Abstract
ME-344 [(3R,4S)-3,4-bis(4-hydroxyphenyl)-8-methyl-3,4-dihydro-2H-chromen-7-ol] is a second-generation derivative natural product isoflavone presently under clinical development. ME-344 effects were compared in lung cancer cell lines that are either intrinsically sensitive or resistant to the drug and in primary immortalized human lung embryonic fibroblasts (IHLEF). Cytotoxicity at low micromolar concentrations occurred only in sensitive cell lines, causing redox stress, decreased mitochondrial ATP production, and subsequent disruption of mitochondrial function. In a dose-dependent manner the drug caused instantaneous and pronounced inhibition of oxygen consumption rates (OCR) in drug-sensitive cells (quantitatively significantly less in drug-resistant cells). This was consistent with targeting of mitochondria by ME-344, with specific effects on the respiratory chain (resistance correlated with higher glycolytic indexes). OCR inhibition did not occur in primary IHLEF. ME-344 increased extracellular acidification rates in drug-resistant cells (significantly less in drug-sensitive cells), implying that ME-344 targets mitochondrial proton pumps. Only in drug-sensitive cells did ME-344 dose-dependently increase the intracellular generation of reactive oxygen species and cause oxidation of total (mainly glutathione) and protein thiols and the concomitant immediate increases in NADPH levels. We conclude that ME-344 causes complex, redox-specific, and mitochondria-targeted effects in lung cancer cells, which differ in extent from normal cells, correlate with drug sensitivity, and provide indications of a beneficial in vitro therapeutic index.
Introduction
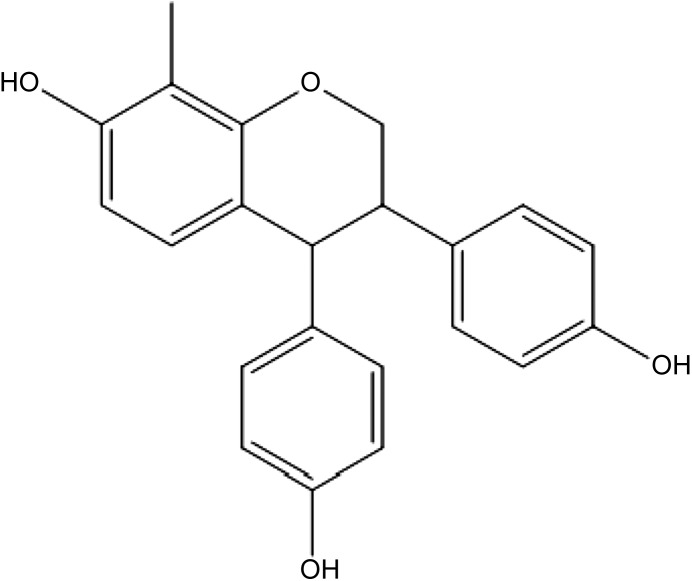
Natural product isoflavones have been studied as agents useful in cancer prevention (de Souza et al., 2010; Leclercq and Jacquot, 2014). However, one isoflavone, phenoxodiol, has cytotoxic antitumor activities more characteristic of a therapeutic agent; consequently, consideration has been given as to how this agent and some of its derivatives might induce cell death (Sapi et al., 2004; Aguero et al., 2005; Silasi et al., 2009). Mechanistically, proapoptotic effects may result from interference with mitochondrial functions (Kamsteeg et al., 2003) and interference with plasma membrane electron transport and/or cell surface thiol: disulfide exchange reactions (Morre and Morre, 2003; Herst et al., 2007). Downstream of such events, phenoxodiol derivatives can activate caspase-dependent and independent apoptotic pathways, perhaps through protein kinase B (Akt) signaling events (Wang et al., 2011; Pant et al., 2014). Related isoflavones alter mitochondrial membrane potential, deplete ATP, reduce steady-state levels of cytochrome c oxidase (complex IV of electron transport chain), and might eventually inhibit mammalian target of rapamycin (mTOR) activity (Alvero et al., 2011). One commonality of these effects is interference with mitochondrial energy production. Related to phenoxodiol, ME-344 [(3R,4S)-3,4-bis(4-hydroxyphenyl)-8-methyl-3,4-dihydro-2H-chromen-7-ol] (Fig. 1) is a second-generation derivative of triphendiol currently under development as a clinical therapeutic drug candidate in both small-cell lung and ovarian cancers. ME-344 is the lead compound selected from a number of analogs, where structure–activity relationship analyses have revealed that the ring hydroxyl groups provides characteristics that are prerequisite for cytotoxicity (Silasi et al., 2009).
Fig. 1.
Structure of ME-344.
Initial in vitro cytotoxicity screens demonstrated that only 20 of 240 human tumor cell lines (mostly solid tumor origins) were intrinsically (naturally) resistant to the effects of ME-344. Within the context of the present study, this provided the opportunity to compare the effects of ME-344 in these cell lines and compare with an immortalized cell line of lung fibroblast origin. Such an approach obviates the complexity of using drug selection conditions to derive acquired resistant clones and facilitates the identification of traits that might be important to the mechanism of action of ME-344. In turn, such information should have relevance to how the drug is clinically applied. Our results indicate that ME-344 impacts redox homeostasis and may act in mitochondria through interference with pathways that regulate switching from oxidative phosphorylation to glycolysis. Our present data implicate certain proteins and pathways that may be linked with the drug’s mechanism of action.
Materials and Methods
Tissue Culture.
SHP-77–small cell lung carcinoma (epithelial cells, sensitive) and H-460 large cell lung cancer (epithelial cells, sensitive) cells were purchased from the National Cancer Institute (Frederick, MD) and cultured according to supplier recommendations. Primary immortalized human lung embryonic fibroblasts (MRC-5), SW 900 grade IV squamous cell lung carcinoma (epithelial cells, resistant), and NCI-H596 adenosquamous lung carcinoma (epithelial cells, resistant) were purchased from American Type Culture Collection (Manassas, VA) and cultured according to the supplier recommendations. A panel of 240 cell lines was initially screened by MEI Pharma (San Diego, CA), and the present lines were chosen to represent natural (intrinsic) sensitivity or resistance to ME-344.
Seahorse Bioscience Experiments.
For standard and glycolytic mitochondrial stress test experiments, 96-well plates (XF96; Seahorse Bioscience/Agilent, Santa Clara, CA) with attached confluent cells were used in the Seahorse Bioscience apparatus in the MUSC COBRE Metabolomics Core. Standard protocols for OCR and extracellular acidification rates (ECAR) recording were used. Incubation time and ME-344 concentration dependence of OCR and ECAR were analyzed. The results were normalized for cell number in each well and presented as mean ± S.E., for n = 8.
Cell Viabilities and Fluorometric Detection of Caspase-3 Protease.
We used a standard Caspase-3/CPP32 Fluorometric Assay Kit from BioVision (Milpitas, CA) to study the effects of ME-344 induction of apoptosis in lung cancer cells and immortalized lung fibroblasts. Cells were grown in standard Petri dishes (60 mm) to ∼80% confluence, and the attached cells (control: dimethylsulfoxide [DMSO], or ME-344 treated) were washed with phosphate-buffered saline (PBS) 3 times, lyzed, combined in reaction buffer and fluorescent substrate DEVD-AFC (caspase-3 substrate) and incubated at 37°C for 1.5 hours. Finally, samples were placed in a quartz cuvette (5 × 5 × 40 mm), and the intensity of the DEVD-AFC cleavage product (AFC: 7-amino-4-trifluoromethyl coumarin, Ex/Em = 400/505 nm) was recorded using a standard kinetic mode (resolution, 0.1 second) of the QM-4 fluorometer for 200 seconds. The resultant emissions were averaged and plotted as mean ± S.D., n = 400. We recorded the kinetics of caspase-3 activation during incubation of cells with ME-344 for 0, 2, 4, and 6 hours. Each time point was measured in triplicate, and the final results are presented as mean ± S.D.
Viability after incubation with ME-344 for 0, 2, 4, and 6 hours at 37°C were assessed. After incubation, the cells were washed with PBS and detached from Petri dishes with trypsin, optimized for each cell type. After washing cells with fresh media, their counts/viabilities were determined using a Cellometer Auto T4 (Nexcelom Bioscience, Lawrence, MA) and trypan blue exclusion. All experiments were in triplicate and results presented as mean ± S.D.
Real-Time Kinetic Detection of Intracellular Reactive Oxygen Species and NADPH.
For reactive oxygen species (ROS)/NADPH fluorescent analyses, cells were grown as confluent monolayers adherent to Aclar plastic slides (14 × 25 mm). These slides were placed in a quartz cuvette (10 × 10 × 40 mm) with PBS (with 100 µM of CaCl2, 37°C) at a 45° angle to excitation light and the dynamics of NADPH emission (under constant stirring) were detected (Ex 340, Em 460 nm) before and after ME-344 (or other effectors) in real-time kinetics with a resolution of 0.1 second using a QM-4 fluorometer (PTI, Edison, NJ) (Manevich et al., 2002). In other experiments, adhered cells were labeled with specific dyes (ROS measurements) and similarly analyzed in the QM-4 fluorometer. Intracellular O2*− was monitored with CellTracker Deep Red fluorescent dye (Ex/Em, 630/650 nm; Life Technologies, Carlsbad, CA), intracellular H2O2 generation with H2DCF-DA dye (Sigma-Aldrich, St. Louis, MO), and *OH radical with 3-CCA-DA dye (Manevich et al., 1997). Live cells attached to Aclar slides were incubated in complete media with 5 µM of the appropriate dye for 45 minutes at 37°C. Slides with labeled cells were washed 3 times with PBS to eliminate excess dye before placing them in a quartz cuvette for data recording.
High-Performance Liquid Chromatography Analyses of ATP, ADP, and AMP.
Incubation medium from cell cultures in six-well plastic plates was replaced with 5% metaphosphoric acid (MPA) at 4°C. After 3 minutes, the plates were scraped, and the extracts centrifuged at 15,000g for 5 minutes at 4°C. The supernatants were then neutralized with 0.1 M Tris-acetate buffer. Adenine nucleotides in the extracts were separated using a Model 1525 Binary Breeze high-performance liquid chromatography (HPLC) pump equipped with a Model 717 Plus Autosampler and detected using a Model 2487 UV-Vis Detector (all from Waters, Milford, MA). Experiments were performed using a C18, 5 µm, 4.6 × 150-mm reverse phase column (SunFire; Waters), isocratic elution (1 ml/min) with 100 mM Na-phosphate buffer (pH 5.5) and UV detection at 260 nm.
The MPA lysates (25 µl) were injected into the column and ATP, ADP, and AMP were detected at approximate retention times of 4.1, 4.9, and 11.5 minutes, respectively (Maldonado et al., 2013). Authenticity of detection was confirmed by spiking actual samples with known amounts of ATP, ADP, and AMP as standards. Appropriate peak areas were quantified using Empower 2 software (Waters), and quantification of individual compounds was achieved with the use of particular calibration curves with purified ATP, ADP, and AMP standards.
HPLC Analysis of Glutathione Disulfide.
Attached cells were incubated with ice-cold 5% (final concentration) MPA. After 3 to 5 minutes, the cells were scraped, and the extracts centrifuged at 15,000g for 5 minutes at 4°C. The supernatants were collected and used for analysis. Glutathione disulfide (GSSG) in the extracts was detected using an HPLC method similar to that described for ATP (ADP, AMP) analyses. The supernatants from cell lysates treated with MPA (25 µl) were injected into the column, and the GSSG peak was detected (absorbance at 260 nm) at a retention time of ∼2.85 minutes. Authenticity of detection was confirmed by spiking actual samples with known amounts of GSSG as standards. Peak areas were quantified using Empower 2 software (Waters), and quantification of GSSG was achieved using calibration curves with purified GSSG.
Fluorescent Analysis of Low-Molecular-Weight and Protein Reduced Thiols.
Detection and quantification of sulfhydryls is based on their specific reaction with the maleimide probe TG-1 (Calbiochem, San Diego, CA), which becomes fluorescent (Bowers et al., 2012). Cell lysates were passed through a size-exclusion chromatography microcolumn (Bio-Spin 6, cutoff ∼6 kDa; Bio-Rad Laboratories, Hercules, CA) to separate proteins from lower molecular weight thiols. Both size-exclusion chromatography processed and intact samples were diluted in a quartz cuvette 1:100 (by volume) with 20 mM PB (pH 7.4) at 37°C under constant stirring in a sample holder of the QM-4 fluorometer. After addition of TG-1 (DMSO solution, final concentration 5 μM), the fluorescent detection (Ex 379 nm, Em 513 nm) of labeled sulfhydryls was performed using a standard kinetic mode of the QM-4 to reach saturation. Where pertinent, background fluorescence’s of cell lysates, effects of DMSO and background fluorescence of TG-1 in 20 mM PB (pH 7.4 at 37°C) were subtracted from the final results. The final emission values were averaged for 50 seconds (500 time points) using Sigma Plot 10.0 software (Systat Software, San Jose, CA) and presented as the mean ± S.D.
Fluorescent Analysis of Cell Surface Thiols.
Either lung cancer or immortalized primary (MRC-5) cells attached to Aclar plastic slides (∼85% confluent) were treated with ME-344 for 30 minutes in complete media. After washing these slides in PBS (with 100 μM of CaCl2) they were placed in a quartz cuvette with PBS (with 100 μM CaCl2) at 4°C (to prevent TG-1 endocytosis) similar to that for NADPH detection. Both before and after addition of 5 µM TG-1, emissions were detected in real-time kinetics until saturation. Background fluorescence for the cells and TG-1 were each subtracted from the final results. Samples were also corrected for DMSO and the final TG-1 fluorescence was normalized for cell number and presented as mean ± S.D. for three independent experiments. After those measurements, cells were detached from the slides and their counts/viabilities estimated using Cellometer Auto T4 (Nexcelom Bioscience LLC, Lawrence, MA) and trypan blue exclusion. Cell viabilities under our experimental conditions were very close (98% to 99%) before and after treatment with ME-344 (30 minutes) and TG-1.
Mitochondrial Membrane Depolarization Measurements in Real-Time Kinetics.
Cells on the Aclar plastic slides (∼85% confluent) were incubated with 5.0 µM of JC-9 fluorescent dye (Life Technology, CA) in complete media for 45 minutes at 37°C. Labeled cells were washed 3 times with PBS containing 100 µM CaCl2 and placed in a quartz cuvette similar to the above measurements for NADPH dynamics. JC-9 fluorescence was measured as a ratio of (485/529 nm monomeric dye fluorescence) to (535/590 nm J-aggregate fluorescence) in real-time kinetics before and after addition of indicated amounts of those agents shown (ME-344 and glucose).
Statistical Analysis.
We used SigmaStat 3.5 (Systat Software) for statistical analysis of data. One-way analysis of variance (ANOVA) was used for statistical relevance of data comparison.
Results
Recent work has implicated inhibition of mitochondrial complex I as a possible contributory mechanism for this class of drugs (Lim et al., 2015). We now provide further interrogation through SeaHorse technology and mitochondrial-stress testing measurements of the effects of ME-344 (Fig. 1) on oxygen consumption and extracellular acidification rates (OCR and ECAR) in drug-sensitive (SHP-77 and H460) and resistant (H596 and SW900) lung cancer and IHLEF (MRC-5) cell lines.
Mitochondrial Stress in Lung Cancer Cells.
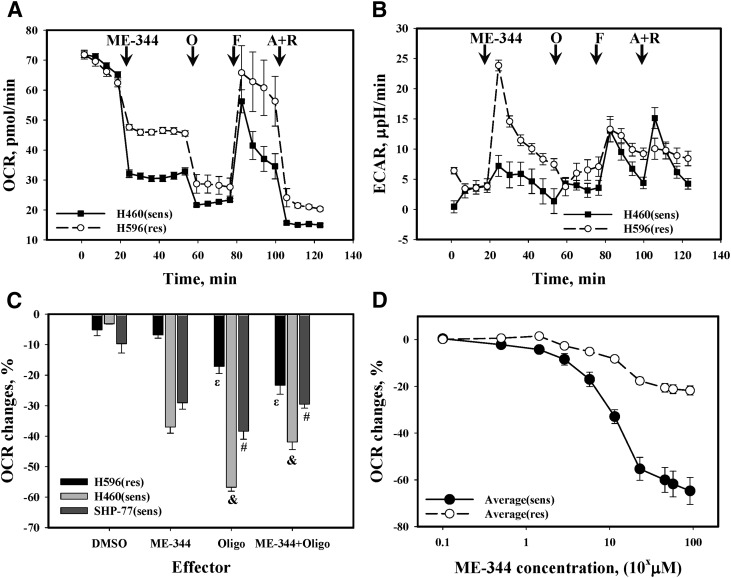
ME-344 caused a substantial and essentially instantaneous inhibition of OCR (similar in extent to oligomycin) in the lung cancer cell lines, correlating with the natural sensitivity of cells to the drug. Application of an uncoupler p-triflouromethoxyphenylhydrazone (FCCP) caused a fast recovery in the drug-resistant lung cancer cells, but was substantially retarded in the sensitive counterpart (Fig. 2A). Subsequent addition of antimycin A and rotenone completely inhibited respiration, with the effect more pronounced in resistant cells. Mitochondrial stress testing of the same cells revealed a substantial increase of ECAR in resistant cells after ME-344 (Fig. 2B), an effect that was barely detectable in sensitive cells. The impact of other standard stressors on ECAR was similar for both sensitive and resistant cell lines (Fig. 2B).
Fig. 2.
Standard mitochondrial stress test traces for lung cancer cells. (A) OCR and (B) ECAR changes upon addition of: ME-344 (4.0 µg/ml), O (oligomycin, 0.5 µM), F (FCCP [p-triflouromethoxyphenylhydrazone], 1.0 µM), and A + R (antimycin A, 1.0 µM + rotenone, 1.0 µM) to lung cancer H460 (sensitive) and H596 (resistant) cells in standard Seahorse 96-well microplates. The traces are representative of six independent experiments. (The results were essentially the same for SHP-77 [sensitive] and SW900 [resistant] lung cancer cells.) Data were normalized for cell numbers. (C) ME-344 effects on ATP synthase inhibition by oligomycin. Presented as a difference (%) between OCR with or without ME-344 before and after oligomycin. Data are normalized for values before oligomycin addition, and they represent the mean ± S.D. for three independent experiments. &,ε,#Statistical significance P ≤ 0.05. (D) ME-344 dose effects on OCR changes (% of initial OCR without effector). The data were averaged for either sensitive or resistant cells and are presented as mean ± S.E. for six independent experiments, P ≤ 0.009.
A further interesting difference was found between the resistant and sensitive cells. While comparing the effects of oligomycin on cells treated with either DMSO or ME-344, we measured OCR inhibition and normalized it for the OCR value before oligomycin addition. Pretreatment of cells with ME-344 inhibited the effects of subsequent oligomycin, but only in the sensitive cell lines. In the resistant cells, ME-344 was shown to potentiate the effects of oligomycin. (Fig. 2C). The effects of ME-344 on OCR were concentration-dependent and reached saturation at ∼57.4 µM (ED50 ∼11.5 µM, Fig. 2D).
Glycolytic Mitochondrial Stress Test.
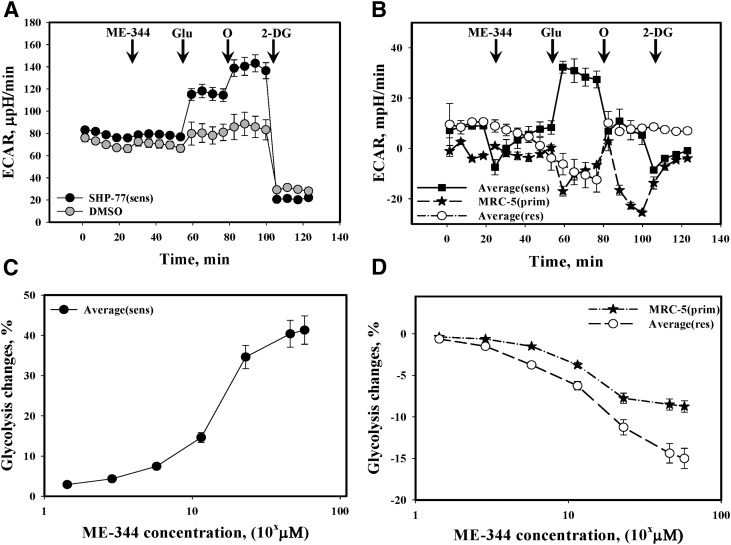
We next studied the effect of ME-344 on glycolysis in the lung cancer cells. Pretreatment of sensitive lung cancer cells resulted in an increase of ECAR (Fig. 3A) after addition of glucose. Interestingly the effects of oligomycin treatment were suppressed in sensitive lung cancer cells following pretreatment with ME-344 (Fig. 3B).
Fig. 3.
Effects of ME-344 on glycolysis in lung cancer and IHLEF cells. (A) Glycolysis stress tests were presented as ECAR changes upon addition of: ME-344 (11.48 μM), Glu (glucose, 10.0 mM), O (oligomycin, 1.0 µM), and 2-DG (2-deoxy-glucose, 10.0 mM) using SHP-77 lung cancer cells (sensitive). The trace is representative of six independent experiments. (B) Averaged differences between ME-344 (11.48 µM) and DMSO (same volume)-treated sensitive (H460 and SHP-77), IHLEF MRC-5, and resistant (H596 and SW900) cells. The differences are statistically significant only where sensitive cells are compared with either resistant or IHLEF cells (P ≤ 0.001). Data were normalized for cell numbers. (C) ME-344 dose effects on glycolysis (percentage of initial ECAR changes after glucose addition, averaged for sensitive [H460, SHP-77] cells). (D) ME-344 dose effects on glycolysis (percentage of initial ECAR changes after addition of glucose) averaged for IHLEF (MRC-5) and resistant (H596, SW900) cells. All data are presented as mean ± S.D. for six independent experiments and are statistically different (P ≤ 0.001).
Initial ECAR values under conditions of glucose deprivation were lower in IHLEF (MRC-5) cells (Fig. 3B) compared with resistant (averaged H596 and SW900) lung cancer cell lines (Fig. 3B). While ME-344 decreased the glycolytic response in IHLEF MRC-5 cells (Fig. 3B), in resistant cells, there was a greater decrease (Fig. 3B). Subsequent treatments with oligomycin caused a pronounced inhibitory response in both sensitive and the primary MRC-5 cells and an increased response in resistant cells (Fig. 3B). The effects of ME-344 on glycolysis were concentration dependent (Fig. 3C) in the sensitive cells and reached saturation at ∼57.4 µM (ED50 ∼11.5 µM) a value similar to that for OCR. Our data also suggest that ME-344 had dose dependent effects upon glycolytic stress that were different between the immortalized primary cells and drug-resistant cell lines (ED50 ∼11.5 µM, Fig. 3D).
Bioenergetic and Redox Effects in Cancer and Immortalized Primary Cells.
Following ME-344, ATP levels were decreased ∼12% in sensitive cells, with essentially no depletion in resistant cells (Fig. 4A). ME-344 also mildly altered redox stress in sensitive compared with resistant cells (as detected by GSSG levels; Fig. 4B). Initial ATP to ADP hydrolysis was 2-fold lower in primary cells (Fig. 4C) when compared with sensitive cells (Fig. 4D). ATP to ADP hydrolysis and redox stress (GSSG) were practically unaffected by ME-344 in primary cells (Fig. 4C) but the former was dose-dependently facilitated in sensitive lung cancer cells (Fig. 4D). These data again imply a degree of specificity for ME-344 effects in sensitive lung cancer cells.
Fig. 4.
Effects of ME-344 on bioenergetics and redox stress in IHLEF and lung cancer cells. HPLC analyses of (A) ATP and (B) GSSG in intact (DMSO, black bars) and ME-344 treated (11.48 µM, white bars) sensitive (SENS) or resistant (RES) cell lysates. Data were averaged for sensitive (H460, SHP-77) or resistant (H596, SW900) cells and presented as mean ± S.D. for three independent experiments. HPLC analyses of ME-344 dose effects on ATP/ADP ratios (black bars) and GSSG (normalized for its value before treatment, gray bars) in (C) IHLEF (MRC-5) or (D) sensitive cells (H460) are presented as mean ± S.E. for three independent experiments. ATP/ADP changes in sensitive (H460) cells were statistically different (P ≤ 0.05).
Cytotoxicity and Apoptosis.
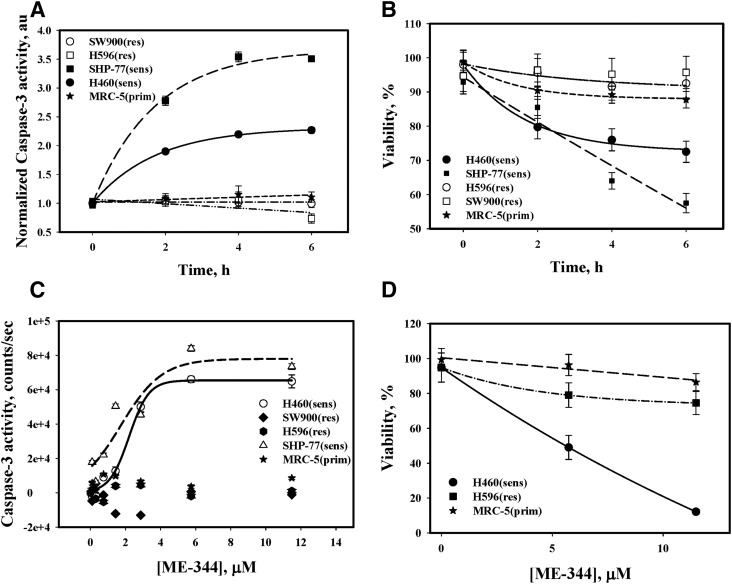
Cytotoxicity of ME-344 was assessed through its effects on apoptosis and viability. The drug rapidly (2 hours) induced apoptosis in the sensitive cell lines (H460 and SHP-77; Fig. 5A) through an activation of caspase-3 in a time dependent manner. In contrast, there was minimal (if any) induction of apoptosis in the IHLEF (MRC-5) and resistant cells. Figure 5B shows that progressive death of sensitive cells occurred after ME-344, but in resistant cells and IHLEF (MRC-5) these effects were minimal. Figure 5C shows the dose effects of ME-344 on caspase-3 activation in all cell types. The ED50 of ME-344-mediated caspase-3 activation occurred at 2.13 µM. Figure 5D shows viability of sensitive, resistant lung cancer and IHLEF after 24 hours incubation with 5.74 and 11.48 µM of ME-344. Approximately 90% of the sensitive cells were killed, but only minimal cytotoxicity was apparent in resistant and IHLEF cells.
Fig. 5.
Me-344 effects on caspase-3 activation and cell viability. (A) The kinetics of ME-344 (11.4 μM) inhibition of caspase-3 activity (normalized for DMSO) in H596 and SW-900 (resistant: □ and ○), H460 and SHP-77 (sensitive: ▪ and ●) and MRC-5 (IHLEF, ★). (B) Temporal viability of sensitive (▪ and ●) and resistant (□ and ○) cells and of IHLEF (MRC-5, ★) after ME-344 (11.48 μM). (C) Dose effects of ME-344 on caspase-3 activity in all cell types. (D) Viabilities of sensitive (H460) and resistant (H596) cells and IHLEF (MRC-5) after 24 hours’ incubation with ME-344 (5.74 and 11.48 µM). Data are mean ± S.D. for three independent experiments.
ME-344 Effects on Intracellular Oxidant Stress.
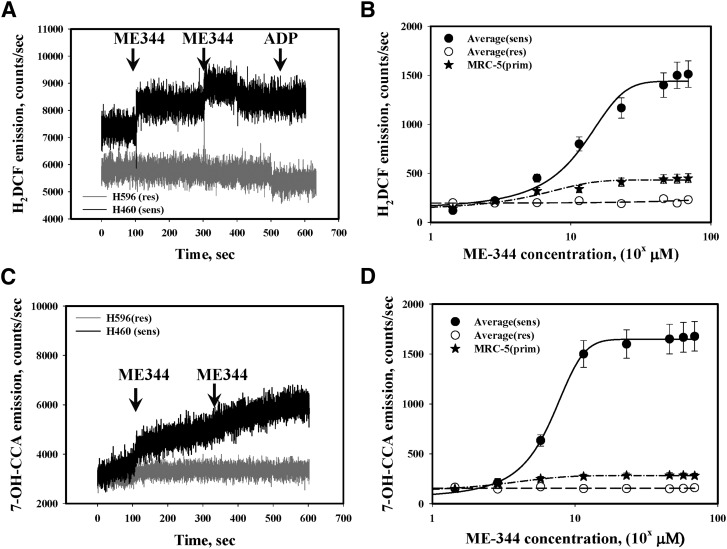
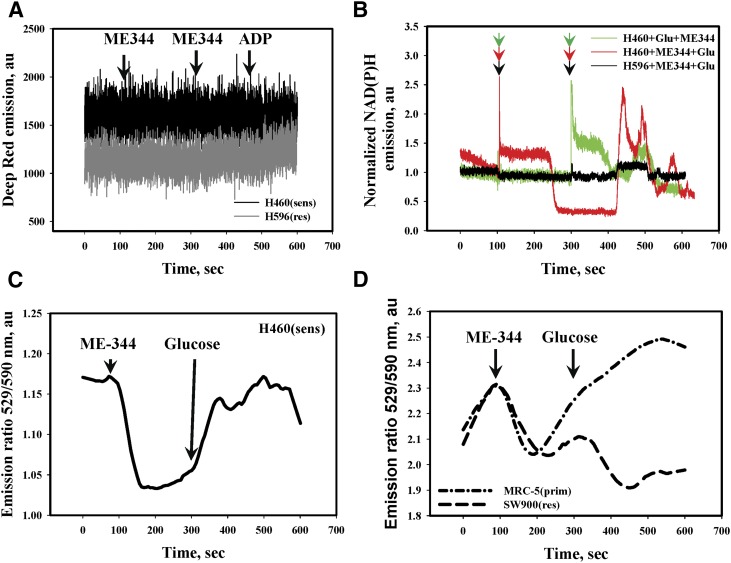
ME-344-mediated inhibition of OCR may contribute to oxidative stress through impairment of the mitochondrial electron transfer chain. ME-344 induced pronounced, concentration-dependent generation of H2O2 and *OH in sensitive cells (SHP-77 and H460; ED50 ∼11.5 µM, Fig. 6, A–C). This effect was not observed in resistant cells and was minimal in IHLEF (Fig. 6, B and D), implying some degree of specificity of ME-344 toward the sensitive lung cancer cells. ME-344 treatments did not lead to superoxide anion radical generation in either lung cancer cells or IHLEF (Fig. 7A).
Fig. 6.
ME-344 treatments linked with hydrogen peroxide and *OH generation. (A, C) ME-344 (11.4 μM) effects on intracellular H2O2 (*OH) generation in H460 sensitive (black traces) and H596 resistant (gray traces) cells in real-time kinetics. The traces are representative of three independent experiments and similar for SHP-77 (sensitive) and SW900 (resistant) cells. (B, D) Averaged dose effects of ME-344 on intracellular generation of averaged H2O2 (*OH) in sensitive (H460 and SHP-77, ●) and resistant (H596 and SW900, ○) cells and IHLEF (MRC-5, ★) as mean ± S.D. for three independent experiments, where the effects are statistically different (P ≤ 0.001). The lines represent sigmoid fit (three parameters) using Sigma Plot software. The arrows indicate points where effector was added.
Fig. 7.
ME-344 effects on the dynamics of superoxide radical generation, NADPH redox, and inner mitochondrial membrane potentials (Ψin). (A) ME-344 (11.4 μM) addition does not affect intracellular superoxide radical generation (detected with Deep Red fluorescent dye in real-time kinetics) in H460 (sensitive: black trace) and H596 (resistant: gray trace) cells. (B) ME-344 (11.48 μM) effects on NADPH emission either before (red trace) or after (green trace) glucose (Glu, 2.0 mM) as indicated by arrows in sensitive (H460) cells or in IHLEF (MRC-5, black trace). Traces are representative of three independent experiments and similar for SHP-77 (sensitive, not shown) cells. (C, D) Effects of ME-344 (11.48 μM) and glucose (2.0 mM, arrows) on the IMM potentials of sensitive (H460, solid line) cells, IHLEF (MRC-5, dotted and dashed line) and resistant (SW900, dashed line) cells; detected with JC-9 ratiometric fluorescent dye in real-time kinetics. Traces are representative of three independent experiments and were similar for SHP-77 sensitive and H596 resistant cells.
ME-344 Effects on General Redox Stress.
NADPH is a universal intracellular source of electrons for redox reactions. Both NADH (mainly mitochondrial) and NADPH (mainly cytosolic) have indistinguishable biofluorescences. Dynamic changes in NADPH fluorescence frequently accompany active redox signaling and have been used previously for evaluation of drugs that impact intermediate metabolism (Biaglow et al., 1998). Our present data indicate that under conditions of glucose deprivation, ME-344 treatment of sensitive cells (H460) caused a rapid increase and subsequent slow decrease of NADPH to below normal resting levels (Fig. 7B, red trace). As detected by fluorescence measurements, subsequent addition of glucose to the same cells resulted in a rapid increase in the generation and cycling of NADPH (Fig. 7B, red trace). A similar dose of glucose given before ME-344 did not alter NADPH, but further addition of ME-344 induced a similar rapid increase and subsequent cycling of NADPH fluorescence (Fig. 7B, green trace). Such effects did not occur in resistant (H596, Fig. 7B, black trace) or IHLEF (not shown).
Because proton fluxes control mitochondrial functions, we measured the dynamics of inner mitochondrial membrane potential (IMM) changes (ΔΨmit) in sensitive (H460), resistant (SW900) cells and IHLEF using a ratiometric fluorescent dye JC-9 (Fig. 7, C and D). Our data showed rapid hyperpolarization of IMM immediately after ME-344 addition (under glucose deprivation conditions), which was significant (∼12%) and sustained in sensitive (H460) cells (Fig. 7C). In IHLEF and resistant cells the effects were smaller (∼5%), and hyperpolarization was followed by rapid depolarization of IMM, which was slower and of smaller intensity in resistant cells (Fig. 7D).
In the cancer cells the addition of glucose after ME-344 caused a fast, sustained depolarization of IMM almost up to resting levels (Fig. 7C). Similar treatments of IHLEF caused slower depolarization of IMM, exceeding resting levels by ∼6% and further hyperpolarization of IMM in resistant cells (Fig. 7D). These results again suggest that ME-344 has some degree of specificity for effecting mitochondrial proton fluxes in sensitive cells.
ME-344 Effects on Intra- and Extracellular Redox States.
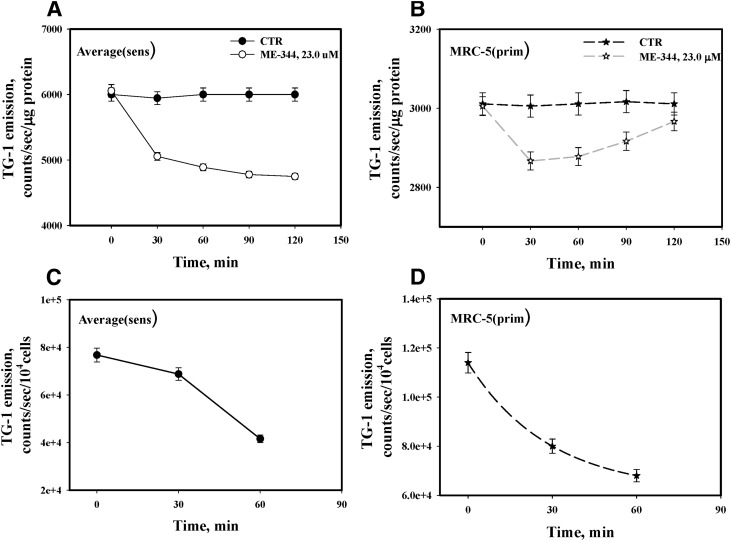
The dynamics of ME-344-mediated induction of oxidant stress (H2O2 and *OH, Fig. 6, A and C) were further investigated using fluorescent detection of intracellular thiol oxidation. The data show a progressive increase of redox stress (corresponding with decreases in reduced thiols) in lung cancer cells with saturation approximately 1 hour after drug treatment (Fig. 8). Maximal decreases in reduced thiols were seen in sensitive cells (Fig. 8A). Interestingly, ME-344 induced redox stress was reversible in IHLEF (Fig. 8B), inferring a plausible therapeutic advantage. ME-344 triggered pronounced oxidation of cell surface thiols with similar kinetics and magnitude (∼40%–50% in 1 hour) in both sensitive cells and IHLEF (Fig. 8, C and D).
Fig. 8.
ME-344 effects on dynamics of intra- and extracellular thiol redox status in lung cancer and IHLEF cells. Averaged reduced thiol (sulfhydryls) dynamics in lysates of (A) sensitive (H460 and SHP-77), and (B) IHLEF (MRC-5) before (DMSO control [CTR], ● and ★) and after incubation with ME-344 (22.8 μM, ○ and ☆), analyzed with sulfhydryl-specific fluorescent dye TG-1. Emissions were normalized for protein concentrations and presented as mean ± S.E. for three independent experiments. (C) Sensitive (H460 and SHP-77 cells) and (D) IHLEF (MRC-5) were treated with ME-344 (22.8 μM) for the indicated times, and cell surface thiol (sulfhydryls) were detected with TG-1. Emissions were normalized for cell numbers, and are averaged and presented as mean ± S.E. for three independent experiments.
Discussion
Because of the hydroxyl constituents on its phenol rings (Silasi et al., 2009), ME-344 has some properties that distinguish it from most of this family of chemicals. Of particular importance, our present results demonstrate that at low micromolar concentrations, cytotoxic effects are not induced in either immortalized normal lung fibroblasts or in lung cancer cell lines that have intrinsic resistance to the drug. Because ME-344 is in early stage clinical trials, there would be benefit in defining the mechanism(s) of action. In the present study, we describe a series of drug effects, which may be associated with a beneficial therapeutic index and can be linked with the unusual cytotoxic properties of the drug. Previous work has identified inhibition of energy production downstream of isoflavone effects, so we focused our attention on redox signaling and its consequent impact on bioenergetics in lung cancer cell lines sensitive and resistant to the drug. The cell lines were chosen from a preliminary screen of over 200 where those identified as resistant have not been previously exposed to drug selection.
Energy production through the mitochondrial respiratory chain requires an intensive flux of protons from the matrix into the inner-membrane space. Inhibition of respiration through impairment of electron flow diminishes this proton flux and causes IMM depolarization (Alberts, 2002). ME-344 can inhibit complex I of the respiratory chain and cause dissipation of inner mitochondrial membrane potential (Lim et al., 2015). Our OCR inhibition data indicate that sensitive lung cancer cells follow a similar response pattern. Instead of depolarization, ME-344 causes hyperpolarization of the inner mitochondrial membrane, serving to minimize the subsequent impact of oligomycin on OCR and ECAR. In this context, oligomycin is a known inhibitor of the F0 component (proton pump) of ATP synthase, which regulates proton efflux into the matrix from the intermembrane space (Shchepina et al., 2002). In the sensitive cells, ME-344 may be targeting the oligomycin-sensitive F0 component of ATP synthase, and some degree specificity is implied because ATP levels and redox signaling were affected only in these sensitive cells.
On the basis of dose, the efficacy of ME-344 is less than that of oligomycin (compare 11.5 µM to 1.0 µM), but the specificity of this effect may be beneficial for the eventual therapeutic index of ME-344. Also, in sensitive lung cancer cells ME-344 dose dependently and effectively (∼50% at 23.0 µM, 30 minutes’ incubation) facilitated ATP hydrolysis. This did not occur in IHLEF cells, perhaps again indicating some degree of selectivity for transformed cells. Taken together, these data are consistent with one interpretation to suggest that ME-344 targets the inner mitochondrial membrane proton channel (F0 component of ATP synthase), causes reduced levels of ATP production, alters proton fluxes, and initiates inner membrane hyperpolarization. This would provide a rational explanation for the unusually rapid and immediate effects of ME-344 on bioenergetics and redox signaling in these cells. In this regard, there are existing examples of other isoflavones that target ATP synthase (Zheng and Ramirez, 2000; Chinnam et al., 2010).
Under hypoxic conditions glycolysis is a complementary pathway for cancer cells to generate ATP. We showed that resting ECAR in immortalized primary cells was ∼2-fold lower than in cancer cells. With glucose deprivation, ME-344 had little effect on ECAR in any of the cells. Glycolytic rates in sensitive lung cancer cells were ∼54.0 µpH/min, but because of the Warburg effect (Vander Heiden et al., 2009) the rates were expectedly lower in IHLEF cells (∼12.0 µpH/min with nonglycolytic acidification at ∼4.0 µpH/min). In resistant lung cancer cells ME-344 did not evince any glycolytic response. Although this could be interpreted to suggest a direct impact of ME-344 on glycolysis, at this stage, such a conclusion would be premature. In addition, the results for estimating glycolysis (Fig. 3B) indicate that the drug may selectively inhibit ATP synthase (following oligomycin addition) only in the sensitive cells and IHLEF.
Two other effects of ME-344 in sensitive cancer cells are described. Drug treatment was linked with increases in intracellular hydroxyl radicals (*OH) and NADPH. Generation of *OH is surprising because traditionally isoflavones are not viewed as pro-oxidants. Biologic formation of *OH radicals can occur through Fenton chemistry, where transition metals react with H2O2 or other organic peroxides (Winterbourn, 1995). In our present study we show that ME-344 increases H2O2 generation in cytosol, and such a process would be consistent with interference with the mitochondrial respiratory chain and subsequent leakage of electrons (Halliwell et al., 1992) causing cytosolic redox stress. Again, this would be consistent with our data showing progressive thiol oxidation. Moreover, this process has been previously shown to induce heme oxygenase (HO-1; Choi and Alam, 1996).
In addition, Park et al. (2011) showed that certain isoflavones can induce HO-1 in primary astrocytes and this can be accompanied by its translocation from the ER to mitochondria (Bindu et al., 2011; Bansal et al., 2013). The first intermediate step of HO-1 catalysis produces Fe2+, an effective component of the Fenton reaction. It is plausible that this sequence of events could be involved in the mechanism by which ME-344 induces cytotoxicity. The precise chain of events by which ME-344 impacts HO-1 induction and/or translocation to mitochondria requires further investigation, but it is perhaps pertinent that HO-1 has been suggested as a possible anticancer target (Choi and Alam, 1996).
The rapid drug-induced increases in NADPH could also result from direct or indirect effects on the F0 component of ATP synthase. In turn, this could be linked with IMM hyperpolarization and compensatory proton fluxes, which could influence nicotinamide nucleotide transhydrogenase (NNT), an enzyme localized in the inner mitochondrial membrane catalyzing NADH-mediated reduction of NADP+ to NADPH (Mather et al., 2004; Gameiro et al., 2013). NNT-controlled catalysis is dynamic, efficient, and does seem to parallel the instantaneous effects of ME-344 (Gameiro et al., 2013). In concert with some of the other results, the immediacy of the effects of ME-344 on NADPH in the sensitive lung cancer cells could also imply that the drug has a direct or indirect impact on this enzyme, although further experimentation in this area will be required.
Earlier genetic characterizations of the lung cancer cell lines used here shed a little light on targeting redox pathways and their plausible influence drug response data. For example, SHP-77 cells express the cell surface markers that are reminiscent of HLA-DR macrophages and hematopoietic cells, and these may influence cell attachment (Ruff et al., 1986). These are not found in H460 (NSCLC) cells and may help to explain cell surface thiol differences (Davidson et al., 2004) and perhaps influence ME-344 sensitivities (likely only the magnitude of such responses). In the resistant cells, SW900 (Wesley et al., 2004) and H596 cells (Nguyen et al., 2015) are each characterized by the Notch1 transmembrane receptor, linked with the malignant phenotype and absent from the sensitive cell lines. In theory, this difference might also influence drug response. In general, however, the quantitative aspects of the differences in response to ME-344 are unlikely to be a function of simple monogenic differences between the cell lines.
In summary, our study shows that ME-344 is an unusual isoflavone with mitochondria-centered activities engendering pronounced and specific inhibition of bioenergetics and redox signaling pathways in lung cancer cells that are sensitive to the drug. At equivalent concentrations these effects do not occur in naturally resistant or immortalized primary cells. Two major mitochondrial enzymes: ATP-synthase and NNT are potentially direct or indirect targets for ME-344. Hypothetically, translocation of HO-1 into mitochondria as well as its activity could also be influenced by ME-344 and further studies in this area are warranted. The clinical use of ME-344 either as a single agent or in drug combinations may be contingent upon the cancer cell’s balance between oxidative phosphorylation and glycolysis. As a prediction, patient tumors more dependent upon glycolysis would be more responsive to the drug. In addition, the drug has lower toxicities in immortalized primary cells, indicating that at least in vitro an advantageous therapeutic index may be attainable.
Acknowledgments
The authors thank Dr. C. Beeson, Ms. G. Beeson, and Mr. B. Hoover of the Metabolomics Core of the COBRE for their help and advice in conducting SeaHorse experiments.
Abbreviations
- AFC
7-amino-4-trifluoromethyl coumarin
- DMSO
dimethylsulfoxide
- ECAR
extracellular acidification rates
- GSSG
glutathione disulfide
- HPLC
high-performance liquid chromatography
- IHLEF
immortalized human lung embryonic fibroblasts
- IMM
inner mitochondrial membrane
- ME-344
(3R,4S)-3,4-bis(4-hydroxyphenyl)-8-methyl-3,4-dihydro-2H-chromen-7-ol
- MPA
metaphosphoric acid
- NNT
nicotinamide nucleotide transhydrogenase
- OCR
oxygen consumption rates
- PBS
phosphate-buffered saline
- ROS
reactive oxygen species
Authorship Contributions
Participated in research design: Manevich, Britten, Townsend, Tew.
Conducted experiments: Manevich, Reyes, Townsend.
Performed data analysis: Manevich, Townsend, Tew.
Wrote or contributed to the writing of the manuscript: Manevich, Tew.
Footnotes
This work was supported by grants from the National Institutes of Health National Cancer Institute [Grants CA08660, CA117259] and National Center for Research Resources [Grant NCRR P20RR024485], COBRE in Oxidants, Redox Balance and Stress Signaling] and support from the South Carolina Centers of Excellence program and was conducted in a facility constructed with the support from the National Institutes of Health [Grant RR015455] from the Extramural Research Facilities Program of the National Center for Research Resources. C.D.B. and K.D.T. hold endowed Chairs from the South Carolina SmartState program. Supported in part by the Drug Metabolism and Clinical Pharmacology shared Resource, Hollings Cancer Center, Medical University of South Carolina. Further financial support was through an unrestricted grant from MEI Pharma Inc.
References
- Aguero MF, Facchinetti MM, Sheleg Z, Senderowicz AM. (2005) Phenoxodiol, a novel isoflavone, induces G1 arrest by specific loss in cyclin-dependent kinase 2 activity by p53-independent induction of p21WAF1/CIP1. Cancer Res 65:3364–3373. [DOI] [PubMed] [Google Scholar]
- Alberts B. (2002) Molecular Biology of the Cell, Garland Sciences, New York. [Google Scholar]
- Alvero AB, Montagna MK, Holmberg JC, Craveiro V, Brown D, Mor G. (2011) Targeting the mitochondria activates two independent cell death pathways in ovarian cancer stem cells. Mol Cancer Ther 10:1385–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S, Biswas G, Avadhani NG. (2013) Mitochondria-targeted heme oxygenase-1 induces oxidative stress and mitochondrial dysfunction in macrophages, kidney fibroblasts and in chronic alcohol hepatotoxicity. Redox Biol 2:273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biaglow JE, Manevich Y, Leeper D, Chance B, Dewhirst MW, Jenkins WT, Tuttle SW, Wroblewski K, Glickson JD, Stevens C, et al. (1998) MIBG inhibits respiration: potential for radio- and hyperthermic sensitization. Int J Radiat Oncol Biol Phys 42:871–876. [DOI] [PubMed] [Google Scholar]
- Bindu S, Pal C, Dey S, Goyal M, Alam A, Iqbal MS, Dutta S, Sarkar S, Kumar R, Maity P, et al. (2011) Translocation of heme oxygenase-1 to mitochondria is a novel cytoprotective mechanism against non-steroidal anti-inflammatory drug-induced mitochondrial oxidative stress, apoptosis, and gastric mucosal injury. J Biol Chem 286:39387–39402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers RR, Manevich Y, Townsend DM, Tew KD. (2012) Sulfiredoxin redox-sensitive interaction with S100A4 and non-muscle myosin IIA regulates cancer cell motility. Biochemistry 51:7740–7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnam N, Dadi PK, Sabri SA, Ahmad M, Kabir MA, Ahmad Z. (2010) Dietary bioflavonoids inhibit Escherichia coli ATP synthase in a differential manner. Int J Biol Macromol 46:478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi AM, Alam J. (1996) Heme oxygenase-1: function, regulation, and implication of a novel stress-inducible protein in oxidant-induced lung injury. Am J Respir Cell Mol Biol 15:9–19. [DOI] [PubMed] [Google Scholar]
- Davidson JD, Ma L, Flagella M, Geeganage S, Gelbert LM, Slapak CA. (2004) An increase in the expression of ribonucleotide reductase large subunit 1 is associated with gemcitabine resistance in non-small cell lung cancer cell lines. Cancer Res 64:3761–3766. [DOI] [PubMed] [Google Scholar]
- de Souza PL, Russell PJ, Kearsley JH, Howes LG. (2010) Clinical pharmacology of isoflavones and its relevance for potential prevention of prostate cancer. Nutr Rev 68:542–555. [DOI] [PubMed] [Google Scholar]
- Gameiro PA, Laviolette LA, Kelleher JK, Iliopoulos O, Stephanopoulos G. (2013) Cofactor balance by nicotinamide nucleotide transhydrogenase (NNT) coordinates reductive carboxylation and glucose catabolism in the tricarboxylic acid (TCA) cycle. J Biol Chem 288:12967–12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM, Cross CE. (1992) Free radicals, antioxidants, and human disease: where are we now? J Lab Clin Med 119:598–620. [PubMed] [Google Scholar]
- Herst PM, Petersen T, Jerram P, Baty J, Berridge MV. (2007) The antiproliferative effects of phenoxodiol are associated with inhibition of plasma membrane electron transport in tumour cell lines and primary immune cells. Biochem Pharmacol 74:1587–1595. [DOI] [PubMed] [Google Scholar]
- Kamsteeg M, Rutherford T, Sapi E, Hanczaruk B, Shahabi S, Flick M, Brown D, Mor G. (2003) Phenoxodiol--an isoflavone analog—induces apoptosis in chemoresistant ovarian cancer cells. Oncogene 22:2611–2620. [DOI] [PubMed] [Google Scholar]
- Leclercq G, Jacquot Y. (2014) Interactions of isoflavones and other plant derived estrogens with estrogen receptors for prevention and treatment of breast cancer-considerations concerning related efficacy and safety. J Steroid Biochem Mol Biol 139:237–244. [DOI] [PubMed] [Google Scholar]
- Lim SC, Carey KT, McKenzie M. (2015) Anti-cancer analogues ME-143 and ME-344 exert toxicity by directly inhibiting mitochondrial NADH: ubiquinone oxidoreductase (complex I). Am J Cancer Res 5:689–701. [PMC free article] [PubMed] [Google Scholar]
- Maldonado EN, Sheldon KL, DeHart DN, Patnaik J, Manevich Y, Townsend DM, Bezrukov SM, Rostovtseva TK, Lemasters JJ. (2013) Voltage-dependent anion channels modulate mitochondrial metabolism in cancer cells: regulation by free tubulin and erastin. J Biol Chem 288:11920–11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manevich Y, Held KD, Biaglow JE. (1997) Coumarin-3-carboxylic acid as a detector for hydroxyl radicals generated chemically and by gamma radiation. Radiat Res 148:580–591. [PubMed] [Google Scholar]
- Manevich Y, Sweitzer T, Pak JH, Feinstein SI, Muzykantov V, Fisher AB. (2002) 1-Cys peroxiredoxin overexpression protects cells against phospholipid peroxidation-mediated membrane damage. Proc Natl Acad Sci USA 99:11599–11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather OC, Singh A, van Boxel GI, White SA, Jackson JB. (2004) Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase. Biochemistry 43:10952–10964. [DOI] [PubMed] [Google Scholar]
- Morré DJ, Morré DM. (2003) Cell surface NADH oxidases (ECTO-NOX proteins) with roles in cancer, cellular time-keeping, growth, aging and neurodegenerative diseases. Free Radic Res 37:795–808. [DOI] [PubMed] [Google Scholar]
- Nguyen D, Rubinstein L, Takebe N, Miele L, Tomaszewski JE, Ivy P, Doroshow JH, Yang SX. (2015) Notch1 phenotype and clinical stage progression in non-small cell lung cancer. J Hematol Oncol 8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S, Burris HA, 3rd, Moore K, Bendell JC, Kurkjian C, Jones SF, Moreno O, Kuhn JG, McMeekin S, Infante JR. (2014) A first-in-human dose-escalation study of ME-143, a second generation NADH oxidase inhibitor, in patients with advanced solid tumors. Invest New Drugs 32:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Jung JS, Jeong YH, Hyun JW, Le TK, Kim DH, Choi EC, Kim HS. (2011) Antioxidant mechanism of isoflavone metabolites in hydrogen peroxide-stimulated rat primary astrocytes: critical role of hemeoxygenase-1 and NQO1 expression. J Neurochem 119:909–919. [DOI] [PubMed] [Google Scholar]
- Ruff MR, Farrar WL, Pert CB. (1986) Interferon gamma and granulocyte/macrophage colony-stimulating factor inhibit growth and induce antigens characteristic of myeloid differentiation in small-cell lung cancer cell lines. Proc Natl Acad Sci USA 83:6613–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapi E, Alvero AB, Chen W, O’Malley D, Hao XY, Dwipoyono B, Garg M, Kamsteeg M, Rutherford T, Mor G. (2004) Resistance of ovarian carcinoma cells to docetaxel is XIAP dependent and reversible by phenoxodiol. Oncol Res 14:567–578. [DOI] [PubMed] [Google Scholar]
- Shchepina LA, Pletjushkina OY, Avetisyan AV, Bakeeva LE, Fetisova EK, Izyumov DS, Saprunova VB, Vyssokikh MY, Chernyak BV, Skulachev VP. (2002) Oligomycin, inhibitor of the F0 part of H+-ATP-synthase, suppresses the TNF-induced apoptosis. Oncogene 21:8149–8157. [DOI] [PubMed] [Google Scholar]
- Silasi DA, Alvero AB, Rutherford TJ, Brown D, Mor G. (2009) Phenoxodiol: pharmacology and clinical experience in cancer monotherapy and in combination with chemotherapeutic drugs. Expert Opin Pharmacother 10:1059–1067. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McKernan R, Kim KH, Alvero AB, Whiting A, Thompson JA, Mor G, Saif MW, Husband AJ, Brown DM, et al. (2011) Triphendiol (NV-196), development of a novel therapy for pancreatic cancer. Anticancer Drugs 22:719–731. [DOI] [PubMed] [Google Scholar]
- Wesley UV, Tiwari S, Houghton AN. (2004) Role for dipeptidyl peptidase IV in tumor suppression of human non small cell lung carcinoma cells. Int J Cancer 109:855–866. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC. (1995) Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol Lett 82-83:969–974. [DOI] [PubMed] [Google Scholar]
- Zheng J, Ramirez VD. (2000) Inhibition of mitochondrial proton F0F1-ATPase/ATP synthase by polyphenolic phytochemicals. Br J Pharmacol 130:1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]