ABSTRACT
Magnetotactic bacteria biosynthesize specific organelles, the magnetosomes, which are membrane-enclosed crystals of a magnetic iron mineral that are aligned in a linear chain. The number and size of magnetosome particles have to be critically controlled to build a sensor sufficiently strong to ensure the efficient alignment of cells within Earth's weak magnetic field while at the same time minimizing the metabolic costs imposed by excessive magnetosome biosynthesis. Apart from their biological function, bacterial magnetosomes have gained considerable interest since they provide a highly useful model for prokaryotic organelle formation and represent biogenic magnetic nanoparticles with exceptional properties. However, potential applications have been hampered by the difficult cultivation of these fastidious bacteria and their poor yields of magnetosomes. In this study, we found that the size and number of magnetosomes within the cell are controlled by many different Mam and Mms proteins. We present a strategy for the overexpression of magnetosome biosynthesis genes in the alphaproteobacterium Magnetospirillum gryphiswaldense by chromosomal multiplication of individual and multiple magnetosome gene clusters via transposition. While stepwise amplification of the mms6 operon resulted in the formation of increasingly larger crystals (increase of ∼35%), the duplication of all major magnetosome operons (mamGFDC, mamAB, mms6, and mamXY, comprising 29 genes in total) yielded an overproducing strain in which magnetosome numbers were 2.2-fold increased. We demonstrate that the tuned expression of the mam and mms clusters provides a powerful strategy for the control of magnetosome size and number, thereby setting the stage for high-yield production of tailored magnetic nanoparticles by synthetic biology approaches.
IMPORTANCE Before our study, it had remained unknown how the upper sizes and numbers of magnetosomes are genetically regulated, and overproduction of magnetosome biosynthesis had not been achieved, owing to the difficulties of large-scale genome engineering in the recalcitrant magnetotactic bacteria. In this study, we established and systematically explored a strategy for the overexpression of magnetosome biosynthesis genes by genomic amplification of single and multiple magnetosome gene clusters via sequential chromosomal insertion by transposition. Our findings also indicate that the expression levels of magnetosome proteins together limit the upper size and number of magnetosomes within the cell. We demonstrate that tuned overexpression of magnetosome gene clusters provides a powerful strategy for the precise control of magnetosome size and number.
INTRODUCTION
Magnetotactic bacteria (MTB) biosynthesize specific organelles, the magnetosomes, which are membrane-enclosed crystals of a magnetic iron mineral. To most efficiently function as magnetic sensors for navigation within Earth's weak magnetic field, the size, shape, number, and intracellular arrangement of magnetosome particles are evolutionarily optimized while at the same time minimizing the metabolic burden imposed by excessive magnetosome biosynthesis (1). In accordance with these constraints, the alphaproteobacterium Magnetospirillum gryphiswaldense MSR-1 (here referred to as MSR) and related magnetospirilla under environmental conditions typically form ∼20 cuboctahedral crystals of magnetite (Fe3O4) with a mean diameter of ∼42 nm (2, 3). Magnetosome biosynthesis proceeds in several steps, which include the formation of magnetosome membrane (MM) vesicles by invagination from the cytoplasmic membrane, the sorting of MM-specific proteins, magnetosomal iron uptake, and redox-controlled magnetite crystallization, which is followed by the assembly of nascent crystals into a single regular chain along an actin-like magnetosome cytoskeleton (4–8).
The high degree of biological control over their synthesis results in unprecedented properties of the nanoscaled magnetite crystals, such as high crystallinity; strong magnetization; and uniform composition, shape, and size, that cannot be replicated by abiogenic fabrication (8). Their structural perfection, exceptional magnetic properties, and biocompatibility make magnetosomes highly attractive for many bionanotechnological applications (9), such as magnetic drug targeting, immunoassays, magnetic resonance imaging, and hyperthermia (10–14). However, all applications using magnetosome particles so far have been hampered by their poor availability due to the fastidious growth requirements of MTB and the low magnetosome content of biomass. Therefore, an attractive possibility to increase magnetosome bioproduction would be genetic engineering or overexpression of biosynthesis pathway genes in native or foreign hosts.
However, until recently, magnetosome biosynthesis has remained poorly understood at the molecular level. We previously discovered that genes controlling magnetosome synthesis in MSR are clustered into several operons within a large genomic magnetosome island (MAI) (15, 16). These genes (∼30) also proved sufficient to induce the biosynthesis of well-ordered magnetosome chains upon transfer into the photosynthetic bacterium Rhodospirillum rubrum as a foreign expression host (17). In the native host MSR, the large mamAB operon alone is sufficient to sustain rudimentary magnetite biomineralization in the absence of all other magnetosome genes (16). From the 17 mamAB operon genes, a subset of only a few (mamE, mamM, mamO, mamQ, and mamB) is essential for magnetite biomineralization, while in single-deletion mutants of the other genes, biomineralization was impaired to different degrees (18). The smaller mamGFDC, mms6, and mamXY operons are not essential but have accessory roles in controlling the biomineralization of properly sized and shaped crystals (2, 16, 19). For instance, the combined deletion of mamG, mamF, mamD, and mamC, encoding some of the most abundant magnetosome membrane proteins, resulted in somewhat smaller and less regular magnetite particles (2), whereas deletion of the entire 3.7-kb (5-gene) mms6 operon caused the synthesis of substantially smaller and misaligned crystals (16). Consistent with these findings, the MmsF and Mms6 proteins were predicted to be major regulators of crystal size and shape in the closely related bacterium Magnetospirillum magneticum (20, 21). In MSR, deletion of the entire mamXY operon that encodes various functions in redox and transport of iron also caused a mixed phenotype, characterized by smaller, regular-shaped magnetite crystals that were flanked by irregular-shaped tiny flake-like hematite crystals. Unlike deletions of all other magnetosome genes, which consistently resulted in crystal size reductions of various degrees, the loss of mms36 and mms48 (within the mms6 operon) intriguingly caused the synthesis of fewer but larger magnetite crystals (18). Plasmid expression of the entire mamGFDC operon in the wild type (WT) also resulted in the formation of slightly enlarged (by 14.7%) magnetite particles (2). From these observations, it was concluded that magnetite biomineralization is affected in a cumulative manner by various different magnetosome proteins (18). However, how the maximum size and number of magnetosomes are precisely regulated has remained unknown, and controlled overproduction of magnetosome biosynthesis genes has not been achieved, due mostly to the difficulties in large-scale genome engineering in native MTB.
In this study, we established and systematically explored a strategy for the overexpression of magnetosome biosynthesis genes by genomic multiplication of single and multiple magnetosome gene clusters via sequential chromosomal insertion by transposition. While stepwise insertion of additional mms6 operon copies caused the formation of increasingly larger (up to 80 nm) regularly shaped magnetite crystals, duplication of all four known magnetosome operons resulted in an overproducing strain in which numbers of magnetosomes were increased >2-fold on average. Our findings also indicate that the expression levels of many Mam and Mms proteins together limit the upper size and number of magnetosomes within the cell. We demonstrate that genetically tuned expression of the mam and mms clusters provides a powerful strategy for the control of the size and number of magnetosomes, thereby setting the stage for high-yield production of tailored magnetic nanoparticles in various hosts by synthetic biology approaches.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
MSR and its mutant strains (see Table S1 in the supplemental material) were grown in liquid modified flask standard medium (FSM) or low-iron medium (LIM) at 30°C under microaerobic conditions if not otherwise specified (22). Therefore, cells were cultivated in gassed flasks with a mixture of 2% O2 and 98% N2 or in purged jars. For anaerobic cultivation, O2 was excluded from the gas mixture, while aerobic conditions were generated by free gas exchange to air. Single colonies were transferred into 100 μl FSM in 96-deep-well plates (Eppendorf, Hamburg, Germany) and incubated in anaerobic jars for 5 to 6 days. The liquid cultures were scaled up to a final volume of 10 ml. Culture conditions for Escherichia coli strains (see Table S1 in the supplemental material) were described previously (23). For strains BW29427 and WM3064, lysogeny broth medium was supplemented with 1 mM dl-α,ε-diaminopimelic acid (DAP). For selection of antibiotic-resistant strains, the following antibiotics were used: 25 μg ml−1 kanamycin (Km), 12 μg ml−1 tetracycline (Tet), and 15 μg ml−1 gentamicin (Gm) for E. coli strains and 5 μg ml−1 kanamycin, 5 μg ml−1 tetracycline, and 20 μg ml−1 gentamicin for MSR strains. Magnetosomes were isolated as described previously by Grünberg et al. after microaerobic cultivation of 5-liter cultures (24). The optical density and magnetic response (Cmag) were analyzed photometrically at 565 nm (25).
Molecular and genetic techniques.
Oligonucleotide sequences for amplification of DNA fragments (Table 1) were deduced from the working draft genome sequence of MSR (GenBank accession number CU459003) and were purchased from Sigma-Aldrich (Steinheim, Germany). Standard PCR procedures were used to amplify genetic fragments, and plasmids were sequenced by using BigDye Terminator v3.1 chemistry on an in-house ABI 3730 capillary sequencer (Applied Biosystems, Darmstadt, Germany). Sequences were analyzed with Vector NTI Advance 11.5 software (Invitrogen, Darmstadt, Germany). For genomic sequencing of overexpression strains, tagged libraries (∼200- to 300-bp insert size) were constructed from 1 ng of genomic DNA with the Nextera XT DNA kit (Illumina) according to the manufacturer's protocol. The eight libraries were sequenced in a multiplex format by using the Illumina MiSeq technology. The obtained sequences were assembled de novo as well as onto the reference genome with the commercial software CLC Genomics Workbench 5.5.
TABLE 1.
DNA oligonucleotides used in this work
Analytical methods.
The iron content of magnetosomes or whole cells was measured three times in triplicates by a ferrozine assay (26). After 16 h of cultivation, cells were washed with a solution containing 20 mM Tris-HCl and 5 mM EDTA (pH 7.4) to remove extracellular iron. One-milliliter cultures were centrifuged for 1 min at 11,000 × g and resuspended in 90 μl HNO3 (65%) for 3 h at 99°C. Afterwards, the lysate was cleared by centrifugation and resuspended in 1 ml H2O, and a ferrozine assay was performed as previously described (26, 27).
Construction of plasmids for overexpression and conjugative transfer.
Plasmids pTps_AB and pTps_XYZ were constructed in a previous study by Kolinko et al. (17). For cloning of plasmid Gm-pTps_AB, the Km resistance gene on plasmid pTps_AB was exchanged with the gentamicin gene via recombinogenic cloning (28, 29). To this end, a cloning cassette comprising the gentamicin gene and the respective promoter was PCR amplified (oligonucleotide pair IB173/IB174) and transferred into electrocompetent E. coli cells (DH10β/pTps_AB) expressing phage-derived recombinases from a plasmid (pSC101-BAD-gbaA). After transfer of the cassette, recombination between homologous regions on the linear fragment and plasmid pTps_AB occurred.
For overexpression of the mms6 and mamGFDC operons, a modified pBam-1 vector was designed. To this end, the EGFP (enhanced green fluorescent protein) gene was amplified with oligonucleotides IB102 and IB103 and integrated into pBam-1 after digestion with KpnI and EcoRI, resulting in pBam-gfp. The mamGFDC (2,185-bp) and mms6 (3,638-bp) operons were amplified by PCR from the genome of MSR (primer pair AL179/AL301) and were inserted into XbaI- and KpnI-digested pBam-gfp, resulting in pBam_mamGFDC and pBam_mms6 1×, respectively, with a C-terminal fusion of the EGFP gene to mamC or mms48. For the generation of pBam_mms6 2× and pBam_GFDC/mms6, the mms6 operon of pBam_mms6 1× was amplified with oligonucleotide pair AL377/AL379 and integrated into pBam_mamGFDC as well as pBam_mms6 1× after digestion with EcoRI. To realize the insertion of four mms6 operons, the gentamicin gene, flanked by lox71 and lox66 sequences, was generated by amplification from pBBR-MCS5 with oligonucleotide pair AL300/AL303 and cloned into the SanDI/AatII site of pBam_mamGFDC, resulting in pBam_GFDC/Gm. The mamGFDC operon was exchanged with the mms6 operon after digestion of pBam_GFDC/Gm with XbaI and KpnI, generating pBam_mms6/Gm. The generated plasmids were examined by restriction analysis with a set of different enzymes or PCR and transferred into different recipients via conjugation as described previously (27).
Fluorescence microscopy.
For localization studies of the EGFP fusion proteins and cell length measurements, the generated mutant strains of MSR were immobilized on agarose pads (FSM salts in H2O, supplemented with 1% agarose) and analyzed with an Olympus BX81 microscope provided with a 100 UPLSAPO100XO objective (numerical aperture of 1.40) and a Hamamatsu Orca AG camera. Data were evaluated with Olympus cell software.
Electron microscopy techniques.
Cells or magnetosomes were concentrated and adsorbed onto carbon-coated copper grids for transmission electron microscopy (TEM) analyses. Isolated magnetosomes were treated with 1% (vol/vol) uranyl acetate for staining of the magnetosome membrane. Cells and vesicles were imaged with a Morgagni 268 instrument (FEI, Eindhoven, Netherlands) at an accelerating voltage of 200 kV.
For cryo-electron tomography (CET) analysis, cells were cultivated anaerobically in FSM or aerobically in LIM and treated with formaldehyde (Fluka, Switzerland) at a final concentration of 0.1% (vol/vol) after 16 h of cultivation. A Tecnai F30 Polara transmission electron microscope (FEI, Eindhoven, Netherlands), equipped with a 300-kV field emission gun, a Gatan GIF 2002 Post-Column Energy filter, and a 2,048- by 2,048-pixel charge-coupled-device (CCD) camera (Gatan, Pleasanton, CA), was used for data generation, whereby image acquisition was performed at 300 kV, with the energy filter operated in the zero-loss mode (slit width of 20 eV). Sample preparation and the acquisition scheme were implemented as described previously (5). Tilt series were acquired with Serial EM software, and three-dimensional reconstructions were performed with the weighted back-projection method by using the TOM toolbox (30).
Statistical analysis of magnetosome sizes and numbers.
The t test (http://www.socscistatistics.com/tests/studentttest/Default.aspx) was used to determine the significance of differences in crystal size distributions. For statistical analysis of the differences in crystal numbers, the U test was performed (https://ccb-compute2.cs.uni-saarland.de/wtest/?id=www/www-ccb/html/wtest).
Cell fractionation.
Mutant strains IK-1, 3×mms6op, and 2×ABG6Xop were grown in 5 liters of FSM under microaerobic conditions. After centrifugation at 9,000 × g, cells were resuspended in a solution containing 20 mM Tris-HCl (pH 7.4) and 5 mM EDTA and stored at 4°C. Cell fractionation and magnetosome isolation were executed as described previously (31).
Gel electrophoresis and Western blot experiments.
A bicinchoninic acid (BCA) protein microassay kit (Pierce) was used for the determination of protein concentrations, according to the manufacturer's recommendations. Protein samples from the magnetosome membrane fraction were resuspended in electrophoresis sample buffer and denatured at 98°C for 5 min. Polyacrylamide gels were prepared according to procedures described previously by Laemmli (32). Ten micrograms of protein extracts was separated on a 15% SDS-polyacrylamide gel. Protein bands were visualized by Coomassie brilliant blue staining. Western blot analysis for the detection of MamM, MamA, and MamC was performed as previously described (24). The intensities of the protein bands of MamM, MamA, and MamC were compared by using ImageJ software.
RESULTS
Magnetosome gene clusters can be multiplied by sequential chromosomal transposition.
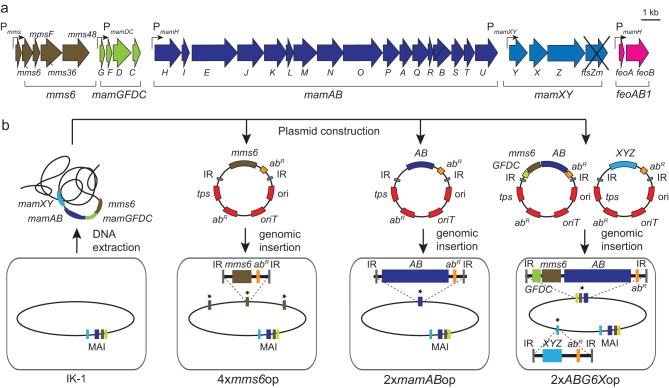
In the absence of appropriate promoters for transcriptional overexpression (13), we explored the effects of genetic multiplication of magnetosome operons by sequential transposition. For the stabilization of multiple identical gene copies, we chose MSR recA mutant strain IK-1 (27) as the chassis, which is otherwise wild type (WT) with respect to magnetosome biosynthesis (Table 2). As described below, multiple copies of various magnetosome gene clusters of between 2 and 17 kb cloned on nonreplicative vectors were inserted into the chromosome of parental strain IK-1. Therefore, sequential rounds of mariner or Tn5 transposon-driven random chromosomal insertions were selected by multiple antibiotic resistances (Fig. 1), as previously applied for genetic transfer of the entire magnetosome biosynthesis pathway into a foreign host (17). Transconjugants were typically obtained at frequencies of 10−7 and 10−8 and found inserted in single copies, as revealed by genome sequencing (see Table S2 in the supplemental material). As tested for several of the resulting strains, antibiotic resistance and magnetosome phenotypes remained stable for >100 generations in the absence of selective pressure.
TABLE 2.
Characteristics of generated overexpression strains compared to those of parental strain IK-1c
| Strain | Genotypeb | No. of measured crystals | Mean crystal size (nm) ± SD | Mean Cmag ± SD | Increase in crystal size (%)a | P value for statistical analysis of differences in crystal sizea | Mean no. of crystals per cell ± SD | Increase in no. of crystals (%)a | P value for statistical analysis of difference in crystal no.a | Mean iron content (% of dry wt) ± SDa | Mean cell length (μm) ± SD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IK-1 | 1× MAI | 390 | 36.2 ± 11.0 | 0.8 ± 0.2 | 33.9 ± 10.3 | 3.9 ± 0.2d | 4.4 ± 1.3 | ||||
| 2×GFDCop | 2× mamGFDC operon | 424 | 44.9 ± 13.5 | 0.9 ± 0.03 | 24.0 | <1.0E−5 | 36.3 ± 12.4 | 7.1 | <0.303 | 7.4 ± 1.1 | ND |
| 2×mms6op | 2× mms6 operon | 616 | 45.7 ± 14.2 | 0.9 ± 0.03 | 26.2 | <1.0E−5 | 46.5 ± 14.3 | 36.8 | <1.3E−6 | 14.9 ± 2.9 | 4.5 ± 1.6 |
| 2×mms6op+2×GFDC | 2× mms6 2× mamGFDC operon | 483 | 45.1 ± 12.2 | 0.9 ± 0.01 | 24.6 | <3.4E−5 | 45.1 ± 14.3 | 32.7 | <8.2E−6 | 14.1 ± 1.9 | ND |
| 3×mms6op | 3× mms6 operon | 1,183 | 47.9 ± 12.8 | 1.0 ± 0.2 | 32.3 | <1.0E−5 | 54.3 ± 29.9 | 59.5 | <7.2E−7 | 34.8 ± 2.5 | 4.6 ± 1.5 |
| 4×mms6op | 4× mms6 operon | 665 | 44.4 ± 13.2 | 1.0 ± 0.03 | 22.6 | <1.0E−5 | 57.8 ± 26.9 | 69.7 | <4.2E−10 | 38.8 ± 2.5 | 5.1 ± 1.9 |
| 5×mms6op | 5× mms6 operon | 545 | 41.9 ± 12.0 | 0.9 ± 0.1 | 15.7 | <1.0E−5 | 46.0 ± 14.8 | 35.3 | <9.5E−7 | ND | 5.3 ± 1.7 |
| 2×mamABop | 2× mamAB operon | 436 | 34.0 ± 17.6 | 1.0 ± 0.04 | −6.1 | <0.05 | 73.4 ± 43.1 | 115.2 | <1.5E−9 | 0.4 ± 0.5 | 4.8 ± 1.8 |
| 3×mamABop | 3× mamAB operon | 477 | 35.6 ± 15.4 | 0.5 ± 0.1 | −1.6 | <0.05 | 68.8 ± 13.2 | 102.9 | <1.5E−9 | 9.4 ± 0.5 | 6.0 ± 2.6 |
| 2×ABG6Xop | 2× mamGFDC 2× mms6 2× mamAB 2× mamXYZ operon | 708 | 38.7 ± 11.9 | 0.8 ± 0.1 | 6.9 | <1.0E−5 | 74.5 ± 34.9 | 118.3 | <9.8E−15 | 140.7 ± 2.4 | 5.1 ± 2.1 |
| 2×AG6X+2×feo | 2× mamGFDC 2× mms6 2× mamAB 2× mamXYZ operon 2× feoAB operon | 697 | 41.1 ± 8.4 | 0.7 ± 0.2 | 13.5 | <0.00001 | 69.3 ± 13.4 | 104.4 | <1.5E−15 | 135.9 ± 6.4 | ND |
Compared to IK-1.
With respect to magnetosome genes.
ND, not determined.
Compared to WT.
FIG 1.
(a) Schematic overview of cloned magnetosome operons (brown, mms6; green, mamGFDC; dark blue, mamAB; pale blue, mamXY; pink, feoAB1) and the respective promoters (angled arrows). (b) Strategy for construction of overexpression strains by amplification of different magnetosome operons. Genomic fragments from M. gryphiswaldense were cloned into transposon-based vectors. Plasmids contained the magnetosome operons mamAB (AB) (blue), mamGFDC (GFDC) (green), and mms6 (brown) and the mamXY operon lacking ftsZm (XYZ) (pale blue). The vector backbone (genes are indicated in red) contains a transposase gene (tps), inverted repeats (IR), the origin of transfer (oriT), an R6K or p15A origin of replication (ori), and an antibiotic resistance cassette (abR). After conjugative transfer of the plasmids, the transposase recognizes inverted repeat sequences and catalyzes the chromosomal insertion of the target sequence. Additional copies of the respective magnetosome operons in the chromosome (oval shape) are marked with asterisks.
Multiplication of mms6 and mamGFDC operons causes enlargement of magnetite crystals.
First, we tested the approach by attempting to insert one, two, three, and four additional copies of the mms6 operon by consecutive transposition of pBam_mms6 1×, pBam_mms6 2×, and pBam_mms6/Gm, resulting in strains 2×mms6op, 3×mms6op, 4×mms6op, and 5×mms6op, respectively. The merodiploid strain 2×mms6op, which now harbored one additional copy of the mms6 operon, displayed a slightly increased magnetic response and increased intracellular iron accumulation (+15%), and the number of magnetosome crystals per cell (47 particles; P < 1.3E−6) was higher (+37%) than that in IK-1, with the mean particle size being increased by 26% (46 nm; P < 1.0E−5) (Table 2). The insertion of two and three additional copies resulted in strains 3×mms6op and 4×mms6op, respectively, and caused further increases in crystal size and number, by ∼32% and 70%, respectively, compared to those of IK-1. However, the insertion of an additional mms6 operon copy (yielding strain 5×mms6op) did not further enhance biomineralization but caused reductions of particle size and number compared to those of its parent strain, 4×mms6op (Table 2). Also, increasing extracellular iron concentrations up to 250 μM had no effect on biomineralization in the overproducing strains (data not shown).
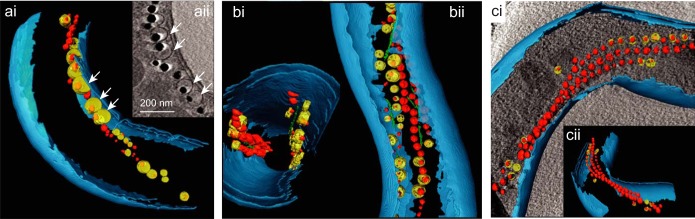
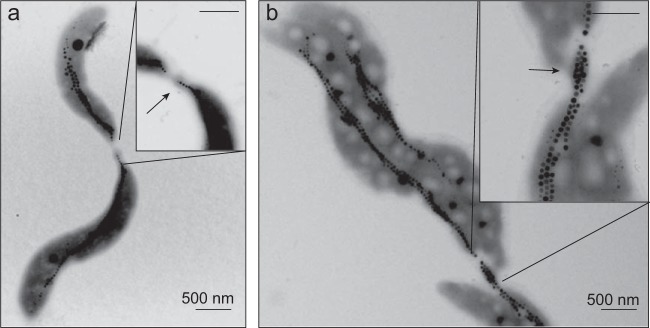
Within some cells of 2×mms6op and 2×mamABop, variable proportions of enlarged vesicles were visible in cryo-electron tomograms (Fig. 2a). These “giant” vesicles appeared spherically shaped, as in the WT, but their size was increased up to nearly 110 nm (2×mms6op) or 81 nm (2×mamABop), whereas vesicles of the WT had a maximum size of 63 nm. In addition, the increase in the mms6 operon copy number caused slight, gradual cell elongation, with a maximum mean cell length of 5.3 μm for strain 5×mms6op, (IK-1, 4.4 μm) (Table 2 and Fig. 3a). The cell length seemed to be correlated with the magnetosome number within the same strain. For example, shorter cells of strain 2×mms6op contained fewer (43 particles) and smaller (43-nm) crystals. On the other hand, highly elongated cells (>10 μm) contained significantly more (53 to 138 particles; mean, 104) and larger (49 nm) magnetosome crystals, with a maximum size of 80 nm (see Fig. S1 in the supplemental material). While magnetosome chains in cells of strains 2×mms6op and 3×mms6op were persistently located at midcell and split during cell division as in the WT (33), daughter cells of 2×mms6op and 3×mms6op remained frequently connected by deformed separation sites, sometimes even extending into tubular structures connecting the daughter cells, which probably resulted from the incomplete separation of the cells during or after division (Fig. 4). Within these structures, often up to 20 magnetosome particles were found to be enclosed (Fig. 4b).
FIG 2.
Cryo-electron tomographs of 3×mms6op, 2×mamABop, and 2×ABG6Xop. (ai) Segmented tomogram of a 3×mms6op cell illustrating the intracellular position of the magnetosome chain as well as the membrane vesicles. Slightly enlarged vesicles are indicated by white arrows. (aii) Tomogram section of the same 3×mms6op cell with enlarged vesicles (white arrows). (b) Tomograms of a 2×mamABop mutant cell from different angles, which illustrate two independent magnetosome chains aligned along two parallel filaments at opposite sites of the cell. (c) Segmented tomograms of a 2×ABG6Xop cell shown from different angles, which contains three intertwined magnetosome chains and regularly sized vesicles. Different colors indicate the outer and inner membranes (blue), magnetosome membrane vesicles (yellow), magnetosome filament (green), and magnetite crystals (red).
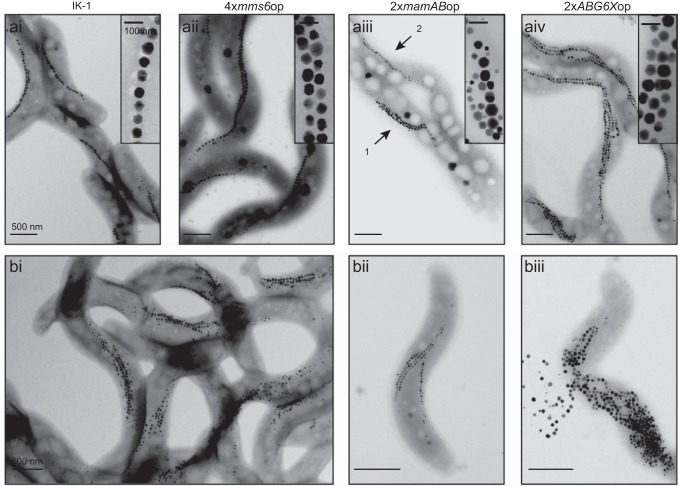
FIG 3.
TEM analysis of overexpression strains compared to the parental strain IK-1. (a) TEM of the overexpression strains 4×mms6op (ii), 2×mamABop (iii), and 2×ABG6Xop (iv) compared to IK-1 (i). Arrows 1 and 2 in panel iii illustrate two different morphotypes found for 2×mamABop. (b) Transmission electron micrographs of 2×ABG6Xop. (ii) Cells with one chain located at the inner convex cell curvature and up to three magnetosome chains with ectopic localization. (iii) Cells that lack a clearly ordered chain-like alignment of the produced particles and tend to burst during TEM grid preparation.
FIG 4.
TEM micrographs of dividing cells of 4×mms6op (a) and 2×ABG6X+2×feo (b), which remained connected by deformed separation sites with enclosed magnetosomes (arrows).
Next, we explored the effects of overexpressing the mamGFDC operon, which is adjacent to the mms6 operon and which was previously shown to be involved in size control of magnetosomes (2). While duplication of the mamGFDC operon (strain 2×GFDCop) alone resulted in only moderate increases in the number (+7%; P < 0.32) (Table 2) and size (+24%; P < 1.0E−5) (Table 2) of crystals, the simultaneous duplication of both the mms6 and mamGFDC operons (2×mms6op+2×GFDCop) caused the synthesis of 33% more crystals per cell (P < 8.2E−6) (Table 2), consistent with the iron content being increased by 7.4% in 2×GFDCop and even further by 14.1% in strain 2×mms6op+2×GFDCop (Table 2). Magnetosome crystals of strains 2×GFDCop and 2×mms6op+2×GFDCop were larger than those of IK-1 by ∼25% (Table 2).
Multiplication of the mamAB operon results in heterogeneous magnetosome morphotypes.
We next attempted to multiply the large mamAB operon that was shown previously to encode all magnetosome proteins essential for magnetosome formation (16, 18). The merodiploid strain 2×mamABop (i.e., harboring one native and one additional mamAB operon copy) showed a magnetic response and iron content similar to those of the parental strain (Table 2). Remarkably, the mean number of crystals per cell was increased by 115%. However, closer inspection by TEM revealed that cultures of strain 2×mamABop were heterogeneous with respect to magnetosome phenotypes. Three distinct morphotypes were present in variable proportions: (i) ∼50% were cells in which the number of regular-sized magnetosomes was increased to 77 particles per cell, with a regular chain-like organization; (ii) ∼40% were cells with increased numbers of magnetosomes (68 per cell) but with smaller crystal sizes and an aberrant intracellular localization; and (iii) 10% were WT-like cells (see Fig. S2 in the supplemental material). This pattern remained constant even after repeated clonal selection by colony purification of the strain. Compared to the single chain typically found in WT cells, we frequently observed two distinct magnetosome chains within cells of 2×mamABop, which were aligned along two separate bundles of cytoskeletal filaments located at opposite sides of the cell, as detected by CET (Fig. 2b). Purified magnetosome crystals from strain 2×mamABop were enveloped by a magnetosome membrane (MM) with the same appearance and thickness as those of MMs of the WT. Coomassie-stained SDS-PAGE profiles of the MM from strains 2×mamABop and 3×mms6op also displayed band patterns similar to those of the MM of strain IK-1. However, the intensity of the bands at 33 and 24 kDa (corresponding to magnetosome proteins MamA and MamM, which were among the duplicated genes) was increased, and quantitative Western blot analysis confirmed the higher abundance of these proteins by ∼130% and 145%, respectively. On the contrary, the abundance of MamC, which is encoded outside the mamAB operon, was not significantly affected (<10% variation) (see Fig. S3 in the supplemental material).
The merotriploid insertion mutant 3×mamABop (obtained by transfer of pTps-mamAB-Gm into strain 2×mamABop) showed a phenotype similar to that of strain 2×mamABop, and the number of magnetosomes did not further increase, despite a slightly increased intracellular iron content (+9.4%). The Cmag (magnetic response) of strain 3×mamABop was even lower than that of the parental strain (0.5 ± 0.2), possibly caused by the altered cell dimensions (Table 2). In summary, overexpression of the mamAB operon alone caused a heterogeneous phenotype but overall did not consistently enhance magnetosome biosynthesis.
Chromosomal duplication of all magnetosome gene clusters causes magnetosome overproduction.
The above-described experiments revealed that chromosomal insertion of up to three additional mms6 operons gradually enhanced the biosynthesis of magnetosomes. However, neither the introduction of additional mms6 operon copies nor the combined overexpression of the mamGFDC and mamAB operons alone further increased biomineralization, suggesting that magnetosome synthesis in these strains was likely limited by different factors encoded outside the multiplied operons. Therefore, we next attempted the combined overexpression of all four major operons (mms6, mamGFDC, mamAB, and mamXY operons). To this end, the mamAB, mms6, and mamGFDC operons were altogether inserted into the chromosome of IK-1 by transposition of pTps_ABG6. Next, in addition, we inserted the mamXYZ operon (lacking ftsZm to avoid interference with cell division during cloning [17]), resulting in strain 2×ABG6Xop, which is merodiploidic for all four key magnetosome operons (Fig. 1).
Transconjugants of 2×ABG6Xop were obtained at frequencies of only 2 × 10−9 to 5 × 10−9. Compared to strains in which the copy number of a single operon was increased and which showed essentially WT-like growth under all tested conditions, growth of strain 2×ABG6Xop was severely impaired at temperatures of ≥30°C. However, since WT-like growth was observed at 23°C, cells were cultivated at this temperature in all subsequent experiments. Cells on average were more elongated (5.1 μm) than cells of the parent strain IK-1, and we also observed deformed separation sites during or after cell division that were similar to those in 4×mms6op (data not shown). While the magnetic response of strain 2×ABG6Xop was similar to that of IK-1, the intracellular iron content was 2.4-fold increased. Cells contained 74 electron-dense particles on average, equivalent to an increase in number by 118% (Table 2 and Fig. 1).
Magnetosome yields (milligrams of Fe3O4 per gram [wet weight] of biomass) were also substantially increased during anaerobic, nitrate-fed growth experiments at a larger scale in a 30-liter fermentor, in which strain 2×ABG6Xop showed cell yields (cells per milliliter) similar to those of the WT (optical density at 565 nm [OD565] of 0.5 after 36 h), and 2.8 mg Fe3O4 per g (wet weight) of biomass was isolated, compared to the ∼1.20 mg/g obtained from fermentor-grown WT cells.
As we recently found that coexpression of the ferrous iron uptake system FeoAB1 encoded next to the MAI together with the four key magnetosome operons further enhanced heterologous magnetosome biosynthesis (17), we tested whether the overexpression of these proteins may also enhance biomineralization in overproducing MSR. To this end, the feoA and feoB genes (carried on Tet-pBam_feoAB1) were inserted into 2×ABG6Xop by Tn5 transposition. Whereas crystal sizes were not significantly increased in the resulting strain, 2×ABG6X+2×feo (41 nm versus 39 nm in 2×ABG6Xop), the number of crystals per cell was slightly decreased (69 particles instead of 74 for strain 2×ABG6Xop) (Table 2).
About 28% of the cells from the overproducer strain 2×ABG6Xop had >100 magnetosomes (maximum of 170), whereas IK-1 cells were found to never contain more than ∼60 magnetosomes. However, the mean size of crystals was only slightly increased, to 39 nm (Table 2; see also Fig. S4 in the supplemental material), and empty and partially filled MM vesicles (∼54 nm in size) were not significantly enlarged compared to those of the WT (Fig. 2c). Most cells contained multiple (two to four) magnetosome chains (9% contained one chain, 23% contained two chains, 34% contained three chains, and 18% contained four chains) (Fig. 3), whereas the WT typically exhibited not more than 2 chains per cell (76% contained one chain, 20% contained two chains, 4% contained three chains, and 0% contained four chains) (33). Besides cells with proper WT-like chains (localized near the inner curvature of the cell), we frequently observed cells with ectopically positioned chains (Fig. 3bi). The plentiful particles present in some cells often lacked any clearly ordered chain-like alignment but appeared “stuffed” into compact bundles or large irregular clusters (Fig. 3bii). Cells exhibited a tendency to spontaneously disintegrate, as indicated by the presence of released magnetosome particles in the vicinity of partially lysed cell bodies in TEM micrographs (Fig. 3biii).
DISCUSSION
Despite some progress by growth optimization (22, 34), previous attempts to increase magnetosome yields at a large scale were met by only limited success. In this work, we established a strategy to enhance magnetosome production by genetic engineering. In the absence of well-characterized promoters for very strong and inducible transcription, we attempted overexpression by increasing the copy numbers of several or all mam and mms operons comprising the genomic magnetosome island. Since we previously found that unbalanced multicopy expression of the large mamAB operon from replicative plasmids was unstable in MSR and caused spontaneous deletions and rearrangements (data not shown), we employed chromosomal insertion via transposition. In striking contrast to plasmid expression, chromosomally multiplied gene clusters apparently remained stable, at least within the >100 tested generations grown under laboratory-scale conditions.
Similar amplification via chromosomal multicopy insertion of gene clusters has been applied in several other studies. For instance, Tang et al. recently increased the production of the secondary metabolite spinosyn in the native host by partial gene cluster duplication (35). Using a chemically inducible chromosomal evolution approach, 40 copies of a poly-3-hydroxybutyrate gene cluster were consecutively inserted into the chromosome of E. coli, thereby causing a significant increase in the production of this biopolymer (36).
In our study, duplication or triplication of the mms6 operon alone caused a gradual strong enlargement of magnetite crystals but only a moderate increase in their numbers. In contrast, duplication of all mam and mms gene clusters yielded an overproducing strain (2×ABG6Xop) in which average magnetosome numbers per cell were greatly increased, by at least 120%, compared to those reported in previous studies (10, 16). This strain is also highly promising for future magnetosome production at a larger scale, as it continued to produce large numbers of magnetosomes per cell (up to 170 magnetosomes cell−1), even during mass cultivation, while the cell yield remained wild-type-like. Thus, magnetosome yields from the overproducing strain (2×ABG6Xop) were 2.3-fold increased compared to those of the WT.
On the other hand, the mean size of magnetite crystals was only weakly increased by duplication of all MAI clusters, compared to the strong effect of mms6 operon multiplication alone. This suggests that the engineering of distinct determinants can control both magnetosome size and number independently. However, the fact that the iron contents of less magnetic strains with fewer particles were similar to those of strains with more particles, such as 2×mms6op and 2×ABG6Xop, raises the question of whether the less magnetic strains might have accumulated iron in a form other than magnetite or other substantial iron pools. The existence of such variable iron pools could have been affected to different degrees by inserted mutations in an as-yet-unknown way and will be the subject to future studies.
It is well established that various physical characteristics of magnetic nanoparticles, such as sedimentation stability and magnetic remanence, are functions of their size (14). While particles smaller than ∼30 nm are superparamagnetic (i.e., no permanent magnetic signal at room temperature in the absence of an external field), particles between 30 and ∼60 to 100 nm have stable single domains (i.e., remanent magnetization), and the large single-domain particles generated in this study can be expected to be close to the maximum with respect to coercivity (37). Although full characterization of their magnetic properties remains to be completed, an initial analysis of some of the engineered large particles indeed indicated stable single-domain behavior and a higher magnetic coercivity than that of WT magnetosome particles (38). Since no synthetic routes for the abiogenic fabrication of such large magnetic single-domain nanoparticles are available so far, our findings will be of particular interest in applications that depend on the specific magnetic properties of such large and size-adjustable particles, such as magnetic resonance imaging (39) or hyperthermia (14).
The size, shape, number, and intracellular organization of magnetosomes also critically affect the strength of the cellular dipole moment that aligns cells by magnetic torque (1). Under environmental conditions, magnetosome biosynthesis has to be precisely regulated to be strong enough to orient the cell within the weak geomagnetic field with sufficient efficiency. On the other hand, increasing the number and size beyond the rather narrow evolutionary optimum that has been calculated for magnetic spirilla with ∼20 to 25 particles per cell (40) would not significantly improve the orientation because of the asymptotic Langevin function for alignment of magnetic dipoles (1) but would increase the metabolic burden and energetic costs of excessive magnetosome biosynthesis as well as potential harmful effects related to increased iron uptake (1). However, how the maximum number and upper size limit of magnetic single-domain particles are controlled at the molecular and structural levels has remained unknown. One important factor spatially constraining the growth of crystals can be expected to be the size of magnetosome vesicles (2). In fact, we sometimes observed significantly enlarged vesicles in the mms6 insertion strains by CET (Fig. 2a). This indicates that the overexpression of a subset of proteins might directly influence the vesicle diameter prior to crystallization, thereby defining the increase in crystal size. Besides vesicle biogenesis, magnetosomal iron import into the vesicles is also required to sustain the growth of magnetite (Fe3O4) crystals. While none of the proteins encoded by the mms6 operon likely function as iron transport proteins, amplification of putative iron transporters (MamB, MamM, MamH, and MamZ) might explain the increase in the number of magnetosomes in 2×ABG6Xop. In fact, in strain 2×mamABop, expression of MamM in the MM was markedly increased by ∼130% compared to that of the parent strain. Overexpression of a subset of magnetosome genes might also influence the expression or recruitment of other proteins controlling accessory processes during magnetosome formation (41, 42).
Altogether, our findings suggest that the balanced expression level of magnetosome proteins is one important factor for the determination of the number and size of magnetite crystals. However, this also raises the intriguing question of what might potentially define the upper limit of the number or size of magnetosomes in overexpression strains. For instance, the extracellular supply is known to be critical for magnetosome formation in MSR, and iron concentrations of 100 μM were found to be already saturating for cell yield and magnetism (43). Surprisingly, increasing the extracellular iron levels beyond this concentration had no effect on biomineralization in the overproducing strains, and chromosomal duplication of genes encoding the ferrous iron transporter FeoAB1 also had only minor effects (Table 2), indicating that the iron supply is not a limiting factor for magnetite overproduction under the tested conditions. However, biomineralization of the mixed-valence iron oxide magnetite was found to be critically dependent on a proper ferric-to-ferrous iron ratio (44–46), and thus, regulation by general metabolic pathways such as aerobic and anaerobic respiration processes might become limiting with increasing expression levels of the magnetosome synthesis machinery.
It might also be possible that further increases in magnetosome overproduction might be limited simply by spatial constraints imposed by the shape and confined size of the cell lumen. We also found that magnetosome overexpression increasingly impaired the growth of overproducing cells. For instance, strain 2×ABG6Xop eventually reached similar cell yields, and it no longer grew at the usual temperature of 30°C but required incubation at lower temperatures of ∼23°C, possibly due to an accumulation of misfolded overexpressed proteins or effects on lipid biosynthesis required for increased MM biosynthesis. In addition, we observed that cell division was slightly impaired in cells, as indicated by the presence of conspicuously elongated cells that often remained connected by deformed separation sites at advanced stages of constriction. This phenotype might be caused by the stronger magnetostatic interactions between the larger and more abundant particles within the chains, which require higher mechanical forces to be overcome (33). Altogether, in addition to the maximum cell size, there are likely multiple factors that set an upper limit for future overexpression approaches that may require engineering of the native host or the use of alternative foreign expression hosts.
In conclusion, our results are very promising for the construction of further engineered strains of magnetotactic bacteria, overproducing, for example, genetically engineered magnetosomes with enhanced functionalities and even higher magnetosome yields (9, 13). Furthermore, overexpression of selected magnetosome genes and clusters in homologous and heterologous hosts might be exploited for the design and mass production of size-adjusted nanocrystals with tuned magnetic properties.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful to A. Nieto, R. Susen, and A. Hähle for expert technical and experimental assistance.
We declare that we have no competing interests.
Funding Statement
We thank the Human Frontier Science Program (grant RGP0052/2012 to D.S.), Deutsche Forschungsgemeinschaft (DFG SCHU 1080/12-1 and 1080/15-3 to D.S.), and Konrad-Adenauer-Stiftung (A.L.) for financial support of this work.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03860-15.
REFERENCES
- 1.Frankel RB, Bazylinski DA. 1995. Structure and function of the bacterial magnetosome. ASM News 61:337–343. [Google Scholar]
- 2.Scheffel A, Gardes A, Grünberg K, Wanner G, Schüler D. 2008. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J Bacteriol 190:377–386. doi: 10.1128/JB.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schleifer KH, Schüler D, Spring S, Weizenegger M, Amann R, Ludwig W, Köhler M. 1991. The genus Magnetospirillum gen. nov. Description of Magnetospirillum gryphiswaldense sp. nov. and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb. nov. Syst Appl Microbiol 14:379–385. [Google Scholar]
- 4.Schüler D. 2008. Genetics and cell biology of magnetosome formation in magnetotactic bacteria. FEMS Microbiol Rev 32:654–672. doi: 10.1111/j.1574-6976.2008.00116.x. [DOI] [PubMed] [Google Scholar]
- 5.Katzmann E, Scheffel A, Gruska M, Plitzko JM, Schüler D. 2010. Loss of the actin-like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense. Mol Microbiol 77:208–224. doi: 10.1111/j.1365-2958.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- 6.Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schüler D. 2006. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- 7.Uebe R, Voigt B, Schweder T, Albrecht D, Katzmann E, Lang C, Böttger L, Matzanke B, Schüler D. 2010. Deletion of a fur-like gene affects iron homeostasis and magnetosome formation in Magnetospirillum gryphiswaldense. J Bacteriol 192:4192–4204. doi: 10.1128/JB.00319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faivre D, Schüler D. 2008. Magnetotactic bacteria and magnetosomes. Chem Rev 108:4875–4898. doi: 10.1021/cr078258w. [DOI] [PubMed] [Google Scholar]
- 9.Lang C, Schüler D. 2006. Biogenic nanoparticles: production, characterization, and application of bacterial magnetosomes. J Phys Condens Matter 18:2815–2828. [Google Scholar]
- 10.Sun JB, Duan JH, Dai SL, Ren J, Guo L, Jiang W, Li Y. 2008. Preparation and anti-tumor efficiency evaluation of doxorubicin-loaded bacterial magnetosomes: magnetic nanoparticles as drug carriers isolated from Magnetospirillum gryphiswaldense. Biotechnol Bioeng 101:1313–1320. doi: 10.1002/bit.22011. [DOI] [PubMed] [Google Scholar]
- 11.Lang C, Schüler D, Faivre D. 2007. Synthesis of magnetite nanoparticles for bio- and nanotechnology: genetic engineering and biomimetics of bacterial magnetosomes. Macromol Biosci 7:144–151. doi: 10.1002/mabi.200600235. [DOI] [PubMed] [Google Scholar]
- 12.Lang C, Schüler D. 2008. Expression of green fluorescent protein fused to magnetosome proteins in microaerophilic magnetotactic bacteria. Appl Environ Microbiol 74:4944–4953. doi: 10.1128/AEM.00231-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borg S, Hofmann J, Pollithy A, Lang C, Schüler D. 2014. New vectors for chromosomal integration enable high-level constitutive or inducible magnetosome expression of fusion proteins in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 80:2609–2616. doi: 10.1128/AEM.00192-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hergt R, Hiergeist R, Zeisberger M, Schüler D, Heyen U, Hilger I, Kaiser WA. 2005. Magnetic properties of bacterial magnetosomes as diagnostic and therapeutic tools. J Magn Magn Mater 293:80–86. doi: 10.1016/j.jmmm.2005.01.047. [DOI] [Google Scholar]
- 15.Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D. 2005. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J Bacteriol 187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohsse A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, Voigt B, Schweder T, Schüler D. 2011. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS One 6:e25561. doi: 10.1371/journal.pone.0025561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolinko I, Lohsse A, Borg S, Raschdorf O, Jogler C, Tu Q, Posfai M, Tompa E, Plitzko JM, Brachmann A, Wanner G, Müller R, Zhang Y, Schüler D. 2014. Biosynthesis of magnetic nanostructures in a foreign organism by transfer of bacterial magnetosome gene clusters. Nat Nanotechnol 9:193–197. doi: 10.1038/nnano.2014.13. [DOI] [PubMed] [Google Scholar]
- 18.Lohsse A, Borg S, Raschdorf O, Kolinko I, Tompa É, Pósfai M, Faivre D, Baumgartner J, Schüler D. 2014. Genetic dissection of the mamAB and mms6 operons reveals a gene set essential for magnetosome biogenesis in Magnetospirillum gryphiswaldense. J Bacteriol 196:2658–2669. doi: 10.1128/JB.01716-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raschdorf O, Müller FD, Pósfai M, Plitzko JM, Schüler D. 2013. The magnetosome proteins MamX, MamZ and MamH are involved in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. Mol Microbiol 89:872–886. doi: 10.1111/mmi.12317. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka M, Mazuyama E, Arakaki A, Matsunaga T. 2011. MMS6 protein regulates crystal morphology during nano-sized magnetite biomineralization in vivo. J Biol Chem 286:6386–6392. doi: 10.1074/jbc.M110.183434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murat D, Falahati V, Bertinetti L, Csencsits R, Kornig A, Downing K, Faivre D, Komeili A. 2012. The magnetosome membrane protein, MmsF, is a major regulator of magnetite biomineralization in Magnetospirillum magneticum AMB-1. Mol Microbiol 85:684–699. doi: 10.1111/j.1365-2958.2012.08132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heyen U, Schüler D. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl Microbiol Biotechnol 61:536–544. doi: 10.1007/s00253-002-1219-x. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 24.Grünberg K, Müller EC, Otto A, Reszka R, Linder D, Kube M, Reinhardt R, Schüler D. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl Environ Microbiol 70:1040–1050. doi: 10.1128/AEM.70.2.1040-1050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schüler D, Uhl R, Bäuerlein E. 1995. A simple light-scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol Lett 132:139–145. doi: 10.1111/j.1574-6968.1995.tb07823.x. [DOI] [Google Scholar]
- 26.Viollier E, Inglett PW, Hunter K, Roychoudhury AN, Van Cappellen P. 2000. The ferrozine method revisited: Fe(II)/Fe(III) determination in natural waters. Appl Geochem 15:785–790. doi: 10.1016/S0883-2927(99)00097-9. [DOI] [Google Scholar]
- 27.Kolinko I, Jogler C, Katzmann E, Schüler D. 2011. Frequent mutations within the genomic magnetosome island of Magnetospirillum gryphiswaldense are mediated by RecA. J Bacteriol 193:5328–5334. doi: 10.1128/JB.05491-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Sarov M, Rientjes J, Fu J, Hollak H, Kranz H, Xie W, Stewart AF, Zhang Y. 2006. An improved recombineering approach by adding RecA to λ Red recombination. Mol Biotechnol 32:43–53. doi: 10.1385/MB:32:1:043. [DOI] [PubMed] [Google Scholar]
- 29.Zhang YM, Buchholz F, Muyrers JPP, Stewart AF. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet 20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 30.Nickell S, Förster F, Linaroudis A, Net WD, Beck F, Hegerl R, Baumeister W, Plitzko JM. 2005. TOM software toolbox: acquisition and analysis for electron tomography. J Struct Biol 149:227–234. doi: 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Uebe R, Junge K, Henn V, Poxleitner G, Katzmann E, Plitzko JM, Zarivach R, Kasama T, Wanner G, Posfai M, Böttger L, Matzanke B, Schüler D. 2011. The cation diffusion facilitator proteins MamB and MamM of Magnetospirillum gryphiswaldense have distinct and complex functions, and are involved in magnetite biomineralization and magnetosome membrane assembly. Mol Microbiol 82:818–835. doi: 10.1111/j.1365-2958.2011.07863.x. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Katzmann E, Müller FD, Lang C, Messerer M, Winklhofer M, Plitzko JM, Schüler D. 2011. Magnetosome chains are recruited to cellular division sites and split by asymmetric septation. Mol Microbiol 82:1316–1329. doi: 10.1111/j.1365-2958.2011.07874.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Zhang X, Jiang W, Li Y, Li J. 2011. Semicontinuous culture of Magnetospirillum gryphiswaldense MSR-1 cells in an autofermentor by nutrient-balanced and isosmotic feeding strategies. Appl Environ Microbiol 77:5851–5856. doi: 10.1128/AEM.05962-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Y, Xia L, Ding X, Luo Y, Huang F, Jiang Y. 2011. Duplication of partial spinosyn biosynthetic gene cluster in Saccharopolyspora spinosa enhances spinosyn production. FEMS Microbiol Lett 325:22–29. doi: 10.1111/j.1574-6968.2011.02405.x. [DOI] [PubMed] [Google Scholar]
- 36.Tyo KE, Ajikumar PK, Stephanopoulos G. 2009. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol 27:760–765. doi: 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- 37.Bennet M, Bertinetti L, Neely RK, Schertel A, Kornig A, Flors C, Müller FD, Schüler D, Klumpp S, Faivre D. 2015. Biologically controlled synthesis and assembly of magnetite nanoparticles. Faraday Discuss 181:71–83. doi: 10.1039/C4FD00240G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumari M, Widdrat M, Tompa E, Uebe R, Schüler D, Pósfai M, Faivre D, Hirt AM. 2014. Distinguishing magnetic particle size of iron oxide nanoparticles with first-order reversal curves. J Appl Phys 116:12304. [Google Scholar]
- 39.Sun C, Lee JS, Zhang M. 2008. Magnetic nanoparticles in MR imaging and drug delivery. Adv Drug Deliv Rev 60:1252–1265. doi: 10.1016/j.addr.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frankel RB. 1984. Magnetic guidance of organisms. Annu Rev Biophys Bioeng 13:85–103. doi: 10.1146/annurev.bb.13.060184.000505. [DOI] [PubMed] [Google Scholar]
- 41.Zeytuni N, Ozyamak E, Ben-Harush K, Davidov G, Levin M, Gat Y, Moyal T, Brik A, Komeili A, Zarivach R. 2011. Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly. Proc Natl Acad Sci U S A 108:E480–E487. doi: 10.1073/pnas.1103367108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murat D, Quinlan A, Vali H, Komeili A. 2010. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc Natl Acad Sci U S A 107:5593–5598. doi: 10.1073/pnas.0914439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schüler D, Bäuerlein E. 1996. Iron-limited growth and kinetics of iron uptake in Magnetospirillum gryphiswaldense. Arch Microbiol 166:301–307. doi: 10.1007/s002030050387. [DOI] [PubMed] [Google Scholar]
- 44.Li YJ, Katzmann E, Borg S, Schüler D. 2012. The periplasmic nitrate reductase Nap is required for anaerobic growth and involved in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. J Bacteriol 194:4847–4856. doi: 10.1128/JB.00903-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li YJ, Bali S, Borg S, Katzmann E, Ferguson SJ, Schüler D. 2013. Cytochrome cd1 nitrite reductase NirS is involved in anaerobic magnetite biomineralization in Magnetospirillum gryphiswaldense and requires NirN for proper d1 heme assembly. J Bacteriol 195:4297–4309. doi: 10.1128/JB.00686-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Raschdorf O, Silva KT, Schüler D. 2014. The terminal oxidase cbb3 functions in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. J Bacteriol 196:2552–2562. doi: 10.1128/JB.01652-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.