ABSTRACT
Efficient microbial utilization of cellulosic sugars is essential for the economic production of biofuels and chemicals. Although the yeast Saccharomyces cerevisiae is a robust microbial platform widely used in ethanol plants using sugar cane and corn starch in large-scale operations, glucose repression is one of the significant barriers to the efficient fermentation of cellulosic sugar mixtures. A recent study demonstrated that intracellular utilization of cellobiose by engineered yeast expressing a cellobiose transporter (encoded by cdt-1) and an intracellular β-glucosidase (encoded by gh1-1) can alleviate glucose repression, resulting in the simultaneous cofermentation of cellobiose and nonglucose sugars. Here we report enhanced cellobiose fermentation by engineered yeast expressing cdt-1 and gh1-1 through laboratory evolution. When cdt-1 and gh1-1 were integrated into the genome of yeast, the single copy integrant showed a low cellobiose consumption rate. However, cellobiose fermentation rates by engineered yeast increased gradually during serial subcultures on cellobiose. Finally, an evolved strain exhibited a 15-fold-higher cellobiose fermentation rate. To identify the responsible mutations in the evolved strain, genome sequencing was performed. Interestingly, no mutations affecting cellobiose fermentation were identified, but the evolved strain contained 9 copies of cdt-1 and 23 copies of gh1-1. We also traced the copy numbers of cdt-1 and gh1-1 of mixed populations during the serial subcultures. The copy numbers of cdt-1 and gh1-1 in the cultures increased gradually with similar ratios as cellobiose fermentation rates of the cultures increased. These results suggest that the cellobiose assimilation pathway (transport and hydrolysis) might be a rate-limiting step in engineered yeast and copies of genes coding for metabolic enzymes might be amplified in yeast if there is a growth advantage. This study indicates that on-demand gene amplification might be an efficient strategy for yeast metabolic engineering.
IMPORTANCE In order to enable rapid and efficient fermentation of cellulosic hydrolysates by engineered yeast, we delve into the limiting factors of cellobiose fermentation by engineered yeast expressing a cellobiose transporter (encoded by cdt-1) and an intracellular β-glucosidase (encoded by gh1-1). Through laboratory evolution, we isolated mutant strains capable of fermenting cellobiose much faster than a parental strain. Genome sequencing of the fast cellobiose-fermenting mutant reveals that there are massive amplifications of cdt-1 and gh1-1 in the yeast genome. We also found positive and quantitative relationships between the rates of cellobiose consumption and the copy numbers of cdt-1 and gh1-1 in the evolved strains. Our results suggest that the cellobiose assimilation pathway (transport and hydrolysis) might be a rate-limiting step for efficient cellobiose fermentation. We demonstrate the feasibility of optimizing not only heterologous metabolic pathways in yeast through laboratory evolution but also on-demand gene amplification in yeast, which can be broadly applicable for metabolic engineering.
INTRODUCTION
The efficient utilization of sugars prevalent in cellulosic hydrolysates by microorganisms is essential to develop a bioeconomy through the cost-effective production of biofuels and value-added chemicals (1). Sugarcane and corn starch have been used as sugar sources for value-added biotransformation based on microbial fermentation, but the “food versus biofuel” issue has been a concern (2). Cellulosic biomass could provide an ethical and sustainable solution (3). Sugarcane and starch contain only hexoses, but cellulosic biomass consists of various hexoses and pentoses. Glucose and xylose are the primary sugars in cellulosic hydrolysates (4). Therefore, efficient microbial fermentation of both hexose and pentose sugars is necessary to produce biofuels such as ethanol (5).
The hexose-fermenting Saccharomyces cerevisiae has been employed for the large-scale fermentation of sugarcane and corn starch, as well as for many other industrial fermentations for the production of value-added chemicals (6, 7). In addition, genetic tools of S. cerevisiae have been well developed so that sophisticated genetic modifications can be readily performed (8). Nonetheless, S. cerevisiae cannot natively ferment xylose, which is the second most abundant sugar in cellulosic hydrolysates. Therefore, metabolic engineering efforts to construct xylose-fermenting S. cerevisiae have been made for over 2 decades (9). A common strategy for the construction is to introduce either the oxidoreductase pathway consisting of xylose reductase (XR) and xylitol dehydrogenase (XDH) or the single-step isomerization pathway catalyzed by xylose isomerase (XI) (10–17). After introducing the xylose assimilation pathways, engineered S. cerevisiae was able to convert xylose into ethanol but utilized glucose preferentially from mixtures of glucose and xylose (18, 19). This “glucose repression” problem is a major barrier to efficient fermentation using sugar mixtures from terrestrial and marine biomass (20, 21). To mitigate this issue, direct fermentation of cellobiose, a dimer of glucose, by engineered yeast has been proposed (22, 23). Wild-type S. cerevisiae cannot utilize cellobiose, but the introduction of a cellodextrin transporter gene (cdt-1) and an intracellular β-glucosidase gene (gh1-1) from Neurospora crassa allowed cellobiose assimilation in S. cerevisiae (23). The extracellular glucose can inhibit xylose uptake, while the intracellular glucose does not impede xylose transport. As cellobiose is hydrolyzed into glucose intracellularly in engineered yeast, glucose repression on xylose can be alleviated so that engineered yeast with both xylose- and cellobiose-assimilating pathways was able to ferment cellobiose and xylose simultaneously. Moreover, the simultaneous cofermentation of cellobiose and xylose led to a higher ethanol yield and productivity than those obtained with the sequential fermentation of glucose and xylose (22).
There are two potential problems with cellobiose fermentation in prior studies (22, 24). First, the engineered yeast contained the episomal plasmids that can result in plasmid loss in nonselective medium due to a metabolic burden on the host (25). Second, the engineered yeast accumulated substantial amounts of extracellular cellodextrin due to the transglycosylation activity of β-glucosidase when there is a mismatch between glucose production and consumption rates during the cellobiose fermentation. While the accumulated cellodextrin can be reassimilated later, it might decrease ethanol productivity (22). In this study, we aimed to solve these two problems in the cellobiose-fermenting yeast constructed using episomal plasmids. To alleviate these issues, we integrated the cellobiose assimilation pathway into the genome and performed laboratory evolution using cellobiose as the sole carbon source. When we introduced the cdt-1 and gh1-1 genes into the genome of S. cerevisiae, the engineered strain showed a low cellobiose consumption rate. However, we observed that the cellobiose fermentation rate increased significantly after serial subcultures on cellobiose. After isolating the evolved strains, we identified the genetic changes responsible for the improved phenotype of the evolved strain through genome sequencing. Interestingly, the improved cellobiose fermentation with reduced cellodextrin accumulation was not caused by mutations in endogenous genes of S. cerevisiae but instead by greatly increasing the copy numbers of cdt-1 and gh1-1 with a presumably optimized copy number ratio of 1:2, respectively. These results suggest that copies of genes coding for metabolic enzymes might be amplified in yeast if there is a growth advantage and the cellobiose assimilation pathway (transport and hydrolysis) might be a rate-limiting step of cellobiose fermentation by engineered S. cerevisiae.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The yeast plasmids and strains used in this study are summarized in Table 1. Escherichia coli DH5 [F− recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA relA1] (Invitrogen, Gaithersburg, MD) was used for gene cloning and manipulation. E. coli was grown in Luria-Bertani medium; 50 μg/ml of ampicillin was added as required. Yeast strains were routinely cultivated at 30°C in YP medium (10 g/liter of yeast extract and 20 g/liter of peptone) with 20 g/liter of glucose or 20 g/liter of cellobiose. To select the transformants using an auxotrophic marker, yeast synthetic complete (YSC) medium was used, which contained 6.7 g/liter of yeast nitrogen base (YNB) plus 20 g/liter of glucose, 20 g/liter of agar, and CSM-Leu-Trp-Ura-His (MP Biomedicals, Santa Ana, CA) with a supply of appropriate nucleotides and amino acids.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| S. cerevisiae D452-2 | MATα leu2 his3 ura3 can1 | 43 |
| S. cerevisiae CEN.PK2-1D | MATα ura3-52 trp1-289 leu2-3,112 his3Δ1 MAL2-8C SUC2 | 44 |
| S. cerevisiae EJ1 | D452-2 leu2::LEU2 pRS405-gh1-1 ura3::URA3 pRS406-cdt-1 | This study |
| S. cerevisiae EJ2 | Evolved strain of EJ1 | This study |
| S. cerevisiae CEN-C1B1 | CEN.PK2-1D/pRS424-gh1-1/pRS426-cdt-1 | This study |
| S. cerevisiae CEN-C1B2 | CEN.PK2-1D/pRS424-gh1-1/pRS425-gh1-1/pRS426-cdt-1 | This study |
| S. cerevisiae CEN-C1B3 | CEN.PK2-1D/pRS423-gh1-1/pRS424-gh1-1/pRS425-gh1-1/pRS426-cdt-1 | This study |
| S. cerevisiae CEN-C2B1 | CEN.PK2-1D/pRS425-gh1-1/pRS424-cdt-1/pRS426-cdt-1 | This study |
| S. cerevisiae CEN-C3B1 | CEN.PK2-1D/pRS425-gh1-1/pRS423-cdt-1/pRS424-cdt-1/pRS426-cdt-1 | This study |
| S. cerevisiae EJ1 svl3Δ | EJ1svl3::KanMX4 | This study |
| S. cerevisiae EJ1 svl3Δ-1S | EJ1 svl3Δ/pRS423-EJ1 SVL3 | This study |
| S. cerevisiae EJ1 svl3Δ-2S | EJ1 svl3Δ/pRS423-EJ2 SVL3 | This study |
| S. cerevisiae DCDT-1G | D452-2/pRS425-gh1-1/pRS426-cdt-1 | 24 |
| Plasmids | ||
| pRS405 | Integration plasmid, LEU2 | 45 |
| pRS406 | Integration plasmid, URA3 | 45 |
| pRS423 | 2μm origin, HIS3 | 46 |
| pRS424 | 2μm origin, TRP1 | 46 |
| pRS425 | 2μm origin, LEU2 | 46 |
| pRS426 | 2μm origin, URA3 | 46 |
| pRS405-gh1-1 | pRS405 PGK1p-gh1-1-CYC1t | This study |
| pRS423-gh1-1 | pRS423 PGK1p-gh1-1-CYC1t | This study |
| pRS424-gh1-1 | pRS424 PGK1p-gh1-1-CYC1t | This study |
| pRS425-gh1-1 | pRS425 PGK1p-gh1-1-CYC1t | 23 |
| pRS406-cdt-1 | pRS406 PGK1p-cdt-1-CYC1t | This study |
| pRS423-cdt-1 | pRS423 PGK1p-cdt-1-CYC1t | This study |
| pRS424-cdt-1 | pRS424 PGK1p-cdt-1-CYC1t | This study |
| pRS426-cdt-1 | pRS426 PGK1p-cdt-1-CYC1t | 23 |
| pRS423-EJ1 SVL3 | pRS423 TDH3p-SVL3 from EJ1-CYC1t | This study |
| pRS423-EJ2 SVL3 | pRS423 TDH3p-SVL3 from EJ2-CYC1t | This study |
Strains and plasmid construction.
All of the primers used for constructing plasmids are listed in Table S1 in the supplemental material. We used the CloneEZ PCR cloning kit (Genscript, Piscataway, NJ) to construct plasmids expressing β-glucosidase (encoded by gh1-1) and cellodextrin transporter (encoded by cdt-1) from N. crassa. The plasmids pRS425-gh1-1 and pRS426-cdt-1 were constructed in a previous study to overexpress β-glucosidase (gh1-1) and cellodextrin transporter (cdt-1) under the control of the PGK1 promoter and CYC1 terminator (23). Each expression cassette of β-glucosidase and cellodextrin transporter was amplified by PCR between the T3 and T7 sites on both pRS425-gh1-1 and pRS426-cdt-1. Linearized vectors were prepared by PCR, amplifying the backbone region (including the auxotrophic markers and replicons) between the T3 and T7 primer binding sites of templates pRS405, pRS406, pRS423, pRS424, pRS425, and pRS426. The expression cassettes for the T3-to-T7 section were cloned to the linearized vectors by a recombination procedure. The resulting plasmids pRS405-gh1-1 and pRS406-cdt-1 were integrated into the LEU2 and URA3 loci of S. cerevisiae strain D452-2, yielding strain EJ1. S. cerevisiae strain CEN.PK2-1D was used as a host strain to express β-glucosidase (gh1-1) and cellodextrin transporter (cdt-1) using multicopy plasmids. The yeast EZ-Transformation kit (MP Biomedicals, Santa Ana, CA) was used to transform the expression cassettes. The transformants were selected on YSC agar medium containing 20 g/liter of glucose. Amino acids and nucleotides were added as necessary.
To construct the SVL3 deletion mutants, the svl3Δ::KanMX4 cassette was amplified by PCR from the genomic DNA of the BY4742 svl3Δ strain in the Yeast Knockout Collection (Open Biosystems, Pittsburgh, PA). The PCR product was purified and integrated into strain EJ1, and the deletion mutant was selected on YP medium (10 g/liter of yeast extract and 20 g/liter of peptone) containing 20 g/liter of glucose with 500 μg/ml of G418. The S. cerevisiae SVL3 gene was PCR amplified from the genomic DNA of strains EJ1 and EJ2 using the primers SVL3 cloning-F and SVL3 cloning-R. Each amplified DNA fragment was cloned into the plasmid pRS423 under the control of the TDH3 promoter and CYC1 terminator. The transformation of the constructed plasmids was performed using the yeast EZ-Transformation kit (MP Biomedicals, Santa Ana, CA).
Fermentation experiments.
The yeast cells were grown in YP medium containing 20 g/liter of cellobiose to prepare inoculums for the fermentation experiments. The cells were harvested in the mid-exponential phase (optical density at 600 nm [OD600], 1.0) and inoculated into the experimental flasks after washing them twice with sterilized water. The 125-ml flasks for fermentation contained 25 ml of YP medium with the appropriate sugars at 30°C with an initial OD600 of 1.0 or 0.1 under oxygen-limited conditions.
Preparation of crude extract and β-glucosidase activity assay.
Yeast cells grown to mid-log phase (OD600 = 1.0) at 30°C in YP medium with 20 g/liter of cellobiose were harvested by centrifugation at 3,000 × g for 5 min. The cell pellets were washed and suspended in Y-PER solution (Pierce, Rockford, IL). After incubation at room temperature for 20 min, the cell suspension was centrifuged at 13,000 rpm for 10 min to completely remove the cell debris. The supernatant (crude enzyme extract) was kept on ice for the enzyme assay. The β-glucosidase activity was measured according to the previous method (23) in a reaction solution containing 50 mM sodium acetate buffer (pH 4.8) and 6.7 mM para-nitrophenyl β-d-glucopyranoside (pNPG). The release of p-nitrophenol (NP) was monitored by a microplate reader (Synergy 2; Biotek, Winooski, VT) at 405 nm during 1 h of incubation at 30°C. The p-nitrophenol absorption coefficient was 18.4 mM−1 cm−1. One unit of enzyme activity is defined as the amount of enzyme that catalyzes 1 μmol of substrate per min at 30°C. The protein concentration was determined by the bicinchoninic acid (BCA) method (Pierce, Rockford, IL).
Quantitative PCR for determining genomic copy numbers of gh1-1 and cdt-1.
The genomic DNA of the strain from each subculture was prepared with the YeaStar Genomic DNA kit (Zymo Research, Orange, CA) and quantified by NanoDrop ND-1000 (Thermo Fisher Scientific, Wilmington, DE). Real-time PCR was performed using SYBR green I Master (Roche) on a Lightcycler 480 instrument (Roche Applied Science, Indianapolis, IN) according to the manufacturer's directions. As shown in Table S1 in the supplemental material, the primers were designed to detect the gh1-1 and cdt-1 genes. The concentrations of the genes were quantified by standard curves generated by the gh1-1 and cdt-1 fragments purified from pRS425-gh1-1 and pRS426-cdt-1, respectively. The calculations of the genomic copy numbers were determined by a comparison of the relative concentrations of the genes obtained by quantitative PCR, as described by Kim et al. (26).
Genome sequencing, SNP discovery, and estimation of copy numbers.
The genomic DNA from S. cerevisiae EJ1 and EJ2 was prepared using the YeaStar Genomic DNA kit (Zymo Research, Irvine, CA). The genome sequencing was performed using an Illumina HiSeq2000 machine in the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign. The barcoded shotgun genomic DNA libraries were constructed with the TruSeq DNAseq Sample Prep kit (Illumina, San Diego, CA). Diluted libraries were mixed (approximately equimolar) to a final total concentration of 10 pM for loading. The diluted equimolar libraries (10 nM) were multiplexed on a lane and sequenced using single reads for 100 nucleotides (nt) by the Illumina HiSeq2000, with SBS chemistry version 2. The raw data were processed by Casava 1.7. Single nucleotide polymorphism (SNP) discovery and the estimation of copy numbers were performed using CLC Genomics Workbench 7.0.2 (CLC Bio, Aarhus, Denmark). The sequence reads were trimmed with a quality score limit of 0.05 and assembled into reference sequences. As reference sequences, S. cerevisiae S288C was used for the SNP discovery, as were the combinations of the RT-qPCR reference genes commonly used for quantitative gene expression (27). The average read depth values for ACT1, IPP1, and PDA1 were used to estimate the copy numbers of gh1-1 and cdt-1 to get the average coverage for them. Probabilistic variant detection was performed with a minimum coverage of 10 and a probability of 90, and these variants were compared to the reference variants to discover SNPs.
PFGE.
Chromosomal DNA was prepared with the contour-clamped homogeneous electric field (CHEF) genomic DNA plug kit (Bio-Rad, Hercules, CA). The agarose plugs containing yeast chromosomes were loaded to a 0.8% agarose gel in a 0.5× Tris-borate-EDTA (TBE) buffer. The Bio-Rad CHEF Mapper XA pulsed-field gel electrophoresis (PFGE) system was used to separate chromosomes under the following conditions: a voltage of 6 V/cm, a run time of 24 h, and an angle of 120° at 14°C. The gel was stained with ethidium bromide after electrophoresis.
Analytical methods.
Cell growth was monitored by OD600 using a UV-visible spectrophotometer (Biomate 5; Thermo, NY). The glucose, cellobiose, xylose, xylitol, glycerol, acetate, and ethanol concentrations were determined by a high-performance liquid chromatography (HPLC) instrument (Agilent Technologies 1200 Series) equipped with a refractive index detector using a Rezex ROA-Organic Acid H+ (8%) column (Phenomenex Inc., Torrance, CA). The column was eluted with 0.005 N H2SO4 at a flow rate of 0.6 ml/min at 50°C.
RESULTS
Construction of cellobiose-utilizing strains via integration of cdt-1 and gh1-1.
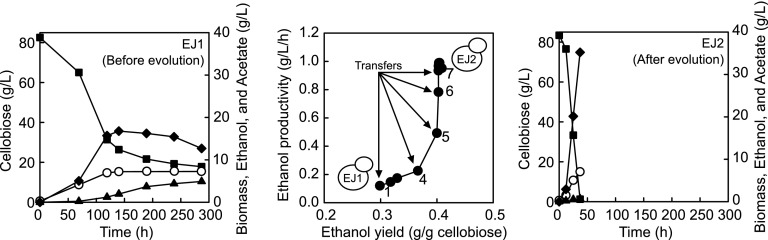
In contrast to prior studies that employed episomal plasmids for expressing cdt-1 and gh1-1, we integrated cdt-1 and gh1-1 into the genome of S. cerevisiae. The resulting strain, EJ1, harboring both cdt-1 and gh1-1 in the genome showed a limited ability to metabolize cellobiose as the sole carbon source (Fig. 1). When 80 g/liter of cellobiose was provided, the EJ1 strain consumed 51 g/liter of cellobiose and produced 16 g/liter of ethanol during 120 h, but further metabolism of cellobiose was not significant until ∼300 h. The volumetric productivity (PEthanol) of the cellobiose fermentation was 0.037 g/(liter · h), and the ethanol yield (YEthanol) was 0.18 g ethanol/g cellobiose. In addition to the low values for PEthanol and YEthanol, the cellobiose consumption rate of strain EJ1 decreased during the fermentation. The initial rate was 0.40 g/(liter · h), and after 140 h, when acetate concentration had increased to 1.94 g/liter, it became much lower. Thus, acetate accumulation might have inhibited cellobiose consumption. Compared to a strain expressing episomal multicopies of the same genes (24), strain EJ1, expressing the single chromosomal copy of cdt-1 and gh1-1, assimilated cellobiose at an extremely low rate (2.22 versus 0.18 g/[liter · h]).
FIG 1.
Improvement of cellobiose fermentation capability of engineered S. cerevisiae EJ1 during adaptive evolution on a high concentration of cellobiose. The subcultures were performed under oxygen-limited conditions (100 rpm) in YP medium with 80 g/liter of cellobiose. The initial cell density of each subculture was adjusted to 0.29 g/liter (OD600 = 1), and the cells were transferred to fresh medium when they reached the stationary phase. Open circle, biomass; solid square, cellobiose; solid diamond, ethanol; solid triangle, acetate.
Improved cellobiose-fermenting capability through laboratory evolution.
To improve the cellobiose fermentation rate, strain EJ1 was subjected to laboratory evolution, which consisted of serial subcultures in YP medium containing 80 g/liter of cellobiose as the sole carbon source. We hypothesized that repeated subculturing on 80 g/liter of cellobiose would select for spontaneous mutants with an increased growth rate. As expected, strain EJ1 showed gradual but noticeable improvement in its cellobiose consumption rate during the serial subcultures (Fig. 1; see also Fig. S1 in the supplemental material). The ethanol concentration was 11.6 g/liter at the end of the first culture, but the ethanol production was 35.2 g/liter at the end of the last culture. Both yields and productivities of ethanol production from cellobiose were improved substantially as the serial subcultures progressed (Fig. 1). As the 12th subculture might be a heterogenous population of various evolved strains, we plated and selected nine colonies from the last subculture, and their cellobiose fermentation capabilities were evaluated using the same culture medium (80 g/liter of cellobiose) as that used for the serial subcultures (see Fig. S2 in the supplemental material). The colony that exhibited the highest ethanol productivity was named EJ2. The cellobiose fermentation profile by strain EJ2 was similar to those of the last serial subculture. These results indicate that the majority of the population in the last subculture had phenotypes similar to those of strain EJ2. It was evident that strain EJ2 can ferment cellobiose much faster than strain EJ1, possibly as a result of strain EJ2 having accumulated beneficial mutations in the genome during the serial subcultures.
Identification of genetic mutations in the evolved cellobiose-fermenting strain.
Laboratory evolution under a particular selection condition and subsequent genome sequence analysis of a selected colony with the desired phenotype constitute a proven approach of inverse metabolic engineering (26, 28). After comparing the genome sequences of strains EJ2 and EJ1, we identified only one nonsynonymous change in strain EJ2 compared to strain EJ1. A protein of unknown function encoded by SVL3 in strain EJ2 had a mutation from methionine to isoleucine at position 685 (M685I) compared to strain EJ1. The SNP (G2005A) in the SVL3 gene was confirmed by Sanger sequencing of a PCR product amplified from the genomic DNA of strain EJ2. As a previous report discovered that SVL3 might be involved in vacuolar function (29), we reasoned that the alteration of vacuole function might increase the stabilities of the cellobiose transporter and β-glucosidase and improve the cellobiose fermentation. To test this idea, we both deleted and overexpressed the SVL3 gene in the slow cellobiose-fermenting strain EJ1 and examined cellobiose fermentation rates of strain EJ1, strain EJ1 svl3Δ, and the EJ1 SVL3-overexpressing strains. However, no changes in cellobiose fermentation by those strains were observed. In addition, we overexpressed the mutant allele of SVL3 (M685I) that was present in EJ2, but there was no significant difference in the cellobiose fermentation rate (see Fig. S3 in the supplemental material). These results suggest that the mutation in the SVL3 gene is not directly associated with the improved cellobiose fermentation capability of the EJ2 strain.
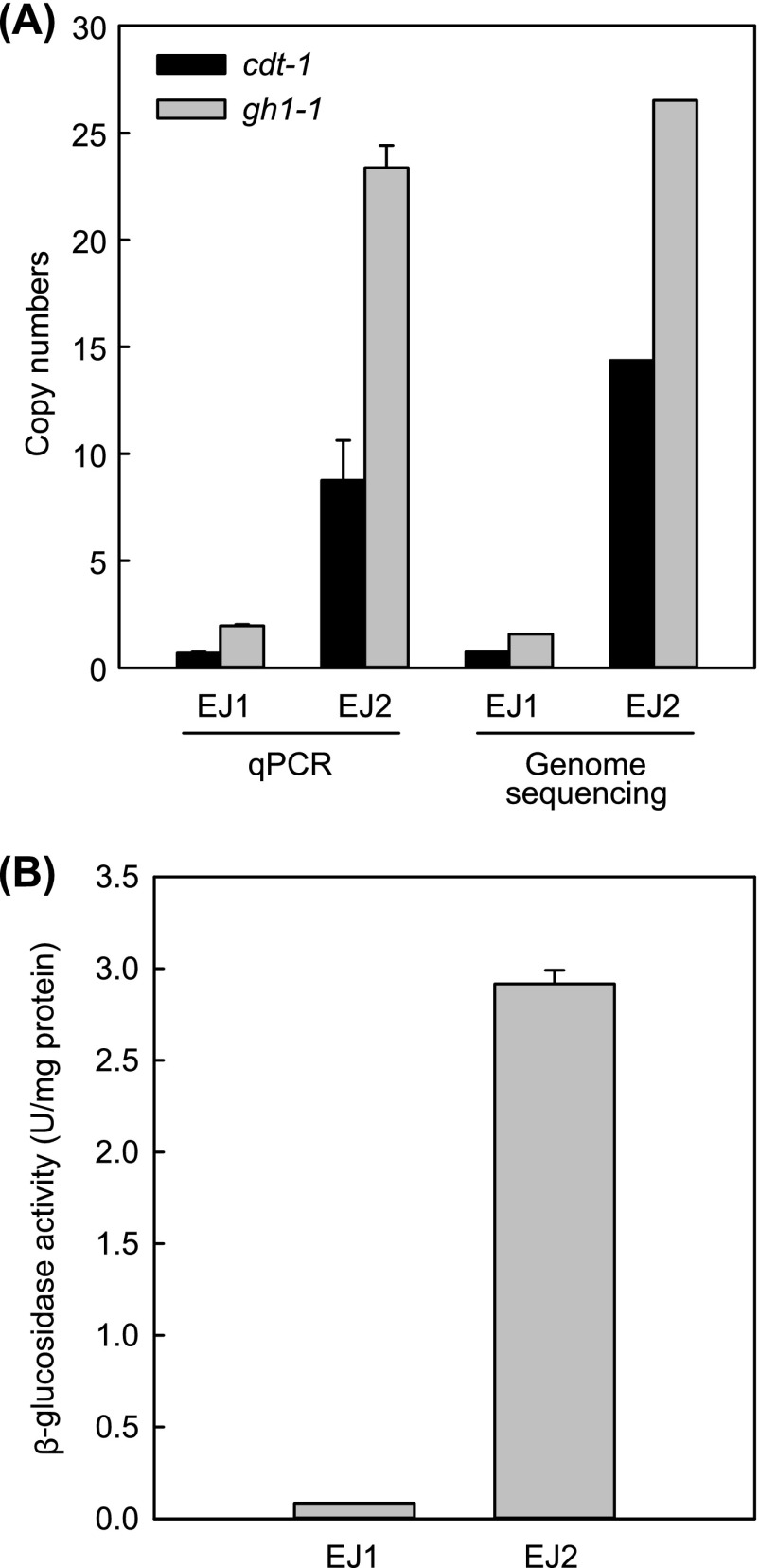
As there were no additional nonsynonymous variants in coding regions, we then investigated the copy number variations in EJ2 compared to EJ1. The average read depth, which is calculated from the number of short sequences mapped to each target from the Illumina sequence reads, was used to estimate the copy number of the integrated cdt-1 and gh1-1 genes. Therefore, we examined the average read depths of cdt-1 and gh1-1 and all open reading frames in strains EJ1 and EJ2. We could not find any copy number variations of endogenous genes in S. cerevisiae, but 19-fold and 17-fold increases in average read depths of cdt-1 and gh1-1, respectively, in strain EJ2 compared to strain EJ1 were observed (Fig. 2A). We also determined the copy numbers of cdt-1 and gh1-1 in strains EJ1 and EJ2 using real-time quantitative PCR (RT-qPCR). As expected, strain EJ2 showed much higher copy numbers for cdt-1 and gh1-1 (9 and 23, respectively) than for EJ1 (1 and 2, respectively) (Fig. 2A). This result suggested that the gene amplification of cdt-1 and gh1-1 in the genome might be responsible for the improved phenotype of the evolved strain (EJ2). We also measured the β-glucosidase activities in strains EJ1 and EJ2 and observed that the crude extract of strain EJ2 exhibited a higher β-glucosidase activity than strain EJ1, suggesting that the increased copy numbers of gh1-1 in strain EJ2 resulted in increased GH1-1 enzyme (Fig. 2B).
FIG 2.
Copy numbers of cellodextrin transporter gene (cdt-1) and β-glucosidase gene (gh1-1) in the parental (EJ1) and evolved (EJ2) strains. (A) Copy numbers of cdt-1 and gh1-1 estimated by genome sequencing data and qPCR. (B) The β-glucosidase activity of engineered S. cerevisiae EJ1 and evolved strain EJ2. The results are the means from triplicate experiments, and the error bars indicate standard deviations. Cells grown to mid-log phase in YP medium containing cellobiose (20 g/liter) were harvested for the enzymatic assay.
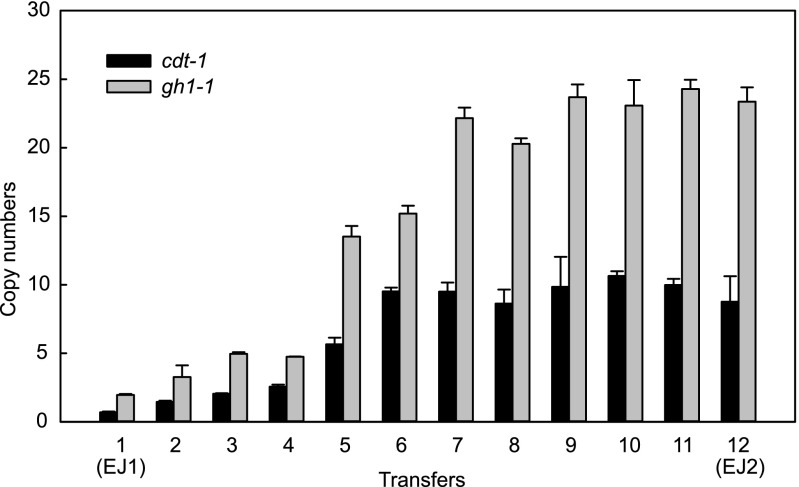
Amplification of cdt-1 and gh1-1 during serial subcultures on cellobiose.
As we identified the increased copy numbers of cdt-1 and gh1-1 in the evolved strain EJ2, which is a strain isolated from the final serial subculture, we reasoned that the copy numbers of cdt-1 and gh1-1 in the evolving strains might increase during the serial subcultures. Interestingly, the copy numbers of cdt-1 and gh1-1 in the evolving strains increased gradually, with good correlations of cellobiose consumption rates and ethanol yields (Fig. 1 and 3). Notably, there was a significant improvement of the cellobiose consumption rate at the fifth subculture with a substantial increase in the copy numbers of cdt-1 and gh1-1 (from 3 copies of cdt-1 and 5 copies of gh1-1 into 6 copies of cdt-1 and 14 copies of gh1-1). Whereas it took more than 140 h to consume 80 g/liter of cellobiose in the initial EJ1 strain, it took only 71 h to ferment 80 g/liter of cellobiose after the fifth subculture. This suggests that amplification of the cdt-1 and gh1-1 genes during laboratory evolution was the cause of improved cellobiose fermentation.
FIG 3.
Gradual increase in the copy numbers of cellodextrin transporter gene (cdt-1) and β-glucosidase gene (gh1-1) during serial subcultures of EJ1. Copy numbers of cdt-1 and gh1-1 in each transfer were estimated by qPCR. The results are the means from triplicate experiments, and the error bars indicate standard deviations.
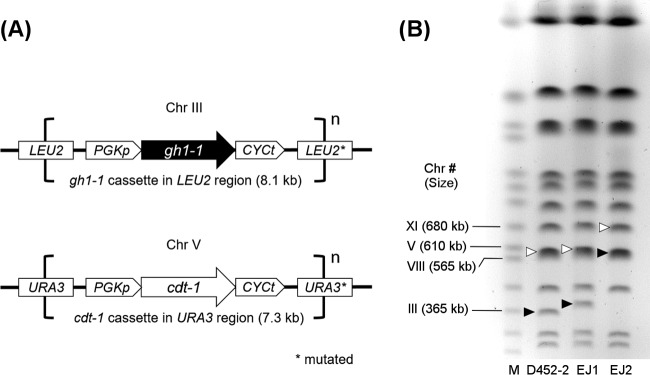
The massive gene amplifications of cdt-1 and gh1-1 in the genome of strain EJ2 were also confirmed by PFGE. The expression cassettes for cdt-1 and gh1-1 were integrated into the URA3 and LEU2 loci in chromosomes V and III, respectively, in strain EJ1 (Fig. 4A). Therefore, the sizes of chromosomes V and III of strain EJ2 should be greater than those of strain EJ1. As the size of the cdt-1 cassette was 7.3 kb and the EJ2 chromosome V was ∼100 kb bigger, this is consistent with a copy number of 14 (7.3 kb/copy × 14 copies) (Fig. 4B). The size of chromosome III of strain EJ2 appeared to be ∼220 kb greater than that of EJ1, which is consistent with a gh1-1 cassette copy number of 27 (8.1 kb/copy × 27 copies) (Fig. 4B). The sizes of the chromosomes in the EJ1 and EJ2 strains obtained from PFGE were entirely consistent with the copy number estimates determined by Illumina read depth analysis and qPCR. Together, all three methods demonstrate that massive gene amplification in the chromosomes occurred during laboratory evolution and likely contributed to substantial improvements in cellobiose fermentation.
FIG 4.
Amplification of integrative cassettes for cdt-1 and gh1-1 in chromosomes. (A) Locations of expression cassettes in chromosomes (Chr) of EJ1 and EJ2. (B) Pulsed-field gel electrophoresis of chromosomes from parental (D452-2) and engineered (EJ1 and EJ2) strains. The chromosome numbers and sizes are supplied. M, yeast chromosome markers; solid arrow, chromosome III; open arrow, chromosome V.
Effects of copy number variations of cdt-1 and gh1-1 on fermentation of cellobiose.
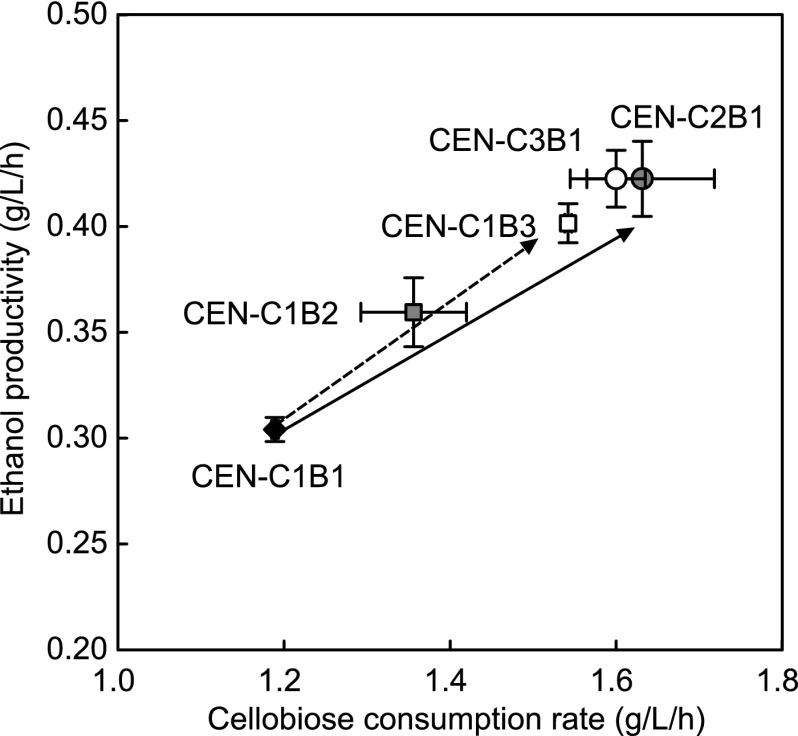
To determine whether increased copy numbers of the cdt-1 and gh1-1 genes were responsible for the enhanced cellobiose fermentation that we observed in the evolved strains, we constructed a series of engineered yeast strains with varied numbers of the cdt-1 and gh1-1 cassettes. We used a different strain background (S. cerevisiae CEN.PK2-1D) not only because the strain has quadruple autotrophic markers but also to examine whether or not the copy number effect is strain independent. A total of five engineered strains that contained different numbers of plasmids with either the cdt-1 or gh1-1 expression cassettes were constructed: CEN-C1B1 had one for the cdt-1 expression cassette and one plasmid for the gh1-1 expression cassette, CEN-C1B2 had one plasmid containing the cdt-1 expression cassette and two plasmids containing the gh1-1 expression cassette, CEN-C1B3 had one plasmid containing the cdt-1 expression cassette and three plasmids containing the gh1-1 expression cassette, CEN-C2B1 had two plasmids containing the cdt-1 expression cassette and one plasmid containing the gh1-1 expression cassette, and CEN-C3B1 had three plasmids containing the cdt-1 expression cassette and one plasmid containing the gh1-1 expression cassette transformed into the CEN.PK2-1D strain.
Using the above-described five strains, we examined the effects of varied expression levels of the cellodextrin transporter and β-glucosidase on cellobiose fermentation rates. As shown in Fig. 5, S. cerevisiae CEN.PK2-1D strains harboring higher copy numbers of cdt-1 or gh1-1 exhibited higher cellobiose consumption rates and ethanol productivity than ones containing lower copy numbers of the genes. When cultured in 80 g/liter cellobiose as the sole carbon source, the cellobiose consumption rate and ethanol productivity by strain CEN-C3B1 were 35% and 39% higher, respectively, than those of strain CEN-C1B1. However, the differences in the fermentation parameters between strains CEN-C2B1 and CEN-C3B1 were not significant, suggesting that strain CEN-C2B1 was already saturated with cdt-1 copies (Fig. 5). We also observed that an increase in the gh1-1 copy number improved the cellobiose fermentation. The ethanol productivity of strain CEN-C1B3 (0.402 ± 0.009 g/[liter · h]) was higher than that of strain CEN-C1B1 (0.304 ± 0.006, P < 0.05), corresponding to a higher (30%) cellobiose consumption rate. These results indicate that the key fermentation parameters, cellobiose consumption rate and ethanol productivity, improve by increasing the copy numbers of gh1-1 and cdt-1, regardless of the strain background.
FIG 5.
Effect of copy number variations of cdt-1 and gh1-1 in episomal plasmids on cellobiose fermentation capability of engineered S. cerevisiae CEN.PK2-1D. The effects of increasing episomal plasmids with cdt-1 (CEN-C2B1 and CEN-C3B1; solid arrow) and gh1-1 (CEN-C1B2 and CEN-C1B3; dashed arrow) in YP medium containing 80 g/liter of cellobiose were compared to those obtained with engineered S. cerevisiae CEN-C1B1. The initial cell density was adjusted to 0.029 g/liter, and all fermentations were performed under oxygen-limited conditions. The results are the means from duplicate experiments, and the error bars indicate standard deviations.
Stability of the integrative cassettes of cdt-1 and gh1-1 in chromosomes of strain EJ2.
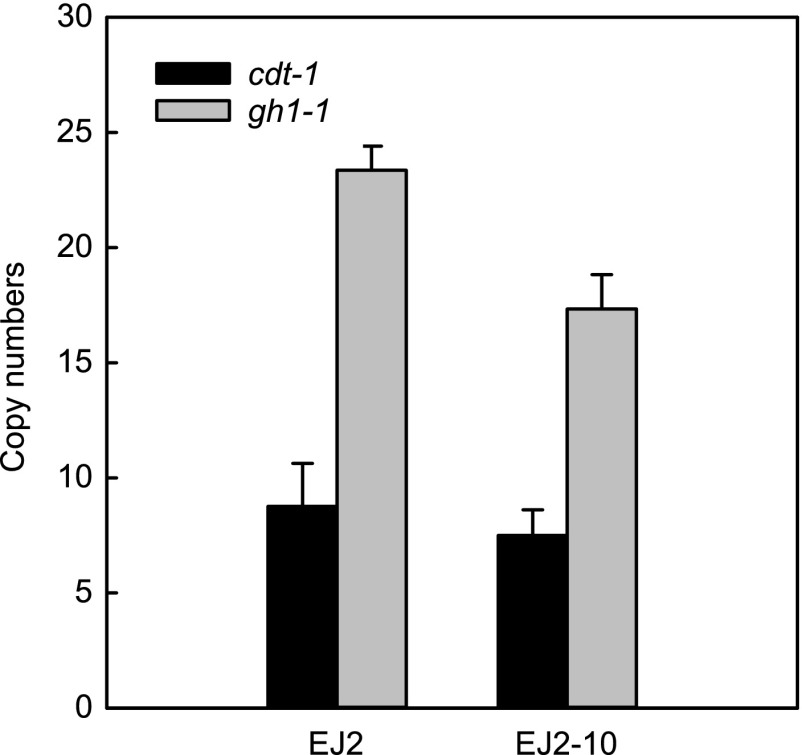
It is evident that the copy number increases of the cdt-1 or gh1-1 genes in engineered yeast, either through gene amplification or increased episomal plasmid copy number, can accelerate cellobiose utilization. One obvious advantage of gene amplification over episomal plasmids is long-term stability under nonselective culture conditions, which are mostly used in industrial fermentations. Therefore, we examined the long-term stability of the amplified cassettes of cdt-1 and gh1-1 without any selective pressure, i.e., without using cellobiose as a carbon source. We performed 10 serial transfers of the EJ2 strain in 20 g/liter glucose. The cells from the 10th transfer (EJ2-10) were evaluated in medium containing cellobiose as the sole carbon source. Interestingly, EJ2-10 exhibited cellobiose consumption rate and ethanol productivity similar to those of strain EJ2 (Table 2). Strain EJ2-10 yielded a specific cellobiose consumption rate (0.46 ± 0.01 g cellobiose/[g cell · h]) comparable to that of EJ2 (0.49 ± 0.00 g cellobiose/[g cell · h]). Using real-time PCR (qPCR), the copy numbers for cdt-1 and gh1-1 (7 and 17) in EJ2-10 were found to be lower than in EJ2 (9 and 23) (Fig. 6). These results showed that the copy numbers of cdt-1 and gh1-1 in engineered yeast might be variable, depending on culture conditions, but did not change drastically. While subculturing on glucose 10 times is an exaggerated condition, strain EJ2 was able to keep the rapid cellobiose-fermenting phenotype. Here, we show that cellobiose can be used as an efficient selection pressure for gene amplification and the amplified cassettes in the genome are stable.
TABLE 2.
Changes in cellobiose fermentation parameters after serial subculturesa
| Parameter (unit) | Value (mean ± SD) |
||
|---|---|---|---|
| EJ1 | EJ2 | EJ2-10 | |
| Cellobiose consumption rate (g/[liter · h]) | 0.18 ± 0.00 | 2.73 ± 0.01 | 2.62 ± 0.10 |
| Specific cellobiose consumption rate (g/[g cell · h]) | 0.09 ± 0.00 | 0.49 ± 0.00 | 0.46 ± 0.01 |
| Ethanol yield (g/g cellobiose) | 0.40 ± 0.00 | 0.40 ± 0.00 | |
| Volumetric ethanol productivity (g/[liter · h]) | 1.09 ± 0.00 | 1.04 ± 0.03 | |
| Specific ethanol productivity (g/[g cell · h]) | 0.19 ± 0.00 | 0.18 ± 0.00 | |
Fermentation experiments were performed under oxygen-limited conditions (100 rpm) in YP medium with 80 g/liter of cellobiose, and the initial cell density was adjusted to 0.29 g/liter (OD600 = 1). The parameters were measured at 29 h. The results are the means from duplicate experiments. Serial subcultures were performed under oxygen-limited conditions (100 rpm) in YP medium with 80 g/liter of cellobiose or 20 g/liter of glucose, and the initial cell density of each subculture was adjusted to 0.29 g/liter (OD600 = 1).
FIG 6.
Changes in copy numbers of cellodextrin transporter gene (cdt-1) and β-glucosidase gene (gh1-1) during serial subcultures of EJ2 on glucose. The results are the means from triplicate experiments, and the error bars indicate standard deviations. The subcultures were performed under oxygen-limited conditions (100 rpm) in YP medium with 20 g/liter of glucose. The initial cell density of each subculture was adjusted to 0.29 g/liter (OD600 = 1), and the cells were transferred to fresh medium when the cells reached the stationary phase. EJ2-10, cells from the 10th transfer.
DISCUSSION
Production of biofuels and chemicals from mixed sugars present in the hydrolysates of marine and terrestrial biomass has been anticipated for decades, but both yields and productivities of biofuels and chemicals are not currently feasible for commercialization (30–32). Among many bottlenecks, glucose repression on utilizing nonglucose sugars is a significant problem for fermenting mixed sugars by engineered S. cerevisiae. Direct fermentation of cellobiose by engineered yeast opened the possibility of simultaneous utilization of cellobiose and nonglucose sugars (22, 23), and it is proven that cellobiose fermentation by engineered yeast can be applied for fermenting pretreated cellulosic biomass (33, 34).
To ferment cellobiose in cellulosic hydrolysates, it is necessary for the engineered S. cerevisiae strain to keep the plasmids for expressing the β-glucosidase and the cellodextrin transporter. Therefore, we introduced cellodextrin transporter and β-glucosidase genes into the genome of S. cerevisiae to enable their stable expression under nonselective conditions. Also, we applied an evolutionary engineering strategy to improve the cellobiose fermentation rate of the integrant strain. In this study, the evolved strain harbored higher copy numbers of the cellodextrin transporter gene (cdt-1) and β-glucosidase gene (gh1-1) in the genome than the parental strain did. The fast-cellobiose-consuming strain showed high enzyme activity with gh1-1, indicating that the β-glucosidase activity corresponded to the copy number variations. The tandem amplification of genes might be initiated by the unequal crossover of the ura3::URA3 PGK1p-cdt-1-CYC1t or leu2::LEU2 PGK1p-gh1-1-CYC1t construct in the genome (Fig. 4A). This difference suggests that amplification of integrated cdt-1 and gh1-1 in the genome during laboratory evolution might be the primary genetic event. In a prior study, xylose utilization by engineered S. cerevisiae harboring xylose isomerase (xylA) was also significantly improved by tandem gene amplification of the xylA in the chromosome, resulting in an increase in the expression level of the xylose isomerase during evolutionary engineering (12). Taken together, these findings indicate that the increase by gene amplification in the copy numbers of the essential genes for sugar utilization (cdt-1 and gh1-1 or xylA) might be a key factor in improving the fermentation performance.
The effect of the copy number variations of cdt-1 and gh1-1 on cellobiose fermentation in the recombinant S. cerevisiae was also tested using a different host strain. The increase of the cdt-1 and gh1-1 plasmid copy numbers in S. cerevisiae CEN.PK2-1D resulted in an improvement of the ethanol productivity and cellobiose consumption rate. In the case of the copy number variations of cdt-1 with the limited, constant expression of gh1-1 (CEN-C1B1, CEN-C2B1, and CEN-C3B1), the cellobiose consumption rate and ethanol productivity were saturated with two multicopy plasmids expressing cdt-1. This saturation suggests that there might be optimized levels for cellodextrin transporter and β-glucosidase in efficient cellobiose-fermenting S. cerevisiae. Interestingly, the copy number ratio of cdt-1 and gh1-1 converged to 1:2 as the engineered strains evolved to ferment the cellobiose faster (Fig. 3). In another study, directed evolution using the screening of promoter libraries was applied to optimize the cellobiose-utilizing pathway. Similarly, the efficient cellobiose-utilizing strain showed a higher consumption rate at the optimized ratio of the expression levels of cdt-1 and gh1-1 (1:2.5) (35). Although our approach for improving the cellobiose fermentation was different from that of Yuan and Zhao (35), we found that the copy numbers of cdt-1 and gh1-1 in the engineered S. cerevisiae increased to the optimized ratio. These results indicated that not only the absolute increase of the copy numbers of cdt-1 and gh1-1 but also their ratio is important for the large improvement in cellobiose fermentation that we observed in the evolved strain EJ2.
Despite the promising results from the cellobiose fermentation experiments, transient cellodextrin accumulation by the transglycosylation of β-glucosidase was observed in the medium during the processes (24). The accumulated cellodextrin might decrease the ethanol productivity because the cellodextrin transport and reconsumption might not be as rapid as the cellobiose fermentation (22, 24). S. cerevisiae strain D452-2 expressing the cellodextrin transporter and β-glucosidase from episomal plasmids (DCDT-1G) accumulated 15.5 g/liter of cellodextrin in the fermentation experiments using YP medium with 80 g/liter cellobiose under oxygen-limited conditions. In contrast, strain EJ2, which is originated from the same host (S. cerevisiae D452-2), showed only a 2.6-g/liter cellodextrin accumulation and a higher ethanol productivity (0.52 versus 1.02 g/[liter · h]) at 24 h under the same conditions (see Fig. S4 in the supplemental material). When the engineered yeast strains uptake cellobiose rapidly, the accumulated intracellular cellobiose can be used as a substrate for producing cellodextrin by the transglycosylation activity of β-glucosidase (gh1-1). It is well known that high cellobiose concentrations cause cellodextrin production by β-glucosidase in vitro (36), and the intracellular cellobiose concentration is determined by the rates of cellobiose transport and hydrolysis. Optimization of the cellodextrin transporter and β-glucosidase expression levels in the genome by evolutionary engineering may minimize the accumulation of intracellular cellobiose and its transglycosylation by β-glucosidase.
By genome integration of the cellobiose-utilizing pathway followed by evolutionary engineering, we improved the cellobiose fermentation performance of the parental strain impressively. Based on the data presented in this study, the evolved strain showed a significantly improved cellobiose consumption rate and ethanol productivity without directed evolution (35) or using an industrial strain as the host strain (37). In addition, our approach of integration followed by evolution could be applied to construct stable systems in S. cerevisiae that require high gene copy number. It is hard to implement induction of antibiotic systems in industrial fermentation processes; therefore, stable strains based on chromosomal integration are desirable (38–41). Here we used cellobiose, a sugar derived from cellulosic biomass, as a strong selective pressure to produce massive gene amplification. Since the approach did not utilize antibiotics as a selective pressure for gene amplification or need additional genetic manipulations to keep the amplified gene copy numbers after the laboratory evolution (42), it demonstrates an industrially applicable and efficient metabolic engineering strategy. Also, we observed that loss in the copy numbers of cdt-1 and gh1-1 in the genome of strain EJ2 was marginal after serial subcultures on glucose for 240 h (Fig. 6 and Table 2). The results suggested that the stability of integrated cassettes of cdt-1 and gh1-1 in chromosomes can be maintained even in the absence of the selection pressure. We envision that the construction of a stable industrial strain using our engineering strategy that can coferment cellobiose and xylose will enable a commercially viable simultaneous saccharification and cofermentation (SSCF) process.
Supplementary Material
Funding Statement
This work was supported by funding from the Energy Biosciences Institute (EBI).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00410-16.
REFERENCES
- 1.Wyman CE. 2007. What is (and is not) vital to advancing cellulosic ethanol. Trends Biotechnol 25:153–157. doi: 10.1016/j.tibtech.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Pimentel D, Marklein A, Toth MA, Karpoff MN, Paul GS, McCormack R, Kyriazis J, Krueger T. 2009. Food versus biofuels: environmental and economic costs. Hum Ecol 37:1–12. doi: 10.1007/s10745-009-9215-8. [DOI] [Google Scholar]
- 3.Schmer MR, Vogel KP, Mitchell RB, Perrin RK. 2008. Net energy of cellulosic ethanol from switchgrass. Proc Natl Acad Sci U S A 105:464–469. doi: 10.1073/pnas.0704767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim SR, Ha SJ, Wei N, Oh EJ, Jin YS. 2012. Simultaneous co-fermentation of mixed sugars: a promising strategy for producing cellulosic ethanol. Trends Biotechnol 30:274–282. doi: 10.1016/j.tibtech.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Rubin EM. 2008. Genomics of cellulosic biofuels. Nature 454:841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- 6.Romanos MA, Scorer CA, Clare JJ. 1992. Foreign gene expression in yeast: a review. Yeast 8:423–488. doi: 10.1002/yea.320080602. [DOI] [PubMed] [Google Scholar]
- 7.Kuyper M, Toirkens MJ, Diderich JA, Winkler AA, van Dijken JP, Pronk JT. 2005. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res 5:925–934. doi: 10.1016/j.femsyr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Forsburg SL. 2001. The art and design of genetic screens: yeast. Nat Rev Genet 2:659–668. [DOI] [PubMed] [Google Scholar]
- 9.Kötter P, Ciriacy M. 1993. Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 38:776–783. doi: 10.1007/BF00167144. [DOI] [Google Scholar]
- 10.Jeffries TW. 2006. Engineering yeasts for xylose metabolism. Curr Opin Biotechnol 17:320–326. doi: 10.1016/j.copbio.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Matsushika A, Inoue H, Kodaki T, Sawayama S. 2009. Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl Microbiol Biotechnol 84:37–53. doi: 10.1007/s00253-009-2101-x. [DOI] [PubMed] [Google Scholar]
- 12.Zhou H, Cheng JS, Wang B, Fink GR, Stephanopoulos G. 2012. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng 14:611–622. doi: 10.1016/j.ymben.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 13.Kim SR, Park YC, Jin YS, Seo JH. 2013. Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnol Adv 31:851–861. doi: 10.1016/j.biotechadv.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Kötter P, Amore R, Hollenberg CP, Ciriacy M. 1990. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr Genet 18:493–500. doi: 10.1007/BF00327019. [DOI] [PubMed] [Google Scholar]
- 15.Bruinenberg PM, de Bot PH, van Dijken JP, Scheffers WA. 1983. The role of redox balances in the anaerobic fermentation of xylose by yeasts. Eur J Appl Microbiol Biotechnol 18:287–292. doi: 10.1007/BF00500493. [DOI] [Google Scholar]
- 16.Walfridsson M, Hallborn J, Penttilä M, Keränen S, Hahn-Hägerdal B. 1995. Xylose-metabolizing Saccharomyces cerevisiae strains overexpressing the TKL1 and TAL1 genes encoding the pentose phosphate pathway enzymes transketolase and transaldolase. Appl Environ Microbiol 61:4184–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho NW, Chen Z, Brainard AP. 1998. Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol 64:1852–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trumbly R. 1992. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol 6:15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- 19.Carlson M. 1999. Glucose repression in yeast. Curr Opin Microbiol 2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 20.Hahn-Hägerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. 2007. Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol 74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- 21.Wei N, Quarterman J, Jin YS. 2013. Marine macroalgae: an untapped resource for producing fuels and chemicals. Trends Biotechnol 31:70–77. doi: 10.1016/j.tibtech.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Ha SJ, Galazka JM, Kim SR, Choi JH, Yang X, Seo JH, Glass NL, Cate JH, Jin YS. 2011. Engineered Saccharomyces cerevisiae capable of simultaneous cellobiose and xylose fermentation. Proc Natl Acad Sci U S A 108:504–509. doi: 10.1073/pnas.1010456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galazka JM, Tian C, Beeson WT, Martinez B, Glass NL, Cate JH. 2010. Cellodextrin transport in yeast for improved biofuel production. Science 330:84–86. doi: 10.1126/science.1192838. [DOI] [PubMed] [Google Scholar]
- 24.Kim H, Lee WH, Galazka JM, Cate JH, Jin YS. 2014. Analysis of cellodextrin transporters from Neurospora crassa in Saccharomyces cerevisiae for cellobiose fermentation. Appl Microbiol Biotechnol 98:1087–1094. doi: 10.1007/s00253-013-5339-2. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Moo-Young M, Chisti Y. 1996. Plasmid stability in recombinant Saccharomyces cerevisiae. Biotechnol Adv 14:401–435. doi: 10.1016/S0734-9750(96)00033-X. [DOI] [PubMed] [Google Scholar]
- 26.Kim SR, Skerker JM, Kang W, Lesmana A, Wei N, Arkin AP, Jin YS. 2013. Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS One 8:e57048. doi: 10.1371/journal.pone.0057048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teste MA, Duquenne M, François J, Parrou JL. 2009. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol 10:99. doi: 10.1186/1471-2199-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oud B, Maris AJ, Daran JM, Pronk JT. 2012. Genome-wide analytical approaches for reverse metabolic engineering of industrially relevant phenotypes in yeast. FEMS Yeast Res 12:183–196. doi: 10.1111/j.1567-1364.2011.00776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng B, Wu JN, Schober W, Lewis DE, Vida T. 1998. Isolation of yeast mutants defective for localization of vacuolar vital dyes. Proc Natl Acad Sci U S A 95:11721–11726. doi: 10.1073/pnas.95.20.11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fortman J, Chhabra S, Mukhopadhyay A, Chou H, Lee TS, Steen E, Keasling JD. 2008. Biofuel alternatives to ethanol: pumping the microbial well. Trends Biotechnol 26:375–381. doi: 10.1016/j.tibtech.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 31.Dellomonaco C, Fava F, Gonzalez R. 2010. The path to next generation biofuels: successes and challenges in the era of synthetic biology. Microb Cell Fact 9:1–15. doi: 10.1186/1475-2859-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young E, Lee SM, Alper H. 2010. Optimizing pentose utilization in yeast: the need for novel tools and approaches. Biotechnol Biofuels 3:24–24. doi: 10.1186/1754-6834-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee WH, Nan H, Kim HJ, Jin YS. 2013. Simultaneous saccharification and fermentation by engineered Saccharomyces cerevisiae without supplementing extracellular β-glucosidase. J Biotechnol 167:316–322. doi: 10.1016/j.jbiotec.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 34.Jin YS, Cate JH. 2012. Model-guided strain improvement: simultaneous hydrolysis and co-fermentation of cellulosic sugars. Biotechnol J 7:328–329. doi: 10.1002/biot.201100489. [DOI] [PubMed] [Google Scholar]
- 35.Yuan Y, Zhao H. 2013. Directed evolution of a highly efficient cellobiose utilizing pathway in an industrial Saccharomyces cerevisiae strain. Biotechnol Bioeng 110:2874–2881. doi: 10.1002/bit.24946. [DOI] [PubMed] [Google Scholar]
- 36.Bohlin C, Praestgaard E, Baumann MJ, Borch K, Praestgaard J, Monrad RN, Westh P. 2013. A comparative study of hydrolysis and transglycosylation activities of fungal β-glucosidases. Appl Microbiol Biotechnol 97:159–169. doi: 10.1007/s00253-012-3875-9. [DOI] [PubMed] [Google Scholar]
- 37.Eriksen DT, Hsieh PCH, Lynn P, Zhao H. 2013. Directed evolution of a cellobiose utilization pathway in Saccharomyces cerevisiae by simultaneously engineering multiple proteins. Microb Cell Fact 12:61. doi: 10.1186/1475-2859-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neubauer P, Lin H, Mathiszik B. 2003. Metabolic load of recombinant protein production: inhibition of cellular capacities for glucose uptake and respiration after induction of a heterologous gene in Escherichia coli. Biotechnol Bioeng 83:53–64. doi: 10.1002/bit.10645. [DOI] [PubMed] [Google Scholar]
- 39.Westfall PJ, Gardner TS. 2011. Industrial fermentation of renewable diesel fuels. Curr Opin Biotechnol 22:344–350. doi: 10.1016/j.copbio.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 40.Olmstead J, Wallinga D. 2010. Antimicrobial alternatives: public health risks call into question continued antibiotic use in ethanol production. Foodborne Pathog Dis 7:871. doi: 10.1089/fpd.2010.0620. [DOI] [PubMed] [Google Scholar]
- 41.Rich JO, Leathers TD, Nunnally MS, Bischoff KM. 2011. Rapid evaluation of the antibiotic susceptibility of fuel ethanol contaminant biofilms. Bioresour Technol 102:1124–1130. doi: 10.1016/j.biortech.2010.08.118. [DOI] [PubMed] [Google Scholar]
- 42.Tyo KE, Ajikumar PK, Stephanopoulos G. 2009. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol 27:760–765. doi: 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- 43.Hosaka K, Nikawa J, Kodaki T, Yamashita S. 1992. A dominant mutation that alters the regulation of INO1 expression in Saccharomyces cerevisiae. J Biochem 111:352–358. [DOI] [PubMed] [Google Scholar]
- 44.Entian KD, Kötter P. 1998. 23 yeast mutant and plasmid collections. Methods Microbiol 26:431–449. doi: 10.1016/S0580-9517(08)70344-1. [DOI] [Google Scholar]
- 45.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119–122. doi: 10.1016/0378-1119(92)90454-W. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.