Abstract
Acute myeloid leukemia (AML) is a heterogenous disease with differential oncogene association, outcome and treatment regimens. Treatment strategies for AML have improved outcome but despite increased molecular biological information AML is still associated with poor prognosis. Proteomic analysis on the effects of a range of leukemogenic oncogenes showed that the protein transglutaminase 2 (TG2) is expressed at greater levels as a consequence of oncogenic transformation. Further analysis of this observation was performed with 511 AML samples using reverse phase proteomic arrays, demonstrating that TG2 expression was higher at relapse than diagnosis in many cases. In addition elevated TG2 expression correlated with increased expression of numerous adhesion proteins and many apoptosis regulating proteins, two processes related to leukemogenesis. TG2 has previously been linked to drug resistance in cancer and given the negative correlation between TG2 levels and peripheral blasts observed increased TG2 levels may lead to the protection of the leukemic stem cell due to increased adhesion/reduced motility. TG2 may therefore form part of a network of proteins that define poor outcome in AML patients and potentially offer a target to sensitize AML stem cells to drug treatment.
Keywords: Acute myeloid leukemia, Biomedicine, Reverse phase protein array, Transglutaminase2
1 Introduction
Major progress has been made in defining the molecular pathogenesis of specific leukemias. However, as survival has not improved significantly in the last 2 decades, there remains a requirement for novel therapies in the treatment of the acute myeloid leukemias (AMLs). The leukemias have been characterized as diseases in which complementary action of mutated signal transduction proteins and transcription factors produce an overt acute disease [1, 2]. There is evidence that the effects of leukemogenic proteins are regulated by posttranslational governance of protein expression levels via process such as ubiquitination [3, 4]. Thus proteomic analyses of events downstream from both transcription factor and signal transduction oncogenes will be a key feature of the determination of transformation processes. Identifying common effectors essential in leukemogenesis can be achieved effectively with proteomic analysis and is able to discover those that are regulated posttranslationally. This is a necessary pursuit in the acute myeloid leukemias where there is wide diversity in the molecular pathology of the disease.
Measurement of protein entities in a systematic fashion in the primitive hematopoietic cells offers technical challenges due to their extremely low number. While enrichment strategies exist there is an inevitable diminution in the quantity of cells available that means there is a need to have a sensitive means of assaying the analyte [5]. For this reason, an antibody-based technique requiring relatively few cells for protein relative quantification has been developed to pursue targets that have been identified through discovery proteomics mass spectrometry screens. There is data (https://brd.nci.nih.gov/BRN/search.seam) showing that technical variation in clinical sample handling leads to changes in the characteristics of the samples for DNA and RNA; this is certainly also true for protein. This can lead to varied results in profiling arrays and biomarker studies. We have therefore used a well validated and documented [6–9] reverse phase protein array (RPPA) method on primary material to investigate the expression of proteins implicated in leukemic transformation from our model systems. We have previously shown that RPPA has applicability in leukemia research screening a large number of primary cell samples [7,8].
Our previous proteomic screen of multiple leukemogenic oncogenes identified transglutaminase2 (TG2) as a protein whose expression was enhanced by a number of different oncogenes [10]. While some of the roles of TG2 under normal physiological conditions remain obscure, the protein is believed to participate in the pathogenesis of several unrelated diseases, including celiac sprue, neurodegenerative diseases, and some cancers [11–13]. TG2 expression has also been associated with resistance to chemotherapy and apoptosis in some cancers [14–16]. TG2 has been reported to be an anti-apoptotic mediator of hypoxia inducible factor conferring a growth advantage to tumor cells. As such it has been considered a potential target in cancer therapy [17]. In addition it has been shown that TG2 overexpression is an adverse prognostic factor in ovarian carcinoma and TG2 targeting may be an attractive therapeutic approach in this disease [18].
In this study, we show that TG2 is expressed in AML blasts and has higher expression at relapse compared to diagnosis. Further analysis of proteins that correlated with changes in TG2 expression revealed an abundance of adhesive and motility proteins. Thus TG2 may form a part of a network of proteins that can potentially define poor outcome in AML patients.
2 Materials and methods
2.1 Cell lines and Western blot analysis
Ba/F3 cells were transfected with either an empty MSCV retroviral vector, or MSCV containing BCR/ABL, TEL/PDGFRβ, FIP1/PDGFRα, KIT D816V, NPM/ALK, or FLT3ITD as previously described [10]. Western blotting was performed using standard protocols. Antibodies used were actin (Sigma, UK) and Transglutaminase 2 (Abcam, UK).
2.2 Primary cell samples and preparations
Patient samples were collected between September 1999 and March 2007 from peripheral blood and bone marrow of newly diagnosed AML evaluated at the University of Texas MD Anderson Cancer Center. Samples were acquired during routine diagnostic assessments in accordance with the regulations and protocols approved by the Investigational Review Board of the University of Texas MD Anderson Cancer Center. Informed consent was obtained in accordance with the Declaration of Helsinki. Samples were analyzed under the Institutional Review Board-approved laboratory protocol. Collected material was enriched for leukemic cells by performing Ficoll separation to yield a mononuclear fraction followed by CD3/CD19 depletion to remove contaminating T and B cells, if they were calculated to be >5% based on the differential. The samples were normalized to a concentration of 1 × 104 cells/μL and a whole cell lysate prepared as reported [8].
2.3 Reverse phase protein array
The study employed a RPPA dataset that was generated by using samples from patients with acute lymphoblastic leukemia (ALL) and AML as described elsewhere [7, 8]. In brief, patient samples were printed in five serial dilutions onto slides along with normalization and expression controls. Slides were probed with a strictly validated TG2 primary antibody (Abcam, ab2386, UK) and a secondary antibody followed by colorimetric assay [9]. Stained slides were analyzed using Microvigene® software (Vigene Tech, Carlisle, MA) to produce quantified data. Supercurve algorithms were used to generate a single value from the five serial dilutions. Loading control and topographical normalization procedures accounted for protein concentration and background staining variations. Protein associations were investigated using Ingenuity pathway analysis software (Ingenuity Systems, USA).
2.4 Statistical assessments
Statistical approaches have been described in detail elsewhere [8]. In brief, we used SuperCurve software (http://bioinformatics.mdanderson.org/Software/OOMPA) to define robust confidence intervals for the signal strength (protein expression) of each sample dilution series on the slide. Differences in protein expression between leukemic cases and normal CD34+ cells were assessed using a standard t-test. Comparison of the protein levels between paired samples (e.g. cryopreserved and fresh material, or diagnosis and relapse material) was carried out by performing paired t-tests. We used ANOVA to assess the association of protein level with categorical clinical variables and used linear regression for continuous clinical variables. We calculated both Pearson and Spearman correlation coefficients (R) between protein level and continuous variables. For time-to-event analyses (overall survival, progression-free survival, and complete remission duration), we divided protein expression into sixths based on the range of expression of all 511 samples. We then applied the log-rank test to assess survival differences among the groups of subjects with different protein levels. The Kaplan–Meier method was used to generate the survival curves. Univariate and multivariate Cox proportional hazard modeling was carried out to investigate the association with survival with protein levels as categorized variables. Bonferroni corrections were performed to account for multiple statistical parameters for calculating statistical significance. Statistical analyses were performed using R version 2.15.0 and Statistica version 10 software (StatSoft, Tulsa, USA).
3 Results
3.1 The effect of leukemogenic oncogenes on TG2 expression
In a systems biology project using isobaric tagging for relative quantification technology we have recently reported the effects of six leukemogenic oncogenes on the proteome of hematopoietic cells [10]. One protein shown to be altered by all six oncogenes in this model was TG2. Here, we have validated this observation by western blot analysis. Figure 1 clearly demonstrates that the six oncogenes induce the expression of TG2 albeit to a different extent.
Figure 1.
TG2 expression. Ba/F3 cells transfected with the oncogenes shown were lysed and the expression of TG2 assessed by Western blot analysis. Actin levels were used as a loading control.
3.2 Workflow for relative analysis of TG2 expression
In a systems biology approach based in model cells it is important to develop high throughput assays to confirm findings in primary material. Here, we describe such a system, investigating TG2 expression in 719 AML samples from 511 AML patients, along with an additional 28 samples from 21 acute promyelocytic leukemia patients. There were ten peripheral blood leucocyte controls, 11 normal, unstimulated bone marrow derived CD34 positive (primitive normal) cell controls, and ten G-CSF primed peripheral blood derived CD34+ cell controls. In total there were 80 clinical and laboratory parameters or assessments associated with the samples gathered for this study including the demographics, cytogenetic information, treatment, disease information, and flow cytometry data. The expression observed in the normal CD34+ bone marrow samples was used to establish a control value of normal expression for ratiometric analyses for the data described below.
3.3 Comparison of the protein levels for paired cryopreserved and fresh cell samples or bone marrow compared to peripheral blood cells
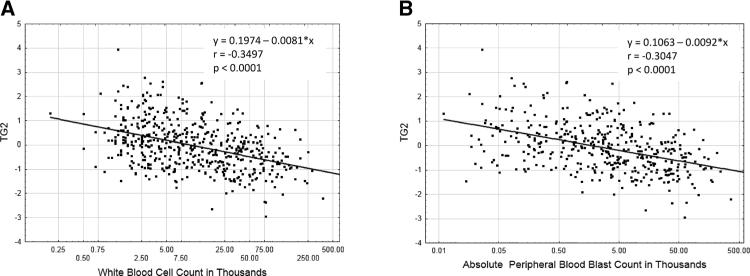
A potential issue with clinical material analyses is whether cryopreservation affects the stability of the specific protein in question. We have observed that lengthy delays can affect the quality of data accrued [6]. In order to assess this possibility, we compared, expression in 58 paired samples, with a protein preparation made immediately on the day of collection and another prepared later from cryopreserved cells. The level of TG2 expression in fresh and cryopreserved samples was statistically similar (p = 0.71) as shown in Supporting Information Fig. 1. There were 140 paired collections of samples from bone marrow and blood enabling a comparison of TG2 expression in each compartment. As shown in Supporting Information Fig. 2 expression of TG2 in blood and marrow was statistically similar (p = 0.43). This suggests that the data from either time of preparation (freshly prepared or cryop-reserved) or sample source (blood versus marrow) could be used in the analysis. An initial investigation into TG2 levels and peripheral blood cell counts in the 719 samples revealed that there was a negative correlation between TG2 level and both white blood cell count (Fig. 2A) and the absolute number of blasts in the peripheral blood (Fig. 2B). Thus further analysis of the protein was viewed as appropriate.
Figure 2.
Correlation between TG2 levels and number of white cells and peripheral blasts. Scatterplot and correlation data from 719 patient samples between TG2 levels and white blood cell count (A) or number of peripheral blood blasts (B). The shape of the curve on a y = mx + c straight line fit and the correlation coefficient (R square) are shown in the panel.
3.4 Comparison of AML samples and normal primitive hematopoietic cells
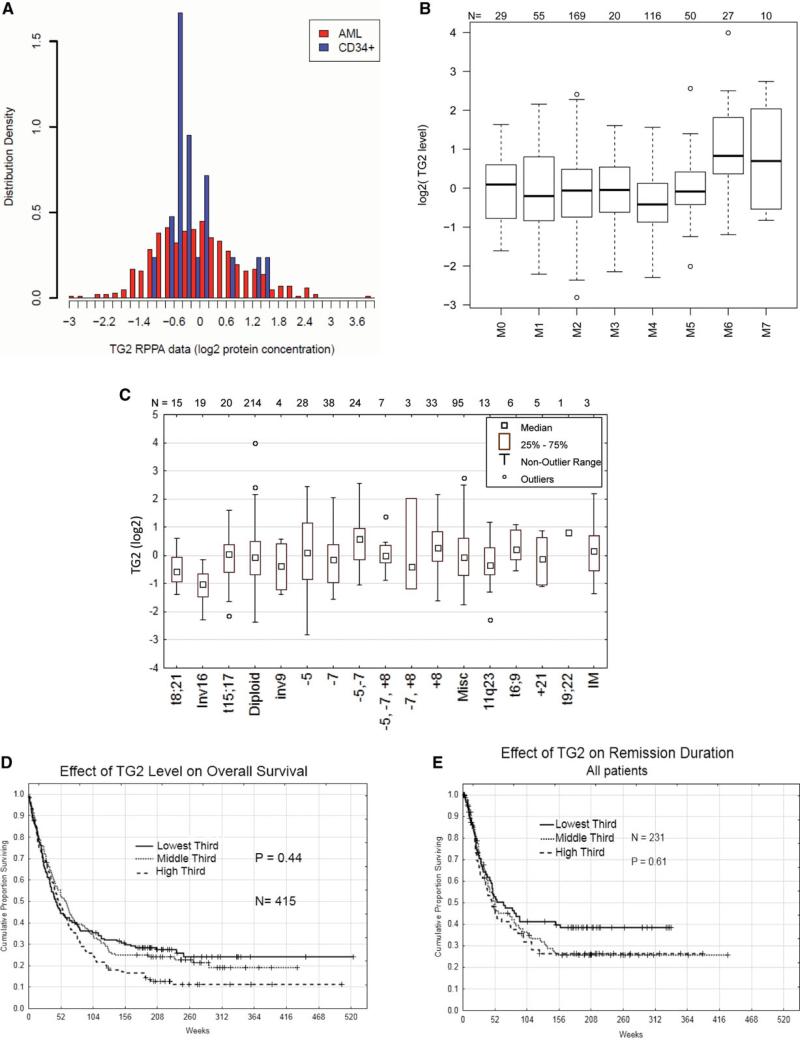
We first asked whether TG2 was expressed in AML and observed that the majority of patients did express TG2 and at levels that were similar to that of normal CD34+ bone marrow cells (Fig. 3A). We next assessed the correlation of protein level and clinical data of newly diagnosed patients. The F-statistics (8.03) from an ANOVA test with a p-value of less than 0.00001 suggests expression was similar (with respect to median expression or the range of expression) for French American British AML categorizations M0 through M5, but levels were higher in erythrocytic (M6) and megakaryocytic (M7) subtypes (Fig. 3B). Using a pair wise analysis in a Tukey HSD test M6 and M7 are statistically higher than M0–5 (p-value < 0.00001 and < 0.0075, respectively). Likewise TG2 expression was not associated with cytogenetic alterations with the exception that expression in patients with inversion 16 (inv16) AML were lower (Fig. 3C).
Figure 3.
Correlation of TG2 protein status to clinical data. (A) Comparison of protein levels in newly diagnosed AML samples and normal CD34+ samples. (B) Correlation of French American British leukemia classification and TG2 protein expression status using Box plot analysis. (C) Cytogenetic analysis data for the AML patient set and its correlation to TG2 protein level measurements in Box plot form. For survival analysis, 256 newly diagnosed AML patients were divided equally into three groups according to the TG2 protein level. Kaplan–Meier survival estimates for AML patients with different protein levels are shown for each group (low, middle, and high levels of TG2 expression) (D) and all three groups in terms of remission duration (E).
Next, we examined whether the level of TG2 was correlated with outcome. The Martingale residual plot suggested a threshold effect with survival worsening between the 4th and 5th sextiles and a lesser suggestion for another threshold somewhere in the second sextile (data not shown). Based on this, the cohort of patients was divided into thirds for comparison. The effect of TG2 level on overall survival was then examined (Fig. 3D). Those with higher levels of TG2 had an inferior overall survival with a plateau of ~11% survival versus 19% and 25% but neither achieved statistical significance. The potential effect on overall survival was also examined in subsets of patients with favorable, intermediate, or unfavorable cytogenetics, but TG2 level was not significantly prognostic in any subset. The lone inv16 patient with high TG2 relapsed and ultimately died at 140 weeks (from diagnosis), compared to five deaths among the other 17 Inv16 cases. Having observed a late divergence between the survival curves of those with higher TG2 we queried whether this might be associated with a difference in remission duration (Fig. 3E). Once again, those with higher TG2 levels had a somewhat inferior remission duration experience, but this was not statistically significant regardless of the cut off point used or whether all cases, or those with favorable, intermediate or unfavorable cytogenetics were examined.
3.5 Comparison of TG2 protein levels for paired “newly diagnosed” and “relapse” samples
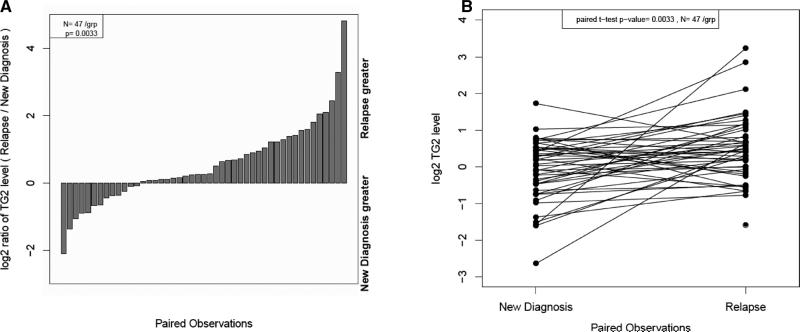
This dataset has 47 paired diagnosis-relapse samples taken from the same patient at different disease states, enabling an assessment of whether TG2 levels change with disease progression. As shown in Fig. 4A and B levels were markedly higher at relapse compared to diagnosis (p = 0.003).
Figure 4.
The distributions of TG2 protein levels for AML patients where paired relapse and new diagnosis samples are available. (A) Paired sample data are shown in a relative quantification plot demonstrating samples where relapse sample or material at diagnosis is greater. (B) Paired samples from relapse or new diagnosis material were analyzed for TG2 expression. The cryopreserved samples are shown connected to the fresh samples for each of 46 pairs.
3.6 TG2 expression correlates with proteins associated with apoptosis or cell adhesion
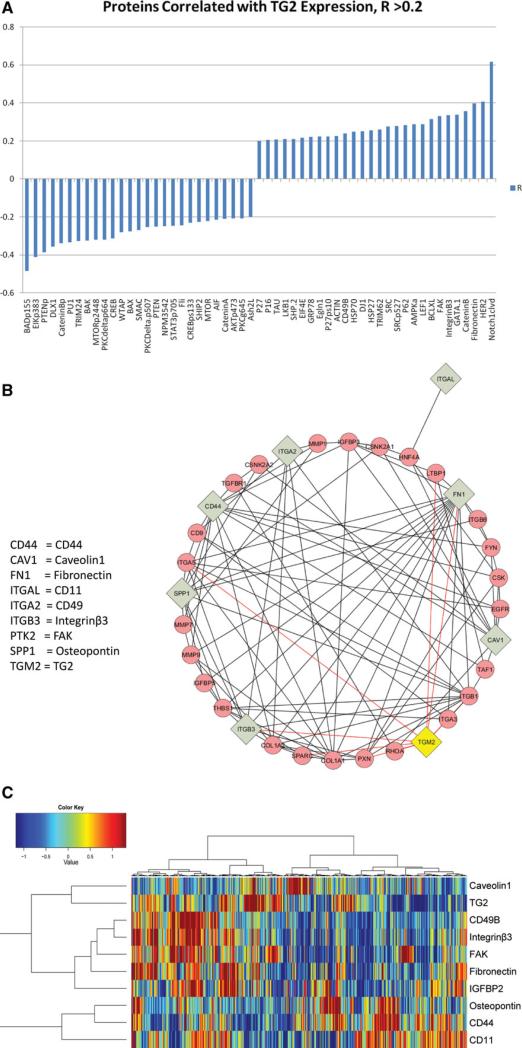
The RPPA method employed in this study has been used to assess the expression of 203 other protein entities on the samples described here. This is, of course, a subjective set chosen for potential interest in AML. Nonetheless it is striking that 55 proteins from the 203 correlated with TG2 expression (Fig. 5A). Analysis of online databases of known protein–protein interactions revealed that TG2 has known interactions with integrinβ3 and fibronectin as shown in Supporting Information Fig. 3. TG2 is known to bind and posttranslationally modify fibronectin while it can act as a coreceptor for fibronectin with Integrinβ3 [19]. TG2 expression also positively correlates with focal adhesion kinase (FAK) expression, a key protein in extracellular matrix/intracellular signaling. The known interactions between TG2 and the nine adhesion/integrin proteins analyzed in this dataset is shown in the Cytoscape interaction plot (Fig. 5B). In this dataset, TG2 expression was strongly associated (positively and negatively) with multiple proteins involved in adhesion and mobility, such as fibronectin, Integrinβ3, FAK, and Actin, replicating the Cytoscape interactome. Protein changes were examined further using a hierarchical clustering (Fig. 5C). The analysis illustrated that these integrin/adhesion high cluster into two groups of tightly associated proteins: Group 1: CD49, FAK, Fibronectin, IGFBP3, and Integrinβ3 and Group 2: CD11, CD44, and Ostepontin with Caveolin 1 (CAV1) expression associated with neither. As shown in the dendogram along the top of Fig. 5C, AML divides into cases with high levels of group 1 and low levels of group 2 proteins (far left) another with the inverse (high group 2, low group 1) and that very few cases have neither group elevated (middle group with only high CAV1), elevated TG2 expression was tightly correlated with expression of group 1 proteins and inversely correlated with that of Group 2 proteins, although not all group 1 high expressors had high TG2, and not all high TG2 cases had high group 1 protein expression. In combination this suggests that there is canonical interaction between TG2 and integrin and adhesion proteins active in AML.
Figure 5.
Proteins correlated with TG2 expression. (A) Negative and positive correlation from a list of 201 proteins also assayed using the same reverse phase proteomics analysis on the same samples; Pearson correlation coefficient R > 0.2, p < 0.001 or < 0.001. (B) Cytoscape software analysis of TG2 and the nine adhesion/integrin proteins assayed on the RPPA illustrating protein interactions. The adhesion/integrin used in the analysis are shown in gray boxes and pink circles are used for proteins included as nearest neighbors interacting with two or more components. TG2 is highlighted in yellow. Lines between nodes are red for those associated with TG2 and black for all other protein interactions. The adhesion/integrin proteins on the array and TG2 were entered as a MIMI query limited to human proteins with known direct interactions. Names appear as gene symbols with the corresponding protein name given for the nine proteins used in the analysis. (C) Unbiased hierarchical clustering was performed in an analysis including the nine adhesion/integrin proteins on the array known to be correlated with TG2 expression. A heat map is shown of the reverse phase protein array data for the expression of adhesion/integrin proteins identified linked to TG2 expression. Blue indicates low expression and red the highest expression as shown by the scale. The dendogram on the right side shows that proteins divided into two groups with Caveolin 1 not being correlated with the other adhesion/integrin proteins. The dendogram along the top demonstrates that AML divides into two groups, high for of the protein groups and that few cases had low expression of both group 1 and group 2 proteins. TG2 expression was strongly positively correlated with group 1 proteins and inversely correlated with group 2 proteins.
4 Discussion
Our cell line models clearly demonstrated that increased TG2 expression is a common consequence of many different leukemogenic oncogenes. There are other data that indicate TG2 involvement in leukemia. It has been reported to play an important role in neutrophil granulocyte differentiation [20] and through this role it has been linked to acute promyelocytic leukemia [21]. Our observations, emanating from a discovery proteomics platform therefore warranted validation in primary material. Here, we demonstrate that TG2 expression is present in the majority of cases of AML at levels comparable to normal bone marrow CD34+ cells at initial diagnosis and that levels became higher at relapse. This suggests that the protein expression signature associated with high TG2 levels may be selected for, or confer a subtle survival advantage to leukemic blasts with higher levels. In support of this, while the level of TG2 was not statistically significantly prognostic for either overall survival or remission duration, patients with higher levels were somewhat more likely to relapse, and less likely to live beyond three years. We are presently investigating whether a combination of CAV1, ITGA2, TG2, and CD44 offer the potential to identify the cohort of patients at risk of relapse. In addition to identifying patients at risk of relapse we believe that the utility of these biomarkers would be more in selecting what integrin/adhesion interfering agents to use in which patient. This would however, require the development of such anti-integrin agents and also the demonstration that triaging based on this panel offers increased cell death in vitro with primary cells from patients with AML before any assessment in clinical trials.
Our further analysis concerned correlative expression patterns of the TG2 with other proteins. With respect to the work of Satpathy et al. in which a decrease in TG2 expression was noted to accompany increased Protein Phosphatase 2a levels [22] a trend for decreased Protein Phosphatase 2a with increasing TG2 (R = −.153, p = 0.0005) that is in agreement with their findings, but in our hands this resulted in less, not more, CREBpSer133. However, the stand out observation from the dataset as a whole was the correlation with adhesion/motility protein expression seen with TG2.
Many reports have demonstrated elevated expression of TG2 in multiple tumors and those resistant to chemotherapy suggesting a link between TG2 expression and metastatic and drug resistance in cancer [16,23,24]. TG2 has been shown to activate FAK in breast cancer cells leading to increased invasion [15]. In gene expression profiling experiments of breast cancer stromal cells, Mehta et al. (personal communication) demonstrated that upregulation of TG2 expression resulted in increased expression of several of the same proteins shown to be correlated in our study: fibronectin (41-fold) LEF1(29-fold) Trim62 (7.8-fold), Notch1 (6.8-fold). We observed a positive correlation of FAK, integrinβ3, fibronectin, and TG2. The activation of integrinβ3 is known to potentiate apoptosis in certain cell types and TG2 expression positively correlated with antiapoptotic Bcl-XL (BCL2L1) expression and negatively correlates with both the BAX proapoptotic protein and the BAD protein phosphorylated on serine 155, which inactivates BAD, resulting in a decrease in proapoptotic forces within the cell. This phosphorylation on BAD governs dissociation with BCL2L1 and enhanced cell death. Thus this phosphosite correlation was contraindicatory when compared with that seen with apoptotic proteins. Furthermore, the proapoptotic protein SMAC/DIABLO negatively correlated with TG2 expression. The LKB1/AMPK proteins also positively correlated with TG2 expression, these proteins pathway can function in cellular metabolic stress sensing and regulate autophagy.
The question arises how could TG2 levels be linked to a higher probability of relapse? Interaction of cells with the surrounding extra cellular matrix is critical for cell adhesion and migration. TG2 is known to modify fibronectin by interacting with integrinβ3 thus increases cell adhesion [19]. Given the negative correlation between TG2 levels and peripheral blasts it can be hypothesized that the increase in TG2 levels results in the protection of the leukemic stem cell due to increased adhesion hence reduced motility. In support of the concept that TG2 promotes the expression of integrins and adhesion markers, thereby increasing leukemic—stromal interaction is the observation that both the white blood cell count and the absolute peripheral blood blast count were inversely correlated with TG2 levels. This TG2 induced stabilization of cell–cell and cell–niche interaction could also be responsible for the reported relationship between CD44 expression and the relapse of AML [25]. Thus there is a positively reinforced pathway in many cases of AML studied here, with respect to extracellular matrix association. Metastasis of primary tumors involves many steps including successful intravasation via cell/matrix interactions, survival during circulation extravasation, and colonization by the tumor cells. As well as true in solid tumors these processes are important in the leukemias. We have recently demonstrated this to be true in relapse of childhood ALL [26]. We reported that ALL cells acquire the ability to populate the central nervous system via changes in expression of proteins involved in adhesion and invasion. Similarly here TG2 expression correlated to relapse of AML patients and the expression of proteins associated with survival (negative correlation with proapoptotic BAK and BAX and positive correlation with BCLxl) adhesion and motility (fibronectin, integrinβ3, and FAK). Strategies to disrupt TG2 expression or function might decrease these inhibitory leukemic blast-stromal cell interactions thereby sensitizing AML blasts to chemotherapy or targeted therapies.
In summary, we have extended observations from a discovery proteomics approach to validation of potential targets via reverse phase protein arrays. In the process, we have demonstrated that higher levels of TG2 are seen in the more advanced state of disease in relapsed patients. We were also able to correlate TG2 expression with altered expression of proteins involved in apoptosis and motility/extracellular matrix association, two processes closely related to leukemogenesis and disease progression. Thus discovery proteomics can direct insights into leukemias via highlighting proteins of interest, which in turn leads to identification of pathways and associated proteins that define primary cell transformation events.
Supplementary Material
Acknowledgments
This work was supported by Leukaemia Lymphoma Research UK (A.D.W., S.M.), Cancer Research UK (A.D.W., S.M.), and Leukemia Lymphoma Society (S.K.).
Thanks to Dr. Kapil Mehta for sharing his GEP data with us.
Abbreviations
- ALL
acute lymphoblastic leukemia
- AML
acute myeloid leukemia
- CAV1
Caveolin1
- FAK
focal adhesion kinase
- TG2
transglutaminase2
- RPPA
reverse phase protein array
Footnotes
Additional supporting information may be found in the online version of this article at the publisher's web-site
The authors have declared no conflict of interest.
References
- 1.Kelly LM, Gilliland DG. Genetic's of myeloid leukemias. Annu. Rev. Genomics Hum. Genet. 2002;3:179–198. doi: 10.1146/annurev.genom.3.032802.115046. [DOI] [PubMed] [Google Scholar]
- 2.Tefferi A, Gilliland DG. Oncogenes in myeloproliferative disorders. Cell Cycle. 2007;6:550–566. doi: 10.4161/cc.6.5.3919. [DOI] [PubMed] [Google Scholar]
- 3.Naujokat C, Šarić T. Concise review: role and function of the ubiquitin-proteasome system in mammalian stem and progenitor cells. Stem cells. 2007;25:2408–2418. doi: 10.1634/stemcells.2007-0255. [DOI] [PubMed] [Google Scholar]
- 4.Vink J, Cloos J, Kaspers GJL. Proteasome inhibition as novel treatment strategy in leukaemia. Br. J. Haematol. 2006;134:253–262. doi: 10.1111/j.1365-2141.2006.06170.x. [DOI] [PubMed] [Google Scholar]
- 5.Williamson AJK, Whetton AD. The requirement for proteomics to unravel stem cell regulatory mechanisms. J. Cell. Physiol. 2011;226:2478–2483. doi: 10.1002/jcp.22610. [DOI] [PubMed] [Google Scholar]
- 6.Kornblau SM, Coombes KR. Use of reverse phase protein microarrays to study protein expression in leukemia: technical and methodological lessons learned. MethodsMol. Biol. 2011;785:141–155. doi: 10.1007/978-1-61779-286-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kornblau SM, Qiu YH, Zhang N, Singh N, et al. Abnormal expression of friend leukemia virus integration 1 protein is an adverse prognostic factor in acute myeloid leukemia. Blood. 2011;118:5604–5612. doi: 10.1182/blood-2011-04-348052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kornblau SM, Tibes R, Qiu YH, Chen W, et al. Functional proteomic profiling of AML predicts response and survival. Blood. 2009;113:154–164. doi: 10.1182/blood-2007-10-119438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tibes R, Qiu Y, Lu Y, Hennessy B, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 10.Pierce A, Unwin RD, Evans CA, Griffiths S, et al. Eight-channel iTRAQ enables comparison of the activity of six leukemogenic tyrosine kinases. Mol. Cell. Proteomics. 2008;7:853–863. doi: 10.1074/mcp.M700251-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Fok JY, Ekmekcioglu S, Mehta K. Implications of tissue transglutaminase expression in malignant melanoma. Mol. Cancer Ther. 2006;5:1493–1503. doi: 10.1158/1535-7163.MCT-06-0083. [DOI] [PubMed] [Google Scholar]
- 12.Verma A, Wang H, Manavathi B, Fok JY, et al. Increased expression of tissue transglutaminase in pancreatic ductal adenocarcinoma and its implications in drug resistance and metastasis. Cancer Res. 2006;66:10525–10533. doi: 10.1158/0008-5472.CAN-06-2387. [DOI] [PubMed] [Google Scholar]
- 13.Yuan L, Siegel M, Choi K, Khosla C, et al. Transglutaminase 2 inhibitor, KCC009, disrupts fibronectin assembly in the extracellular matrix and sensitizes orthotopic glioblastomas to chemotherapy. Oncogene. 2006;26:2563–2573. doi: 10.1038/sj.onc.1210048. [DOI] [PubMed] [Google Scholar]
- 14.Antonyak MA, Miller AM, Jansen JM, Boehm JE, et al. Augmentation of tissue transglutaminase expression and activation by epidermal growth factor inhibit doxorubicin-induced apoptosis in human breast cancer cells. J. Biol. Chem. 2004;279:41461–41467. doi: 10.1074/jbc.M404976200. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Xu J, Brady S, Gao H, et al. Tissue transglutaminase promotes drug resistance and invasion by inducing mesenchymal transition in mammary epithelial cells. PLoS One. 2010;5:e13390. doi: 10.1371/journal.pone.0013390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehta K, Fok J, Miller FR, Koul D, Sahin AA. Prognostic significance of tissue transglutaminase in drug resistant and metastatic breast cancer. Clin. Cancer Res. 2004;10:8068–8076. doi: 10.1158/1078-0432.CCR-04-1107. [DOI] [PubMed] [Google Scholar]
- 17.Jang GY, Jeon JH, Cho SY, Shin DM, et al. Transglutaminase 2 suppresses apoptosis by modulating caspase 3 and NF-[kappa]B activity in hypoxic tumor cells. Oncogene. 2010;29:356–367. doi: 10.1038/onc.2009.342. [DOI] [PubMed] [Google Scholar]
- 18.Hwang JY, Mangala LS, Fok JY, Lin YG, et al. Clinical and biological significance of tissue transglutaminase in ovarian carcinoma. Cancer Res. 2008;68:5849–5858. doi: 10.1158/0008-5472.CAN-07-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akimov SS, Krylov D, Fleischman LF, Belkin AM. Tissue transglutaminase is an integrin-binding adhesion coreceptor for fibronectin. J. Cell Biol. 2000;148:825–838. doi: 10.1083/jcb.148.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balajthy Z, Csomós K, Vámosi G, Szántó A, et al. Tissue-transglutaminase contributes to neutrophil granulocyte differentiation and functions. Blood. 2006;108:2045–2054. doi: 10.1182/blood-2004-02-007948. [DOI] [PubMed] [Google Scholar]
- 21.Csomoós K, Német I, Fésüs L, Balajthy Z. Tissue transglutaminase contributes to the all-trans-retinoic acid—induced differentiation syndrome phenotype in the NB4 model of acute promyelocytic leukemia. Blood. 2010;116:3933–3943. doi: 10.1182/blood-2010-01-266064. [DOI] [PubMed] [Google Scholar]
- 22.Satpathy M, Shao M, Emerson R, Donner DB, Matei D. Tissue transglutaminase regulates matrix metalloproteinase-2 in ovarian cancer by modulating cAMP-response element-binding protein activity. J. Biol. Chem. 2009;284:15390–15399. doi: 10.1074/jbc.M808331200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao L, Petrusca DN, Satpathy M, Nakshatri H, et al. Tissue transglutaminase protects epithelial ovarian cancer cells from cisplatin-induced apoptosis by promoting cell survival signaling. Carcinogenesis. 2008;29:1893–1900. doi: 10.1093/carcin/bgn158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangala LS, Fok JY, Zorrilla-Calancha IR, Verma A, Mehta K. Tissue transglutaminase expression promotes cell attachment, invasion and survival in breast cancer cells. Oncogene. 2006;26:2459–2470. doi: 10.1038/sj.onc.1210035. [DOI] [PubMed] [Google Scholar]
- 25.Quere R, Andradottir S, Brun ACM, Zubarev RA, et al. High levels of the adhesion molecule CD44 on leukemic cells generate acute myeloid leukemia relapse after withdrawal of the initial transforming event. Leukemia. 2011;25:515–526. doi: 10.1038/leu.2010.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland M, Castro FV, Alexander S, Smith D, et al. RAC2, AEP, and ICAM1 expression are associated with CNS disease in a mouse model of pre-B childhood acute lymphoblastic leukemia. Blood. 2011;118:638–649. doi: 10.1182/blood-2010-09-307330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.