella as seen at 5 hrs post-infection, but LAMP-2 protein localized distinctly away from Salmonella (Fig 3, bottom row).
Fig. 3.
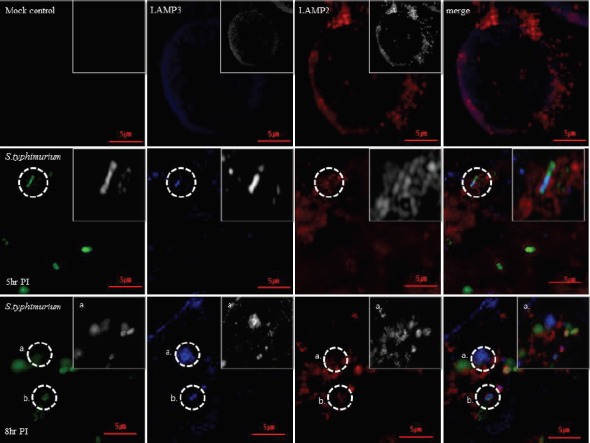
LAMP-3 co-localizes with Salmonella typhimurium. N-SIM super resolution microscope was used to investigate the co-localization of LAMP3 with LAMP2 and Salmonella typhimurium (eGFP) in Hela cells. HeLa cells were infected with PBS buffer (mock control) or the Salmonella bacteria for 5 or 8 h, and were processed as indicated in the materials and methods. After fixation, permeabilization and FBS blocking, the infected cells were reacted with primary LAMP-2 and LAMP-3 antibodies and with secondary antibodies (Alexa-594 or Alexa-633) before PBS washing and mounting on slide glass for N-SIM microscope analysis with NIS-Elements Viewer.
LAMP-3 knockdown interferes with the intracellular proliferation of Salmonella Typhimurium
To investigate the relationship between LAMP-3 expression and intracellular proliferation of Salmonella, a stable HeLa cell line was transfected with a cocktail of two shRNA plasmids that knock down LAMP-3. Compared to the control cells transfected with a scrambled shRNA plasmid, LAMP-3 protein expression was decreased in LAMP-3 knockdown cells by 40% (Figs. 4A and 4B). Next, cells were infected with Salmonella for 2, 6, and 12 h. Cells were lysed to recover live bacteria that were cultured on bacterial growth media. After culturing overnight, the number of bacterial colonies from LAMP-3 knockdown cells were counted and compared to those recovered from control cells (scrambled shRNA). Knockdown of LAMP-3 decreased the number of Salmonella by 47%, 55%, and 54% at each time point respectively. These data suggest that LAMP-3 may contribute to the intracellular proliferation of Salmonella. Down-regulation of LAMP-2 yielded similar results by decreasing the numbers of recovered live bacteria (Figs. 4D, 4E and 4F).
Fig. 4.
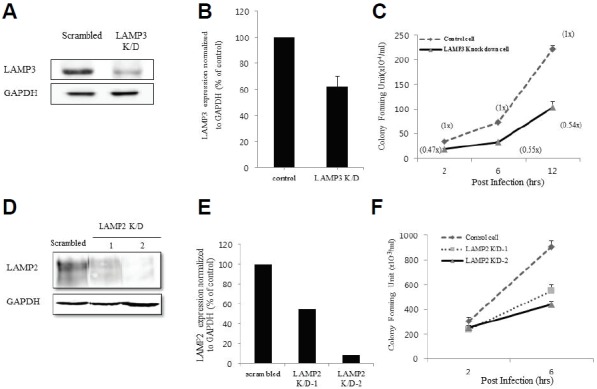
Interference of LAMP-3 expression inhibits intracellular proliferation of Salmonella typhimurium. (A) Stable control (Scrambled) or LAMP-3 knockdown HeLa cell line was checked for the level of down-regulation of LAMP-3 expression by western blotting using antibody against LAMP-3. GAPDH expression was checked as a loading control. (B) Intracellular proliferation of Salmonella was assayed according to the procedure in the materials and methods. The 2, 6 and 12 h after infection, the cells were lyzed with lysis buffer to isolate live bacteria in the LB agar plate containing 100 ug/ml of Ampicillin. After, overnight, the number of bacteria colonies was counted and compared to that of control cell line. Data are the average of three independent experiments. Error bars represent standard deviation. (C) LAMP-2 knockdown HeLa cell line was established and the level of LAMP-2 down-regulation was checked by Western blotting with anti-LAMP-2 antibody. (D) Intra cellular survival assay was performed with LAMP-2 K/D cell line and scrambled control cell line at 2 and 6 h after the Salmonella infection.
DISCUSSION
Pathogenic Salmonella isolates, like mycobacteria, have evolved to avoid intracellular targeting to lysosomes and subsequent killing by establishing intracellular niches such as SCVs and Sifs (Bakowski et al., 2008; Wileman, 2013). About 20% of Salmonella are estimated to be outside of SCVs and SIFs or have damaged SCV membranes inside infected cells (Birmingham et al., 2006).
For intracellular survival, Salmonella recruits LAMP-1 and LAMP-2 from the cell surface (Baldeon et al, 2001) to the SCV, and later to Sifs with the help of the SPI-1 effector protein SpoI (Bakowski et al., 2008). LAMP-2 appears to be more important than LAMP-1 in establishing and maintaining SCVs and Sifs. RNAi mediated silencing of LAMP-2 results in a greater reduction in the number of Salmonella than silencing of LAMP-1 (Roark et al., 2008). Despite their abundance, it is not clear how LAMP-1 and LAMP-2 proteins play a role in the biogenesis and maintenance of SCVs and Sifs. From a screening of LPS responsive genes in THP-1 cells, we identified LAMP-3 as one of the target genes. The human LAMP-3 amino acid sequence was similar in size, but distinctively different from LAMP-1 and LAMP-2 (data not shown).
Although LAMP-3 expression is documented in many cell types including lung, dendritic, and cancer cells, as well as influenza virus-infected cells (Barois et al., 2002; Bechetoille et al., 2006; Honess and Simmons, 1993; Kanao et al., 2005; Kleijmeer et al., 2001; de Saint-Vis et al., 1998; Zhou et al., 2011), little is known about its expression in Salmonella infected cells. In contrast to constitutively and abundantly expressed LAMP-1 and LAMP-2 (Furuta et al., 1999), we show that LAMP-3 expression is inducible by both LPS and Salmonella at the transcriptional and translational levels in THP-1 and HeLa cells. Salmonella infection had a more profound effect on the expression levels of LAMPs than did LPS stimulation. LAMP-3 contains a greater number of potential glycosylation than seen in LAMP-1 and LAMP-2, and LAMP-3 protein migrated more frequently as multiple bands, as reported in previous studies (Dominguez-Bautista et al., 2015; Nagelkerke et al., 2011).
LAMP-3 is also known to be a novel hypoxia-inducible gene under the control of the PERK/eIF2α/ATF4 arm of the unfolded protein response (UPR) and is independent of HIF-1α (Mujcic et al., 2009; Nagelkerke et al., 2011). In epithelial cells infected with either influenza A virus or Pseudomonas aeruginosa, increased LAMP-3 expression is observed endosomal or vacuolar compartments (Angus et al., 2008; Zhou et al., 2011). As reported for the TLR4 signaling pathway in Salmonella-infected cells (Arpaia et al., 2011), we found that the signaling route to LAMP-3 expression in THP-1 cells was dependent on NF-κB and p38 MAPK activity. In this study, NF-κB may be regulated by translocation of the p65 subunit into nucleus rather than by phosphorylation, whereas p38 MAPK is phosphorylated in response to LPS-mediated signaling. Similar to LPS stimulation, Salmonella infection appears to elicit similar signaling to activate NF-κB and p38 MAPK to a greater extent than the other MAPKs, although TLR2 is also known to be activated by Salmonella infection in mice (Arpaia et al., 2011).
N-SIM super-resolution imaging data revealed that LAMP-3 localized at the periphery of HeLa cells before infection, but localized with Salmonella in the cytosol by 5–8 h after infection. Before infection, LAMP-3 partially colocalized at the cell surface with LAMP-2, but 5 h after infection, some LAMP-2 protein surrounded the Salmonella, as reported (Roark and Haldar, 2008). It is not clear why LAMP-2 and LAMP-3 colocalized before infection and exhibited distinct localization patterns after infection. Recruitment of LAMP-1 to SCVs is known to be controlled by SifA-mediated translocation. The absence of SifA protein in Salmonella-infected cells results in independent LAMP-1 localization away from SCV (Beuzón et al., 2000). In this regard, it remains to be investigated whether SifA may control LAMP-3 localization.
LAMP-3 knockdown by specific RNAi expression in a stable cell line inhibited the intracellular proliferation of Salmonella, resulting in fewer recovered Salmonella bacterial colonies than seen in the case of control cells transfected with scrambled RNAi. LAMP-2 knockdown also resulted in a similar suppression of intracellular proliferation of Salmonella. Given that the LAMP-3 localizes primarily at the cell surface before Salmonella infection and subsequently with intracellular Salmonella, there is a possibility that the downregulation of LAMP-3 and LAMP-2 may actually interfere with the Salmonella infection process at the cell surface, resulting in fewer numbers of infected bacteria per cell. The reduced number of Salmonella bacteria found at the 2-h time point, in LAMP-3 or LAMP-2 knockdown cells versus scrambled controls, may support this interpretation. Thus, LAMP-3 or LAMP-2 deficiency may impede the early Salmonella infection process at the cell surface through phagocytosis. In this regard, it will be useful to use a LAMP-3 knockout cell line to test this possibility.
Acknowledgments
This work was carried out with the support of Next Generation BioGreen21 project (Project No.PJ01104401 and PJ01110901) of Rural Development Administration, Republic of Korea. This paper was also supported by research funds for newly appointed professors of Chonbuk National University in 2015. We appreciate Professor Heo, Won-Do and Dr. Kim, Nury at the Center for Cognition and Sociality, IBS, Korea for help with N-SIM Imaging.
REFERENCES
- Agarwal A.K., Srinivasan N., Godbole R., More S.K., Budnar S., Gude R.P., Kalraiya R.D. Role of tumor cell surface lysosome-associated membrane protein-1(LAMP1) and its associated carbohydrates in lung metastasis. J. Cancer Res. Clin. Oncol. 2015;141:1563–1574. doi: 10.1007/s00432-015-1917-2. [DOI] [PubMed] [Google Scholar]
- Andrejewski N., Punnonen E.L., Guhde G., Tanaka Y., Lullmann-Rauch R., Hartmann D., von Figura K., Saftig P. Normal lysosomal morphology and function in LAMP-1-deficient mice. J. Biol. Chem. 1999;274:12692–12701. doi: 10.1074/jbc.274.18.12692. [DOI] [PubMed] [Google Scholar]
- Angus A.A., Lee A.A., Augustin D.K., Lee E.J., Evans D.J., Fleiszig S.M. Pseudomonas aeruginosa induces membrane blebs in epithelial cells, which are utilized as a niche for intracellular replication and motility. Infect. Immun. 2008;76:1992–2001. doi: 10.1128/IAI.01221-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelqvist H., Waster P., Kagedal K., Ollinger K. The lysosome: from waste bag to potential therapeutic target. J. Mol. Cell Biol. 2013;5:214–226. doi: 10.1093/jmcb/mjt022. [DOI] [PubMed] [Google Scholar]
- Arpaia N., Godec J., Lau L., Sivick K.E., McLaughlin L.M., Jones M.B., Dracheva T., Peterson S.N., Monack D.M., Barton G.M. TLR signaling is required for Salmonella typhimurium virulence. Cell. 2011;144:675–688. doi: 10.1016/j.cell.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowski M.A., Braun V., Brumell J.H. Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic. 2008;9:2022–2031. doi: 10.1111/j.1600-0854.2008.00827.x. [DOI] [PubMed] [Google Scholar]
- Baldeon M.E., Ceresa B.P., Casanova J.E. Expression of constitutively active Rab5 uncouples maturation of the Salmonella-containing vacuole from intracellular replication. Cell. Microbiol. 2001;3:473–486. doi: 10.1046/j.1462-5822.2001.00130.x. [DOI] [PubMed] [Google Scholar]
- Barois N., de Saint-Vis B., Lebecque S., Geuze H.J., Kleijmeer M.J. MHC class II compartments in human dendritic cells undergo profound structural changes upon activation. Traffic. 2002;3:894–905. doi: 10.1034/j.1600-0854.2002.31205.x. [DOI] [PubMed] [Google Scholar]
- Bechetoille N., Andre V., Valladeau J., Perrier E., Dezutter-Dambuyant C. Mixed Langerhans cell and interstitial/dermal dendritic cell subsets emanating from monocytes in Th2-mediated inflammatory conditions respond differently to pro-inflammatory stimuli. J. Leukocyte Biol. 2006;80:45–58. doi: 10.1189/jlb.0205109. [DOI] [PubMed] [Google Scholar]
- Beuzon C.R., Salcedo S.P., Holden D.W. Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology. 2002;148:2705–2715. doi: 10.1099/00221287-148-9-2705. [DOI] [PubMed] [Google Scholar]
- Birmingham C.L., Smith A.C., Bakowski M.A., Yoshimori T., Brumell J.H. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- Castillo E.F., Dekonenko A., Arko-Mensah J., Mandell M.A., Dupont N., Jiang S., Delgado-Vargas M., Timmins G.S., Bhattacharya D., Yang H., et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc. Natl. Acad. Sci. USA. 2012;109:E3168–3176. doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravortty D., Hansen-Wester I., Hensel M. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J. Exp. Med. 2002;195:1155–1166. doi: 10.1084/jem.20011547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn B., Grassl G.A., Finlay B.B. Salmonella, the host and disease: a brief review. Immunol. Cell Biol. 2007;85:112–118. doi: 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- Cox T.M., Cachon-Gonzalez M.B. The cellular pathology of lysosomal diseases. J. Pathol. 2012;226:241–254. doi: 10.1002/path.3021. [DOI] [PubMed] [Google Scholar]
- De Saint-Vis B., Vincent J., Vandenabeele S., Vanbervliet B., Pin J.J., Ait-Yahia S., Patel S., Mattei M.G., Banchereau J., Zurawski S., et al. A novel lysosome-associated membrane glycoprotein, DC-LAMP, induced upon DC maturation, is transiently expressed in MHC class II compartment. Immunity. 1998;9:325–336. doi: 10.1016/s1074-7613(00)80615-9. [DOI] [PubMed] [Google Scholar]
- Dominguez-Bautista J.A., Klinkenberg M., Brehm N., Subramaniam M., Kern B., Roeper J., Auburger G., Jendrach M. Loss of lysosome-associated membrane protein 3 (LAMP3) enhances cellular vulnerability against proteasomal inhibition. Eur. J. Cell Biol. 2015;94:148–161. doi: 10.1016/j.ejcb.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Eskelinen E.L. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Aspects Med. 2006;27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Eskelinen E.L., Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim. Biophys. Acta. 2009;1793:664–673. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Fabrega A., Vila J. Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 2013;26:308–341. doi: 10.1128/CMR.00066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K., Yang X.L., Chen J.S., Hamilton S.R., August J.T. Differential expression of the lysosome-associated membrane proteins in normal human tissues. Arch. Biochem. Biophysics. 1999;365:75–82. doi: 10.1006/abbi.1999.1147. [DOI] [PubMed] [Google Scholar]
- Guha M., Mackman N. The phosphatidylinositol 3-kinase-Akt pathway limits lipopolysaccharide activation of signaling pathways and expression of inflammatory mediators in human monocytic cells. J. Biol. Chem. 2002;277:32124–32132. doi: 10.1074/jbc.M203298200. [DOI] [PubMed] [Google Scholar]
- Holness C.L., Simmons D.L. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood. 1993;81:1607–1613. [PubMed] [Google Scholar]
- Jo E.K., Yuk J.M., Shin D.M., Sasakawa C. Roles of autophagy in elimination of intracellular bacterial pathogens. Front. Immunol. 2013;4:97. doi: 10.3389/fimmu.2013.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanao H., Enomoto T., Kimura T., Fujita M., Nakashima R., Ueda Y., Ueno Y., Miyatake T., Yoshizaki T., Buzard G.S., et al. Overexpression of LAMP3/TSC403/DC-LAMP promotes metastasis in uterine cervical cancer. Cancer Res. 2005;65:8640–8645. doi: 10.1158/0008-5472.CAN-04-4112. [DOI] [PubMed] [Google Scholar]
- Kleijmeer M., Ramm G., Schuurhuis D., Griffith J., Rescigno M., Ricciardi-Castagnoli P., Rudensky A.Y., Ossendorp F., Melief C.J., Stoorvogel W., et al. Reorganization of multivesicular bodies regulates MHC class II antigen presentation by dendritic cells. J. Cell Biol. 2001;155:53–63. doi: 10.1083/jcb.200103071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler L.A., Celli J. Eating the strangers within: host control of intracellular bacteria via xenophagy. Cell. Microbiol. 2011;13:1319–1327. doi: 10.1111/j.1462-5822.2011.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X., Chen Y., Liu D., Li F., Li X., Jia W. High expression of LAMP3 is a novel biomarker of poor prognosis in patients with esophageal squamous cell carcinoma. Int. J. Mol. Sci. 2015;16:17655–17667. doi: 10.3390/ijms160817655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujcic H., Rzymski T., Rouschop K.M., Koritzinsky M., Milani M., Harris A.L., Wouters B.G. Hypoxic activation of the unfolded protein response (UPR) induces expression of the metastasis-associated gene LAMP3. Radiother. Oncol. 2009;92:450–459. doi: 10.1016/j.radonc.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Nagelkerke A., Mujcic H., Bussink J., Wouters B.G., van Laarhoven H.W., Sweep F.C., Span P.N. Hypoxic regulation and prognostic value of LAMP3 expression in breast cancer. Cancer. 2011;117:3670–3681. doi: 10.1002/cncr.25938. [DOI] [PubMed] [Google Scholar]
- Nagelkerke A., Bussink J., Mujcic H., Wouters B.G., Lehmann S., Sweep F.C., Span P.N. Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response. Breast Cancer Res. 2013;15:R2. doi: 10.1186/bcr3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagelkerke A., Sieuwerts A.M., Bussink J., Sweep F.C., Look M.P., Foekens J.A., Martens J.W., Span P.N. LAMP3 is involved in tamoxifen resistance in breast cancer cells through the modulation of autophagy. Endocr. Relat. Cancer. 2014;21:101–112. doi: 10.1530/ERC-13-0183. [DOI] [PubMed] [Google Scholar]
- Nishino I., Fu J., Tanji K., Yamada T., Shimojo S., Koori T., Mora M., Riggs J.E., Oh S.J., Koga Y., et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- Roark E.A., Haldar K. Effects of lysosomal membrane protein depletion on the Salmonella-containing vacuole. PLoS One. 2008;3:e3538. doi: 10.1371/journal.pone.0003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter S.W., Cloonan S.M., Choi A.M. Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol. Cells. 2013;36:7–16. doi: 10.1007/s10059-013-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy. 2012;8:1643–1656. doi: 10.4161/auto.21654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 2004;17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff A, Ayyar K, Saha D, Reddy KVR. Host autophagy response: friend or foe in reproductive tract infections. SOJ Microbiol. Infect Dis. 2014;2:1–9. [Google Scholar]
- Vazquez-Torres A., Xu Y., Jones-Carson J., Holden D.W., Lucia S.M., Dinauer M.C., Mastroeni P., Fang F.C. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science. 2000;287:1655–1658. doi: 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- Wileman T. Autophagy as a defence against intracellular pathogens. Essays Biochem. 2013;55:153–163. doi: 10.1042/bse0550153. [DOI] [PubMed] [Google Scholar]
- Wilke S., Krausze J., Büssow K. Crystal structure of the conserved domain of the DC lysosomal associated membrane protein: implications for the lysosomal glycocalyx. BMC Biol. 2012;10:62. doi: 10.1186/1741-7007-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Xue Q., Wan Y., Yang Y., Wang J., Hung T. Lysosome-associated membrane glycoprotein 3 is involved in influenza A virus replication in human lung epithelial (A549) cells. Virology J. 2011;8:384. doi: 10.1186/1743-422X-8-384. [DOI] [PMC free article] [PubMed] [Google Scholar]


