Abstract
Squalene synthase inhibitors (SSIs), such as squalestatin 1 (SQ1), reduce cholesterol biosynthesis but cause the accumulation of isoprenoids derived from farnesyl pyrophosphate (FPP), which can modulate the activity of nuclear receptors, including the constitutive androstane receptor (CAR), farnesoid X receptor, and peroxisome proliferator-activated receptors (PPARs). In comparison, 3-hydroxy-3-methylglutaryl-coenzyme A reductase inhibitors (e.g., pravastatin) inhibit production of both cholesterol and nonsterol isoprenoids. To characterize the effects of isoprenoids on hepatocellular physiology, microarrays were used to compare orthologous gene expression from primary cultured mouse and rat hepatocytes that were treated with either SQ1 or pravastatin. Compared with controls, 47 orthologs were affected by both inhibitors, 90 were affected only by SQ1, and 51 were unique to pravastatin treatment (P < 0.05, ≥1.5-fold change). When the effects of SQ1 and pravastatin were compared directly, 162 orthologs were found to be differentially coregulated between the two treatments. Genes involved in cholesterol and unsaturated fatty acid biosynthesis were up-regulated by both inhibitors, consistent with cholesterol depletion; however, the extent of induction was greater in rat than in mouse hepatocytes. SQ1 induced several orthologs associated with microsomal, peroxisomal, and mitochondrial fatty acid oxidation and repressed orthologs involved in cell cycle regulation. By comparison, pravastatin repressed the expression of orthologs involved in retinol and xenobiotic metabolism. Several of the metabolic genes altered by isoprenoids were inducible by a PPARα agonist, whereas cytochrome P450 isoform 2B was inducible by activators of CAR. Our findings indicate that SSIs uniquely influence cellular lipid metabolism and cell cycle regulation, probably due to FPP catabolism through the farnesol pathway.
Introduction
Cardiovascular disease (CVD) is the leading cause of mortality worldwide, and elevated low-density lipoprotein (LDL) cholesterol is a major risk factor for CVD development (Mathers et al., 2009; Goldstein and Brown, 2015). Inhibitors of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR, i.e., statins), the rate-limiting enzyme in cholesterol biosynthesis (Fig. 1), are the most widely used class of anticholesterol drugs. Various clinical trials have demonstrated the efficacy of statin therapy in lowering LDL cholesterol and reducing both CVD morbidity and mortality (Baigent et al., 2010; Fulcher et al., 2015). However, statin use in some individuals is limited due to the development of adverse reactions such as myopathies (Tomaszewski et al., 2011). Additionally, despite aggressive statin therapy, certain populations have a high residual risk for CVD (Campbell et al., 2007; Sampson et al., 2012), so alternative lipid-modifying agents are needed.
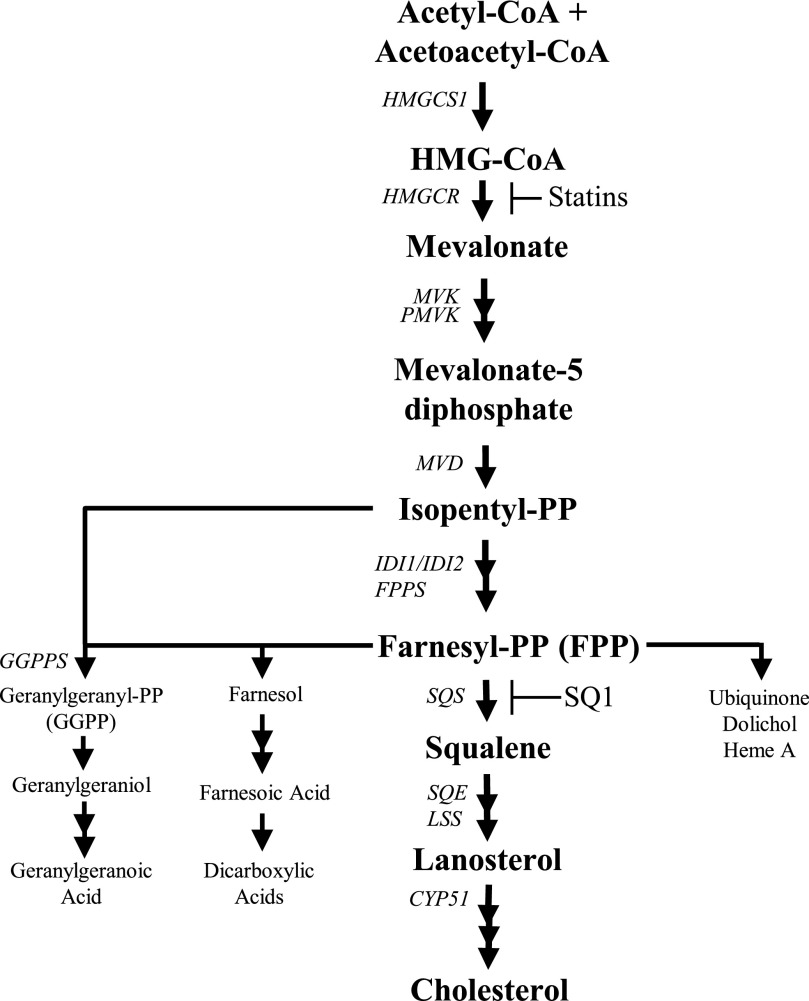
Fig. 1.
Overview of the cholesterol biosynthetic pathway. Cholesterol synthesis inhibitors and their enzymatic targets are indicated. CYP51, lanosterol 14α-demethylase; FPPS, farnesyl pyrophosphate synthase; GGPP, geranylgeranyl pyrophosphate; GGPPS, geranylgeranyl pyrophosphate synthase; HMGCS1, 3-hydroxy-3-methylglutaryl coenzyme A (CoA) synthase 1; IDI1/2, isopentenyl-diphosphate delta isomerase 1 and 2; LSS, lanosterol synthase; MVD, mevalonate diphosphate decarboxylase; MVK, mevalonate kinase; PMVK, phosphomevalonate kinase; SQS, squalene synthase; SQE, squalene epoxidase.
Squalene synthase inhibitors [SSIs such as squalestatin 1 (SQ1)] are a class of anticholesterol drugs that block the first committed step in sterol synthesis, in which farnesyl pyrophosphate (FPP) is converted to squalene (Fig. 1) (Do et al., 2009). Compared with inhibitors of HMGCR, SSIs preserve the synthesis of nonsterol isoprenoids that are used in the posttranslational modification of proteins, in glycoprotein synthesis, and for components of aerobic metabolism (Krag, 1998; Edwards and Ericsson, 1999; Dallner and Sindelar, 2000; McTaggart, 2006). Additionally, FPP can be dephosphorylated to farnesol and then further metabolized to produce farnesoic acid and a number of chain-shortened dicarboxylic acids (DCAs), which are detectable in urine (Bostedor et al., 1997; Vaidya et al., 1998). Several of these isoprenoid compounds function as signaling molecules that can modulate the activities of nuclear receptors, including peroxisome proliferator-activated receptors α and γ (PPARα and PPARγ), constitutive androstane receptor (CAR), and farnesoid X receptor (FXR) (Forman et al., 1995; Kocarek and Mercer-Haines, 2002; Takahashi et al., 2002; Rondini et al., 2016). Therefore, compared with statins, SSIs likely have distinctive effects on hepatic metabolism.
Similar to statins, treatment with SSIs has been shown to lower non–high-density lipoprotein cholesterol in preclinical models (Hiyoshi et al., 2000, 2001; Nishimoto et al., 2003). SSIs also markedly reduce serum triglycerides through an LDL receptor-independent mechanism (Hiyoshi et al., 2001). The effect on triglyceride levels is thought to be mediated through farnesol and/or a farnesol metabolite (Hiyoshi et al., 2003), as farnesol treatment also decreased triglyceride biosynthesis and reduced hepatic steatosis, in part through PPARα activation (Duncan and Archer, 2008; Goto et al., 2011a). Elevated triglycerides are a known risk factor for CVD and are commonly associated with metabolic dyslipidemia (Eckel et al., 2005; Boullart et al., 2012). Although the SSI lapaquistat (TAK-475) showed promising effects on serum lipid profiles in primates (Nishimoto et al., 2003), its development was terminated during phase III clinical trials because of safety concerns and the lack of commercial viability at lower doses (Stein et al., 2011). Nonetheless, there is still significant interest in SSIs and the isoprenoid pathway with respect to hepatic lipid metabolism (Goto et al., 2011a; Nagashima et al., 2015) and as a potential therapeutic target for a variety of other conditions (Shang et al., 2014; Yang et al., 2014; Saito et al., 2015; Healey et al., 2016).
The liver is central to cholesterol metabolism and is a major target for hypolipidemic drugs. Previous studies have compared the effect of different statins on hepatocellular gene expression (Hafner et al., 2011; Leszczynska et al., 2011; Schroder et al., 2011). However, a detailed evaluation of SSI treatment on global gene responses has not yet been performed, which is important for understanding both the beneficial as well as the potentially adverse effects of isoprenoids on hepatocellular physiology. Thus, our current investigation used microarrays to evaluate the effects of SQ1 on orthologous gene expression changes in primary cultured rodent hepatocytes. Both mouse and rat hepatocytes were included to focus our analysis on conserved responses that are likely indicative of isoprenoid signaling mechanisms across species. Additionally, because SSIs also reduce cholesterol biosynthesis, the effects of SQ1 were compared with those of the HMGCR inhibitor pravastatin (Prav). Prav was selected because, unlike other statins, it is not extensively metabolized (Hatanaka, 2000), and it does not produce off-target effects such as activation of xenobiotic-sensing receptor(s) (Kocarek and Reddy, 1996; Kocarek et al., 2002). Therefore, Prav was used to distinguish the gene expression changes that were due to hepatic sterol depletion (Prav and SQ1) from those due to endogenous isoprenoid accumulation (SQ1). Treatment-induced changes were further validated using more sensitive methods, and informative signaling pathways are discussed.
Materials and Methods
Materials.
SQ1 was generously supplied from GlaxoSmithKline (Research Triangle Park, NC) and Prav from Bristol-Myers Squibb (Wallingford, CT). Cell culture medium and reagents were purchased from Invitrogen (Carlsbad, CA), primers from Integrated DNA Technologies (Coralville, IA), PureCol from Advanced Biomatrix (San Diego, CA), and phenobarbital (PB), dimethyl sulfoxide (DMSO), 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP), and ciprofibrate (Cipro) from Sigma-Aldrich (St. Louis, MO). Matrigel was obtained from Corning (Tewksbury, MA), and the cDNA synthesis kit and SYBR green master mix from Life Technologies (Carlsbad, CA). Other sources of reagents are provided throughout the article.
Primary Culture of Mouse and Rat Hepatocytes.
Animal procedures were conducted in accordance with the regulatory guidelines of the Division of Laboratory Animal Resources at Wayne State University (Detroit, MI). Adult male and female C57BL/6 mouse breeder pairs, generously donated by Dr. Masahiko Negishi (National Institute of Environmental Health Sciences, Research Triangle Park, NC), were used for colony generation. Male offspring (7 to 8 weeks of age) produced from this colony were then used for primary hepatocyte isolation. Adult male Sprague-Dawley rats (175–200 g) were purchased from Harlan Sprague Dawley (Indianapolis, IN); upon their receipt, they were allowed 1 week to acclimatize before use. All animals were housed in an Association for Assessment and Accreditation of Laboratory Animal Care–approved animal facility in temperature-controlled (23 ± 2°C) and humidity-controlled rooms with a 12-hour light/dark cycle and were allowed free access to chow and house-distilled water.
Primary hepatocytes were isolated with a two-step collagen perfusion using methods described in detail elsewhere (Kocarek and Reddy, 1996; Wu et al., 2001). Immediately after isolation, 1.2 million (mouse) or 1.6 million (rat) viable hepatocytes per well were plated onto collagen-coated six-well plates. Hepatocyte cultures were maintained in Williams’ E medium supplemented with 0.25 U/ml insulin, 0.1 μM triamcinolone acetonide, 100 U/ml penicillin, and 100 μg/ml streptomycin. Twenty-four hours after plating, the medium was replaced with fresh medium containing Matrigel (1 to 50 dilution). The next day, cholesterol synthesis inhibitors (0.1 μM SQ1 or 30 μM Prav) or nuclear receptor activators (100 μM PB, 0.25 μM TCPOBOP, or 100 μM Cipro) were added to the culture medium from concentrated stock solutions dissolved in either water (SQ1, Prav, PB) or DMSO (TCPOBOP, Cipro). Culture medium containing drugs was replaced once after 24 hours. Forty-eight hours after the initial treatment, total RNA was extracted from the hepatocytes and processed for microarray or quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis as will be described in more detail.
Microarrays of Mouse and Rat Hepatocytes Treated with SQ1 or Prav.
Two independent preparations of primary cultured mouse or rat hepatocytes were treated with either medium alone (i.e., untreated control) or medium containing SQ1 or Prav (eight wells/treatment/hepatocyte preparation; 16 wells/treatment/species total). Forty-eight hours after the initial drug treatment, total RNA was extracted from the hepatocytes and column purified with RNeasy columns (Qiagen, Valencia, CA). After isolation, RNA quality was assessed using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), and only high-quality RNA (RNA integrity number >9) was used in the subsequent steps.
Total RNA was pooled from two wells/treatment group, providing eight RNA samples/treatment group/species for microarray analysis. The RNA samples (500 ng) with Agilent spike-in controls were amplified using the TargetAMP 1-Round Aminoallyl-aRNA Amplification Kit 101 according to manufacturers’ instructions (Epicentre, Madison, WI). Five µg of each aminoallyl-aRNA sample was then fluorescently labeled with Alexa 647 or Alexa 555. Microarrays were performed using Mouse GE 4x44K v2 and Rat GE 4x44K v3 whole genome arrays (Agilent), with a basic two-color hybridization design to compare relative gene expression levels in SQ1 versus untreated (control) and Prav versus control samples.
To minimize the potential effect of transcript-dependent dye bias on fluorescent intensity signals, for each set of eight arrays we hybridized four of the arrays with Alexa 647-labeled RNA from control samples and Alexa 555-labeled RNA from drug-treated samples, while in the other four arrays the dye orientation was reversed. Microarrays were scanned using the Agilent dual-laser DNA microarray scanner (Model G2565AA), and image analysis was performed using Agilent Feature Extraction software. Outlier features having aberrant image characteristics were flagged and excluded from subsequent analyses.
The data were further processed and analyzed as described in detail within the Statistical Analyses section. All microarray data have been deposited and are available on the GEO database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81659).
Quantitative Reverse Transcription Polymerase Chain Reaction.
Forty-eight hours after the initial drug treatment, total RNA was extracted from cells, and cDNA was synthesized from total RNA (2 µg) using the High Capacity Reverse Transcription cDNA Kit (Life Technologies). Quantitative determination of gene expression was performed using the StepOne Plus Real Time PCR System (Applied Biosystems, Foster City, CA), gene-specific primers (75–250 nM), and SYBR green master mix. Primers were designed using the Primer-BLAST program (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Ye et al., 2012). A complete listing of the mouse and rat primer pairs used for gene expression analysis is available in Supplemental Tables 1 and 2.
For each assay, a commercially designed primer set to detect mouse (P/N Mm.PT.39a.22214839) or rat (P/N Rn.PT.51.24118050) TATA box binding protein was purchased from Integrated DNA Technologies and used as the endogenous control. The polymerase chain reaction cycling conditions have been described in detail elsewhere (Rondini et al., 2016). All assays were performed in duplicate (n = 3–6/treatment), and the relative fold changes were then quantitated using the comparative cycle threshold (ΔΔCT) method.
Statistical Analyses.
The statistical analysis for qRT-PCR was performed using SigmaStat Statistical Software (Version 3.5; Point Richmond, CA). Normalized expression values were analyzed using a one-way or two-way analysis of variance as appropriate; when statistically significant differences were detected (P < 0.05), group comparisons were made using the Student-Newman-Keuls test. The results are presented as mean ± S.E.M.
For the microarray analysis, raw fluorescence intensities were normalized using the Lowess method in Feature Extraction Software (Agilent). Normalized values were then imported into Genespring version 12.6.1 (Agilent) for further filtering and statistical analyses. Differentially expressed transcripts (SQ1 versus untreated controls, Prav versus control, SQ1 versus Prav) were detected using a t test against zero and the Benjamini and Hochberg false discovery rate post-test to correct for multiple comparisons. Results were filtered to exclude genes with a false discovery rate of >5% and those displaying less than a ±1.5-fold change between treatments. Differentially expressed transcripts were then exported into Excel and Access (Microsoft, Redmond, WA) for further sorting and processing. When present, the values from replicate transcripts were averaged.
For interspecies comparisons, orthologous genes were retrieved using Homologene (http://www.ncbi.nlm.nih.gov/homologene) and the rat genome databases (http://rgd.mcw.edu/). The PANTHER database (http://www.pantherdb.org/) was used to classify transcripts into biologic categories and the Web-based Gene Set Analysis Toolkit (http://bioinfo.vanderbilt.edu/webgestalt/option.php) for Pathway enrichment analysis. Pathway enrichment P values were calculated using a hypergeometric distribution test followed by the Benjamini and Hochberg post-test. Heat maps displaying differential gene changes were generated using GENE-E software version 3.0.204 (http://www.broadinstitute.org/cancer/software/GENE-E/index.html), and the data are displayed on a log2 scale.
Results
Profile of Global Gene Expression Changes Significantly Altered in Primary Cultured Mouse and Rat Hepatocytes After Treatment with SQ1 or Prav.
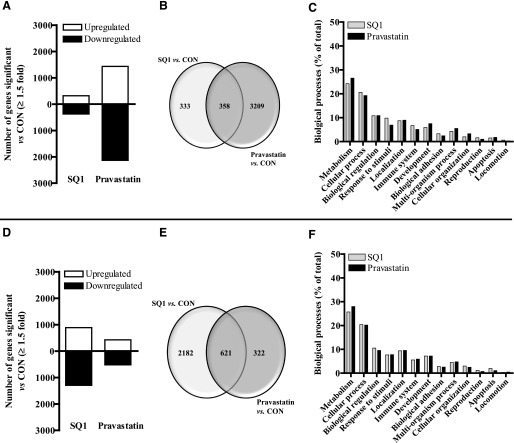
The number and distribution of RNA transcripts significantly altered by either SQ1 or Prav in primary cultured mouse hepatocytes are presented in Fig. 2. Among the 39,430 genes represented on the mouse Agilent microarray slides, 691 transcripts (318 up-regulated and 373 down-regulated) were significantly affected by SQ1 and 3567 transcripts by Prav (1433 up-regulated, 2134 down-regulated) when compared with untreated, control hepatocytes (Fig. 2A). Among these, 333 genes (166 up-regulated, 167 down-regulated) were unique to SQ1 treatment, 3209 genes (1303 up, 1906 down) were unique to Prav, and 358 genes were significantly affected by both treatments (111 co-up-regulated, 188 co-down-regulated, 59 differentially regulated; ≥1.5-fold, P < 0.05; Fig. 2B). As shown in Fig. 2C, a large portion of the known genes that were significantly affected by either treatment were associated with metabolic (lipid, protein, nucleobase-containing) and cellular processes (cell communication, movement, and cell cycle), representing ∼45% of the total gene expression changes.
Fig. 2.
Characterization of gene expression changes in primary cultured mouse (A–C) and rat (D–F) hepatocytes after treatment with SQ1 or pravastatin identified using microarrays. Primary mouse and rat hepatocytes were freshly isolated and plated onto six-well plates as described in the Materials and Methods. Forty-eight hours after initial plating, hepatocytes were treated with Williams’ E medium either alone (control, CON) or containing SQ1 (0.1 μM) or pravastatin (30 μM). The culture medium was replaced once after 24 hours. Forty-eight hours after the initial drug treatment, total RNA was isolated from the hepatocytes, and fluorescent-labeled RNA was synthesized and hybridized onto Agilent whole-genome microarrays as described in Materials and Methods (n = 8/treatment group/species). Significant differences in the normalized fluorescence intensities (≥1.5-fold) among the different drug treatments (SQ1 versus CON, pravastatin versus CON) were detected using GeneSpring. (A, D) The total number of transcripts significantly affected by either SQ1 or pravastatin in mouse (A) and rat (D) hepatocytes. (B, E) Venn diagrams displaying the number of common and unique transcripts significantly affected among the different drug treatments in mouse (B) and rat (E) hepatocytes. (C, F) Functional distribution of known gene changes from mouse (C) and rat (F) hepatocytes grouped into biologic categories. Data are presented as percentage of total gene changes.
RNA transcripts significantly affected in primary cultured rat hepatocytes are presented in Fig. 2D. Among the 30,003 Entrez gene probes represented on the rat arrays, 2803 transcripts (1196 up-regulated and 1607 down-regulated) were significantly affected by SQ1 and 943 transcripts by Prav (425 up-regulated, 518 down-regulated) compared with the untreated controls (Fig. 2D). SQ1 treatment uniquely affected 2182 genes (888 up-regulated, 1294 down-regulated), Prav affected 322 (126 up-regulated, 196 down-regulated), and 621 genes were commonly affected by both cholesterol synthesis inhibitors (294 co-up-regulated, 308 co-down-regulated, 19 differentially regulated; Fig. 2E). Similar to the effects observed in mouse hepatocytes, a large share of the genes affected by either treatment were associated with metabolic and cellular processes, representing ∼45% of total known gene changes (Fig. 2F).
Comparative Ortholog Analysis to Identify Conserved Common and Drug-Specific Gene Expression Changes in Primary Mouse and Rat Hepatocytes.
Species-specific responses to pharmacologic and toxicologic agents are generally accepted and have been well reported (Forgacs et al., 2013). To understand the key biologic pathways affected by isoprenoid accumulation that are conserved across species and that therefore likely represent core functions of isoprenoids on hepatocellular physiology, we further processed lists of regulated genes among species to identify treatment-specific orthologs, as depicted in Fig. 3A. To identify orthologs uniquely affected by SQ1 treatment, we evaluated 313 of the SQ1-regulated mouse genes with known orthologs (264 of the 333 genes originally identified as unique to SQ1 treatment [Fig. 2B] and 49 differentially regulated transcripts from the 358 genes originally identified as regulated by both SQ1 and Prav [Fig. 2B]) and 1561 SQ1-regulated rat genes with known orthologs (1552 of the 2182 genes originally identified as unique to SQ1 and 9 differentially regulated transcripts from the 621 genes originally identified to be regulated by both SQ1 and Prav [Fig. 2E]).
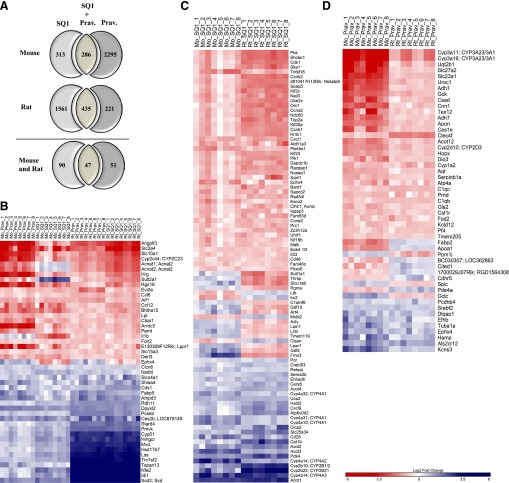
Fig. 3.
Orthologous genes affected by both SQ1 and Prav treatment in primary cultured mouse and rat hepatocytes identified using microarrays. Treatment-specific orthologs were retrieved using Homologene and the rat genome database as described in Materials and Methods. (A) Venn diagram displaying the number of putative orthologs that were significantly affected by either SQ1 alone, both SQ1 and Prav, or Prav alone in mouse (upper panel) and rat hepatocytes (middle panel) and used to screen for conserved gene changes. The lower panel displays the number of treatment-specific orthologs that were commonly and uniquely affected in both species. (B–D) Heat maps displaying the normalized (log2) expression values of orthologs (B) commonly affected by both SQ1 and Prav, (C) unique to SQ1-treated hepatocytes, and (D) unique to Prav-treated hepatocytes (n = 8/treatment group/species). When ortholog names differed between species, the mouse name is provided first followed by the rat name. Blue-colored bars indicate up-regulated genes, and red-colored bars indicate down-regulated genes.
To identify those uniquely affected by Prav treatment, we evaluated 2295 Prav-regulated mouse genes with known orthologs (2246 of the 3209 genes originally identified as unique to Prav [Fig. 2B] and 49 genes differentially regulated by SQ1 and Prav) and 221 Prav-regulated rat genes (212 of the 322 genes originally identified as unique to Prav in Fig. 2E, and nine differentially expressed transcripts). Differentially expressed transcripts (SQ1 versus control compared with Prav versus control) are defined as genes significantly affected by both SQ1 and Prav and thus originally included in the intersection (Fig. 2, B and E), but displaying the opposite direction of change. Orthologs significantly coregulated by both SQ1 and Prav treatment (286 genes in mice and 435 genes in rat hepatocytes) were also evaluated to identify conserved, drug-independent gene expression changes responsive to cellular cholesterol depletion.
Orthologous Genes Commonly Affected by Both SQ1 and Prav in Primary Cultured Mouse and Rat Hepatocytes.
As shown in Fig. 3, A and B, we identified 47 orthologous genes commonly affected by both SQ1 and Prav in mouse and rat primary hepatocytes. Among these, 42 genes showed similar responses in both species with 23 genes coinduced by cholesterol synthesis inhibition and 19 genes corepressed (Fig. 3B). However, the overall magnitude of the coinduced genes was species dependent, with rats generally having responses of up to ∼10-fold greater than those observed in mice for the most highly induced genes.
Among the orthologs coinduced were several involved in cholesterol biosynthesis (Cyp51, Hmgcr, Mvd, Lss, Idi1, Pmvk, Nsdhl, Tm7sf2, and Hsd17b7), whereas those corepressed were associated with chemokine signaling (Ccl6, Ccl12), adipogenesis (Lpl, Slc2a4), and sodium/bile acid transport (Slc10a1). Other coinduced genes include the proprotein convertase subtilisin/kexin type 9 (Pcsk9), which targets the LDL receptor for degradation (Horton et al., 2007); Stard4, a sterol-responsive gene that regulates cholesterol transport between organelles (Rodriguez-Agudo et al., 2008; Calderon-Dominguez et al., 2014); the transcription factor Nfe2; and Scd2 (Scd in rats), which is involved in the synthesis of monounsaturated fatty acids.
Orthologous Genes Uniquely Affected by SQ1 in Primary Cultured Mouse and Rat Hepatocytes.
A heat map displaying orthologs unique to SQ1 treatment compared with untreated controls is displayed in Fig. 3C. Ninety orthologs were found to be unique to SQ1 treatment, with 72 genes (26 coinduced and 46 corepressed; 80% of genes) displaying the same direction of change whereas 18 genes were differentially expressed in mouse when compared with rat hepatocytes.
Previous studies have indicated that treatment with either an SSI or farnesol increased the expression of select PPAR-regulated genes (Takahashi et al., 2002; Goto et al., 2011b).
Consistent with these findings, we found that many of the genes coinduced by SQ1 were strongly associated with lipid/fatty acid metabolism and are also identified as PPAR target genes, including Cyp4a10, Cyp4a14 (CYP4A2 and 4A3 in rats), Cyp4a31 and Cyp4a32 (CYP4A1 in rats), Cpt1b, and Ehhadh. Additional classes of genes coinduced included those involved in drug/xenobiotic metabolism (Cyp2b10, Cyp2b23 [CYP2B1/2, and CYP2B21, respectively, in rats]) as well as several acyl-CoA thioesterases (Acot1, Acot2, Acot3, Acot4), which play a role in lipid metabolism by regulating intracellular CoA levels and levels of activated substrates for peroxisomal and mitochondrial fatty acid oxidation (Hunt et al., 2012, 2014) (Fig. 3C). Among the genes uniquely corepressed by SQ1-treated when compared with control hepatocytes were those involved in cell cycle regulation, including several cyclins (Ccna2, Ccnb1, Ccnb2, Ccne2), the cyclin-dependent kinase Cdk1, and genes associated with DNA replication/chromosome segregation (Orc1, Plk1, and Top2a) (Fig. 3C).
Orthologous Genes Uniquely Affected by Prav in Primary Cultured Mouse and Rat Hepatocytes.
A heat map displaying orthologs unique to Prav treatment compared with untreated controls is displayed in Fig. 3D. A total of 51 orthologous genes were identified, among which 41 (9 coinduced and 32 corepressed; 80% of genes) displayed the same direction of change across species. A majority of the conserved genes specific for Prav treatment were corepressed, including several involved in retinol and xenobiotic metabolism (Adh1, Adh7, Cyp1a2, Ugtb1) as well as in metabolic pathways (Ces1e, Uroc1, Adh7, Gck, Gls2, Adh1, Csad). Orthologs that were coinduced include the sterol-sensitive transcription factor sterol regulatory element-binding factor-2 (Srebf2), epoxide hydrolase 4 (Ephx4), the iron-regulatory and type II acute phase protein hepcidin (Hamp), and the amyotrophic lateral sclerosis 2 gene (Als2cr12), which encodes a putative GTPase regulator (Hadano et al., 2001) (Fig. 3D).
Orthologous Genes Differentially Affected by SQ1 Compared with Prav in Primary Cultured Mouse and Rat Hepatocytes.
Obvious species-specific differences in the magnitude of fold changes were observed after treatment with SQ1 or Prav, most notably for cholesterol biosynthetic enzymes (Fig. 3B). To gain further insight into effects that may have been more subtly regulated by isoprenoid depletion (Prav) or accumulation (SQ1) across species but are nonetheless important in isoprenoid signaling and physiology, we additionally compared orthologous expression between treatments (SQ1 versus Prav) using a 1.5-fold cutoff for biologic significance.
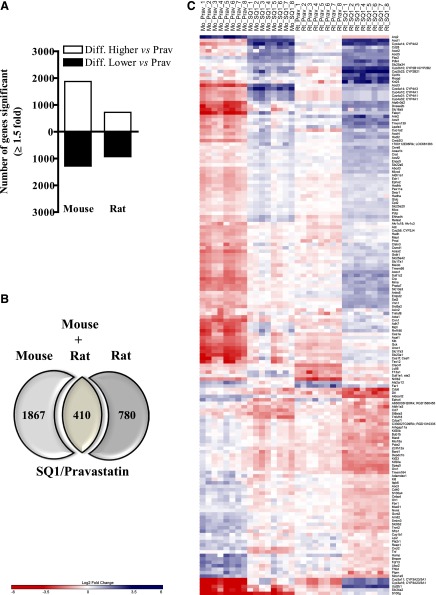
As shown in Fig. 4A, we identified 3154 transcripts (1873 higher, 1281 lower) that were differentially affected in the mouse and 1652 (723 higher, 929 lower) in the rat when comparing SQ1- versus Prav-treated hepatocytes (Fig. 4A). Among these, 410 orthologs were affected in both species (Fig. 4, B and C), whereas 1867 (mouse) and 780 (rat) orthologs exhibited species-specific expression patterns (Fig. 4B). Within the 410 orthologs that were commonly affected, 162 orthologs displayed similar expression patterns in both species (104 genes that were differentially higher and 58 differentially lower in SQ1-treated relative to Prav-treated hepatocytes) (Fig. 4, B and C).
Fig. 4.
Hepatic expression of orthologs differentially affected by SQ1 compared with Prav in primary cultured mouse and rat hepatocytes identified using microarrays. Treatment-specific orthologs were retrieved using Homologene and the rat genome database as described in Materials and Methods. (A) The total number of transcripts differentially affected in SQ1-treated compared with Prav-treated mouse and rat hepatocytes. (B) Venn diagram displaying the number of orthologs differentially affected by SQ1 compared with Prav treatment (≥1.5-fold) in primary cultured mouse and rat primary hepatocytes. (C) A heat map displaying the normalized (log2) expression values of orthologs differentially expressed in SQ1 compared with Prav-treated hepatocytes isolated from mice and rats (n = 8/treatment group/species). When ortholog names differed between species, the mouse name is provided first followed by the rat name. Blue-colored bars indicate up-regulated genes, and red-colored bars indicate down-regulated genes.
Orthologs that were differentially lower in SQ1-treated hepatocytes included several genes involved in cell cycle regulation (Orc1, Cdc6, Pole2, Bub1b) as well as cytokine signaling (Tnf, Cd40, Ccl7, Tnfsf18, Kitl, Cxcl2) (Fig. 4C). Nr0b2, the prototypical target gene for FXR activation was also differentially regulated by the treatments, being strongly repressed by Prav and more moderately reduced by SQ1.
As observed when comparing SQ1 to controls, several genes that were differentially higher in SQ1-treated compared with Prav-treated hepatocytes included a cluster of PPAR-regulated genes (Cyp4a10 [CYP4A1 in rats], Fabp1, Acox1, Cpt1b, Cyp4a31, and 4a32 [CYP4A1 in rats], Cyp4a14 [CYP4A2 and 4A3 in rats], Acaa1b, Acsl1, Cpt2, and Ehhadh), and genes involved in fatty acid β-oxidation (Cpt1b, Acat1, Ehhadh, Ech1, Hadh, Hadha, Hadhb, Decr1, Crat, Acsl1, Slc25a20, Cpt2, Abcd3), fatty acid ω-oxidation (Aldh1a1, Adh7, Cyp1a2), drug and xenobiotic metabolism (Cyp2b10 [CYP2B1 in rats]), Cyp2b23 [CYP2B21 in rats], Cyp3a11 [CYP3A23/3A1 in rats], Cyp3a16 [CYP3A23/3A1 in rats], Adh7, Cyp1a2, Ugt2b1, Gstk1]), and various ACOT enzymes such as Acot1 (cytosolic), Acot2 (mitochondrial), and Acot3, 4, and 5 (peroxisomal).
Validation of Select Gene Expression Changes by qRT-PCR.
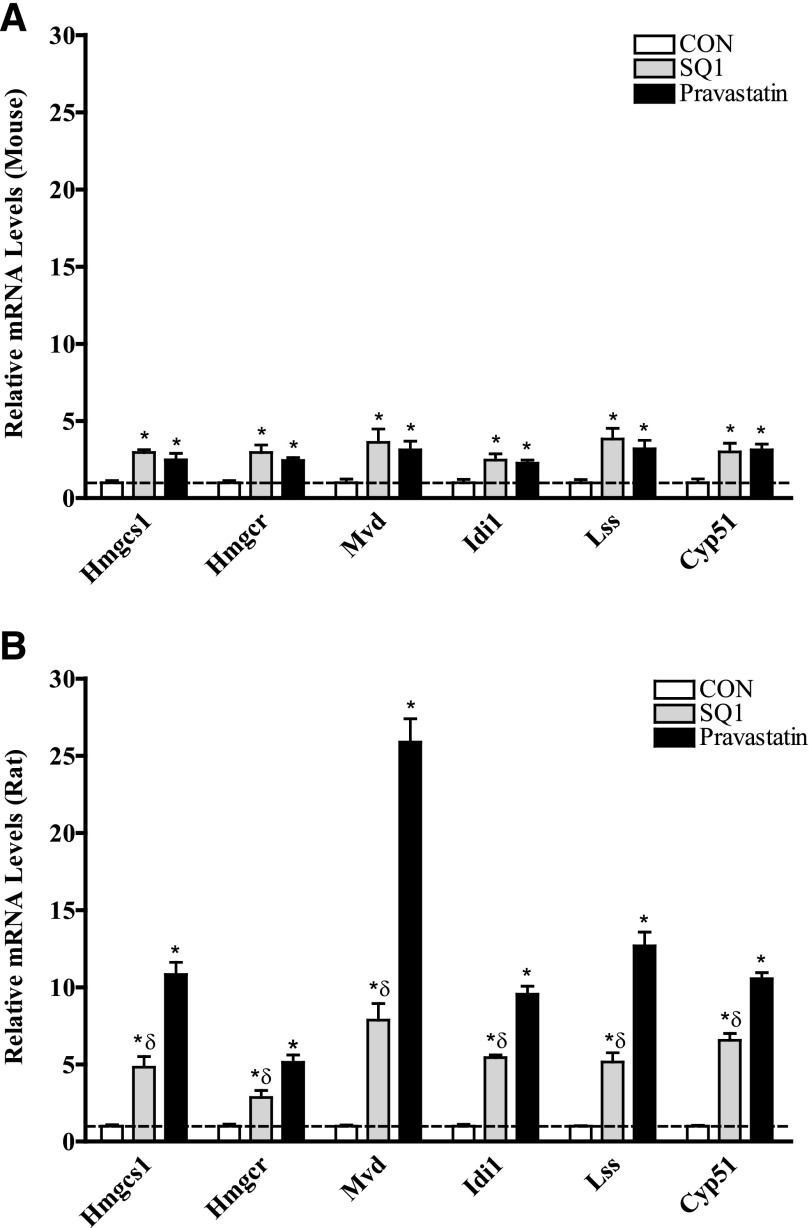
The relative expression of several genes involved in cholesterogenesis and lipid metabolism in response to SQ1 and Prav treatment was further validated by qRT-PCR, and the results are presented in Figs. 5 and 6. In general, the expression changes observed using microarrays were in accordance with the changes measured by qRT-PCR. As shown in Fig. 5, A and B, treatment with either drug, as expected, resulted in transcriptional up-regulation of several cholesterol biosynthetic enzymes including Hmgcs1, Hmgcr, Mvd, Idi1, Lss, and Cyp51; however, the relative fold changes were generally lower in mouse hepatocytes (Fig. 5A) than those observed in rat hepatocytes (Fig. 5B), reflecting species differences in hepatocellular responses to cholesterol depletion.
Fig. 5.
Effect of SQ1 and pravastatin on the mRNA expression of cholesterol biosynthetic enzymes in primary cultured mouse (A) and rat (B) hepatocytes using qRT-PCR. Primary mouse (A) and rat (B) hepatocytes were freshly isolated, plated onto six-well plates, and 48 hours later were treated with Williams’ E medium either alone (control, CON) or containing SQ1 (0.1 μM) or Prav (30 μM). The medium was replaced once after 24 hours, and 48 hours after the initial treatment, cDNA was synthesized from total RNA as described in Material and Methods. Relative changes in mRNA expression levels were quantified using qRT-PCR. Each bar represents the normalized values (mean ± S.E.M.) from five to six hepatocyte preparations (two combined wells/treatment group/preparation). *Statistically significant compared with untreated (CON) hepatocytes (P < 0.05). δStatistically significant compared with pravastatin-treated hepatocytes (P < 0.05).
Fig. 6.
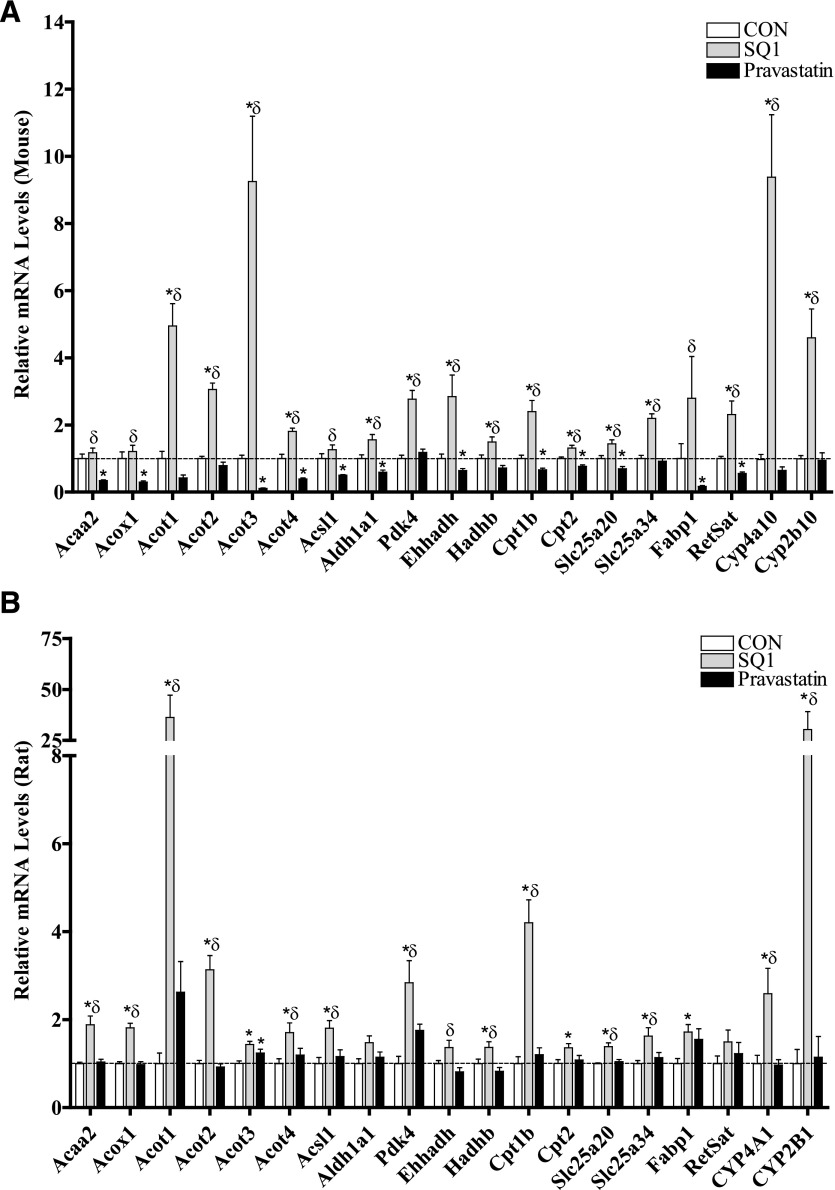
Validation of select regulated genes identified through microarrays by qRT-PCR in primary cultured mouse (A) and rat (B) hepatocytes. Primary mouse (A) and rat (B) hepatocytes were freshly isolated and plated onto six-well plates, and 48 hours later were treated with Williams’ E medium either alone (control, CON) or containing SQ1 (0.1 μM) or Prav (30 μM). The medium was replaced once after 24 hours, and 48 hours after the initial treatment total RNA was isolated and cDNA synthesized as described in Materials and Methods. Relative changes in mRNA expression levels were determined using qRT-PCR. Each bar represents the normalized values (mean ± S.E.M.) from five to six hepatocyte preparations (two combined wells/treatment group/preparation). *Statistically significant compared with untreated (CON) hepatocytes (P < 0.05). δStatistically significant compared with Prav-treated hepatocytes (P < 0.05).
Select genes differentially regulated by SQ1 and Prav were also analyzed by qRT-PCR (Fig. 6, A and B). Many of the differentially regulated transcripts were identified as putative PPAR-target or CAR-target genes with identified roles in lipid and xenobiotic metabolism, respectively, so we focused on orthologs within this cluster for further confirmation (Fig. 6A, 6B). As shown in Fig. 6, A and B, the prototypical PPAR and CAR target genes, Cyp4a10 (CYP4A1 in rats) and Cyp2b10 (CYP2B1 in rats), respectively, were strongly induced (>3-fold) by SQ1 across species, although CAR-mediated responses tended to be higher in rat hepatocytes whereas PPAR-mediated responses were greater in mouse hepatocytes (Fig. 6, A and B). Genes associated with peroxisomal (Acox1, Acot3, Acot4, Ehhadh) and mitochondrial β-oxidation (Acaa2, Acot2, Hadhb, Cpt1b, Cpt2, Slc25a20) were also generally expressed at higher levels in SQ1-treated compared with Prav-treated hepatocytes. However, in the mouse, endogenous isoprenoids appeared to influence basal expression of these genes as Prav treatment down-regulated their mRNA levels, whereas SQ1 either did not change or increased the mRNA levels compared with untreated hepatocytes (Fig. 6A). In comparison, in rat hepatocytes the expression levels were much less affected by Prav and generally induced after SQ1 treatment (Fig. 6B).
Other genes differentially regulated by SQ1 included the mitochondrial solute transporter Slc25a34, the pyruvate dehydrogenase inhibitor Pdk4, liver fatty acid binding protein (Fabp1), Aldh1a1, an aldehyde dehydrogenase enzyme involved in ω-oxidation and retinol metabolism, as well as Acsl1 and Acot1, which are enzymes involved in regulating activation and deactivation of fatty acids, respectively (Fig. 6, A and B).
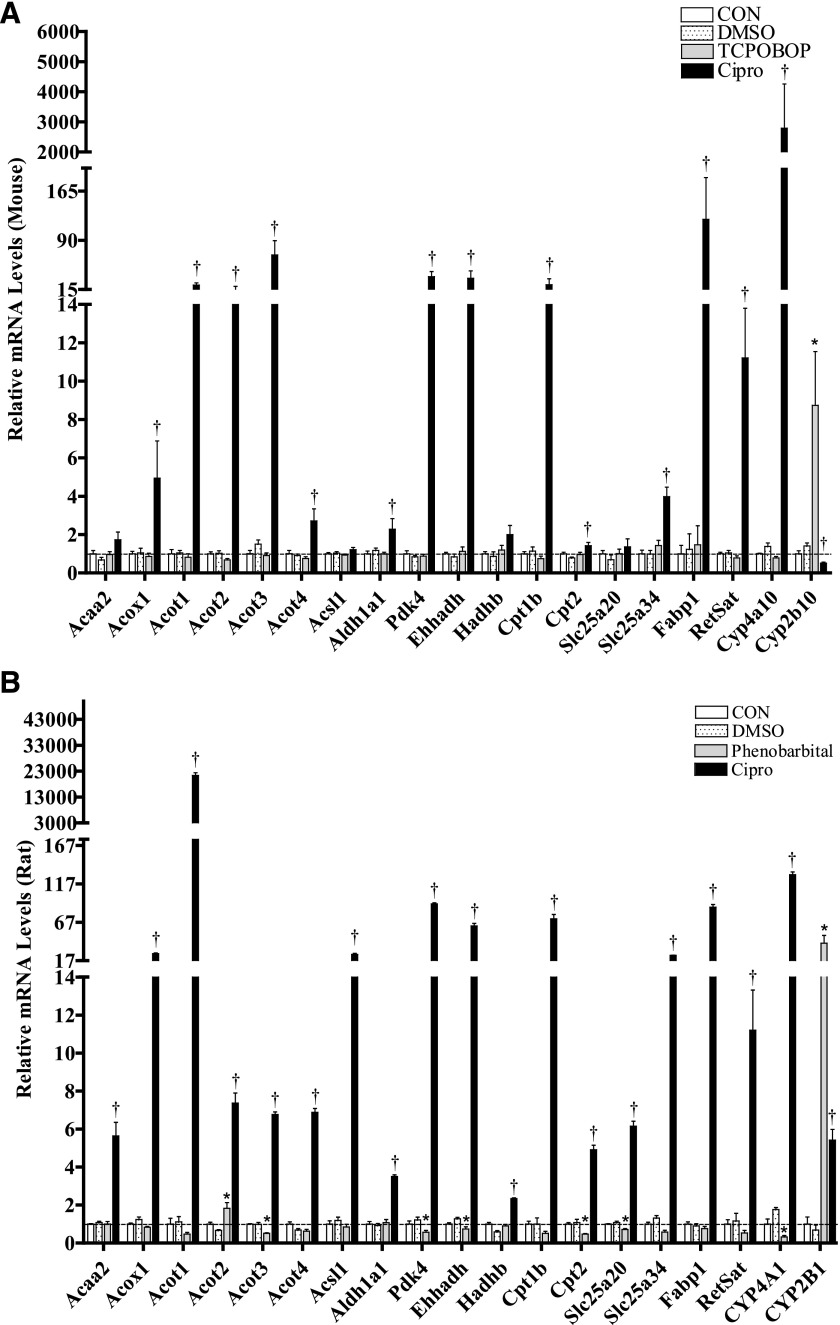
As shown in Fig. 7, A and B, most of the metabolic enzymes that were surveyed were also inducible by the prototypical PPARα agonist Cipro in both species, whereas Cipro-mediated induction of the mRNA for Acaa2, Acsl1, Hadhb, and Slc25a20 was significant in rats only. Comparably, despite a suggested role for CAR in lipid metabolism in rodents, only CYP2B expression was induced by the prototypical CAR agonists TCPOBOP and PB in mouse and rat hepatocytes, respectively (Fig. 7, A and B). A species-dependent interaction was also observed among treatments with respect to gene regulation by the nuclear receptors CAR and PPAR. These changes may reflect species-dependent regulation of CAR responses after PPAR activation as discussed elsewhere (Guo et al., 2006; Wieneke et al., 2007; Saito et al., 2010; Rondini et al., 2016). In mouse hepatocytes, PPAR activation by the prototypical agonist Cipro decreased expression of the CAR target gene Cyp2b10 by ∼50%, whereas in the rat hepatocytes, PPAR activation increased CYP2B1 expression by ∼5-fold. CAR activation in rat hepatocytes also partially suppressed expression of the PPAR target genes Cyp4a1, Pdk4, Ehhadh, Cpt2, and Slc25a20, but this effect was not observed in mouse hepatocytes (Fig. 7, A and B).
Fig. 7.
Effects of nuclear receptor activators on the mRNA levels of select target genes in primary cultured mouse (A) and rat (B) hepatocytes. Primary mouse (A) and rat (B) hepatocytes were freshly isolated, plated onto six-well plates, and after 48 hours treated with Williams’ E medium either alone (CON) or containing DMSO (0.1%), the PPAR-activator, Cipro (100 μM), or one of the CAR activators, TCPOBOP (0.25 µM, mouse) or PB (100 μM, rats). Medium was replaced once after 24 hours, and 48 hours after the initial treatment, total RNA was extracted from hepatocytes and used to synthesize cDNA as described in the Materials and Methods. Relative changes in mRNA levels were determined using qRT-PCR. Each bar represents the normalized values (mean ± S.E.M.) from three to four hepatocyte preparations (two combined wells/treatment group/preparation). *Statistically significant compared with untreated (CON) hepatocytes (P < 0.05). †Statistically significant compared with DMSO-treated hepatocytes (P < 0.05).
Discussion
The current investigation was designed to elucidate the effects of endogenous isoprenoids on gene expression in primary mouse and rat hepatocytes. Using microarrays, we compared orthologous gene expression produced by two cholesterol synthesis inhibitors: SQ1, which causes isoprenoid accumulation, and Prav, which blocks isoprenoid production. Although not the primary focus of this study, orthologs commonly affected by both cholesterol synthesis inhibitors were also evaluated to differentiate the effects of sterol depletion from those due to isoprenoid accumulation.
Both statins and SSIs reduce non–high-density lipoprotein cholesterol by inhibiting de novo cholesterol biosynthesis, leading to a compensatory up-regulation of LDL receptors on the surface of the hepatocytes and enhanced clearance of LDL-cholesterol (Bilheimer et al., 1983; Nishimoto et al., 2003). This transcriptional pathway is regulated through activation of the sterol regulatory element-binding proteins (SREBPs) (Horton et al., 2002). Under reduced sterol conditions, inactive SREBP precursors are transported from the endoplasmic reticulum to the Golgi apparatus for proteolytic processing, resulting in nuclear translocation and activation of responsive genes (Goldstein and Brown, 2015). In the current study, many of the orthologs coinduced by SQ1 and Prav have been previously identified as SREBP2 targets (Horton et al., 2003) and are consistent with changes identified in tissues of SREBP2-transgenic mice (Ma et al., 2014). For example, the most highly induced class of genes was associated with cholesterol and unsaturated fatty acid biosynthesis. The level of up-regulation was similar between drugs, indicating comparable effects on cholesterol inhibition. However, the most striking difference was in the magnitude of change across species. In general, the fold changes were several-fold higher in rat than in mouse hepatocytes. The basal hepatic cholesterol synthesis rate is reportedly 4 to 12 times higher in rats compared with other species (Spady and Dietschy, 1983), suggesting that this difference may influence the magnitude of SREBP2-mediated responses under conditions of reduced sterol synthesis.
SSIs markedly reduce triglyceride biosynthesis, which is independent from effects on LDL cholesterol (Hiyoshi et al., 2001; Amano et al., 2003) and is dependent on metabolism through the farnesol pathway (Hiyoshi et al., 2003). Isoprenoids are known to modulate nuclear receptor signaling pathways, including PPAR, CAR, and FXR (Forman et al., 1995; O’Brien et al., 1996; Kocarek and Mercer-Haines, 2002; Takahashi et al., 2002; Goto et al., 2011a), which is suggested to partially account for some of these effects (Goto et al., 2011a). In the current study, many of the orthologs uniquely affected by SQ1 treatment were associated with fatty acid metabolism and are also transcriptional targets of PPARα. Therefore, at least some of these gene expression changes are likely attributable to isoprenoid-mediated PPAR activation. The CAR-target gene CYP2B was also induced, although more strongly in rat than in mouse hepatocytes. Although CAR is suggested to influence hepatocellular lipid metabolism (Dong et al., 2009; Goto et al., 2011a), the panel of fatty acid-metabolizing enzymes altered by isoprenoids in this study did not appear to be strongly regulated through CAR activation.
The metabolic route for FPP metabolism following squalene synthase inhibition is proposed to occur through the farnesol–farnesoic acid–DCA pathway described by Gonzalez-Pacanowska et al. (1988), although an alternative pathway has been suggested (DeBarber et al., 2004). Farnesol-derived DCAs are then partially catabolized from the ω-carbon, producing a family of C12 and C10 DCAs that can be detected in the urine (Bostedor et al., 1997; Vaidya et al., 1998). The enzymatic steps involved in this pathway have not been fully elucidated; however, based on the gene expression changes observed in this study, they may involve components of microsomal, peroxisomal, and mitochondrial oxidation. For example, oxidation of farnesol to farnesoic acid was recently shown to be catalyzed by alcohol dehydrogenases 1 and 7 followed by a microsomal aldehyde dehydrogenase (Endo et al., 2011), and we found Adh7 to be regulated by SQ1 treatment. Several CYP4A orthologs were also highly induced by SQ1 compared with control or Prav-treated cells. CYP4A enzymes catalyze the ω-hydroxylation of various fatty acid substrates (Okita and Okita, 2001), including the branched fatty acid phytanic acid (Xu et al., 2006), and based on the extent of up-regulation in this study may potentially be involved in catalyzing the ω-hydroxylation of farnesoic acid.
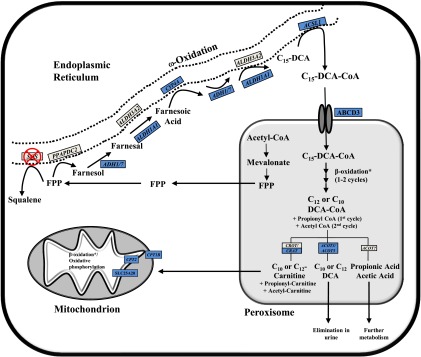
Peroxisomal β-oxidation is involved in chain-length shortening of fatty acids and is the primary pathway for metabolism of bile acids, and very-long-chain and branched-chain fatty acids (Reddy and Hashimoto, 2001). In the current study, several genes associated with peroxisomal and mitochondrial β-oxidation were regulated by SQ1. Additionally, Acot genes were also up-regulated after SQ1 treatment. ACOTs are auxiliary enzymes catalyzing the hydrolysis of fatty acyl-CoAs with suggested roles in regulating intracellular CoA levels and in fatty acid metabolism (Hunt and Alexson, 2008), potentially by generating ligands for PPAR activation (Gachon et al., 2011; Hunt et al., 2014). Many of these changes are consistent with farnesol-derived DCAs undergoing partial oxidation in the peroxisome, producing chain-shortened DCAs that can then either be excreted or further metabolized in the mitochondria (Hiyoshi et al., 2003). Based on substrate specificity, ACOT3 or ACOT5 (ACOT4 in humans) would likely terminate peroxisomal β-oxidation, producing the DCAs identified by Bostedor et al. (1997), although additional steps involved in regulating the extent of oxidation and degree of saturation are not currently known. Building on previous findings and the SQ1-mediated gene expression changes detected in the current study, a proposed model of FPP metabolism is depicted in Fig. 8.
Fig. 8.
Proposed pathway of FPP metabolism after squalene synthase inhibition. Enzymes involved in the cholesterol biosynthetic pathway are localized to different subcellular compartments, with the production of FPP from mevalonate occurring in peroxisomes (Krisans et al., 1994). FPP is used either for the production of squalene by squalene synthase (SQS) or for the synthesis of nonsterol isoprenoids. In the presence of squalene synthase inhibitors (SSI), FPP accumulates and is thought to be metabolized primarily through the farnesol–farnesoic acid–dicarboxylic acid (DCA) pathway (Gonzalez-Pacanowska et al., 1988). Farnesol-derived DCAs are then partially oxidized from the ω-carbon, producing chain-shortened (C12, C10) DCAs that are detectable in urine (Bostedor et al., 1997; Vaidya et al., 1998). The enzymes involved in FPP catabolism have not been fully elucidated; however, based on mRNA gene expression changes observed after SQ1 treatment in the current study, they probably involve components of microsomal, peroxisomal, and mitochondrial oxidation. Shown is a proposed model of FPP catabolism in the presence of squalene synthase inhibitors, which extends on findings from others (Gonzalez-Pacanowska et al., 1988; Bergstrom et al., 1993; Endo et al., 2011; Pant and Kocarek, 2016). FPP can be dephosphorylated by phosphatidic acid phosphatase domain containing 2 (PPAPDC2) to produce farnesol, which is then sequentially oxidized to farnesal and then farnesoic acid. ADH1 and ADH7 were recently identified to catalyze the first oxidation step whereas ALDH3A2 was proposed to catalyze the second (Endo et al., 2011), although ALDH1A1 is another possible candidate based on the expression changes observed in this study. Farnesoic acid is then oxidized from the ω-carbon to produce a C15 dicarboxylic acid. We identified several CYP4A orthologs induced by SQ1, which we propose catalyzes the ω-hydroxylation of farnesoic acid followed by sequential oxidation at the ω-carbon, possibly by the same enzymes involved in farnesol oxidation. The C15-DCA could then be activated by ACSL1 to its CoA derivative for transport into the peroxisome via the dicarboxylic acid transporter ABCD3 (van Roermund et al., 2014), and then undergo one or two rounds of β-oxidation, producing C10 and C12 DCAs as well as propionyl-CoA and acetyl-CoA. The chain-shortened DCAs could either be conjugated with carnitine and exported to the mitochondria for further oxidation or processed by ACOT enzymes, producing free acids that are then eliminated from the cell. For brevity, not all of the enzyme names and/or cofactors are shown for each step. Blue-colored boxes represent orthologous genes that were identified through microarrays to be differentially higher in SQ1-treated compared with untreated controls or to Prav-treated cells (≥1.5-fold). *A number of enzymes involved in peroxisomal and mitochondrial β-oxidation were differentially higher in SQ1-treated compared with control or Prav-treated cells but were excluded for clarity (see Figs. 3 and 4 and text).
The most enriched gene class repressed by SQ1 when compared with either control or Prav-treated hepatocytes was associated with cell cycle regulation and DNA synthesis. These effects were not observed with Prav. Farnesol has been reported to induce apoptosis and cell cycle arrest in a number of different cell lines, with cancer cells more sensitive to the effects of farnesol (Wiseman et al., 2007; Joo and Jetten, 2010). Although SSIs had good hepatic safety in preclinical trials (Nishimoto et al., 2003), Nagashima et al. (2015) recently described a transient liver dysfunction and mild hepatomegaly in hepatic-specific squalene synthase knockout mice (Nagashima et al., 2015). This was associated with elevated serum alanine aminotransferase levels and an increase in apoptotic and proliferative markers at a time point that coincided with higher farnesol production. Farnesol is hypothesized to be rapidly metabolized and difficult to detect even after treatment with SSI in vivo (Bergstrom et al., 1993; Vaidya et al., 1998). However, whether individual variations in the metabolic flux of FPP-derived isoprenoids could partially account for the hepatotoxicity that was observed in some individuals at the highest dose of TAK-475 (Stein et al., 2011) is worthy of future consideration.
Conserved gene expression changes unique to Prav treatment are likely secondary to depletion of the pool of isoprenoids available for protein prenylation and other signaling pathways. For example, Prav uniquely repressed genes involved in xenobiotic and retinol metabolism, some of which are regulated by the nuclear receptors CAR and PXR (Xie et al., 2000; Yoshinari et al., 2010). Adh1 and 7 were also lower in Prav-treated cells, and these have been identified as enzymes involved in metabolizing farnesol and geranylgeraniol to their aldehyde derivatives (Endo et al., 2011). Additionally, some genes associated with fatty acid oxidation tended to be repressed in Prav-treated mouse, but not rat, hepatocytes, suggesting a greater sensitivity of PPAR target genes to isoprenoid depletion in mice. Conversely, Prav treatment increased expression of several orthologs, including a putative GTPase regulator (Als2cr12), iron regulatory peptide (Hamp), potassium channel protein (Kcn), and epoxide hydrolase 4 (Ephx), suggesting that their transcription is normally repressed by endogenous isoprenoids although the mechanism underlying this regulation is currently unknown.
Squalene synthase plays a pivotal role in cholesterol biosynthesis by regulating the flux of mevalonate-derived intermediates used for either sterol synthesis or production of nonsterol isoprenoids (Do et al., 2009). The current study provides insight into some of the conserved effects of SSIs on hepatocellular gene expression. The most profound effect of SQ1 was induction of several PPARα-regulated genes associated with fatty acid oxidation, which may at least partially explain the suppressive effects of SSIs on triglyceride biosynthesis (Ugawa et al., 2000; Hiyoshi et al., 2003). SQ1 treatment also influenced CAR targets involved in drug metabolism and repressed several orthologs associated with cell cycle regulation. Limitations to this study are that SQ1- and Prav-mediated effects on gene expression may not always correspond to changes in protein levels or enzyme activities, and that the drug effects do not identify which specific isoprenoids mediate individual gene changes. Also, although we are unaware of evidence that Prav and SQ1 have any pharmacologic targets other than HMGCR and squalene synthase, respectively, it remains possible that some effects on gene expression could have been produced by an off-target mechanism(s). Additional studies are needed to address the functional role(s) of identified gene expression changes on hepatocellular physiology. Notwithstanding these limitations, as there is continued interest in SSIs and the isoprenoid pathway (Goto et al., 2011a; Ichikawa et al., 2013; Nagashima et al., 2015; Saito et al., 2015), our findings provide a contextual framework for more mechanistic studies.
Supplementary Material
Acknowledgments
The authors thank Mary Garagano for isolating the primary hepatocytes, and Dr. Masahiko Negishi (National Institute of Environmental Health Sciences, Research Triangle Park, NC) for providing the breeding mice that were essential to this study.
Abbreviations
- ACOT
acetyl-coenzyme A thioesterase
- CAR
constitutive androstane receptor (NR1I3)
- Cipro
ciprofibrate
- CoA
coenzyme A
- CVD
cardiovascular disease
- DCA
dicarboxylic acid
- DMSO
dimethylsulfoxide
- FPP
farnesyl pyrophosphate
- FXR
farnesoid X receptor (NR1H4)
- HMGCR
3-hydroxy-3-methylglutaryl-CoA reductase
- LDL
low-density lipoprotein
- PB
phenobarbital
- P450
cytochrome P450
- PPARα
peroxisome proliferator-activated receptor α (NR1C1)
- PPARγ
peroxisome proliferator-activated receptor γ (NR1C3)
- Prav
pravastatin
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
- SQ1
squalestatin 1 (zaragozic acid A)
- SREBP
sterol regulatory element-binding protein
- SSI
squalene synthase inhibitor
- TAK-475
lapaquistat
- TCPOBOP
1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene
Authorship Contributions
Participated in research design: Rondini, Dombkowski, Kocarek.
Conducted experiments: Rondini, Duniec-Dmuchowski, Cukovic.
Performed data analysis: Rondini, Dombkowski.
Wrote or contributed to the writing of the manuscript: Rondini, Kocarek.
Footnotes
This work was supported by the National Institutes of Health National Heart, Lung, and Blood Institute [Grant R01 HL050710] and National Institute of Environmental Health Sciences [Center Grant P30 ES020957]. Dr. Rondini was funded in part through a post-doctoral fellowship awarded through the Office of Vice President of Research (Wayne State University, Detroit, MI).
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Amano Y, Nishimoto T, Tozawa Ri, Ishikawa E, Imura Y, Sugiyama Y. (2003) Lipid-lowering effects of TAK-475, a squalene synthase inhibitor, in animal models of familial hypercholesterolemia. Eur J Pharmacol 466:155–161. [DOI] [PubMed] [Google Scholar]
- Bergstrom JD, Kurtz MM, Rew DJ, Amend AM, Karkas JD, Bostedor RG, Bansal VS, Dufresne C, VanMiddlesworth FL, Hensens OD, et al. (1993) Zaragozic acids: a family of fungal metabolites that are picomolar competitive inhibitors of squalene synthase. Proc Natl Acad Sci USA 90:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilheimer DW, Grundy SM, Brown MS, Goldstein JL. (1983) Mevinolin and colestipol stimulate receptor-mediated clearance of low density lipoprotein from plasma in familial hypercholesterolemia heterozygotes. Proc Natl Acad Sci USA 80:4124–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostedor RG, Karkas JD, Arison BH, Bansal VS, Vaidya S, Germershausen JI, Kurtz MM, Bergstrom JD. (1997) Farnesol-derived dicarboxylic acids in the urine of animals treated with zaragozic acid A or with farnesol. J Biol Chem 272:9197–9203. [DOI] [PubMed] [Google Scholar]
- Boullart AC, de Graaf J, Stalenhoef AF. (2012) Serum triglycerides and risk of cardiovascular disease. Biochim Biophys Acta 1821:867–875. [DOI] [PubMed] [Google Scholar]
- Calderon-Dominguez M, Gil G, Medina MA, Pandak WM, Rodríguez-Agudo D. (2014) The StarD4 subfamily of steroidogenic acute regulatory-related lipid transfer (START) domain proteins: new players in cholesterol metabolism. Int J Biochem Cell Biol 49:64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CY, Rivera JJ, Blumenthal RS. (2007) Residual risk in statin-treated patients: future therapeutic options. Curr Cardiol Rep 9:499–505. [DOI] [PubMed] [Google Scholar]
- Baigent C, Blackwell L, Emberson J, Holland LE, Reith C, Bhala N, Peto R, Barnes EH, Keech A, Simes J, et al. Cholesterol Treatment Trialists’ (CTT) Collaboration (2010) Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet 376:1670–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher J, O’Connell R, Voysey M, Emberson J, Blackwell L, Mihaylova B, Simes J, Collins R, Kirby A, Colhoun H, et al. Cholesterol Treatment Trialists’ (CTT) Collaboration (2015) Efficacy and safety of LDL-lowering therapy among men and women: meta-analysis of individual data from 174,000 participants in 27 randomised trials. Lancet 385:1397–1405. [DOI] [PubMed] [Google Scholar]
- Dallner G, Sindelar PJ. (2000) Regulation of ubiquinone metabolism. Free Radic Biol Med 29:285–294. [DOI] [PubMed] [Google Scholar]
- DeBarber AE, Bleyle LA, Roullet JB, Koop DR. (2004) Omega-hydroxylation of farnesol by mammalian cytochromes p450. Biochim Biophys Acta 1682:18–27. [DOI] [PubMed] [Google Scholar]
- Do R, Kiss RS, Gaudet D, Engert JC. (2009) Squalene synthase: a critical enzyme in the cholesterol biosynthesis pathway. Clin Genet 75:19–29. [DOI] [PubMed] [Google Scholar]
- Dong B, Saha PK, Huang W, Chen W, Abu-Elheiga LA, Wakil SJ, Stevens RD, Ilkayeva O, Newgard CB, Chan L, et al. (2009) Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proc Natl Acad Sci USA 106:18831–18836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan RE, Archer MC. (2008) Farnesol decreases serum triglycerides in rats: identification of mechanisms including up-regulation of PPARα and down-regulation of fatty acid synthase in hepatocytes. Lipids 43:619–627. [DOI] [PubMed] [Google Scholar]
- Eckel RH, Grundy SM, Zimmet PZ. (2005) The metabolic syndrome. Lancet 365:1415–1428. [DOI] [PubMed] [Google Scholar]
- Edwards PA, Ericsson J. (1999) Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu Rev Biochem 68:157–185. [DOI] [PubMed] [Google Scholar]
- Endo S, Matsunaga T, Ohta C, Soda M, Kanamori A, Kitade Y, Ohno S, Tajima K, El-Kabbani O, Hara A. (2011) Roles of rat and human aldo-keto reductases in metabolism of farnesol and geranylgeraniol. Chem Biol Interact 191:261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgacs AL, Dere E, Angrish MM, Zacharewski TR. (2013) Comparative analysis of temporal and dose-dependent TCDD-elicited gene expression in human, mouse, and rat primary hepatocytes. Toxicol Sci 133:54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, et al. (1995) Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81:687–693. [DOI] [PubMed] [Google Scholar]
- Gachon F, Leuenberger N, Claudel T, Gos P, Jouffe C, Fleury Olela F, de Mollerat du Jeu X, Wahli W, Schibler U. (2011) Proline- and acidic amino acid-rich basic leucine zipper proteins modulate peroxisome proliferator-activated receptor alpha (PPARα) activity. Proc Natl Acad Sci USA 108:4794–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. (2015) A century of cholesterol and coronaries: from plaques to genes to statins. Cell 161:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pacanowska D, Arison B, Havel CM, Watson JA. (1988) Isopentenoid synthesis in isolated embryonic Drosophila cells. Farnesol catabolism and omega-oxidation. J Biol Chem 263:1301–1306. [PubMed] [Google Scholar]
- Goto T, Kim YI, Funakoshi K, Teraminami A, Uemura T, Hirai S, Lee JY, Makishima M, Nakata R, Inoue H, et al. (2011a) Farnesol, an isoprenoid, improves metabolic abnormalities in mice via both PPARα-dependent and -independent pathways. Am J Physiol Endocrinol Metab 301:E1022–E1032. [DOI] [PubMed] [Google Scholar]
- Goto T, Nagai H, Egawa K, Kim YI, Kato S, Taimatsu A, Sakamoto T, Ebisu S, Hohsaka T, Miyagawa H, et al. (2011b) Farnesyl pyrophosphate regulates adipocyte functions as an endogenous PPARγ agonist. Biochem J 438:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Sarkar J, Ahmed MR, Viswakarma N, Jia Y, Yu S, Sambasiva Rao M, Reddy JK. (2006) Peroxisome proliferator-activated receptor (PPAR)-binding protein (PBP) but not PPAR-interacting protein (PRIP) is required for nuclear translocation of constitutive androstane receptor in mouse liver. Biochem Biophys Res Commun 347:485–495. [DOI] [PubMed] [Google Scholar]
- Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, et al. (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29:166–173. [DOI] [PubMed] [Google Scholar]
- Hafner M, Juvan P, Rezen T, Monostory K, Pascussi JM, Rozman D. (2011) The human primary hepatocyte transcriptome reveals novel insights into atorvastatin and rosuvastatin action. Pharmacogenet Genomics 21:741–750. [DOI] [PubMed] [Google Scholar]
- Hatanaka T. (2000) Clinical pharmacokinetics of pravastatin: mechanisms of pharmacokinetic events. Clin Pharmacokinet 39:397–412. [DOI] [PubMed] [Google Scholar]
- Healey GD, Collier C, Griffin S, Schuberth HJ, Sandra O, Smith DG, Mahan S, Dieuzy-Labaye I, Sheldon IM. (2016) Mevalonate biosynthesis intermediates are key regulators of innate immunity in bovine endometritis. J Immunol 196:823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyoshi H, Yanagimachi M, Ito M, Ohtsuka I, Yoshida I, Saeki T, Tanaka H. (2000) Effect of ER-27856, a novel squalene synthase inhibitor, on plasma cholesterol in rhesus monkeys: comparison with 3-hydroxy-3-methylglutaryl-CoA reductase inhibitors. J Lipid Res 41:1136–1144. [PubMed] [Google Scholar]
- Hiyoshi H, Yanagimachi M, Ito M, Saeki T, Yoshida I, Okada T, Ikuta H, Shinmyo D, Tanaka K, Kurusu N, et al. (2001) Squalene synthase inhibitors reduce plasma triglyceride through a low-density lipoprotein receptor-independent mechanism. Eur J Pharmacol 431:345–352. [DOI] [PubMed] [Google Scholar]
- Hiyoshi H, Yanagimachi M, Ito M, Yasuda N, Okada T, Ikuta H, Shinmyo D, Tanaka K, Kurusu N, Yoshida I, et al. (2003) Squalene synthase inhibitors suppress triglyceride biosynthesis through the farnesol pathway in rat hepatocytes. J Lipid Res 44:128–135. [DOI] [PubMed] [Google Scholar]
- Horton JD, Cohen JC, Hobbs HH. (2007) Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem Sci 32:71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109:1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA 100:12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt MC, Alexson SE. (2008) Novel functions of acyl-CoA thioesterases and acyltransferases as auxiliary enzymes in peroxisomal lipid metabolism. Prog Lipid Res 47:405–421. [DOI] [PubMed] [Google Scholar]
- Hunt MC, Siponen MI, Alexson SE. (2012) The emerging role of acyl-CoA thioesterases and acyltransferases in regulating peroxisomal lipid metabolism. Biochim Biophys Acta 1822:1397–1410. [DOI] [PubMed] [Google Scholar]
- Hunt MC, Tillander V, Alexson SE. (2014) Regulation of peroxisomal lipid metabolism: the role of acyl-CoA and coenzyme A metabolizing enzymes. Biochimie 98:45–55. [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Ohtsuka M, Ohki H, Ota M, Haginoya N, Itoh M, Shibata Y, Ishigai Y, Terayama K, Kanda A, et al. (2013) Discovery of DF-461, a potent squalene synthase inhibitor. ACS Med Chem Lett 4:932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JH, Jetten AM. (2010) Molecular mechanisms involved in farnesol-induced apoptosis. Cancer Lett 287:123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocarek TA, Dahn MS, Cai H, Strom SC, Mercer-Haines NA. (2002) Regulation of CYP2B6 and CYP3A expression by hydroxymethylglutaryl coenzyme A inhibitors in primary cultured human hepatocytes. Drug Metab Dispos 30:1400–1405. [DOI] [PubMed] [Google Scholar]
- Kocarek TA, Mercer-Haines NA. (2002) Squalestatin 1-inducible expression of rat CYP2B: evidence that an endogenous isoprenoid is an activator of the constitutive androstane receptor. Mol Pharmacol 62:1177–1186. [DOI] [PubMed] [Google Scholar]
- Kocarek TA, Reddy AB. (1996) Regulation of cytochrome P450 expression by inhibitors of hydroxymethylglutaryl-coenzyme A reductase in primary cultured rat hepatocytes and in rat liver. Drug Metab Dispos 24:1197–1204. [PubMed] [Google Scholar]
- Krag SS. (1998) The importance of being dolichol. Biochem Biophys Res Commun 243:1–5. [DOI] [PubMed] [Google Scholar]
- Krisans SK, Ericsson J, Edwards PA, Keller GA. (1994) Farnesyl-diphosphate synthase is localized in peroxisomes. J Biol Chem 269:14165–14169. [PubMed] [Google Scholar]
- Leszczynska A, Gora M, Plochocka D, Hoser G, Szkopinska A, Koblowska M, Iwanicka-Nowicka R, Kotlinski M, Rawa K, Kiliszek M, et al. (2011) Different statins produce highly divergent changes in gene expression profiles of human hepatoma cells: a pilot study. Acta Biochim Pol 58:635–639. [PubMed] [Google Scholar]
- Ma K, Malhotra P, Soni V, Hedroug O, Annaba F, Dudeja A, Shen L, Turner JR, Khramtsova EA, Saksena S, et al. (2014) Overactivation of intestinal SREBP2 in mice increases serum cholesterol. PLoS One 9:e84221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers CD, Boerma T, Ma Fat D. (2009) Global and regional causes of death. Br Med Bull 92:7–32. [DOI] [PubMed] [Google Scholar]
- McTaggart SJ. (2006) Isoprenylated proteins. Cell Mol Life Sci 63:255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima S, Yagyu H, Tozawa R, Tazoe F, Takahashi M, Kitamine T, Yamamuro D, Sakai K, Sekiya M, Okazaki H, et al. (2015) Plasma cholesterol-lowering and transient liver dysfunction in mice lacking squalene synthase in the liver. J Lipid Res 56:998–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto T, Amano Y, Tozawa R, Ishikawa E, Imura Y, Yukimasa H, Sugiyama Y. (2003) Lipid-lowering properties of TAK-475, a squalene synthase inhibitor, in vivo and in vitro. Br J Pharmacol 139:911–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien ML, Rangwala SM, Henry KW, Weinberger C, Crick DC, Waechter CJ, Feller DR, Noonan DJ. (1996) Convergence of three steroid receptor pathways in the mediation of nongenotoxic hepatocarcinogenesis. Carcinogenesis 17:185–190. [DOI] [PubMed] [Google Scholar]
- Okita RT, Okita JR. (2001) Cytochrome P450 4A fatty acid omega hydroxylases. Curr Drug Metab 2:265–281. [DOI] [PubMed] [Google Scholar]
- Pant A, Kocarek TA. (2016) Role of phosphatidic acid phosphatase domain containing 2 in squalestatin 1-mediated activation of the constitutive androstane receptor in primary cultured rat hepatocytes. Drug Metab Dispos 44:352–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy JK, Hashimoto T. (2001) Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu Rev Nutr 21:193–230. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Agudo D, Ren S, Wong E, Marques D, Redford K, Gil G, Hylemon P, Pandak WM. (2008) Intracellular cholesterol transporter StarD4 binds free cholesterol and increases cholesteryl ester formation. J Lipid Res 49:1409–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondini EA, Duniec-Dmuchowski Z, Kocarek TA. (2016) Nonsterol isoprenoids activate human constitutive androstane receptor in an isoform-selective manner in primary cultured mouse hepatocytes. Drug Metab Dispos 44:595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Kobayashi K, Mizuno Y, Fukuchi Y, Furihata T, Chiba K. (2010) Peroxisome proliferator-activated receptor alpha (PPARα) agonists induce constitutive androstane receptor (CAR) and cytochrome P450 2B in rat primary hepatocytes. Drug Metab Pharmacokinet 25:108–111. [DOI] [PubMed] [Google Scholar]
- Saito K, Shirasago Y, Suzuki T, Aizaki H, Hanada K, Wakita T, Nishijima M, Fukasawa M. (2015) Targeting cellular squalene synthase, an enzyme essential for cholesterol biosynthesis, is a potential antiviral strategy against hepatitis C virus. J Virol 89:2220–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson UK, Fazio S, Linton MF. (2012) Residual cardiovascular risk despite optimal LDL cholesterol reduction with statins: the evidence, etiology, and therapeutic challenges. Curr Atheroscler Rep 14:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder A, Wollnik J, Wrzodek C, Dräger A, Bonin M, Burk O, Thomas M, Thasler WE, Zanger UM, Zell A. (2011) Inferring statin-induced gene regulatory relationships in primary human hepatocytes. Bioinformatics 27:2473–2477. [DOI] [PubMed] [Google Scholar]
- Shang N, Li Q, Ko TP, Chan HC, Li J, Zheng Y, Huang CH, Ren F, Chen CC, Zhu Z, et al. (2014) Squalene synthase as a target for Chagas disease therapeutics. PLoS Pathog 10:e1004114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spady DK, Dietschy JM. (1983) Sterol synthesis in vivo in 18 tissues of the squirrel monkey, guinea pig, rabbit, hamster, and rat. J Lipid Res 24:303–315. [PubMed] [Google Scholar]
- Stein EA, Bays H, O’Brien D, Pedicano J, Piper E, Spezzi A. (2011) Lapaquistat acetate: development of a squalene synthase inhibitor for the treatment of hypercholesterolemia. Circulation 123:1974–1985. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Kawada T, Goto T, Yamamoto T, Taimatsu A, Matsui N, Kimura K, Saito M, Hosokawa M, Miyashita K, et al. (2002) Dual action of isoprenols from herbal medicines on both PPARγ and PPARα in 3T3-L1 adipocytes and HepG2 hepatocytes. FEBS Lett 514:315–322. [DOI] [PubMed] [Google Scholar]
- Tomaszewski M, Stepień KM, Tomaszewska J, Czuczwar SJ. (2011) Statin-induced myopathies. Pharmacol Rep 63:859–866. [DOI] [PubMed] [Google Scholar]
- Ugawa T, Kakuta H, Moritani H, Matsuda K, Ishihara T, Yamaguchi M, Naganuma S, Iizumi Y, Shikama H. (2000) YM-53601, a novel squalene synthase inhibitor, reduces plasma cholesterol and triglyceride levels in several animal species. Br J Pharmacol 131:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya S, Bostedor R, Kurtz MM, Bergstrom JD, Bansal VS. (1998) Massive production of farnesol-derived dicarboxylic acids in mice treated with the squalene synthase inhibitor zaragozic acid A. Arch Biochem Biophys 355:84–92. [DOI] [PubMed] [Google Scholar]
- van Roermund CW, Ijlst L, Wagemans T, Wanders RJ, Waterham HR. (2014) A role for the human peroxisomal half-transporter ABCD3 in the oxidation of dicarboxylic acids. Biochim Biophys Acta 1841:563–568. [DOI] [PubMed] [Google Scholar]
- Wieneke N, Hirsch-Ernst KI, Kuna M, Kersten S, Püschel GP. (2007) PPARα-dependent induction of the energy homeostasis-regulating nuclear receptor NR1i3 (CAR) in rat hepatocytes: potential role in starvation adaptation. FEBS Lett 581:5617–5626. [DOI] [PubMed] [Google Scholar]
- Wiseman DA, Werner SR, Crowell PL. (2007) Cell cycle arrest by the isoprenoids perillyl alcohol, geraniol, and farnesol is mediated by p21(Cip1) and p27(Kip1) in human pancreatic adenocarcinoma cells. J Pharmacol Exp Ther 320:1163–1170. [DOI] [PubMed] [Google Scholar]
- Wu W, Kocarek TA, Runge-Morris M. (2001) Sex-dependent regulation by dexamethasone of murine hydroxysteroid sulfotransferase gene expression. Toxicol Lett 119:235–246. [DOI] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, Neuschwander-Tetri BA, Brunt EM, Guzelian PS, Evans RM. (2000) Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature 406:435–439. [DOI] [PubMed] [Google Scholar]
- Xu F, Ng VY, Kroetz DL, de Montellano PR. (2006) CYP4 isoform specificity in the omega-hydroxylation of phytanic acid, a potential route to elimination of the causative agent of Refsum’s disease. J Pharmacol Exp Ther 318:835–839. [DOI] [PubMed] [Google Scholar]
- Yang YF, Jan YH, Liu YP, Yang CJ, Su CY, Chang YC, Lai TC, Chiou J, Tsai HY, Lu J, et al. (2014) Squalene synthase induces tumor necrosis factor receptor 1 enrichment in lipid rafts to promote lung cancer metastasis. Am J Respir Crit Care Med 190:675–687. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari K, Yoda N, Toriyabe T, Yamazoe Y. (2010) Constitutive androstane receptor transcriptionally activates human CYP1A1 and CYP1A2 genes through a common regulatory element in the 5′-flanking region. Biochem Pharmacol 79:261–269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.