Abstract
Fungal mycelia serve as effective dispersal networks for bacteria in water-unsaturated environments, thereby allowing bacteria to maintain important functions, such as biodegradation. However, poor knowledge exists on the effects of dispersal networks at various osmotic (Ψo) and matric (Ψm) potentials, which contribute to the water potential mainly in terrestrial soil environments. Here we studied the effects of artificial mycelium-like dispersal networks on bacterial dispersal dynamics and subsequent effects on growth and benzoate biodegradation at ΔΨo and ΔΨm values between 0 and −1.5 MPa. In a multiple-microcosm approach, we used a green fluorescent protein (GFP)-tagged derivative of the soil bacterium Pseudomonas putida KT2440 as a model organism and sodium benzoate as a representative of polar aromatic contaminants. We found that decreasing ΔΨo and ΔΨm values slowed bacterial dispersal in the system, leading to decelerated growth and benzoate degradation. In contrast, dispersal networks facilitated bacterial movement at ΔΨo and ΔΨm values between 0 and −0.5 MPa and thus improved the absolute biodegradation performance by up to 52 and 119% for ΔΨo and ΔΨm, respectively. This strong functional interrelationship was further emphasized by a high positive correlation between population dispersal, population growth, and degradation. We propose that dispersal networks may sustain the functionality of microbial ecosystems at low osmotic and matric potentials.
INTRODUCTION
Population dispersal is considered a key factor of ecosystem functioning, enabling bacteria to invade new habitats and to harness new pools of resources (1). Consequently, it is also a prerequisite for efficient biodegradation in soils because it increases the contact probability for bacteria and contaminants (2). However, in terrestrial environments, bacterial surface motility is usually restricted due to limited water availability, which is anticipated to become an even more important constraint in future due to prolonged drought periods (3, 4). Water availability is described by the concept of the water potential (Ψw), which is a measure of the energetic state of water in a system (5). In water-saturated soils, Ψw is determined almost exclusively by the osmotic potential (Ψo), which refers to the amount of solutes in the aqueous phase. In unsaturated environments, the matric potential (Ψm) also becomes important due to increasing capillary and adsorptive forces between water and the soil matrix (5, 6). Several studies evaluated the effects of different water potentials on the biodegradation performance of bacteria, mainly over time, without consideration of spatial processes (7, 8). However, both the spatial and temporal dynamics are crucial for a system's biodegradation performance, as substrates and bacteria in soil are typically distributed heterogeneously (2, 9). In a sand matrix adjusted to different Ψm values, degradation has been shown to depend strongly on the initial distribution and the dispersal of degrading bacteria (10). Researchers have revealed various bacterial strategies to cope with the direct physiological effects of low water potentials by, for example, accumulating compatible solutes or changing the lipid content of the membranes (11), but mechanisms to overcome the motility restrictions in thin water films are still poorly understood.
In contrast to bacteria, fungi do not rely on continuous water pathways to grow through the soil matrix. They can bridge air-water interfaces due to efficient resource translocation in the mycelium, thus connecting fragmented soil habitats (12, 13). Flagellated bacteria can actively move in the liquid films surrounding hydrophilic hyphae and thus overcome motility restrictions in unsaturated environments (14). Different studies using soil column experiments and simulation modeling showed that this phenomenon can enhance biodegradation of phenanthrene and glucose, respectively (15–17). However, studies to assess the benefits of dispersal networks were carried out with culture plates with different agar concentrations routinely used in standard bacterial motility assays (18–20). Indeed, these concentrations represent only a narrow range of Ψm values, which is insufficient for studying relevant environmental conditions, in particular in soils, where Ψm often falls to −1.5 MPa (21). Salt accumulation caused by irrigation or fertilization is a major threat in agricultural soils, and globally, 108 ha (5%) of arable land are affected by salt at a level that causes osmotic stress (22). Furthermore, soil drying and soil surface proximity are often accompanied by high solute concentrations (9), which have been shown to cause a repression of motility genes, leading to an impairment of movement in Pseudomonas and Bacillus strains (23, 24). However, no study has yet evaluated the effects of dispersal networks at different Ψo values.
In the present study, we tested the hypothesis that dispersal networks facilitate bacterial dispersal over a broad range of environmentally relevant Ψw values and hence improve the ecosystem's functional performance in terms of bacterial biodegradation. To obtain a mechanistic understanding of the effects of different water potentials and dispersal networks on bacterial population dispersal, we developed highly controlled laboratory microcosms in which we measured the radial colony expansion at different Ψw values in both the presence and absence of an artificial dispersal network. By varying either Ψo or Ψm, we aimed to disentangle the different modes of action as well as the capability of dispersal networks to improve the systems' performance for the two main descriptors of Ψw in soil. The systems' performance was assessed by following population growth of a green fluorescent protein (GFP)-tagged derivative of the well-studied soil bacterium Pseudomonas putida KT2440 and the biodegradation of benzoate as a model for water-soluble and therefore well-accessible substrates.
MATERIALS AND METHODS
Organisms and culture conditions.
A benzoate-degrading, GFP-tagged derivative of the soil bacterium Pseudomonas putida KT2440 was chosen for the experiments because of its well-known motility behavior (4, 25). It was cultivated in FAB minimal medium (26) supplemented with 50 mM sodium benzoate (FAB-50; Sigma-Aldrich, Munich, Germany) at room temperature with rotary culture flask movement at 150 rpm. For long-term cultivation, bacteria were transferred weekly to FAB-50 plates containing 1.5% (wt/vol) agar and incubated at room temperature.
Multiple-microcosm approach.
Laboratory systems were designed to investigate bacterial motility behavior at five water potentials in a miniaturized system allowing parallel and semicontinuous microscopic observation of 24 microcosms. Therefore, we used clear, sterile, flat-bottom 24-well microtiter plates covered with low-evaporation lids (Orange Scientific, Braine-l'Alleud, Belgium) and filled the wells of each column with 0.3% (wt/vol) FAB agar. The whole system contained four replicates per ΔΨw treatment and for the abiotic control (Fig. 1).
FIG 1.
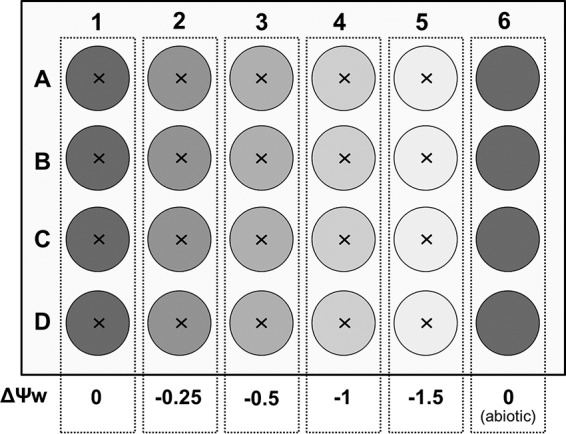
Scheme of the multiple-microcosm setup to assess the effects of five different Ψw values on bacterial dispersal, growth, and biodegradation. The water potential in each column was adjusted with different concentrations of NaCl or PEG 8000. P. putida KT2440-gfp was inoculated into the center (×) of each well, and one column served as an abiotic control.
Adjustment of ΔΨo and ΔΨm.
To change the Ψo value of the FAB-50 medium by −0.25, −0.5, −1, or −1.5 MPa, we added 3.2, 6.4, 12.8, or 19.2 g liter−1 of sodium chloride, respectively, to the agar prior to autoclaving (7). Agar without sodium chloride served as a control treatment (ΔΨo = 0 MPa). Each well was then filled with 1 ml molten agar in a laminar flow cabinet. Comparable changes of the Ψm value were obtained by overlaying 1 ml FAB double-strength agar containing 100 mM sodium benzoate directly in the well with 1.5 ml polyethylene glycol (PEG) solution containing 250, 392, 584, or 704 g PEG 8000 (Carl Roth, Karlsruhe, Germany) per liter of distilled water (27). For controls (ΔΨm = 0 MPa), the agar was overlaid only with distilled water. After 72 h of equilibration, solutions were discarded and residues were carefully removed by pipetting. Before use, all plates were dried with open lids in a laminar flow cabinet for 5 min.
Microcosm inoculation.
Cells were harvested from liquid culture by centrifugation at 8,000 × g for 10 min after 16 h of cultivation. The pellet was washed once with 10 mM potassium phosphate buffer (PB) at pH 7.2 and adjusted to an optical density of 50. Microcosms were inoculated with a 0.2-μl bacterial suspension (approximately 2.44 × 106 CFU) in the center of each well by use of a microliter syringe. Abiotic controls were left uninoculated. In experiments with the abiotic dispersal networks, a glass fiber mat (Mühlmeier Composite, Bärnau, Germany) with an area weight of 14 g m−2 was cut into circular pieces with a diameter of 1.2 mm, heat sterilized at 450°C for 4 h in a muffle furnace, and placed on top of the agar in the microcosms before inoculation with bacteria. The addition of the glass fiber mat had no effect on water volumes in the microcosms. Plates were incubated in plastic containers at room temperature in the dark.
Bacterial dispersal measurement.
Directly after inoculation, a plate was placed under a microscope equipped with an Hg vapor lamp and a black-and-white camera (AZ 100 Multizoom; Nikon, Amsterdam, Netherlands). Colony images were captured at intervals of 30 min for 24 h at respective x-y positions, using the 4D module and GFP filter settings. Picture stacks were imported into ImageJ (28) and converted to binary images after applying an intensity threshold of 40. Original image size information was included via the set scale command before the colony area was measured.
Bacterial growth measurement.
Cell numbers in replicate plates were determined after 6, 24, 30, and 48 h. The agar in each well was suspended in 2 ml PB and transferred to a sterile Falcon tube. Detachment was carried out by vortexing and subsequent ultrasonication treatment for 1 min. Culturable cells were analyzed by the number of CFU. Therefore, bacteria were spread on FAB-50 plates by using the drop plate method as described earlier (29). Briefly, 10-fold dilution series of the supernatants were prepared directly in 96-well microtiter plates, and 5-μl samples of consecutive dilutions were dropped on an agar plate. Plates were incubated at 25°C for 48 h. Droplets containing between 5 and 30 single colonies were used to determine CFU numbers.
For further analyses, 1.5 ml of supernatant obtained from cell detachment was fixed by incubation with 500 μl of a 12% (wt/vol) formaldehyde solution for 24 h at 4°C. To verify the results obtained from the CFU counts, we tested whether the change in Ψo or Ψm had an influence on the culturability of P. putida KT2440. Therefore, 0.5-μl droplets of the fixed samples obtained from treatments giving Δ0 and Δ−1.5 MPa after 48 h were spotted on individual boxes of a cellulose acetate grid filter (0.45 μm; Sartorius, Goettingen, Germany). Afterwards, cells were stained by embedding the filter in 4′-6-diamidino-2-phenylindole (DAPI)-amended mountant containing 9 parts Citifluor mountant (Citifluor, Leicester, United Kingdom), 1 part PB, and 1 μg ml−1 DAPI (30). Filters were stored at −20°C until analysis. For cell number determination, the size of each sample drop was first measured under transmission light by using a 1× objective. Subsequently, images of the bacteria were recorded at 5 random positions within the same drop by using a 20× objective and DAPI filter settings. Cells were counted using NIS Elements software (Nikon, Amsterdam, Netherlands). Therefore, an appropriate threshold was chosen to separate cells from background fluorescence. The ratios of the numbers of cells and CFU were calculated and compared by performing a two-sample t test for the two different water potentials and for Ψo and Ψm.
Bacterial degradation measurement.
One milliliter of fixed supernatant obtained from the detachment procedure was filtered through a 0.22-μm syringe filter (Carl Roth, Karlsruhe, Germany) to remove bacteria. The benzoate concentration was determined with a high-pressure liquid chromatography (HPLC) system equipped with a C18 reverse-phase column (250 by 4 mm) and a photodiode array detector (PDA) set at 271 nm. The system was operated at a flow rate of 1.2 ml min−1, with a 10-μl injection volume and a mobile phase consisting of 80% sodium acetate (50 mM; pH 4.5) and 20% methanol (MeOH) (31). Benzoate had a retention time of 13.8 min under these operation conditions.
Evaluation of dispersal, growth, and degradation data.
For evaluation of the dispersal, growth, and degradation data for P. putida KT2440-gfp, we calculated the areas under the curve (AUC) for the time span over which the respective data were experimentally obtained (24 h for dispersal and 48 h for growth and degradation). The AUC rises when a curve increases either earlier or to a higher level. Hence, it serves as an aggregate measure of the extent and temporal performance of the respective characteristics. The AUC values were calculated using the trapezoidal method. This numerical approximation of the time integral allows for nonequidistant time data and thus is not hampered by missing measurements for certain points in time. Relative AUC values were calculated by dividing the respective value by the ΔΨo (0 MPa) or ΔΨm (0 MPa) AUC value for the control treatment without network presence. To evaluate the correlation strength between population dispersal, population growth, and benzoate degradation, we calculated pairwise Pearson's product moment correlation coefficients (ρ) between AUC values at the different ΔΨo and ΔΨm values. Therefore, we combined the AUC values for the experiments with and without dispersal networks.
RESULTS
Population dispersal at different ΔΨo and ΔΨm values in the presence and absence of dispersal networks.
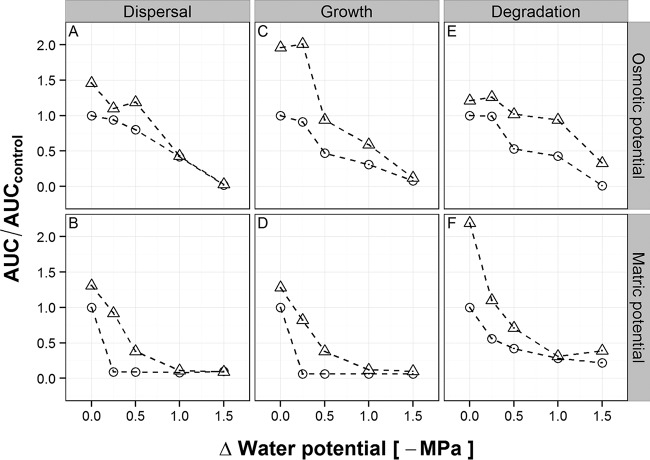
The radial colony expansion graph revealed two-phasic behavior for most of the ΔΨw treatments without a network presence. The initial phase is a result of the accumulation of cells due to growth taking place within the clearly delimited droplet on the agar surface formed during the inoculation procedure. The second phase reflects the swimming of the cells through the pores of the agar matrix away from the injection site. In the presence of dispersal networks, the biphasic behavior vanished because bacterial cells did not accumulate in a colony on the surface but immediately used the network to move away from the injection site. This dispersal, however, was not measured during the first hours of the experiments because we needed to adjust the detection threshold for image analysis to a constant value for all experiments, and we selected a value optimized for measuring the bulk of cells dispersing, which we consider to be responsible for efficient degradation (see Fig. S1 in the supplemental material). The population dispersal performance characterized by the AUC remained nearly constant (94% of the control) at a ΔΨo of −0.25 MPa but decreased continuously at lower ΔΨo values (Fig. 2A). At a ΔΨo of −1.5 MPa, only 2% of the AUC compared to that of the control (ΔΨo = 0 MPa) remained. In contrast, changing the ΔΨm diminished the AUC for population dispersal by at least 90% for all treatments, due to a complete repression of bacterial movement through the agar matrix (Fig. 2B). The remaining 8 to 10% of the AUC for the ΔΨm treatments stemmed from bacterial colony growth on the surface of the agar resulting in a passive push-forward effect.
FIG 2.
Relative dispersal (A and B), growth (C and D), and degradation (E and F) of P. putida KT2440-gfp for the different ΔΨo (A, C, and E) and ΔΨm (B, D, and F) values in the presence (triangles) and absence (circles) of dispersal networks, as determined by AUC calculation (see Material and Methods for details). Dispersal was measured microscopically by following the colony expansion based on the GFP signal in the single wells every 30 min for 24 h. Growth and degradation were assessed by the increase in the number of CFU per milliliter and the decrease in benzoate concentration after 6, 24, 30, and 48 h. Performance is given as the AUC relative to the AUC for the control (ΔΨw = 0 MPa) without dispersal networks.
A glass fiber mat was used as an abiotic model of mycelium-like dispersal networks to test whether they could maintain bacterial dispersal processes. This glass fiber network accelerated the dispersal of P. putida KT2440 between ΔΨo and ΔΨm values of 0 and −0.5 MPa (Fig. 2A and B). The highest benefits compared to the treatments without network presence were found at a ΔΨo value of 0 MPa and a ΔΨm value of −0.25 MPa, with 46% and 83% absolute differences of the AUC, respectively.
Population growth at different ΔΨo and ΔΨm values in the presence and absence of dispersal networks.
Bacterial population growth was analyzed after detachment of the cells from the agar matrix at the four different time points. The AUC for CFU-based population growth of P. putida KT2440 remained nearly stable at a ΔΨo value of −0.25 MPa but dropped at lower water potentials, by up to 92% at a ΔΨo value of −1.5 MPa relative to the control (0 MPa) (Fig. 2C). For ΔΨm, population growth was reduced by 94% relative to the control treatment at all potentials tested (Fig. 2D). The presence of dispersal networks resulted in an increase of the AUC for population growth for all treatments, with an absolute difference of 110% for a ΔΨo value of −0.25 MPa and 76% for a ΔΨm value of −0.25 MPa compared to the corresponding treatments without network presence (Fig. 2C and D). However, the networks' effects at a ΔΨo value of −1.5 MPa and a ΔΨm value of −1 MPa and below were negligible (between 4 and 6% differences in the AUC values) (see Fig. S2 in the supplemental material).
We further tested if a change of the water potentials led to different proportions of culturable cells in all cells. We did this in order to exclude biases potentially occurring due to a viable but not culturable state of the bacteria, which is often related to the occurrence of environmental stressors (32). Therefore, we microscopically counted cells obtained from the incubations at ΔΨo and ΔΨm values of 0 MPa and −1.5 MPa and compared the proportions of CFU in all cells for the two water potentials. There was no significant difference in these proportions for ΔΨo (32.4% ± 4.6% at a ΔΨo value of 0 MPa and 30.1% ± 4.6% at a ΔΨo value of −1.5 MPa) and ΔΨm (36.7% ± 3.3% at a ΔΨm value of 0 MPa and 33.2% ± 8.6% at a ΔΨm value of −1.5 MPa), which supports the results obtained from the measurement of culturable cells.
Benzoate degradation at different ΔΨo and ΔΨm values in the presence and absence of dispersal networks.
Sodium benzoate was the sole carbon and energy source in the agar, and its decrease in concentration was monitored by HPLC analysis. Degradation performance was not affected at a ΔΨo value of −0.25 MPa (99% of the AUC remained), decreased by 47% at a ΔΨo value of −0.5 MPa, and almost disappeared at a ΔΨo value of −1.5 MPa (1% of the AUC remained) (Fig. 2E) relative to the performance at a ΔΨo value of 0 MPa. In contrast, at a ΔΨm value of −0.25 MPa, the degradation performance had already decreased by 44%, but at a ΔΨm value of −1.5 MPa, it decreased by only 78% (Fig. 2F). The presence of dispersal networks was beneficial for biodegradation for all treatments. For ΔΨo treatments, absolute AUC improvements ranged from 21% at 0 MPa to 52% at −0.5 MPa compared to the corresponding treatments without network presence (Fig. 2E). For ΔΨm treatments, AUC values varied much more, leading to increases of 2% at −1 MPa and 119% at 0 MPa (Fig. 2F). Population dispersal, population growth, and benzoate degradation were found to be positively correlated for both ΔΨo (ρ > 0.86) and ΔΨm (ρ > 0.92) (Table 1).
TABLE 1.
Pairwise Pearson's product moment correlation coefficients (ρ) and corresponding P values
| Pairwise combination | Data for Ψo |
Data for Ψm |
||
|---|---|---|---|---|
| ρ value | P value | ρ value | P value | |
| Growth vs degradation | 0.86 | 0.002 | 1 | <0.001 |
| Growth vs dispersal | 0.87 | 0.001 | 0.93 | <0.001 |
| Degradation vs dispersal | 0.88 | <0.001 | 0.92 | <0.001 |
DISCUSSION
Effects of dispersal networks on bacterial dispersal at different ΔΨo and ΔΨm values.
In this study, we tested the hypothesis that network-based dispersal increases the functional performance of bacterial population growth and biodegradation by facilitating bacterial motility at environmentally relevant ΔΨo and ΔΨm values. The motile soil bacterium P. putida KT2440 was chosen for the experiments because its motility under unsaturated conditions has been studied extensively (4, 33).
In the absence of the dispersal networks, colony expansion was found to decrease with lowering ΔΨo values, down to −1.5 MPa. A similar effect was reported for Pseudomonas and Bacillus strains at salt concentrations which correspond to ΔΨo values between −1.8 and −2.3 MPa (23, 24), likely as a result of downregulation of motility genes to avoid energetic disadvantages due to flagellar system formation. In contrast to ΔΨo, a reduction of ΔΨm to −0.25 MPa (as induced by PEG 8000 addition) already resulted in drastically reduced dispersal, which is in good agreement with several studies pointing out that bacterial motility is restricted to a narrow range of high matric water potentials (4, 9, 33, 34). Dechesne et al. also reported a drastically reduced lateral colony expansion in response to insufficient water film thickness for P. putida KT2440 at Ψm values as low as −0.0036 MPa on a porous surface model system (25).
The use of PEG 8000 to change the matric component of ΔΨw is still debated, as different gene expression profiles were observed, unlike the case with directly applied ΔΨm (35). However, for plant cells, PEG 8000 was shown to evoke the same effects as soil drying, causing cytorrhysis rather than plasmolysis, without toxic effects of the PEG itself (27). Furthermore, the method of adjusting the Ψm by adding PEG 8000 has a distinct advantage over the use of different agar concentrations (36), as the latter imposes a physical hindrance rather than a decrease of the Ψm (37).
The presence of glass fiber networks clearly improved bacterial dispersal until the ΔΨw and ΔΨo values reached −0.5 MPa. Glass fibers were used to simulate hyphae surrounded by liquid films (16, 18) and to exclude effects of hyphal activities on bacterial growth and nutrition (38–41) to avoid additional complexity that might mask effects attributable to the promotion of bacterial dispersal at a lowered Ψm.
From the experimental data, we cannot infer that −0.5 MPa is a critical threshold below which dispersal along the glass fibers is completely restricted. Population dispersal was observed for 24 h because the microscopic observation area for the control treatments without network presence was completely colonized in this time span. Most, but not all, scenarios with lower ΔΨw values showed an improvement by use of dispersal networks within this time span. However, the shape of the population dispersal curves for the remaining three scenarios (see Fig. S1 in the supplemental material) suggests that benefits from the dispersal networks would probably occur soon after 24 h. We hypothesize that the dispersal network-mediated benefits for bacteria may compensate the energetic costs of flagellar maintenance and hence prevent the downregulation of motility genes. Moreover, our results indicate that bacterial dispersal in soil in the presence of dispersal-enabling mycelia is probably more dynamic than previously assumed and may not be restricted to the generally accepted Ψw of −0.05 MPa, a water potential which corresponds to high soil humidity (21).
Effects on bacterial growth and biodegradation.
The presence of dispersal networks resulted in an increase in population growth relative to the situation without network presence for ΔΨw values down to −1 MPa. This points to a dispersal network-mediated benefit also occurring at values below −0.5 MPa, which occurred with a delay of more than 24 h but within the 48-h observation period used for growth analysis. Such an observation is consistent with a previous study showing that faster dispersal is accompanied by increased bacterial abundance (10).
Biodegradation activity was assessed by following the consumption of sodium benzoate in the microcosms (see Fig. S3 in the supplemental material). The good water solubility of benzoate makes it a suitable model compound for highly accessible and bioavailable contaminants. With the developed microcosm system, we could clearly demonstrate that the network-mediated benefit for bacterial dispersal also led to an accelerated biodegradation over a wide range of ΔΨo and ΔΨm values. This is highly relevant for soil, for which bacterial dispersal was identified as one of the key factors for efficient biodegradation (17, 42–44). Interestingly, the observed negative influence of salt exposure on biodegradation is inconsistent with a study showing no influence of Ψo on degrading activity in liquid culture experiments (7). This discrepancy is presumably a consequence of the completely different conditions of the experimental test systems. Shaken liquid culture systems are virtually homogeneous regarding bacterial cell and substrate distribution, leading to short diffusion pathways. In contrast, in the agar systems applied here, bacteria are initially concentrated at the inoculation spot (Fig. 1), and restricted dispersal causes diffusion-limited degradation. Moreover, differences may arise from the use of degradation rates to compare effects at different ΔΨw rates for liquid culture experiments. Degradation rates are probably not a robust measure for comparing effects at different ΔΨw values because abrupt osmotic shifts are known to increase lag times (45), which are not considered for rate calculation but are well represented by the AUC.
Relevance for soil ecological functions.
The dispersal of microorganisms is recognized as a key factor for soil ecological functions (1, 46–48), such as the promotion of microbial diversity or the turnover of chemicals. Bacterial dispersal along fungal mycelia has been shown for numerous bacterial species and fungi (18, 49, 50) and hence may be of special relevance in unsaturated soil systems. Fungi constitute up to 75% of the soil microbial biomass, with a length of 102 to 104 m hyphae per g of soil (12, 51). Moreover, fungi have a unique lifestyle that is adapted to environmental changes, and they were shown to possess a marked resistance to desiccation in the field (52, 53). Because of the ubiquity, high abundance, and adaptive capacity of fungi, interactions between fungi and bacteria may substantially affect soil environmental dynamics and should therefore not be neglected (54).
The overall level and spatiotemporal variability of soil moisture have long been described as some of the primary environmental regulators of soil microbial activity (55, 56). Indeed, desiccation is a frequent physiological stress for soil microbial communities and is anticipated to gain further importance during future climate change (21). Simulation models predict an increased risk of drought in the 21st century (57). Furthermore, soil salinization is also a rising problem, especially in agriculture, due to irrigation and fertilizer amendment (58). However, the steadily increasing global population necessitates land use changes toward agriculture, which will also increase the amount of pesticides applied to the soil. Thus, to remain efficient in future, bioremediation approaches have to consider the effects of drought and salts on bacterial dispersal, growth, and degradation performance, which were shown to react drastically to changes in Ψo and Ψm in this study.
Obviously, the developed microcosm system reflects natural conditions in a highly simplified manner. However, such simplifications are also a strength of microbial model systems, which are necessary to better understand single aspects of natural systems (59, 60), such as the observed beneficial effects of dispersal networks on bacterial degradation performance at various water potentials.
The use of gel media to study bacterial dispersal in soil is discussed controversially (33). Scanning electron microscopy pictures revealed that soil surfaces, on top of which bacterial dispersal occurs, are not just bare mineral surfaces but rather are covered by patchy materials most likely originating from broken cell envelopes (61). The role of these sponge-like structures on bacterial motility in soils is completely neglected. However, they are probably better represented by agar matrices than by ceramic or quartz surfaces. Further investigations, in particular with fungi in a real soil system, are advisable to extrapolate our findings and to further elucidate the combined effects of fungal-bacterial interactions on contaminant degradation. Nevertheless, our findings strongly indicate that fungi may act as promoters of biodegradation under low-hydration and high-salinity conditions, thus improving the functional performance of microbial ecosystems.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Helmholtz Association via the integrated project Controlling Chemicals' Fate for the research topic Chemicals in the Environment (CITE) within the research program Terrestrial Environment.
We thank Rita Remer, Jana Reichenbach, and Birgit Würz for skilled experimental help, Karin Johst for valuable discussions, and three anonymous reviewers for comments on earlier versions of the article.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03901-15.
REFERENCES
- 1.Lindstrom ES, Ostman O. 2011. The importance of dispersal for bacterial community composition and functioning. PLoS One 6:e25883. doi: 10.1371/journal.pone.0025883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semple KT, Doick KJ, Wick LY, Harms H. 2007. Microbial interactions with organic contaminants in soil: definitions, processes and measurement. Environ Pollut 150:166–176. doi: 10.1016/j.envpol.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 3.Rijsberman FR. 2006. Water scarcity: fact or fiction? Agric Water Manag 80:5–22. doi: 10.1016/j.agwat.2005.07.001. [DOI] [Google Scholar]
- 4.Dechesne A, Wang G, Gulez G, Or D, Smets BF. 2010. Hydration-controlled bacterial motility and dispersal on surfaces. Proc Natl Acad Sci U S A 107:14369–14372. doi: 10.1073/pnas.1008392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris R. 1981. Effect of water potential on microbial growth and activity, p 23–95. In Parr JF, Gardner WR, Elliott LF (ed), Water potential relations in soil microbiology. Soil Science Society of America special publication no. 9. Soil Science Society of America, Madison, WI. [Google Scholar]
- 6.Papendick RI, Campbell GS. 1981. Theory and measurement of water potential, p 1–22. In Parr JF, Gardner WR, Elliott LF (ed), Water potential relations in soil microbiology. Soil Science Society of America special publication no. 9. Soil Science Society of America, Madison, WI. [Google Scholar]
- 7.Holden PA, Halverson LJ, Firestone MK. 1997. Water stress effects on toluene biodegradation by Pseudomonas putida. Biodegradation 8:143–151. doi: 10.1023/A:1008237819089. [DOI] [PubMed] [Google Scholar]
- 8.Stott DE, Elliott LF, Papendick RI, Campbell GS. 1986. Low temperature or low water potential effects on the microbial decomposition of wheat residue. Soil Biol Biochem 18:577–582. doi: 10.1016/0038-0717(86)90078-7. [DOI] [Google Scholar]
- 9.Or D, Smets BF, Wraith JM, Dechesne A, Friedman SP. 2007. Physical constraints affecting bacterial habitats and activity in unsaturated porous media—a review. Adv Water Resour 30:1505–1527. doi: 10.1016/j.advwatres.2006.05.025. [DOI] [Google Scholar]
- 10.Dechesne A, Owsianiak M, Bazire A, Grundmann GL, Binning PJ, Smets BF. 2010. Biodegradation in a partially saturated sand matrix: compounding effects of water content, bacterial spatial distribution, and motility. Environ Sci Technol 44:2386–2392. doi: 10.1021/es902760y. [DOI] [PubMed] [Google Scholar]
- 11.Potts M. 1994. Desiccation tolerance of prokaryotes. Microbiol Rev 58:755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harms H, Schlosser D, Wick LY. 2011. Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat Rev Microbiol 9:177–192. doi: 10.1038/nrmicro2519. [DOI] [PubMed] [Google Scholar]
- 13.Guhr A, Borken W, Spohn M, Matzner E. 2015. Redistribution of soil water by a saprotrophic fungus enhances carbon mineralization. Proc Natl Acad Sci U S A 112:14647–14651. doi: 10.1073/pnas.1514435112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohlmeier S, Smits TH, Ford RM, Keel C, Harms H, Wick LY. 2005. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ Sci Technol 39:4640–4646. doi: 10.1021/es047979z. [DOI] [PubMed] [Google Scholar]
- 15.Wick LY, Remer R, Würz B, Reichenbach J, Braun S, Schäfer F, Harms H. 2007. Effect of fungal hyphae on the access of bacteria to phenanthrene in soil. Environ Sci Technol 41:500–505. doi: 10.1021/es061407s. [DOI] [PubMed] [Google Scholar]
- 16.Banitz T, Fetzer I, Johst K, Wick LY, Harms H, Frank K. 2011. Assessing biodegradation benefits from dispersal networks. Ecol Model 222:2552–2560. doi: 10.1016/j.ecolmodel.2010.07.005. [DOI] [Google Scholar]
- 17.Banitz T, Wick LY, Fetzer I, Frank K, Harms H, Johst K. 2011. Dispersal networks for enhancing bacterial degradation in heterogeneous environments. Environ Pollut 159:2781–2788. doi: 10.1016/j.envpol.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Pion M, Bshary R, Bindschedler S, Filippidou S, Wick LY, Job D, Junier P. 2013. Gains of bacterial flagellar motility in a fungal world. Appl Environ Microbiol 79:6862–6867. doi: 10.1128/AEM.01393-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 20.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gülez G, Dechesne A, Smets BF. 2010. The pressurized porous surface model: an improved tool to study bacterial behavior under a wide range of environmentally relevant matric potentials. J Microbiol Methods 82:324–326. doi: 10.1016/j.mimet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Lambers H. 2003. Introduction, dryland salinity: a key environmental issue in southern Australia. Plant Soil 257:5–7. [Google Scholar]
- 23.Kristoffersen SM, Ravnum S, Tourasse NJ, Økstad OA, Kolstø A-B, Davies W. 2007. Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14570. J Bacteriol 189:5302–5313. doi: 10.1128/JB.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soutourina OA, Semenova EA, Parfenova VV, Danchin A, Bertin P. 2001. Control of bacterial motility by environmental factors in polarly flagellated and peritrichous bacteria isolated from Lake Baikal. Appl Environ Microbiol 67:3852–3859. doi: 10.1128/AEM.67.9.3852-3859.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dechesne A, Or D, Gülez G, Smets BF. 2008. The porous surface model, a novel experimental system for online quantitative observation of microbial processes under unsaturated conditions. Appl Environ Microbiol 74:5195–5200. doi: 10.1128/AEM.00313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen SK, Haagensen JA, Gjermansen M, Jorgensen TM, Tolker-Nielsen T, Molin S. 2007. Characterization of a Pseudomonas putida rough variant evolved in a mixed-species biofilm with Acinetobacter sp. strain C6. J Bacteriol 189:4932–4943. doi: 10.1128/JB.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verslues PE, Bray EA. 2004. LWR1 and LWR2 are required for osmoregulation and osmotic adjustment in Arabidopsis. Plant Physiol 136:2831–2842. doi: 10.1104/pp.104.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int 11:36–42. [Google Scholar]
- 29.Chen C-Y, Nace GW, Irwin PL. 2003. A 6×6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli. J Microbiol Methods 55:475–479. doi: 10.1016/S0167-7012(03)00194-5. [DOI] [PubMed] [Google Scholar]
- 30.Wendeberg A. 2010. Fluorescence in situ hybridization for the identification of environmental microbes. Cold Spring Harb Protoc 2010:pdb.prot5366. doi: 10.1101/pdb.prot5366. [DOI] [PubMed] [Google Scholar]
- 31.Warikoo V, McInerney MJ, Robinson JA, Suflita JM. 1996. Interspecies acetate transfer influences the extent of anaerobic benzoate degradation by syntrophic consortia. Appl Environ Microbiol 62:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver JD. 2005. The viable but nonculturable state in bacteria. J Microbiol 43:93–100. [PubMed] [Google Scholar]
- 33.Dechesne A, Smets BF. 2012. Pseudomonad swarming motility is restricted to a narrow range of high matric water potentials. Appl Environ Microbiol 78:2936–2940. doi: 10.1128/AEM.06833-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G, Or D. 2010. Aqueous films limit bacterial cell motility and colony expansion on partially saturated rough surfaces. Environ Microbiol 12:1363–1373. doi: 10.1111/j.1462-2920.2010.02180.x. [DOI] [PubMed] [Google Scholar]
- 35.Gulez G, Dechesne A, Workman CT, Smets BF. 2012. Transcriptome dynamics of Pseudomonas putida KT2440 under water stress. Appl Environ Microbiol 78:676–683. doi: 10.1128/AEM.06150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jost D, Winter J, Gallert C. 2011. Water and oxygen dependence of growing in silica sand capillary fringes. Vadose Zone J 10:532. doi: 10.2136/vzj2010.0092. [DOI] [Google Scholar]
- 37.Pernodet N, Maaloum M, Tinland B. 1997. Pore size of agarose gels by atomic force microscopy. Electrophoresis 18:55–58. doi: 10.1002/elps.1150180111. [DOI] [PubMed] [Google Scholar]
- 38.Furuno S, Foss S, Wild E, Jones KC, Semple KT, Harms H, Wick LY. 2012. Mycelia promote active transport and spatial dispersion of polycyclic aromatic hydrocarbons. Environ Sci Technol 46:5463–5470. doi: 10.1021/es300810b. [DOI] [PubMed] [Google Scholar]
- 39.Schamfuss S, Neu TR, van der Meer JR, Tecon R, Harms H, Wick LY. 2013. Impact of mycelia on the accessibility of fluorene to PAH-degrading bacteria. Environ Sci Technol 47:6908–6915. doi: 10.1021/es304378d. [DOI] [PubMed] [Google Scholar]
- 40.Banitz T, Johst K, Wick LY, Schamfuss S, Harms H, Frank K. 2013. Highways versus pipelines: contributions of two fungal transport mechanisms to efficient bioremediation. Environ Microbiol Rep 5:211–218. doi: 10.1111/1758-2229.12002. [DOI] [PubMed] [Google Scholar]
- 41.Pion M, Spangenberg JE, Simon A, Bindschedler S, Flury C, Chatelain A, Bshary R, Job D, Junier P. 2013. Bacterial farming by the fungus Morchella crassipes. Proc Biol Sci 280:20132242. doi: 10.1098/rspb.2013.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harms H, Wick LY. 2006. Dispersing pollutant-degrading bacteria in contaminated soil without touching it. Eng Life Sci 6:252–260. doi: 10.1002/elsc.200620122. [DOI] [Google Scholar]
- 43.Vieublé Gonod L, Chadoeuf J, Chenu C. 2006. Spatial distribution of microbial 2,4-dichlorophenoxy acetic acid mineralization from field to microhabitat scales. Soil Sci Soc Am J 70:64. doi: 10.2136/sssaj2004.0034. [DOI] [Google Scholar]
- 44.Dechesne A, Badawi N, Aamand J, Smets BF. 2014. Fine scale spatial variability of microbial pesticide degradation in soil: scales, controlling factors, and implications. Front Microbiol 5:667. doi: 10.3389/fmicb.2014.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mellefont L, McMeekin T, Ross T. 2003. The effect of abrupt osmotic shifts on the lag phase duration of foodborne bacteria. Int J Food Microbiol 83:281–293. doi: 10.1016/S0168-1605(02)00377-X. [DOI] [PubMed] [Google Scholar]
- 46.Martiny JBH, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR. 2006. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 47.Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH. 2010. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J 4:337–345. doi: 10.1038/ismej.2009.122. [DOI] [PubMed] [Google Scholar]
- 48.Kerr B, Riley MA, Feldman MW, Bohannan BJM. 2002. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 49.Warmink J, Nazir R, Corten B, Van Elsas J. 2011. Hitchhikers on the fungal highway: the helper effect for bacterial migration via fungal hyphae. Soil Biol Biochem 43:760–765. doi: 10.1016/j.soilbio.2010.12.009. [DOI] [Google Scholar]
- 50.Warmink J, Van Elsas J. 2009. Migratory response of soil bacteria to Lyophyllum sp. strain Karsten in soil microcosms. Appl Environ Microbiol 75:2820–2830. doi: 10.1128/AEM.02110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ritz K, Young IM. 2004. Interactions between soil structure and fungi. Mycologist 18:52–59. doi: 10.1017/S0269915X04002010. [DOI] [Google Scholar]
- 52.Barnard RL, Osborne CA, Firestone MK. 2013. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J 7:2229–2241. doi: 10.1038/ismej.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heaton L, Obara B, Grau V, Jones N, Nakagaki T, Boddy L, Fricker MD. 2012. Analysis of fungal networks. Fungal Biol Rev 26:12–29. doi: 10.1016/j.fbr.2012.02.001. [DOI] [Google Scholar]
- 54.Frey-Klett P, Burlinson P, Deveau A, Barret M, Tarkka M, Sarniguet A. 2011. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev 75:583–609. doi: 10.1128/MMBR.00020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waksman SA, Tenney FG, Stevens KR. 1928. The role of microorganisms in the transformation of organic matter in forest soils. Ecology 9:126–144. doi: 10.2307/1929350. [DOI] [Google Scholar]
- 56.Waksman SA, Gerretsen FC. 1931. Influence of temperature and moisture upon the nature and extent of decomposition of plant residues by microorganisms. Ecology 12:33–60. doi: 10.2307/1932933. [DOI] [Google Scholar]
- 57.Dai A. 2013. Increasing drought under global warming in observations and models. Nat Clim Change 3:52–58. doi: 10.1038/nclimate1633. [DOI] [Google Scholar]
- 58.Egamberdieva D, Renella G, Wirth S, Islam R. 2010. Secondary salinity effects on soil microbial biomass. Biol Fertil Soils 46:445–449. doi: 10.1007/s00374-010-0452-1. [DOI] [Google Scholar]
- 59.Drake JA, Huxel GR, Hewitt CL. 1996. Microcosms as models for generating and testing community theory. Ecology 77:670–677. [Google Scholar]
- 60.Lawton JH. 1995. Ecological experiments with model systems. Science 269:328–331. doi: 10.1126/science.269.5222.328. [DOI] [PubMed] [Google Scholar]
- 61.Miltner A, Bombach P, Schmidt-Brücken B, Kästner M. 2012. SOM genesis: microbial biomass as a significant source. Biogeochemistry 111:41–55. doi: 10.1007/s10533-011-9658-z. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.