ABSTRACT
Microbicides are broad-spectrum antimicrobial agents that generally interact with multiple pharmacological targets. While they are widely deployed in disinfectant, antiseptic, and preservative formulations, data relating to their potential to select for microbicide or antibiotic resistance have been generated mainly by testing the compounds in much simpler aqueous solutions. In the current investigation, antibiotic susceptibility was determined for bacteria that had previously exhibited decreased microbicide susceptibility following repeated exposure to microbicides either in formulation with sequestrants and surfactants or in simple aqueous solution. Statistically significant increases in antibiotic susceptibility occurred for 12% of bacteria after exposure to microbicides in formulation and 20% of bacteria after exposure to microbicides in aqueous solutions, while 22% became significantly less susceptible to the antibiotics, regardless of formulation. Of the combinations of a bacterium and an antibiotic for which British Society for Antimicrobial Chemotherapy breakpoints are available, none became resistant. Linear modeling taking into account phylogeny, microbicide, antibiotic, and formulation identified small but significant effects of formulation that varied depending on the bacterium and microbicide. Adaptation to formulated benzalkonium chloride in particular was more likely to increase antibiotic susceptibility than adaptation to the simple aqueous solution. In conclusion, bacterial adaptation through repeated microbicide exposure was associated with both increases and decreases in antibiotic susceptibility. Formulation of the microbicide to which the bacteria had previously adapted had an identifiable effect on antibiotic susceptibility, but it effect was typically small relative to the differences observed among microbicides. Susceptibility changes resulting in resistance were not observed.
IMPORTANCE The safety of certain microbicide applications has been questioned due to the possibility that microbicide exposure could select for microbicide and antibiotic resistance. Evidence that this may happen is based mainly on in vitro experiments where bacteria have been exposed to microbicides in aqueous solution. Microbicides are, however, normally deployed in products formulated with surfactants, sequestrants, and other compounds. While this may influence the frequency and extent of susceptibility changes, few studies reported in the literature have assessed this. In the current investigation, therefore, we have investigated changes in antibiotic susceptibility in bacteria which exhibited decreased microbicide susceptibility following repeated exposure to microbicides in simple aqueous solutions and in formulation. We report that the microbicide formulation had an identifiable effect on antibiotic susceptibility, but it was typically small relative to the differences observed among microbicides. We did not observe susceptibility changes resulting in resistance.
INTRODUCTION
Microbicides are broad-spectrum antimicrobial compounds that are widely deployed to control the growth of microorganisms or eliminate them. Applications include the control of biofouling and microbial contamination in industry (1) as well as clinical antisepsis (2–4). They are also used extensively in the domestic environment as hygiene adjuncts and preservatives in a range of formulations, including oral care products (5), hand sanitizers (6), and hard-surface cleaners (7).
The safety of certain microbicide applications has been questioned due to the possibility that long-term microbicide exposure could select for reduced antimicrobial susceptibility in bacteria (8–10). Reduced microbicide susceptibility has been reported for some combinations of a bacterium and a microbicide (11), and changes in bacterial susceptibility to chemically unrelated antimicrobials, such as antibiotics or other microbicides, following laboratory microbicide exposure have been reported (12, 13). The mechanisms involved in such cross-resistance include selection for mutations in shared cellular target sites, upregulation of efflux pumps (14), reductions in cell permeability (15), and in some cases, sporulation (16).
Evidence that microbicides can select for reduced microbicide susceptibility in the environment is limited, with the majority of reports relating to in vitro exposure (17). Similarly, little evidence to firmly link microbicide/antibiotic cross-resistance to microbicide use has emerged (18–21). The majority of studies aiming to better understand the potential risks of resistance development through microbicide exposure have exposed bacteria to microbicides in aqueous solution with or without the addition of cosolvents, such as dimethyl sulfoxide (22) or ethanol (23). In real use, however, microbicides are deployed in products formulated with surfactants, sequestrants, and other compounds that can interact with cellular targets to influence antimicrobial potency. As previously reported, such formulations can decrease the frequency and extent of the acquisition of reduced microbicide susceptibility in bacteria (24). Accordingly, evaluation of the effects of bacterial exposure to microbicides within a formulation chassis containing surfactants and sequestrants may generate more realistic data on which to base assessments of the risk of induction of changes in bacterial susceptibility. In the current investigation, we have therefore assessed changes in antibiotic susceptibility in bacteria which have previously exhibited decreases in microbicide susceptibility following repeated exposure to a range of microbicides in simple aqueous solutions and in formulations containing commonly used nonionic surfactants and sequestrants (24). The microbicides tested reflect those frequently used in consumer products, such as laundry detergents, hard-surface disinfectants, and personal care products. The antibiotics were selected on the basis of their common therapeutic use and their inclusion in a U.S. investigation of links between domestic microbicide use and antibiotic resistance (25).
MATERIALS AND METHODS
Bacteria.
Pseudomonas aeruginosa ATCC 9027, Staphylococcus aureus ATCC 6538, and Escherichia coli ATCC 25922 were obtained from Oxoid (Basingstoke, United Kingdom). Acinetobacter baumannii MBRG15.1, Pseudomonas putida MBRG15.2, Escherichia coli MBRG15.4, and Cronobacter sakazakii MBRG15.5 were isolated from a domestic kitchen drain biofilm. Enterococcus faecalis MRBG15.6 is a wound isolate provided by Angela Oates, The University of Manchester.
Chemical reagents and growth media.
Bacteriological growth media were purchased from Oxoid (Basingstoke, United Kingdom). All other chemical reagents were purchased from Sigma-Aldrich (Dorset, United Kingdom) unless otherwise stated. Bacterial growth media were sterilized at 121°C and 15 lb/in2 for 15 min prior to use. Pseudomonas aeruginosa, Staphylococcus aureus, Escherichia coli, and Enterococcus faecalis were cultured on tryptone soy agar and broth. Acinetobacter baumannii, Pseudomonas putida, and Cronobacter sakazakii were grown on Wilkins-Chalgren agar and broth containing 2% sucrose. All bacteria were incubated aerobically at 37°C for 18 h unless stated otherwise.
Antimicrobials.
The microbicides benzalkonium chloride (BAC), chlorhexidine digluconate (CHX; 20%, vol/vol), thymol, and triclosan were purchased from Sigma-Aldrich (Dorset, United Kingdom). Didecyldimethyl ammonium chloride (DDAC; 50%, vol/vol) was purchased from Merck Millipore (Durham, United Kingdom). A 20% (vol/vol) aqueous solution of polyhexamethylene biguanide (PHMB; Vantocil) was obtained from Arch Chemicals Inc. (Manchester, United Kingdom). 1,3-Dimethylol-5,5-dimethylhydantoin (DMDM hydantoin; Glydant) at 54% (vol/vol) was obtained from Lonza (Bishops Stortford, United Kingdom), while benzisothiazolinone (BIT) was supplied by Unilever (Port Sunlight, United Kingdom). All microbicides were prepared in aqueous solution or added to a microbicide-free formulation chassis containing sequestrants and surfactants as previously described (24) at concentrations reflective of their normal deployment in consumer products. BAC, CHX, DDAC, DMDM hydantoin, PHMB, and thymol were prepared at 1% (vol/vol) in a general-purpose cleaner. Triclosan was added to a laundry detergent at 0.0066% (wt/vol). Benzisothiazolinone was formulated into a laundry detergent at 0.02% (vol/vol). Ampicillin (10 μg), cephalothin (20 μg), ciprofloxacin (1 μg), kanamycin (5 μg), and tetracycline (10 μg) antibiotic discs were obtained from Oxoid (Basingstoke, United Kingdom).
16S rRNA gene sequencing.
Single bacterial colonies were dispersed in 100 μl of nanopure water, vortexed for 30 s, and boiled at 100°C for 15 min to lyse the cells. Microcentrifuge tubes were centrifuged at 16,000 × g for 1 min to remove cellular debris, and the resulting supernatant was retained as the DNA template. PCR was performed using the primers 8FLP (5′-GAG TTT GAT CCT GGS TCA G-3′) and 806R (5′-GGA CTA CCA GGG TAT CTA AT-3′) at 5 μM per reaction mixture. PCR was conducted using a Biometra TGradient thermocycler (Analytik Jena, Jena, Germany) and run for 35 thermal cycles of 94°C (1 min), 53°C (1 min), and 72°C (1 min). A 15-min elongation step was included in the final cycle. The PCR products were purified using a QIAquick PCR purification kit (Qiagen, West Sussex, United Kingdom) according to the manufacturer's instructions, and the resulting DNA yield was quantified using a NanoDrop 2000c UV-visible spectrophotometer (Thermo Scientific, Wilmington, DE, USA). A reaction mixture containing 4 pM forward or reverse primer and 40 to 50 ng of DNA in a 10-μl total volume was used for DNA sequencing. DNA sequencing was performed using an Applied Biosystems 3730 DNA analyzer (Thermo Fisher, Paisley, United Kingdom).
Microbicide exposure in aqueous solution and formulation.
A system previously validated to be highly selective for changes in antimicrobial susceptibility (26, 27) was used. Reproducible ca. 100-fold-concentration gradients of the antimicrobial compounds were generated on tryptone soy or Wilkins-Chalgren agar plates using an automated spiral plater (Don Whitley Scientific, Shipley, United Kingdom). Antimicrobials in aqueous solution or in formulation (50 μl) were deposited on the agar surface. The plates were dried for 1 h at room temperature prior to radial deposition of bacterial pure cultures and then incubated (4 days, 37°C) in an aerobic incubator. After incubation, the growth observed at the highest microbicide concentration was aseptically removed and streaked onto a fresh plate containing the same antimicrobial compound concentration gradient. Where growth was observed across the whole antimicrobial gradient, a new plate produced with a 5-fold higher microbicide concentration was used. This process was repeated until 14 passages (P14) had occurred. The bacteria obtained before passage (P0) and at P14 were archived for subsequent susceptibility testing.
Determination of antibiotic susceptibility.
Bacteria showing ≥4-fold increases in the minimum bactericidal concentration (MBC) after microbicide/formulation exposure were investigated for changes in antibiotic susceptibility. Antibiotic susceptibilities were determined for ciprofloxacin (1 μg), cephalothin (20 μg), ampicillin (10 μg), kanamycin (5 μg), and tetracycline (10 μg). Disc diffusion assays were performed according to the British Society for Antimicrobial Chemotherapy (BSAC) disc diffusion method for antimicrobial susceptibility testing (28).
Statistical analyses.
Antibiotic zone-of-inhibition sizes were compared before and after adaptation to microbicides using Mann-Whitney U tests, and those in the cross-resistance assays were compared using linear mixed-effect models (LMMs). LMMs were required to simultaneously compare and account for the effects on the inhibition zone of (i) the microbicidal environment to which the bacterium was adapted, (ii) the antibiotic against which it was tested, and (iii) the interaction of the microbicidal environment and antibiotic (each of which was fitted as a fixed effect) plus (iv) the different bacteria (which was fitted as a random effect), allowing the variation among bacteria to differ for different antibiotics. Initial models with this structure violated the statistical assumptions of the normality of residuals and the homogeneity of variance. Box-Cox transformation indicated that a transformation with a power of 0.5 (square root) was approximately optimal to address the nonnormality and was therefore used. A wide range of different models accounting for the nonhomogeneity of variance in response to different variables was tested. Models allowing different variances for different bacteria and different variances for different microbicidal environments were superior to all others tested (they had the lowest Akaike information criterion). To account for the fact that closely related bacteria are likely to respond more similarly than others just through having a more recent common ancestor, a correlation term was included on the basis of the 16S rRNA sequence-based phylogenetic tree of the strains used. Testing of different weightings on this correlation term (Pagel's λ [29]) determined that a Brownian model (i.e., Pagel's λ = 1) was the best. In addition, an LMM was fitted for the subset of data involving microbicides where bacteria that had adapted to both formulated and unformulated versions of the microbicidal environment were tested. In this case, accounting for nonhomogeneous variance was best done by allowing different variances for different microbicidal environments and for variance to increase at higher values according to the formula e(0.65 · zone-of-clearance value). All models were fitted using the NLME package (version 3.1) (30) in R (version 3.2) (31), with phylogenetic correlation structures being created using the APE package (version 3.3) (32). Where P values are not explicitly given, statistical significance was deemed to be a P value of <0.05.
RESULTS
After exposure to microbicides in simple aqueous solution, out of 90 possible combinations of a bacterium and an antibiotic, the bacteria in 22% of the combinations had significantly (P < 0.05) reduced antibiotic susceptibility (the bacteria in 8%, 6%, 4%, 2%, and 2% of the combinations had reduced susceptibility to ciprofloxacin, ampicillin, kanamycin, tetracycline, and cephalothin, respectively). In comparison, the bacteria in 20% of the combinations had significantly increased antibiotic susceptibility (the bacteria in 6%, 4%, 4%, 3%, and 2% of the combinations had increased susceptibility to ciprofloxacin, kanamycin, tetracycline, cephalothin, and ampicillin, respectively). After exposure to the formulated microbicides, out of 50 possible combinations of a bacterium and an antibiotic, the bacteria in 22% of the combinations had significantly decreased antibiotic susceptibility (the bacteria in 6%, 6%, 4%, 4%, and 2% of the combinations had decreased susceptibility to ciprofloxacin, kanamycin, cephalothin, tetracycline, and ampicillin, respectively). In comparison, the bacteria in 12% of the combinations had significantly increased antibiotic susceptibility (the bacteria in 8%, 2%, and 2% of the combinations had increased susceptibility to ciprofloxacin, kanamycin, and tetracycline, respectively). Importantly, while statistically significant increases and decreases in antibiotic susceptibility occurred, the generation of resistance, as defined by BSAC breakpoints, was not observed in any previously susceptible bacterium.
The frequency of the reduction in antibiotic susceptibility was the highest in organisms exhibiting previously reduced susceptibility to DMDM hydantoin (80%), followed by those exhibiting previously reduced susceptibility to BAC (20%), CHX (20%), DDAC (20%), triclosan (20%), and PHMB (16%). Bacteria with reduced susceptibility to triclosan showed the highest frequency of increased antibiotic susceptibility (45%), followed by bacteria with reduced susceptibility to CHX (30%), DDAC (27%), DMDM hydantoin (20%), and PHMB (4%). In comparison, after exposure to the formulations, 27% of thymol formulation-adapted isolates and 20% of DDAC formulation-adapted isolates exhibited increased antibiotic susceptibility, while 40% of DDAC formulation-adapted bacteria, 33% of thymol formulation-adapted bacteria, 10% of BAC formulation-adapted bacteria, and 7% of PHMB formulation-adapted bacteria had significantly decreased antibiotic susceptibility. The following sections detail the effects of each microbicide on antibiotic susceptibility.
Benzalkonium chloride.
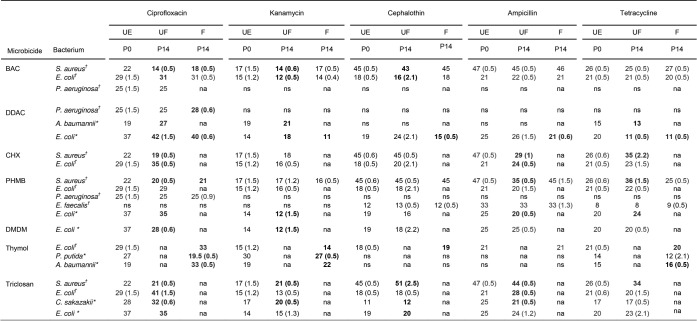
When comparing unexposed organisms to BAC-adapted organisms, there was a significant decrease in the susceptibility of S. aureus to ciprofloxacin and kanamycin (Table 1). E. coli also showed a significant reduction in kanamycin susceptibility after exposure to BAC. After repeated exposure to the BAC formulation, S. aureus showed significantly decreased susceptibility to ciprofloxacin (Table 1).
TABLE 1.
Antibiotic susceptibility of bacterial isolates that showed a ≥4-fold decrease in microbicide/formulation susceptibility following exposure to microbicides in simple aqueous solution or formulated with surfactants and sequestrantsa
Data show growth inhibition zones (in millimeters) representative of antibiotic susceptibility before passage (P0) and after 14 passages (P14) in the presence of a microbicide/formulation. Antibiotic zones of inhibition were determined before antimicrobial exposure (unexposed [UE]) and after antimicrobial exposure to both unformulated (UF) microbicides (i.e., microbicides in a simple aqueous solution) and formulated (F) microbicides (i.e., microbicides with surfactants and sequestrants). †, nondrain isolates; *, drain isolates. Statistically significant changes are in bold text (P < 0.05). Bacteria that did not undergo a ≥4-fold change in MBC were not assessed for changes in antibiotic susceptibility. Where data varied between biological replicates, standard deviations are given in parentheses (n = 6). Combinations of a bacterium and an antibiotic for which BSAC breakpoints are available are indicated in blue text. According to these data, no susceptible bacterium became antibiotic resistant following microbicide adaptation. na, not applicable (i.e., the bacterium did not exhibit a ≥4-fold decrease in microbicide susceptibility following previous exposure); ns, nonsusceptible; BAC, benzalkonium chloride; DDAC, didecyldimethyl ammonium chloride; CHX, chlorhexidine digluconate; PHMB, polyhexamethylene biguanide; DMDM (hydantoin), 1,3-dimethylol-5,5-dimethylhydantoin.
Chlorhexidine.
S. aureus showed a significant decrease in susceptibility to ampicillin and ciprofloxacin after CHX exposure as well as an increase in susceptibility to tetracycline (Table 1). E. coli demonstrated increased susceptibility to ciprofloxacin and ampicillin after repeated exposure to chlorhexidine.
Didecyldimethyl ammonium chloride.
After exposure to DDAC, A. baumannii showed a significant increase in susceptibility to ciprofloxacin and kanamycin and decreased susceptibility to tetracycline compared to the susceptibility of the bacterium before microbicide exposure (Table 1). Increased susceptibility to ciprofloxacin, kanamycin, and cephalothin was observed for the E. coli drain isolate, while a significant reduction in tetracycline susceptibility was also evident in this bacterium. After exposure to the DDAC formulation, the E. coli drain isolate underwent a significant reduction in kanamycin, cephalothin, tetracycline, and ampicillin susceptibility and an increase in susceptibility to ciprofloxacin. P. aeruginosa showed a significant increase in ciprofloxacin susceptibility after long-term exposure to the DDAC formulation (Table 1).
DMDM hydantoin.
After repeated exposure to DMDM hydantoin, the E. coli drain isolate demonstrated a significant reduction in ciprofloxacin, kanamycin, cephalothin, and ampicillin susceptibility and an increase in tetracycline susceptibility compared to the susceptibility of its preexposed counterpart (Table 1).
Polyhexamethylene biguanide.
Following adaptation to PHMB, the E. coli drain isolate exhibited a decrease in kanamycin and ciprofloxacin susceptibility (Table 1). S. aureus developed a significantly reduced susceptibility to ampicillin and ciprofloxacin after repeated PHMB exposure but higher tetracycline susceptibility than the unexposed parent strain. After exposure to the PHMB formulation, S. aureus also showed a significant reduction in ciprofloxacin susceptibility.
Thymol.
None of the test bacteria demonstrated a significant change in antibiotic susceptibility after exposure to thymol in aqueous solution. Following exposure to the thymol-containing formulation, however, P. putida underwent significant decreases in susceptibility to ciprofloxacin and kanamycin (Table 1), while E. coli showed significant increases in ciprofloxacin and cephalothin susceptibility but decreases in susceptibility to kanamycin and tetracycline. The susceptibility of A. baumannii to ciprofloxacin, kanamycin, and tetracycline increased compared to that of its unexposed counterpart (Table 1).
Triclosan.
Following exposure to triclosan, S. aureus exhibited significant reductions in ciprofloxacin and ampicillin susceptibility, while its susceptibility to kanamycin, tetracycline, and cephalothin increased (Table 1). E. coli showed increased susceptibility to ampicillin and ciprofloxacin after triclosan exposure, while the E. coli drain isolate showed decreased ciprofloxacin susceptibility but increased cephalothin susceptibility compared to that of the parent strain. Comparatively, C. sakazakii showed a significant increase in ciprofloxacin, cephalothin, and kanamycin susceptibility and a decrease in ampicillin susceptibility after repeated triclosan exposure (Table 1).
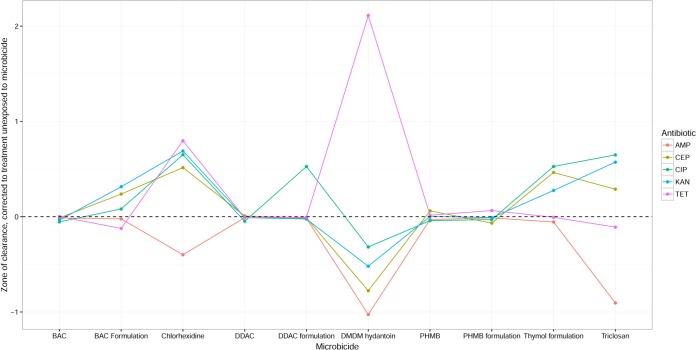
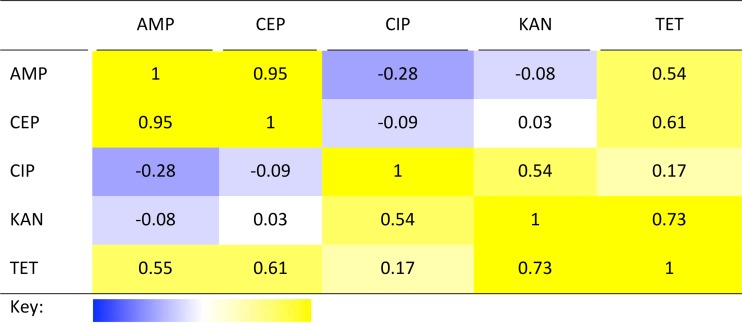
To gain an overview of the statistical significance of the observed changes in antibiotic susceptibility and ask whether it was possible to identify consistent patterns in susceptibility, linear mixed-effects models for how the susceptibility to particular antibiotics varied depending on the antibiotic in question, the bacterium, and the microbicidal environment that the bacterium was previously adapted to were fitted. A highly significant interaction (F40, 298 = 15, P < 2 × 10−16) indicative of different responses to particular antibiotics depending on the microbicidal environment to which the organism had previously adapted (Fig. 1) was observed. Bacterial strains differed most in their response to ampicillin (standard deviation among strains = 5.1) and least in their response to tetracycline (standard deviation among strains = 2.7), with the responses of different strains to some antibiotics being associated either positively (cephalothin and ampicillin, r = 0.95) or negatively (ciprofloxacin and ampicillin, r = −0.28) (Table 2).
FIG 1.
Antibiotic susceptibility of strains adapted to different microbicides. The values plotted are the differences in the average zone of clearance across strains before and after adaptation to the given microbicide, as estimated by the linear mixed-effects model (arbitrary scale; see Materials and Methods); i.e., values above 0 indicate antibiotic cross-susceptibility arising from adaptation to the microbicide, and values below 0 indicate cross-resistance. Points are connected for ease of comparison only. For abbreviations on the x axis, see footnote a to Table 1. AMP, ampicillin; CEP, cephalothin; CIP, ciprofloxacin; KAN, kanamycin; TET, tetracycline.
TABLE 2.
Correlation of responses to different antibiotics across strains in LMMa
A value of 1 (deep yellow in the key) indicates that all organisms responded in a perfectly correlated way to the two antibiotics indicated (they were either more or less sensitive to both); a value of −1 (deep blue in the key) indicates a perfect negative correlation, i.e., organisms that are more sensitive to one antibiotic being less sensitive to the other and vice versa. For abbreviations, see the legend to Fig. 1.
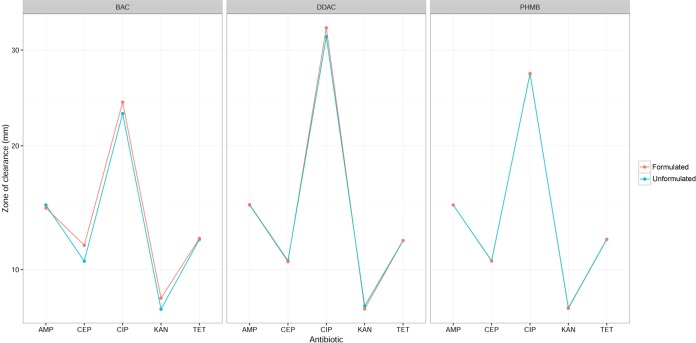
Data presented in Fig. 1 indicate differences in the antibiotic susceptibilities of organisms previously adapted to either formulated or unformulated microbicides. The differences in susceptibility changes observed between microbicides in simple aqueous solution or in a complex formulation were highly significant (in a likelihood ratio [LR] test of the full model against a model treating formulated and unformulated versions of microbicides as equivalent, LR88, 70 = 61 and P = 8.6 × 10−10). To test whether there was any consistent effect of formulation, a second linear mixed-effects model was created for the subset of the data where strains had adapted to both formulated and unformulated versions of the same microbicide (PHMB, BAC, and DDAC). This indicated that the way in which bacteria adapted to formulated versus nonformulated versions of a microbicide depended on the microbicide in question (F2, 145 = 4.5, P = 0.012), although that did not vary significantly among the antibiotics (F8, 145 = 0.70, P = 0.69). The effect of formulation was specific to BAC, with formulation giving a small increase in the antibiotic susceptibility of microbes adapted to it (Fig. 2).
FIG 2.
Antibiotic susceptibility of strains exposed to different microbicides in formulation with surfactants and sequestrants and simple aqueous solution (unformulated). A significant difference is apparent only for BAC. The values plotted are the average zone of clearance (in millimeters) estimated in the linear mixed-effects model (note the transformed scale used by the model; see Materials and Methods). For abbreviations, see the legend to Fig. 1.
DISCUSSION
Investigations into the potential of microbicides to select for reduced microbicide susceptibility in bacteria and induce cross-resistance to antibiotics have largely been conducted by evaluating susceptibility changes following the exposure of bacteria to microbicides in simple aqueous solution (17). In such experiments, the susceptibility of the exposed bacteria has been reported to decrease for certain combinations of a bacterium and a microbicide either transiently or stably (26). In the real world, however, microbicides are deployed in complex formulations containing sequestrants, surfactants, and other compounds. Recent investigations indicate that the formulation of microbicides can significantly enhance antibacterial potency and that decreases in microbicide susceptibility after sublethal microbicide exposure may be significantly lower in frequency and extent when the microbicides are incorporated into formulations reflecting application in the real world (24, 33). This highlights the value of risk assessments that more accurately reflect the way in which microbicides are deployed. In the current investigation, we evaluated whether the formulation of microbicides additionally mitigates the development of antibiotic insusceptibility in bacteria.
In order to investigate whether the formulation of microbicides affects cross-resistance to antibiotics, we studied the induction of changes in antibiotic susceptibility in bacteria that had been repeatedly exposed, using a highly selective system arguably representing a worst-case scenario, to microbicides in simple aqueous solution and in formulation with ingredients that are used in consumer products, such as laundry detergents, hard-surface disinfectants, and personal care products (24). It should be noted that while the majority of microbicides tested are widely used in domestic cleaning products, the use of triclosan in Europe is generally restricted to applications where its utility is the greatest, such as oral care.
Out of 288 microbicide-exposed bacteria, 28 organisms previously demonstrated a ≥4-fold decrease in microbicide susceptibility (18 organisms adapted to microbicides following exposure to simple aqueous solutions and 10 adapted to microbicides in formulation). These were further evaluated for changes in antibiotic susceptibility in the current study. The difference in the numbers of test bacteria between treatment groups results from the mitigating effects that the formulation of microbicides had on the development of microbicide insusceptibility. Increases in antibiotic susceptibility occurred at a higher frequency following exposure to simple solutions than following exposure to formulations (20% versus 12% of the bacteria), while 22% of the bacteria became significantly less susceptible to the antibiotics regardless of formulation. While both increases and decreases in the antibiotic susceptibility of the test bacteria were observed after exposure to a microbicide/formulation, no bacterium became resistant according to published BSAC breakpoints.
Changes in antibiotic susceptibility varied between the test antibiotics, the bacteria, and the microbicides that the bacteria had been previously adapted to, suggesting little correlative effect between the different variables. One positive correlation was, however, observed between the β-lactam antibiotics ampicillin and cephalothin (Table 2). In this case, microbicide exposure could have altered transpeptidase expression or otherwise influenced cell wall permeability, subsequently impacting the efficacy of these antibiotics, which target cell wall synthesis.
In some cases, bacterial antibiotic susceptibility was increased following microbicide exposure. It is notable that such cross-susceptibility was associated with adaptation to at least some microbicides for all antibiotics except ampicillin (Fig. 1). The phenomenon of cross-susceptibility has been observed in several previous investigations (17, 22, 34, 35), where links between antibiotics and decreased microbicide susceptibility in bacteria have been demonstrated in vitro (14, 17). In a recent study, exposure of Burkholderia cepacia to low concentrations of either CHX or BAC resulted in variable reductions in antibiotic susceptibility (36). CHX exposure was reportedly associated with significant decreases in susceptibility to ceftazidime, ciprofloxacin, and imipenem, while short-term exposure to BAC resulted in significant decreases in ceftazidime, ciprofloxacin, and meropenem susceptibility. These effects were, however, highly variable between biological replicates in a manner suggestive of stochastic effects. In another recent investigation, six S. aureus strains, including methicillin-resistant S. aureus, were repeatedly exposed to triclosan. Susceptibility to triclosan was significantly decreased in all exposed bacteria, whereas antibiotic susceptibility was significantly increased in the majority of cases. While the reasons for cross-susceptibility have not been elucidated, they are likely to include the general fitness costs of adaptation and transient cellular damage, as previously hypothesized (37).
The mechanisms of cross-resistance have been more extensively investigated and include nonspecific reductions in cell permeability, active efflux of the compound from the bacterial cell, or the acquisition of mutations in shared target sites (14, 17). Antibiotics such as aminoglycosides enter the cell through a mechanism of self-promoted uptake (38), whereby they displace cations in the bacterial cell envelope, leading to the reorganization of lipopolysaccharide, which may facilitate antibiotic entry. This mechanism of self-promoted uptake mirrors that of polymeric biguanides, such as PHMB and CHX (39), which has led to the question as to whether any adaptation to reduce biguanide uptake may have a resulting effect on the uptake of aminoglycosides into the bacterial cell. The current investigation included the evaluation of any changes in susceptibility to the aminoglycoside antibiotic kanamycin in bacteria that had previously shown to have reduced susceptibility to both CHX and PHMB. However, we found no evidence of a systematic effect of this sort (indeed, adaptation to CHX typically led to an increase in susceptibility to kanamycin; Fig. 1), and only the PHMB-adapted E. coli drain isolate showed any significant reduction in antibiotic susceptibility (Table 1).
Cross-resistance between quaternary ammonium compounds (QACs), such as BAC and DDAC, and antibiotics has been attributed to the expression of broad-range efflux systems capable of removing both the microbicide and certain antibiotics from the bacterial cell (40–42). It has additionally been noted that genes encoding QAC-specific efflux pumps, such as qacA and qacB, may be detected on plasmids bearing β-lactamases in certain clinical isolates, suggesting another cause for cross-resistance between QACs and penicillins (43). Furthermore, the qacE gene has been detected in the 3′ conserved structure (3′-CS) of certain class 1 integrons found in many Gram-negative bacteria. Class 1 integrons often contain multiple antibiotic resistance genes, and since they are commonly located on plasmids, this makes them highly mobile via the action of plasmid-mediated conjugation. This consequentially facilitates the dissemination of both QAC and antibiotic resistance genes through a population via horizontal gene transfer (44). Our data indicate that 20% of bacterial isolates with reduced BAC and DDAC susceptibility, in addition to 40% and 10% of isolates with reduced DDAC or BAC formulation susceptibility, respectively, were also significantly reduced in their antibiotic susceptibility. Linear mixed-effect modeling revealed that the formulation of BAC conferred a moderate protective effect on the development of antibiotic cross-resistance (Fig. 2), possibly suggesting a regulatory impact of the formulation excipients on the induction of the aforementioned efflux mechanisms due to nonspecific effects on cell permeability or through other cellular changes.
Triclosan exposure may select for mutations in the gene that encodes the target enzyme FabI, an enoyl-acyl carrier protein reductase that participates in bacterial fatty acid synthesis (45). There has been concern over the induction of cross-resistance between triclosan and therapeutic agents that also share this target enzyme, such as isoniazid, which is used to treat Mycobacterium tuberculosis infections. Cross-resistance between triclosan and certain antibiotics has been reported in P. aeruginosa and is largely believed to be due to increased expression of the MexAB-OprM efflux system (14). In the current investigation, the data showed reductions in ciprofloxacin susceptibility in S. aureus and the E. coli drain isolate together with reductions in ampicillin susceptibility in S. aureus and C. sakazakii after repeated triclosan exposure, which may potentially be mediated through the regulation of efflux or cell permeability.
While the induction of cross-resistance between microbicides and antibiotics has been previously investigated, little information concerning any effect of incorporation of microbicides into formulations containing surfactants and sequestrants on antibiotic susceptibility in adapted bacteria is available. The data presented here indicate that both decreases and increases in antibiotic susceptibility can occur in bacteria following exposure to microbicides in simple solution and in formulations using a highly selective system. A rigorous statistical analysis demonstrated that formulation significantly affected the development of cross-resistance but that this was variable, with the only consistently identified formulation effect being a small increase in susceptibility across antibiotics in strains adapted to the formulated relative to the unformulated version of the microbicide benzalkonium chloride.
In conclusion, while both increases and decreases in antibiotic susceptibility were observed in microbicide- and formulation-adapted bacteria, these were not sufficient to confer clinical resistance according to published BSAC breakpoints.
ACKNOWLEDGMENTS
We thank Joanne O'Keeffe and Andrew Jamieson from Unilever R&D, Port Sunlight, United Kingdom, for their advice regarding the selection of microbicides and formulations.
This project was funded by Unilever's Safety & Environmental Assurance Centre (SEAC).
Alejandro Amézquita is an employee of Unilever. Peter McClure was an employee of Unilever when this project was initiated. None of the other authors has a conflict to declare.
REFERENCES
- 1.Pereira M, Vieira M, Beleza V, Melo L. 2001. Comparison of two biocides—carbamate and glutaraldehyde—in the control of fouling in pulp and paper industry. Environ Technol 22:781–790. doi: 10.1080/095933322086180318. [DOI] [PubMed] [Google Scholar]
- 2.Barbolt TA. 2002. Chemistry and safety of triclosan, and its use as an antimicrobial coating on coated VICRYL* Plus antibacterial suture (coated polyglactin 910 suture with triclosan). Surg Infect (Larchmt) 3(Suppl 1):S45–S53. doi: 10.1089/sur.2002.3.s1-45. [DOI] [PubMed] [Google Scholar]
- 3.Bibbo C, Patel D, Gehrmann R, Sheldon L. 2005. Chlorhexidine provides superior skin decontamination in foot and ankle surgery: a prospective randomized study. Clin Orthop Relat Res 438:204–208. [DOI] [PubMed] [Google Scholar]
- 4.Abreu AC, Tavares RR, Borges A, Mergulhão F, Simões M. 2013. Current and emergent strategies for disinfection of hospital environments. J Antimicrob Chemother 68:2718–2732. doi: 10.1093/jac/dkt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBain AJ, Bartolo RG, Catrenich CE, Charbonneau D, Ledder RG, Gilbert P. 2003. Effects of a chlorhexidine gluconate-containing mouthwash on the vitality and antimicrobial susceptibility of in vitro oral bacterial ecosystems. Appl Environ Microbiol 69:4770–4776. doi: 10.1128/AEM.69.8.4770-4776.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koburger T, Hubner NO, Braun M, Siebert J, Kramer A. 2010. Standardized comparison of antiseptic efficacy of triclosan, PVP-iodine, octenidine dihydrochloride, polyhexanide and chlorhexidine digluconate. J Antimicrob Chemother 65:1712–1719. doi: 10.1093/jac/dkq212. [DOI] [PubMed] [Google Scholar]
- 7.Best M, Kennedy M, Coates F. 1990. Efficacy of a variety of disinfectants against Listeria spp. Appl Environ Microbiol 56:377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McBain A, Gilbert P. 2001. Biocide tolerance and the harbingers of doom. Int Biodeterior Biodegrad 47:55–61. doi: 10.1016/S0964-8305(01)00037-3. [DOI] [Google Scholar]
- 9.Maillard J-Y. 2010. Emergence of bacterial resistance to microbicides and antibiotics. Microbiol Aust 31:159–164. [Google Scholar]
- 10.Maillard J-Y. 2007. Bacterial resistance to biocides in the healthcare environment: should it be of genuine concern? J Hosp Infect 65:60–72. doi: 10.1016/S0195-6701(07)60018-8. [DOI] [PubMed] [Google Scholar]
- 11.Karatzas KA, Webber MA, Jorgensen F, Woodward MJ, Piddock LJ, Humphrey TJ. 2007. Prolonged treatment of Salmonella enterica serovar Typhimurium with commercial disinfectants selects for multiple antibiotic resistance, increased efflux and reduced invasiveness. J Antimicrob Chemother 60:947–955. doi: 10.1093/jac/dkm314. [DOI] [PubMed] [Google Scholar]
- 12.Tattawasart U, Maillard JY, Furr JR, Russell AD. 1999. Development of resistance to chlorhexidine diacetate and cetylpyridinium chloride in Pseudomonas stutzeri and changes in antibiotic susceptibility. J Hosp Infect 42:219–229. doi: 10.1053/jhin.1999.0591. [DOI] [PubMed] [Google Scholar]
- 13.Webber MA, Whitehead RN, Mount M, Loman NJ, Pallen MJ, Piddock LJ. 2015. Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J Antimicrob Chemother 70:2241–2248. doi: 10.1093/jac/dkv109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuanchuen R, Beinlich K, Hoang TT, Becher A, Karkhoff-Schweizer RR, Schweizer HP. 2001. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob Agents Chemother 45:428–432. doi: 10.1128/AAC.45.2.428-432.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winder CL, Al-Adham ISI, Abdel Malek SMA, Buultjens TEJ. 2000. Outer membrane protein shifts in biocide resistant Pseudomonas aeruginosa PAO1. J Appl Microbiol 89:289–295. doi: 10.1046/j.1365-2672.2000.01119.x. [DOI] [PubMed] [Google Scholar]
- 16.Bloomfield SF, Arthur M. 1994. Mechanisms of inactivation and resistance of spores to chemical biocides. J Appl Microbiol 76:91S–104S. [DOI] [PubMed] [Google Scholar]
- 17.Walsh SE, Maillard J-Y, Russell A, Catrenich C, Charbonneau D, Bartolo R. 2003. Development of bacterial resistance to several biocides and effects on antibiotic susceptibility. J Hosp Infect 55:98–107. doi: 10.1016/S0195-6701(03)00240-8. [DOI] [PubMed] [Google Scholar]
- 18.Oggioni MR, Furi L, Coelho JR, Maillard J-Y, Martínez JL. 2013. Recent advances in the potential interconnection between antimicrobial resistance to biocides and antibiotics. Exp Rev Anti Infect Ther 11:363–366. doi: 10.1586/eri.13.16. [DOI] [PubMed] [Google Scholar]
- 19.Cottell A, Denyer S, Hanlon G, Ochs D, Maillard J-Y. 2009. Triclosan-tolerant bacteria: changes in susceptibility to antibiotics. J Hosp Infect 72:71–76. doi: 10.1016/j.jhin.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 20.Maillard J-Y. 2005. Antimicrobial biocides in the healthcare environment: efficacy, usage, policies, and perceived problems. Ther Clin Risk Manag 1:307–320. [PMC free article] [PubMed] [Google Scholar]
- 21.Morrissey I, Oggioni MR, Knight D, Curiao T, Coque T, Kalkanci A, Martinez JL, BIOHYPO Consortium. 2014. Evaluation of epidemiological cut-off values indicates that biocide resistant subpopulations are uncommon in natural isolates of clinically-relevant microorganisms. PLoS One 9:e86669. doi: 10.1371/journal.pone.0086669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forbes S, McBain AJ, Felton-Smith S, Jowitt TA, Birchenough HL, Dobson CB. 2013. Comparative surface antimicrobial properties of synthetic biocides and novel human apolipoprotein E derived antimicrobial peptides. Biomaterials 34:5453–5464. doi: 10.1016/j.biomaterials.2013.03.087. [DOI] [PubMed] [Google Scholar]
- 23.Ledder RG, Gilbert P, Willis C, McBain AJ. 2006. Effects of chronic triclosan exposure upon the antimicrobial susceptibility of 40 ex-situ environmental and human isolates. J Appl Microbiol 100:1132–1140. doi: 10.1111/j.1365-2672.2006.02811.x. [DOI] [PubMed] [Google Scholar]
- 24.Cowley N, Forbes S, Amézquita A, McClure P, Humphreys G, McBain AJ. 2015. The effect of formulation on microbicide potency and mitigation of the development of bacterial insusceptibility. Appl Environ Microbiol 81:7330–7338. doi: 10.1128/AEM.01985-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall BM, Robleto E, Dumont T, Levy SB. 2012. The frequency of antibiotic-resistant bacteria in homes differing in their use of surface antibacterial agents. Curr Microbiol 65:407–415. doi: 10.1007/s00284-012-0172-x. [DOI] [PubMed] [Google Scholar]
- 26.Forbes S, Dobson CB, Humphreys GJ, McBain AJ. 2014. Transient and sustained bacterial adaptation following repeated sublethal exposure to microbicides and a novel human antimicrobial peptide. Antimicrob Agents Chemother 58:5809–5817. doi: 10.1128/AAC.03364-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore LE, Ledder RG, Gilbert P, McBain AJ. 2008. In vitro study of the effect of cationic biocides on bacterial population dynamics and susceptibility. Appl Environ Microbiol 74:4825–4834. doi: 10.1128/AEM.00573-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews JM. 2001. BSAC standardized disc susceptibility testing method. J Antimicrob Chemother 48:43–57. doi: 10.1093/jac/48.suppl_1.43. [DOI] [PubMed] [Google Scholar]
- 29.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 30.Pinheiro J, Bates D. 2006. Mixed-effects models in S and S-PLUS. Springer Science & Business Media, New York, NY. [Google Scholar]
- 31.R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.r-project.org. [Google Scholar]
- 32.Paradis E. 2011. Analysis of phylogenetics and evolution with R. Springer Science & Business Media, New York, NY. [Google Scholar]
- 33.Knapp L, Amézquita A, McClure P, Stewart S, Maillard J-Y. 2015. Development of a protocol for predicting bacterial resistance to microbicides. Appl Environ Microbiol 81:2652–2659. doi: 10.1128/AEM.03843-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krašovec R, Belavkin RV, Aston JA, Channon A, Aston E, Rash BM, Kadirvel M, Forbes S, Knight CG. 2014. Mutation rate plasticity in rifampicin resistance depends on Escherichia coli cell-cell interactions. Nat Commun 5:3742. doi: 10.1038/ncomms4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forbes S, Latimer J, Bazaid A, McBain AJ. 2015. Altered competitive fitness, antimicrobial susceptibility, and cellular morphology in a triclosan-induced small-colony variant of Staphylococcus aureus. Antimicrob Agents Chemother 59:4809–4816. doi: 10.1128/AAC.00352-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knapp L, Rushton L, Stapleton H, Sass A, Stewart S, Amezquita A, McClure P, Mahenthiralingam E, Maillard JY. 2013. The effect of cationic microbicide exposure against Burkholderia cepacia complex (Bcc); the use of Burkholderia lata strain 383 as a model bacterium. J Appl Microbiol 115:1117–1126. doi: 10.1111/jam.12320. [DOI] [PubMed] [Google Scholar]
- 37.McBain AJ, Ledder RG, Sreenivasan P, Gilbert P. 2004. Selection for high-level resistance by chronic triclosan exposure is not universal. J Antimicrob Chemother 53:772–777. doi: 10.1093/jac/dkh168. [DOI] [PubMed] [Google Scholar]
- 38.Hancock RE. 1981. Aminoglycoside uptake and mode of action—with special reference to streptomycin and gentamicin. I. Antagonists and mutants. J Antimicrob Chemother 8:249–276. doi: 10.1093/jac/8.4.249. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert P, Pemberton D, Wilkinson DE. 1990. Synergism within polyhexamethylene biguanide biocide formulations. J Appl Microbiol 69:593–598. [DOI] [PubMed] [Google Scholar]
- 40.Chen J, Kuroda T, Huda MN, Mizushima T, Tsuchiya T. 2003. An RND-type multidrug efflux pump SdeXY from Serratia marcescens. J Antimicrob Chemother 52:176–179. doi: 10.1093/jac/dkg308. [DOI] [PubMed] [Google Scholar]
- 41.Levy SB. 2002. Active efflux, a common mechanism for biocide and antibiotic resistance. J Appl Microbiol 92:65S–71S. doi: 10.1046/j.1365-2672.92.5s1.4.x. [DOI] [PubMed] [Google Scholar]
- 42.Maseda H, Hashida Y, Konaka R, Shirai A. 2009. Mutational upregulation of a resistance-nodulation-cell division-type multidrug efflux pump, SdeAB, upon exposure to a biocide, cetylpyridinium chloride, and antibiotic resistance in Serratia marcescens. Antimicrob Agents Chemother 53:5230–5235. doi: 10.1128/AAC.00631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lyon BR, Skurray R. 1987. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev 51:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paulsen IT, Littlejohn TG, Rådström P, Sundström L, Sköld O, Swedberg G, Skurray RA. 1993. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother 37:761–768. doi: 10.1128/AAC.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMurry LM, Oethinger M, Levy SB. 1998. Triclosan targets lipid synthesis. Nature 394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]