ABSTRACT
Agrobacterium tumefaciens has a cluster of genes (Atu3178, Atu3179, and Atu3180) encoding an ABC-type transporter, here named troA, troB, and troC, respectively, which is shown here to be a zinc-specific uptake system. Reverse transcription (RT)-PCR analysis confirmed that troA, troB, and troC are cotranscribed, with troC as the first gene of the operon. The yciC (Atu3181) gene is transcribed in the opposite orientation to that of the troCBA operon and belongs to a metal-binding GTPase family. Expression of troCBA and yciC was inducible under zinc-limiting conditions and was controlled by the zinc uptake regulator, Zur. Compared to the wild type, the mutant strain lacking troC was hypersensitive to a metal chelator, EDTA, and the phenotype could be rescued by the addition of zinc, while the strain with a single yciC mutation showed no phenotype. However, yciC was important for survival under zinc limitation when either troC or zinT was inactivated. The periplasmic zinc-binding protein, ZinT, could not function when TroC was inactivated, suggesting that ZinT may interact with TroCBA in zinc uptake. Unlike many other bacteria, the ABC-type transporter ZnuABC was not the major zinc uptake system in A. tumefaciens. However, the important role of A. tumefaciens ZnuABC was revealed when TroCBA was impaired. The strain containing double mutations in the znuA and troC genes exhibited a growth defect in minimal medium. A. tumefaciens requires cooperation of zinc uptake systems and zinc chaperones, including TroCBA, ZnuABC, ZinT, and YciC, for survival under a wide range of zinc-limiting conditions.
IMPORTANCE Both host and pathogen battle over access to essential metals, including zinc. In low-zinc environments, physiological responses that make it possible to acquire enough zinc are important for bacterial survival and could determine the outcome of host-pathogen interactions. A. tumefaciens was found to operate a novel pathway for zinc uptake in which ZinT functions in concert with the high-affinity zinc importer TroCBA.
INTRODUCTION
Zinc is an essential metal for bacteria because it is required for the functions of many enzymes and proteins (1, 2). However, zinc overload is toxic to cells (3–7). Bacteria have mechanisms to maintain zinc homeostasis via the coordinated response of genes involved in zinc uptake, efflux, and storage (8–13). The zinc uptake regulator Zur is a transcriptional regulator belonging to the Fur family and functions as a repressor of zinc uptake genes, including znuABC and zinT (10). To prevent excessive amounts of zinc in cells under high-zinc conditions, Zur uses Zn2+ as its cofactor to bind to a conserved AT-rich sequence, called the Zur box (14), found in the promoter region of the zinc uptake genes, leading to inhibition of gene expression (15–17). ZnuA is a periplasmic protein that binds zinc and transfers it to the membrane permease ZnuB and the ATPase ZnuC (15). The ZinT protein is a periplasmic zinc-binding protein (18–21) that has been shown to directly interact with and assist ZnuABC in transporting zinc in Salmonella enterica (22, 23).
Agrobacterium tumefaciens is an alphaproteobacterium that causes crown gall tumor in plants (24). Negative regulation of zinc uptake genes, such as znuABC and zinT, by A. tumefaciens Zur (ZurAt) has been reported previously (25). Expression of A. tumefaciens znuABC and zinT was inducible with zinc depletion and was repressed in response to increased zinc concentrations (25). Loss of ZurAt led to derepression of the znuABC and zinT genes and increased accumulation of intracellular zinc content (25). The roles of the periplasmic zinc-binding proteins A. tumefaciens ZnuA and ZinT (ZnuAAt and ZinTAt, respectively) have been investigated. It was found that ZinTAt played an important role in A. tumefaciens survival under severe zinc shortage, whereas ZnuAAt did not show an apparent role under the tested conditions (25). Disruption of both znuAAt and zinTAt slightly affected the total cellular zinc content (25), implying the existence of other, unidentified zinc uptake genes in A. tumefaciens.
The A. tumefaciens C58 genome contains a cluster of genes consisting of Atu3178, Atu3179, and Atu3180 that are annotated as a putative zinc/manganese ABC transport system (http://www.genome.jp/kegg-bin/show_organism?org=atu). Atu3178 encodes a periplasmic substrate-binding protein belonging to the TroA (transport-related operon) superfamily. Atu3179 encodes a permease, and Atu3180 encodes an ATP-binding protein. Here, we have named these genes troA, troB, and troC (the first gene of the operon), respectively. Atu3181 is a gene that has a transcription orientation opposite to that of the troCBA operon and encodes a putative metal chaperone, YciC, belonging to the COG0523 family of GTPases (26). In Bacillus subtilis, YciC was proposed to be a zinc chaperone that participates in an unidentified low-affinity zinc transport pathway (27, 28). B. subtilis yciC is regulated by Zur (28). The A. tumefaciens troCBA operon and yciC were predicted to be controlled by Zur due to the presence of a Zur box in their promoter regions (26). However, their physiological functions and metal regulation have not been experimentally verified.
The role of TroABCD in zinc transport was first reported in Treponema pallidum (29, 30). TroA is a substrate-binding protein, and TroB is an ATPase, while TroC and TroD form a heterodimeric cytoplasmic membrane permease. T. pallidum has the ZnuABC and TroABCD systems for zinc uptake (30). While the T. pallidum ZnuABC transporter is specific to zinc, T. pallidum TroABCD can transport zinc, manganese, and iron (29, 30). T. pallidum troABCD was shown to be negatively regulated by a zinc-responsive transcriptional regulator, TroR, a DtxR-like repressor (29). However, a later study showed that T. pallidum TroR is a manganese-dependent rather than a zinc-dependent regulator (31). Unlike many other bacteria, T. pallidum does not contain Zur, and the regulator of T. pallidum znuABC has not been reported.
Both ZnuA and TroA are substrate-binding proteins that belong to the cluster A-1 family (32). While ZnuA has been shown to respond specifically to zinc, the metal selectivity of TroA proteins and regulation of tro operons are different among bacteria (29, 33–36). Streptococcus suis TroA is involved in the uptake of manganese, not zinc (36), even though it can bind both Mn2+ and Zn2+ with high affinity (35). A gene encoding a protein that has high similarity to the manganese-responsive transcriptional regulator ScaR, located directly downstream of the S. suis troA gene, may be involved in the manganese regulation of the troA gene (36). Treponema denticola troABCD is negatively regulated by Mn2+ and Fe2+ via TroR (33). In contrast, Corynebacterium diphtheriae troA is under the control of Zur in a zinc-dependent manner (34).
Here, the physiological functions of the A. tumefaciens troCBA operon and yciC are characterized. The metal regulation of A. tumefaciens troCBA and yciC was investigated, and their regulator was identified. Important roles for A. tumefaciens TroCBA and YciC in zinc acquisition under zinc-limiting conditions were demonstrated. A. tumefaciens ZinT may interact with the TroCBA system. In addition, a role for A. tumefaciens ZnuA under zinc starvation conditions was revealed when the TroCBA system was disrupted. Our findings provide a step toward understanding how A. tumefaciens controls zinc homeostasis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. The growth conditions, media, and antibiotic concentrations that were used for A. tumefaciens and E. coli were reported previously (25, 44).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Agrobacterium tumefaciens | ||
| NTL4 | WT strain: a Ti plasmid-cured derivative of strain C58 | 37 |
| PC135 | zinT::pKNOCK-Gm (zinT::Gm); Gmr | 25 |
| PS132 | znuA::pKNOCK-Gm (znuA::Gm); Gmr | 25 |
| SPP12 | zur::pKNOCK-Gm (zur::Gm); Gmr | 25 |
| TC142 | troC::pKNOCK-Gm (troC::Gm); Gmr | This study |
| TCZA15 | troC::pKNOCK-Gm and znuA::pKNOCK-Km (troC::Gm znuA::Km); Gmr and Kmr | This study |
| TCZT14 | troC::pKNOCK-Gm and zinT::pKNOCK-Km (troC::Gm zinT::Km); Gmr Kmr | This study |
| TCYC15 | troC::pKNOCK-Gm and yciC::pKNOCK-Km (troC::Gm yciC::Km); Gmr Kmr | This study |
| YC154 | yciC::pKNOCK-Km (yciC::Km); Kmr | This study |
| ZAYC15 | znuA::pKNOCK-Gm and yciC::pKNOCK-Km (znuA::Gm yciC::Km); Gmr Kmr | This study |
| ZTYC15 | zinT::pKNOCK-Gm and yciC::pKNOCK-Km (zinT::Gm yciC::Km); Gmr Kmr | This study |
| ZURYC15 | zur::pKNOCK-Gm and yciC::pKNOCK-Km (zur::Gm yciC::Km); Gmr Kmr | This study |
| Escherichia coli | ||
| DH5α | Host for general DNA cloning | 38 |
| BW20767 | Host for plasmid pKNOCK-Gm and pKNOCK-Km | 39 |
| Plasmids for gene inactivation | ||
| pKNOCK-Gm | Suicide vector; Gmr | 40 |
| pKNOCK-Km | Suicide vector; Kmr | 40 |
| pKNOCKTROC | Internal coding region of troC cloned into pKNOCK-Gm; Gmr | This study |
| pKNOCKmYCIC | Internal coding region of yciC cloned into pKNOCK-Km; Kmr | This study |
| pKNOCKmZINT | Internal coding region of zinT cloned into pKNOCK-Km; Kmr | This study |
| pKNOCKmZNUA | Internal coding region of znuA cloned into pKNOCK-Km; Kmr | 25 |
| Plasmids for complementation | ||
| pBBR1MCS-4 | Expression vector; Apr (pBBR) | 41 |
| pTROC | Full-length troB cloned into pBBR1MCS-4; Apr | This study |
| pTROCBA | Full-length troCBA operon cloned into pBBR1MCS-4; Apr | This study |
| pYCIC | Full-length yciC cloned into pBBR1MCS-4; Apr | This study |
| pZINT | Full-length zinT cloned into pBBR1MCS-4; Apr | 25 |
| pZNUA | Full-length znuA cloned into pBBR1MCS-4; Apr | 25 |
| pZUR | Full-length zur cloned into pBBR1MCS-4; Apr | 25 |
| Promoter-lacZ transcriptional fusions | ||
| pUFR027lacZ | Promoter probe vector; Tcr | 42 |
| p027troC-lacZ | 266-bp DNA fragment containing troC promoter fused to lacZ of pUFR027lacZ | This study |
| p027yciC-lacZ | 274-bp DNA fragment containing yciC promoter fused to lacZ of pUFR027lacZ | This study |
| YciC-LacZ and YciC-PhoA translational fusions | ||
| pPR9TT | Reporter vector, LacZ as a reporter; Apr | 43 |
| pYCLacZ | YciC amino acid residues 1–400 fused to LacZ of pPR9TT; Apr | This study |
| p'PhoA | 5′-truncated phoA gene lacking its signal peptide sequence cloned into pBBR; Apr | 44 |
| pYCPhoA | YciC amino acid residues 1–400 fused to PhoA of p′PhoA; Apr | This study |
| Plasmid for tumor assay | ||
| pCMA1 | pTiC58traM::nptII; Kmr | 45 |
Apr, ampicillin resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance.
Molecular techniques.
General molecular techniques were performed according to standard protocols (46). The nucleotide sequence of the cloned DNA fragment was confirmed by DNA sequencing (Macrogen). Plasmids were transferred into A. tumefaciens by electroporation (47). All the A. tumefaciens mutant strains were confirmed by Southern blotting.
Construction of the A. tumefaciens troC mutant and yciC mutant strains.
The A. tumefaciens troC and yciC mutant strains were constructed using a single homologous-recombination method as described previously (48). The internal coding regions of troC (BT3751 [5′-TGAAACCGCTCGGCGGTGAG-3′] and BT3752 [5′-CTGCGCATCCTGCAACATGG-3′]; 293 bp) and yciC (BT5027 [5′-GTGATGGCGACAACCGCACC-3′] and BT5028 [5′-GCCCTGGCGAAATCGAAGCG-3′]; 223 bp) were amplified by PCR using the specific primers indicated. The PCR products were cloned into the unique SmaI sites of pKNOCK-Gm and pKNOCK-Km, respectively (40). The resulting plasmids, pKNOCKTROC and pKNOCKmYCIC, were electroporated into wild-type (WT) NTL4 (a Ti plasmid-cured derivative of strain C58 with an internal deletion of the tetA-tetR locus) (37). The troC mutant (TC142) and yciC mutant (YC154) were selected on Luria-Bertani (LB) agar plates containing 60 μg/ml gentamicin (Gm) and 30 μg/ml kanamycin (Km), respectively.
Construction of the double-mutant (TCYC15, ZAYC15, ZTYC15, and ZURYC15) strains.
The plasmid pKNOCKmZNUA (25) was electroporated into the TC142 strain, generating the TCZA15 strain (with disruptions of both the troC and znuA genes), which was selected on LB agar (LA) plates containing 60 μg/ml Gm and 30 μg/ml Km. The plasmid pKNOCKmZINT was constructed by PCR amplification of the internal coding region of zinT (238 bp) using primers BT3747 (5′-TGACGGACTGGGAAGGCGAC-3′) and BT3748 (5′-ATCTCCTGGCCGTCGCTGAC-3′), which was then cloned into SmaI-digested pKNOCK-Km (40). The plasmid pKNOCKmZINT was electroporated into the TC142 strain, generating the TCZT14 strain (troC and zinT mutations). The plasmid pKNOCKmYCIC was electroporated into TC142, PS132 (25), PC135 (25), and SPP12 (25) to generate the double-mutant strains TCYC15 (troC and yciC mutations), ZAYC15 (znuA and yciC mutations), ZTYC15 (zinT and yciC mutations), and ZURYC15 (zur and yciC mutations), respectively.
Construction of plasmids expressing functional troC, troCBA, and yciC for complementation.
DNA fragments of full-length troC (BT3809 [5′-ATGAACGATCCGTGCCTCAC-3′] and BT3810 [5′-AAATCAGCCATGGGCATTCG-3′]; 837 bp), troCBA (BT3809 and BT4627 [5′-GAAACTGTGCGTTTTACTGG-3′]; 2,747 bp), and yciC (BT5033 [5′-ATGAAAAAACTCCCTGTCAC-3′] and BT5034 [5′-AATCAGGCCGCCTGTCTATC-3′]; 1,205 bp) were amplified by PCR using Pfu DNA polymerase (Fermentas) and the specific primers. The PCR products were cloned into SmaI-digested pBBR1MCS-4 (41), generating pTROC, pTROCBA, and pYCIC, respectively.
RT-PCR.
Total RNA was extracted from cells grown in LB at 28°C for 4 h and then treated with 1 mM EDTA for 15 min using a modified hot-phenol method (49). Reverse transcription (RT)-PCR was performed as previously described (49). Primer sets were used for amplifying the junction of troC-troB (BT4265 [5′-AGGCGCGTCATTTCCACGAG-3′] and BT4266 [5′-TGAGGCTCATGCGCCGCAAC-3′]; 280 bp) and troB-troA (BT4267 [5′-GCATGGTCTCCTGCTTTGCC-3′] and BT4268 [5′-CGATGATCGAGAAGCTGGCC-3′]; 310 bp). The PCR products were visualized using gel electrophoresis on a 1.8% agarose gel with ethidium bromide staining.
qRT-PCR analysis.
Quantitative real-time (qRT)-PCR was performed as previously described (50). Log-phase cells grown in LB were not treated or were treated with metal and a metal chelator for 15 min prior to harvest. The metals CdCl2, CoCl2, CuSO4, FeCl3, MgCl2, MnCl2, NiCl2, and ZnCl2 were used at a final concentration of 0.45 mM. The metal chelator used was EDTA (1 mM). Gene-specific primers for troC (BT3751 [5′-TGAAACCGCTCGGCGGTGAG-3′] and BT3752 [5′-CTGCGCATCCTGCAACATGG-3′]; 293 bp), yciC (BT5027 [5′-GTGATGGCGACAACCGCACC-3′] and BT5028 [5′-GCCCTGGCGAAATCGAAGCG-3′]; 223 bp), and the 16S rRNA gene (BT1421 [5′-GAATCTACCCATCTCTGCGG-3′] and BT1422 [5′-AAGGCCTTCATCACTCACGC-3′]; 280 bp) were used.
The amount of a specific mRNA target was normalized to the amount of a housekeeping gene 16S rRNA. Fold changes in gene expression are relative to untreated samples from wild-type NTL4 using the 2−ΔΔCt method (51). The data are reported as the means of biological triplicates and standard deviations (SD).
Determination of the transcriptional start site for troC and yciC using 5′ RACE.
RNA samples were isolated from wild-type NTL4 log-phase cells grown in LB and treated with 1 mM EDTA for 15 min. 5′ Rapid amplification cDNA ends (RACE) (Roche) was performed according to the manufacturer's instructions. The specific primers SP1 and SP2 for troC are BT3752 (5′-CTGCGCATCCTGCAACATGG-3′) and BT3758 (5′-TAGCGCCATCCAGATGATGC-3′), respectively. The specific primers SP1 and SP2 for yciC are BT5028 (5′-GCCCTGGCGAAATCGAAGCG-3′) and BT5042 (5′-CGAGGATGTGGTTGAGAAGG-3′), respectively.
Construction of promoter-lacZ transcriptional fusions.
DNA fragments containing the promoter region of troC (BT3757 [5′-TATGTGCGACATGTCAACGG-3′] and BT3758 [5′-TAGCGCCATCCAGATGATGC-3′]; 269 bp) and yciC (BT5041 [5′-TTAGAGTGGCGCGCTGTTGG-3′] and BT5042 [5′-CGAGGATGTGGTTGAGAAGG-3′]; 274 bp) were amplified from A. tumefaciens NTL4 genomic DNA using PCR. The PCR products were cloned into a unique HindIII site (and end-gap filled with Klenow enzyme) of the promoter probe vector pUFR027lacZ, a derivative of pUFR027 (42), to generate the plasmids p027troC-lacZ and p027yciC-lacZ, respectively.
β-Galactosidase activity assay.
β-Galactosidase (β-Gal) activity was measured as described by Miller (52). Log-phase cells grown in LB were not treated or were treated with 1 mM EDTA for 1 h. The cells were then harvested. Crude bacterial cell lysates were prepared as previously described (48). Protein concentrations were determined using the Bradford Bio-Rad protein assay. Specific activity was calculated in units per milligram of protein. Data are reported as the means of biological triplicates ± SD.
EDTA sensitivity test.
Log-phase cells grown in LB were adjusted, serially diluted, and spotted onto plates containing AB medium (47) and AB plus EDTA (0.3, 0.6, 0.9, 1, 1.1, 1.2, 1.3, 1.4, and 1.5 mM) according to a protocol previously described (25). In some experiments, AB plates containing 1.2 mM EDTA were individually supplemented with 50 μM ZnCl2, CdCl2, CoCl2, CuSO4, FeCl3, MgCl2, MnCl2, or NiCl2. The plates were then incubated at 28°C for 48 h. Each strain was tested in duplicate, and the experiment was repeated at least twice.
Construction of yciC-lacZ and yciC-phoA translational fusions.
Primers BT5041 (5′-TTAGAGTGGCGCGCTGTTGG-3′) and BT5309 (5′-GCCGCCTGTCTATCCCAGT-3′) were used to amplify the yciC promoter region and sequences encoding the entire 400 amino acids of YciC. The DNA fragments were cloned into the SmaI sites of the plasmid vectors pPR9TT (43) and p′PhoA (44) to generate plasmids pYCLacZ and pYCPhoA, respectively. The β-galactosidase and alkaline phosphatase activities were assayed as previously described (44), using wild-type NTL4 carrying plasmids pYCLacZ and pYCPhoA, which express the hybrid proteins YciC-LacZ and YciC-PhoA, respectively.
Measurement of total cellular zinc content.
Zinc ions were measured in parts per billion using an inductively coupled plasma mass spectrometer (ICP-MS), as previously described (44). Samples were prepared from cells grown in LB plus 0.5 mM EDTA at 28°C for 24 h. The data are reported as the means of biological triplicates and SD.
Virulence assay.
A. tumefaciens strains carrying plasmid pCMA1 were used to infect young Nicotiana benthamiana plants as previously described (25). Cells grown in LB plus 0.5 mM EDTA at 28°C for 24 h were washed and resuspended in induction broth (47), pH 5.5 (IB 5.5), plus 300 μM acetosyringone (AS). The cells were incubated at 28°C with shaking for 20 min, harvested, and adjusted to an optical density at 600 nm (OD600) of 0.1 in IB 5.5 plus 300 μM AS. A 5-μl aliquot of the cell suspension was inoculated into a wounded N. benthamiana petiole. Each bacterial strain was used to infect 15 petioles. Tumor formation at 4 weeks after infection was assessed.
RESULTS
The A. tumefaciens troCBA operon and yciC are negatively regulated by Zur.
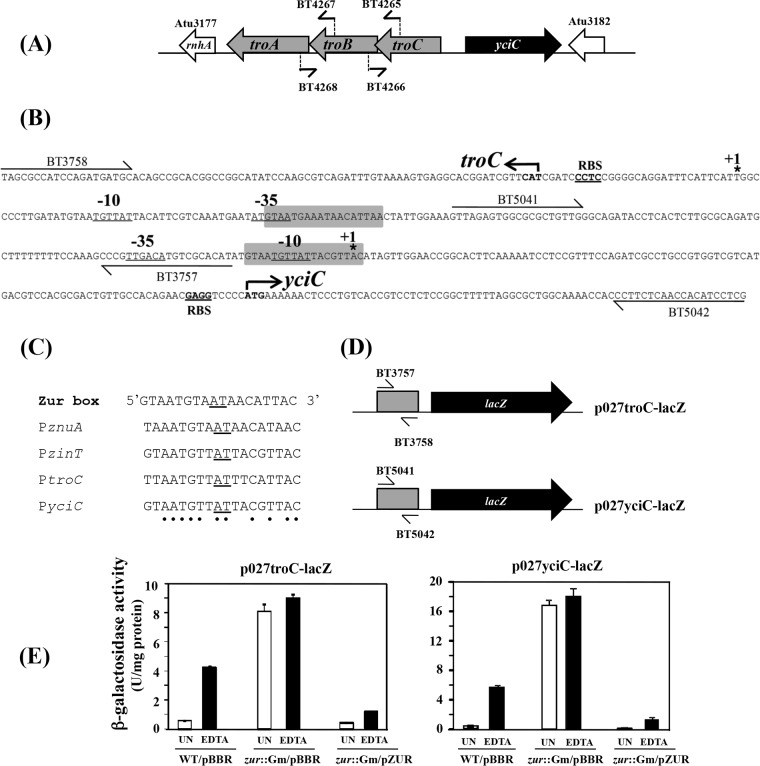
A. tumefaciens genome sequence analysis (53) revealed that the gene arrangement in the tro operon starts with troC (Atu3180), followed by troB (Atu3179) and troA (Atu3178) (Fig. 1A), and these genes may be cotranscribed. RT-PCR analysis was performed, confirming that troC, troB, and troA are cotranscribed (see Fig. S1 in the supplemental material). The A. tumefaciens troCBA operon and yciC are divergently oriented. The translational start codons of troC and yciC are separated by 300 bp (Fig. 1B). The transcriptional start sites of troC (at the A residue) and of yciC (at the A residue) were determined by 5′ RACE, and the −10 and −35 sequences were predicted using BPROM (Softberry) (Fig. 1B). A Zur-binding site (5′-TTAATGTTATTTCATTAC-3′; underlined nucleotides are the center of symmetry for the inverted palindrome) was identified previously in the intergenic region between A. tumefaciens troCBA and yciC (26) and overlaps the predicted −35 site of troC (Fig. 1B). Furthermore, we identified another Zur-binding site (5′-GTAATGTTATTACGTTAC-3′) that overlaps the predicted −10 site of yciC (Fig. 1B). An alignment of A. tumefaciens Zur boxes found in the Zur-regulated genes is shown in Fig. 1C.
FIG 1.
(A) Genomic context of the A. tumefaciens troCBA operon and yciC. The rnhA (Atu3177) gene encoding RNase H is located downstream of the troCBA operon. The Atu3182 gene encoding a hypothetical protein with unknown function is located downstream of the yciC gene. The primer sets used to amplify the junctions between troC and troB (BT4265 and BT4266) and between troB and troA (BT4267 and BT4268) with RT-PCR analysis are indicated. (B) troC and yciC promoters. The ATG start codons for troC and yciC are shown in boldface with bent arrows. The transcriptional +1 start site of troC and yciC was determined using 5′ RACE and is indicated by asterisks. The predicted −10 and −35 sequences are underlined. The Zur-binding sites are shaded. The putative ribosome-binding sites (RBS) are underlined. (C) Conserved Zur-binding site (Zur box) for A. tumefaciens (14). The sequences of Zur boxes found in the promoter regions of A. tumefaciens znuA, zinT, troC, and yciC (PznuA, PzinT, PtroC, and PyciC, respectively) are shown. The conserved residues in all the Zur boxes are marked with dots. (D) Schematic representation of the promoter-lacZ fusions (not drawn to scale) from plasmids p027troC-lacZ and p027yciC-lacZ. (E) β-Galactosidase activity assay. Wild-type NTL4 (WT/pBBR), the zur mutant strain SPP12 (zur::Gm/pBBR), and the complemented strain (zur::Gm/pZUR) contain either p027troC-lacZ or p027yciC-lacZ. pBBR is a plasmid vector, while pZur contains a functional zur gene. Cells were grown in LB for 4 h and then were left untreated (UN) or treated with 1 mM EDTA for 1 h. The results are the means and SD of triplicate samples.
To assess the promoter activity of troC and yciC, the DNA fragments upstream of the translational start codon, predicted −10 and −35 sequences, and a potential Zur-binding site for the troC promoter or for the yciC promoter were fused to a promoterless lacZ reporter gene (transcriptional fusions) (plasmids p027troC-lacZ and p027yciC-lacZ, respectively) (Fig. 1D), and β-Gal activity was measured. In wild-type NTL4 (the WT), β-Gal activities from both troC-lacZ and yciC-lacZ fusions were increased when cells were treated with EDTA (WT/pBBR) (Fig. 1E). β-Gal in the zur mutant strain, SPP12, was constitutively expressed at high levels under all the tested conditions (zur::Gm/pBBR) (Fig. 1E); however, expression of the zur gene from a multicopy plasmid, pZUR, could suppress this phenotype (zur::Gm/pZUR) (Fig. 1E).
The A. tumefaciens troCBA operon and yciC are inducible with zinc depletion.
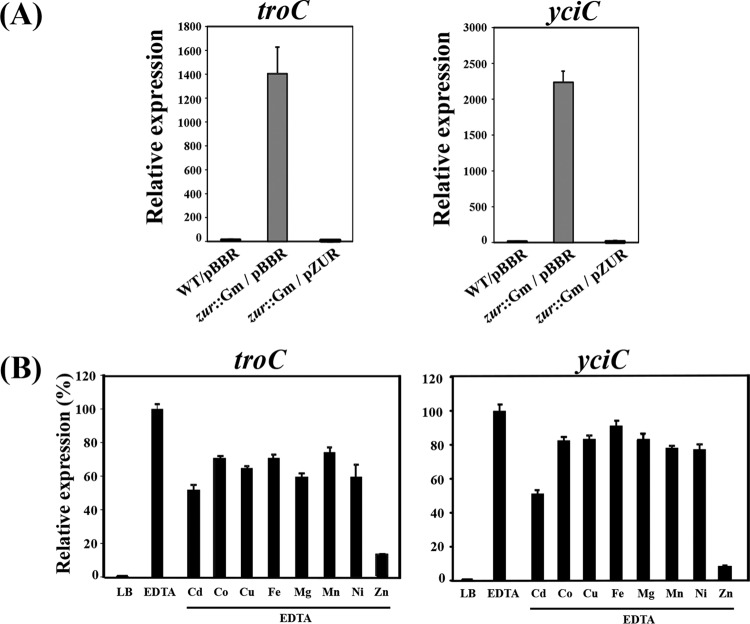
Expression of the troC and yciC genes was determined using qRT-PCR. As expected for genes involved in metal acquisition, expression of the troC and yciC genes in the WT was inducible (∼103-fold) by the metal chelator EDTA compared to untreated cells. Both genes were constitutively expressed at high levels in the zur mutant but were suppressed when complemented with pZUR (Fig. 2A). These results further supported the view that A. tumefaciens Zur negatively regulates troCBA and yciC. To investigate the metal-specific responses of troC and yciC, repression of the EDTA-induced expression by various metals was determined. In the wild type, Zn2+ was the most potent metal ion that could repress the EDTA-induced expression of troC and yciC (Fig. 2B) compared to other metals, including cadmium, cobalt, copper, iron, magnesium, manganese, and nickel. These results demonstrated that the A. tumefaciens troCBA operon and yciC are inducible, specifically by zinc depletion, which supports the view that A. tumefaciens TroCBA and YciC are involved in zinc acquisition.
FIG 2.
(A) Expression analysis of troC and yciC using qRT-PCR. (A) RNA samples were isolated from log-phase cells of the wild type (WT/pBBR), the zur mutant (zur::Gm/pBBR), and the complemented strain (zur::Gm/pZUR) grown in LB medium. pBBR is the plasmid vector. pZUR is the plasmid containing a functional zur gene. The expression of the target genes was normalized to that of 16S rRNA, and the fold changes in gene expression in zur::Gm/pBBR and zur::Gm/pZUR are relative to those in WT/pBBR (regarded as 1). The experiment was performed in biological triplicate, and the error bars indicate the standard deviations. (B) Induction of troC and yciC is specific to zinc limitation. Shown is qRT-PCR analysis of troC and yciC expression. Wild-type NTL4 cells were grown in LB medium and under metal-limiting conditions (LB plus 1 mM EDTA). Metals (CdCl2, CoCl2, CuSO4, FeCl3, MnCl2, NiCl2, and ZnCl2) were supplemented at a final concentration of 0.45 mM. The expression levels are presented as percentages and are relative to those in cells grown in LB plus 1 mM EDTA (100%).
The troC mutant is hypersensitive to EDTA treatment, and inactivation of yciC further increases the sensitivity; however, the TroCBA transporter can function independently of YciC.
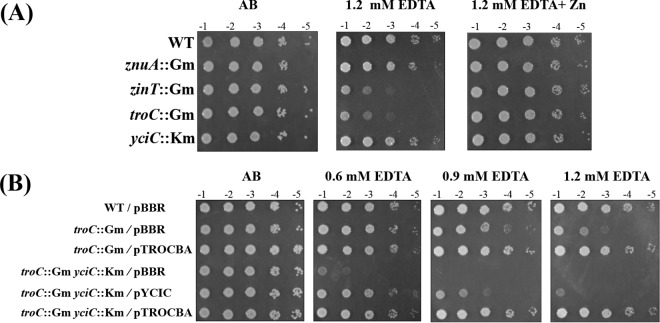
A. tumefaciens mutant strains were constructed to assess the physiological functions of troC (TC142) and yciC (YC154). Growth was determined under metal limitation in the presence of EDTA (1, 1.1, 1.2, 1.3, 1.4, and 1.5 mM [see Fig. S2 in the supplemental material]). Inactivation of troC, but not yciC, caused cells to become hypersensitive to EDTA. Consistent with a previous report (25), the znuA mutant (PS132) showed no phenotype, while the zinT mutant (PC135) was hypersensitive to EDTA (Fig. 3A). Similar to the zinT mutant, the troC mutant was ∼102-fold more sensitive to 1.2 mM EDTA than the WT (Fig. 3A) and could be fully rescued by the addition of 50 μM ZnCl2 (Fig. 3A, 1.2 mM EDTA plus Zn) but not by other metals (see Fig. S3A in the supplemental material).
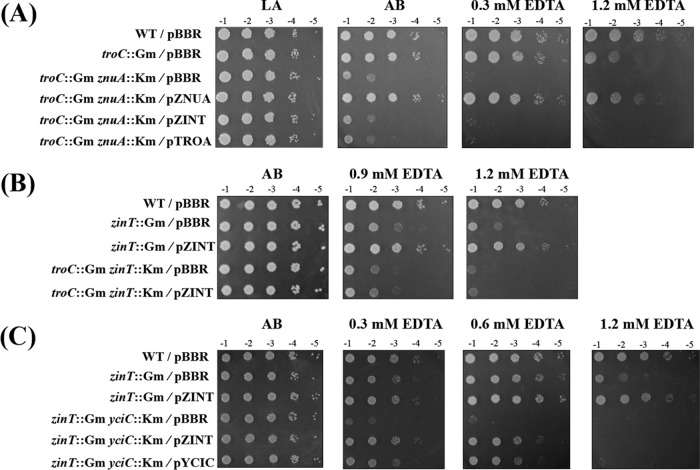
FIG 3.
Effects of a single mutation (A) and double mutations (B) in the troC and yciC genes on the sensitivity of A. tumefaciens to EDTA. (A) The strains are wild-type NTL4 (WT), PS132 (znuA::Gm), PC135 (zinT::Gm), TC142 (troC::Gm), and YC154 (yciC::Km). (B) The WT, TC142 (troC::Gm), and TCYC15 (troC::Gm yciC::Km) strains carry the plasmid vector (pBBR). The mutant strains were complemented with functional troCBA or yciC from the multicopy plasmids pTROCBA and pYciC, respectively. Cells were adjusted, serially diluted, and spotted onto plates containing AB and AB plus EDTA (0.6, 0.9, and 1.2 mM) with or without 50 μM ZnCl2. Tenfold serial dilutions are indicated. The plates were incubated at 28°C for 48 h.
The EDTA-sensitive phenotype of the troC mutant (TC142) could not be reversed by the plasmid pTROC (see Fig. S3B in the supplemental material), suggesting that the troC gene knockout by insertional inactivation used to generate the TC142 strain may have a polar effect on other downstream genes in the troCBA operon. This notion was supported by the observation that the plasmid pTROCBA could fully restore the growth of the troC mutant (Fig. 3B; see Fig. S3B in the supplemental material).
Although the yciC mutant strain showed no phenotype (Fig. 3A; see Fig. S2 and S4 in the supplemental material), the inactivation of yciC further increased the sensitivity of the troC mutant to EDTA (Fig. 3B). Growth of the troC mutant strain TC142 (troC::Gm/pBBR) on the AB plate containing 0.6 mM EDTA was similar to that of the WT (WT/pBBR), while the TCYC15 strain lacking both troC and yciC (troC::Gm yciC::Km/pBBR) was ∼103-fold more sensitive to 0.6 mM EDTA than the WT (Fig. 3B). Expressing the functional yciC gene from the plasmid pYCIC could restore the growth defect of TCYC15 (troC and yciC mutations) to levels similar to that of TC142 (troC mutation) and the WT at 0.6 mM EDTA, but not at higher levels of EDTA (0.9 and 1.2 mM) (troC::Gm yciC::Km/pYCIC) (Fig. 3B). In contrast, at 0.6, 0.9, and 1.2 mM EDTA, the EDTA-hypersensitive phenotype of TCYC15 could be fully reversed to WT by complementation with pTROCBA (troC::Gm yciC::Km/pTROCBA) (Fig. 3B), demonstrating that A. tumefaciens requires the TroCBA transporter to survive under severe metal-limiting conditions and that TroCBA can function even in the absence of YciC.
The important role of ZnuA is revealed when TroCBA is disrupted.
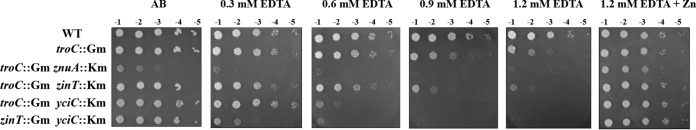
Inactivation of either znuA or znuAB had no detectable effect on the growth of A. tumefaciens on the AB plate containing 1.4 mM EDTA compared to the WT (see Fig. S2 in the supplemental material) (25). However, disruption of znuA in combination with troC (TCZA15) led to a growth defect of A. tumefaciens in a minimal AB medium and hypersensitivity to EDTA (troC::Gm znuA::Km) (Fig. 4). The growth defect of TCZA15 could be rescued by zinc supplementation (1.2 mM EDTA plus Zn) (Fig. 4). Furthermore, the EDTA sensitivity of TCZA15 (troC and znuA mutations) could be reversed to that of TC142 (troC mutation) by complementation with the plasmid pZNUA (troC::Gm znuA::Km/pZNUA) (Fig. 5A), but not by the plasmid pZINT or pTROA. Although TCZA15 showed a growth defect on the AB plate (a minimal medium), it could grow on the LA plate (a rich medium) similarly to the WT (troC::Gm znuA::Km/pBBR) (Fig. 5A). These results demonstrated the role of A. tumefaciens ZnuA (a periplasmic zinc-binding protein) in zinc acquisition under zinc limitation and that A. tumefaciens ZnuA function could not be substituted for by other periplasmic zinc-binding proteins, such as ZinT and TroA.
FIG 4.
EDTA-sensitive phenotypes of strains containing double mutations of genes encoding zinc uptake proteins and zinc chaperones. The EDTA sensitivities of TCZA15 (troC::Gm znuA::Km), TCZT14 (troC::Gm zinT::Km), TCYC15 (troC::Gm yciC::Km), and ZTYC15 (zinT::Gm yciC::Km) were compared to that of wild-type NTL4 (WT) and TC142 (troC::Gm). Sensitivity to EDTA on plates containing AB and AB plus EDTA (0.3, 0.6, 0.9, and 1.2 mM) with or without 50 μM ZnCl2 was assessed as described for Fig. 3.
FIG 5.
(A) ZnuA under metal limitation conditions could not be replaced by the periplasmic zinc-binding proteins ZinT and TroA. The TCZA15 (troC::Gm znuA::Km) strain was complemented with functional znuA, zinT, or troA from the plasmids pZNUA, pZINT, and pTROA, respectively. The cells were tested on plates containing LA (rich medium) and AB (minimal medium). Sensitivity to EDTA on AB plates containing EDTA (0.3, 0.6, 0.9, and 1.2 mM) was assessed as described for Fig. 3. (B) ZinT could not function when TroCBA was inactivated. The PC135 (zinT::Gm) and TCZT14 (troC::Gm zinT::Km) strains were complemented with functional zinT from the plasmid pZINT. (C) Important role of zinc chaperones ZinT and YciC. The ZTYC15 (zinT::Gm yciC::Km) strain was complemented with functional zinT or yciC from the plasmids pZINT and pYCIC, respectively.
ZinT may interact with TroCBA.
It has been reported previously that A. tumefaciens ZinT functions independently of ZnuABC (25). The inactivation of zinT caused a reduction in the total zinc content and hypersensitivity to EDTA (25). To test the possibility of interaction between A. tumefaciens ZinT (a periplasmic protein) and TroCBA in zinc transport, a strain (TCZT14) containing mutations in troC and zinT was generated, and its sensitivity to EDTA was determined. The TC142 (troC mutation) and TCZT14 (troC and zinT mutations) strains were ∼102-fold and ∼103-fold, respectively, more sensitive to 1.2 mM EDTA than the WT (Fig. 4). Moreover, the EDTA-hypersensitive phenotype of TCZT14 (troC and zinT mutations) could not be reversed to that of TC142 (troC mutation) by complementation with the plasmid pZINT (Fig. 5B), but the EDTA-hypersensitive phenotype of PC135 (zinT mutation) could be fully restored by pZINT (Fig. 5B). The results suggested that TroCBA is required for the function of A. tumefaciens ZinT.
Loss of the zinc chaperones ZinT and YciC causes a severe EDTA-mediated growth defect.
Double mutations in zinT and yciC (ZTYC15) led to hypersensitivity to EDTA, and this EDTA-dependent growth defect could be detected at 0.3 mM EDTA (∼103-fold more sensitive than the wild type) (Fig. 4). At 0.3 and 0.6 mM EDTA, the growth defect of ZTYC15 could be fully restored to PC135 (zinT mutation) by complementation with the plasmid pZINT or pYCIC (Fig. 5C). At 1.2 mM EDTA, complementation with either pZINT or pYCIC did not rescue ZTYC15 (Fig. 5C), but addition of zinc did (1.2 mM EDTA plus Zn) (Fig. 4). These results demonstrated that A. tumefaciens requires both zinc chaperones, ZinT and YciC, to cope with severe zinc-limiting conditions.
YciCAt is a cytoplasmic protein.
B. subtilis YciC (YciCBs) is a membrane protein (27). Unlike YciCBs, A. tumefaciens YciC (YciCAt) is likely to reside in the cytoplasm. The hydropathy of YciCAt was determined using the Kyte-Doolittle hydropathy plot (54). The hydropathy plot and other programs, including HMMTOP (55) and Phobius (56), predicted that there would be no transmembrane segment in the YciCAt protein. To determine the localization of YciCAt, the entire 400 amino acids of YciCAt was fused to PhoA (alkaline phosphatase) and LacZ (β-galactosidase) to generate YciCAt-PhoA and YciCAt-LacZ fusions using a previously described protocol (44). The alkaline phosphatase and β-galactosidase activities of the hybrid proteins could be used as indicators for the periplasmic and cytoplasmic locations, respectively, of the target protein fusion sites (57, 58). The YciCAt-PhoA fusion displayed low levels of alkaline phosphatase activity (∼0.22 U), while the YciCAt-LacZ fusion exhibited high levels of β-galactosidase activity (∼240 U) compared to the negative-control vectors, p′PhoA (∼0.04 U) and pPR9TT (∼8 U), which showed low activity of alkaline phosphatase and β-galactosidase, respectively. The results suggest that the YciCAt protein is a cytoplasmic protein.
YciCAt may possibly catalyze GTP hydrolysis to drive the TroCBAAt transporter. However, YciCAt is not essential for the TroCBAAt transporter (Fig. 3B). Another possible function of YciCAt is as a zinc chaperone that may transfer zinc ions to the zinc sensor ZurAt. To test this idea, expression of the znuA, zinT, and troC genes in the yciC::Km strain was determined by qRT-PCR. Similarly to the WT, EDTA induction and zinc repression of znuA, zinT, and troC were observed in the yciC::Km strain (data not shown). Therefore, YciCAt is unlikely to function in zinc loading to ZurAt for mediating repression of the ZurAt regulon.
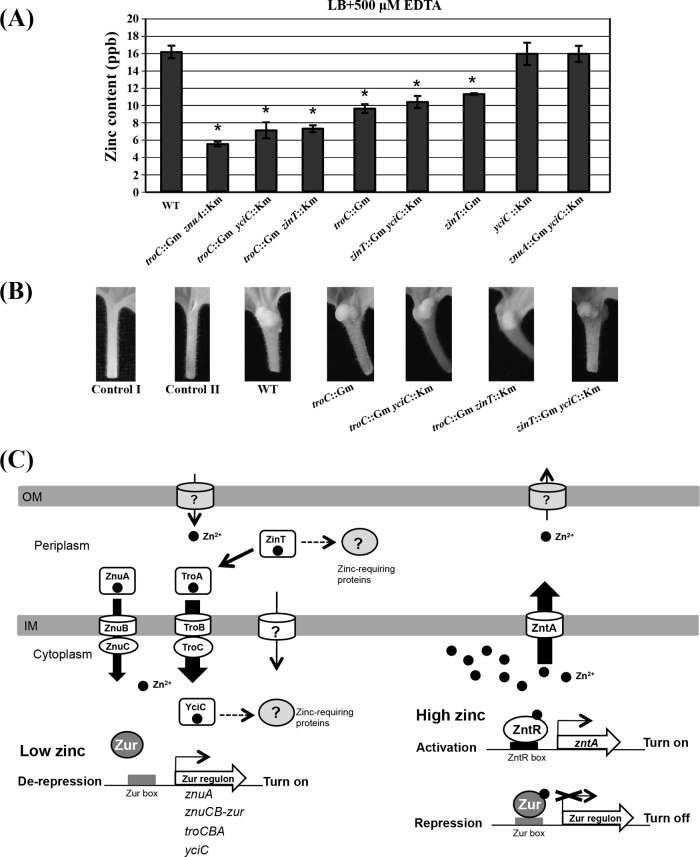
Decreased zinc levels in the EDTA-hypersensitive mutants.
Total zinc content in the mutant strains compared to the WT was determined using ICP-MS (Fig. 6A). Because the troC znuA double-mutant strain has a growth defect in the minimal AB medium (Fig. 4), ICP-MS was performed using cells grown in LB (rich medium) for 24 h; however, the zinc levels in the mutant strains were not dramatically different from those in the WT (data not shown). When cells were grown in LB plus 0.5 mM EDTA for 24 h, all the EDTA-hypersensitive mutants showed an obvious reduction in zinc content compared to the WT (Fig. 6A). It should be noted that 0.5 mM EDTA in LB did not inhibit the growth of the WT and all the tested mutant strains (data not shown). The lowest levels of intracellular zinc were detected in the troC::Gm znuA::Km strain (troC::Gm yciC::Km and troC::Gm zinT::Km strains < troC::Gm and zinT::Gm yciC::Km strains < zinT::Gm strain) (Fig. 6A). The WT and the yciC::Km and znuA::Gm yciC::Km mutant strains showed similar levels of EDTA sensitivity (see Fig. S4 in the supplemental material) and intracellular zinc levels (Fig. 6A).
FIG 6.
(A) Intracellular zinc content was determined using ICP-MS. The WT and the mutant strains TCZA15 (troC::Gm znuA::Km), TCYC15 (troC::Gm yciC::Km), TCZT15 (troC::Gm zinT::Km), TC142 (troC::Gm), ZTYC15 (zinT::Gm yciC::Km), PC135 (zinT::Gm), YC154 (yciC::Km), and ZAYC15 (znuA::Gm yciC::Km) were grown in LB plus 0.5 mM EDTA for 24 h. The results are shown as the means of samples in triplicate, and the error bars indicate the standard deviations. The bars marked with asterisks are significantly different from the WT (P < 0.05 in an unpaired Student's t test). (B) Virulence assay. Shown is tumor formation on the petioles of N. benthamiana plants infected with the WT and the mutant strains TC142 (troC::Gm), TCYC15 (troC::Gm yciC::Km), TCZT15 (troC::Gm zinT::Km), and ZTYC15 (zinT::Gm yciC::Km) grown in IB 5.5 medium containing 300 μM AS. Control I, without inoculation; control II, inoculation of the wounded petiole with IB 5.5 plus 300 μM AS. (C) Proposed model of zinc homeostasis. A. tumefaciens contains two zinc sensors, Zur and ZntR, which control zinc homeostasis. Zur is the transcriptional repressor of zinc acquisition genes, including znuA, znuBC, zinT, troCBA, and yciC. ZntR is a transcriptional activator of a zinc efflux gene, zntA. Under low-zinc conditions, Zur is in the apo form and dissociates from DNA to trigger expression of the Zur regulon. The TroABC system is the major zinc uptake system (thick solid arrow) compared to ZnuABC. ZinT functions in association with TroCBA. ZinT and YciC may function as zinc chaperones that supply Zn2+ (black dots) to zinc-requiring proteins in the periplasm and cytoplasm, respectively. When the intracellular zinc levels are high, the complexes ZntR-Zn2+ and Zur-Zn2+ are formed, which in turn activates zinc efflux by ZntA and represses zinc uptake systems, respectively. The outer membrane (OM) channels for zinc import and zinc extrusion have not been identified. Zinc ions may also be transported into the cytoplasm via unknown inner membrane (IM) transporters. ?, unknown; dashed arrows, possible mechanisms.
Virulence assay.
N. benthamiana plants were infected with the WT and the mutant strains carrying pCMA1 (a tumor-inducing plasmid). The mutant strains TC142 (troC mutation), TCYC15 (troC and yciC mutations), TCZT14 (troC and zinT mutations), and ZTYC15 (zinT and yciC mutations) were tested. All A. tumefaciens strains grown in LB for 24 h caused similar tumor formation on infected plants (data not shown). Next, the A. tumefaciens strains were grown under metal limitation in LB plus 0.5 mM EDTA for 24 h and used to infect plants. However, TC142, TCYC15, TCZT14, and ZTYC15 all caused tumors on the infected plants and were similar to the WT (Fig. 6B). Unfortunately, several attempts were made, but we could not select the TCZA15 (troC and znuA mutations) strain carrying pCMA1; thus, we were unable to determine the virulence of TCZA15.
DISCUSSION
The high-affinity zinc importer ZnuABC is found widely in many bacteria. Rarely, some bacteria have a second high-affinity zinc uptake system in addition to ZnuABC, such as T. pallidum TroABCD (TroABCDTp) (29, 30) and Haemophilus influenzae ZevAB (59). Listeria monocytogenes contains two zinc permease systems, ZurAM and ZinABC, which are regulated by Zur (60). The presence of two high-affinity zinc uptake systems helps these bacteria to survive in diverse zinc-limiting environments and during infection. In addition to ZnuABC, A. tumefaciens has a cluster of genes (Atu3178, Atu3179, and Atu3180) encoding an ABC-type transporter. The Atu3178 gene encodes a protein belonging to the TroA superfamily that has amino acid identity to TroATp (28%), ZinA (26%), and A. tumefaciens ZnuA (25%). Therefore, we named Atu3178, Atu3179, and Atu3180 troA, troB, and troC, respectively. In other bacteria, tro operons are involved in metal uptake and can transport either zinc or manganese and possibly iron (29, 33–36). The A. tumefaciens troCBA operon (troCBAAt) was shown here to respond specifically to zinc limitation.
To survive under a wide range of zinc-deficient conditions, A. tumefaciens requires the cooperation of Zur-regulated zinc acquisition systems, including two ABC-type zinc importers, TroCBAAt and ZnuABCAt, and two zinc chaperones, ZinTAt and YciCAt. Zinc uptake in A. tumefaciens differs from the systems in many other bacteria in that TroCBAAt is the major zinc importer while ZnuABCAt plays a lesser role. ZnuABCAt is very important for supporting growth under zinc limitation in the absence of TroCBAAt. This indicates that ZnuABCAt may function as a backup system for high-affinity zinc uptake. It is also possible that the two transporters function optimally under different conditions, such as different pHs. Consequently, TroCBAAt may play a dominant role in zinc uptake under laboratory test conditions and ZnuABCAt may play a more crucial role under other, as yet unidentified conditions. The severe growth defect of the troC::Gm znuA::Km strain could be rescued by supplementation with zinc, suggesting that zinc can be taken up through additional low-affinity zinc transporters.
ZinT proteins in Escherichia coli O157 (20) and S. enterica (22, 23) (ZinTEc and ZinTSe, respectively) are accessory components of the ZnuABC transporters that enhance the ability of cells to recruit zinc under severe zinc shortage. In contrast to ZinTEc and ZinTSe, ZinTAt plays a critical role for survival under zinc depletion and can function in the absence of ZnuABCAt (25). It was found that ZinTAt may interact with TroCBAAt. It is likely that ZinTAt is an additional component of the TroCBAAt transporter, and it may help increase the rate of zinc delivery to TroCBAAt when A. tumefaciens faces severe zinc deprivation. ZinTAt may also supply zinc ions to other periplasmic zinc-requiring proteins.
The members of the COG0523 subfamily of G3E-GTPases have been reported to function as metal insertases by using GTP hydrolysis to drive the incorporation of a metal cofactor into the catalytic site of the target enzyme and/or as metal chaperones to allocate a metal cofactor to the target zinc-requiring proteins (26). B. subtilis YciC (YciCBs) belongs to a subgroup, Zur-regulated COG0523 proteins. YciCBs is a membrane protein, and it was proposed to be a component of a low-affinity zinc transporter and possibly a zinc chaperone (27, 28). Unlike YciCBs, YciCAt is a cytoplasmic protein. Inactivation of yciCAt in combination with the mutation of a gene encoding the major zinc importer, troCAt, or the periplasmic zinc chaperone, zinTAt, caused a growth defect on LA plates containing EDTA. When A. tumefaciens encounters conditions of severe zinc shortage, even if the high-affinity zinc uptake TroCBAAt and ZnuABCAt systems are still functioning, it may need ZinTAt and YciCAt to effectively shuttle zinc ions to essential zinc-dependent proteins whose functions are required to cope with low-zinc stress.
Zinc uptake systems are known to be important for virulence in many bacteria (59, 60). However, in virulence assays using N. benthamiana as the host plant, growth either in LB (a zinc-replete condition) or in LB plus 0.5 mM EDTA (a zinc-depleted condition) had no effect on the virulence of A. tumefaciens mutant strains lacking zinc uptake genes compared to the wild type. Zinc availability at the infection site may differ between different types of plants, and this may alter the sensitivity of the assay in different host plants.
A model of zinc homeostasis in A. tumefaciens is proposed in Fig. 6C. Intracellular zinc levels are sensed by two transcriptional regulators, Zur and ZntR (25, 61). Zur is a repressor in the Fur family, whereas ZntR is an activator belonging to the MerR family. Under low-zinc conditions, Zur is in the apo form, which in turn triggers the expression of zinc acquisition genes, including znuABC, troCBA, zinT, and yciC. The TroCBA transporter plays a dominant role in zinc uptake compared to the ZnuABC system. ZinT may help enhance the efficiency of zinc recruitment to the TroCBA transporter. When TroCBA is impaired, the ZnuABC system plays an essential role in zinc uptake to support bacterial growth. It is likely that zinc can also be transported into the cytoplasm by an unidentified low-affinity zinc importer(s). The ZinT and YciC proteins may act as metallochaperones to allocate zinc to their target zinc-dependent proteins in the periplasm and cytoplasm, respectively. When intracellular zinc is overloaded, the complexes of Zn2+ with the zinc sensors, Zur-Zn2+ and ZntR-Zn2+, are formed. The Zur-Zn2+ complex represses the zinc uptake genes, whereas the ZntR-Zn2+ complex activates the zinc efflux gene, zntA. Consequently, the high-affinity zinc uptake systems, TroCBA and ZnuABC, are shut down, and excess zinc can be pumped out of the cytoplasm by a P1B-type ATPase, ZntA. However, the outer-membrane channels for zinc import and zinc extrusion in A. tumefaciens remain to be identified.
A. tumefaciens was shown here to operate a novel pathway for zinc uptake in which ZinTAt is part of a high-affinity zinc importer, TroCBAAt. Whether ZinTAt interacts with TroAAt or directly interacts with the inner-membrane permease, TroBAt, during the process of zinc transport awaits further study. High-affinity zinc uptake systems, such as TroCBAAt and ZnuABCAt, are necessary for A. tumefaciens to survive under low-zinc stress, but zinc allocation to zinc-dependent proteins facilitated by the zinc chaperones ZinTAt and YciCAt is also essential. Nevertheless, the exact molecular mechanisms of ZinTAt and YciCAt, as well as their target proteins, have yet to be elucidated.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. K. Farrand for the plasmid pCMA1. We also thank P. Srifah Huehne and K. Bhinija for technical assistance with the virulence assay.
This work was supported by the Chulabhorn Research Institute and Thailand Research Fund grant RSA5880010.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00299-16.
REFERENCES
- 1.Vallee BL, Falchuk KH. 1993. The biochemical basis of zinc physiology. Physiol Rev 73:79–118. [DOI] [PubMed] [Google Scholar]
- 2.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. 2008. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem 13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 3.Kasahara M, Anraku Y. 1974. Succinate- and NADH oxidase systems of Escherichia coli membrane vesicles. Mechanism of selective inhibition of the systems by zinc ions. J Biochem 76:967–976. [PubMed] [Google Scholar]
- 4.Singh AP, Bragg PD. 1974. Inhibition of energization of Salmonella typhimurium membrane by zinc ions. FEBS Lett 40:200–202. doi: 10.1016/0014-5793(74)80927-0. [DOI] [PubMed] [Google Scholar]
- 5.Beard SJ, Hughes MN, Poole RK. 1995. Inhibition of the cytochrome bd-terminated NADH oxidase system in Escherichia coli K-12 by divalent metal cations. FEMS Microbiol Lett 131:205–210. doi: 10.1111/j.1574-6968.1995.tb07778.x. [DOI] [PubMed] [Google Scholar]
- 6.Aagaard A, Brzezinski P. 2001. Zinc ions inhibit oxidation of cytochrome c oxidase by oxygen. FEBS Lett 494:157–160. doi: 10.1016/S0014-5793(01)02299-2. [DOI] [PubMed] [Google Scholar]
- 7.Xu FF, Imlay JA. 2012. Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl Environ Microbiol 78:3614–3621. doi: 10.1128/AEM.07368-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blencowe DK, Morby AP. 2003. Zn(II) metabolism in prokaryotes. FEMS Microbiol Rev 27:291–311. doi: 10.1016/S0168-6445(03)00041-X. [DOI] [PubMed] [Google Scholar]
- 9.Nies DH. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 10.Hantke K. 2005. Bacterial zinc uptake and regulators. Curr Opin Microbiol 8:196–202. doi: 10.1016/j.mib.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Nies DH. 2007. Biochemistry. How cells control zinc homeostasis. Science 317:1695–1696. [DOI] [PubMed] [Google Scholar]
- 12.Shin JH, Oh SY, Kim SJ, Roe JH. 2007. The zinc-responsive regulator Zur controls a zinc uptake system and some ribosomal proteins in Streptomyces coelicolor A3(2). J Bacteriol 189:4070–4077. doi: 10.1128/JB.01851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabriel SE, Helmann JD. 2009. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J Bacteriol 191:6116–6122. doi: 10.1128/JB.00802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panina EM, Mironov AA, Gelfand MS. 2003. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc Natl Acad Sci U S A 100:9912–9917. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patzer SI, Hantke K. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol 28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 16.Patzer SI, Hantke K. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J Biol Chem 275:24321–24332. doi: 10.1074/jbc.M001775200. [DOI] [PubMed] [Google Scholar]
- 17.Outten CE, O'Halloran TV. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 18.Kershaw CJ, Brown NL, Hobman JL. 2007. Zinc dependence of zinT (yodA) mutants and binding of zinc, cadmium and mercury by ZinT. Biochem Biophys Res Commun 364:66–71. doi: 10.1016/j.bbrc.2007.09.094. [DOI] [PubMed] [Google Scholar]
- 19.Graham AI, Hunt S, Stokes SL, Bramall N, Bunch J, Cox AG, McLeod CW, Poole RK. 2009. Severe zinc depletion of Escherichia coli: roles for high affinity zinc binding by ZinT, zinc transport and zinc-independent proteins. J Biol Chem 284:18377–18389. doi: 10.1074/jbc.M109.001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabbianelli R, Scotti R, Ammendola S, Petrarca P, Nicolini L, Battistoni A. 2011. Role of ZnuABC and ZinT in Escherichia coli O157:H7 zinc acquisition and interaction with epithelial cells. BMC Microbiol 11:36. doi: 10.1186/1471-2180-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim J, Lee KM, Kim SH, Kim Y, Kim SH, Park W, Park S. 2011. YkgM and ZinT proteins are required for maintaining intracellular zinc concentration and producing curli in enterohemorrhagic Escherichia coli (EHEC) O157:H7 under zinc deficient conditions. Int J Food Microbiol 149:159–170. doi: 10.1016/j.ijfoodmicro.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 22.Petrarca P, Ammendola S, Pasquali P, Battistoni A. 2010. The Zur-regulated ZinT protein is an auxiliary component of the high-affinity ZnuABC zinc transporter that facilitates metal recruitment during severe zinc shortage. J Bacteriol 192:1553–1564. doi: 10.1128/JB.01310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ilari A, Alaleona F, Tria G, Petrarca P, Battistoni A, Zamparelli C, Verzili D, Falconi M, Chiancone E. 2014. The Salmonella enterica ZinT structure, zinc affinity and interaction with the high-affinity uptake protein ZnuA provide insight into the management of periplasmic zinc. Biochim Biophys Acta 1840:535–544. doi: 10.1016/j.bbagen.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Ziemienowicz A. 2001. Odyssey of Agrobacterium T-DNA. Acta Biochim Pol 48:623–635. [PubMed] [Google Scholar]
- 25.Bhubhanil S, Sittipo P, Chaoprasid P, Nookabkaew S, Sukchawalit R, Mongkolsuk S. 2014. Control of zinc homeostasis in Agrobacterium tumefaciens via zur and the zinc uptake genes znuABC and zinT. Microbiology 160:2452–2463. doi: 10.1099/mic.0.082446-0. [DOI] [PubMed] [Google Scholar]
- 26.Haas CE, Rodionov DA, Kropat J, Malasarn D, Merchant SS, de Crécy-Lagard V. 2009. A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10:470. doi: 10.1186/1471-2164-10-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaballa A, Helmann JD. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol 180:5815–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabriel SE, Miyagi F, Gaballa A, Helmann JD. 2008. Regulation of the Bacillus subtilis yciC gene and insights into the DNA-binding specificity of the zinc-sensing metalloregulator Zur. J Bacteriol 190:3482–3488. doi: 10.1128/JB.01978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hazlett KR, Rusnak F, Kehres DG, Bearden SW, La Vake CJ, La Vake ME, Maguire ME, Perry RD, Radolf JD. 2003. The Treponema pallidum tro operon encodes a multiple metal transporter, a zinc-dependent transcriptional repressor, and a semi-autonomously expressed phosphoglycerate mutase. J Biol Chem 278:20687–20694. doi: 10.1074/jbc.M300781200. [DOI] [PubMed] [Google Scholar]
- 30.Desrosiers DC, Sun YC, Zaidi AA, Eggers CH, Cox DL, Radolf JD. 2007. The general transition metal (Tro) and Zn2+ (Znu) transporters in Treponema pallidum: analysis of metal specificities and expression profiles. Mol Microbiol 65:137–152. doi: 10.1111/j.1365-2958.2007.05771.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Li W, Wei Y, Jiang Y, Tan X. 2013. Efficient preparation and metal specificity of the regulatory protein TroR from the human pathogen Treponema pallidum. Metallomics 5:1448–1457. doi: 10.1039/c3mt00163f. [DOI] [PubMed] [Google Scholar]
- 32.Berntsson RP, Smits SH, Schmitt L, Slotboom DJ, Poolman B. 2010. A structural classification of substrate-binding proteins. FEBS Lett 584:2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 33.Brett PJ, Burtnick MN, Fenno JC, Gherardini FC. 2008. Treponema denticola TroR is a manganese- and iron-dependent transcriptional repressor. Mol Microbiol 70:396–409. doi: 10.1111/j.1365-2958.2008.06418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith KF, Bibb LA, Schmitt MP, Oram DM. 2009. Regulation and activity of a zinc uptake regulator, Zur, in Corynebacterium diphtheriae. J Bacteriol 191:1595–1603. doi: 10.1128/JB.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng B, Zhang Q, Gao J, Han H, Li M, Zhang J, Qi J, Yan J, Gao GF. 2011. Insight into the interaction of metal ions with TroA from Streptococcus suis. PLoS One 6:e19510. doi: 10.1371/journal.pone.0019510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wichgers Schreur PJ, Rebel JM, Smits MA, van Putten JP, Smith HE. 2011. TroA of Streptococcus suis is required for manganese acquisition and full virulence. J Bacteriol 193:5073–5080. doi: 10.1128/JB.05305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo ZQ, Clemente TE, Farrand SK. 2001. Construction of a derivative of Agrobacterium tumefaciens C58 that does not mutate to tetracycline resistance. Mol Plant Microbe Interact 14:98–103. doi: 10.1094/MPMI.2001.14.1.98. [DOI] [PubMed] [Google Scholar]
- 38.Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metcalf WW, Jiang W, Daniels LL, Kim SK, Haldimann A, Wanner BL. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 40.Alexeyev MF. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques 26:824–826, 828. [DOI] [PubMed] [Google Scholar]
- 41.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 42.DeFeyter R, Kado CI, Gabriel DW. 1990. Small, stable shuttle vectors for use in Xanthomonas. Gene 88:65–72. doi: 10.1016/0378-1119(90)90060-5. [DOI] [PubMed] [Google Scholar]
- 43.Santos PM, Di Bartolo I, Blatny JM, Zennar E, Valla S. 2001. New broad-host-range promoter probe vectors based on the plasmid RK2 replicon. FEMS Microbiol Lett 195:91–96. doi: 10.1111/j.1574-6968.2001.tb10503.x. [DOI] [PubMed] [Google Scholar]
- 44.Bhubhanil S, Chamsing J, Sittipo P, Chaoprasid P, Sukchawalit R, Mongkolsuk S. 2014. Roles of Agrobacterium tumefaciens membrane-bound ferritin (MbfA) in iron transport and resistance to iron under acidic conditions. Microbiology 160:863–871. doi: 10.1099/mic.0.076802-0. [DOI] [PubMed] [Google Scholar]
- 45.Hwang I, Cook DM, Farrand SK. 1995. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J Bacteriol 177:449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 47.Cangelosi GA, Best EA, Martinetti G, Nester EW. 1991. Genetic analysis of Agrobacterium. Methods Enzymol 204:384–397. doi: 10.1016/0076-6879(91)04020-O. [DOI] [PubMed] [Google Scholar]
- 48.Kitphati W, Ngok-Ngam P, Suwanmaneerat S, Sukchawalit R, Mongkolsuk S. 2007. Agrobacterium tumefaciens fur has important physiological roles in iron and manganese homeostasis, the oxidative stress response, and full virulence. Appl Environ Microbiol 73:4760–4768. doi: 10.1128/AEM.00531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ngok-Ngam P, Ruangkiattikul N, Mahavihakanont A, Virgem SS, Sukchawalit R, Mongkolsuk S. 2009. Roles of Agrobacterium tumefaciens RirA in iron regulation, oxidative stress response and virulence. J Bacteriol 191:2083–2090. doi: 10.1128/JB.01380-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhubhanil S, Niamyim P, Sukchawalit R, Mongkolsuk S. 2014. Cysteine desulphurase-encoding gene sufS2 is required for the repressor function of RirA and oxidative resistance in Agrobacterium tumefaciens. Microbiology 160:79–90. doi: 10.1099/mic.0.068643-0. [DOI] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 53.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF Jr, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee D Sr, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, Nester EW. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 54.Kyte J, Doolittle R. 1982. A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 55.Tusnady GE, Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 56.Käll L, Krogh A, Sonnhammer EL. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 57.Manoil C, Beckwith J. 1986. A genetic approach to analyzing membrane protein topology. Science 233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 58.Manoil C. 1990. Analysis of protein localization by use of gene fusions with complementary properties. J Bacteriol 172:1035–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosadini CV, Gawronski JD, Raimunda D, Argüello JM, Akerley BJ. 2011. A novel zinc binding system, ZevAB, is critical for survival of nontypeable Haemophilus influenzae in a murine lung infection model. Infect Immun 79:3366–3376. doi: 10.1128/IAI.05135-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corbett D, Wang J, Schuler S, Lopez-Castejon G, Glenn S, Brough D, Andrew PW, Cavet JS, Roberts IS. 2012. Two zinc uptake systems contribute to the full virulence of Listeria monocytogenes during growth in vitro and in vivo. Infect Immun 80:14–21. doi: 10.1128/IAI.05904-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaoprasid P, Nookabkaew S, Sukchawalit R, Mongkolsuk S. 2015. Roles of Agrobacterium tumefaciens C58 ZntA and ZntB and the transcriptional regulator ZntR in controlling Cd2+/Zn2+/Co2+ resistance and the peroxide stress response. Microbiology 161:1730–1740. doi: 10.1099/mic.0.000135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.