ABSTRACT
Animal development and physiology depend on beneficial interactions with microbial symbionts. In many cases, the microbial symbionts are horizontally transmitted among hosts, thereby making the acquisition of these microbes from the environment an important event within the life history of each host. The light organ symbiosis established between the Hawaiian squid Euprymna scolopes and the bioluminescent bacterium Vibrio fischeri is a model system for examining how hosts acquire horizontally transmitted microbial symbionts. Recent studies have revealed that the light organ of wild-caught E. scolopes squid contains polyclonal populations of V. fischeri bacteria; however, the function and development of such strain diversity in the symbiosis are unknown. Here, we report our phenotypic and phylogenetic characterizations of FQ-A001, which is a V. fischeri strain isolated directly from the light organ of an E. scolopes individual. Relative to the type strain ES114, FQ-A001 exhibits similar growth in rich medium but displays increased bioluminescence and decreased motility in soft agar. FQ-A001 outcompetes ES114 in colonizing the crypt spaces of the light organs. Remarkably, we find that animals cocolonized with FQ-A001 and ES114 harbor singly colonized crypts, in contrast to the cocolonized crypts observed from competition experiments involving single genotypes. The results with our two-strain system suggest that strain diversity within the squid light organ is a consequence of diversity in the single-strain colonization of individual crypt spaces.
IMPORTANCE The developmental programs and overall physiologies of most animals depend on diverse microbial symbionts that are acquired from the environment. However, the basic principles underlying how microbes colonize their hosts remain poorly understood. Here, we report our findings of bacterial strain competition within the coevolved animal-microbe symbiosis composed of the Hawaiian squid and bioluminescent bacterium Vibrio fischeri. Using fluorescent proteins to differentially label two distinct V. fischeri strains, we find that the strains are unable to coexist in the same niche within the host. Our results suggest that strain competition for distinct colonization sites dictates the strain diversity associated with the host. Our study provides a platform for studying how strain diversity develops within a host.
INTRODUCTION
Microbes directly contribute to the physiology, development, and evolution of metazoans (1). Many metazoan-microbe symbioses are established through horizontal transmission, i.e., animals acquire microbial symbionts from their environment (2). Coevolution of host-microbe pairs can result in genetic factors that promote remarkably high specificity between the partners, thereby assisting in the acquisition of symbionts from typically unpredictable environments (3). An important but understudied topic in coevolved metazoan-microbe symbioses is how strain diversity impacts the establishment of these associations.
A particularly powerful model system to explore the molecular mechanisms underlying host-microbe interactions is the symbiosis established between the Hawaiian bobtail squid Euprymna scolopes and the marine bacterium Vibrio fischeri. V. fischeri populations persist within a host structure called the light organ, where they produce bioluminescence that is used by the squid for a camouflaging behavior known as counterillumination (4). Bioluminescence is the primary function of V. fischeri during symbiosis and is a product of luciferase, which is the heterodimeric enzyme encoded by luxA and luxB (5). Regulation of the bioluminescence genes (luxICDABEG) in V. fischeri occurs primarily at the transcriptional level via the LuxI/LuxR quorum-sensing system, as previously reviewed (6). The synthase LuxI produces the signaling molecule N-oxohexanoyl-homoserine lactone (3-oxo-C6 HSL), which binds to and activates the transcription factor LuxR. Both bioluminescence and a functional LuxI/LuxR signaling system are required for V. fischeri populations to persist within the light organ (7–9).
The initial stages of bacterial colonization in the squid-Vibrio symbiosis are well characterized, in large part due to the numerous studies that couple cell biology and microbial genetic approaches, which were recently reviewed (10). Environmental V. fischeri cells first associate with cilia located on the light organ surface (11) and then aggregate for a few hours outside three pores present on either side of the light organ (12). Using flagellum-based motility and chemotaxis, V. fischeri cells migrate up a gradient of chitin-derived oligosaccharides (13). Host chitinases are hypothesized to generate these chitin-derived sugars (14). At the end of each duct, approximately 1 to 2 cells successfully pass through a physical bottleneck to enter deep crypt spaces, where the ensuing infections persist for the lifetime of the animal (15, 16).
While a single strain of V. fischeri is sufficient to fully colonize the juvenile light organ, the V. fischeri infections within the light organs of wild-caught adult animals are polyclonal (16). V. fischeri strains isolated from adult light organs show significant phenotypic variability, including in bioluminescence, colony pigmentation, motility, growth rates, and siderophore production (16). Phylogenetic analyses created from concatenated housekeeping genes have revealed that several well-supported clades of strains exist within the greater V. fischeri clade. Interestingly, one clade, referred to as group A strains, exhibits competitive dominance when competed against non-group A strains in colonization experiments with juvenile squid (17). Among V. fischeri strains, the luxIR intergenic region was also shown to be hypervariable and to contribute to the bioluminescence profile of each strain, suggesting that bioluminescence and its regulation are under selection (18, 19). How such V. fischeri strain diversity develops within the squid light organ is unclear.
In this study, we tested the hypothesis that the initial colonization of the squid light organ contributes to strain diversity associated with the host. We investigated the light organ colonization patterns of two phylogenetically distinct symbiotic strains of V. fischeri. We found that animals cocolonized with both strains contain crypts that are singly colonized, which suggests that strain competition occurs for each crypt space. We present here a model of how strain competition can impact the V. fischeri diversity established during the initial period of light organ colonization.
MATERIALS AND METHODS
Media and growth conditions.
V. fischeri strains were grown aerobically at 28°C in LBS medium (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 2% [wt/vol] NaCl, 50 mM Tris-HCl [pH 7.5]). For the maintenance of plasmids, chloramphenicol was added to the medium at a final concentration of 2.5 μg/ml. Tryptone-based Instant Ocean (TB-IO) medium for motility assays contained 0.5% tryptone (wt/vol) and 0.3% yeast extract (wt/vol) in 70% Instant Ocean (Spectrum Brands, Blacksburg, VA, USA). Filter-sterilized seawater (FSSW) was prepared by filtering 100% Instant Ocean through a 0.2-μm-pore-size surfactant-free filter.
Strains.
The V. fischeri strains used in this study are listed in Table 1. To isolate FQ-A001, the light organ of an adult female squid was homogenized, and serial dilutions of the homogenate were plated onto LBS agar. An isolated colony was restreaked and named FQ-A001.
TABLE 1.
Strains and plasmids used in this study
| Strain/plasmid | Characteristics | Reference |
|---|---|---|
| Strains | ||
| ES114 | Symbiotic V. fischeri type strain isolated from E. scolopes collected in Kaneohe Bay, Oahu, HI, in 1989 | 29 |
| EVS102 | ES114 ΔluxCDABEG | 8 |
| FQ-A001 | Symbiotic V. fischeri strain isolated from a female E. scolopes squid collected in Kaneohe Bay, HI, in 2015 | This study |
| Plasmids | ||
| pVSV105 | R6Kori ori(pES213) RP4 oriT cat | 46 |
| pTM267 | pVSV105 kan gfp PtetA-mCherry | 20 |
| pTM280 | pVSV105 PluxIES114-gfp PtetA-mCherry | 20 |
| pEDL01 | pVSV105 PluxIFQ-A001-gfp PtetA-mCherry | This study |
| pSCV38 | pVSV105 PtetA-yfp PtetA-mCherry | Lab stock |
| pYS112 | pVSV105 PproD-cfp PtetA-mCherry | This study |
| proD | Reporter plasmid with proD promoter | 21 |
Plasmids.
The V. fischeri-specific plasmids used in this study are listed in Table 1. To construct pEDL01, the luxI promoter region was amplified from a colony of FQ-A001 using the primers luxI-prom-XbaI-u1 (5′-TGCCCGGGGTTCACCTAGCTTATTGTTATGTT-3′)/luxI-prom-XmaI-l1 (5′-TTCTAGATACAGCCATGCAACCTCTCT-3′) and cloned into the vector pCR-Blunt (Life Technologies, Carlsbad, CA, USA). The pCR-Blunt clone was confirmed and subcloned via XmaI/XbaI restriction digestion into the XmaI/XbaI vector fragment of pTM267 (20) to yield pEDL01.
Plasmid pYS112 was constructed by amplifying the insulated promoter proD from a pSB3C5-derived plasmid (21) using primers pro-XmaI-u1 (5′-TATACCCGGGCACAGCTAACACCACGTCGTCC-3′)/pro-XbaI-l1 (5′-CTTTCCTGTGTGACTCTAGACTCTAGTAAAAG-3′) and cloned into pCR-Blunt. The pCR-Blunt clone was verified by sequencing and cloned into an XbaI/XmaI-generated backbone of a pTM267-derived vector that contains cfp in place of gfp.
Squid colonization assays.
For each indicated strain, single-strain colonization assays were performed in FSSW using an inoculum of approximately 5,000 CFU/ml, as previously described (20). For each competition experiment, freshly hatched E. scolopes juveniles were exposed for 18 h to a mixed inoculum containing a yellow fluorescent protein (YFP)-labeled strain (ES114 or FQ-A001) and a cyan fluorescent protein (CFP)-labeled strain (ES114) and then transferred to fresh FSSW. At 48 h postinoculation (p.i.), luminescent animals were either prepared for crypt colonization assays, as described below, or homogenized for serial dilutions that were plated onto LBS. The resulting CFU were scored for YFP or CFP using an Olympus SX16 fluorescence dissecting microscope (Olympus Corp., Tokyo, Japan) equipped with YFP and CFP filter sets. For the head start competition assays, initial and mixed inoculums were prepared in FSSW. Animals were exposed to either the initial inoculum or FSSW. After 3.5 h, animals were separately transferred as groups to a mixed inoculum. At 18 h p.i., the animals were transferred to fresh FSSW. At 48 h p.i., luminescent animals were prepared for crypt colonization assays, as described below. Pennsylvania State University does not require IACUC approval for invertebrate research.
Motility assay.
Motility assays were performed using TB-IO plates containing 0.25% (wt/vol) agar, as previously described (22). Motility rates were determined by linear regression using the GraphPad Prism software version 6.04 (GraphPad Software, Inc., La Jolla, CA, USA).
Bioluminescence assay.
Bioluminescence assays were performed in LBS medium, as previously reported (23), with the following slight modifications. Each strain was grown in the presence or absence of 120 nM (final concentration) 3-oxo-C6 HSL, and measurements were taken when the cultures were at an optical density at 600 nm (OD600) of ∼1.0.
Fluorescence assay.
Fluorescence assays to measure luxI gene expression were performed with liquid cultures, as previously reported (23), with the following slight modifications. Overnight LBS cultures were diluted 1:100 into LBS medium in the presence or absence of 120 nM (final concentration) 3-oxo-C6 HSL (Sigma-Aldrich, St. Louis, MO, USA). Samples of ES114 harboring the nonfluorescent plasmid pVSV105 were used to account for autofluorescence levels.
Statistical analyses.
Two-way analysis of variance (ANOVA), as performed with GraphPad Prism software, was used to analyze variation among strain identities and HSL treatments for the bioluminescence and fluorescence assays.
Phylogenetics.
The phylogenetics analysis was completed using partial sequences of four concatenated loci (recA, mdh, katA, and pyrC) from previously published data and newly amplified sequences from FQ-A001 in a manner similar to that of previously published methods (17). Concatenations of these partial sequences for 48 Vibrio isolates were aligned with Clustal X 2.1 (24) and analyzed via phylogenetic reconstruction methods in the programs ClonalFrame 1.2 (25) and SplitsTree 4.12.2 (26). For the data set, ClonalFrame generated three independent runs; in each run, the 50% majority rule consensus genealogy was estimated from the posterior distribution of 200,000 generations (with a thinning interval of 100) following burn-in of 100,000 generations. For each run, the Markov chain's Monte Carlo convergence was judged to be satisfactory via use of the Gelman-Rubin test (27). The resulting posterior distribution of 6,000 samples (3 runs of 2,000 samples each) from each concatenated data set was analyzed using a consensus network (28), with mean edge weights at a threshold of 0.2 and an equal-angle splits transformation, as implemented via the SplitsTree software. Nodes with 95% posterior probability (from the 6,000 original ClonalFrame samples) were identified and highlighted in the network graph.
Crypt colonization assay.
Luminescent animals were prepared for confocal microscopy using 4% paraformaldehyde fixative, as previously described (22). Differential interference contrast (DIC), CFP, and YFP images were collected using a Zeiss 780 confocal microscope (Carl Zeiss AG, Jena, Germany) equipped with a 10× water lens. Light organ crypts were scored for CFP and YFP fluorescence.
Aggregation assay.
Juvenile squid were exposed to a 1:1 mixed inoculum of YFP-labeled FQ-A001 and CFP-labeled ES114 (total, ∼10,000 CFU/ml). At 3.5 h p.i., the animals were stained with CellTracker orange CMRA (Life Technologies), according to the manufacturer's instructions, and anesthetized in 5% ethanol-FSSW. The anesthetized animals were dissected to expose their light organs and mounted for examination with a Zeiss 780 confocal microscope equipped with a 63× water lens. CellTracker orange fluorescence was used to locate light organ pores, and z-stacks of CFP, YFP, and CellTracker orange fluorescence were collected around each region. Each pore was examined for aggregates, and the corresponding aggregates were scored for CFP- and YFP-labeled cells.
Growth competition assay.
The growth competition assays were performed in LBS medium, as previously described (22), with the following modification. Where indicated, 120 nM (final concentration) 3-oxo-C6 HSL was added to the medium.
Nucleotide sequence accession numbers.
The sequence of the region upstream of luxI in FQ-A001 has been submitted to GenBank under the accession no. KU851258. Sequences associated with the phylogenetics analysis of the four concatenated loci have been submitted to the GenBank database under the accession numbers KU756584 to KU756587.
RESULTS
Isolation and initial characterization of FQ-A001.
FQ-A001 was isolated directly from the light organ of an adult female E. scolopes squid that had been collected in February 2015 along the coastal area of Kaneohe Bay, HI. At the time of dissection, the individual had a normal mantle length (2.1 cm) and appeared healthy relative to other animals collected in the same cohort. On rich medium, FQ-A001 colonies appeared opaque and smooth, which are features comparable to those of ES114, which is the sequenced type strain of V. fischeri used to study the light organ symbiosis (29).
To determine whether FQ-A001 can initiate a functional symbiosis with E. scolopes, squid colonization assays were conducted by exposing naive juvenile squid to approximately 5,000 CFU/ml of the FQ-A001 culture. At 48 h p.i., the average luminescence level of juveniles was 4.0 × 105 relative light units (RLU), which was 5.2-fold higher than the corresponding luminescence in animals exposed to ES114 (Fig. 1A). As expected, juveniles that were not exposed to a V. fischeri inoculum (i.e., were aposymbiotic) remained nonluminous (Fig. 1A). However, the CFU level of individual squid exposed to FQ-A001 (1.9 × 105 CFU) was comparable to that in juveniles exposed to ES114 (1.8 × 105 CFU) (Fig. 1B). Thus, the specific luminescence, i.e., the luminescence production per CFU, is higher for FQ-A001 than for ES114 within the light organ. The results of these experiments demonstrate that FQ-A001 is able to colonize E. scolopes juveniles and produce luminescence.
FIG 1.
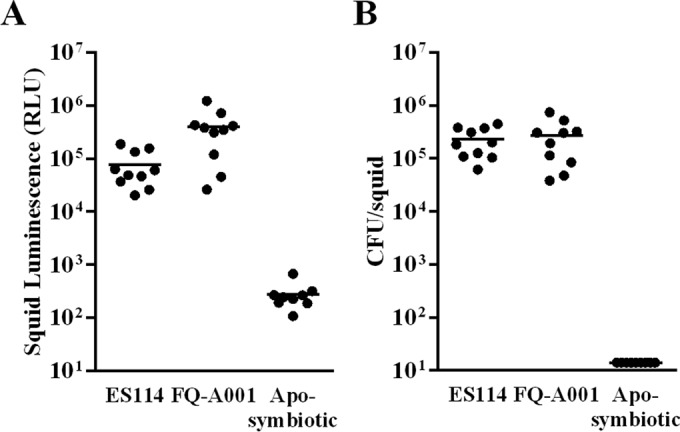
Squid colonization assays with V. fischeri strains. (A) Luminescence of individual E. scolopes juveniles (n = 10 per group) 48 h after exposure to the ES114 or FQ-A001 inoculum. Each point represents an individual animal, and the horizontal bars represent geometric means. (B) CFU levels of squid in panel A. The limit of detection is 14 CFU/squid. The experiment was repeated twice, with similar results. Aposymbiotic, no bacteria were added.
Because flagellum-based motility and chemotaxis are important bacterial behaviors for V. fischeri cells to colonize the squid light organ (13, 30), we examined the ability of FQ-A001 to swim through soft agar containing tryptone, which leads to the chemotaxis-directed motility of V. fischeri (22, 31). Consistent with our previous report (22), ES114 formed two motility rings, with the rate of motility for the outer ring being 0.78 ± 0.04 cm/h (data not shown). Like ES114, FQ-A001 also formed two motility rings but swam at a rate of 0.27 ± 0.01 cm/h, which is 2.9-fold lower than the rate of ES114 (P < 0.0001, Student's t test). These results, which were repeated twice with similar outcomes, demonstrate that FQ-A001 is a motile V. fischeri strain.
Induction of lux gene expression in FQ-A001.
The results from the squid colonization assays described above suggest that the specific luminescence of FQ-A001 in symbiosis is elevated relative to that of ES114 (Fig. 1). To further investigate this finding, we examined the bioluminescence response of FQ-A001 to 3-oxo-C6 HSL, which increases transcription of the lux genes by activating the transcription factor LuxR (32). In response to 120 nM 3-oxo-C6 HSL, FQ-A001 showed 2.1 × 103-fold higher levels of luminescence (Fig. 2A). This response was higher than the 58-fold response displayed by ES114. As expected, EVS102, which is an ES114-derived strain that lacks the luxCDABEG genes (8), remained nonluminous under both conditions. Using two-way ANOVA on log-transformed data, we found that strain identity (F2,12 = 3,726), HSL treatment (F1,12 = 9,808), and their interaction (F2,12 = 2,925) were significant at a P value of <0.001. Post hoc Tukey tests found that ES114 ΔluxCDABEG groups treated and untreated with HSL were not significantly different (P < 0.05); likewise, ES114 and FQ-A001 groups untreated with HSL were not significantly different (P < 0.05); all other group comparisons were found to be significant (P < 0.05). These results demonstrate that FQ-A001 exhibits a stronger luminescence response to 3-oxo-C6 HSL than that of ES114.
FIG 2.
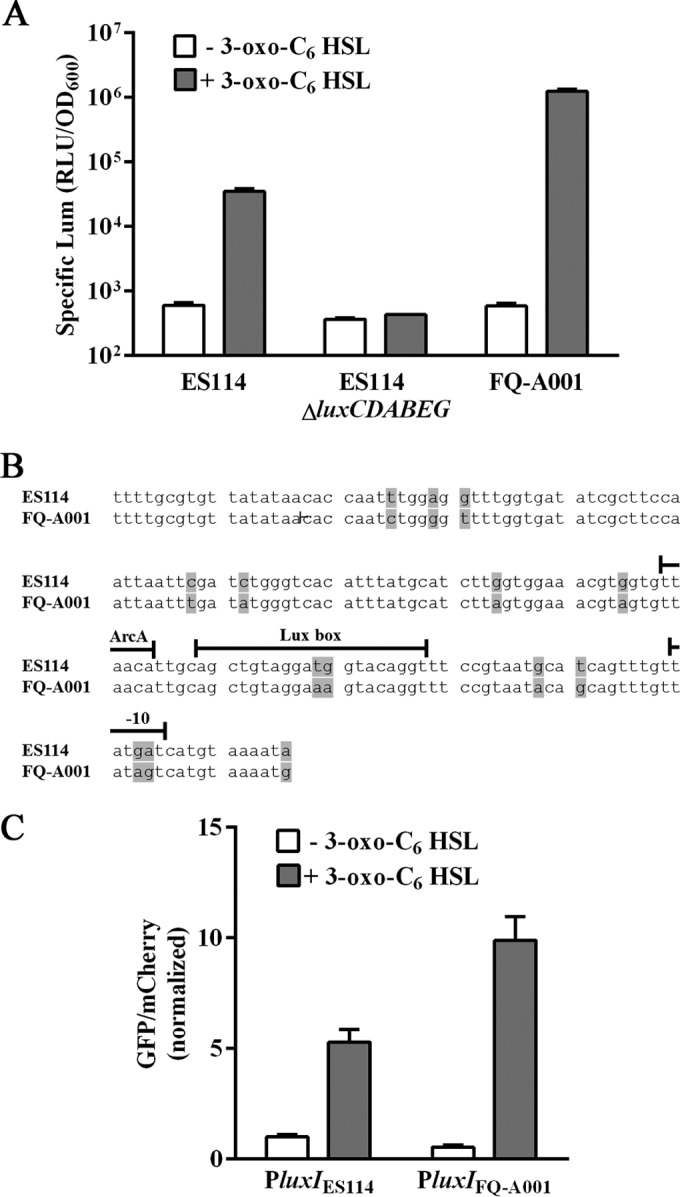
Quorum signaling in V. fischeri strains. (A) Luminescence of ES114, EVS102 (ES114 ΔluxCDABEG), and FQ-A001 grown in LBS medium without (white) or with (gray) 120 nM 3-oxo-C6 HSL. The bars and error bars represent the means and standard deviations of the results from three biological replicates, respectively. The experiment was performed twice, with similar results. See Results for two-way ANOVA statistical analysis. Lum, luminescence. (B) luxIR intergenic region in ES114 and FQ-A001. Bases highlighted in gray indicate differences between strains. The binding sites for ArcA and LuxR and the −10 region are shown above the corresponding sequences. (C) Transcriptional response of luxI in ES114 grown in LBS medium without (white) or with (gray) 120 nM 3-oxo-C6 HSL. The luxI transcriptional reporters are pTM280 (PluxIES114) and pEDL01 (PluxIFQ-A001). The bars and error bars represent the means and standard deviations of the results from three biological replicates, normalized to pTM280/ES114 grown in the absence of 3-oxo-C6 HSL. See Results for two-way ANOVA statistical analysis. GFP, green fluorescent protein.
Sequencing of 166 bp of the luxIR intergenic region in FQ-A001 revealed differences in 14 positions (Fig. 2B). While two of these differences occur at positions 11 and 12 within the Lux box, which is the 20-bp site to which LuxR binds, these positions were previously shown not to impact the induction of bioluminescence by 3-oxo-C6 HSL (33). However, five differences are located at positions downstream of the Lux box, two of which are associated with the −10 region (Fig. 2B). To study transcription of the FQ-A001-specific lux genes, we constructed pEDL001 by cloning the luxI promoter of FQ-A001 (PluxIFQ-A001) upstream of gfp in the plasmid pTM267 (20). Plasmid pTM267 contains a V. fischeri-specific origin of replication and also constitutively expresses the red fluorescent protein mCherry. When measured in ES114 cells harboring pEDL001, the level of luxI expression in response to 3-oxo-C6 HSL was 1.9-fold higher than that of the luxI promoter of ES114 (PluxIES114) (Fig. 2C). Two-way ANOVA found that HSL treatment (F1,8 = 378.6) was significant at a P value of 0.004 and that plasmid identity (F1,8 = 34.89) and their interaction (F1,9 = 52.11) were significant at a P value of <0.001. Post hoc Tukey tests revealed that the PluxIES114 and PluxIFQ-A100 groups treated with HSL were significantly different (P < 0.05), but untreated groups were not (P < 0.05). These results suggest that the differences within the luxIR intergenic region of FQ-A001 promote increased lux transcription in the presence of 3-oxo-C6 HSL.
Phylogenetic relationship of FQ-A001 to other V. fischeri strains.
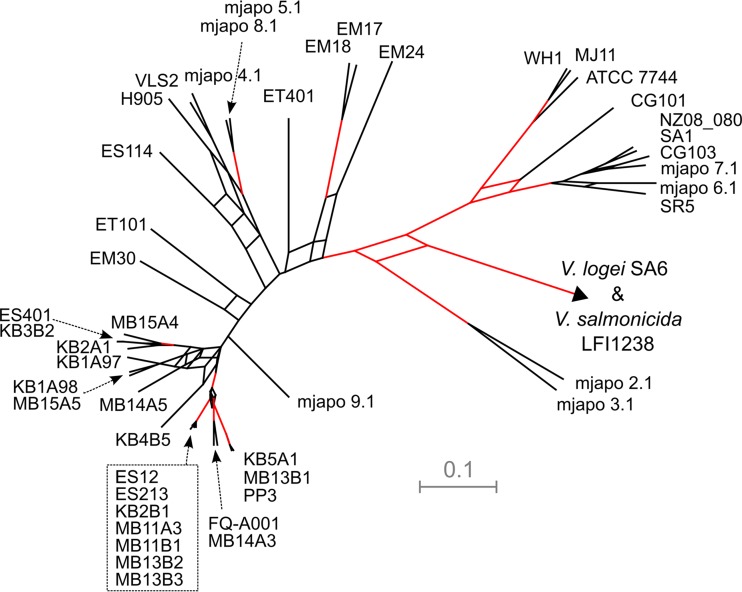
Previous studies have revealed significant phenotypic, genetic, and phylogenetic diversity among symbiotic V. fischeri strains (16, 17). To explore the evolutionary relationship of FQ-A001 to other V. fischeri strains, we conducted a phylogenetic analysis. This phylogenetic analysis used a previously described methodology of constructing a phylogenetic hypothesis using concatenated partial sequences of four housekeeping loci (recA, mdh, katA, and pyrC) in V. fischeri and several closely related Vibrio species (17). Our phylogenetic analysis strongly supported a very close evolutionary relationship of FQ-A001 and MB14A3 (Fig. 3). MB14A3 is a V. fischeri strain that was isolated from an E. scolopes adult collected in Maunalua Bay, HI (16). In addition, the phylogenetic analysis of these four loci did not support the hypotheses that FQ-A001 is a member of the monophyletic group A clade and that FQ-A001 is more similar to ES114 than other V. fischeri strains. However, FQ-A001 was contained in a strongly supported (>95% posterior probability support from ClonalFrame trees) extended monophyletic clade that contains the group A strains as well as KB5A1, MB13B1, PP3, and MB14A3.
FIG 3.
Phylogenetic relationship of FQ-A001 with V. fischeri strains. A consensus network of 6,000 trees was derived from ClonalFrame analyses using concatenated fragments of recA, mdh, katA, and pyrC. Strains highlighted by the stippled box are group A strains. The scale bar represents a branch length of 0.1 substitution per site; outgroups are derived from the closely related species Vibrio logei and Vibrio salmonicida, with respective branch lengths of 0.51 and 1.59 substitutions per site extended beyond the arrowhead; these two outgroups are located at the end of different branches that share a node at the arrowhead in the figure. Clades with ≥95% posterior probability from the 6,000 samples generated by ClonalFrame are highlighted with red lines.
Competitiveness of FQ-A001 during colonization of the squid light organ.
The culture assays and phylogenetic analysis described above suggest that FQ-A001 is both phenotypically and genetically distinct from ES114. To determine the relative efficiency of FQ-A001 in colonizing the squid light organ, we conducted cocolonization experiments involving ES114. To distinguish between strains in each experiment, one strain harbored a plasmid that constitutively produces cyan fluorescent protein (CFP), and the other strain contained a plasmid that constitutively produces yellow fluorescent protein (YFP).
Squid exposed to a mixed inoculum with differentially labeled ES114 strains yielded primarily cocolonized animals (73 to 93%) (Table 2). As expected, the competitive index, i.e., the ratio of YFP-labeled CFU to CFP-labeled CFU, was approximately 1.0 for three independent trials, indicating that the efficiency of ES114 to colonize the animals is independent of the fluorescent label.
TABLE 2.
Light organ cocolonization assays
| Test straina | Trial no. | Inoculum ratio (YFP/CFP) | Total inoculum (CFU/ml × 103) | No. of animalsb |
Avg log CI ± 95% confidence intervalc | ||
|---|---|---|---|---|---|---|---|
| Control only | Test only | Cocolonized | |||||
| ES114 | 1 | 1.24 | 9.89 | 4 | 0 | 11 | −0.164 ± 0.309 |
| 2 | 1.72 | 6.91 | 1 | 0 | 13 | 0.175 ± 0.436 | |
| 3 | 1.42 | 1.84 | 0 | 1 | 13 | 0.119 ± 0.112 | |
| FQA001 | 1 | 1.31 | 12.0 | 0 | 14 | 1 | 1.45 |
| 2 | 0.44 | 4.60 | 0 | 14 | 1 | 1.37 | |
| 3 | 0.26 | 3.34 | 0 | 5 | 10 | 1.18 ± 0.497 | |
For each trial, newly hatched E. scolopes juveniles were exposed for approximately 18 h to a mixed inoculum containing the indicated test strain harboring a YFP+ plasmid (pSCV38) and the control strain ES114 harboring a CFP+ plasmid (pYS112).
After 48 h p.i., E. scolopes juveniles were homogenized and plated onto LBS medium. The squid were scored via the resulting fluorescent CFU counts as singly colonized by either the control (CFP) or test (YFP) strain or as cocolonized with both the control and test strains.
For cocolonized animals, the average competitive index (CI) was determined using log-transformed data. Also shown for trials with >1 cocolonized animal are the 95% confidence intervals of the log-transformed data.
Using this approach, we conducted cocolonization experiments with FQ-A001 labeled with YFP. In two trials, the majority of animals (93%) were colonized by FQ-A001 only (Table 2). A third trial, in which the initial inoculum ratio was biased toward ES114, yielded 67% cocolonized animals. However, the competitive indices associated with these cocolonized animals remained elevated (10-fold higher FQ-A001 CFU than ES114 CFU), indicating that even in cocolonized animals, FQ-A001 appears to have an advantage in establishing symbiosis within the light organ. Together, these CFU-based results indicate that FQ-A001 is a competitively dominant strain.
Crypt colonization profiles of FQ-A001 and ES114.
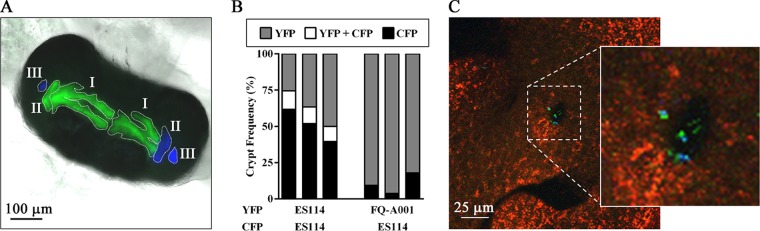
The nascent light organ has six distinct crypt spaces that serve as the primary colonization sites for V. fischeri cells (Fig. 4A). Previous work with ES114 has suggested that only 1 to 2 cells enter each crypt space and serve as founder cells of the resulting crypt populations (16). To determine the crypt colonization profiles of FQ-A001, we conducted a series of cocolonization experiments with the fluorescently labeled strains described above. However, instead of homogenizing bacterium-containing tissue, we prepared samples for fluorescence microscopy by fixing and dissecting out light organs from luminescent animals, which permits each colonized crypt to be scored for specific strains.
FIG 4.
Colonization profiles of V. fischeri strains. (A) Light organ colonized in each crypt with either YFP-labeled FQ-A001 (green) or CFP-labeled ES114 (blue) cells. The distinct crypt spaces, which are indicated by the dashed lines, are labeled I, II, or III. (B) Frequency of crypts with each strain type. Each bar represents the results from a single trial, in which at least 100 crypt spaces were scored. (C) Coaggregate of YFP-labeled FQ-A001 (green) and CFP-labeled ES114 (blue) cells associated with light organ pore. Host tissue is stained with CellTracker orange (red). The image was generated as the maximum projection of a z-stack.
When ES114 was differentially labeled and mixed evenly in an inoculum, the frequency of animals containing both YFP- and CFP-fluorescent populations was 77 to 89%, which is comparable to the frequency of cocolonization, as determined by CFU plating (Table 2). Across all animals in each trial, the frequencies of crypts singly colonized by YFP and CFP ranged from 25 to 50% and 40 to 62%, respectively (Fig. 4B). In addition, 10 to 13% of the resulting crypts were cocolonized with CFP- and YFP-labeled cells. Together, these results indicate that the majority of crypts were singly colonized and that cocolonized animals arose primarily due to distinct populations of each type.
When YFP-labeled FQ-A001 cells were combined with CFP-labeled ES114 cells as mixed inoculums for cocolonization experiments, we found that the majority of the crypts (82 to 96%) were colonized by FQ-A001, suggesting that the competitive dominance of FQ-A001 observed from CFU plating was due to the prevalence of crypts containing FQ-A001 (Fig. 4B). Remarkably, among over 100 crypts scored in each trial, no cocolonized crypts were observed, which is in stark contrast to the occurrence of cocolonized crypts in the experiments using only ES114. This striking result led to our investigation of the aggregation stage of host colonization, during which V. fischeri cells could compete to access the pores that lead to crypt spaces. However, 3-dimensional confocal microscopy scans of the pore regions of animals exposed to a mixed FQ-A001-ES114 inoculum revealed that coaggregation occurred in 38% of the observed aggregates (8 coaggregates/21 aggregates) (Fig. 4C), suggesting that the observed crypt incompatibility of FQ-A001 and ES114 is independent of aggregate formation.
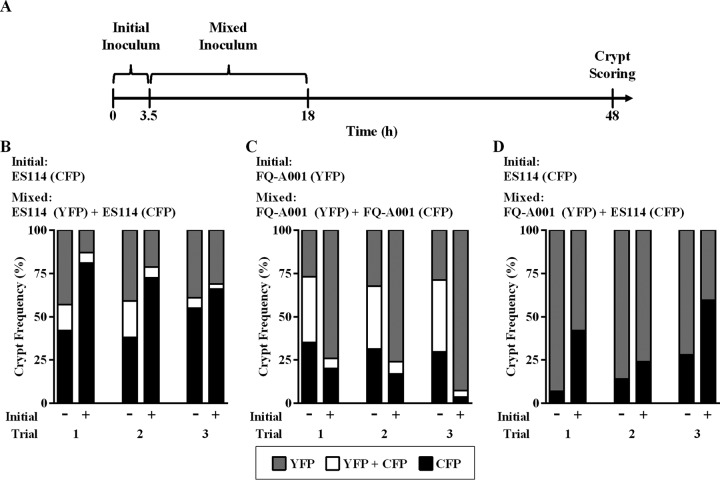
To further characterize the competitive dominance of FQ-A001, we conducted cocolonization experiments designed to provide one strain a temporal advantage, i.e., a head start, during initial colonization of the light organ (Fig. 5A). We first investigated the competition between differentially labeled ES114 strains. In the absence of a head start, there were similar frequencies of crypts singly colonized by YFP- and CFP-producing populations (Fig. 5B). In addition, 6 to 21% of the crypts were cocolonized, which is a frequency range similar to that obtained from the standard cocolonization assay described above. When CFP-labeled ES114 cells were given a head start, the frequency of crypts singly colonized with CFP-labeled cells increased, concomitant with lower frequencies of crypts either singly colonized with YFP-labeled cells or cocolonized (Fig. 5B). Notably, in this experiment, as well as in the other head start experiments shown in Fig. 5, the total number of crypts colonized was independent of whether an initial inoculum was present. These results demonstrate that the preexposure of animals to one strain will bias the crypt-specific colonization patterns toward that strain.
FIG 5.
Crypt colonization profiles in response to strain preexposure. (A) Timeline of a head start cocolonization experiment. Each experiment consisted of a mixed inoculum with CFP- and YFP-labeled strains with (+) or without (−) the initial fluorescently labeled single-strain inoculum. The frequency of crypts with each strain type is shown. Each experiment was performed three times and is presented as an individual trial. (B) Mixed inoculum, CFP- and YFP-labeled ES114; initial inoculum, CFP-labeled ES114. (C) Mixed inoculum, CFP- and YFP-labeled FQ-A001; initial inoculum, YFP-labeled FQ-A001. (D) Mixed inoculum, CFP-labeled ES114 and YFP-labeled FQ-A001; initial inoculum, CFP-labeled ES114.
For the competitions involving differentially labeled strains of FQ-A001, the percentages of crypts singly colonized by YFP or CFP were similar to one another (Fig. 5C). Interestingly, the percentage of cocolonized crypts observed for FQ-A001 (36 to 42%) was higher than that found for the competitions involving ES114 (6 to 21%), suggesting that the likelihood of two founder FQ-A001 cells is higher than that of two founder ES114 cells. The impact of a head start by YFP-labeled FQ-A001 was an increased abundance of YFP-labeled crypts (74 to 93%) and a decreased abundance of CFP-labeled or cocolonized crypts (4 to 20% and 4 to 7%, respectively). Overall, this outcome was similar to that in the experiments involving ES114, demonstrating that earlier access to the light organ by a strain can increase the likelihood of that strain successfully colonizing the host.
For the final set of head start experiments, we conducted three trials with the mixed inoculum of YFP-labeled FQ-A001 and CFP-labeled ES114 strains. The frequency of crypts singly colonized by FQ-A001 exceeded that of crypts singly colonized by ES114 (Fig. 5D), which is consistent with results shown in Fig. 4B. Remarkably, no cocolonized crypts were observed in any of the trials (Fig. 5D), which each involved scoring approximately 100 crypts. In the animals initially exposed to CFP-labeled ES114 cells, the frequency of CFP-labeled crypts increased; however, cocolonized crypts were not observed.
In summary, the results from the head start experiments, which involves strain competitions during the initial colonization of the squid light organ, demonstrate the following: (i) preexposure to a strain increases the frequency of crypt occupancy by that strain, (ii) preexposure to a strain decreases the incidence of cocolonized crypts, and (iii) ES114 and FQ-A001 are unable to cocolonize the same crypt.
Fitness of FQ-A001 and ES114 in culture.
Our unexpected finding of only singly colonized crypts with animals exposed to FQ-A001 and ES114 suggests the possibility of strain incompatibility within crypt spaces. To determine whether such incompatibility is a property solely associated with these two strains, we conducted culture-based fitness assays. For each assay, we combined cultures containing a CFP-labeled strain with a YFP-labeled strain and tracked the relative competitive index, i.e., the ratio of the CFU of the YFP-labeled strain to the CFU of the CFP-labeled strain normalized by the initial CFU ratio. For LBS, we found that the relative competitive index (RCI) of each competition (ES114 versus ES114, FQ-A001 versus FQ-A001, and ES114 versus FQ-A001) remained near 1.0 for 20 generations (Fig. 6A), which exceeds the number of generations required to establish a light-producing infection of V. fischeri within the juvenile light organ. We also considered the possibility that high 3-oxo-C6 HSL levels within the crypt space may induce the strain incompatibility complex, but the RCI remained comparable to 1.0 for each competition grown in the presence of 120 nM 3-oxo-C6 HSL (Fig. 6B).
FIG 6.
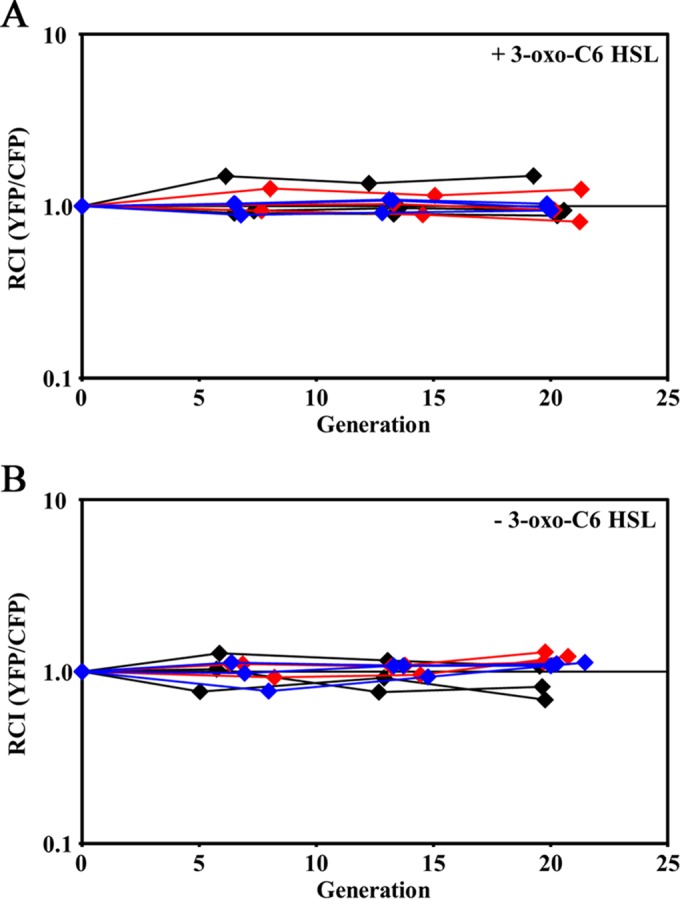
Impact of autoinducer on fitness in rich medium. Differentially labeled strains (ES114 or FQ-A001) were grown in LBS without (A) or with (B) 120 nM 3-oxo-C6 HSL for the indicated number of generations of the CFP-labeled strain. The relative competitive index (RCI) was calculated by normalizing the YFP/CFP CFU ratio at the indicated generation by the initial YFP/CFP CFU ratio. Each line represents an independent culture.
DISCUSSION
In this study, we examined whether the initial stages of colonization could contribute to the previously reported V. fischeri strain diversity associated with the light organs of wild-caught squid (16, 17). Microbiological assays and phylogenetic analyses demonstrated that the strain FQ-A001, which was isolated directly from an adult light organ, is physiologically and genetically distinct from the V. fischeri type strain ES114. Relative to ES114, FQ-A001 displays a stronger bioluminescence response to 3-oxo-C6 HSL and reduced motility in soft agar. In addition, FQ-A001 outcompetes ES114 in squid cocolonization experiments. Phylogenetic analysis of concatenated fragments of housekeeping genes revealed that FQ-A001 falls into a clade that also contains a subclade denoted “group A” (17). Group A is comprised of competitively dominant symbiotic strains, and the proximity of FQ-A001 to this clade suggests that FQ-A001 may contain host specificity factors that enhance light organ colonization.
Using FQ-A001 and ES114, we developed a two-strain symbiosis model and found that the individual crypt spaces of the light organ represent distinct niches for V. fischeri cells initiating host colonization (Fig. 7). This model highlights that the higher abundance of FQ-A001 in the juvenile light organ relative to that in ES114 is due to more crypt spaces being colonized by FQ-A001. The results from our head start experiments show that a temporal advantage for one strain to access the host results in more crypts harboring that strain type; i.e., sampling of potential symbionts within the seawater environment by the juvenile host is a strong determinant of the resulting crypt composition. However, if two different strains coexist within the aggregates that develop outside the light organ pores, they subsequently compete to singly colonize the crypt space, indicating that a bottleneck exists after the host samples the environment. While there is a physical bottleneck located at each crypt entrance that reduces in size within 2 days after colonization (15), two lines of evidence from our study suggest that this physical bottleneck does not dictate which strain type colonizes the corresponding crypt space. First, FQ-A001 exhibits lower motility rates than those of ES114, which suggests that a race between cells of each strain type through the ducts and physical bottleneck is insufficient to explain the overall dominance of FQ-A001 in crypt colonization. Notably, there is a caveat to this interpretation, because the soft-agar plate assays used to study bacterial motility in vitro may not accurately reflect how V. fischeri uses motility to colonize the light organ. Second, cocolonized crypts were detected when differentially labeled strains of an identical genotype (with either FQ-A001 or ES114) were used, suggesting that at least two cells are able to pass the physical bottleneck and enter the crypt spaces. Instead of the physical bottleneck, differences in the abilities of cells to escape from the aggregate, the formation of which is strongly linked to the process of biofilm development (34), may contribute to the dominance of certain strains when presented as mixed inoculums. Finally, because a competitive advantage for either FQ-A001 or ES114 failed to emerge in culture (Fig. 6), we speculate that the crypt environment itself may also limit strain diversity either directly through the preferential selection of strain type (e.g., bacterial adhesion to crypt epithelial cells or utilization of crypt-specific nutrients) or indirectly by activating strain-specific genes that target and kill other strain types. Imaging-based approaches that track individual V. fischeri cells associated with the host are necessary to understand the dynamics of how crypt colonization is determined in the presence of multiple strain types.
FIG 7.
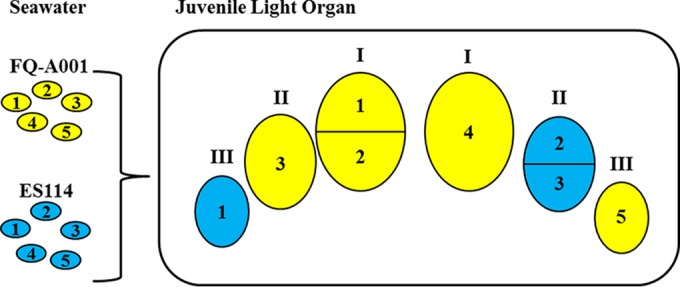
Model for intraspecific competition during initial colonization of the E. scolopes light organ. The cartoon shows the six crypts (indicated by Roman numerals) of a juvenile light organ cocolonized with FQ-A001 (yellow) and ES114 (blue). FQ-A001-ES114 symbiont diversity can arise in the nascent light organ only through colonization of distinct crypts. Multiple cells of the same type can serve as founder cells for the resulting populations, e.g., FQ-A001 cells 1 and 2 or ES114 cells 2 and 3. FQ-A001 outcompetes ES114 during crypt colonization, which leads to a higher abundance of FQ-A001 than of ES114 in whole animals. (ES114 cells 4 and 5 fail to colonize.)
For this case of FQ-A001/ES114 strain diversity, our results appear to refute a model based strictly on niche partitioning within a light organ crypt, in which different strains coexist within the same crypt by accessing different nutrients. Niche partitioning is an important process that stabilizes the interspecific diversity of microbial consortia, in which individual members occupy specific niches that are generated by the host environment and the metabolic activities of other consortium members. However, this process can lead to intraspecific competition for unique niches within a host. For instance, germfree mouse models have revealed that colonization of the gut by Bacteroides fragilis, which is a member of the highly abundant Bacteroides family found within the mammalian gut microbiota, prevents the colonization of the same, but not other, Bacteroides species (35). Remarkably, a single polysaccharide utilization locus enables B. fragilis to occupy intestinal crypt spaces, and these bacterial cells are thought to serve as a reservoir for recolonization events when the structure of the microbial consortium is perturbed by antibiotic treatment. More recently, a two-species (Aeromonas veronii and Vibrio sp.) gut microbiota model was developed with zebrafish larvae; this model used transposon-associated genomic insertion sites to differentially label non-wild-type cells of each species (36). While some interspecific competition was detected between Aeromonas and Vibrio, the level of intraspecific competition was profound for each species, with the transposon-labeled cells unable to establish infections within intestines already colonized with the corresponding wild-type strains. Our study suggests that niche partitioning does not occur with FQ-A001 and ES114 and, furthermore, demonstrates that the squid-Vibrio symbiosis can serve as a general platform for more-comprehensive investigations of niche partitioning and intraspecific competition using additional strains.
The V. fischeri strain that successfully colonizes a crypt space is predicted to influence the abundance of symbiotic strains in the seawater environment and, as a consequence, future generations of the symbiosis. Each day at dawn, squid will expel up to 95% of the symbiont population, which then regrows from the remaining 5%, thereby enriching squid-containing seawater environments with symbiotic strains (37). Recent work has shown that group A strains also exhibit lower fitness within local Hawaiian seawater than that of non-group A strains (17), which highlights the fact that host-independent factors also impact the ecology of symbiotic V. fischeri strains. Interestingly, this result suggests that the stability of diverse strains within the seawater environment, in contrast to within the crypt spaces, may be subject to niche partitioning. Sequencing-based approaches that can monitor V. fischeri populations at the strain level will be necessary to explore the impact of daily expulsion on the abundance of V. fischeri strains within the environment.
At the organismal level, sequencing-based approaches have begun to reveal the physiological significance of symbiont strain diversity, especially in the context of gut microbiota. For instance, genetic diversity was shown among strains of Gilliamella gut symbionts within the honey bee Apis mellifera (38), which contains a gut microbiome that is acquired through the eusocial behavior of the host. Genetically different Gilliamella isolates displayed different levels of pectin degradation, suggesting unique niche adaptation for each strain within the honey bee gut. Within the human gut microbiome, significant intraspecies copy number variation was detected among genes that function in transport, sugar metabolism, and antibiotic resistance (39), supporting the notion that strain diversity can provide novel functions within the community. Interestingly, the population structures for many of the species were unique in each sample, indicating the individualized nature of the human gut microbiota. These population structures are particularly dynamic during the initial colonization of the infant gut, as revealed by a 10-day study of the gut microbiome in a 1-month-old infant, which found that the abundance profiles of three distinct Staphylococcus epidermidis strains were unique for each strain (40).
Strain diversity among gut symbionts can also impact the ability of pathogens to colonize the gut. For instance, colonization by the enterohemorrhagic Escherichia coli strain EDL933 was blocked by a complementary mixture of commensal strains that effectively occupy the various intestinal niches that serve as colonization sites for EDL933 (41). Subsequent work revealed that other E. coli pathotypes target other intestinal sites for colonization (42), indicating that further investigation of the link between strain diversity and niche specificity is necessary to develop broad-spectrum probiotics. Interestingly, in some cases, strain diversity of pathogens within a mixed infection may augment the corresponding disease. This outcome was demonstrated in a necrotizing fasciitis model with two Aeromonas hydrophila strain types that occupied distinct niches within the host, with one strain exhibiting increased virulence due to the secretion of exotoxin A by the other strain (43).
Our study suggests that FQ-A001/ES114 strain diversity in the juvenile light organ is a consequence of the multiple colonization sites harboring distinct strain types. However, because the infections associated with juvenile squid can persist over the lifetime of the host (9), we speculate that the populations also evolve over time, thereby contributing to the strain diversity observed in the light organs of adult animals (16). Recent experimental evolution studies using juvenile squid to propagate nonsymbiotic V. fischeri strains resulted in evolved lines that exhibit phenotypes that differ from that of the parental strain (44). For instance, the passage of the highly visible fish symbiont MJ11 through multiple juvenile squid resulted in strains with in vitro luminescence profiles comparable to that of the control strain ES114 (44). Similar diversification has been reported in the persistent infections of Pseudomonas aeruginosa within the lungs of patients with cystic fibrosis (45). The ability to monitor individual V. fischeri strains within the light organ throughout the lifetime of the host will increase our understanding of the microbial diversity associated with infectious diseases. Future studies with other natural isolates will reveal how strain phenotype and genome content contribute to the strain diversity established during the initial stages of light organ colonization.
Our two-strain system also underscores the importance of incorporating strain diversity in experiments that involve horizontally acquired microbes. The majority of studies of the squid-Vibrio symbiosis have used the type strain ES114, which falls outside the competitively dominant group A clade and is a relatively dim strain of V. fischeri (18). Many squid-symbiotic strains exhibit levels of bioluminescence that are at least an order of magnitude higher than that of ES114 (e.g., FQ-A001). While the role of bacterial bioluminescence is involved in the counterillumination behavior of the squid host (4), the impact of light organ infections containing strains with different bioluminescence capacities on this behavior is unclear. We anticipate that the recent success in rearing juvenile squid to adulthood (9) will make long-term behavioral studies of individuals harboring polyclonal infections much more feasible.
ACKNOWLEDGMENTS
A.G.C. caught adult squid, performed dissection for FQ-A001 isolation, and contributed to the design of culture-based experiments. J.L.-V. scored and analyzed crypt colonization profiles. Y.S., E.D.L., and K.B.L. conducted culture competition experiments. M.S.W. conducted phylogenetic analysis of FQ-A001. Y.S., E.D.L., and T.M. designed, performed, analyzed, and interpreted all other experiments. All authors contributed to the drafting of the article.
We thank the Sauer lab for the proD plasmid. We also thank three anonymous reviewers for their comments, which significantly improved the final manuscript.
Funding Statement
This work, including the efforts of Tim Miyashiro, was funded by HHS | National Institutes of Health (NIH) (097032) and the Eberly College of Science at Penn State University. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Gilbert SF, Bosch TC, Ledón-Rettig C. 2015. Eco-Evo-Devo: developmental symbiosis and developmental plasticity as evolutionary agents. Nat Rev Genet 16:611–622. doi: 10.1038/nrg3982. [DOI] [PubMed] [Google Scholar]
- 2.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandel MJ. 2010. Models and approaches to dissect host-symbiont specificity. Trends Microbiol 18:504–511. doi: 10.1016/j.tim.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Jones BW, Nishiguchi MK. 2004. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar Biol 144:1151–1155. doi: 10.1007/s00227-003-1285-3. [DOI] [Google Scholar]
- 5.Meighen EA. 1993. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J 7:1016–1022. [DOI] [PubMed] [Google Scholar]
- 6.Miyashiro T, Ruby EG. 2012. Shedding light on bioluminescence regulation in Vibrio fischeri. Mol Microbiol 84:795–806. doi: 10.1111/j.1365-2958.2012.08065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol 182:4578–4586. doi: 10.1128/JB.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bose JL, Rosenberg CS, Stabb EV. 2008. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol 190:169–183. doi: 10.1007/s00203-008-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch EJ, Miyashiro T, McFall-Ngai MJ, Ruby EG. 2014. Features governing symbiont persistence in the squid-Vibrio association. Mol Ecol 23:1624–1634. doi: 10.1111/mec.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McFall-Ngai MJ. 2014. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol 68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altura MA, Heath-Heckman EA, Gillette A, Kremer N, Krachler AM, Brennan C, Ruby EG, Orth K, McFall-Ngai MJ. 2013. The first engagement of partners in the Euprymna scolopes-Vibrio fischeri symbiosis is a two-step process initiated by a few environmental symbiont cells. Environ Microbiol 15:2937–2950. doi: 10.1111/1462-2920.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyholm SV, McFall-Ngai MJ. 2003. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl Environ Microbiol 69:3932–3937. doi: 10.1128/AEM.69.7.3932-3937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandel MJ, Schaefer AL, Brennan CA, Heath-Heckman EA, Deloney-Marino CR, McFall-Ngai MJ, Ruby EG. 2012. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol 78:4620–4626. doi: 10.1128/AEM.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heath-Heckman EA, McFall-Ngai MJ. 2011. The occurrence of chitin in the hemocytes of invertebrates. Zoology (Jena) 114:191–198. doi: 10.1016/j.zool.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sycuro LK, Ruby EG, McFall-Ngai M. 2006. Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their morphological complexity and dynamic function in symbiosis. J Morphol 267:555–568. doi: 10.1002/jmor.10422. [DOI] [PubMed] [Google Scholar]
- 16.Wollenberg MS, Ruby EG. 2009. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl Environ Microbiol 75:193–202. doi: 10.1128/AEM.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wollenberg MS, Ruby EG. 2012. Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME J 6:352–362. doi: 10.1038/ismej.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bose JL, Wollenberg MS, Colton DM, Mandel MJ, Septer AN, Dunn AK, Stabb EV. 2011. Contribution of rapid evolution of the luxR-luxI intergenic region to the diverse bioluminescence outputs of Vibrio fischeri strains isolated from different environments. Appl Environ Microbiol 77:2445–2457. doi: 10.1128/AEM.02643-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. 2009. A single regulatory gene is sufficient to alter bacterial host range. Nature 458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. 2010. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol 77:1556–1567. doi: 10.1111/j.1365-2958.2010.07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis JH, Rubin AJ, Sauer RT. 2011. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res 39:1131–1141. doi: 10.1093/nar/gkq810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Y, Verma SC, Bogale H, Miyashiro T. 2015. NagC represses N-acetyl-glucosamine utilization genes in Vibrio fischeri within the light organ of Euprymna scolopes. Front Microbiol 6:741. doi: 10.3389/fmicb.2015.00741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyashiro T, Oehlert D, Ray VA, Visick KL, Ruby EG. 2014. The putative oligosaccharide translocase SypK connects biofilm formation with quorum signaling in Vibrio fischeri. Microbiologyopen 3:836–848. doi: 10.1002/mbo3.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 27.Gelman A, Rubin DB. 1992. Inference from iterative simulation using multiple sequences. Stat Sci 7:457–472. doi: 10.1214/ss/1177011136. [DOI] [Google Scholar]
- 28.Holland BR, Huber KT, Moulton V, Lockhart PJ. 2004. Using consensus networks to visualize contradictory evidence for species phylogeny. Mol Biol Evol 21:1459–1461. doi: 10.1093/molbev/msh145. [DOI] [PubMed] [Google Scholar]
- 29.Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg EP. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci U S A 102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brennan CA, Mandel MJ, Gyllborg MC, Thomasgard KA, Ruby EG. 2013. Genetic determinants of swimming motility in the squid light-organ symbiont Vibrio fischeri. Microbiologyopen 2:576–594. doi: 10.1002/mbo3.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deloney-Marino CR. 2013. Observing chemotaxis in Vibrio fischeri using soft agar assays in an undergraduate microbiology laboratory. J Microbiol Biol Educ 14:271–272. doi: 10.1128/jmbe.v14i2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens AM, Dolan KM, Greenberg EP. 1994. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc Natl Acad Sci U S A 91:12619–12623. doi: 10.1073/pnas.91.26.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antunes LC, Ferreira RB, Lostroh CP, Greenberg EP. 2008. A mutational analysis defines Vibrio fischeri LuxR binding sites. J Bacteriol 190:4392–4397. doi: 10.1128/JB.01443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visick KL. 2009. An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri. Mol Microbiol 74:782–789. doi: 10.1111/j.1365-2958.2009.06899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. 2013. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephens WZ, Wiles TJ, Martinez ES, Jemielita M, Burns AR, Parthasarathy R, Bohannan BJ, Guillemin K. 2015. Identification of population bottlenecks and colonization factors during assembly of bacterial communities within the zebrafish intestine. mBio 6(6):e01163-15. doi: 10.1128/mBio.01163-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee K-H, Ruby EG. 1994. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol 60:1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engel P, Martinson VG, Moran NA. 2012. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci U S A 109:11002–11007. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenblum S, Carr R, Borenstein E. 2015. Extensive strain-level copy-number variation across human gut microbiome species. Cell 160:583–594. doi: 10.1016/j.cell.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. 2013. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res 23:111–120. doi: 10.1101/gr.142315.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leatham MP, Banerjee S, Autieri SM, Mercado-Lubo R, Conway T, Cohen PS. 2009. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect Immun 77:2876–2886. doi: 10.1128/IAI.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meador JP, Caldwell ME, Cohen PS, Conway T. 2014. Escherichia coli pathotypes occupy distinct niches in the mouse intestine. Infect Immun 82:1931–1938. doi: 10.1128/IAI.01435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ponnusamy D, Kozlova EV, Sha J, Erova TE, Azar SR, Fitts EC, Kirtley ML, Tiner BL, Andersson JA, Grim CJ, Isom RP, Hasan NA, Colwell RR, Chopra AK. 2016. Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc Natl Acad Sci U S A 113:722–727. doi: 10.1073/pnas.1523817113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuster BM, Perry LA, Cooper VS, Whistler CA. 2010. Breaking the language barrier: experimental evolution of non-native Vibrio fischeri in squid tailors luminescence to the host. Symbiosis 51:85–96. doi: 10.1007/s13199-010-0074-2. [DOI] [Google Scholar]
- 45.Jorth P, Staudinger BJ, Wu X, Hisert KB, Hayden H, Garudathri J, Harding CL, Radey MC, Rezayat A, Bautista G, Berrington WR, Goddard AF, Zheng C, Angermeyer A, Brittnacher MJ, Kitzman J, Shendure J, Fligner CL, Mittler J, Aitken ML, Manoil C, Bruce JE, Yahr TL, Singh PK. 2015. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe 18:307–319. doi: 10.1016/j.chom.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol 72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]