Abstract
Asthma is a heterogeneous disease comprising a number of subtypes which may be caused by different pathophysiologic mechanisms (sometimes referred to as endotypes) but may share similar observed characteristics (phenotypes). The use of unsupervised clustering in adult and paediatric populations has identified subtypes of asthma based on observable characteristics such as symptoms, lung function, atopy, eosinophilia, obesity, and age of onset. Here we describe different clustering methods and demonstrate their contributions to our understanding of the spectrum of asthma syndrome. Precise identification of asthma subtypes and their pathophysiological mechanisms may lead to stratification of patients, thus enabling more precise therapeutic and prevention approaches.
Keywords: Adult asthma, Asthma, Clustering, Endotypes, Paediatric asthma, Phenotypes
Introduction
Asthma is a heterogeneous disease, defined by the most recent Global Initiative for Asthma (GINA) global strategy for asthma management and prevention consensus as a condition characterised by the presence of respiratory symptoms such as wheeze, shortness of breath, chest tightness and cough that vary over time and in intensity, together with variable airflow obstruction [1]. However, various definitions of asthma do not capture the heterogeneity of this common complex condition. It is becoming increasingly clear that asthma is not a single disease, but a syndrome which consists of a number of disease subtypes with similar observable clinical characteristics [2]. These observable characteristics of the disease are often referred to as asthma phenotypes. The term 'asthma endotype' is not synonymous with phenotype, and it should be used to refer to the distinct disease entity under the umbrella diagnosis of asthma, which has defined pathophysiological mechanisms that give rise to clinical symptoms [3]. It should be emphasised that the same observable characteristic (i.e. phenotype) can arise as a consequence of different underlying pathologies (i.e. endotypes), which is consistent with observations showing that there are subtypes of asthma that share similar clinical symptoms but have differing underlying pathophysiological mechanisms [4]. There are numerous examples in other disease areas of a similar or identical clinical presentation arising as a consequence of different pathology (e.g. fever in childhood can be caused by numerous different mechanisms).
The traditional constructs of ‘asthma phenotypes’ have been largely descriptive, with little uniformity, and usually informed by subjective observations of single dimensions of the disease, such as triggering factors (e.g. extrinsic and intrinsic asthma [5], exercise-induced asthma [6]), patterns of airway obstruction (e.g. reversible and irreversible asthma [7]), or pathology (e.g. eosinophilic and non-eosinophilic asthma [8]).
In paediatric asthma, changes over time in symptoms such as wheeze have been used to define phenotypes of wheezing illness during childhood [9]. For example, based on clinical observation of changes in the temporal pattern of wheezing illness during childhood, as confirmed in the birth cohort study (Tucson Children’s Respiratory Study), Martinez et al. divided children into three groups (or phenotypes) of wheezing: transient early wheezers, late-onset wheezers, and persistent wheezers [10]. Although these phenotypes are clinically meaningful in their association with lung function and subsequent development of asthma [11], their distinct underlying pathophysiological mechanisms have not been elucidated or confirmed—they cannot be considered as endotypes.
Based on expert opinion and consensus, Lotvall et al. [4] suggested the existence of six asthma endotypes: aspirin-sensitive asthma, allergic bronchopulmonary mycosis, allergic asthma, asthma predictive index-positive preschool wheezers, severe late-onset hypereosinophilic asthma, and asthma in cross-country skiers. However, the well-defined pathophysiological mechanisms and biomarkers which differentiate these proposed endotypes have not been discovered, and there is no universal agreement that these subtypes of asthma represent true endotypes [12]. At this time, the endotype concept remains largely hypothetical, but may have a tangible value in helping us to formulate strategies to better understand the mechanisms underlying different asthma-related diseases, and thus to identify more effective stratified treatment strategies [13].
In recent years, approaches to subtyping asthma have evolved from subjective expert opinion to more data-driven methodologies such as machine learning [14, 15]. Statistical machine-learning methods facilitate the efficient exploration of data for the identification and analysis of disease patterns. These methods are able to draw upon the vast array of data generated from birth and patient cohorts in order to cluster, classify, regress, and make predictions from data based on inherent patterns within the large complex data set. This is in contrast to the traditional methods based on human observation and testing of hypotheses using prior knowledge. Within the context of asthma subtyping, methods such as unsupervised clustering approaches, factor analysis, and principal component analysis come into wide use within the last decade. These are hypothesis-generating, with the overarching notion that the inherent patterns within the data may be a reflection of different underlying aetiologies, genetic basis, and/or immunopathophysiologies, and that identified clusters may represent distinct asthma endotypes. If this assumption is correct, clustering methodologies could facilitate better understanding of the disease mechanisms, identification of novel therapeutic targets, and better clinical trial design incorporating group-specific targeted treatment, all of which are essential steps towards delivery of stratified medicine in asthma.
Here we present a review of the different clustering methodologies—model-free and model-based—and their applications in asthma subtyping. We provide an overview of the major studies and discuss the implications and approaches used.
What Is Clustering?
Cluster analysis is a popular unsupervised machine-learning method that seeks to identify similar characteristics in subjects (or variables) and to group them together on that basis. In selecting groups, the primary aim is to minimize intra-group variance while simultaneously maximizing inter-group variance. Clustering ‘classifies’ data by labelling objects with cluster ‘labels’ or giving each object a probability of belonging to a certain cluster. Cluster labels are not known a priori, and are derived solely from the data. This is in contrast to supervised methods such as logistic regression and support vector machines, which seek to derive rules for classifying new objects based on a set of previously classified objects.
Selection of Variables/Features and Dimension Reduction
Cluster analysis lacks the ability to differentiate between clinically relevant and irrelevant variables; thus the choice of variables to input into the clustering algorithm is one of the most important considerations. Variable or feature selection can be performed subjectively or objectively. Subjective methods involve choosing relevant variables based on expert advice and published work. In contrast, objective methods use data-driven approaches to variable/feature selection, the most common of which are stepwise methods (such as backward and forward selection) and dimension reduction techniques (such as principal components analysis [PCA] and factor analysis [FA]). Forward selection progressively adds variables of greatest significance (based on pre-set p values) to the model. Backward selection starts with all variables and progressively drops the least significant ones until all the remaining variables are statistically significant.
To reduce the large number of variables, the majority of studies we reviewed employed manual extraction based on expert advice. For example, Moore et al. [16] manually reduced the number of variables from 600 to 34 by excluding variables with missing data and those that were either deemed redundant because information was captured by another variable (multicollinearity) or considered not clinically relevant. Other studies used dimension reduction techniques such as PCA and FA, which reduce data by generating small subsets of generally uncorrelated variables from a large data set of potentially correlated variables. It is useful when we assume that there are underlying latent (unobserved) constructs (factors/components) in the data which cannot be measured directly but which can influence responses on measured variables. Although these two methods were used almost interchangeably in the literature we reviewed, there are differences between them. As a general rule, PCA is used to reduce data into smaller subsets, while FA is used to determine the unobserved factors which explain the data.
Clustering Methods
Three main clustering methods have generally been used in asthma subtyping: hierarchical approaches, non-hierarchical or partitioning-based approaches, and model-based or probabilistic approaches.
Hierarchical Clustering
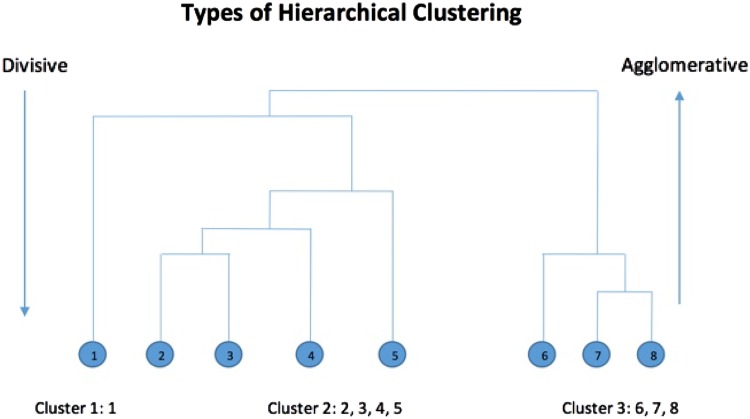
Hierarchical clustering aims to create a pyramidal or (as its name implies) ‘hierarchical’ grouping of homogeneous clusters that can be displayed in a tree-like graph (dendrogram). It does not require the number of clusters to be specified a priori, and cluster assignment is based on similarity of measured characteristics. Within hierarchical clustering there are two subcategories: agglomerative and divisive methods (Fig. 1).
Fig. 1.
Overview of the difference between agglomerative and divisive hierarchical clustering
Agglomerative Method
The agglomerative method is a bottom-up approach that starts with each data point assigned to its own cluster, and iteratively merges the two closest clusters until all the data belong to a single cluster [17]. Once clusters are formed, there is no inter-cluster switching. The choice of which clusters to combine is determined by measuring distances, similarities/dissimilarities, and/or using linkage criteria.
This method formulates decisions based on the pattern of variables used, without accounting for the overall distribution.
Divisive Method
This variant is a top-down approach whereby all objects initially belong to one cluster, which is then recursively divided into sub-clusters until the desired number of clusters is obtained [18]. By initially having a single cluster, the model gains insight into the spread and type of data, and subsequently makes decisions on when and how to divide the sub-clusters.
Similarity/Dissimilarity Measures
To determine whether objects within the same clustered group are similar or dissimilar, distance measures and linkage criteria (Table 1) are used. Distance metrics measure the distance between observations, while linkage criteria measure the distance between clusters. In order to define a similarity measure, the actual similarities between objects can be evaluated using a distance measure. Choosing a measure for calculating the distance between data can sometimes be arbitrary, as there are no general theoretical guidelines. The Euclidean distance measure, which is the default method in most statistical packages, was used in all but one of the studies reviewed here [19].
Table 1.
Most commonly used linkage criteria
| Linkage criteria | |
|---|---|
| Centroid | Measures distance between the central points of each cluster |
| Ward’s method | Measures the distance between clusters as the ANOVA sum of squares—i.e. combining information over all cluster members |
| Complete | Measures the distance between the members of clusters farthest apart |
Non-Hierarchical Clustering
The prototype of non-hierarchical clustering is k-means (Fig. 2), which is a partitioning method in which the number of clusters is specified a priori and the optimal solution is chosen. It is a variance-minimizing algorithm whereby each subject is assigned to its nearest cluster based on the minimum squared Euclidean distance. This method is sensitive to outliers and is generally limited to numeric attributes.
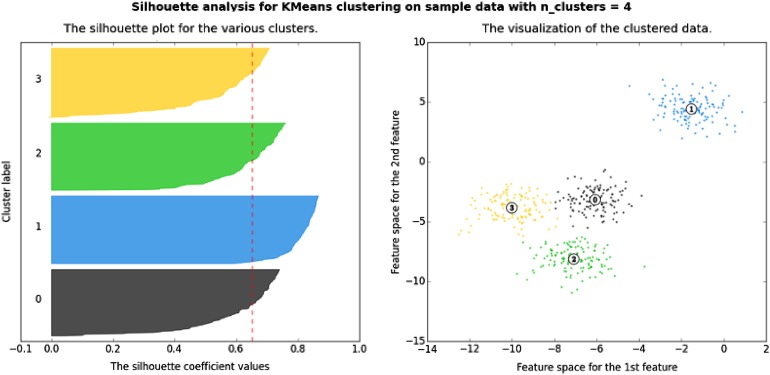
Fig. 2.
A silhouette plot used for non-hierarchical clustering (k-means) (from [20], with permission). A silhouette plot shows how close observations from neighbouring clusters are to each other using a measure of −1 to +1. A value of +1 indicates that observations are far away, 0 indicates that the observations are very close to the boundary of deciding exactly which cluster they belong to, and −1 indicates that the observations may be assigned to the wrong cluster
Model-Based Clustering
Model-based clustering (also known as latent class analysis or mixture modelling), is based on the assumption that the observed data are generated by a collection of models, with each cluster corresponding to a different model. Each resulting cluster is represented by a (most commonly) parametric distribution, and can be either spherical or ellipsoidal of varying sizes and variance. The advantage of model-based clustering is that it can produce probabilistic cluster assignments for individuals—i.e. it captures the uncertainty in assigning individuals to clusters. Bayesian extensions (e.g. Markov chain Monte Carlo [MCMC], expectation-maximisation [EM]) of model-based clustering can also be used to incorporate prior distributions to reflect uncertainty around model assumptions.
A major challenge in model-based clustering is identifying and representing the underlying model assumptions with reasonable complexity. However, unlike a model-free approach, log-likelihood-based statistics such as the Bayesian information criteria (BIC) and model evidence allow us to select the most parsimonious set of assumptions by penalising model complexity for accuracy. This is in contrast to model-free clustering, where an arbitrary distance measure is used to find clusters. Importantly, choosing the best statistically fitting model is not enough; there must be an element of expert input into choosing the number of clusters to maximise the potential clinical relevance of the identified subgroups.
Stability of Resulting Clusters
Cluster stability is an important aspect of validity, because cluster methods can generate groups in fairly homogenous data sets. Furthermore, there is always a risk of identifying less meaningful clusters. Stability in this context refers to clusters not disappearing when, for example, outliers are added, data is sub-set, or random error is introduced to every point to simulate measurement error [21]. The most common means of doing this is to apply the same cluster method to a sample data set taken from the original one (also termed bootstrapping), and identifying similar clusters using similarity measures. The similarity values are then compared, and stability is taken to be the mean similarity in the new data set [21].
Clustering Methods in Asthma Subtyping
The Use of Principal Components Analysis/Factor Analysis in Asthma Subtyping
Studies which used PCA/FA as stand-alone analyses for demonstrating the heterogeneity of asthma syndrome and its risk factors are summarised in Table 2 [22–40]. Sample sizes ranged from 69 to 16,635, and the number of variables used initially ranged from 5 to 97. The number of resulting components/factors ranged from one to six.
Table 2.
Studies using principal components analysis/factor analysis in asthma subtyping
| Cohort/data setting | Year | Age group | Sample size | No. variables | Method, % variance | Resulting components (PCA)/factors (FA) | Author group references |
|---|---|---|---|---|---|---|---|
| Manchester Asthma and Allergy Study | 2008 | 3 | 946 | 21 | PCA | [22] | |
| 5 | 904 | 32 | Age 3: 47.5% | Age 3: 4 | |||
| Age 5: 49.8% | Age 5: 5 | ||||||
| 59 rural communities in Ecuador | 2011 | 7–15 | Mean 73 | 29 | PCA | 2 | [23] |
| Component 1: 54.4% | |||||||
| Component 2: 50.1% | |||||||
| Component 3: 50.7% | |||||||
| Three clinical trials | 2012 | 15–79 | 1114 | 21 | PCA | 6 | [24] |
| 76% cumulative | |||||||
| Generation R study | 2012 | ≤4 | 2173 | 21 | PCA | 2 | [25] |
| Component 1: 16.3% | |||||||
| 8.2% | |||||||
| Education department Sao Francisco do Conde, Brazil | 2013 | 6–12 | 1307 | 22 | PCA | 2 | [26] |
| 45.7% cumulative | |||||||
| COREA | 2013 | Avg age 70.2 | 434 | 11 | PCA | Elderly: 4 | [27] |
| 53.5% cumulative | |||||||
| Non-elderly: 4 | |||||||
| Avg age 44.2 | 1633 | ||||||
| Manchester Asthma and Allergy Study | 2014 | Children | 1051 | 97 | PCA | 3 | [28] |
| 15.3% cumulative | |||||||
| Riyadh Cohort Study | 2014 | 7–17 | 195 | 6 | PCA | 2 | [29] |
| 57.3% cumulative | |||||||
| COPSAC | 2015 | Neonates | 411 | 5 | PCA | 1 | [30] |
| 41% cumulative | |||||||
| CAMP, CARE, PACT, ACRN, IMPACT, SOCS | 2015 | Children | 327 | 6 | PCA | 6 | [31] |
| 100% cumulative | |||||||
| University of Alabama at Birmingham Pulmonary Medicine Clinic | 1992 | Adults | 199 | 10 | FA | 3 | [32] |
| Institute of Immunoallergology, Florence IT | 1999 | 16–75 | 99 | 8 | FA | 3 | [33] |
| 74.8% cumulative | |||||||
| European Community Respiratory Health Study | 2000 | 20–44 | 16,635 | 18 | FA | 4 | [34] |
| 58% cumulative | |||||||
| Tucson Children’s Respiratory Study | 2001 | 6–11 | 877 | 25 | FA | 2 | [35] |
| 22.6% cumulative | |||||||
| Stable chronic asthmatics | 2001 | Adults | 69 | – | FA | 3 | [36] |
| 78% cumulative | |||||||
| Salmeterol Quality of Life Study Group | 2004 | >12 | 763 | 21 | FA | 4 | [37] |
| 80.8% cumulative | |||||||
| Health Maintenance Organisation, Kaiser-Permanente, US | 2005 | 18–56 | 2854 | 53 | FA | 5 | [38] |
| 59% cumulative | |||||||
| Paediatric outpatients, Chinese University of Hong Kong | 2005 | 7–18 | 92 | 12 | FA | 5 | [39] |
| 64.6% cumulative | |||||||
| Childhood Asthma Management Program, clinical trial, Boston, USA | 2008 | 5–12 | 990 | 17 | FA | 5 | [40] |
| 51.2% cumulative |
Avg average, COREA (Korea) Cohort for Reality and Evolution of Adult Asthma in Korea, COPSAC (Denmark) Copenhagen Prospective Study on Asthma in Childhood, CAMP (US) Childhood Asthma Management Program, IMPACT (US) Improving Asthma Control Trial, PACT (US) Pediatric Asthma Controller Trial, CARE (US) Childhood Asthma Research and Education Network, SOCS (US) Salmeterol or Corticosteroids Study, ACRN (US) Asthma Clinical Research Network, MAAS Manchester Asthma and Allergy Study
The PCA was first used in the context of asthma by Smith et al. to examine whether syndromes of coexisting respiratory symptoms could be discovered using the response to a large number of questions (>100) from validated questionnaires administered in a birth cohort (Manchester Asthma and Allergy Study [MAAS]) [22]. The analysis demonstrated that symptom components (wheeze, cough, wheeze with allergens, wheeze with irritants, chest congestion) were better indicators of the presence and developmental changes in observable secondary asthma phenotypes (such as lung function, airway reactivity, and immunoglobulin E (IgE)-mediated sensitisation) than the presence of individual symptoms such as wheeze.
Using factor analysis, Bailey et al. [32] found that the intensity of asthma symptoms, asthma management, and airflow impairment (forced expiratory volume [FEV1]) were independent components of the disease. This was also seen in the study by Grazzini et al. [36], where lung function (FEV1) was a factor independent from asthma symptoms in a mixed teenager-adult population of 69 asthmatics. Lung function was also independent of inflammatory markers (fraction of exhaled nitric oxide [FeNO], sputum eosinophils) in other studies [33, 39, 40]. The study by Juniper et al. [37], which included 763 patients older than 12 years who participated in clinical trials, showed that, despite medication, daytime and nighttime symptoms were distinct and independent factors of asthma. Clemmer et al. [31] used PCA to demonstrate that a clinical ‘endophenotype’ relating to corticosteroid responsiveness best predicted corticosteroid response in all replication populations. Other studies in Brazilian [26], British [28], and Japanese [41] children have shown that ‘Western diets’ were independently associated with an increased risk of wheezing by school age.
More recently, both PCA and FA have been used as dimension reduction techniques to generate small subsets from a large number of variables; these small subsets (components/factors) were then used for further clustering. For example, Just et al. used PCA to reduce 40 variables to 19, characterising age and body mass index (BMI), asthma duration, medication use, hospitalisation, atopy, and lung function [42], which were then used in hierarchical clustering. This approach acts as feature extraction in that it can initially visualize/reveal clusters prior to the cluster analysis.
Asthma Subtype Classification with Model-Free Approaches
The studies identified from our literature search which used model-free approaches for subtyping asthma are shown in Table 3 [16, 19, 43–61]. Of 22 studies, 12 were carried out in adult populations. Population sample sizes ranged from 57 to 1843. The approach of choice was Ward’s hierarchical method with some form of data reduction, whether with PCA, multiple regression analysis, discriminant analysis, factor analysis, or decision trees. k-means clustering was performed in 9 of 22 studies, but always as a supplementary method. The resulting numbers of clusters ranged from two to six.
Table 3.
Studies using model-free approaches for subtyping asthma
| Cohort/data setting | Year | Age group (years) | Sample size, N | Data reduction technique | Method of clustering | Number of clusters | Author group reference |
|---|---|---|---|---|---|---|---|
| Glenfield Hospital Difficult Asthma Clinic GLAD Glenfield Hospital Clinical Trial |
2008 | Avg age: 49.2 | 184 | PCA | Two-step: | 3 | [43] |
| Clustering | 4 | ||||||
| Avg age: 43.4 | 187 | ||||||
| k-means | 3 | ||||||
| Avg age: 52.4 | 68 | ||||||
| Random selection of patients in Wellington, NZ | 2009 | 25–75 | 175 | Two-step: Agglomerative ‘agnes’ clustering Divisive ‘Diana’ Gower’s distance measure Clusters chosen subjectively from tree diagram to include ≥10 subjects per cluster |
Agnes: 5 | [44] | |
| Diana: 4 | |||||||
| SARP | 2010 | 12–80 | 726 | Ward’s hierarchical clustering | 5 | [16] | |
| Post hoc | |||||||
| Discriminant analysis for tree analysis | |||||||
| Asthma Severity Modifying Polymorphisms Project, USA | 2010 | 6–20 | 154 | PCA | Two-step: | 3 | [45] |
| Hierarchical clustering | |||||||
| k-means clustering | |||||||
| SARP | 2011 | 6–17 | 161 | Ward’s hierarchical clustering | 4 | [46] | |
| Centroid linkage | |||||||
| Post hoc | |||||||
| Fisher discriminant analysis-predictors of cluster assignment | |||||||
| John Hunter Hospital Ambulatory Care Clinic, Newcastle, Australia | 2011 | 19–75 | 72 | Hierarchical clustering | 3 | [47] | |
| Complete linkage | |||||||
| TAP | 2012 | 6–12 | 315 | PCA | Two-step: | 3 | [42] |
| k-means clustering | |||||||
| Ward’s hierarchical clustering | |||||||
| Korean Genome Research Centre for Allergy and Respiratory Diseases cohort | 2012 | Adults | 86 | Two-step: | 4 | [48] | |
| Hierarchical cluster analysis | |||||||
| k-means clustering | |||||||
| NYUBAR, New York City, Bellevue Hospital Center Asthma Clinic | 2012 | 18–75 | 471 | Ward’s hierarchical clustering | 5 | [49] | |
| TAP | 2012 | 0–3 | 551 | Ward’s hierarchical clustering | 3 | [50] | |
| Post hoc | |||||||
| Classification and regression trees | |||||||
| Random forest for predictors of cluster assignment | |||||||
| TAP | 2012 | <36 mos | 79 | Ward’s hierarchical clustering | 3 | [51] | |
| TALC and BASALT trials, USA | 2012 | Avg age: 37.6 | 250 | Ward’s hierarchical clustering | 4 | [52] | |
| Post hoc | |||||||
| Discriminant analysis for predicting cluster membership | |||||||
| TAP | 2013 | 5 | 150 | Ward’s hierarchical clustering | 4 | [53] | |
| COREA Soonchunhyang University Asthma Genome Research Centre cohort |
2013 | >18 | 724 | Two-step: | 4 4 |
[54] | |
| 1843 | Ward’s hierarchical clustering | ||||||
| k-means | |||||||
| University of Tsukuba Hospital, Hokkaido University Hospital | 2013 | 16–84 | 800 | Ward’s hierarchical clustering | 6 | [55] | |
| Post hoc Classification and regression trees | |||||||
| Random forest for predictors of cluster assignment | |||||||
| Quebec City Case–Control Asthma Cohort | 2013 | Avg age: 35.7 | 377 | Factor analysis | Two-step: Ward’s hierarchical clustering |
4 | [56] |
| k-means | |||||||
| Niigata University Hospital, Japan | 2013 | Avg age: 59.8 | 86 | Step-wise multiple regression | Ward’s hierarchical clustering | 3 | [57] |
| Decision tree analysis for cluster assignment | |||||||
| Paediatric Asthma Clinic, Hacettepe University, Ankara, Turkey | 2013 | 6–18 | 383 | Factor analysis PCA |
Hierarchical clustering | 4 | [19] |
| Gower, Jaccard distances | |||||||
| Logistic models | |||||||
| Dutch multi-centre study | 2013 | Adults | 200 | Factor analysis | Wards hierarchical clustering | 3 | [58] |
| k-means | |||||||
| The epidemiology and natural history of asthma: outcomes and treatment regimens, San Diego, USA | 2014 | 6–11 >12 |
518 3612 |
Ward’s hierarchical clustering | Children: 5 Adults: 5 |
[59] | |
| SARP | 2014 | Adults | 378 | InfoGain [96] for relevant variables >0.2 Redundant variables removed by Markov blanket algorithm |
k-means to partition subjects | 6 | [60] |
| Ward’s hierarchical clustering | |||||||
| Outpatient clinics, Portugal | 2015 | Avg age: 45.6 | 57 | Ward’s hierarchical clustering | 5 clusters | [61] |
PCA principal components analysis, FA factor analysis, Avg average, SARP Severe Asthma Research Program (USA), GLAD (UK) GPIAG [General Practitioners in Asthma Group] and Leicester Asthma and Dysfunctional breathing study, TAP Trousseau Asthma Program (Paris, FR), COREA (Korea) Cohort for Reality and Evolution of Adult Asthma in Korea
Paediatric Studies
The Trousseau Asthma Program (TAP) in France used Ward’s hierarchical clustering as the method of choice [42, 50, 53]. In the TAP preschool population of 551 wheezers, ‘three clusters of wheezing’ were identified: mild episodic viral wheeze, atopic multiple-trigger wheeze, and non-atopic uncontrolled wheeze [50]. The mild episodic viral wheeze class was identified in one British [62] and one French cohort [63] using model-based approaches (see below), and the non-atopic uncontrolled wheeze cluster was reproduced in a separate TAP cohort [53]. The multiple-trigger wheeze was previously identified using supervised methods in the Avon Longitudinal Study of Parents and Children (ALSPAC) [64]. This cluster described children with either early- or late-onset persistent wheezing characterised by atopy and poor lung function. A similar description of wheezing was used in the MAAS cohort to demonstrate that persistent wheezing and multiple early atopy were associated with diminished lung function by age 11 years [65].
The clusters of wheezing described in the TAP cohort remained stable at age 5 years [53]. However, at school age, the clusters were different: ‘asthma with severe exacerbations and multiple allergies’, ‘severe asthma with bronchial obstruction’, and ‘mild asthma’ [42]. These accounted for two ‘phenotypes’: asthma with severe exacerbations, and multiple allergic severe asthma with bronchial obstruction [42]. It is important to note, however, that not only were the children from a separate cohort within the TAP, but the clustering methodology was also different; PCA was used for data reduction and a two-step clustering approach including k-means [42]. Furthermore, differing post hoc analyses were used.
The Severe Asthma Research Program (SARP) is a US multi-centre study comprising both children and adults with persistent asthma. The study by Fitzpatrick et al. [46] included 161 children aged 6–17 years. Variables were selected subjectively with no data reduction technique, and the authors derived ‘composite variables’ from binary and questionnaire data discerned by physicians. After Ward’s hierarchical clustering, four clusters were identified: ‘late-onset symptomatic asthma’, ‘early-onset atopic asthma and normal lung function’, ‘early-onset atopic asthma with mild airflow limitation and comorbidities’, and ‘early-onset atopic asthma with advanced airflow limitation’. These results and the accompanying clinical characteristics exhibited by the children were consistent with previously reported data from clinical observations [66–68]. However, these results differed from findings in a Turkish cohort of children aged 6–18 years with moderate–severe asthma [19]. In contrast to previous studies, the predictive ability of clusters and of original variables in relation to asthma severity in this population was relatively poor [19]. The authors concluded that the search for asthma subtypes needs careful selection of variables, which should be consistent across studies, and that a cautious interpretation of results is warranted [19].
Studies in Adults
The initial work that sparked further interest in clustering methodology was the study conducted by Haldar et al. in Leicester, UK [43]. A two-step Ward’s hierarchical and subsequent k-means cluster analysis was performed in three different data sets (refractory asthmatics from secondary care, primary care data, refractory asthmatics from clinical trial). After variable selection to identify ‘most clinically relevant’, PCA was performed, which reduced the variables into five components. Results of the subsequent cluster analysis revealed three clusters in the primary care data set and four clusters in the secondary care data. Two clusters were identified in each data set: ‘early-onset atopic asthma’ and ‘obese female with no eosinophilic inflammation’. The primary care data set identified a third ‘benign asthma’ cluster, while the secondary care set identified an ‘early-onset, symptom-predominant group with minimal eosinophils’ cluster as well as a ‘late-onset, male predominant, eosinophilic inflammation with few symptoms’ cluster. These results were then validated in the clinical trial data set, which revealed a three-cluster model similar to that in the secondary care set.
Expanding on Haldar’s findings, the SARP study [16], which included 726 patients older than 12 years, began with 628 variables, which were reduced to 34 by excluding missing data, text data, and redundant and ‘irrelevant’ variables. Half of the variables were composite. Ward’s method and post hoc discriminant analysis for tree analysis was performed to describe five clusters highly determined by frequency of symptoms, medication use, and lung function. Both studies identified a group of obese women with adult-onset asthma and less atopy, as well as a group of severe late-onset atopic asthmatics with poor lung function. However, SARP did not use sputum eosinophilia, which was an important feature in the Leicester study. A few years later, the SARP group used a different approach, and identified six clusters [60]. k-means clustering partitioned the 378 subjects, while Ward’s method clustered the 112 variables into 10 InfoGain (information gain—measures how well variables predict clusters)-ranked variable clusters based on symptoms, atopy, medication use, lung function, corticosteroid use and cause, Th2 inflammation, inflammatory markers, and demographics. Preprocessing of the data included imputing variables with less than 5% missing data while excluding those with more than 5%. Markov blanket algorithms identified redundant variables. Three clusters overlapped with previous results (severe asthmatics, female late-onset with normal lung function), while two were novel (late-onset severe eosinophilic asthmatics with nasal polyps, severe atopic Hispanics). It is interesting to note that similar clusters were seen in children from SARP and the Asthma Severity Modifying Polymorphisms (AsthMaP) Project [45], though the degree of lung function impairment was less.
Patrawalla et al. [49] based their clustering and variable selection technique on SARP, and identified clusters similar to those found by Wu et al. [60], though the Hispanic women had milder disease. This was explained by the fact that the sample was from an urban New York City population with a higher proportion of Hispanics.
The results obtained in the Leicester and SARP populations were reproduced in part in a Dutch cohort of patients with severe asthma that included more thorough inflammatory markers [58]. The resulting three clusters confirmed the existence of two previously reported clusters: ‘severe eosinophilic inflammation-predominant asthma with few symptoms and poor lung function’, and ‘obese late-onset asthma with low eosinophils additionally provoked by comorbidities such as gastrointestinal oesophageal reflux disease (GORD)’. The third cluster in the Dutch cohort (‘mild adult-onset well-controlled asthma’), which was not found in Leicester or SARP, had been seen in studies in Asian populations which included smoking status in their analysis [54, 55].
The recurring obesity-related subtypes were explored in more detail in two US trials comprising 250 adults [52]. With the incorporation of detailed data on inflammation, major differences were found between the obese and non-obese populations. Non-obese asthmatics had significantly better lung function. Obese patients with early-onset asthma and poor lung function had greater degrees of systemic inflammation (represented by the inverse association between hsCRP and GCRα); this was directly associated with increased glucocorticoid resistance (measured by reduced MKP-1 expression via dexamethasone).
Asthma Subtyping and Model-Based Approaches
Latent Variable Modelling
This topic was recently discussed in detail in another review article, which identified a total of 36 studies within the last 5 years that used model-based approaches to asthma subtyping (four in adult populations, 32 in children) [69]. Sample sizes in these studies ranged from 201 to 11,632. Methods included latent class analysis (14 studies), longitudinal latent class analysis (11 studies), latent class growth analysis (one study), latent growth mixture modelling (eight studies), and mixture models (two studies). The number of resulting classes ranged from three to eight, and were in most cases characterised by physician-diagnosed asthma, atopy, and/or FeNO. The most common outcome was ‘wheeze phenotype’ [64, 71–82], followed by ‘atopy class’ [64, 76, 81–86].
In these studies, the wheeze classes (often referred to as ‘phenotypes’, although by definition these were not observable, but latent) were described as either early-onset (transient [78, 87, 88] or prolonged [70]), late-onset (characterised as wheeze after age 3 years, persisting into later childhood) [70, 74, 78, 80, 83], or persistent (controlled and troublesome, characterised by diminished lung function by school age) [9, 74]. Early-onset wheeze was found to be predictive of poor lung function, but not atopy, eczema, or rhinitis at age 6–8 years [87]. Late-onset wheeze was associated with bronchial hyperresponsiveness and, in some cohorts, poorer lung function at age 6 years [64]. The persistent wheeze phenotype was consistently characterized by diminished lung function by school age [9, 74].
Atopic sensitisation was the second most common phenotype investigated by latent variable modelling, based on the hypothesis that distinct subtypes may be present. Simpson et al. applied a hidden Markov chain model to cluster children in MAAS into five sensitization classes using skin tests and specific IgE data at ages 1, 3, 5, and 8 years [83]. The underlying assumption was that children in each class had the same probability of becoming sensitized or resolving sensitization at each age (and to a similar panel of inhalant and food allergens), and that this differed between classes. Children in one of the four classes (comprising ~25% of sensitised participants), which the authors assigned as ‘multiple early atopy’, were much more likely to have asthma and worse lung function than children in any of the other classes [65, 83]. An almost-identical five-class model was identified by extending the analysis in MAAS through to age 11 years and, in another British birth cohort (Isle of Wight study), indicating stability over time and across different populations [84, 89]. However, these classes of sensitisation can be identified only by using statistical inference on longitudinal data, and differentiation between classes at any single cross-sectional point is currently not possible. This underscores the need to develop diagnostic tools that delineate different classes at any cross-sectional time point among the patient population, in order to facilitate the application of these findings in clinical practice [89–92].
In the adult population, Newby et al. performed a cluster analysis using mixture models on a multi-centre longitudinal observational study of 349 asthma patients in the British Thoracic Society Severe Refractory Asthma Registry [93]. Variables were initially restricted to those with less than 30% missing data that were non-categorical, and factor analysis was then applied. The resulting five factors (airflow obstruction, exacerbation frequency, IgE/BMI, treatment scaling, blood eosinophilia) were used in the cluster analysis to describe five clusters: (1) ‘early-onset atopic’, (2) ‘obese, late-onset’, (3) ‘normal lung function least severe asthma’, (4) ‘late-onset, eosinophilic’, and (5) ‘airflow obstruction’. The best-fitting models were chosen by the Akaike information criterion (AIC) or BIC, and the clusters were validated using a classifier on a separate data set from the same registry. Cluster stability for the whole group was only 52%, with cluster 2 accounting for 71% as the highest, while cluster 4 accounted for only 25%. A significant proportion of subjects in clusters 1, 4, and 5 moved to clusters 2 and 3 at follow-up, indicating greater obesity, lower blood eosinophilia, better lung function, and fewer exacerbations. Taking into account small differences in variables used, the results were broadly in accordance with previously reported clusters derived using model-free approaches [16, 43]. Gaussian mixture model clustering was also used to investigate cytokine response patterns of peripheral blood mononuclear cells to mite allergens, with results suggesting that asthma was associated with a broad range of immunophenotypes [94]. Various machine-learning approaches were also used to identify patterns of IgE responses to a large number of individual allergen molecules in component-resolved diagnostics microarrays and to associate these with asthma and allergic diseases [14].
Challenges in Asthma Clustering
Mixed Types of Data
Medicine generates many different types of data, including binary, numerical, and categorical variables, non-normal distributions, missing values, and outliers, and applying a model that combines these is challenging. One solution may be to transform the raw variables into a single type (i.e. all binary variables). Prosperi et al. [19] showed that, although results were vastly different when comparing the raw and binary variables, they were still clinically consistent with each other. However, in certain instances, changing continuous variables into binary variables would require the creation of categories. For example, if we take FEV1 and categorise it based on levels of obstruction (e.g. 80%, 60–80%, below 60%, above 80%), we assume that an FEV1 of 60% has the same clinical significance as an FEV1 of 79%, which is not necessarily true. Other issues with dichotomizing variables include a loss of information, leading to a reduction in statistical power, a loss of linear relationships between two groups, and underestimation of outcome variability between groups [95]. Another way to minimize this problem is to create clinically meaningful categories, but this will likely introduce an element of subjectivity.
Lack of Robustness to Choice of Variables and Clustering Methods
Different input parameters, even within the same data set, may produce different results. For example, in the SARP, the same hierarchical clustering techniques on the same data set produced different clusters [16, 46]. The major differences were in the preprocessing of the data and the cluster input. Wu et al. also included inflammatory markers in their analysis, which would account for better atopy delineation [60].
As mentioned previously, the choice of variables has been generally limited to consideration of expert opinion based on previous work. Furthermore, there is a practical consideration involved in that the variables chosen must correspond to the type of data in the cohort, given that some studies included all variables [58, 60, 61] in the data set, while others chose those that were ‘most relevant’ [42, 43, 48, 54, 55, 57]. This resulted in patient exclusion, particularly when there was a requirement to remove variables with missing data. Although some studies implemented imputation techniques in order to overcome this [60, 93], the impact on clinical outcome was not fully explored, which should be taken into account when interpreting the results.
In most studies, the choice of distance measure was not specified, and so it was assumed that the default measures in statistical packages were used (i.e. Euclidean distance). Only two studies [19, 44] specified varying the distance measures (Gower and/or Jaccard) to observe the effect. One study group used centroid linkage as their similarity measure, whereas the rest were based on Ward. Consequently, we cannot say whether the methods employed were the most reliable, as there is a repository containing hundreds of options.
Prosperi et al. hypothesized that clusters resulting from various studies differed because of variation in investigator choice of factors, encoding/categorization/transformation of variables, and methodology [19]. They proceeded to verify this using different hierarchical clustering and data reduction approaches on a cohort of children aged 6–18 years from the Paediatric Asthma Clinic in Ankara, Turkey. Data reduction was performed by both FA and PCA, resulting in five ‘dimensions’ of variables accounting for 35% of the variance. Multiple hierarchical clustering analyses were performed by varying the variable encoding scheme, distance linkages, feature selection, and dimensionality reduction space. Although the authors demonstrated that small variations in linkage-distance functions did not affect the resulting clusters [19], they tested only two, and it is possible that other linkage criteria may have influenced the results. Most significant was the fact that changes in variable encoding schemes and transformations resulted in different clusters [19]. While it is possible to test the strength of the methods employed by bootstrapping and/or multiple repetitions, this does not necessarily translate into more plausible results overall.
This is where model-free clustering runs into issues, and where a model-based approach might provide more structured methods, as MCMC and EM algorithms are applicable to all modelled distributions. However, in latent class analysis, there is no agreement on the optimal way to determine the number of classes. The most common method is the BIC, though other methods such as the AIC, likelihood tests, bootstrapping, and entropy have been used extensively, which may account for the different classes across populations.
Differing Subtypes Across Populations
It is clear that different clusters are identified across different populations (see Table 3). Other than differences in statistical methodologies, these disparities may be due to differences in features/variables selected to inform the mode (for example, the choice of lung function variables differed among studies, and post-bronchodilator FEV1 was included in only a few of these [43]). Of note, in addition to influencing heterogeneity in identified clusters, the non-inclusion of some of the potentially important variables (e.g. post-bronchodilator lung function) may result in failure to capture some important underlying mechanisms. Additionally, most studies were conducted in patients with severe or moderate–severe asthma, and the same subtypes may not be seen in the mild asthma population (Table 3).
It is also important to note that clusters identified cross-sectionally at a specific time point may not always be seen at different time points. Further longitudinal analysis is required to visualize how the clusters vary over time.
Conclusion
Our understanding of asthma has come a long way, and data-driven hypothesis-generating clustering methods have aided in identifying distinct subtypes. However, we must exercise caution when translating these results into clinical practice, as statistical inference on a large data set is needed to identify disease subtypes, and biomarkers that would allow differentiation of such subtypes at any cross-sectional time point are in most cases not available. Further challenges to the optimal use of clustering methodologies include tailoring models to individual data sets and incorporating genetic, epigenetic, and more detailed molecular-level data. The resulting models should then be able to accommodate large volumes of data in order to discern the developmental profiles of each individual, facilitating a genuinely personalised approach to asthma management.
Acknowledgments
No funding or sponsorship was received for this study or publication of this article. All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this manuscript, take responsibility for the integrity of the work as a whole, and have given final approval for the version to be published.
Disclosures
A. Custovic reports grants from the Medical Research Council, the JP Moulton Charitable Foundation, and the North West Lung Research Centre Charity, and personal fees from AstraZeneca, Novartis, ThermoFisher, Regeneron/Sanofi, and Novartis, outside the submitted work. M. Deliu is supported in part by UK Medical Research Council (MRC) grant MR/K002449/1 and MRC Health eResearch Centre (HeRC) grant MR/K006665/1. D. Belgrave is supported by MRC Career Development Award in Biostatistics grant MR/M015181/1. M. Sperrin has nothing to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not involve any new studies of human or animal subjects performed by any of the authors.
Footnotes
Enhanced Content
To view enhanced content for this article, go to http://www.medengine.com/Redeem/48D4F06023BDDEA2.
References
- 1.From the Global Strategy for Asthma Management and Prevention (GINA 2015). http://www.ginasthma.org/ . Accessed 21 Mar 2016.
- 2.Wenzel SE. Asthma: defining of the persistent adult phenotypes. Lancet. 2006;368(9537):804–813. doi: 10.1016/S0140-6736(06)69290-8. [DOI] [PubMed] [Google Scholar]
- 3.Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008;372(9643):1107–1119. doi: 10.1016/S0140-6736(08)61452-X. [DOI] [PubMed] [Google Scholar]
- 4.Lotvall J, et al. Asthma endotypes: a new approach to classification of disease entities within the asthma syndrome. J Allergy Clin Immunol. 2011;127(2):355–360. doi: 10.1016/j.jaci.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 5.Rackemann FM. A clinical survey of 1074 patients with asthma followed for 2 years. J Lab Clin Med. 1927;12:1185–1197. [Google Scholar]
- 6.Custovic A, et al. Exercise testing revisited. The response to exercise in normal and atopic children. Chest. 1994;105(4):1127–1132. doi: 10.1378/chest.105.4.1127. [DOI] [PubMed] [Google Scholar]
- 7.Vonk JM, et al. Risk factors associated with the presence of irreversible airflow limitation and reduced transfer coefficient in patients with asthma after 26 years of follow up. Thorax. 2003;58(4):322–327. doi: 10.1136/thorax.58.4.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavord ID, Agusti A. Blood eosinophil count: a biomarker of an important treatable trait in patients with airway disease. Eur Respir J. 2016;47(5):1299–1303. doi: 10.1183/13993003.00055-2016. [DOI] [PubMed] [Google Scholar]
- 9.Belgrave DC, Custovic A, Simpson A. Characterizing wheeze phenotypes to identify endotypes of childhood asthma, and the implications for future management. Expert Rev Clin Immunol. 2013;9(10):921–936. doi: 10.1586/1744666X.2013.836450. [DOI] [PubMed] [Google Scholar]
- 10.Martinez FD, et al. Asthma and wheezing in the first 6 years of life. The Group Health Medical Associates. N Engl J Med. 1995;332(3):133–138. doi: 10.1056/NEJM199501193320301. [DOI] [PubMed] [Google Scholar]
- 11.Lowe LA, et al. Wheeze phenotypes and lung function in preschool children. Am J Respir Crit Care Med. 2005;171(3):231–237. doi: 10.1164/rccm.200406-695OC. [DOI] [PubMed] [Google Scholar]
- 12.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18(5):716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 13.Custovic A, et al. The Study Team for Early Life Asthma Research (STELAR) consortium ‘Asthma e-lab’: team science bringing data, methods and investigators together. Thorax. 2015;70(8):799–801. doi: 10.1136/thoraxjnl-2015-206781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prosperi MC, et al. Challenges in interpreting allergen microarrays in relation to clinical symptoms: a machine learning approach. Pediatr Allergy Immunol. 2014;25(1):71–79. doi: 10.1111/pai.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prosperi MC, et al. Predicting phenotypes of asthma and eczema with machine learning. BMC Med Genom. 2014;7(Suppl 1):S7. doi: 10.1186/1755-8794-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore WC, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181(4):315–323. doi: 10.1164/rccm.200906-0896OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duda R, Hart P. Pattern classification and scene analysis. NY: Wiley; 1973. [Google Scholar]
- 18.Rokach L, Oded M. Clustering methods, in data mining and knowledge discovery handbook. NY:Springer; 2005. P.321–52.
- 19.Prosperi MC, et al. Challenges in identifying asthma subgroups using unsupervised statistical learning techniques. Am J Respir Crit Care Med. 2013;188(11):1303–1312. doi: 10.1164/rccm.201304-0694OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedregosa F, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 21.Hennig C. Cluster-wise assessment of cluster stability. London: University College London; 2006. [Google Scholar]
- 22.Smith JA, et al. Dimensions of respiratory symptoms in preschool children: population-based birth cohort study. Am J Respir Crit Care Med. 2008;177(12):1358–1363. doi: 10.1164/rccm.200709-1419OC. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez A, et al. Urbanisation is associated with prevalence of childhood asthma in diverse, small rural communities in Ecuador. Thorax. 2011;66(12):1043–1050. doi: 10.1136/thoraxjnl-2011-200225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenberg S, et al. Airway obstruction lability helps distinguish levels of disease activity in asthma. Respir Med. 2012;106(4):500–507. doi: 10.1016/j.rmed.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 25.Tromp II, et al. Dietary patterns and respiratory symptoms in pre-school children: the Generation R Study. Eur Respir J. 2012;40(3):681–689. doi: 10.1183/09031936.00119111. [DOI] [PubMed] [Google Scholar]
- 26.de Cassia Ribeiro Silva R, et al. Dietary patterns and wheezing in the midst of nutritional transition: a study in Brazil. Pediatr Allergy Immunol Pulmonol. 2013;26(1):18–24. doi: 10.1089/ped.2012.0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park HW, et al. Differences between asthma in young and elderly: results from the COREA study. Respir Med. 2013;107(10):1509–1514. doi: 10.1016/j.rmed.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 28.Patel S, et al. Cross-sectional association of dietary patterns with asthma and atopic sensitization in childhood—in a cohort study. Pediatr Allergy Immunol. 2014;25(6):565–571. doi: 10.1111/pai.12276. [DOI] [PubMed] [Google Scholar]
- 29.Al-Daghri NM, et al. Th1/Th2 cytokine pattern in Arab children with severe asthma. Int J Clin Exp Med. 2014;7(8):2286–2291. [PMC free article] [PubMed] [Google Scholar]
- 30.Chawes BL, et al. Neonates with reduced neonatal lung function have systemic low-grade inflammation. J Allergy Clin Immunol. 2015;135(6):1450–1456. doi: 10.1016/j.jaci.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clemmer GL, et al. Measuring the corticosteroid responsiveness endophenotype in asthmatic patients. J Allergy Clin Immunol. 2015;136(2):274–281. doi: 10.1016/j.jaci.2015.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey WC, et al. Asthma severity: a factor analytic investigation. Am J Med. 1992;93(3):263–269. doi: 10.1016/0002-9343(92)90231-Y. [DOI] [PubMed] [Google Scholar]
- 33.Rosi E, et al. Sputum analysis, bronchial hyperresponsiveness, and airway function in asthma: results of a factor analysis. J Allergy Clin Immunol. 1999;103(2 Pt 1):232–237. doi: 10.1016/S0091-6749(99)70496-3. [DOI] [PubMed] [Google Scholar]
- 34.Sunyer J, et al. International assessment of the internal consistency of respiratory symptoms. European Community Respiratory Health Study (ECRHS) Am J Respir Crit Care Med. 2000;162(3 Pt 1):930–935. doi: 10.1164/ajrccm.162.3.9911062. [DOI] [PubMed] [Google Scholar]
- 35.Holberg CJ, et al. Factor analysis of asthma and atopy traits shows 2 major components, one of which is linked to markers on chromosome 5q. J Allergy Clin Immunol. 2001;108(5):772–780. doi: 10.1067/mai.2001.119158. [DOI] [PubMed] [Google Scholar]
- 36.Grazzini M, et al. Relevance of dyspnoea and respiratory function measurements in monitoring of asthma: a factor analysis. Respir Med. 2001;95(4):246–250. doi: 10.1053/rmed.2000.1017. [DOI] [PubMed] [Google Scholar]
- 37.Juniper EF, et al. Relationship between quality of life and clinical status in asthma: a factor analysis. Eur Respir J. 2004;23(2):287–291. doi: 10.1183/09031936.04.00064204. [DOI] [PubMed] [Google Scholar]
- 38.Schatz M, et al. Relationships among quality of life, severity, and control measures in asthma: an evaluation using factor analysis. J Allergy Clin Immunol. 2005;115(5):1049–1055. doi: 10.1016/j.jaci.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 39.Leung TF, et al. Clinical and atopic parameters and airway inflammatory markers in childhood asthma: a factor analysis. Thorax. 2005;60(10):822–826. doi: 10.1136/thx.2004.039321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holt EW, et al. Identifying the components of asthma health status in children with mild to moderate asthma. J Allergy Clin Immunol. 2008;121(5):1175–1180. doi: 10.1016/j.jaci.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyake Y, et al. Maternal dietary patterns during pregnancy and risk of wheeze and eczema in Japanese infants aged 16–24 months: the Osaka Maternal and Child Health Study. Pediatr Allergy Immunol. 2011;22(7):734–741. doi: 10.1111/j.1399-3038.2011.01176.x. [DOI] [PubMed] [Google Scholar]
- 42.Just J, et al. Two novel, severe asthma phenotypes identified during childhood using a clustering approach. Eur Respir J. 2012;40(1):55–60. doi: 10.1183/09031936.00123411. [DOI] [PubMed] [Google Scholar]
- 43.Haldar P, et al. Cluster analysis and clinical asthma phenotypes. Am J Respir Crit Care Med. 2008;178(3):218–224. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weatherall M, et al. Distinct clinical phenotypes of airways disease defined by cluster analysis. Eur Respir J. 2009;34(4):812–818. doi: 10.1183/09031936.00174408. [DOI] [PubMed] [Google Scholar]
- 45.Benton AS, et al. Overcoming heterogeneity in pediatric asthma: tobacco smoke and asthma characteristics within phenotypic clusters in an African American cohort. J Asthma. 2010;47(7):728–734. doi: 10.3109/02770903.2010.491142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fitzpatrick AM, et al. Heterogeneity of severe asthma in childhood: confirmation by cluster analysis of children in the National Institutes of Health/National Heart, Lung, and Blood Institute Severe Asthma Research Program. J Allergy Clin Immunol. 2011;127(2):382–389. doi: 10.1016/j.jaci.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baines KJ, et al. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol. 2011;127(1):153–160. doi: 10.1016/j.jaci.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 48.Jang AS, et al. Identification of subtypes of refractory asthma in Korean patients by cluster analysis. Lung. 2013;191(1):87–93. doi: 10.1007/s00408-012-9430-8. [DOI] [PubMed] [Google Scholar]
- 49.Patrawalla P, et al. Application of the asthma phenotype algorithm from the Severe Asthma Research Program to an urban population. PLoS One. 2012;7(9):e44540. doi: 10.1371/journal.pone.0044540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Just J, et al. Novel severe wheezy young children phenotypes: boys atopic multiple-trigger and girls nonatopic uncontrolled wheeze. J Allergy Clin Immunol. 2012;130(1):103–110. doi: 10.1016/j.jaci.2012.02.041. [DOI] [PubMed] [Google Scholar]
- 51.Gouvis-Echraghi R, et al. Exhaled nitric oxide measurement confirms 2 severe wheeze phenotypes in young children from the Trousseau Asthma Program. J Allergy Clin Immunol. 2012;130(4):1005–1007. doi: 10.1016/j.jaci.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 52.Sutherland ER, et al. Cluster analysis of obesity and asthma phenotypes. PLoS One. 2012;7(5):e36631. doi: 10.1371/journal.pone.0036631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Just J, et al. Wheeze phenotypes in young children have different courses during the preschool period. Ann Allergy Asthma Immunol. 2013;111(4):256–261. doi: 10.1016/j.anai.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 54.Kim TB, et al. Identification of asthma clusters in two independent Korean adult asthma cohorts. Eur Respir J. 2013;41(6):1308–1314. doi: 10.1183/09031936.00100811. [DOI] [PubMed] [Google Scholar]
- 55.Kaneko Y, et al. Asthma phenotypes in Japanese adults—their associations with the CCL5 and ADRB2 genotypes. Allergol Int. 2013;62(1):113–121. doi: 10.2332/allergolint.12-OA-0467. [DOI] [PubMed] [Google Scholar]
- 56.Lavoie-Charland E, et al. Multivariate asthma phenotypes in adults: the Quebec City case-control asthma cohort. Open J Respir Dis. 2013;03(04):10. [Google Scholar]
- 57.Sakagami T, et al. Cluster analysis identifies characteristic phenotypes of asthma with accelerated lung function decline. J Asthma. 2014;51(2):113–118. doi: 10.3109/02770903.2013.852201. [DOI] [PubMed] [Google Scholar]
- 58.Amelink M, et al. Three phenotypes of adult-onset asthma. Allergy. 2013;68(5):674–680. doi: 10.1111/all.12136. [DOI] [PubMed] [Google Scholar]
- 59.Schatz M, et al. Phenotypes determined by cluster analysis in severe or difficult-to-treat asthma. J Allergy Clin Immunol. 2014;133(6):1549–1556. doi: 10.1016/j.jaci.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 60.Wu W, et al. Unsupervised phenotyping of Severe Asthma Research Program participants using expanded lung data. J Allergy Clin Immunol. 2014;133(5):1280–1288. doi: 10.1016/j.jaci.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loureiro CC, et al. Cluster analysis in phenotyping a Portuguese population. Rev Port Pneumol (2006). 2015. [DOI] [PubMed]
- 62.Spycher BD, et al. Distinguishing phenotypes of childhood wheeze and cough using latent class analysis. Eur Respir J. 2008;31(5):974–981. doi: 10.1183/09031936.00153507. [DOI] [PubMed] [Google Scholar]
- 63.Herr M, et al. Risk factors and characteristics of respiratory and allergic phenotypes in early childhood. J Allergy Clin Immunol. 2012;130(2):389–396. doi: 10.1016/j.jaci.2012.05.054. [DOI] [PubMed] [Google Scholar]
- 64.Henderson J, et al. Associations of wheezing phenotypes in the first 6 years of life with atopy, lung function and airway responsiveness in mid-childhood. Thorax. 2008;63(11):974–980. doi: 10.1136/thx.2007.093187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Belgrave DCM, et al. Trajectories of lung function during childhood. Am J Respir Crit Care Med. 2014;189(9):1101–1109. doi: 10.1164/rccm.201309-1700OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bossley CJ, et al. Corticosteroid responsiveness and clinical characteristics in childhood difficult asthma. Eur Respir J. 2009;34(5):1052–1059. doi: 10.1183/09031936.00186508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chipps BE, et al. Demographic and clinical characteristics of children and adolescents with severe or difficult-to-treat asthma. J Allergy Clin Immunol. 2007;119(5):1156–1163. doi: 10.1016/j.jaci.2006.12.668. [DOI] [PubMed] [Google Scholar]
- 68.Bacharier LB, et al. Classifying asthma severity in children: mismatch between symptoms, medication use, and lung function. Am J Respir Crit Care Med. 2004;170(4):426–432. doi: 10.1164/rccm.200308-1178OC. [DOI] [PubMed] [Google Scholar]
- 69.Howard R, et al. Distinguishing asthma phenotypes using machine learning approaches. Curr Allergy Asthma Rep. 2015;15(7):38. doi: 10.1007/s11882-015-0542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Savenije OE, et al. Comparison of childhood wheezing phenotypes in 2 birth cohorts: ALSPAC and PIAMA. J Allergy Clin Immunol. 2011;127(6):1505–1512. doi: 10.1016/j.jaci.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 71.Chen Q, et al. Using latent class growth analysis to identify childhood wheeze phenotypes in an urban birth cohort. Ann Allergy Asthma Immunol. 2012;108(5):311–315. doi: 10.1016/j.anai.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weinmayr G, et al. Asthma phenotypes identified by latent class analysis in the ISAAC phase II Spain study. Clin Exp Allergy. 2013;43(2):223–232. doi: 10.1111/cea.12035. [DOI] [PubMed] [Google Scholar]
- 73.Spycher BD, et al. Comparison of phenotypes of childhood wheeze and cough in 2 independent cohorts. J Allergy Clin Immunol. 2013;132(5):1058–1067. doi: 10.1016/j.jaci.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 74.Belgrave DC, et al. Joint modeling of parentally reported and physician-confirmed wheeze identifies children with persistent troublesome wheezing. J Allergy Clin Immunol. 2013;132(3):575–583. doi: 10.1016/j.jaci.2013.05.041. [DOI] [PubMed] [Google Scholar]
- 75.Cano-Garcinuno A, Mora-Gandarillas I, S.S. Group Wheezing phenotypes in young children: an historical cohort study. Prim Care Respir J. 2014;23(1):60–66. doi: 10.4104/pcrj.2014.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Panico L, et al. Asthma trajectories in early childhood: identifying modifiable factors. PLoS One. 2014;9(11):e111922. doi: 10.1371/journal.pone.0111922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Depner M, et al. Clinical and epidemiologic phenotypes of childhood asthma. Am J Respir Crit Care Med. 2014;189(2):129–138. doi: 10.1164/rccm.201307-1198OC. [DOI] [PubMed] [Google Scholar]
- 78.Caudri D, et al. Perinatal risk factors for wheezing phenotypes in the first 8 years of life. Clin Exp Allergy. 2013;43(12):1395–1405. doi: 10.1111/cea.12173. [DOI] [PubMed] [Google Scholar]
- 79.Lodge CJ, et al. Childhood wheeze phenotypes show less than expected growth in FEV1 across adolescence. Am J Respir Crit Care Med. 2014;189(11):1351–1358. doi: 10.1164/rccm.201308-1487OC. [DOI] [PubMed] [Google Scholar]
- 80.Savenije OE, et al. Association of IL33-IL-1 receptor-like 1 (IL1RL1) pathway polymorphisms with wheezing phenotypes and asthma in childhood. J Allergy Clin Immunol. 2014;134(1):170–177. doi: 10.1016/j.jaci.2013.12.1080. [DOI] [PubMed] [Google Scholar]
- 81.Belgrave DC, et al. Developmental profiles of eczema, wheeze, and rhinitis: two population-based birth cohort studies. PLoS Med. 2014;11(10):e1001748. doi: 10.1371/journal.pmed.1001748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Siroux V, et al. Identifying adult asthma phenotypes using a clustering approach. Eur Respir J. 2011;38(2):310–317. doi: 10.1183/09031936.00120810. [DOI] [PubMed] [Google Scholar]
- 83.Simpson A, et al. Beyond atopy: multiple patterns of sensitization in relation to asthma in a birth cohort study. Am J Respir Crit Care Med. 2010;181(11):1200–1206. doi: 10.1164/rccm.200907-1101OC. [DOI] [PubMed] [Google Scholar]
- 84.Lazic N, et al. Multiple atopy phenotypes and their associations with asthma: similar findings from two birth cohorts. Allergy. 2013;68(6):764–770. doi: 10.1111/all.12134. [DOI] [PubMed] [Google Scholar]
- 85.Garden FL, SJ, Marks G. Atopu phenotypes in he Childhood Asthma Prevention Study (CAPS) cohort and the relationship with allergic disease: clinical mechanisms in allergic disease. J Br Soc Allergy Clin Immunol. 2013;43(6):633–41. [DOI] [PubMed]
- 86.Havstad S, et al. Atopic phenotypes identified with latent class analyses at age 2 years. J Allergy Clin Immunol. 2014;134(3):722–727. doi: 10.1016/j.jaci.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Savenije OE, et al. Comparison of childhood wheezing phenotypes in 2 birth cohorts: ALSPAC and PIAMA. J Allergy Clin Immunol. 2011;127(6):1505–1512. doi: 10.1016/j.jaci.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 88.Savenije OE, et al. Association of IL33-IL-1 receptor-like 1 (IL1RL1) pathway polymorphisms with wheezing phenotypes and asthma in childhood. J Allergy Clin Immunol. 2014. [DOI] [PubMed]
- 89.Custovic A, Lazic N, Simpson A. Pediatric asthma and development of atopy. Curr Opin Allergy Clin Immunol. 2013;13(2):173–180. doi: 10.1097/ACI.0b013e32835e82b6. [DOI] [PubMed] [Google Scholar]
- 90.Holt PG, et al. Distinguishing benign from pathologic TH2 immunity in atopic children. J Allergy Clin Immunol. 2016;137(2):379–387. doi: 10.1016/j.jaci.2015.08.044. [DOI] [PubMed] [Google Scholar]
- 91.Custovic A, et al. Evolution pathways of IgE responses to grass and mite allergens throughout childhood. J Allergy Clin Immunol. 2015;136(6):1645–1652. doi: 10.1016/j.jaci.2015.03.041. [DOI] [PubMed] [Google Scholar]
- 92.Simpson A, et al. Patterns of IgE responses to multiple allergen components and clinical symptoms at age 11 years. J Allergy Clin Immunol. 2015;136(5):1224–1231. doi: 10.1016/j.jaci.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Newby C, et al. Statistical cluster analysis of the British Thoracic Society Severe refractory Asthma Registry: clinical outcomes and phenotype stability. PLoS One. 2014;9(7):e102987. doi: 10.1371/journal.pone.0102987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu J, et al. Relationship between cytokine expression patterns and clinical outcomes: two population-based birth cohorts. Clin Exp Allergy. 2015;45(12):1801–1811. doi: 10.1111/cea.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Altman DG, Royston P. The cost of dichotomising continuous variables. BMJ. 2006;332(7549):1080. doi: 10.1136/bmj.332.7549.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mitchell T. Machine learning. Maidenheach: McGraw-Hill Science; 1997. [Google Scholar]