ABSTRACT
The McMurdo Dry Valleys (MCM) of southern Victoria Land, Antarctica, harbor numerous ice-covered bodies of water that provide year-round liquid water oases for isolated food webs dominated by the microbial loop. Single-cell microbial eukaryotes (protists) occupy major trophic positions within this truncated food web, ranging from primary producers (e.g., chlorophytes, haptophytes, and cryptophytes) to tertiary predators (e.g., ciliates, dinoflagellates, and choanoflagellates). To advance the understanding of MCM protist ecology and the roles of MCM protists in nutrient and energy cycling, we investigated potential metabolic strategies and microbial interactions of key MCM protists isolated from a well-described lake (Lake Bonney). Fluorescence-activated cell sorting (FACS) of enrichment cultures, combined with single amplified genome/amplicon sequencing and fluorescence microscopy, revealed that MCM protists possess diverse potential metabolic capabilities and interactions. Two metabolically distinct bacterial clades (Flavobacteria and Methylobacteriaceae) were independently associated with two key MCM lake microalgae (Isochrysis and Chlamydomonas, respectively). We also report on the discovery of two heterotrophic nanoflagellates belonging to the Stramenopila supergroup, one of which lives as a parasite of Chlamydomonas, a dominate primary producer in the shallow, nutrient-poor layers of the lake.
IMPORTANCE Single-cell eukaryotes called protists play critical roles in the cycling of organic matter in aquatic environments. In the ice-covered lakes of Antarctica, protists play key roles in the aquatic food web, providing the majority of organic carbon to the rest of the food web (photosynthetic protists) and acting as the major consumers at the top of the food web (predatory protists). In this study, we utilized a combination of techniques (microscopy, cell sorting, and genomic analysis) to describe the trophic abilities of Antarctic lake protists and their potential interactions with other microbes. Our work reveals that Antarctic lake protists rely on metabolic versatility for their energy and nutrient requirements in this unique and isolated environment.
INTRODUCTION
The microbial loop links the transfer of nutrients and carbon between the microorganisms of the food web and includes complex ecological interactions between microbial eukaryotes, bacteria, archaea, and viruses. Microbial activity is influenced by bottom-up (e.g., availability of nutrient and energy sources) and top-down (e.g., predation, parasitism, and viral lysis) controls, as well as microbe-microbe interactions (e.g., bacterial consortia). Interaction partnerships between microorganisms lead to combinations of win, loss, or neutral outcomes for each microbial partner. Parasitism and predation are examples of win-loss outcomes (1), while mutualistic interactions within biofilms and syntrophic interactions (cross-feeding of metabolic products between two species) between an alga and a heterotrophic bacterium are examples of win-win partnerships (2). Understanding microbial interactions and the influences of environmental factors on microbial distribution patterns is critical for deciphering global nutrient fluxes and biogeochemical cycles.
Single-cell eukaryotic microorganisms (i.e., protists) are ubiquitous in every ecosystem on earth and play critical ecological roles in food web dynamics and global carbon and nutrient cycles (3). Diverse protist lineages possess multiple nutritional modes, playing key roles as producers, decomposers, parasites, and predators. Phototrophic protists are important producers that contribute to global primary production and incorporate a significant proportion of inorganic carbon into the food web (4). Predatory protists are major consumers of planktonic phytoplankton and bacteria, providing control over the abundance of these organisms as well as linking primary producers/consumers with higher trophic levels (i.e., metazoans) (5). Mixotrophic protists (with combined abilities for photosynthesis and phagotrophic ingestion of food particles) are widespread in aquatic ecosystems (6, 7). Recent molecular surveys from marine and freshwater environments have revealed a great diversity of 18S rRNA sequences that are unrelated to existing cultured protists (8, 9). The application of high-throughput sequencing has begun to reveal protist biogeography (10–12), as well as seasonal variability (13, 14).
The McMurdo Dry Valleys (MCM) of southern Victoria Land, Antarctica, represent a polar desert, with average air temperatures of −20°C and precipitation rates of <10 cm per year (15, 16). Numerous marine-derived, perennially ice-covered lakes are the only source of year-round liquid water for life on the Antarctic continent. Permanent ice caps prevent wind mixing and significant nutrient inputs; MCM lake chemistry is vertically stratified in the water column, and lakes exhibit oxygen-rich ultraoligotrophic surface waters that are separated by permanent chemoclines from ancient anoxic saline waters (17). Thus, the MCM lakes represent aquatic environments with strong selective pressures on microbial evolution, including low temperatures, low annual levels of photosynthetically active radiation (PAR), and limited nutrient (nitrogen and phosphorus) availability (18). Salinity, oxygen, and light appear to play important roles in the biogeography of Antarctic lake microorganisms (19). Each lake supports simple food webs that harbor few to no metazoans, with the exception of small numbers of copepods and rotifers in a few lakes (20). Protists play dominating roles in carbon and nutrient cycling in the MCM microbial food web. The majority of dissolved organic carbon is autochthonous (i.e., derived from new photosynthetic activity within the water column). Protists represent the major producers of organic matter in the MCM lake food web (21, 22), while heterotrophic nanoflagellates (HNFs) and ciliates are the top predators of bacteria and smaller protists (23, 24). Despite the energetic costs of maintaining and regulating both photosynthetic and heterotrophic cellular apparatuses, mixotrophy appears to be very prevalent in MCM aquatic food webs (23, 25). Mixotrophic metabolism is a survival strategy for MCM protists to exploit alternative sources of either nutrients (i.e., under oligotrophic conditions) or energy (i.e., in the absence of light) (26).
As part of the NSF Long Term Ecological Research (LTER) Program, several lakes within the MCM have been the focus of more than 2 decades of intensive study. Lake Bonney is one of the most well-studied MCM lakes, and the physical and chemical characteristics of the lake have been thoroughly described (27). Lake Bonney is divided into two basins (the west and east lobes), which are separated by a narrow passage that allows mixing of the upper photic zones for a few weeks in the summer. The shallow waters of both lobes are generally freshwater; however, salinity levels increase steeply at the permanent chemocline (18- to 20-m depth for the east lobe). Below the chemocline, the deep anoxic zone is hypersaline (maximum salinity, 125 to 150 PSU). Photosynthetically active radiation (PAR) is relatively low under the ice (<50 μmol m−2 s−1), due to the attenuation of the ice cover, and declines below detectable levels at depths of >25 m (28). Recently, the diversity and spatial distribution of the planktonic microbial eukaryotic communities residing in Lake Bonney have been reported (28–31). Protist populations residing in the MCM lakes are vertically stratified and exhibit lake-specific differences in distribution patterns. The water column of Lake Bonney is dominated by large chlorophytes (genus Chlamydomonas) in the shallow, nutrient-poor depths, while the dominant photosynthetic protists are nanoplankton (haptophytes and stramenopiles), which exhibit peak abundances within the permanent chemocline, where PAR is extremely limited (29). While these studies have begun to describe the biogeographic distribution of the protist communities residing in these ice-covered polar lakes, a full understanding of the trophic abilities of MCM protists and potential interactions with other MCM microorganisms is currently lacking.
The vast diversity of microbial eukaryotes has been largely inaccessible with conventional cultivation methods. Over the past decade, the technology of single-cell genomics has been exploited to recover genomic information from single uncultivated cells collected from their natural habitats and has revealed cell-specific interactions such as symbiosis, predation, and parasitism (32–34). In this study, single cells of protists originating from an enrichment culture from the zone of maximum primary production in the permanent chemocline of Lake Bonney (east lobe) were isolated on the basis of fluorescence properties, which were related to their potential nutritional mode (photosynthetic, heterotrophic, or mixotrophic). We chose to utilize an enrichment cultivation approach to allow us to work with live organisms and to avoid the logistical issues associated with preserving and shipping natural protist communities from Antarctica to our U.S. laboratory. Our hypothesis was that MCM lake protists possess broad metabolic versatility, which drives nutritional mode-dependent microbial interactions. Specifically, we addressed the following open questions regarding the ecology of the MCM lake microbial eukaryote communities. What is the range of trophic versatility in the MCM planktonic protist communities? Do MCM phototrophic and heterotrophic protists interact with specific microbial partners? In this study, whole-genome amplification (WGA) was performed to produce single amplified genomes (SAGs) from a variety of the sorted eukaryotic cells (with their microbial partners). Combining Illumina sequencing with various microscopic methods, we were able (i) to identify the nutritional mode and metabolic potential of several key MCM protists, (ii) to reveal a diversity of potential microbial interactions, including predation, parasitism, and endosymbiosis, and (iii) to provide new information for understanding microbial metabolic strategies for survival under permanent light-limiting and nutrient-poor conditions.
MATERIALS AND METHODS
Site description, sample collection, and enrichment cultures.
Samples were collected from the water column of the east lobe of Lake Bonney, an ice-covered meromictic lake located in the Taylor Valley, McMurdo Dry Valleys. Samples were collected through the ice hole located in the middle of the lake (77°42.825′S, 162°26.832′E) on 21 December 2012, during the austral summer. A sampling depth (piezometric depth of 13 m) was selected to reflect the depth of maximum phytoplankton biomass (29), which was verified by in situ chlorophyll fluorescence with a submersible FluoroProbe (BBE Moldaenke GmbH) (28). Lake water samples were collected with a 5-liter Niskin bottle (General Oceanics), transferred to 1-liter amber bottles, and stored at 4°C in the dark until processing.
An enrichment cultivation approach was employed to allow for the transport of live cultures with greater biomass to our U.S. laboratory (Miami University, Oxford, OH, USA). To ensure maximum recovery of protist diversity, a series of enrichment cultures was started in the presence of a number of autotrophic medium types (35–37) and growth medium concentrations (10 to 100%). Lake water was inoculated into 25-ml sterile culture flasks and incubated in a photoincubator, at 5°C and 25 μmol photons m−2 s−1, at McMurdo Station, Antarctica, for 2 weeks prior to shipment. Cultures were regularly inspected under the microscope for growth and the presence of microbial eukaryotes. Cultures were then shipped to our U.S. laboratory (Miami University) at 4°C in the dark. Upon arrival, enrichment cultures were transferred to a photoincubator set at the same temperature and light conditions until cell sorting. While cryopreservation in the presence of glycerol buffer works well for preserving bacterial communities (38, 39), preliminary experiments showed that this preservation approach failed to preserve the internal structures of eukaryotic cells and caused lysis of some wall-less protists.
Single-cell sorting.
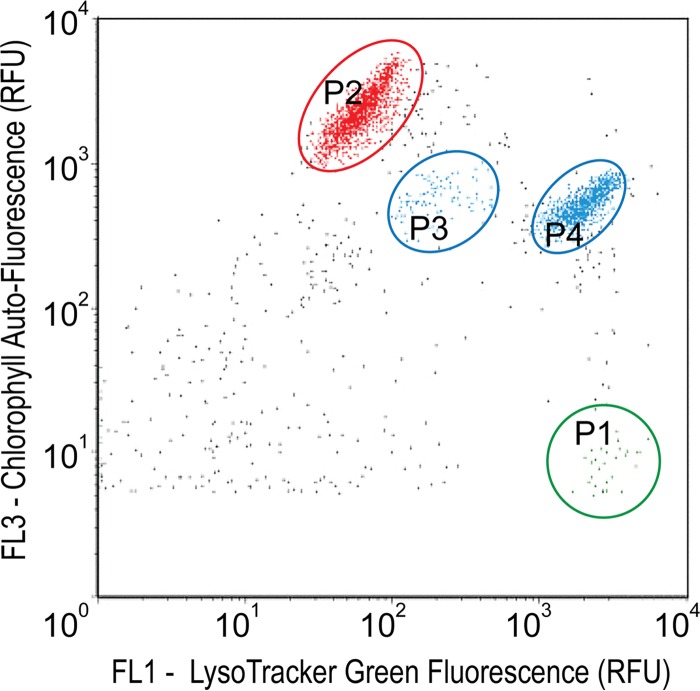
Prior to cell sorting, all enrichment cultures were visually inspected by light microscopy for growth and morphological diversity of the protist communities. Based on these microscopic observations, one enrichment culture (grown in 10% F medium) that exhibited relatively great protist morphological diversity was chosen to be shipped to Oak Ridge National Laboratory for single-cell sorting. Protist food vacuoles were stained using a pH-sensitive probe (LysoTracker Green DND-26; Invitrogen, Carlsbad, CA, USA) (40). A 2-ml aliquot of the culture was gently homogenized with several inversions and incubated for 15 min in the dark on ice in the presence of 75 nmol liter−1 of LysoTracker. Microbial eukaryotic cells of various sizes, morphological types, and trophic abilities (phototrophic, heterotrophic, or mixotrophic) were identified and sorted with a Cytopeia Influx sorter (BD, Franklin Lakes, NJ, USA) in a class 1000 clean room, using a 488-nm argon laser for excitation. Green (528 to 538 nm) and red (670 to 730 nm) fluorescent emission indicated LysoTracker green fluorescence and chlorophyll autofluorescence, respectively. Targeted single eukaryotic cells were sorted into four 96-well plates, containing 3 μl UV-sterilized TE buffer in each well (32). Individual cells were sorted from discrete sectors using three criteria; plate 1 contained cells showing high levels of LysoTracker fluorescence only (i.e., heterotrophic or phagotrophic), plate 2 contained cells exhibiting high levels of red fluorescence (i.e., photosynthetic) and the absence of green fluorescence, and plates 3 and 4 contained smaller cells exhibiting intermediate levels of red and green fluorescence (i.e., mixotrophic) (Fig. 1). Plates were stored at −80°C until whole-genome amplification.
FIG 1.
Sample cytogram showing flow cytometric sorting regions for microbial eukaryotes from an enrichment culture (MCM87) generated from the Antarctic Dry Valley of Lake Bonney. The sample was stained with LysoTracker prior to sorting, and samples were gated on green fluorescence (LysoTracker-stained food vacuoles) versus red fluorescence (chlorophyll autofluorescence). Ovals, groups selected for single-cell sorting. Red, high levels of chlorophyll autofluorescence; green, high levels of LysoTracker fluorescence; blue, intermediate levels of both types of fluorescence. P1 to P4, sorted 96-well plates 1 to 4.
DNA template preparation for PCR.
For DNA preparation, single sorted eukaryotic cells were lysed using alkaline treatment with the addition of 3 μl of buffer consisting of 0.13 M KOH, 3.3 mM EDTA (pH 8.0), and 27.7 mM dithiothreitol (DTT), heated to 95°C for 30 s, and immediately placed on ice for 10 min, followed by the addition of 3 μl of neutralization buffer (0.13 M HCl, 0.42 M Tris [pH 7.0], 0.18 M Tris [pH 8.0]). Genomic DNA from the cell lysates was amplified using multiple displacement amplification (MDA) with the addition of 11 μl of MDA master mix containing 90.9 μM random hexamers with two protective phosporothioate bonds on the 3′ end (Integrated DNA Technologies, Coralville, IA, USA), 1.09 mM deoxynucleoside triphosphates (dNTPs) (Roche, Indianapolis, IN, USA), 1.8× phi29 DNA polymerase buffer (New England BioLabs, Ipswich, MA, USA), 4 mM DTT, and ∼100 U phi29 DNA polymerase (purified in-house) (41). Whole-genome amplification was performed at 30°C for 10 h, followed by inactivation at 80°C for 20 min. MDA products were diluted 100-fold in sterile 1× TE buffer and then screened by PCR and sequencing.
Sanger sequencing.
A fragment of the 18S rRNA gene was amplified from each of the SAGs using the universal eukaryote primers EK-82F (5′-GAAACTGCGAATGGCTC) and EK-1520R (5′-CYGCAGGTTCACCTAC) (42), to generate PCR products for sequencing. PCR was performed in triplicate using 25 cycles of 95°C for 1 min, 52°C for 1 min, and 72°C for 2 min. Sequencing reactions were performed using the BigDye Terminator v3.1 cycle sequencing kit (ABI) with the M13R primer, and the fragments were sequenced on an Applied Biosystems 3730xl DNA analyzer (ABI) located in the Center for Bioinformatics and Functional Genomics (CBFG) at Miami University.
Illumina sequencing.
Following successful identification of the individual eukaryote SAGs, 79 samples were selected for sequencing of the associated bacterial communities on an Illumina MiSeq platform at the CBFG. The 16S rRNA genes from the SAGs were amplified using a primer set (F515/R806) that encoded sequences against the highly variable V4 region of 16S rRNA, barcodes, and linkers. PCRs and MiSeq sequencing reactions strictly followed the protocol provided by the Earth Microbiome Project (http://www.earthmicrobiome.org) (42–45). Samples were sequenced using a 300-cycle MiSeq v2 reagent kit (Illumina), according to the manufacturer's recommendations, in a paired-end run (2 by 150 bp) in the presence of 25% PhiX DNA.
Analysis of sequences.
For 18S rRNA gene sequences derived from Sanger sequencing, representative and closest sequences were selected from GenBank and aligned using MUSCLE. A maximum likelihood tree was generated using MEGA6 software. Bootstrapping was used to estimate the node support of 1,000 replicate trees.
The 16S rRNA gene sequences generated with the MiSeq system were analyzed with QIIME (v1.8.0). Operational taxonomic units (OTUs) were identified at 97% cutoff using the Greengenes database (v13.8) (46). All OTUs with one sequence per sample were discarded. Samples were rarefied to 120 sequences/sample based on the number of sequences in the library with the lowest sequence number. Alpha diversity (number of OTUs, Chao1 index, and Shannon index) was assessed (data not shown). Beta diversity was integrated using the weighted UniFrac distance metric (47, 48). Principal-coordinate analysis (PCoA) and analysis of similarities (ANOSIM) were performed to identify the coexistence of eukaryotic and prokaryotic organisms in the single-cell samples (49–52).
Confocal laser scanning microscopy.
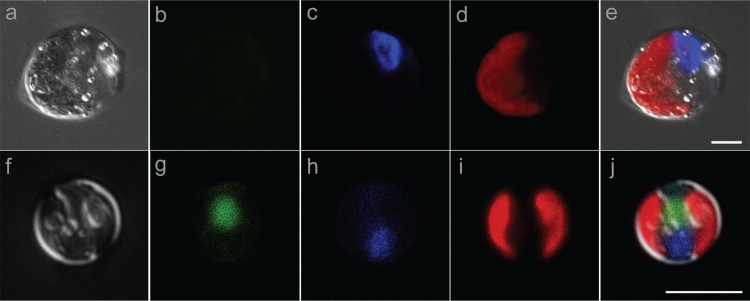
Isochrysis sp. MDV and Chlamydomonas sp. ICE-MDV cultures were treated with the LysoTracker Green DND-26 probe using the same procedure as described above for fluorescence-activated cell sorting (FACS). A Zeiss LSM-710 confocal laser scanning microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) was used for the images in Fig. 3. A 488-nm argon ion laser was used as the elimination/excitation source. LysoTracker fluorescence and chlorophyll fluorescence were detected at 520 to 540 nm and 650 to 750 nm, respectively. Differential interference contrast (DIC) images were also generated, using the transmission light mode. Images were pseudocolored with Zeiss ZEN 2011 software (Carl Zeiss Microscopy GmbH).
FIG 3.
Confocal microscopic images of Chlamydomonas sp. ICE-MDV (a to e) and Isochrysis sp. MDV (f to j) isolates stained with LysoTracker Green and 4′,6-diamidino-2-phenylindole (DAPI). (a and f) DIC images. (b and g) LysoTracker fluorescence. (c and h) DAPI fluorescence. (d and i) Chlorophyll autofluorescence. (e and j) Overlays of a to d and f to i, respectively. Bars, 5 μm.
Scanning electron microscopy.
Samples from the original enrichment culture were fixed with 1% paraformaldehyde and 1.25% glutaraldehyde, and a secondary fixation was performed with 1% osmium tetroxide. Specimens were dehydrated through an ethanol series, critical point dried with CO2, and sputter coated with gold. Micrographs were generated with a Zeiss Supra 35 FEG scanning electron microscope (Carl Zeiss Microscopy GmbH).
Nucleotide sequence accession numbers.
The 79 high-quality partial 18S rRNA sequences (∼600 bp) recovered were submitted to GenBank under accession numbers KU196097 to KU196166 (NCBI BioProject accession no. PRJNA304193).
RESULTS AND DISCUSSION
Identities and trophic modes of sorted eukaryotes.
To overcome the logistical constraints of transporting fresh samples from Antarctica to our U.S. laboratory, as well as low biomass issues associated with MCM planktonic communities, we used an enrichment cultivation-based approach to test FACS/single-cell sorting with McMurdo Dry Valley lake protist communities. A series of enrichment cultures was started in Antarctica using a range of growth media, as described in Materials and Methods, and cultures were incubated for 2 weeks prior to shipment to our U.S. laboratory. When the cultures arrived in our U.S. laboratory, we monitored algal diversity using a BBE FluoroProbe (see Table S2 in the supplemental material) and visually inspected each culture using bright-field and fluorescence microscopy, to assess protist diversity qualitatively. Inspection of one of the cultures (enrichment MCM87) revealed large (>15-μm) biflagellate chlorophytes, several nanophytoplankton identified as haptophytes and cryptophytes (∼5 μm), dinoflagellate cells exhibiting variable pigmentation (owing to the presence or absence of phytoplankton prey), and several nonpigmented heterotrophic nanoflagellates (2 to 5 μm). Based on our recent work on natural protist diversity in Lake Bonney (29), we concluded that MCM87 harbored representatives of several key members of the Lake Bonney protist community, including chlorophytes, haptophytes, choanoflagellates, dinoflagellates, and nanoflagellates, all of which have been detected in environmental sequence libraries generated from the photic zone of the east lobe of Lake Bonney.
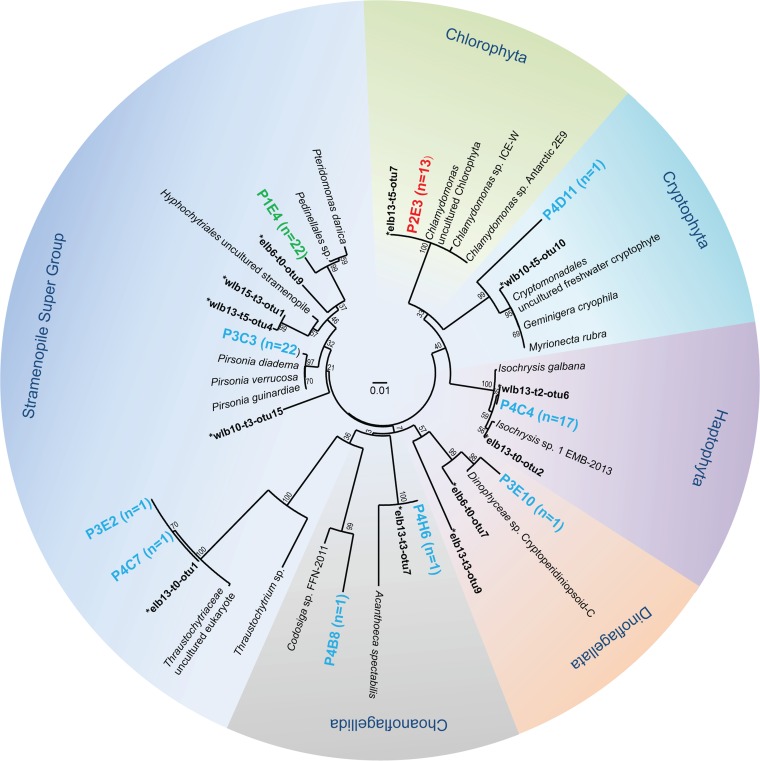
Based on LysoTracker and chlorophyll fluorescence signals combined with cell size information, we gated four eukaryote populations for single-cell sorting, i.e., a group with high levels of LysoTracker fluorescence (i.e., the presence of protist food vacuoles; plate 1) (Fig. 1, green group), a group with high levels of red fluorescence (i.e., the presence of chlorophyll; plate 2) (Fig. 1, red group), and two groups with smaller cells that exhibited intermediate levels of red and green fluorescence (plates 3 and 4) (Fig. 1, blue groups). Targeted single cells were deposited into four 96-well plates according to distinct gates that separated various types of organisms on the basis of the size and fluorescence characteristics mentioned above. Following cell lysis and whole-genome amplification using phi29 DNA polymerase, 65% (250 of 384 samples) of the SAGs were successfully amplified using 18S rRNA primers, based on electrophoretic separation of the PCR products on a 1% agarose gel. All SAGs that exhibited a band of the correct size (∼1,400 bp) were subjected to Sanger sequencing. We recovered 79 high-quality partial 18S rRNA sequences (∼600 bp) for phylogenetic analysis. Representative sequences were aligned with the SILVA and GenBank databases using SINA and BLAST. Phylogenetic analysis revealed that the SAG library harbored a total of 16 OTUs (based on a cutoff value of 97% similarity) related to the Stramenopila supergroup and several other major phyla, including Chlorophyta, Choanoflagellida, Dinoflagellata, Cryptophyta, and Haptophyta. The majority of SAG 18S rRNA sequences were identical to uncultivated eukaryote clones recovered in an earlier study on natural protist populations in Lake Bonney (29).
All SAGs recovered from the photosynthetic group (i.e., high autofluorescence levels and low LysoTracker fluorescence levels; n = 13) (Fig. 1, P2) were related to a large biflagellate chlorophyte, Chlamydomonas, which was closely related (99% identity) to an Antarctic marine species, Chlamydomonas sp. Antarctic 2E9, and two Antarctic ice Chlamydomonas spp., ICE-W and ICE-L (53). The Chlamydomonas SAGs were also closely related to sequences from clone libraries generated from the depth of maximum productivity (13 m depth) in the east lobe of Lake Bonney (29) (Fig. 2). Recent studies on phylogenetic and functional genes indicated that Chlamydomonas sp. ICE is the dominant chlorophyte in the east and west lobes of Lake Bonney (29, 30). Lastly, our FACS results, which suggested that this organism is photoautotrophic, were supported by earlier physiological studies on Chlamydomonas sp. ICE (53), as well as a highly studied Lake Bonney chlorophyte, Chlamydomonas sp. UWO241 (22, 54).
FIG 2.
Maximum likelihood tree (1,000 bootstrap replicates) showing the identity of Lake Bonney microbial eukaryote single amplified genomes (SAGs) recovered from enrichment culture MCM87. The SILVA and GenBank databases were used to assign the phylogenetic positions of eukaryote SAGs. The colors of the SAG sequences are correlated with the sorting parameters in Fig. 1. *, sequences from the natural eukaryote populations in Lake Bonney generated in a previous study (29).
SAG sequences related to a nanoflagellate haptophyte (Isochrysis galbana; 97% identity) represented a significant proportion (80%) of the samples recovered from SAG plate 4 (n = 17) (Fig. 2), which represented one of two plates with intermediate levels of green and red fluorescence (Fig. 1, P4). The 18S rRNA sequences from these SAGs were very similar to sequences recovered from clone libraries generated from 13-m-deep waters from the east and west lobes of Lake Bonney (29), and recent reports suggested that haptophyte communities related to this organism are key primary producers in this lake (28, 30, 55). Communities of Isochrysis dominate the chemoclines of the east and west lobes of Lake Bonney (28). Several recent studies using quantitative PCR analyses to monitor the abundance of photosynthetic functional genes (rbcL and psbA) indicated that, in contrast to chlorophyte populations, the abundance (DNA) and activity (RNA) of haptophyte populations are not correlated with light availability (28, 55). These data suggest that the Dry Valley Isochrysis may rely on alternative metabolic strategies, such as mixotrophy.
Stramenopiles represent a poorly understood supergroup of microbial eukaryotes that exhibit broad trophic abilities (e.g., phototrophy, mixotrophy, predation, and parasitism). The marine stramenopile (MAST) lineage is a diverse group of microbial eukaryotes and represents a large proportion of heterotrophic nanoflagellates (HNFs) in marine environments (56). HNFs are small microbial eukaryotes (2 to 20 μm) that graze on bacteria and picophytoplankton and are recognized as the dominant consumers of picoplanktonic biomass in marine environments (7, 57). Our 18S rRNA gene sequence library contained several SAGs that were related to stramenopiles (Fig. 2). The most abundant stramenopile SAGs were closely related to Pteridomonas danica (99% identification, n = 22; SAG plate 4) and Pirsonia verrucosa (94 to 100% identification, n = 22; SAG plate 3). Sequences related to both stramenopiles were recovered in clone libraries from the west and east lobes of Lake Bonney (29) (Fig. 2). Recent work reported that stramenopiles make up significant proportions of the protist population (up to 60%) in both lobes of Lake Bonney (28, 31). Pirsonia is a phycovorous nanoflagellate that feeds on marine centric diatoms (58). Pteridomonas is a ubiquitous marine heterotrophic nanoflagellate (5). In addition, two OTUs within the Stramenopila supergroup (SAG clones P3E2 and P4C7) exhibited very low levels of similarity (<81%) to the closest related identified sequence in the database (Thraustochytrium sp.). Both OTUs, however, were closely related to the sequences generated for Lake Bonney clone libraries in a previous study (29).
Several other protist groups were represented in the SAG libraries at low abundance. A single SAG was recovered for two choanoflagellates and one dinoflagellate. One SAG related to the psychrophilic marine cryptophyte Geminigera cryophila was also recovered. G. cryophila is abundant in the shallow layers of Lake Bonney (28, 29). Dinoflagellates have been detected in relatively high abundance in the west lobe of Lake Bonney, while sequences related to choanoflagellates were less abundant (29, 31). All low-abundance SAGs were recovered from the two mixotrophic groups (Fig. 1, P3 and P4) and were closely related to sequences recovered from clone libraries generated from the east or west lobe of Lake Bonney, with the exception of one choanoflagellate sequence (SAG P4B8) (Fig. 2).
Isolation and description of two key photosynthetic protists.
Recent work on the natural communities of Lake Bonney suggested that a chlorophyte related to Chlamydomonas sp. ICE-L and a haptophyte related to Isochrysis galbana occupy important but distinct roles in the Antarctic lake food web (28–31). Our FACS analyses indicated that the chlorophyte is a pure photoautotrophic species, while the haptophyte may possess the ability to combine photosynthetic metabolism with heterotrophic activity, such as digestion of captured bacterial prey or particulate carbon. To confirm our hypotheses regarding the trophic capabilities of these key MCM protists, we purified isolates of both organisms from enrichment cultures. A culture of the Chlamydomonas sp. (named Chlamydomonas sp. ICE-MDV) was recovered by plating of an enrichment culture on Bold's basal medium (59) with 1.5% agar. The Isochrysis strain (named Isochrysis sp. MDV) was isolated by dilution to extinction of an enrichment culture in 0.5× seawater supplemented with F2 medium. The identity of both strains was confirmed by 18S rRNA gene sequencing (data not shown). Confocal microscopy showed that Chlamydomonas sp. ICE-MDV had large (15- to 20-μm) biflagellate cells exhibiting high levels of autofluorescence (Fig. 3a, d, and e). The LysoTracker probe is a fluorescent acidotrophic probe; therefore, we wondered whether the acidic compartment (lumen) of chloroplasts (60) might cause false-positive fluorescence signals in flow cytometry studies. We did not detect green fluorescence in the presence of LysoTracker in Chlamydomonas sp. ICE-MDV cells, despite the presence of a large chloroplast (Fig. 3d and e). This finding also confirms that it is unlikely that any of the SAGs sorted in the mixed-SAG plates (Fig. 1, P2 and P3) were products of spurious chloroplast staining. Isochrysis sp. MDV is a small (2- to 5-μm) brown biflagellate alga. It possesses bilobed chloroplasts, which exhibited high levels of autofluorescence (Fig. 3i and j). In contrast to the chlorophyte strain, green fluorescence was localized to a single vacuole when cells of Isochrysis sp. MDV were treated with the LysoTracker probe (Fig. 3g). The presence of green fluorescence was associated only with LysoTracker-stained cells, inasmuch as neither strain exhibited green fluorescence in the absence of LysoTracker (see Fig. S1 and S2 in the supplemental material). We also confirmed in growth experiments that Isochrysis sp. MDV can grow either in the dark (heterotrophic) or in the light (phototrophic) but it grows optimally in the presence of a variety of organic carbon sources and low irradiance (mixotrophic). In contrast, Chlamydomonas ICE-MDV cannot grow in the dark in the presence of organic carbon but exhibits the ability to grow under a broader range of light intensities (for a description of the basic growth physiology, see Table S1 in the supplemental material). Therefore, the chlorophyll and LysoTracker fluorescence information from confocal microscopy, combined with growth physiology, confirmed results from the FACS analyses indicating that Chlamydomonas sp. ICE-MDV is a pure photosynthetic protist, while Isochrysis sp. MDV is likely capable of mixotrophic metabolism. These results fit well with data from functional gene analyses indicating that Lake Bonney chlorophyte populations that occupy the under-ice layers of the water column are strongly correlated with light availability on spatial and seasonal scales (55). In contrast, haptophyte populations dominating planktonic communities in the permanent chemocline are not influenced by light availability but can supplement light-dependent photosynthesis with ingestion of bacterial prey or particulate carbon.
Community composition of organisms cosorted with Lake Bonney eukaryotes.
To gain further insight into the trophic modes and potential microbial partners of the MCM microbial eukaryotes, we selected a number of eukaryote SAGs of different trophic potentials for 16S rRNA amplicon community sequencing. From the population of successfully amplified and sequenced SAGs, we sequenced a fragment of the 16S rRNA gene from 79 eukaryote SAGs using the Illumina MiSeq platform. We recovered both bacterial 16S and plastid small-subunit (SSU) rRNA genes from these sequencing libraries. The plastid sequences were filtered from the 16S rRNA gene libraries of all SAG OTUs known to be photosynthetic, including Chlamydomonas sp. ICE-MDV, Isochrysis sp. MDV, and Geminigera. To assess both the diversity and cooccurrence patterns of organisms that cosorted with the individual eukaryote SAGs, we generated a heat map of 16S rRNA genes associated with each individual SAG sample (see Fig. S3 in the supplemental material). The distribution of recovered 16S rRNA gene sequences was highly variable and was dependent on both the trophic mode and the identity of the microbial eukaryote partner. Bacterial sequences associated with the photosynthetic Chlamydomonas sp. ICE-MDV and the mixotrophic Isochrysis sp. MDV were generally diverse, with a predominance of the phyla Actinobacteria and Bacteroidetes. In contrast, 16S rRNA gene sequences recovered from the two stramenopile SAGs related to Pteridomonas and Pirsonia, which dominated the heterotrophic population (plate 1) and one of the mixotrophic populations (plate 4), respectively, were enriched with plastid sequences (see Fig. S3).
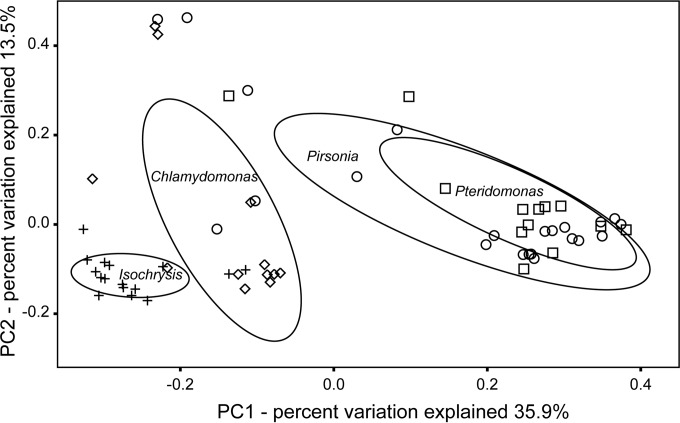
Principal-coordinate analysis (PCoA) based on UniFrac distance metrics of 16S rRNA OTUs from the four most abundant SAGs (Isochrysis sp. MDV, Chlamydomonas sp. ICE-MDV, Pteridomonas, and Pirsonia) demonstrated clustering by protist trophic mode (Fig. 4). Similarity analysis using the ANOSIM method indicated that bacterial OTUs associated with individual eukaryote SAGs were significantly different from each other (P < 0.001, R2 = 0.40). Bacterial OTUs associated with Isochrysis sp. MDV and Chlamydomonas sp. ICE-MDV were clustered separately from each other but were more closely related than either of the stramenopile SAGs. The 16S rRNA gene libraries generated from SAGs of the heterotrophic nanoflagellates (Pteridomonas and Pirsonia) were related more to each other than to either of the photosynthetic protists or the environmental samples (Fig. 4).
FIG 4.
Principle-coordinate analysis (PCoA) of weighted UniFrac distances of 16S rRNA genes associated with eukaryote SAGs of Isochrysis sp. MDV (+), Chlamydomonas sp. ICE-MDV (♢), Pteridomonas (□), and Pirsonia (○). Each data point represents one SAG sample. Ovals, 50% similarities within each sample group.
Potential interactions between Dry Valley protists and bacteria.
Complex ecological interactions between protists and other microorganisms exist, including predation of bacteria by heterotrophic protists and syntrophic interactions between photosynthetic algae and heterotrophic bacteria (2, 61). Traditionally, heterotrophic protists were identified via culture-dependent and microscopic methods (62–65). Recent studies have shown that a single-cell genomic approach can provide direct evidence of specific predator-prey interactions (66, 67). We recovered 22 samples that had high levels of LysoTracker probe-associated fluorescence and were closely related to the stramenopile Pteridomonas danica (Fig. 1 and 2). Pteridomonas is a ubiquitous heterotrophic nanoflagellate in marine systems and is an important link between bacteria and higher trophic levels in the food web (68–70). Interestingly, we observed that more than 90% of 16S rRNA gene sequences from Pteridomonas SAGs were closely related to a stramenopile chloroplast sequence that was neither reported in environmental sequence libraries (29, 31) nor detected in other SAG 16S rRNA libraries in this study. A previous study reported that Pteridomonas danica harbors nonpigmented nonphotosynthetic plastids or vestigial chloroplasts (71); therefore, it is likely that the chloroplast sequences we recovered in 16S rRNA gene sequence libraries generated from Pteridomonas SAGs represent plastid sequences from vestigial chloroplasts. This interpretation was supported by a lack of pigmentation in Pteridomonas cells under light microscopy (data not shown). We removed the stramenopile plastid sequences from the 16S rRNA gene libraries generated from Pteridomonas SAGs and investigated the remaining 16S rRNA gene sequences in the Pteridomonas libraries (Fig. 5a). The most abundant sequences were related to a haptophyte plastid rRNA gene (32% ± 23% of total OTUs) and a firmicute, Streptococcus (13% ± 8% of total OTUs) (Fig. 6a). HNFs represent a diverse ecological group capable of various trophic preferences (e.g., bacteriovorous, phycovorous, or omnivorous). P. danica feeds on both bacteria and picophytoplankton and therefore is an omnivore (72); however, we did not find any reports of this genus preying on haptophytes. Picocyanobacteria are largely absent from the water column of the MCM lakes (28, 30); therefore, we suggest that, in addition to heterotrophic bacteria, the haptophyte population that dominates the photic zone of Lake Bonney could be the natural prey for the MCM Pteridomonas.
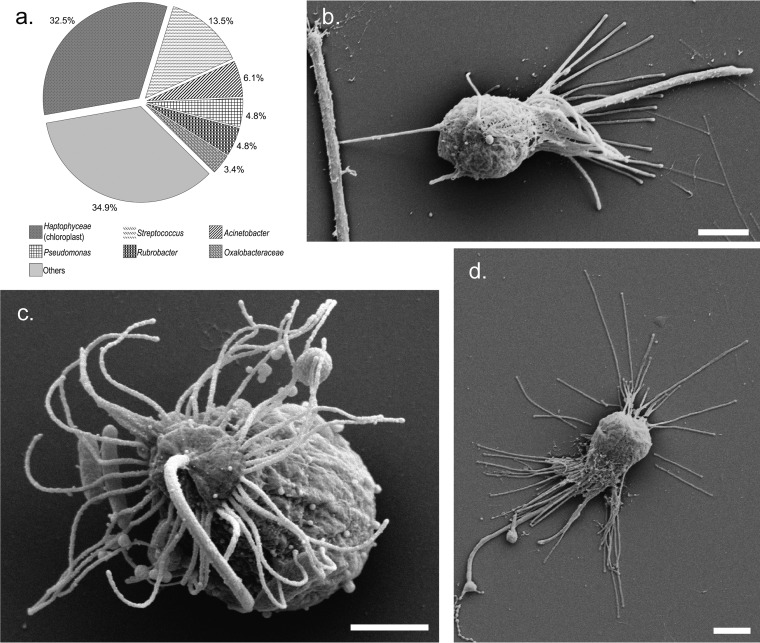
FIG 5.
SEM images and the diversity of microbial partners associated with the heterotrophic nanoflagellate Pteridomonas (n = 22). (a) Chart showing the diversity of 16S rRNA gene OTUs recovered from Pteridomonas eukaryote SAGs. (b to d) SEM images of Pteridomonas sp. cells from the enrichment culture. Bars, 1 μm.
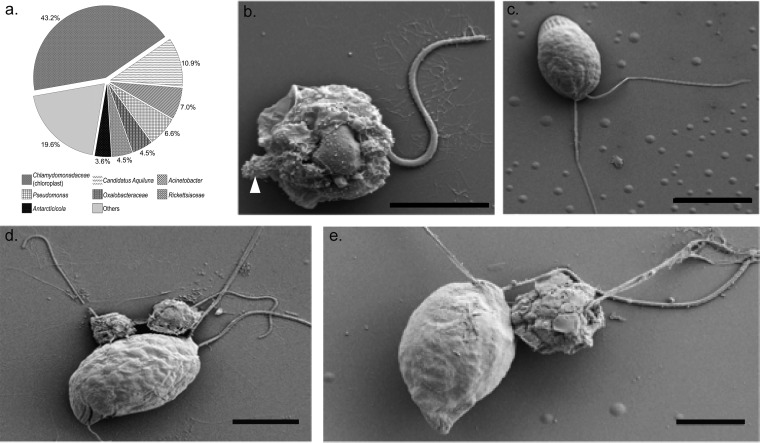
FIG 6.
Evidence of parasite-host interactions among Lake Bonney microbial eukaryotes. (a) Chart showing the diversity of 16S rRNA gene OTUs recovered from Pirsonia eukaryote SAGs (n = 22). (b and c) SEM images showing the sizes of Pirsonia and Chlamydomonas sp. ICE-MDV, respectively. Arrowhead, extrusive organelle of Pirsonia. (d and e) SEM images showing Pirsonia cells attached to Chlamydomonas sp. ICE-MDV cells. Bars, 5 μm.
A number of feeding strategies for heterotrophic flagellates, such as filtration of food particles with an array of radial pseudopodia or tentacles or direct interception of prey by flagella, have been reported. In addition, flagellates are often attached to particles such as marine snow when feeding (73, 74). Examination of the Lake Bonney Pteridomonas by scanning electron microscopy (SEM) revealed the presence of a single long flagellum surrounded by a radial array of tentacles (Fig. 5b to d), which are characteristic morphological features of Pteridomonas spp. (68). In addition, we observed particles attached to the tentacles, and cells were often attached to the surface of a particle by a stalk (Fig. 5b). To our knowledge, our images are some of the first published SEM images of this genus. Thus, the Lake Bonney Pteridomonas appears to rely on a filter feeding mechanism and may be particle attached within the water column. This would be an advantageous strategy within the permanent chemocline, where potential prey would be abundant and particulate organic carbon from the upper photic zone may accumulate. In support of this suggestion, Roberts et al. (24) reported that HNFs dominated heterotrophic protozoa in Lake Bonney and peaked within the permanent chemocline of the west lobe of Lake Bonney.
A second highly abundant SAG (n = 22) within the Stramenopila supergroup exhibited a relatively large proportion of chloroplast sequences within the 16S rRNA gene sequence libraries (Fig. 6; also see Fig. S3 in the supplemental material). These SAGs were recovered from plate 4, which exhibited relatively high levels of green fluorescence and intermediate levels of autofluorescence, and were closely related to a parasitoid nanoflagellate, Pirsonia sp. (Fig. 1 and 2). Parasitoid protists represent a diverse taxonomic group and are thought play major roles in controlling the abundance of their prey; however, their roles in microbial food webs are poorly understood (75). Plastid sequences recovered from the Pirsonia SAGs were all related to Chlamydomonas chloroplast sequences (Fig. 6a); however, unlike Pteridomonas, Pirsonia is not known to harbor a vestigial plastid. Therefore, the presence of plastid sequences within the 16S rRNA gene sequencing libraries suggested that Pirsonia potentially interacts with Chlamydomonas in a predator-prey relationship. However, microscopic examination indicated that the average size of Pirsonia was ∼5 μm, which was noticeable smaller than Chlamydomonas (∼15 μm) (Fig. 6b and c). Previous studies observed that Pirsonia species are host specific for marine centric diatoms. The feeding mechanism involves attachment to a host diatom cell, formation of a trophosome inside the host cell, and transfer of digested material into the parasite cell (58, 76). We observed an extracellular organelle in the Dry Valley Pirsonia (Fig. 6b, arrowhead), which likely functions as part of this feeding mechanism. While diatoms are abundant in the ephemeral streams and microbial mats around the Dry Valleys (77–79), they are rare in MCM lake water columns (28, 29, 31). To our knowledge, there are no reports of Pirsonia interacting with chlorophytes; however, we observed numerous Pirsonia cells attached to Chlamydomonas cells (Fig. 6d and e), which is a critical step for infection of host cells.
Phytoplankton-bacteria interactions in aquatic environments are well known and are largely driven by the dependence of heterotrophic bacteria on the production of alga-derived organic substrates in the form of extracellular phytoplankton products or decaying algal biomass. There is clear evidence that phytoplankton community distribution and dynamics influence specific bacterial assemblages (80–82). The interactions of algae and bacteria involve either free-living, non-particle-attached communities of heterotrophic bacteria or cell-to-cell contact between specific bacteria and algal hosts; bacterial groups associated with the two types of interactions appear to be significantly different from each other (83, 84). Bacterioplankton can exhibit different abilities for incorporation of specific alga-derived substrates (85). The Dry Valley lakes are essentially closed systems for most of the year, and thus heterotrophic bacteria are heavily reliant on newly fixed carbon from phytoplankton production. Recent studies based on environmental sequencing of phytoplanktonic and bacterial communities have shown that spatial distribution of planktonic communities varies greatly within and between lakes (28, 29, 31); however, interactions between MCM phytoplankton and heterotrophic bacteria have not been resolved. In the present study, SAGs of either the chlorophyte Chlamydomonas sp. ICE-MDV or the haptophyte Isochrysis sp. MDV were associated with a number of bacterial OTUs (Fig. 5; also see Fig. S3 in the supplemental material). In general, the evidence for strong interactions between algae and bacteria was lacking. However, we did note two potential bacterial associations with the Lake Bonney phototrophic SAGs. First, a significant proportion of Chlamydomonas ICE-MDV single cells were associated with OTUs related to the Methylobacteriaceae. Members of this group are obligate methylotrophs that can oxidize a number of methylated compounds, including methanol and methylamines. These organisms are relatively abundant in marine environments, particularly during phytoplankton blooms (86, 87). Many marine microalgae produce a number of single-carbon compounds as osmolytes (88); however, less is known about alga-methylotroph interactions in freshwater environments (89). While the presence of methylotrophy in the MCM lakes has not been reported, algal production of osmolytes would be a likely adaptation for survival under harsh conditions such as low temperatures combined with high salinity.
In contrast to the Chlamydomonas sp. ICE-MDV SAGs, Isochrysis sp. MDV SAGs were not generally associated with methylotrophic bacterial sequences. Instead, the majority of haptophyte single cells were associated with a number of bacterial sequences from Flavobacteriaceae (see Fig. S3 in the supplemental material). In the marine environment, Flavobacteria represent dominant Bacteroidetes organisms, which break down complex organic matter and biopolymers by direct attachment to algal cells or alga-derived detritus (90, 91). Haptophyte blooms in the Southern Ocean are associated with peaks in abundance of Flavobacteria (92), and sequences related to Flavobacteria are highly abundant in the bacterioplankton communities in the chemocline of Lake Bonney (31). Our work supports a putative interaction between Lake Bonney haptophytes and Flavobacteria.
The relatively large number of bacterial OTUs associated with the phototrophic SAGs could also represent evidence of low levels of alga-bacterium coupling in the MCM lakes. Phytoplankton exudate quantity and composition are influenced by environmental factors, including nutrient limitations, light availability, and temperature (91, 93). The conditions of the MCM lakes (e.g., limited light and nutrients and low temperatures) might have selected for algal species that excrete a small fraction of alga-derived compounds, and thus bacterioplankton rely more heavily on exudate release during algal lysis or death. In support of the latter hypothesis, we have observed heavy recruitment and colonization of bacteria on dead and dying algal cells in our enrichment cultures, an interaction that would have been excluded in the FACS analysis. There is also evidence that natural bacterioplankton communities obtain a significant fraction of organic carbon from ancient relict pools of organic matter (94).
Conclusions.
Microbial eukaryotes represent the majority of diversity across the eukaryotic tree of life and are capable of complex metabolic modes and interactions. While important in marine ecosystems, their influence reaches profound levels in aquatic systems in which the microbial loop dominates. In the current study, we discovered that the MCM protists possess diverse metabolic capabilities (from pure photosynthesis to mixotrophy, heterotrophy, and parasitism), which likely contributes to the strong vertical layering of key protist communities in the water column of Lake Bonney. The metabolic versatility of MCM protists underpins specific microbe-microbe interactions in some cases (e.g., Pteridomonas-chlorophytes), while interactions with heterotrophic bacteria do not appear to be an important survival strategy for other protists (e.g., haptophytes).
Supplementary Material
ACKNOWLEDGMENTS
We thank the McMurdo LTER, Antarctic Support Contract, and PHI helicopters for logistical assistance in the field. We thank Andor J. Kiss and the Center for Bioinformatics and Functional Genomics at Miami University for assistance with Illumina sequencing. We thank Richard E. Edelmann, Matthew Duley, and the Center for Advanced Microscopy and Imaging at Miami University for assistance with microscopy and image analysis. We thank Steven Allman for technical assistance with flow cytometry.
This work was supported by the NSF Office of Polar Programs (grant OPP-1056396). M.P. has been supported by the Laboratory Directed Research and Development Program of the Oak Ridge National Laboratory; the Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy, under contract DE-AC05-00OR22725.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00478-16.
REFERENCES
- 1.Faust K, Raes J. 2012. Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 2.Amin SA, Parker MS, Armbrust EV. 2012. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev 76:667–684. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montagnes D, Roberts E, Lukeš J, Lowe C. 2012. The rise of model protozoa. Trends Microbiol 20:184–191. doi: 10.1016/j.tim.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Caron DA, Worden AZ, Countway PD, Demir E, Heidelberg KB. 2009. Protists are microbes too: a perspective. ISME J 3:4–12. doi: 10.1038/ismej.2008.101. [DOI] [PubMed] [Google Scholar]
- 5.Sherr EB, Sherr BF. 2002. Significance of predation by protists in aquatic microbial food webs. Antonie Van Leeuwenhoek 81:293–308. doi: 10.1023/A:1020591307260. [DOI] [PubMed] [Google Scholar]
- 6.Moorthi S, Caron DA, Gast RJ, Sanders RW. 2009. Mixotrophy: a widespread and important ecological strategy for planktonic and sea-ice nanoflagellates in the Ross Sea, Antarctica. Aquat Microb Ecol 54:269–277. doi: 10.3354/ame01276. [DOI] [Google Scholar]
- 7.Sanders RW, Berninger U-G, Lim EL, Kemp PF, Caron DA. 2000. Heterotrophic and mixotrophic nanoplankton predation on picoplankton in the Sargasso Sea and on Georges Bank. Mar Ecol Prog Ser 192:103–118. doi: 10.3354/meps192103. [DOI] [Google Scholar]
- 8.Caron DA, Countway PD, Brown MV. 2004. The growing contributions of molecular biology and immunology to protistan ecology: molecular signatures as ecological tools. J Eukaryot Microbiol 51:38–48. doi: 10.1111/j.1550-7408.2004.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 9.Epstein S, Lopez-Garcia P. 2008. Missing protists: a molecular perspective. Biodivers Conserv 17:261–276. doi: 10.1007/s10531-007-9250-y. [DOI] [Google Scholar]
- 10.Lara E, Mitchell EA, Moreira D, Lopez Garcia P. 2011. Highly diverse and seasonally dynamic protist community in a pristine peat bog. Protist 162:14–32. doi: 10.1016/j.protis.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Zinger L, Gobet A, Pommier T. 2012. Two decades of describing the unseen majority of aquatic microbial diversity. Mol Ecol 21:1878–1896. doi: 10.1111/j.1365-294X.2011.05362.x. [DOI] [PubMed] [Google Scholar]
- 12.Simon M, Jardillier L, Deschamps P, Moreira D, Restoux G, Bertolino P, Lopez-Garcia P. 2015. Complex communities of small protists and unexpected occurrence of typical marine lineages in shallow freshwater systems. Environ Microbiol 17:3610–3627. doi: 10.1111/1462-2920.12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon M, Lopez-Garcia P, Deschamps P, Moreira D, Restoux G, Bertolino P, Jardillier L. 2015. Marked seasonality and high spatial variability of protist communities in shallow freshwater systems. ISME J 9:1941–1953. doi: 10.1038/ismej.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nolte V, Pandey RV, Jost S, Medinger R, Ottenwalder B, Boenigk J, Schlotterer C. 2010. Contrasting seasonal niche separation between rare and abundant taxa conceals the extent of protist diversity. Mol Ecol 19:2908–2915. doi: 10.1111/j.1365-294X.2010.04669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds RT, Squyres SW, Colburn DS, McKay CP. 1983. On the habitability of Europa. Icarus 56:246–254. doi: 10.1016/0019-1035(83)90037-4. [DOI] [Google Scholar]
- 16.Chela-Flores J. 2011. The science of astrobiology, p 151–170. Springer, New York, NY. [Google Scholar]
- 17.Green W, Lyons W. 2009. The saline lakes of the McMurdo Dry Valleys, Antarctica. Aquat Geochem 15:321–348. doi: 10.1007/s10498-008-9052-1. [DOI] [Google Scholar]
- 18.Laybourn-Parry J. 2009. No place too cold. Science 324:1521–1522. doi: 10.1126/science.1173645. [DOI] [PubMed] [Google Scholar]
- 19.Cavicchioli R. 2015. Microbial ecology of Antarctic aquatic systems. Nat Rev Microbiol 13:691–706. doi: 10.1038/nrmicro3549. [DOI] [PubMed] [Google Scholar]
- 20.Laybourn-Parry J, Pearce DA. 2007. The biodiversity and ecology of Antarctic lakes: models for evolution. Philos Trans R Soc Lond B Biol Sci 362:2273–2289. doi: 10.1098/rstb.2006.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neale PJ, Priscu JC. 1995. The photosynthetic apparatus of phytoplankton from a perennially ice-covered Antarctic lake: acclimation to an extreme shade environment. Plant Cell Physiol 36:253–263. [Google Scholar]
- 22.Morgan-Kiss RM, Priscu JP, Pocock T, Gudynaite-Savitch L, Hüner NPA. 2006. Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol Mol Biol Rev 70:222–252. doi: 10.1128/MMBR.70.1.222-252.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts EC, Laybourn-Parry J. 1999. Mixotrophic cryptophytes and their predators in the Dry Valley lakes of Antarctica. Freshw Biol 41:737–746. doi: 10.1046/j.1365-2427.1999.00401.x. [DOI] [Google Scholar]
- 24.Roberts EC, Priscu JC, Wolf C, Lyons WB, Laybourn-Parry J. 2004. The distribution of microplankton in the McMurdo Dry Valley Lakes, Antarctica: response to ecosystem legacy or present-day climatic controls? Polar Biol 27:238–249. doi: 10.1007/s00300-003-0582-0. [DOI] [Google Scholar]
- 25.Bell EM, Laybourn-Parry J. 2003. Mixotrophy in the Antarctic phytoflagellate Pyramimonas gelidicola. J Phycol 39:644–649. doi: 10.1046/j.1529-8817.2003.02152.x. [DOI] [Google Scholar]
- 26.Laybourn-Parry J. 2002. Survival mechanisms in Antarctic lakes. Philos Trans R Soc Lond B Biol Sci 357:863–869. doi: 10.1098/rstb.2002.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spigel RH, Priscu JC. 1998. Physical limnology of the McMurdo Dry Valleys lakes, p 153–187. In Priscu JC. (ed), Ecosystem dynamics in a polar desert: the McMurdo Dry Valleys, Antarctica. American Geophysical Union, Washington, DC. [Google Scholar]
- 28.Dolhi JM, Teufel AG, Kong W, Morgan-Kiss RM. 2015. Diversity and spatial distribution of autotrophic communities within and between ice-covered Antarctic lakes (McMurdo Dry Valleys). Limnol Oceanogr 60:977–991. doi: 10.1002/lno.10071. [DOI] [Google Scholar]
- 29.Bielewicz S, Bell E, Kong W, Friedberg I, Priscu JC, Morgan-Kiss RM. 2011. Protist diversity in a permanently ice-covered Antarctic lake during the polar night transition. ISME J 5:1559–1564. doi: 10.1038/ismej.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong W, Ream DC, Priscu JC, Morgan-Kiss RM. 2012. Diversity and expression of RubisCO genes in a perennially ice-covered Antarctic lake during the polar night transition. Appl Environ Microbiol 78:4358–4366. doi: 10.1128/AEM.00029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vick-Majors TJ, Priscu JC, Amaral-Zettler LA. 2014. Modular community structure suggests metabolic plasticity during the transition to polar night in ice-covered Antarctic lakes. ISME J 8:778–789. doi: 10.1038/ismej.2013.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell AG, Campbell JH, Schwientek P, Woyke T, Sczyrba A, Allman S, Beall CJ, Griffen A, Leys E, Podar M. 2013. Multiple single-cell genomes provide insight into functions of uncultured Deltaproteobacteria in the human oral cavity. PLoS One 8:e59361. doi: 10.1371/journal.pone.0059361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon HS, Price DC, Stepanauskas R, Rajah VD, Sieracki ME, Wilson WH, Yang EC, Duffy S, Bhattacharya D. 2011. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 332:714–717. doi: 10.1126/science.1203163. [DOI] [PubMed] [Google Scholar]
- 34.Marcy Y, Ouverney C, Bik EM, Lösekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA. 2007. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A 104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guillard RR, Ryther JH. 1962. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea (cleve). Can J Microbiol 8:229–239. [DOI] [PubMed] [Google Scholar]
- 36.Bischoff HW, Bold HC. 1963. Some soil algae from Enchanted Rock and related algal species. Phycological studies IV. University of Texas Press, Austin, TX. [Google Scholar]
- 37.Stanier R, Kunisawa R, Mandel M, Cohen-Bazire G. 1971. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol Rev 35:171–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rinke C, Lee J, Nath N, Goudeau D, Thompson B, Poulton N, Dmitrieff E, Malmstrom R, Stepanauskas R, Woyke T. 2014. Obtaining genomes from uncultivated environmental microorganisms using FACS-based single-cell genomics. Nat Protoc 9:1038–1048. doi: 10.1038/nprot.2014.067. [DOI] [PubMed] [Google Scholar]
- 39.Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D, Reinthaler T, Poulton NJ, Masland EDP, Gomez ML, Sieracki ME, DeLong EF, Herndl GJ, Stepanauskas R. 2011. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333:1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- 40.Rose JM, Caron DA, Sieracki ME, Poulton N. 2004. Counting heterotrophic nanoplanktonic protists in cultures and aquatic communities by flow cytometry. Aquat Microb Ecol 34:263–277. doi: 10.3354/ame034263. [DOI] [Google Scholar]
- 41.Blainey PC, Quake SR. 2011. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Res 39:e19. doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- 43.Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. 2009. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One 4:e6372. doi: 10.1371/journal.pone.0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamady M, Knight R. 2009. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res 19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. 2012. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol 27:1E.5.1–1E.5.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 52.Anderson MJ, Crist TO, Chase JM, Vellend M, Inouye BD, Freestone AL, Sanders NJ, Cornell HV, Comita LS, Davies KF, Harrison SP, Kraft NJ, Stegen JC, Swenson NG. 2011. Navigating the multiple meanings of β diversity: a roadmap for the practicing ecologist. Ecol Lett 14:19–28. doi: 10.1111/j.1461-0248.2010.01552.x. [DOI] [PubMed] [Google Scholar]
- 53.Liu C, Huang X, Wang X, Zhang X, Li G. 2006. Phylogenetic studies on two strains of Antarctic ice algae based on morphological and molecular characteristics. Phycologia 45:190–198. doi: 10.2216/03-88.1. [DOI] [Google Scholar]
- 54.Dolhi JM, Maxwell DP, Morgan-Kiss RM. 2013. The Antarctic Chlamydomonas raudensis: an emerging model for cold adaptation of photosynthesis. Extremophiles 17:711–722. doi: 10.1007/s00792-013-0571-3. [DOI] [PubMed] [Google Scholar]
- 55.Kong W, Li W, Romancova I, Prasil O, Morgan-Kiss RM. 2014. An integrated study of photochemical function and expression of a key photochemical gene (psbA) in photosynthetic communities of Lake Bonney (McMurdo Dry Valleys, Antarctica). FEMS Microbiol Ecol 89:293–302. doi: 10.1111/1574-6941.12296. [DOI] [PubMed] [Google Scholar]
- 56.Massana R, Castresana J, Balagué V, Guillou L, Romari K, Groisillier A, Valentin K, Pedrós-Alió C. 2004. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl Environ Microbiol 70:3528–3534. doi: 10.1128/AEM.70.6.3528-3534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fenchel T. 1982. Ecology of heterotrophic microflagellates. IV. Quantitative occurrence and importance as bacterial consumers. Mar Ecol Prog Ser 9:35–42. [Google Scholar]
- 58.Kuhn S, Medlin L, Eller G. 2004. Phylogenetic position of the parasitoid nanoflagellate Pirsonia inferred from nuclear-encoded small subunit ribosomal DNA and a description of Pseudopirsonia n. gen. and Pseudopirsonia mucosa (Drebes) comb. nov. Protist 155:143–156. doi: 10.1078/143446104774199556. [DOI] [PubMed] [Google Scholar]
- 59.Nichols HW, Bold HC. 1965. Trichosarcina polymorpha gen. et sp. nov. J Phycol 1:34–38. doi: 10.1111/j.1529-8817.1965.tb04552.x. [DOI] [Google Scholar]
- 60.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2002. Energy conversion: mitochondria and chloroplasts, p 767–829. In Molecular biology of the cell, 4th ed Garland Science, New York, NY. [Google Scholar]
- 61.Buchan A, LeCleir GR, Gulvik CA, Gonzalez JM. 2014. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol 12:686–698. doi: 10.1038/nrmicro3326. [DOI] [PubMed] [Google Scholar]
- 62.Sherr EB, Caron DA, Sherr BF. 1993. Staining of heterotrophic protists for visualization via epifluorescence microscopy, p 213–227. In Kemp PF, Cole JJ, Sherr BF, Sherr EB (ed), Handbook of methods in aquatic microbial ecology. CRC Press, Boca Raton, FL. [Google Scholar]
- 63.Pernthaler J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- 64.Auinger BM, Pfandl K, Boenigk J. 2008. Improved methodology for identification of protists and microalgae from plankton samples preserved in Lugol's iodine solution: combining microscopic analysis with single-cell PCR. Appl Environ Microbiol 74:2505–2510. doi: 10.1128/AEM.01803-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jezbera J, Hornak K, Simek K. 2005. Food selection by bacterivorous protists: insight from the analysis of the food vacuole content by means of fluorescence in situ hybridization. FEMS Microbiol Ecol 52:351–363. doi: 10.1016/j.femsec.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 66.Heywood JL, Sieracki ME, Bellows W, Poulton NJ, Stepanauskas R. 2011. Capturing diversity of marine heterotrophic protists: one cell at a time. ISME J 5:674–684. doi: 10.1038/ismej.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez-Garcia M, Brazel D, Poulton NJ, Swan BK, Gomez ML, Masland D, Sieracki ME, Stepanauskas R. 2012. Unveiling in situ interactions between marine protists and bacteria through single cell sequencing. ISME J 6:703–707. doi: 10.1038/ismej.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caron D, Gast R, Lim E, Dennett M. 1999. Protistan community structure: molecular approaches for answering ecological questions, p 215–227. In Zehr JP, Voytek M (ed), Molecular ecology of aquatic communities. Springer, New York, NY. [Google Scholar]
- 69.Pelegri S, Christaki U, Dolan J, Rassoulzadegan F. 1999. Particulate and dissolved organic carbon production by the heterotrophic nanoflagellate Pteridomonas danica Patterson and Fenchel. Microb Ecol 37:276–284. doi: 10.1007/s002489900150. [DOI] [PubMed] [Google Scholar]
- 70.Zubkov MV, Sleigh MA. 2005. Assimilation efficiency of Vibrio bacterial protein biomass by the flagellate Pteridomonas: assessment using flow cytometric sorting. FEMS Microbiol Ecol 54:281–286. doi: 10.1016/j.femsec.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 71.Sekiguchi H, Moriya M, Nakayama T, Inouye I. 2002. Vestigial chloroplasts in heterotrophic stramenopiles Pteridomonas danica and Ciliophrys infusionum (Dictyochophyceae). Protist 153:157–167. doi: 10.1078/1434-4610-00094. [DOI] [PubMed] [Google Scholar]
- 72.Zwirglmaier K, Spence E, Zubkov MV, Scanlan DJ, Mann NH. 2009. Differential grazing of two heterotrophic nanoflagellates on marine Synechococcus strains. Environ Microbiol 11:1767–1776. doi: 10.1111/j.1462-2920.2009.01902.x. [DOI] [PubMed] [Google Scholar]
- 73.Christensen-Dalsgaard KK, Fenchel T. 2003. Increased filtration efficiency of attached compared to free-swimming flagellates. Aquat Microb Ecol 33:77–86. doi: 10.3354/ame033077. [DOI] [Google Scholar]
- 74.Kiørboe T, Grossart H-P, Ploug H, Tang K, Auer B. 2004. Particle-associated flagellates: swimming patterns, colonization rates, and grazing on attached bacteria. Aquat Microb Ecol 35:141–152. doi: 10.3354/ame035141. [DOI] [Google Scholar]
- 75.Skovgaard A. 2014. Dirty tricks in the plankton: diversity and role of marine parasitic protists. Acta Protozool 53:51–62. [Google Scholar]
- 76.Schweikert M, Schnepf E. 1997. Light and electron microscopical observations on Pirsonia punctigerae spec. nov., a nanoflagellate feeding on the marine centric diatom Thalassiosira punctigera. Eur J Protistol 33:168–177. doi: 10.1016/S0932-4739(97)80033-8. [DOI] [Google Scholar]
- 77.Sumner DY, Hawes I, Mackey TJ, Jungblut AD, Doran PT. 2015. Antarctic microbial mats: a modern analog for Archean lacustrine oxygen oases. Geology 43:887–890. doi: 10.1130/G36966.1. [DOI] [Google Scholar]
- 78.Doran PT, Wharton RA Jr, Lyons WB. 1994. Paleolimnology of the McMurdo Dry Valleys, Antarctica. J Paleolimnol 10:85–114. doi: 10.1007/BF00682507. [DOI] [PubMed] [Google Scholar]
- 79.Spaulding S, McKnight D, Stoermer E, Doran P. 1997. Diatoms in sediments of perennially ice-covered Lake Hoare, and implications for interpreting lake history in the McMurdo Dry Valleys of Antarctica. J Paleolimnol 17:403–420. [Google Scholar]
- 80.Murray A, Arnosti C, De La Rocha C, Grosart H-P, Passow U. 2007. Microbial dynamics in autotrophic and heterotrophic seawater mesocosms. II. Bacterioplankton community structure and hydrolytic enzyme activities. Aquat Microb Ecol 49:123–141. [Google Scholar]
- 81.Paver SF, Hayek KR, Gano KA, Fagen JR, Brown CT, Davis-Richardson AG, Crabb DB, Rosario-Passapera R, Giongo A, Triplett EW. 2013. Interactions between specific phytoplankton and bacteria affect lake bacterial community succession. Environ Microbiol 15:2489–2504. doi: 10.1111/1462-2920.12131. [DOI] [PubMed] [Google Scholar]
- 82.Simek K, Hornak K, Jezbera J, Nedoma J, Znachor P, Hejzlar J, Sed'a J. 2008. Spatio-temporal patterns of bacterioplankton production and community composition related to phytoplankton composition and protistan bacterivory in a dam reservoir. Aquat Microb Ecol 51:249–262. doi: 10.3354/ame01193. [DOI] [Google Scholar]
- 83.Grossart HP, Levold F, Allgaier M, Simon M, Brinkhoff T. 2005. Marine diatom species harbour distinct bacterial communities. Environ Microbiol 7:860–873. doi: 10.1111/j.1462-2920.2005.00759.x. [DOI] [PubMed] [Google Scholar]
- 84.Rösel S, Grossart H-P. 2012. Contrasting dynamics in activity and community composition of free-living and particle-associated bacteria in spring. Aquat Microb Ecol 66:169–181. doi: 10.3354/ame01568. [DOI] [Google Scholar]
- 85.Salcher MM, Posch T, Pernthaler J. 2013. In situ substrate preferences of abundant bacterioplankton populations in a prealpine freshwater lake. ISME J 7:896–907. doi: 10.1038/ismej.2012.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sekar R, Fuchs BM, Amann R, Pernthaler J. 2004. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl Environ Microbiol 70:6210–6219. doi: 10.1128/AEM.70.10.6210-6219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morris RM, Longnecker K, Giovannoni SJ. 2006. Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ Microbiol 8:1361–1370. doi: 10.1111/j.1462-2920.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- 88.Sieburth JM, Keller MD. 1989. Methylaminotrophic bacteria in xenic nanoalgal cultures: incidence, significance, and role of methylated algal osmoprotectants. Biol Oceanogr 6:383–395. [Google Scholar]
- 89.Salcher MM, Neuenschwander SM, Posch T, Pernthaler J. 2015. The ecology of pelagic freshwater methylotrophs assessed by a high-resolution monitoring and isolation campaign. ISME J 9:2442–2453. doi: 10.1038/ismej.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kirchman DL. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol 39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 91.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, Kassabgy M, Huang S, Mann AJ, Waldmann J. 2012. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 92.Williams TJ, Wilkins D, Long E, Evans F, DeMaere MZ, Raftery MJ, Cavicchioli R. 2013. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ Microbiol 15:1302–1317. doi: 10.1111/1462-2920.12017. [DOI] [PubMed] [Google Scholar]
- 93.Eckert EM, Salcher MM, Posch T, Eugster B, Pernthaler J. 2012. Rapid successions affect microbial N-acetyl-glucosamine uptake patterns during a lacustrine spring phytoplankton bloom. Environ Microbiol 14:794–806. doi: 10.1111/j.1462-2920.2011.02639.x. [DOI] [PubMed] [Google Scholar]
- 94.Priscu JC, Wolf CF, Takacs CD, Fritsen CH, Laybourn-Parry J, Roberts EC, Sattler B, Lyons WB. 1999. Carbon transformations in a perennially ice-covered Antarctic lake. Bioscience 49:997–1008. doi: 10.2307/1313733. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.