ABSTRACT
The rhizosphere-inhabiting species Agrobacterium fabrum (genomospecies G8 of the Agrobacterium tumefaciens species complex) is known to degrade hydroxycinnamic acids (HCAs), especially ferulic acid and p-coumaric acid, via the novel A. fabrum HCA degradation pathway. Gene expression profiles of A. fabrum strain C58 were investigated in the presence of HCAs, using a C58 whole-genome oligoarray. Both ferulic acid and p-coumaric acid caused variations in the expression of more than 10% of the C58 genes. Genes of the A. fabrum HCA degradation pathway, together with the genes involved in iron acquisition, were among the most highly induced in the presence of HCAs. Two operons coding for the biosynthesis of a particular siderophore, as well as genes of the A. fabrum HCA degradation pathway, have been described as being specific to the species. We demonstrate here their coordinated expression, emphasizing the interdependence between the iron concentration in the growth medium and the rate at which ferulic acid is degraded by cells. The coordinated expression of these functions may be advantageous in HCA-rich but iron-starved environments in which microorganisms have to compete for both iron and carbon sources, such as in plant roots. The present results confirm that there is cooperation between the A. fabrum-specific genes, defining a particular ecological niche.
IMPORTANCE We previously identified seven genomic regions in Agrobacterium fabrum that were specifically present in all of the members of this species only. Here we demonstrated that two of these regions, encoding the hydroxycinnamic acid degradation pathway and the iron acquisition pathway, were regulated in a coordinated manner. The coexpression of these functions may be advantageous in hydroxycinnamic acid-rich but iron-starved environments in which microorganisms have to compete for both iron and carbon sources, such as in plant roots. These data support the view that bacterial genomic species emerged from a bacterial population by acquiring specific functions that allowed them to outcompete their closest relatives. In conclusion, bacterial species could be defined not only as genomic species but also as ecological species.
INTRODUCTION
The definition of bacterial species is still based on the separation of bacteria into cohesive genomic units (i.e., genomic species) (1). Further investigations are needed in order to understand the biological determinants of this separation. Our hypothesis is that genomic species emerged from a bacterial population by acquiring specific functions that allowed them to outcompete their closest relatives, resulting in their respective progenies forming clearly separate groups with high levels of genomic similarity. To test this idea, we looked for genes that are present in all of the members of one particular species but are absent in the members of closely related species (i.e., species-specific genes). We then determined to which particular niche a given species is specifically adapted by following the genes encoding certain functions. We used the model species Agrobacterium fabrum (a species of the Agrobacterium tumefaciens species complex) (2), which is largely present in soil and in plant root systems. Comparative genomics allowed us to identify seven species-specific genomic regions in A. fabrum (3, 4). One A. fabrum-specific genomic region, called SpG8-1b (spanning atu1409 to atu1423), has been found to encode a novel pathway for the degradation of hydroxycinnamic acids (HCAs) (4).
HCAs are often described as inhibiting the growth of numerous bacteria, such as Pectobacterium carotovorum, Xanthomonas campestris pv. pelargonii, Pseudomonas syringae, Staphylococcus aureus, and Escherichia coli (5–8). HCAs may also be involved in bacterial cell-to-cell signaling, since p-coumaric acid has been linked to a new class of quorum-sensing molecules (9) and ferulic acid has been described as a strong chemoattractant for rhizobia or agrobacteria (10, 11), helping bacteria to move toward phenolic-rich root environments (12). HCAs have also been shown to be carbon sources for several soil bacteria, such as Bacillus subtilis, Pseudomonas fluorescens, Pseudomonas putida, and A. fabrum (4, 13–15). In A. fabrum, ferulic acid and p-coumaric acid are catabolized via a unique coenzyme A (CoA)-dependent β-oxidative deacetylation pathway, i.e., the A. fabrum HCA degradation pathway. In this pathway, coenzyme A is added to ferulic acid by a feruloyl-CoA synthase (Atu1416), and the feruloyl-CoA compound is then converted into 4-hydroxy-3-methoxyphenyl-β-hydroxypropionyl (HMPHP)-CoA by the enoyl-CoA hydratase Atu1417. Two original enzymatic reactions then follow. HMPHP-CoA is first converted into 4-hydroxy-3-methoxyphenyl-β-ketopropionoyl (HMPKP)-CoA by Atu1415, a phenylhydroxypropionyl-CoA dehydrogenase. HMPKP-CoA is then converted into vanillic acid by Atu1421, an uncharacterized protein. Finally, protocatechuic acid is produced from vanillic acid by Atu1420 and Atu1418 and is transformed into acetyl-CoA and succinate, which are known to be carbon and energy sources for A. fabrum (16). The conversion of ferulic acid into protocatechuic acid by six essential enzymes is now well characterized, whereas the impact of this degradation on bacterial cell physiology remains unclear. We suspect that HCAs may play a key role in the regulation of a large number of A. fabrum genes.
In this study, we investigated the transcriptional reprogramming and the cell physiology of A. fabrum in the presence of either ferulic acid or p-coumaric acid, using whole-C58-genome oligoarrays. Because transcriptional analyses highlighted the genes of the A. fabrum HCA degradation pathway and the A. fabrum-specific iron acquisition pathway as being among the most significantly upregulated genes, we focused our study on the coordinated regulation of these two regions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The A. fabrum strains used in this study were C58 (CFBP 1903; Collection Française de Bactéries Associées aux Plantes, INRA, Angers, France), C58ΔSpG8-1b (3), and C58ΔSpG8-3 (constructed in our laboratories). Bacteria were grown at 28°C, with shaking (160 rpm), in YPG-rich medium (5 g/liter yeast extract, 5 g/liter peptone, 55 mM glucose [pH 7.2]) or in minimal AT medium (80 mM KH2PO4, 0.65 mM MgSO4·7H2O, 18 μM FeSO4·7H2O, 70 μM CaCl2·2H2O, 10 μM MnCl2·4H2O [pH 7.2]) supplemented with 10 mM succinate as the carbon source and 10 mM ammonium sulfate as the nitrogen source (17). Media were supplemented, as required, with 500 μM ferulic acid or p-coumaric acid and with the appropriate antibiotics (25 μg/ml gentamicin, 25 μg/ml neomycin, and 25 μg/ml kanamycin) and casamino acids (0.1%).
RNA extractions for oligoarray analyses.
Bacterial cells were precultivated to the stationary phase in AT medium with 10 mM succinate, 10 mM ammonium sulfate, and 0.1% casamino acids. Next, cells were inoculated at 1.5 × 108 cells/ml in AT medium supplemented with 10 mM succinate, 10 mM ammonium sulfate, and 500 μM ferulic acid or p-coumaric acid as required. After 24 h of incubation (stationary phase; 1.5 × 109 cells/ml), cells were harvested by centrifugation and total RNAs were extracted from the pellets by using the frozen acid-phenol method described by Maes and Messens (18). DNA was eliminated with two DNase I treatments (Roche, Mannheim, Germany) followed by a treatment with Ambion DNase (Turbo DNA-free kit). The absence of DNA was checked by PCR with primer pairs for the atu1420 gene (Table 1). Isolated RNAs were quantified and checked for quality using a Bioanalyzer 2100 system, and they were then stored at −80°C for further use.
TABLE 1.
Primer sets used in this study
| Analysis and region | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) |
|---|---|---|
| RT-qPCR analyses | ||
| atu0043 | GCATATCGATGACCACCACA | TTGACGTTCCACACAAGGAA |
| atu0140 | ATGAGCGACTGCCTGATCTT | GCCATAGGTGATGGAGGAGA |
| atu0161 | AAACGGTTCAGCCTTCACAG | TCGGCGTTAACGGTTTCTAC |
| atu0229 | GGTTTGGCAAGATCGTCAAT | CGATGCCGTGAAAATAGACC |
| atu0231 | GCGAACAGAATGCGGTAGAT | AAGATCGGGAAGTTCTGCTG |
| atu0786 | GTTGCCGGTTACGTCTTCAT | TTGTTTTCCCAGACGAGCTT |
| atu1028 | CAAGCTTCTGAAGGCGTTTC | TCCATGTTGCCCTTGATCTT |
| atu1226 | ACGACGCTTGAAAAGCTGAT | CCAGGCTGACAAGGATGATT |
| atu1331 | GTCGGCACGGTCTATCTCTC | GACGATGATTTCCACCACCT |
| atu1413 | GCGAAAAGCTGAAGGTCATC | GGCATGACGAAGTTCCATTT |
| atu1414 | GAGCTTCTGAAGGCAACGTC | GAAAAGCATCGGCAAAACC |
| atu1415 | GGCCTTCGTATTCTCGTCAA | TTGTCTTGGCGACTGTCTTG |
| atu1416 | CCCTATCCGCAGAAATTGAA | ACCGAAAGGTCATGATCGAG |
| atu1417 | CGACATGATGCTGACTGGAC | ACGGCATAGTTGGACAAAGG |
| atu1418 | ATGGGTTGGTCGAGTTTTTG | GGATACCATGTCGGCTATCG |
| atu1419 | GGTGATCGAACTCAGGGAAA | TACTTCAATGGCATCCACGA |
| atu1420 | GCCTATGGCAAGTGGAATGT | CGAGCCGATATTGATCCAGT |
| atu1426 | CGTATGGGTTCGGAAGTGTT | AGCCTTTTCGATCGACTTCA |
| atu1507 | CAGGATGTGGAAAGCCTGAT | TTGACGATCTGCTGGATACG |
| atu1823 | CAAGTCGACGCTGTCCTACA | AACCTTCAGGAACTGCATGG |
| atu1870 | TCATCTGGCAGCTCATCAAG | GGCTCTTCCACATCTGCTTC |
| atu1923 | GAGCGTGGTTTCGGTCTTAC | CATCTTGATGGCGATTTCCT |
| atu1924 | CGACCTTCAACAACACGATG | CGTGTTCCTGAGCCTTCTTC |
| atu1998 | GACACCGTCTTCAACAAGCA | CACCCTCTTTATCCGTGGAA |
| atu2073 | CTGACGATCGAGATGACGAA | TCGACGGATACGAAATAGGC |
| atu2085 | GGACTGCTCGGATTTCAGAC | ACTGACTGGGATAGCGAACG |
| atu2449 | CGGCCAGTGTGTTTCAACTA | AAGACTTACGGATGGCGTTG |
| atu2497 | ACCTATGATCATCGCGGAAC | CAGGCATTGATGAGCACAAG |
| atu2623 | CTGATTTCCGAAGGCAAGAC | GAGCGAGATCACCTTCTTGG |
| atu2829 | ATTTGCTCGGTTTCGAGATG | ACCTTCCAGGGCAAAGAACT |
| atu3327 | TCGGCTGAACAGACACTCAG | TCGGAGAGTTTCACCAGGAC |
| atu3391 | GCCAAGGTCACCGATACACT | GGTTGGAAGCGATGATCTGT |
| atu3564 | GGTGCAGACACCTTCGTTTT | ACGATCTGTCGAGGCTGAGT |
| atu3677 | GTCCTGACGAAAGCAGTTCC | CTTCGCAACTCATGCAGAAA |
| atu3687 | GACGTCCATAACGTCGGACT | GAGGCGATCCATTCCTTGTA |
| atu3916 | ACTGGTCGCTTTCGCATACT | CGAGAACGGTGTTACGGATT |
| atu3918 | GAACTGGGTCTCGTCTTTGC | GCCAGAACAGGCTTGATAGG |
| atu4177 | CTCGATGAATATCGCAAGCA | ATCAGGTCGATTGCCTTGTC |
| atu4333 | GTCGAGATCACCGAGGAAAG | CAGATGTGCGAGGTTGAGAA |
| atu4544 | GTAAACCACCGGATCGATTG | GGTGGTTGGCGTAGATGAGT |
| atu4547 | AGGGACCGAAGAACCTGACT | GGCCAGATACCGGTCTGTAA |
| atu4709 | GGATGAAAAGCTGGTGAGGA | AAACACAAGCGGCAGAAACT |
| atu5311 | GCTGAAACGATCAAGGAAGC | CGCCGATATAGGGCAGATAA |
| atu5449 | TGACTATGCACCGCTGTTTC | ACGGTCAGAAGGTTGGATTG |
| atu6151 | CAGTGATGCGAACGTTTCTG | TTAGCCATGGCCATTCTTTC |
| Inactivation of SpG8-3 gene cluster | ||
| Region upstream of atu3663 | CCGGTTCTACATCCTGGAAA | CCTGCTCAACAGGCTACTCC |
| Region downstream of atu3693 | GACAACATGCCCCTCTCCTA | TCTGGAACGTCACCGACATA |
| nptII gene | TTGCTGCGCGCGACATCAAGGTTTCGACCGAGGAGTAGCCTGTTGAGCAGGTGTGTAGGCTGGAGCTGCTTC | TGAAAATGCCGCTGTATTTCCTCGATCACGTAGGAGAGGGGCATGTTGTCCATATGAATATCCTCCTTA |
| Region for inactivation diagnosis | GAGAGTGACGCTTTGGCTCT | GGTTGATCTGGTGCAGCTTT |
Oligoarray hybridizations and data analyses.
RNA samples were reverse transcribed into cDNA using a cDNA synthesis system (Roche) and then were labeled and amplified with a one-color DNA labeling kit (Roche). A total of 2 μg of purified Cy3-labeled cDNA was hybridized to NimbleGen 4-plex microarrays (Roche) containing probe sets for the 5,345 genes that represent the entire A. fabrum C58 genome. Oligoarrays were scanned in a MS200 NimbleGen scanner (Roche), and fluorescence signals were quantified using NimbleScan software (Roche). Data were normalized using the Robust Multichip Average algorithm (DEVA software; Roche) and then were analyzed for differential gene expression using single-factor analysis of variance (ANOVA) (19). Changes of ≥2-fold were considered to be significant according to the statistical test (false discovery rate [FDR] of ≤0.05). Significant genes were used for hierarchical and principal-component analysis clustering. Transcriptomic assays were performed with three independent biological replicates.
Reverse transcription-quantitative PCR analyses.
RNA extractions, reverse transcriptions, and quantitative PCR analyses were performed as described previously (20). RNAs were extracted by a rapid procedure using the FastPrep-24 cell breaker (MP Biomedicals, Santa Ana, CA) and acid-phenol. DNAs were removed with DNase I treatments (Roche). The absence of DNA was checked by PCR with primer pairs for the atu1420 gene. Reverse transcriptions and quantitative PCR analyses were performed using the primers listed in Table 1. The housekeeping genes atu1028 and atu1823, which displayed constant expression levels in the global transcriptomic analyses performed here, were used for normalization of the reverse transcription-quantitative PCR (qRT-PCR) data (21). The specificity of PCR primers was verified with a melting curve analysis.
Blast2Go annotation.
Genes from A. fabrum C58 were annotated using Blast2Go, with the default settings. They were subjected to a BLAST search against the GenBank database, followed by mapping to Gene Ontology (GO) information for the BLAST hits. Blast2Go functional annotation was analyzed by generating Blast2Go combined graphs of molecular functions and biological processes, with the default settings (22). Enrichment analysis was performed by Blast2Go with Fisher's exact test (FDR of ≤0.05).
Construction of SpG8-3 deletion mutant (siderophore-encoding genes).
A mutant with a deletion from gene atu3663 to gene atu3693 (around 57 kb) was constructed as described previously (3), using the primers listed in Table 1. The deletion event was verified by diagnostic PCR analysis.
Ferulic acid degradation measurements by HPLC.
High-performance liquid chromatography (HPLC) analyses were used to monitor the degradation of ferulic acid. Briefly, cells were cultivated overnight in AT medium supplemented with 10 mM succinate, 10 mM ammonium sulfate, and 0.1% casamino acids. For the induction culture, cells were then inoculated at an optical density at 600 nm (OD600) of 0.1 in minimal AT medium supplemented with nitrogen, a carbon source (10 mM), and ferulic acid (500 μM). As described previously (4), to avoid differential growth effects on the speed of ferulic acid degradation, cells were harvested and suspended at an OD600 of 1 in fresh medium in the presence of 500 μM ferulic acid, a concentration that does not permit cell growth (degradation culture). When necessary, 2,2′-dipyridyl (DIP) was added to the induction and degradation cultures at a concentration (200 μM) that does not prevent C58 growth. As described previously, degradation measurements were performed using an Agilent 1200 series HPLC system (Agilent Technologies, Santa Clara, CA) coupled to a UV-visible diode array detector (Agilent Technologies) equipped with a Kromasil C18 column (Kromasil 100-5-C18, 5-μm particle size, 250 nm by 4.6 mm; AkzoNobel, Amsterdam, Netherlands) (3).
Microarray data accession number.
Data that had been treated according to the MIAME (minimum information about a microarray experiment) guidelines were registered at ArrayExpress under accession number E-MTAB-3589.
RESULTS
General characteristics of the C58 transcriptome in the presence of HCAs.
A whole-genome oligoarray approach was used to identify genes that were differentially expressed in the presence of ferulic acid and p-coumaric acid. For this purpose, C58 RNAs were extracted from bacteria and grown for 24 h in AT medium with succinate, with or without 500 μM ferulic acid or p-coumaric acid. These conditions were chosen because (i) the expression of genes involved in HCA degradation (atu1416, atu1417, and atu1420) was maximal and (ii) no differences in growth rates or growth yields were detected with the addition of HCAs to the medium (see Fig. S1 in the supplemental material) (4).
According to single-factor analysis of variance, the expression levels of 757 genes (14.16% of the C58 genes) were significantly different under conditions with versus without ferulic acid (or p-coumaric acid), while fewer than 10 genes were differentially expressed under conditions with ferulic acid versus p-coumaric acid (see Table S1 in the supplemental material). Among those, 42 genes were chosen, according to the range of their fold changes in expression, for independent validation by qRT-PCR analyses. Significant differences in expression were validated by qRT-PCR analyses for subsets of 40 and 39 genes for ferulic acid regulation and p-coumaric acid regulation, respectively (see Table S2 in the supplemental material). Nonvalidated genes were mostly genes with the smallest observed fold changes in expression (see Table S2). Therefore, we focused subsequent analyses on the 675 genes (12.63%) that displayed ≥2-fold changes in expression in comparisons of ferulic acid medium with minimal medium, p-coumaric acid medium with minimal medium, or p-coumaric acid medium with ferulic acid medium. Clustering analyses revealed that 663/675 genes were regulated in the same way in the presence of succinate and either ferulic acid or p-coumaric acid, compared with minimal medium with succinate but without HCAs. Among those genes, around 59% were upregulated and 41% were downregulated.
Genes involved in the A. fabrum HCA degradation pathway are induced in the presence of ferulic acid and/or p-coumaric acid.
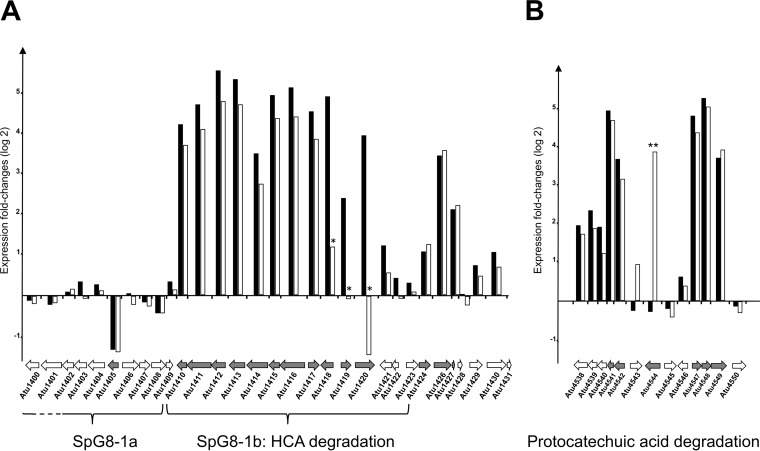
Almost all of the genes in the SpG8-1b genomic region, which is involved in HCA degradation, were induced in the presence of ferulic acid or p-coumaric acid. Upregulations of approximately 30-fold, 34-fold, and 23-fold were observed for atu1415, atu1416, and atu1417, respectively (Fig. 1A; also see Table S1 in the supplemental material). The genes atu4541, atu4542, atu4547, atu4548, and atu4549, which do not belong to the SpG8-1b genomic region but are still known to be necessary for complete HCA degradation through the transformation of protocatechuic acid into acetyl-CoA and succinate (4, 23), were also highly induced, with 30-fold, 10-fold, 25-fold, 35-fold, and 14-fold changes, respectively (Fig. 1B; also see Table S1). All of these inductions were subsequently validated by qRT-PCR analyses (see Table S2 in the supplemental material).
FIG 1.
Genomic regions differentially regulated in the presence of ferulic acid and p-coumaric acid. Each gene is represented by an arrow directed to the right for the gene transcribed from the plus strand and to the left for the gene transcribed from the minus strand. For each gene, histograms represent logarithmic fold changes in expression levels measured from cells cultivated with versus without ferulic acid (black bars) or with versus without p-coumaric acid (white bars). Gray arrows, genes with significant expression differences, according to ANOVA, at the 0.05 level, with the exception of atu1418, atu1419, and atu1420 (*), whose expression differences with p-coumaric acid were not significant. (A) The SpG8-1 region is subdivided into two regions, i.e., the SpG8-1a subregion, spanning atu1398 to atu1408, and the SpG8-1b subregion, spanning atu1409 to atu1423. Genes atu1415, atu1416, atu1417, and atu1421 are essential for the degradation of HCA, whereas atu1418 to atu1420 are essential only for the degradation of ferulic acid (3). (B) The region encompassing pca genes encodes the proteins involved in the second part of the HCA degradation pathway, i.e., the degradation of protocatechuic acid into acetyl-CoA and succinate. The gene atu4544 (pobA) is essential only for the degradation of p-coumaric acid, and its expression was significantly induced only with p-coumaric acid (**) (22).
As expected, the genes coding for proteins that play a role in the degradation of ferulic acid but not in that of p-coumaric acid (atu1418, atu1419, and atu1420) were induced with ferulic acid but not with p-coumaric acid. Conversely, atu4544, encoding PobA, which converts 4-hydroxybenzoate (an intermediate in the p-coumaric acid degradation pathway) into protocatechuic acid (24), was induced only in the presence of p-coumaric acid. This activity is not necessary for the degradation of ferulic acid, and the specific induction of this gene was in accordance with its known functions. The gene atu8021, encoding a hypothetical protein, was also induced only in the presence of p-coumaric acid.
Genes involved in the categories of metabolism and expression are differentially expressed in the presence of ferulic acid or p-coumaric acid.
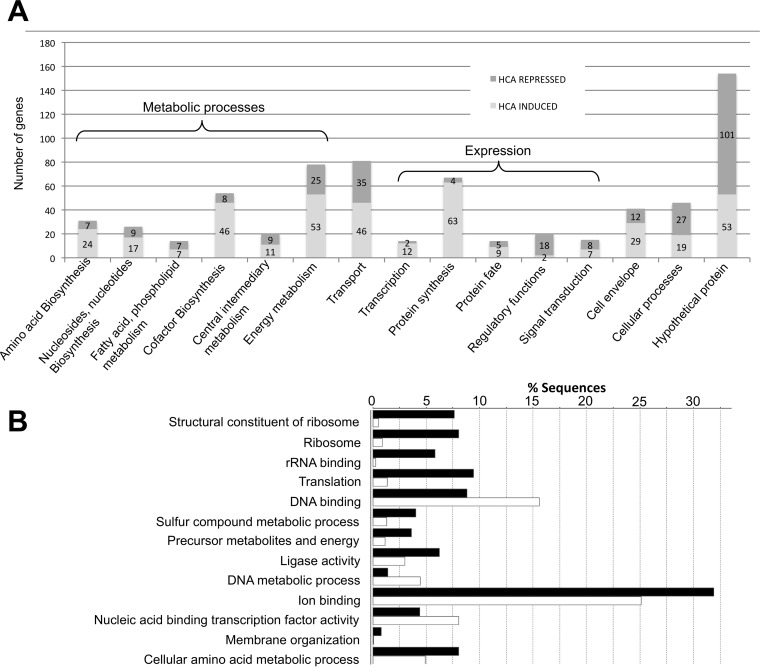
Functional clustering was performed with the 675 genes, according to (i) the 15 KEGG functional categories (http://www.genome.jp/kegg/pathway.html) and (ii) our expert annotations (Fig. 2A). A total of 154 genes (around 20% of the genes in the HCA regulon) encoded hypothetical proteins, with two-thirds being downregulated in the presence of HCAs. Functional enrichment analysis was also performed with Blast2Go (Fig. 2B). Interestingly, genes involved in metabolic processes were significantly enriched in the HCA regulon. Among those, 20 genes were involved in central intermediary metabolism and 78 in energy metabolism, with the upregulation of genes involved in ATP synthase, the tricarboxylic acid (TCA) cycle, and the glycolysis pathway (Fig. 2A). In addition, the expression levels of 31 genes involved in amino acid biosynthesis, 14 genes involved in fatty acid metabolism, and 26 genes involved in nucleotide biosynthesis were significantly different in the presence of HCAs. Genes from the expression category (transcription and protein synthesis) were also significantly enriched in the HCA regulon (Fig. 2A). In conclusion, among the annotated genes regulated by HCAs, a large majority belonged to the categories of metabolic processes or expression.
FIG 2.
Gene function categories of the ferulic acid and p-coumaric acid regulons. (A) Gene abundances in gene function categories. Dark gray, downregulated genes; light gray, upregulated genes. (B) Enrichment analysis of GO terms (FDR of ≤0.05). Black bars, percentage of gene sequences from the ferulic acid and p-coumaric acid regulons (HCA regulon); white bars, percentage of gene sequences from the A. fabrum C58 genome.
Genes involved in iron uptake are induced in the presence of ferulic acid or p-coumaric acid.
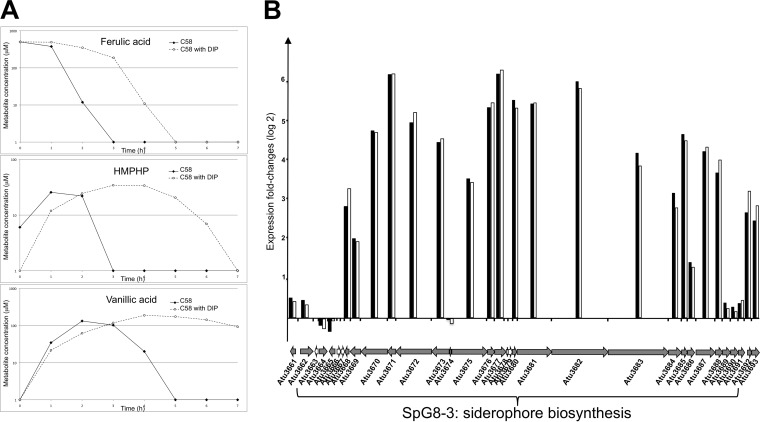
Strikingly, among the 54 genes belonging to the category of biosynthesis of cofactors, including the siderophore and iron storage and iron transporter subcategories, 85% were upregulated in the presence of HCAs (Fig. 2A). The induction of five of those genes was validated independently by qRT-PCR analyses (see Table S2 in the supplemental material). These inductions suggested a possible correlation between the HCA degradation pathway and the siderophore biosynthesis pathway. Therefore, the involvement of iron uptake in HCA degradation efficiency was tested. The rate of ferulic acid degradation was measured in the presence of a low iron concentration, which was obtained with the addition of an iron chelator, namely, 2,2′-dipyridyl (DIP). These degradations were performed using equivalent cellular densities (OD600 of 1) and under conditions without cell growth. As shown in Fig. 3A, degradation of ferulic acid was delayed in the presence of the iron chelator, as was the production and/or consumption of ferulic acid intermediates (HMPHP and vanillic acid). HMPHP was completely consumed after 3 h for C58, compared with 7 h in the presence of DIP. Vanillic acid was completely consumed after 5 h for C58, but consumption did not start until after 7 h in the presence of DIP. These results led us to conclude that the ferulic acid degradation of C58 is significantly slower with low iron concentrations.
FIG 3.
Upregulation of genes in the C58 HCA regulon in the iron uptake category. (A) Ferulic acid degradations by the C58 strain at a low iron concentration. Ferulic acid degradations were performed with cells at an OD600 of 1 in AT medium with 500 μM ferulic acid, with or without DIP (an iron chelator), as described in Materials and Methods. The amounts of ferulic acid and its intermediary compounds (HMPHP and vanillic acid) were monitored using HPLC with a UV-visible diode array detector set at 280 nm, 0 h, 1 h, 2 h, 3 h, 4 h, 5 h, 6 h, and 7 h postinoculation. Results presented are representative of at least three biological replicates. (B) The SpG8-3 region, spanning atu3663 to atu3691, encodes the biosynthesis of a siderophore. It is composed notably of two operons, beginning with atu3674 and atu3675 (24). Histograms represent fold changes in expression levels measured from cells cultivated in AT medium with succinate, with versus without ferulic acid (black bars) or with versus without p-coumaric acid (white bars). Each gene is represented by an arrow directed to the right for the gene transcribed from the plus strand and to the left for the gene transcribed from the minus strand. Gray arrows, genes with significant expression differences, according to ANOVA, at the 0.05 level.
Iron capture and HCA catabolism are species-specific regions interconnected in A. fabrum.
Among the genes that were upregulated in the presence of HCAs and belonged to the iron uptake category, genes from atu3669 to atu3692 were highly induced in the presence of HCAs, with 4-fold to 78-fold changes (Fig. 3B). These genes, which are organized in two divergent operons, are involved in the biosynthesis of a unique siderophore that has not yet been structurally characterized (25) and, remarkably, they are located in the SpG8-3 genomic region, which is known to be specific to A. fabrum (3). Therefore, HCAs induce the expression of two A. fabrum-specific gene clusters, namely, SpG8-1b and SpG8-3.
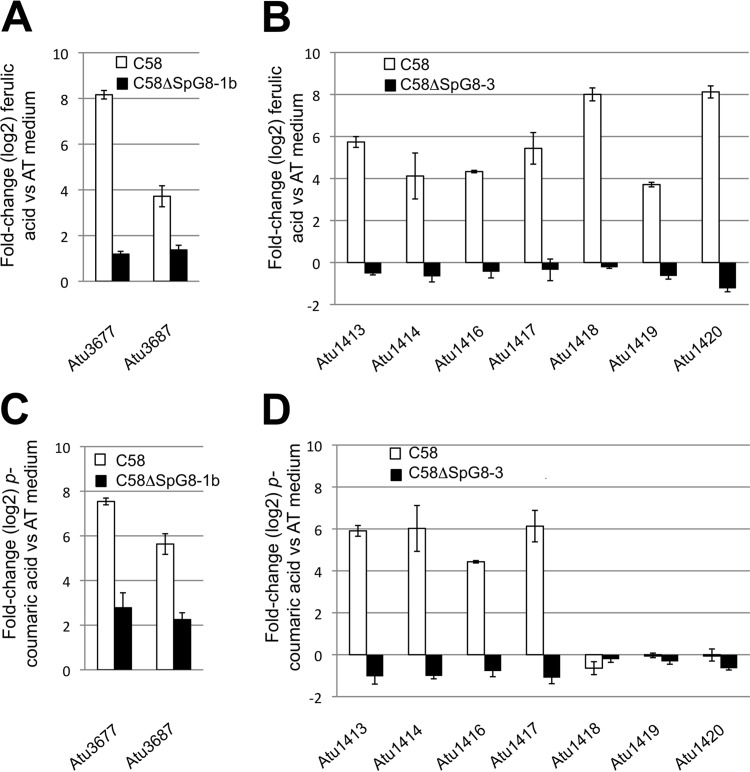
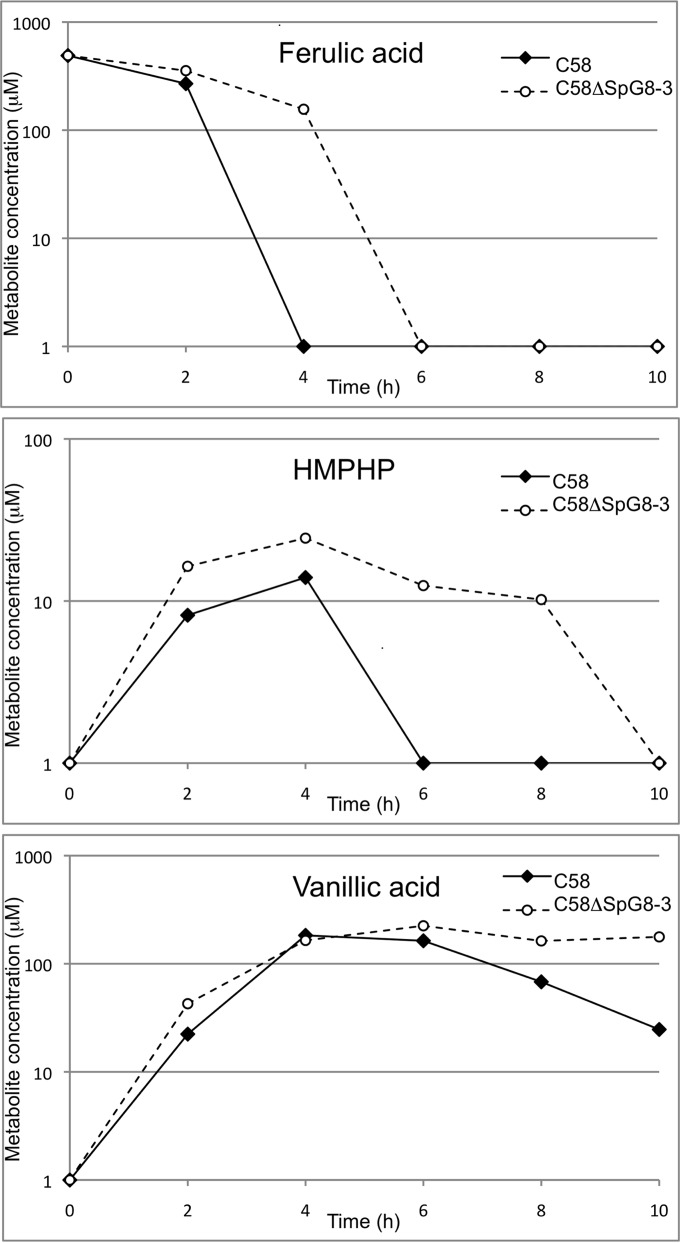
This raised the question of whether the expression level of one of these clusters may influence the expression level of the other. To investigate this, we used qRT-PCR analyses to measure the expression level of one gene cluster in mutants lacking the other cluster. Although neither of the clusters was expressed in the minimal AT medium with succinate, both clusters were induced in C58 in the presence of ferulic acid or p-coumaric acid. In the presence of HCAs, the cluster involved in iron uptake (SpG8-3) was upregulated in the wild-type strain but not in the mutant strain C58ΔSpG8-1b, which lacks the HCA degradation genes (Fig. 4A and C). In a similar way, the genes involved in HCA degradation were not upregulated in the mutant C58ΔSpG8-3 in the presence of HCAs (Fig. 4B and D). At the cellular level, a delay in the consumption of ferulic acid was observed when C58ΔSpG8-3 and C58 were compared. Moreover, the production and/or consumption of intermediates of degradation (HMPHP and vanillic acid) was delayed (Fig. 5). These findings indicate a significant delay of ferulic acid degradation in the iron uptake-deleted mutant.
FIG 4.
Transcript overexpressions in the C58ΔSpG8-1b (A and C) and C58ΔSpG8-3 (B and D) mutant strains, measured in the presence of ferulic acid (A and B) or p-coumaric acid (C and D) versus minimal AT medium, in comparison with the wild-type strain C58. qRT-PCR results for each gene were normalized using the housekeeping genes atu1028 and atu1823.
FIG 5.
Ferulic acid degradations with C58 and C58ΔSpG8-3. Ferulic acid degradations were monitored with cells at equivalent cell densities, in AT medium supplemented with 500 μM ferulic acid. The amounts of ferulic acid, HMPHP, and vanillic acid were monitored using HPLC with a UV-visible diode array detector set at 280 nm. The starting point (0 h) corresponds to the last record before ferulic acid degradation. Results are means of three independent replicates.
The gene clusters SpG8-1 (HCA degradation pathway gene cluster) and SpG8-3 (siderophore biosynthesis gene cluster) could be coregulated.
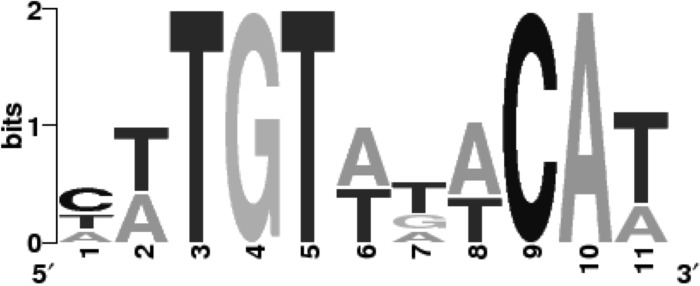
The search for DNA consensus sequences was performed in the promoter regions of SpG8-1b and SpG8-3. The search revealed a consensus sequence, composed of 11 nucleotides, that was present upstream of the six SpG8-1b operons (atu1414, atu1416, atu1417, atu1418, atu1419, and atu1420) and the two SpG8-3 operons (atu3673 and atu3675) (Fig. 6). This consensus sequence was located up to 150 bp upstream of the start codon. Subsequently, a search for the presence of this consensus sequence elsewhere in the A. fabrum genome allowed us to identify the sequence signature in the promoter regions of 12 other genes (atu0043, atu0080, atu0413, atu0428, atu1226, atu1250, atu1765, atu1766, atu3140, atu3356, atu3778, and atu3997). These genes encode proteins involved in various functions, but all of them were regulated by ferulic acid and p-coumaric acid (see Table S1 in the supplemental material). As an indication, none of these genes is specific to A. fabrum, as determined previously using comparative genomic analyses. The significant presence of this DNA consensus sequence in the promoter regions of both the SpG8-1b and SpG8-3 clusters suggests that they may be regulated in the same manner.
FIG 6.
Consensus sequence identified upstream of the coding sequences of genes of the SpG8-1b and SpG8-3 genomic regions, using the SCOPE algorithms (32, 33).
DISCUSSION
Within the framework of research undertaken to elucidate the specific adaptation of species, we investigated the transcriptional reprogramming of A. fabrum cells in the presence of HCAs already known to be involved in the ecological specificity of A. fabrum (3). Our work reveals that ferulic acid and p-coumaric acid have very closely related effects, inducing significant variations in the expression levels of a large number of genes. This defines the HCA regulon of C58, demonstrating major effects of HCAs on cell physiology.
As expected, genes essential for the A. fabrum HCA degradation pathway (atu1416, atu1417, atu1415, atu1418, and atu1420) (4) were among the most significantly differentially expressed genes of the HCA regulon. In agreement with previous results, the Ti plasmid genes virH1 and virH2, which are involved in the transformation of ferulic acid into caffeic acid (8, 26), displayed no or only slight upregulation with HCAs; consequently, they are not part of the HCA regulon. In addition to the genes of the A. fabrum HCA degradation pathway, however, most other SpG8-1b genes are included in the HCA regulon (Fig. 1). Among these genes is the operon atu1410 to atu1413, annotated as a branched-chain amino acid ABC transporter, which might carry HCA or HCA intermediates into cells. Several upregulated genes were previously annotated as putative enzymes. This is the case for atu1414, which encodes a feruloyl-CoA dehydrogenase that is potentially involved in the HCA degradation pathway but is not essential for degradation, since the wild-type strain and the atu1414-deleted mutant showed similar profiles for ferulic acid degradation (data not shown). In addition, atu1405, which is upstream of SpG8-1b in the SpG8-1a region spanning atu1398 to atu1408 (3), was significantly downregulated. This gene encodes a putative GntR family regulator and may be involved in the repression of HCA degradation gene transcriptions.
Cellular functions were overrepresented in the HCA regulon, including those of metabolism, expression processes, and iron uptake. The gene expressions in the categories of metabolism and expression processes were generally increased in the presence of HCAs. Taken together, these results indicate that HCA catabolism via the A. fabrum HCA degradation pathway allows for the maintenance of active metabolism in the cells, reinforcing the fact that ferulic acid and p-coumaric acid are carbon sources for A. fabrum (4). The iron uptake category represents at least 33% of the most highly upregulated genes. We observed upregulations of the genes involved in the categories of biosynthesis of siderophores, TonB-dependent transporters, iron-related ABC transporters, and intracellular iron storage and downregulations of genes from the category of iron-sulfur cluster protein biosynthesis. According to the literature, such regulations are generally observed in cells cultivated under conditions of iron starvation (27). In our experiments, however, these genes were induced in spite of the fact that iron was not a limiting factor in the medium (18 μM FeSO4). This would be expected to lead first to an excess concentration of intracellular iron and then to an oxidative burst, but this has not been recorded in the transcriptional data (see Table S1 in the supplemental material). Hence, we suggest possible control of iron homeostasis in the presence of HCAs. The mechanism by which this iron control operates has yet to be elucidated.
Strikingly, the expression levels of the genes directly involved in HCA degradation and iron uptake were correlated. The presence of SpG8-3 (siderophore biosynthesis gene cluster) is required for the expression of the SpG8-1b genes (HCA degradation pathway gene cluster), and the presence of SpG8-1b is required for the expression of the SpG8-3 genes. Furthermore, iron seems to be essential for ferulic acid degradation. We observed a delay in ferulic acid degradation under iron starvation conditions, either with the mutant strain C58ΔSpG8-3 or in the presence of an iron chelator (Fig. 3). Moreover, the siderophore-free strain C58ΔSpG8-3 displayed growth rates and yields equivalent to those of the wild-type strain regardless of the carbon source tested (data not shown). In our opinion, this indicates that a lack of iron did not lead to general metabolism slowing under these conditions but rather had a specific effect on ferulic acid degradation. Thus, we suspected the presence of a regulatory network coordinating the two functions, either directly or indirectly. Supporting this idea, a consensus sequence has been identified in the promoter regions of almost all of the operons of both SpG8-1b and SpG8-3 and in the region located upstream of other genes of the HCA regulon. This sequence is composed of 11 nucleotides (with eight of them forming a palindrome), suggesting the occurrence of an unknown DNA-binding protein that may bind to this consensus sequence. This putative transcriptional regulator would regulate both HCA degradation and iron uptake functions, in a coordinated manner.
From a physiological point of view, the reason for coordination of the HCA degradation pathway and the iron uptake pathway is intriguing. It could be related to the antioxidative properties of ferulic acid (28) and its ability to reduce iron in vitro in exchange for its oxidation (29). Indeed, the dramatic delay of ferulic acid degradation under conditions of iron starvation suggests that ferulic acid was unable to enter into cells under these conditions. This is perhaps simply due to the fact that the ferulic acid was in a reduced form. An alternative reason is that ferulic acid needs to be coupled with iron to enter into cells, using a suitable transporter. Remarkably, strong coupling between ferulic acid and iron does occur (30), and ferulic acid is suspected to function as a siderophore (31).
From an ecological point of view, the A. fabrum species presents a complex strategy for the sequestration of iron, using (i) the production of siderophores by its own cells and (ii) HCAs originating from the environment, i.e., from the decay of plant cell walls. This could lead to the very specific ability of A. fabrum to capture iron in environments that are rich in HCAs but have extremely low iron levels, such as in decaying lignin-rich plant materials. A. fabrum could have an advantage over other Agrobacterium species in such environments, because the relevant genome regions, SpG8-1 and SpG8-3, have been described as belonging to the set of genes specific to the species A. fabrum (3). We suggest that the coordinated expression of these two species-specific gene clusters reinforces the idea that they are encoding functions that cooperate to determine a species-specific ecological niche for A. fabrum. The present work supports the view that bacterial species are defined according to their genomic coherence, corresponding to ecological species that are adapted to particular niches.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the EcoGenome project of the French Agence Nationale de la Recherche (grant ANR-BLAN-08-0090). T.C. received a doctoral grant from the French Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche.
We thank the ProfilExpert platform for the microarray hybridizations, Vincent Gaillard and Isabelle Kerzaon, from the Centre d'Etude des Substances Naturelles platform, for the HPLC analyses, and Jade Ravent for technical assistance. Quantitative PCR experiments were performed at the DTAMB platform of the BioEnviS Research Federation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00419-16.
REFERENCES
- 1.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kämpfer P, Maiden MC, Nesme X, Rosselló-Mora R, Swings J, Trüper HG, Vauterin L, Ward AC, Whitman WB. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 2.Mousavi SA, Osterman J, Wahlberg N, Nesme X, Lavire C, Vial L, Paulin L, de Lajudie P, Lindström K. 2014. Phylogeny of the Rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Syst Appl Microbiol 37:208–215. doi: 10.1016/j.syapm.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Lassalle F, Campillo T, Vial L, Baude J, Costechareyre D, Chapulliot D, Shams M, Abrouk D, Lavire C, Oger-Desfeux C, Hommais F, Guéguen L, Daubin V, Muller D, Nesme X. 2011. Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens. Genome Biol Evol 3:762–781. doi: 10.1093/gbe/evr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campillo T, Renoud S, Kerzaon I, Vial L, Baude J, Gaillard V, Bellvert F, Chamignon C, Comte G, Nesme X, Lavire C, Hommais F. 2014. Analysis of hydroxycinnamic acid degradation in Agrobacterium fabrum reveals a coenzyme A-dependent, beta-oxidative deacetylation pathway. Appl Environ Microbiol 80:3341–3349. doi: 10.1128/AEM.00475-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalogeraki VS, Zhu J, Eberhard A, Madsen EL, Winans SC. 1999. The phenolic vir gene inducer ferulic acid is O-demethylated by the VirH2 protein of an Agrobacterium tumefaciens Ti plasmid. Mol Microbiol 34:512–522. doi: 10.1046/j.1365-2958.1999.01617.x. [DOI] [PubMed] [Google Scholar]
- 6.Sayadi S, Allouche N, Jaoua M, Aloui F. 2000. Detrimental effects of high molecular-mass polyphenols on olive mill wastewater biotreatment. Process Biochem 35:725–735. doi: 10.1016/S0032-9592(99)00134-X. [DOI] [Google Scholar]
- 7.Seneviratne G, Jayasinghearachchi HS. 2003. Phenolic acids: possible agents of modifying N2-fixing symbiosis through rhizobial alteration? Plant Soil 252:385–395. doi: 10.1023/A:1024725511783. [DOI] [Google Scholar]
- 8.Ravn H, Andary C, Kovacs G, Molgaard P. 1989. Caffeic acid esters as in vitro inhibitors of plant pathogenic bacteria and fungi. Biochem System Ecol 17:175–184. doi: 10.1016/0305-1978(89)90076-8. [DOI] [Google Scholar]
- 9.Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, Harwood CS. 2008. A new class of homoserine lactone quorum-sensing signals. Nature 454:595–599. doi: 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]
- 10.Parke D, Ornston LN, Nester EW. 1987. Chemotaxis to plant phenolic inducers of virulence genes is constitutively expressed in the absence of the Ti plasmid in Agrobacterium tumefaciens. J Bacteriol 169:5336–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kape R, Parniske M, Werner D. 1991. Chemotaxis and nod gene activity of Bradyrhizobium japonicum in response to hydroxycinnamic acids and isoflavonoids. Appl Environ Microbiol 57:316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharya A, Sood P, Citovsky V. 2010. The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection. Mol Plant Pathol 11:705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plaggenborg R, Overhage J, Steinbüchel A, Priefert H. 2003. Functional analyses of genes involved in the metabolism of ferulic acid in Pseudomonas putida KT2440. Appl Microbiol Biotechnol 61:528–535. doi: 10.1007/s00253-003-1260-4. [DOI] [PubMed] [Google Scholar]
- 14.Andreoni V, Bernasconi S, Bestetti G. 1995. Biotransformation of ferulic acid and related compounds by mutant strains of Pseudomonas fluorescens. Appl Microbiol Biotechnol 42:830–835. doi: 10.1007/BF00191177. [DOI] [Google Scholar]
- 15.Gurujeyalakshmi G, Mahadevan A. 1987. Dissimilation of ferulic acid by Bacillus subtilis. Curr Microbiol 16:69–73. doi: 10.1007/BF01588174. [DOI] [Google Scholar]
- 16.Parke D. 1995. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens. J Bacteriol 177:3808–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petit A, Tempe J, Kerr A, Holsters M, Vanmontagu M, Schell J. 1978. Substrate induction of conjugative activity of Agrobacterium tumefaciens Ti plasmids. Nature 271:570–572. doi: 10.1038/271570a0. [DOI] [Google Scholar]
- 18.Maes M, Messens E. 1992. Phenol as grinding material in RNA preparations. Nucleic Acids Res 20:4374. doi: 10.1093/nar/20.16.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharov AA, Dudekula DB, Ko MS. 2005. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics 21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- 20.Dequivre M, Diel B, Villard C, Sismeiro O, Durot M, Coppée JY, Nesme X, Vial L, Hommais F. 2015. Small RNA deep-sequencing analyses reveal a new regulator of virulence in Agrobacterium fabrum C58. Mol Plant Microbe Interact 28:580–589. doi: 10.1094/MPMI-12-14-0380-FI. [DOI] [PubMed] [Google Scholar]
- 21.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 23.Parke D. 2000. Positive selection for mutations affecting bioconversion of aromatic compounds in Agrobacterium tumefaciens: analysis of spontaneous mutations in the protocatechuate 3,4-dioxygenase gene. J Bacteriol 182:6145–6153. doi: 10.1128/JB.182.21.6145-6153.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lah MS, Palfey BA, Schreuder HA, Ludwig ML. 1994. Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants. Biochemistry 33:1555–1564. doi: 10.1021/bi00172a036. [DOI] [PubMed] [Google Scholar]
- 25.Rondon MR, Ballering KS, Thomas MG. 2004. Identification and analysis of a siderophore biosynthetic gene cluster from Agrobacterium tumefaciens C58. Microbiology 150:3857–3866. doi: 10.1099/mic.0.27319-0. [DOI] [PubMed] [Google Scholar]
- 26.Brencic A, Eberhard A, Winans SC. 2004. Signal quenching, detoxification and mineralization of vir gene-inducing phenolics by the VirH2 protein of Agrobacterium tumefaciens. Mol Microbiol 51:1103–1115. doi: 10.1046/j.1365-2958.2003.03887.x. [DOI] [PubMed] [Google Scholar]
- 27.Benjamin JA, Desnoyers G, Morissette A, Salvail H, Massé E. 2010. Dealing with oxidative stress and iron starvation in microorganisms: an overview. Can J Physiol Pharmacol 88:264–272. doi: 10.1139/Y10-014. [DOI] [PubMed] [Google Scholar]
- 28.Butterfield D, Castegna A, Pocernich C, Drake J, Scapagnini G, Calabrese V. 2002. Nutritional approaches to combat oxidative stress in Alzheimer's disease. J Nutr Biochem 13:444. doi: 10.1016/S0955-2863(02)00205-X. [DOI] [PubMed] [Google Scholar]
- 29.Hynes MJ, O'Coinceanainn M. 2004. The kinetics and mechanisms of reactions of iron(III) with caffeic acid, chlorogenic acid, sinapic acid, ferulic acid and naringin. J Inorg Biochem 98:1457–1464. doi: 10.1016/j.jinorgbio.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Angkawijaya AE, Fazary AE, Hernowo E, Taha M, Ju Y-H. 2011. Iron(III), chromium(III), and copper(II) complexes of l-norvaline and ferulic acid. J Chem Eng Data 56:532–540. doi: 10.1021/je101075q. [DOI] [Google Scholar]
- 31.Schrey SD, Erkenbrack E, Früh E, Fengler S, Hommel K, Horlacher N, Schulz D, Ecke M, Kulik A, Fiedler HP, Hampp R, Tarkka MT. 2012. Production of fungal and bacterial growth modulating secondary metabolites is widespread among mycorrhiza-associated streptomycetes. BMC Microbiol 12:164. doi: 10.1186/1471-2180-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakravarty A, Carlson JM, Khetani RS, Gross RH. 2007. A novel ensemble learning method for de novo computational identification of DNA binding sites. BMC Bioinformatics 8:249. doi: 10.1186/1471-2105-8-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlson JM, Chakravarty A, DeZiel CE, Gross RH. 2007. SCOPE: a web server for practical de novo motif discovery. Nucleic Acids Res 35:W259–W264. doi: 10.1093/nar/gkm310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.