ABSTRACT
ArsR is a well-studied transcriptional repressor that regulates microbe-arsenic interactions. Most microorganisms have an arsR gene, but in cases where multiple copies exist, the respective roles or potential functional overlap have not been explored. We examined the repressors encoded by arsR1 and arsR2 (ars1 operon) and by arsR3 and arsR4 (ars2 operon) in Agrobacterium tumefaciens 5A. ArsR1 and ArsR4 are very similar in their primary sequences and diverge phylogenetically from ArsR2 and ArsR3, which are also quite similar to one another. Reporter constructs (lacZ) for arsR1, arsR2, and arsR4 were all inducible by As(III), but expression of arsR3 (monitored by reverse transcriptase PCR) was not influenced by As(III) and appeared to be linked transcriptionally to an upstream lysR-type gene. Experiments using a combination of deletion mutations and additional reporter assays illustrated that the encoded repressors (i) are not all autoregulatory as is typically known for ArsR proteins, (ii) exhibit variable control of each other's encoding genes, and (iii) exert variable control of other genes previously shown to be under the control of ArsR1. Furthermore, ArsR2, ArsR3, and ArsR4 appear to have an activator-like function for some genes otherwise repressed by ArsR1, which deviates from the well-studied repressor role of ArsR proteins. The differential regulatory activities suggest a complex regulatory network not previously observed in ArsR studies. The results indicate that fine-scale ArsR sequence deviations of the reiterated regulatory proteins apparently translate to different regulatory roles.
IMPORTANCE Given the significance of the ArsR repressor in regulating various aspects of microbe-arsenic interactions, it is important to assess potential regulatory overlap and/or interference when a microorganism carries multiple copies of arsR. This study explores this issue and shows that the four arsR genes in A. tumefaciens 5A, associated with two separate ars operons, encode proteins exhibiting various degrees of functional overlap with respect to autoregulation and cross-regulation, as well as control of other functional genes. In some cases, differences in regulatory activity are associated with only limited differences in protein primary structure. The experiments summarized herein also present evidence that ArsR proteins appear to have activator functions, representing novel regulatory activities for ArsR, previously known only to be a repressor.
INTRODUCTION
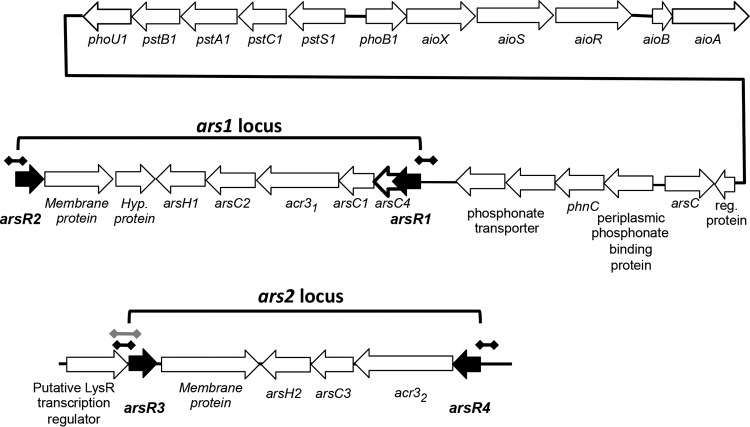
In reaction to arsenic in their environment, microorganisms orchestrate an organized response that may involve arsenite [As(III)] oxidation, arsenate [As(V)] reduction, or both. These redox reactions serve to detoxify or protect the organism or to generate energy, depending on the organism and the genes involved. Current models depict As(III) being taken up into the cell via aquaglyceroporins (e.g., reviewed in references 1, 2, and 3), where it then interacts with a DNA-binding repressor protein, ArsR, resulting in a conformational change in ArsR and causing it to disassociate from the DNA, thereby allowing the DNA to be transcribed (reviewed in references 1, 2, and 3). The arsR gene is autoregulated by its product, ArsR, and is typically part of an operon that contains other ars genes involved in arsenic detoxification. Operon composition usually is comprised of at least arsR, arsC (encoding arsenate reductase), and either an arsB or acr3 gene [coding for different proteins involved in As(III) extrusion from the cytoplasm]. Depending on the organism, additional ars operon elements can include arsA, which codes for an ATPase that associates with ArsB, enabling the latter to use ATP to energize the extrusion of As(III) (4); arsD, coding for a protein that can exhibit weak repressor activity, but with a primary function currently viewed as arsenic metallochaperone activity (5–7); arsH, which was recently shown to encode an organoarsenical oxidase capable of oxidizing trivalent methylated and aromatic arsenicals (8); arsI, encoding an alternative As(V) reductase that differs from ArsC (9); another arsI gene, encoding a C-As lyase (10); arsO, encoding a putative flavin-binding monooxygenase (11); arsP, encoding a putative membrane permease (12, 13); and arsTX, encoding a thioredoxin system transcribed along with an arsRC2 fusion gene (14). Gene duplication within these operons has been observed (15), as is the case with Agrobacterium tumefaciens 5A (Fig. 1). Additionally, though not part of the ars operon, arsM codes for an arsenic methyltransferase that confers As(III) resistance (16, 17).
FIG 1.
Gene composition and organization of the ars1 and ars2 operons in Agrobacterium tumefaciens 5A in relation to the nearby aio operon and the pst/pho gene cluster. Black dumbbell lines indicate the approximate regions of DNA used to construct the different arsR-lacZ fusions. The gray dumbbell line indicates the part of the arsR3-lysR region in the ars2 operon examined in the RT-PCR experiments. The black line connecting phoU1 and “reg. protein” is not drawn to scale and does not depict actual distance but illustrates the physical connectivity of the genes. The open reading frame denoted by a boldly outlined arrow represents a modification from prior depictions of the arsR1 locus. (Modified from reference 20.)
As more genomes are sequenced, it has become increasingly apparent that many ars genes are reiterated, with some microorganisms containing two or more ars operons (18–26). The relative importance of such reiteration has been examined only sparingly. Gene disruption experiments in Corynebacterium glutamicum illustrated the relative contributions of the arsB (18) and acr3 genes to arsenic resistance (27), and the Ramos group explored the functional importance of two ars operons in Pseudomonas putida. Both operons provide protection against arsenic toxicity, although there is a temperature dependence that suggests that there are different ecological niches for different ars operons (15, 18, 28).
In studies aimed at examining the regulatory controls of As(III) oxidation in A. tumefaciens strain 5A, we showed that it has two distinct ars operons (e.g., see reference 29). Each ars operon is bracketed by arsR annotated genes oriented in opposing directions (Fig. 1). We designated these arsR genes the arsR1 and arsR2 genes (bracketing the ars1 operon) and the arsR3 and arsR4 genes (bracketing the ars2 operon). ArsR1 is autoregulatory and exerts control over the operon in which it resides (29). ArsR1 also represses the expression of the nearby phoB1 and pstS1 genes (Fig. 1), which are both required for optimal expression of aioBA [structural genes encoding As(III) oxidase] (20) and are also controlled by the phosphorus stress response (20). In the current study, we examined the contributions of the four ArsR proteins to regulating each arsR gene, phoB1, and pstS1, and we briefly assessed their structural similarities.
MATERIALS AND METHODS
Strains, constructs, plasmids, and primers.
Strains, constructs, plasmids, and primers used in this study are listed in Table 1. The A. tumefaciens strains were cultured at 30°C in a defined minimal mannitol medium (MMNH4) (29) containing a high (1 mM) or low (0.05 mM) phosphate level and mannitol as a carbon and energy source, with aeration by shaking. As(III) (100 μM) was added for induction experiments. Escherichia coli strains were grown at 37°C in lysogeny broth (LB). Bacterial growth was monitored via measurements of the culture optical density by use of a SpectraMax microtiter plate reader (Molecular Devices, CA). Plasmid isolation, gel electrophoresis, transformation, PCR amplification of DNA, and reporter gene assays were conducted as previously described (20, 30). When required, the MMNH4 agar medium was supplemented with 500 μg/ml kanamycin (Km) for selection and maintenance of reporter constructs derived from pLSP-KT2lacZ or with 80 μg/ml gentamicin (Gen), 20 μg/ml tetracycline (Tc), or 15% sucrose to select for/against pJQ200SK for the generation of deletion mutations (see below). E. coli was grown with 50 μg/ml Km, 20 μg/ml Gen, or 20 μg/ml Tc, as required. Constructs were mobilized into A. tumefaciens strains by conjugation with E. coli S17-1 (20, 30).
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant markers and characteristics | Primer use | Reference or source |
|---|---|---|---|
| Bacterial strains | |||
| Agrobacterium tumefaciens strains | |||
| 5A | Wild type, soil isolate, As(III) oxidizer | Lab stock | |
| ΔarsR1 mutant | arsR1 gene deletion mutant | Lab stock | |
| ΔarsR2 mutant | arsR2 gene deletion mutant | This study | |
| ΔarsR3 mutant | arsR3 gene deletion mutant | This study | |
| ΔarsR4 mutant | arsR4 gene deletion mutant | This study | |
| 5A (ParsR1) | Kmr; 5A with pLSP-ParsR1 | This study | |
| 5A (ParsR2) | Kmr; 5A with pLSP-ParsR2 | This study | |
| 5A (ParsR3) | Kmr; 5A with pLSP-ParsR3 | This study | |
| 5A (ParsR4) | Kmr; 5A with pLSP-ParsR4 | This study | |
| ΔarsR1 (ParsR2) | Kmr; ΔarsR1 mutant with pLSP-ParsR2 | This study | |
| ΔarsR1 (ParsR4) | Kmr; ΔarsR1 mutant with pLSP-ParsR4 | This study | |
| ΔarsR1 (PphoB1) | Kmr; ΔarsR1 mutant with pLSP-PphoB1 | This study | |
| ΔarsR1 (PpstS1) | Kmr; ΔarsR1 mutant with pLSP-PpstS1 | This study | |
| ΔarsR2 (ParsR1) | Kmr; ΔarsR2 mutant with pLSP-ParsR1 | This study | |
| ΔarsR2 (ParsR4) | Kmr; ΔarsR2 mutant with pLSP-ParsR4 | This study | |
| ΔarsR2 (PphoB1) | Kmr; ΔarsR2 mutant with pLSP-PphoB1 | This study | |
| ΔarsR2 (PpstS1) | Kmr; ΔarsR2 mutant with pLSP-PpstS1 | This study | |
| ΔarsR3 (ParsR1) | Kmr; ΔarsR3 mutant with pLSP-ParsR1 | This study | |
| ΔarsR3 (ParsR2) | Kmr; ΔarsR3 mutant with pLSP-ParsR2 | This study | |
| ΔarsR3 (ParsR4) | Kmr; ΔarsR3 mutant with pLSP-ParsR4 | This study | |
| ΔarsR3 (PphoB1) | Kmr; ΔarsR3 mutant with pLSP-PphoB1 | This study | |
| ΔarsR3 (PpstS1) | Kmr; ΔarsR3 mutant with pLSP-PpstS1 | This study | |
| ΔarsR4 (ParsR1) | Kmr; ΔarsR4 mutant with pLSP-ParsR1 | This study | |
| ΔarsR4 (ParsR2) | Kmr; ΔarsR4 mutant with pLSP-ParsR2 | This study | |
| ΔarsR4 (PphoB1) | Kmr; ΔarsR4 mutant with pLSP-PphoB1 | This study | |
| ΔarsR4 (PpstS1) | Kmr; ΔarsR4 mutant with pLSP-PpstS1 | This study | |
| Escherichia coli strains | |||
| S17-1 | Pro− Mob+; conjugation donor | Lab stock | |
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm rne-131 (DE3)pLysS Camr | Invitrogen | |
| Plasmids | |||
| pJQ200sk | Genr traJ oriT sacB; suicide vector | Lab stock | |
| pLSP-KT2lacZ | Kmr oriV; lacZ fusion vector used for lacZ fusion constructs | Lab stock | |
| pPROEX Hta | Ampr; His6 N-terminal protein expression vector | Invitrogen | |
| pJQ200sk-arsR2 | pJQ200sk with arsR2-deleted region | This study | |
| pJQ200sk-arsR3 | pJQ200sk with arsR3-deleted region | This study | |
| pJQ200sk-arsR4 | pJQ200sk with arsR4-deleted region | This study | |
| pLSP-ParsR1 | pLSP-KT2lacZ with arsR1 promoter region | This study | |
| pLSP-ParsR2 | pLSP-KT2lacZ with arsR2 promoter region | This study | |
| pLSP-ParsR3 | pLSP-KT2lacZ with arsR3 promoter region | This study | |
| pLSP-ParsR4 | pLSP-KT2lacZ with arsR4 promoter region | This study | |
| Primers | |||
| arsR2-1f/1r (455 bp) | CGCGGATCCAGACGAGGCGCAATAGAGTGACAT/CCCATCCACTAAACTTAAACAGACAGTGCGGCAAAAGAAGTTAGG | Deletion of arsR2 gene | |
| arsR2-2f/2r (469 bp) | TGTTTAAGTTTAGTGGATGGGATTGCTGCTCGGGACATC/CGCTCTAGACGACAAGGGCTGCGAACG | Deletion of arsR2 gene | |
| arsR3-1f/1r (396 bp) | CGCGGATCCTGATGTCCGGCCACTATGTT/CCCATCCACTAAACTTAAACAAGCAAATGCCGAAAGAGCCTGATG | Deletion of arsR3 gene | |
| arsR3-2f/2r (395 bp) | TGTTTAAGTTTAGTGGATGGGGTCCGGCTGGCACTTCGTC/CGCTCTAGAGCGGCCTGATACTGCACCATTCC | Deletion of arsR3 gene | |
| arsR4-1f/1r (352 bp) | CGCGGATCCGCGCGTGAGCCGAACAGAA/CCCATCCACTAAACTTAAACATGCCGCACCAGAAGCCGAAAAG | Deletion of arsR4 gene | |
| arsR4-2f/2r (353 bp) | TGTTTAAGTTTAGTGGATGGGCCACACGCGGGAAAGTC/CGCTCTAGACTGGTCAGCGGGAAGATAGG | Deletion of arsR4 gene | |
| ParsR1-f/r (394 bp) | CGCGAATTCTGTGCCTCAAGTCCTGCCATCGTT/CGCGGATCCAATTGCCTGTTCCTGTTCCATA | Construction of ParsR1-lacZ fusion | |
| ParsR2-f/r (410 bp) | CGCGAATTCCGGAGACCTTGCGAATGATG/CGCGGATCCCAACCACGAGCGCACGCACGATAG | Construction of ParsR2-lacZ fusion | |
| ParsR3-f/r (397 bp) | CGCGAATTCTGATGTCCGGCCACTATGT/CGCGGATCCCAGCAAATGCCGAAAGAGC | Construction of ParsR3-lacZ fusion | |
| ParsR4-f/r (395 bp) | CGCGAATTCAAGGGCGGCGCAACCATCATC/CGCGGATCCGCCAAAGCGAGAATAGCCTGTT | Construction of ParsR4-lacZ fusion |
Deletion mutations were introduced separately into the arsR1, arsR2, arsR3, and arsR4 coding regions by crossover PCR and the levansucrase selection procedure, which we described previously (20, 29, 31). For this purpose, the PCR primers were designed to (i) leave the 5′ and 3′ ends of the deleted gene intact and (ii) avoid polar effects on downstream gene expression by leaving the Shine-Dalgarno sequence and the translational start of the downstream gene untouched. Transcriptional lacZ reporter fusions were also constructed using previously described procedures wherein the promoter regions of the different genes were PCR amplified and then directionally cloned into the multicloning site of pLSP-KT2lacZ containing a promoterless lacZ coding region.
Protein modeling.
Homology modeling and computational studies were undertaken to explore the structural similarities and differences of the four ArsR molecules. As previously described by the Rosen group for the modeling of Acidithiobacillus ferrooxidans or Corynebacterium glutamicum ArsR (32, 33), we used the crystal structure of one of the well-characterized members of the ArsR/SmtB family of proteins (34), SmtB (PDB code 1R1T), as the template to construct homology models of the four ArsR proteins. While Staphylococcus aureus pI258 CadC (35) was also a potential template for the current modeling studies, it was not used owing to the availability of a higher-resolution structure of SmtB (note also that CadC and SmtB share high structural identity). The homology models were constructed using the software tool MODELLER (36) interfaced to Discovery Studio (37). To identify potential DNA binding regions of the ArsR proteins, the electrostatic surfaces of the molecular models were calculated using the APBS (38) plug-in to PyMol (PyMol molecular graphics system, V1.7.2; Schrödinger, LLC). Multiple-sequence alignments were made using the Clustal Omega server and plotted using the ESPRIPT Web server (39; http://espript.ibcp.fr), and phylogenetic analysis was conducted in MEGA 6.0 (40). For construction of the phylogenetic tree, the selected ArsR sequences represent various bacterial phyla in order to provide reasonable phylogenetic information, as well as to compare multiple ArsR proteins within the same organism. Beyond being annotated as an ArsR protein, sequences were also screened for the following three properties: (i) the location of the encoding arsR gene is clearly associated with an ars operon, (ii) the protein contains Cys residues located in positions that have been recognized as likely As(III) binding sites, or (iii) the arsR gene is associated with an lysR-type gene (as with arsR3) (see below). Hydropathy analysis was performed on the basis of the Kyte and Doolittle method (41), with a window size of 3 amino acids, using in-house programs. The hydropathy analysis values were plotted on a scale of 1 to 10, with 1 representing a highly hydrophilic region (white) and 10 indicating a hydrophobic region (black), and were plotted with appropriate gray percentages for intermediate values.
RESULTS
Primary structural features and phylogenetic relatedness of ArsR proteins.
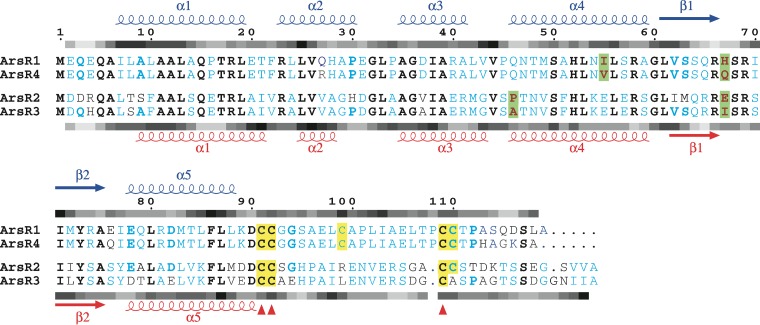
The current study initiated work designed to begin characterizing the expression of multiple arsR genes, structural predictions for the encoded ArsR proteins, and their role in As(III)-linked gene regulation. ArsR1 and ArsR4 are quite similar in terms of amino acid sequence homology, sharing 93% identity and 96% similarity (Fig. 2). ArsR2 and ArsR3 are likewise more similar to each other (78% identity and 86% similarity) than to ArsR1 and ArsR4, e.g., ArsR1 and ArsR3 are 44% identical and 57% similar. As would thus be expected, amino acid alignments clearly distinguish these pairs of proteins (Fig. 2). Sequence differences between ArsR1/ArsR4 and ArsR2/ArsR3 translate to differences in calculated hydrophobicity in a few regions (Fig. 2). For example, the hydrophobicities of the N-terminal and β-turn-β regions of the proteins are quite comparable, while the other regions show significant variations.
FIG 2.
Multiple-sequence alignment of ArsR sequences. Secondary structure information on the top layer corresponds to ArsR1 and ArsR4; that on the bottom layer corresponds to ArsR2 and ArsR3. Strictly conserved sites are represented by bold black letters. Blue letters indicate sequence identity within the similar pairs (ArsR1/ArsR4 and ArsR2/ArsR3). Bold blue letters indicate that three of the four sequences are identical. Cysteine residues are highlighted in yellow, while conserved cysteine residues are indicated by red triangles below the sequence block. The fine changes in the sequences at the potential DNA binding region are highlighted in green. The shaded bars above and below each sequence block show schematic representations of hydropathy analyses of ArsR1 and ArsR3, respectively. Hydropathy indices are shown in various shades of gray, with hydrophilic regions shown in pale gray and hydrophobic regions shown in black.
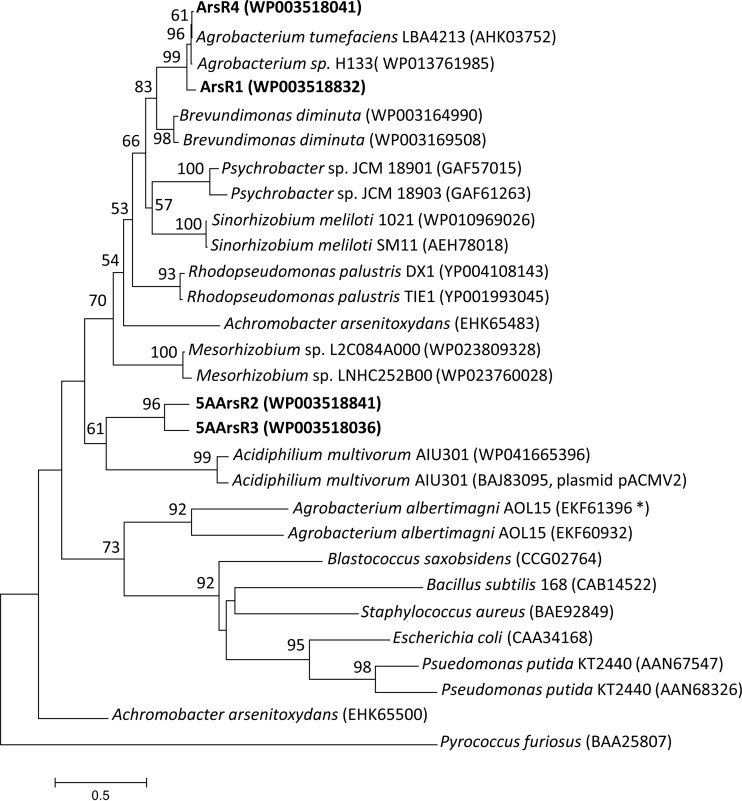
Given these differences, it is not surprising that these proteins also separate phylogenetically (Fig. 3). ArsR1 and ArsR4 cluster closely with ArsR proteins from another A. tumefaciens strain and an Agrobacterium species but are quite distinct from ArsR2 and ArsR3, as well as from two ArsR proteins in Agrobacterium albertimagni AOL15 (Fig. 3). Comparisons with other ArsR proteins from other genera illustrate interesting patterns, ranging from near identity for two ArsR proteins in P. putida and Acidiphilium multivorum (with one encoded by a plasmid-borne gene) to two ArsR proteins in an Achromobacter arsenitoxydans strain that are quite distinct from one another, which implies differing phylogenetic histories (Fig. 3).
FIG 3.
Phylogenetic relatedness of the A. tumefaciens 5A ArsR proteins (shown in bold) and ArsR proteins from other bacteria. The neighbor-joining tree was generated using MEGA (version 6.06). Bootstrap values of >50 are shown and were generated from 1,000 samplings, with the archaean Pyrococcus furiosus ArsR protein designated as the outgroup. The scale bar shows the number of amino acid substitutions per site. Accession numbers are provided in parentheses. The asterisk denotes an ArsR protein encoded adjacent to a gene annotated as an lysR gene, as with arsR3 in A. tumefaciens 5A.
To attain structural insight into the four A. tumefaciens ArsR proteins, as with all other ArsR proteins published to date, homology models were constructed (see Fig. S1A in the supplemental material) by using the crystal structure of the zinc regulatory protein SmtB (34) as the template (26 to 32% identity and 44 to 46% similarity). A few unpublished ArsR-like structures in the Protein Data Bank (PDB codes 2OQG, 3F6V, etc.) that crystallized as dimers or monomers were incomplete or unavailable for viewing or did not have characteristic arsenic binding sites and hence were not considered suitable templates for modeling the A. tumefaciens ArsR structures. All four structures were modeled as homodimers with helix-turn-helix (HTH) winged folds, following the template used. Least-square superposition of the Cα atoms in these structures (see Fig. S1B) showed high structural identity, with only fine-scale changes in the structures. Among the four structures, the maximum root mean square deviation (RMSD) of 2.7 Å was observed between ArsR3 and ArsR1, whereas ArsR2 and ArsR3 showed a low RMSD value (0.873 Å), and ArsR1 and ArsR4 were identical in their Cα positions. Small differences in length and location of the beginning or end of the secondary structures could be artifacts of computational tools and hence are not emphasized here. However, subtle differences in the conserved secondary structure regions may be important in determining the specific DNA sequences that the ArsR proteins recognize and bind (Fig. 2). To aid in identifying the potential DNA binding regions of the ArsR proteins, electrostatic surfaces were calculated (see Fig. S2). As could be expected, the electrostatic surfaces of ArsR1 and ArsR4 were very similar and clearly exhibited a positive surface on one side of each protein depicting the potential DNA binding region of the protein. The charge distributions on ArsR2 and ArsR3 were more discrete across the protein surface, but as with Ars1/ArsR4, the distributions of charge were very similar for the pair. The different patterns suggest that there may be functional differences between the pairs.
An inspection of the location of the helices along the positively charged region of the surfaces is consistent with α4 being the recognition helix in all four of these ArsR proteins, as generally found in HTH winged helix proteins (42), and is in accordance with the nuclear magnetic resonance (NMR)-based structure of the zinc-dependent transcriptional repressor CzrA in the DNA-bound state (43). The α1 and α4 amino acid sequences in ArsR1 and ArsR4 show significant conservation, except for the I55V difference. Similarly, ArsR2 and ArsR3 show high sequence identity; the only change in this region is at the start of α4, with a proline in ArsR2 but an alanine in ArsR3. An examination of the β-turn-β region reveals that position 67 is highly variant: H in ArsR1, Q in ArsR4, E in ArsR2, and I in ArsR3 (Fig. 2). From further inspection, it can be assumed that the arsenite binding sites are comprised of C91, C92, and C109, which are conserved in all four ArsR proteins. However, there are two additional cysteines that exhibit various levels of conservation. C99 is conserved only in ArsR1 and ArsR4, and C110 is conserved in all except ArsR3. The locations of the cysteine residues in these structures indicate that these proteins have simple type 2 arsenic binding sites located at the dimer interface (32, 33).
Autoregulatory and cross-regulatory features of the different ArsR proteins.
We then examined the expression and regulation of the different arsR genes. To construct lacZ reporters, the 5′ region of each coding region was PCR amplified along with at least 166 nucleotides of upstream DNA and directionally cloned into the plasmid pLSP-KT2lacZ carrying a promoterless lacZ gene. Expression profiling of arsR1, arsR2, and arsR4 showed that all were upregulated in response to arsenite (Fig. 4A), illustrating that their regulation is similar to that published for all other arsR-sensitive repressors of which we are aware (32, 44). They did differ with respect to their basal expression levels as well as their induced levels of reporter activity (Fig. 4A). For example, basal expression of the arsR2::lacZ construct was the highest and indeed exceeded the induced level of arsR1::lacZ (Fig. 4A). Maximal As(III)-induced expression levels for arsR2::lacZ and arsR4::lacZ were similar and were 4-fold higher than that for arsR1::lacZ. In contrast to the case for aio gene expression (20), phosphate levels had no effect on arsR expression (results not shown). Also, even though the arsR3::lacZ construct utilized 363 bp of upstream DNA, its expression did not change as a function of added As(III) (results not shown). Consequently, qualitative reverse transcription-PCR (RT-PCR) was then used to assess arsR3 expression in relation to As(III) exposure. There was no detectable change in arsR3 expression when the cells were exposed to As(III) (Fig. 4B), in sharp contrast to the bold upregulation of the arsR1-arsC1 gene region (Fig. 4B). It may be important that background arsR3 expression was evident, as opposed to the case for arsR1-arsC1 (Fig. 4B). The RT-PCR experiments also showed that the arsR3 coding region is cotranscribed with at least the 3′ region of the upstream open reading frame, annotated as an lysR homolog (see Fig. 1 for primer locations).
FIG 4.
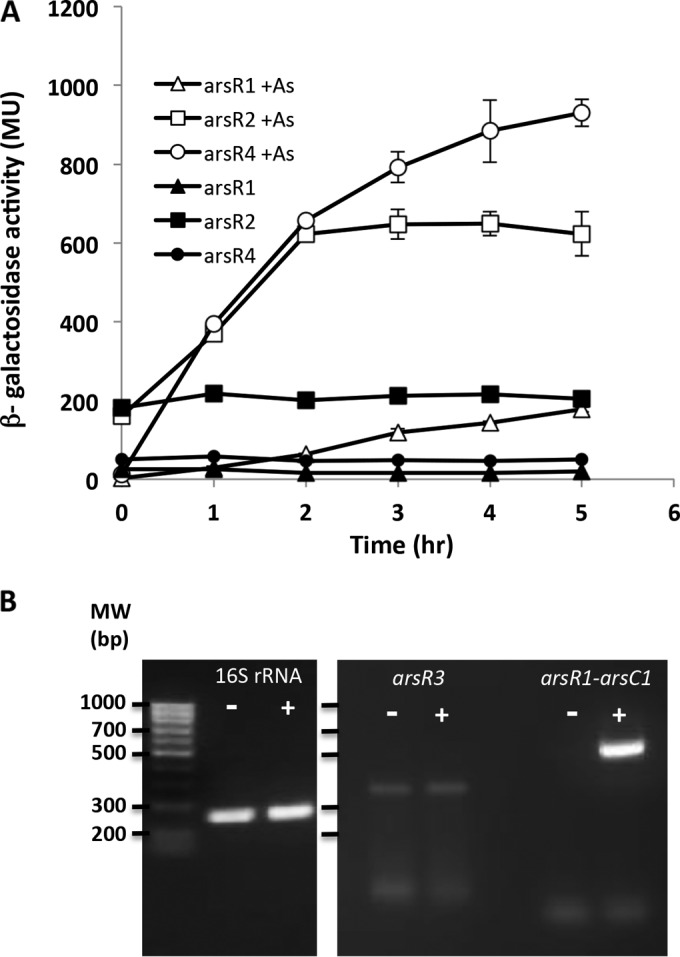
Characterization of arsR gene expression in wild-type A. tumefaciens strain 5A. (A) Expression of arsR1, arsR2, and arsR4 was monitored using lacZ transcriptional fusions. Data are examples from reproducible experiments and represent the means for duplicate cultures, with error bars illustrating the data ranges. (B) Expression of the 16S rRNA and arsR3 genes as monitored using qualitative RT-PCR with an induction period as shown for panel A. Gel images were obtained for separate gels. In all cases for both panels, cultures were incubated with 200 μM Pi, with (+) or without (−) 100 μM As(III).
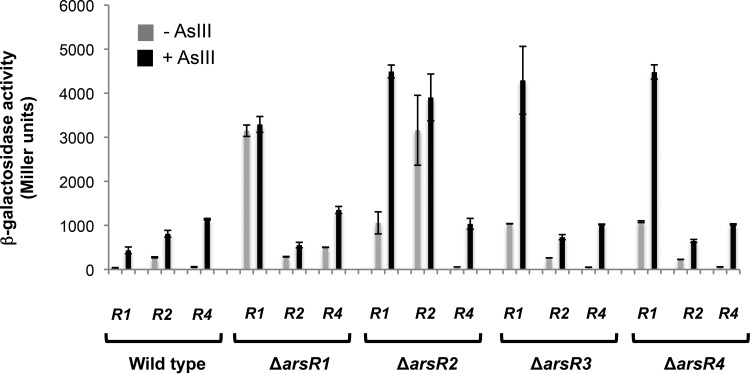
Expression levels were then investigated in different regulatory backgrounds to determine if these ArsR proteins autoregulate their respective genes and/or are capable of regulating one another. When the genes were expressed in wild-type cells, single-time-point assays generated results similar to those observed in the above-described transcriptional profiling (compare the last time point in Fig. 4 with the data in Fig. 5). However, when the genes were expressed in the various ΔarsR mutant backgrounds, important differences were readily apparent. ArsR1 is clearly autoregulatory, as we reported previously (20). When the arsR1::lacZ reporter was carried in the ΔarsR1 mutant, there was essentially no difference in expression levels with or without As(III), but the levels significantly exceeded that in the uninduced wild-type strain (∼8-fold) (Fig. 5). ArsR1 also appears to exert a weak regulatory influence over arsR4, since arsR4::lacZ reporter expression levels in the ΔarsR1 mutant [without As(III)] were >8-fold greater than those in the wild-type cells (503 versus 58 Miller units [MU]) (Fig. 5). ArsR2 is likewise autoregulatory. Expression levels of arsR2::lacZ in the ΔarsR2 mutant were similarly high regardless of As(III) exposure [i.e., lack of As(III) induction] and greatly exceeded expression without As(III) (3,159 versus 275 MU) and As(III)-induced expression (3,906 versus 810 MU) in the wild type. Regulation of arsR2::lacZ reporter activity appeared to be unchanged in all other arsR mutants (Fig. 5), implying that the other ArsR proteins do not participate in arsR2 regulation. Expression of arsR1::lacZ was also very significantly enhanced in the ΔarsR2 mutant, with levels 29- and 10-fold greater than those in wild-type cells without and with As(III), respectively (Fig. 5). Furthermore, enhanced arsR1::lacZ expression was also observed in the ΔarsR3 and ΔarsR4 mutants, similar to that seen in the ΔarsR2 mutant in the presence of As(III). Given the autoregulatory patterns observed for ArsR1/arsR1 and ArsR2/arsR2, we expected greatly enhanced expression of the arsR4::lacZ reporter in the ΔarsR4 mutant; however, this was not the case, even though the effects of the ΔarsR4 mutation could be seen on arsR1::lacZ expression (Fig. 5). In summary, the arsR1, arsR2, and arsR4 reporters exhibited sensitivity to As(III) but displayed a range of expression levels that differed as a function of the regulatory background. In contrast, arsR3 was found to be expressed constitutively at low levels and did not increase in response to As(III).
FIG 5.
Characterization of arsR1 (R1), arsR2 (R2), and arsR4 (R4) expression in wild-type A. tumefaciens strain 5A and in the ΔarsR1, ΔarsR2, ΔarsR3, and ΔarsR4 mutants. Data are examples from reproducible experiments, with the data representing the means for triplicate cultures and the error bars illustrating 1 standard deviation. Cultures were incubated with 200 μM Pi, with or without 100 μM As(III). The induction period was 7 h.
Functional gene regulation.
Given the observed temperature niche potential for the two ars operons in P. putida (15), we examined the transcription of arsR1::lacZ and arsR4::lacZ as proxies for estimating the expression of the ars1 and ars2 operons, respectively, which contain the acr31 and acr32 As(III) antiporter genes (Fig. 1). Expression of arsR1::lacZ at 30°C [1 mM As(III)] was roughly 3-fold greater than that at 15°C (190 ± 15 versus 70 ± 4 MU), whereas expression of arsR4::lacZ was reduced by only about 35% at the lower temperature (477 ± 6 versus 353 ± 30 MU). However, as measured by growth in 1 mM As(III) (24 h at 30°C and 72 h at 15°C), this did not translate to changed As(III) resistance for either the Δacr32 mutant [relies on Acr31 for As(III) resistance] or the Δacr31 mutant [relies on Acr32 for As(III) resistance] in comparison to the wild-type strain, which uses both Acr31 and Acr32 for As(III) resistance (results not shown).
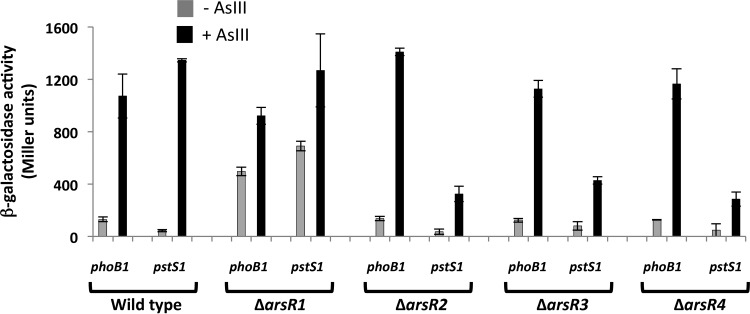
Previously, we reported that ArsR1 behaves as an As(III)-sensitive repressor of the nearby divergently expressed phoB1 and pstS1 genes (Fig. 1) (20), so we were interested in determining whether any of the other ArsR proteins would similarly regulate phoB1 and pstS1. Accordingly, phoB::lacZ and pstS1::lacZ constructs were mobilized into all four arsR deletion mutants and the wild-type parental strain. Significant As(III)-independent expression of phoB1::lacZ was observed in the ΔarsR1 mutant (Fig. 6), whereas phoB1::lacZ reporter profiles in the absence and presence of As(III) for the other arsR mutants were similar to those recorded for the wild-type strain (Fig. 6). ArsR1 control of pstS1 was also evident and consistent with our previous efforts (20). Interestingly, while pstS1::lacZ expression in the ΔarsR2, ΔarsR3, and ΔarsR4 mutants was increased in response to As(III), the degree of induction was 3- to 5-fold lower than that in the wild type (Fig. 6), implying that the corresponding proteins may normally exert some type of activator function for this gene.
FIG 6.
Comparison of levels of regulatory control of phoB1 and pstS1 in the wild-type and ΔarsR mutant strains. Data are examples from reproducible experiments, with the data representing the means ± ranges (n = 2). Cultures were incubated for 7 h with 50 μM Pi, with (black bars) or without (gray bars) 100 μM As(III).
DISCUSSION
The occurrence of multiple ars genes and ars operons in the A. tumefaciens genome is not necessarily novel, as such redundancy has been reported previously (mentioned above), although this is not the most common scenario (15). Thus far, the functional importance of ars operon redundancy has been explored primarily in P. putida KT2440, which has two ars operons. Both are required for optimal arsenic resistance (15, 45), although they differ with respect to temperature optima, with the activity and resistance function of one operon reduced at 15°C (15). In A. tumefaciens strain 5A, the ars operons are not structurally identical (Fig. 1), but they cover the same ground with respect to As(III) resistance, i.e., methylarsenite oxidase (arsH) production, As(V) reduction encoded by one or more arsC genes, and As(III) extrusion via acr3.
Phylogenetic analysis of the four ArsR proteins in strain 5A revealed that ArsR1 and ArsR4 differ from ArsR2 and ArsR3. The consistent orientations of arsR1 relative to arsR2 and of arsR3 relative to arsR4 imply nonrandomness in associating arsR1/arsR4 with arsR2/arsR3. The event(s) encompassing and selecting for the incorporation and orientation of arsR2 and arsR3 in their respective loci is unclear. There is less uncertainty as to how strain 5A acquired one of the ars operons. Prior work reported evidence of the ars1 operon being acquired as a gene island via horizontal transfer (46), as previously suggested for P. putida (15) and annotated as a plasmid acquisition in Acidiphilium multivorum (GenBank accession no. BAJ83095).
Regardless of the origin of the ars operons and their respective gene complements, the occurrence of multiple arsR genes poses interesting questions about regulatory organization, i.e., is there cross-regulation, and if so, is there a regulatory hierarchy? The significant homologies between ArsR1 and ArsR4 and between ArsR2 and ArsR3 spurred our interest in assessing their structural differences and/or similarities in relation to their regulatory function. At present, crystal structures of only a few members of the ArsR/SmtB family are known, but they do not include one for a published ArsR protein known to regulate an ars operon. In an approach similar to that used by the Rosen group (32, 33), modeling of strain 5A ArsR proteins based on SmtB showed them all to be structurally similar. SmtB is one of the best-characterized members of this family of proteins. The Staphylococcus aureus pI258 cadmium repressor CadC, another characterized member of this family of proteins, shares 79% structural identity with SmtB (determined using SABERTOOTH [47]), with an RMSD of 2.3 Å in the positions of common atoms, despite only 48.4% sequence identity (48). Such dissimilar sequences sharing similar structures reinforces the view that the ArsR proteins are homodimeric molecules with an HTH DNA binding motif having a winged helix fold.
Potentially, at least, the fine differences in the A. tumefaciens 5A ArsR amino acid sequences may confer sequence specificity in DNA recognition, and thus it was of interest to determine if the different ArsR proteins might participate in regulating each other's encoding genes, e.g., for cross-regulation between highly similar ArsR proteins, such as ArsR1 and ArsR4. Expression of three of the arsR genes was enhanced by As(III) and thus was consistent with known regulatory responses of arsR genes. This is important in the context of demonstrating the fidelity of the reporters but also to illustrate that expression of these arsR genes indeed responds to cellular As(III) and would be an essential determinant of the arsenic resistance response. The exception concerns arsR3, which exhibited no apparent influence of As(III) (Fig. 4B) and indeed appears to be cotranscribed with the upstream open reading frame, annotated as an lysR gene. This implies that the latter also does not respond to As(III).
Strategic deletion mutations allowed us to further examine the regulatory expression of these genes. The deletion rendering an ArsR1 mutant resulted in high constitutive expression of arsR1, as would be predicted from the significant literature showing that these repressors autoregulate. Surprisingly, the high degree of apparent structural similarity between ArsR1 and ArsR4 did not translate to equivalent cross-regulatory control of each other; constitutive expression levels of arsR4::lacZ and arsR1::lacZ in the ΔarsR1 mutant were not equivalent (Fig. 5). Still, constitutive expression of arsR4::lacZ in the ΔarsR1 mutant was nearly 9-fold higher than that observed in the uninduced wild-type cells (Fig. 5), implying that ArsR1 has some level of affinity for the arsR4 promoter region. Surprisingly, ArsR4 did not exhibit autoregulatory activity, at least as observed with the arsR4::lacZ construct (Fig. 5). There may be an additional DNA binding site further upstream of the region used to construct the arsR4::lacZ construct, but this reporter nevertheless demonstrated sensitivity to As(III) in all regulatory backgrounds, i.e., it otherwise behaved in an expected fashion. These observations are consistent with the suggestion that an ArsR4-specific binding motif is present in the arsR1 promoter region but absent in that of arsR4. If true, this would also argue that these putative DNA binding differences derive from the seemingly subtle amino acid differences between these proteins (Fig. 2).
Greatly enhanced arsR2::lacZ expression [constitutive and with As(III)] was not unexpected for the ΔarsR2 mutant (Fig. 5); however, arsR1 expression in this mutant was also greatly enhanced, even though the reciprocal situation did not hold (see above). Curiously, even though arsR3 did not respond to the addition of As(III) (Fig. 4B), a deletion knockout of ArsR3 also resulted in enhanced arsR1::lacZ expression, but this was not the case for arsR2 or arsR4 (Fig. 5). This implies that the DNA binding motif for ArsR3 differs from that recognized by ArsR2 or ArsR4. The observation that constitutive arsR1::lacZ expression was enhanced in all mutants likewise suggests that the arsR1 promoter region contains multiple DNA binding motifs recognizable by the different proteins, though perhaps with a lower affinity than that exhibited by ArsR1. Given that all four ArsR proteins demonstrated evidence of arsR1 repression (Fig. 5), it may be reasonable to conclude that their combined repression accounts for the lowest relative reporter activity observed for arsR1::lacZ (Fig. 4).
The unexpected regulatory patterns exhibited for the ArsR proteins suggested that there may be differences in the regulatory patterns of functional genes they control. Our prior research found the Acr31 antiporter to be more important than Acr32 in terms of providing Sb/As(III) resistance (29). However, such differences cannot be attributed to gene expression derived from the arsR1 (controls acr31) (28) and arsR4 (controls acr32) promoters, because the arsR4::lacZ reporter activity was nearly an order of magnitude stronger than the arsR1::lacZ reporter activity (Fig. 4A). Relative levels of transcription as influenced by temperature also failed to influence As(III) resistance.
We previously showed that ArsR1 acts as an As(III)-sensitive repressor of phoB1 and pstS1 (Fig. 1) (20). The current study illustrated the same ArsR1 regulatory behavior in that significantly increased constitutive expression of phoB1::lacZ and pstS1::lacZ was observed in the ΔarsR1 mutant (Fig. 6). Use of the same constructs in the other mutants, however, generated different regulatory profiles. Incongruent patterns again emerged in the context of apparent protein structural similarities (e.g., for ArsR1 and ArsR4) (Fig. 2; see Fig. S1 in the supplemental material) failing to translate to similar regulatory activities (Fig. 6). Expression levels and induction ranges of the phoB1::lacZ reporter in response to As(III) were similar for the wild type and the ΔarsR2, ΔarsR3, and ΔarsR4 mutants (basal expression levels ranged from 123 to 137 MU, and upregulation levels ranged from ∼8- to 11-fold) but contrasted sharply with those for the ΔarsR1 mutant (Fig. 6).
Perhaps one of the most significant novel observations derived from this study concerns the behavior of the pstS1::lacZ reporter in the different arsR backgrounds. Expression of pstS1::lacZ in the wild type and the ΔarsR1 mutant was as expected based on our prior efforts (20). As(III)-triggered induction was ∼31-fold in wild-type cells, and the constitutive expression level in the ΔarsR1 mutant exceeded uninduced wild-type expression 5-fold and was approximately half of the fully induced level (Fig. 6). This confirmed the fidelity of the pstS1::lacZ reporter with respect to As(III) sensitivity. In contrast, fully As(III)-induced levels in the ΔarsR2, ΔarsR3, and ΔarsR4 mutants were only 21 to 32% of that in the wild-type cells. This implies that the corresponding ArsR proteins behave in the fashion of a transcription factor, at least with respect to pstS1, departing significantly from the oft-described repressor function of ArsR proteins but possibly similar to the function of AioF, recently described by the Bonnefoy group (49). PhoB is a well-described transcriptional activator of the phosphate stress response (50, 51), and we previously demonstrated PhoB1 to serve this role in strain 5A, activating itself and pstS1 (20).
In summary, based on reporter constructs that should reasonably be viewed as containing relevant promoter DNAs, the auto-/cross-regulatory picture for these arsR genes and encoded proteins appears to be quite complex and is not easily explained by structurally (dis)similar characteristics alone. Other regulatory components are likely involved, with one example being PhoB1 as an activator of phoB1 and pstS1 (20). Equally likely, some of this complexity may be associated with DNA binding sequence variation among the arsR gene promoters. A complete set of double knockouts and DNA footprinting will be required to sort out the various possible scenarios to address this complexity.
Supplementary Material
ACKNOWLEDGMENTS
Support for this research was provided by the U.S. National Science Foundation (grants MCB-0817170 and MCB-1413321 to T.R.M. and B.B.B.) and the Montana Agricultural Experiment Station (project 911310 to T.R.M.). S.T. was supported by an institutional grant to IBAB from the Department of Electronics and Information Technology, Government of India, and the Department of Information Technology, Biotechnology and Science & Technology, Government of Karnataka.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00262-16.
REFERENCES
- 1.Rosen BP. 1999. Families of arsenic transporters. Trends Microbiol 7:207–212. doi: 10.1016/S0966-842X(99)01494-8. [DOI] [PubMed] [Google Scholar]
- 2.Mukhopadhyay R, Rosen BP, Phung LT, Silver S. 2002. Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol Rev 26:311–325. doi: 10.1111/j.1574-6976.2002.tb00617.x. [DOI] [PubMed] [Google Scholar]
- 3.Rosen BP. 2002. Biochemistry of arsenic detoxification. FEBS Lett 529:86–92. doi: 10.1016/S0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- 4.Chen CM, Misra TK, Silver S, Rosen BP. 1986. Nucleotide sequence of the structural genes for an anion pump. J Biol Chem 261:15030–15038. [PubMed] [Google Scholar]
- 5.Chen Y, Rosen BP. 1997. Metalloregulatory properties of the ArsD repressor. J Biol Chem 272:14257–14262. doi: 10.1074/jbc.272.22.14257. [DOI] [PubMed] [Google Scholar]
- 6.Lin YF, Walmsley AR, Rosen BP. 2006. An arsenic metallochaperone for an arsenic detoxification pump. Proc Natl Acad Sci U S A 103:15617–15622. doi: 10.1073/pnas.0603974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Rosen BP. 1993. Metalloregulated expression of the ars operon. J Biol Chem 268:52–58. [PubMed] [Google Scholar]
- 8.Chen J, Bhattacharjee H, Rosen BP. 2015. ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol Microbiol 96:1042–1052. doi: 10.1111/mmi.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.López-Maury L, Sánchez-Riego AM, Reyes JC, Florencio FJ. 2009. The glutathione/glutaredoxin system is essential for arsenate reduction in Synechocystis sp. strain PCC 6803. J Bacteriol 191:3534–3543. doi: 10.1128/JB.01798-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshinaga M, Rosen BP. 2014. A C-As lyase for degradation of environmental organoarsenical herbicides and animal husbandry growth promoters. Proc Natl Acad Sci U S A 111:7701–7706. doi: 10.1073/pnas.1403057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Chen S, Xiao X, Huang X, You D, Zhou X, Deng Z. 2006. arsRBOCT arsenic resistance system encoded by linear plasmid pHZ227 in Streptomyces sp. strain FR-008. Appl Environ Microbiol 72:3738–3742. doi: 10.1128/AEM.72.5.3738-3742.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Jeon B, Sahin O, Zhang Q. 2009. Identification of an arsenic resistance and arsenic-sensing system in Campylobacter jejuni. Appl Environ Microbiol 75:5064–5073. doi: 10.1128/AEM.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakajima T, Hayashi K, Nagatomi R, Matsubara K, Moore JE, Millar BC, Matsuda M. 2013. Molecular identification of an arsenic four-gene operon in Campylobacter lari. Folia Microbiol (Praha) 58:253–260. doi: 10.1007/s12223-012-0207-5. [DOI] [PubMed] [Google Scholar]
- 14.Achour-Rokbani A, Cordi A, Poupin P, Bauda P, Billard P. 2010. Characterization of the ars gene cluster from extremely arsenic-resistant Microbacterium sp. strain A33. Appl Environ Microbiol 76:948–955. doi: 10.1128/AEM.01738-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Páez-Espino AD, Durante-Rodríguez G, de Lorenzo V. 2015. Functional coexistence of twin arsenic resistance systems in Pseudomonas putida KT2440. Environ Microbiol 17:229–238. doi: 10.1111/1462-2920.12464. [DOI] [PubMed] [Google Scholar]
- 16.Qin J, Rosen BP, Zhang Y, Wang G, Franke S, Rensing C. 2006. Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc Natl Acad Sci U S A 103:2075–2080. doi: 10.1073/pnas.0506836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Zhang L, Wang G. 2014. Genomic evidence reveals the extreme diversity and wide distribution of the arsenic-related genes in Burkholderiales. PLoS One 9:e92236. doi: 10.1371/journal.pone.0092236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ordóñez E, Letek M, Valbuena N, Gil JA, Mateos LM. 2005. Analysis of genes involved in arsenic resistance in Corynebacterium glutamicum ATCC 13032. Appl Environ Microbiol 71:6206–6215. doi: 10.1128/AEM.71.10.6206-6215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branco R, Chung AP, Morais PV. 2008. Sequencing and expression of two arsenic resistance operons with different functions in the highly arsenic-resistant strain Ochrobactrum tritici SCII24T. BMC Microbiol 8:95. doi: 10.1186/1471-2180-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang YS, Heinemann J, Bothner B, Rensing C, McDermott TR. 2012. Integrated co-regulation of bacterial arsenic and phosphorus metabolisms. Environ Microbiol 14:3097–3109. doi: 10.1111/j.1462-2920.2012.02881.x. [DOI] [PubMed] [Google Scholar]
- 21.Muller D, Médigue C, Koechler S, Barbe V, Barakat M, Talla E, Bonnefoy V, Krin E, Arsène-Ploetze F, Carapito C, Chandler M, Cournoyer B, Cruveiller S, Dossat C, Duval S, Heymann M, Leize E, Lieutaud A, Lièvremont D, Makita Y, Mangenot S, Nitschke W, Ortet P, Perdrial N, Schoepp B, Siguier P, Simeonova DD, Rouy Z, Segurens B, Turlin E, Vallenet D, Van Dorsselaer A, Weiss S, Weissenbach J, Lett MC, Danchin A, Bertin PN. 2007. A tale of two oxidation states: bacterial colonization of arsenic-rich environments. PLoS Genet 3:e53. doi: 10.1371/journal.pgen.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canovas D, Cases I, de Lorenzo V. 2003. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ Microbiol 5:1242–1256. doi: 10.1111/j.1462-2920.2003.00463.x. [DOI] [PubMed] [Google Scholar]
- 23.Koechler S, Arsene-Ploetze F, Brochier-Armanet C, Goulhen-Chollet F, Heinrich-Salmeron A, Jost B, Lievremont D, Philipps M, Plewniak F, Bertin PN, Lett MC. 2015. Constitutive arsenite oxidase expression detected in arsenic-hypertolerant Pseudomonas xanthomarina S11. Res Microbiol 166:205–214. doi: 10.1016/j.resmic.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Arsène-Ploetze F, Koechler S, Marchal M, Coppée JY, Chandler M, Bonnefoy V, Brochier-Armanet C, Barakat M, Barbe V, Battaglia-Brunet F, Bruneel O, Bryan CG, Cleiss-Arnold J, Cruveiller S, Erhardt M, Heinrich-Salmeron A, Hommais F, Joulian C, Krin E, Lieutaud A, Lièvremont D, Michel C, Muller D, Ortet P, Proux C, Siguier P, Roche D, Rouy Z, Salvignol G, Slyemi D, Talla E, Weiss S, Weissenbach J, Médigue C, Bertin PN. 2010. Structure, function, and evolution of the Thiomonas spp. genome. PLoS Genet 6:e1000859. doi: 10.1371/journal.pgen.1000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, Lin J, Mi S, Lin J. 2010. Arsenic resistance operon structure in Leptospirillum ferriphilum and proteomic response to arsenic stress. Bioresour Technol 101:9811–9814. doi: 10.1016/j.biortech.2010.07.043. [DOI] [PubMed] [Google Scholar]
- 26.Cuebas M, Villafane A, McBridem M, Yeem N, Bini E. 2011. Arsenate reduction and expression of multiple chromosomal ars operons in Geobacillus kaustophilus A1. Microbiology 157:2004–2011. doi: 10.1099/mic.0.048678-0. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q, Qin D, Zhang S, Wang L, Li J, Rensing C, McDermott TR, Wang G. 2015. Fate of arsenate following arsenite oxidation in Agrobacterium tumefaciens GW4. Environ Microbiol 17:1926–1940. doi: 10.1111/1462-2920.12465. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez M, Udaondo Z, Niqui JL, Duque E, Ramos JL. 2014. Synergic role of the two ars operons in arsenic tolerance in Pseudomonas putida KT2440. Environ Microbiol Rep 6:483–489. doi: 10.1111/1758-2229.12167. [DOI] [PubMed] [Google Scholar]
- 29.Kang YS, Shi Z, Bothner B, Wang G, McDermott TR. 2015. Involvement of the Acr3 and DctA anti-porters in arsenite oxidation in Agrobacterium tumefaciens 5A. Environ Microbiol 17:1950–1962. doi: 10.1111/1462-2920.12468. [DOI] [PubMed] [Google Scholar]
- 30.Kang YS, Park W. 2010. Trade-off between antibiotics resistance and biological fitness in Acinetobacter sp. strain DR1. Environ Microbiol 12:1304–1318. doi: 10.1111/j.1462-2920.2010.02175.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu G, Liu M, Kim EH, Matty W, Bothner B, Lei B, Rensing C, Wang G, McDermott TR. 2012. A periplasmic arsenite-binding protein involved in regulating arsenite oxidation. Environ Microbiol 12:1624–1634. doi: 10.1111/j.1462-2920.2011.02672.x. [DOI] [PubMed] [Google Scholar]
- 32.Ordóñez E, Thiyagarajan S, Cook JD, Stemmler TL, Gil JA, Mateos LM, Rosen BP. 2008. Evolution of metal(loid) binding sites in transcriptional regulators. J Biol Chem 283:25706–25714. doi: 10.1074/jbc.M803209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin J, Fu HL, Ye J, Bencze KZ, Stemmler TL, Rawlings DE, Rosen BP. 2007. Convergent evolution of a new arsenic binding site in the ArsR/SmtB family of metalloregulators. J Biol Chem 282:34346–34355. doi: 10.1074/jbc.M706565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eicken C, Pennella MA, Chen X, Koshlap KM, VanZile ML, Sacchettini JC, Giedroc DP. 2003. A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J Mol Biol 333:683–695. doi: 10.1016/j.jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Ye J, Kandegedara A, Martin P, Rosen BP. 2005. Crystal structure of the Staphylococcus aureus pI258 CadC Cd(II)/Pb(II)/Zn(II)-responsive repressor. J Bacteriol 187:4214–4221. doi: 10.1128/JB.187.12.4214-4221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen M, Pieper U, Sali A. 2006. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics Chapter 5:Unit 5.6. doi: 10.1002/0471250953.bi0506s15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Accelrys Software Inc. 2009. Discovery Studio modeling environment, release 2.5.5. Accelrys Software Inc, San Diego, CA. [Google Scholar]
- 38.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. 2011. Integrative analysis of environmental sequences using MEGAN 4. Genome Res 21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kyte J, Doolittle R. 1982. A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 42.Gajiwala KS, Burley SK. 2000. Winged helix proteins. Curr Opin Struct Biol 10:110–116. doi: 10.1016/S0959-440X(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 43.Arunkumar AL, Campanello GC, Giedroc DC. 2009. Solution structure of a paradigm ArsR family zinc sensor in the DNA bound state. Proc Natl Acad Sci U S A 106:18177–18182. doi: 10.1073/pnas.0905558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy JN, Saltikov CW. 2009. The ArsR repressor mediates arsenite-dependent regulation of arsenate respiration and detoxification operons of Shewanella sp. strain ANA-3. J Bacteriol 191:6722–6731. doi: 10.1128/JB.00801-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paez-Espino D, Tamames J, de Lorenzo V, Canovas D. 2009. Microbial responses to environmental arsenic. Biometals 22:117–130. doi: 10.1007/s10534-008-9195-y. [DOI] [PubMed] [Google Scholar]
- 46.Li H, Li M, Huang Y, Rensing C, Wang G. 2013. In silico analysis of bacterial arsenic islands reveals remarkable synteny and functional relatedness between arsenate and phosphate. Front Microbiol 4:347. doi: 10.3389/fmicb.2013.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teichert F, Bastolla U, Porto M. 2007. SABERTOOTH: protein structural alignment based on a vectorial structure representation. BMC Bioinformatics 8:425. doi: 10.1186/1471-2105-8-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kandegadara A, Thiyagarayan S, Kondapalli KC, Stemmler TL, Rosen BP. 2009. Role of bound Zn(II) in the CadC Cd(II)/Pb(II)/Zn(II)-responsive repressor. J Biol Chem 284:14958–14965. doi: 10.1074/jbc.M809179200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moinier D, Slyemi D, Byrne D, Lignon S, Lebrun R, Talla E, Bonnefoy V. 2014. An ArsR/SmtB family member is involved in the regulation by arsenic of the arsenite oxidase operon in Thiomonas arsenitoxydans. Appl Environ Microbiol 80:6413–6426. doi: 10.1128/AEM.01771-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamarche MG, Wanner BL, Crepin S, Harel J. 2008. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol Rev 32:461–473. doi: 10.1111/j.1574-6976.2008.00101.x. [DOI] [PubMed] [Google Scholar]
- 51.Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol 13:198–203. doi: 10.1016/j.mib.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.