ABSTRACT
The antibiotic streptothricin (ST) possesses an amino sugar bound to an l-β-lysine (β-Lys) residue via a peptide bond. The peptide bond formation has been shown to be catalyzed by a nonribosomal peptide synthetase (NRPS) during ST biosynthesis. The focus of this study is the closely related ST analogue BD-12, which carries a glycine-derived side chain rather than a β-Lys residue. Here, in Streptomyces luteocolor NBRC13826, we describe our biosynthetic studies of BD-12, which revealed that the peptide bond between the amino sugar and the glycine residue is catalyzed by a Fem-like enzyme (Orf11) in a tRNA-dependent manner rather than by an NRPS. Although there have been several reports of peptide bond-forming tRNA-dependent enzymes, to our knowledge, Orf11 is the first enzyme that can accept an amino sugar as a substrate. Our findings clearly demonstrate that the structural diversity of the side chains of ST-type compounds in nature is generated in an unusual manner via two distinct peptide bond-forming mechanisms. Moreover, the identification and functional analysis of Orf11 resulted in not only the production of new ST-related compounds, but also the provision of new insights into the structure-activity relationship of the ST-related antibiotics.
IMPORTANCE The antibiotic streptothricin (ST) possesses an amino sugar bound to an l-β-lysine (β-Lys) side chain via a peptide bond formed by a nonribosomal peptide synthetase (NRPS). BD-12, an analogue of ST, carries a glycine-derived side chain rather than β-Lys, and here, we describe the BD-12-biosynthetic gene cluster from Streptomyces luteocolor NBRC13826, which contains the orf11 gene encoding a novel tRNA-dependent peptide bond-forming enzyme. The unique Fem-like enzyme (Orf11) accepts the amino sugar as a substrate and mediates the peptide formation between the amino sugar intermediate and glycine. Our studies demonstrate that the structural diversity of the side chains of ST-related compounds in nature is generated via two distinct peptide bond-forming mechanisms.
INTRODUCTION
Streptothricins (STs) produced by Streptomyces strains are broad-spectrum antibiotics and are chemically characterized by the l-β-lysine (β-Lys) residue and its oligomeric side chains [oligo(β-Lys)]. Since the initial identification, in 1943, of ST-F with one β-Lys residue as the first member of the ST group of antibiotics (1), STs with an oligo(β-Lys) consisting of two to seven residues have been identified (Fig. 1). ST-F inhibits protein biosynthesis in prokaryotic cells (2), and STs carrying the longer oligo(β-Lys) side chains show higher levels of antibacterial activity. Moreover, STs strongly inhibit the growth of eukaryotes, such as yeasts (3–5), fungi (6), protozoa (7), insects (8), plants (9), and mammals (10–13). Although STs have been used effectively as selective agents for recombinant DNA work in some of these organisms, STs are not currently used therapeutically due to their inherent toxicity. In addition to the STs, it has been reported that Streptomyces strains produce ST analogues that possess a glycine-derived side chain rather than the β-Lys residue: BD-12 (14, 15), citromycin (16, 17), glycinothricin (18), A-269A (19), and A-269A′ (19) (Fig. 1). These analogues display potent antibacterial activities, although their molecular targets remain unclear. Also, like the STs, the ST analogues are not used clinically due to their toxicity.
FIG 1.
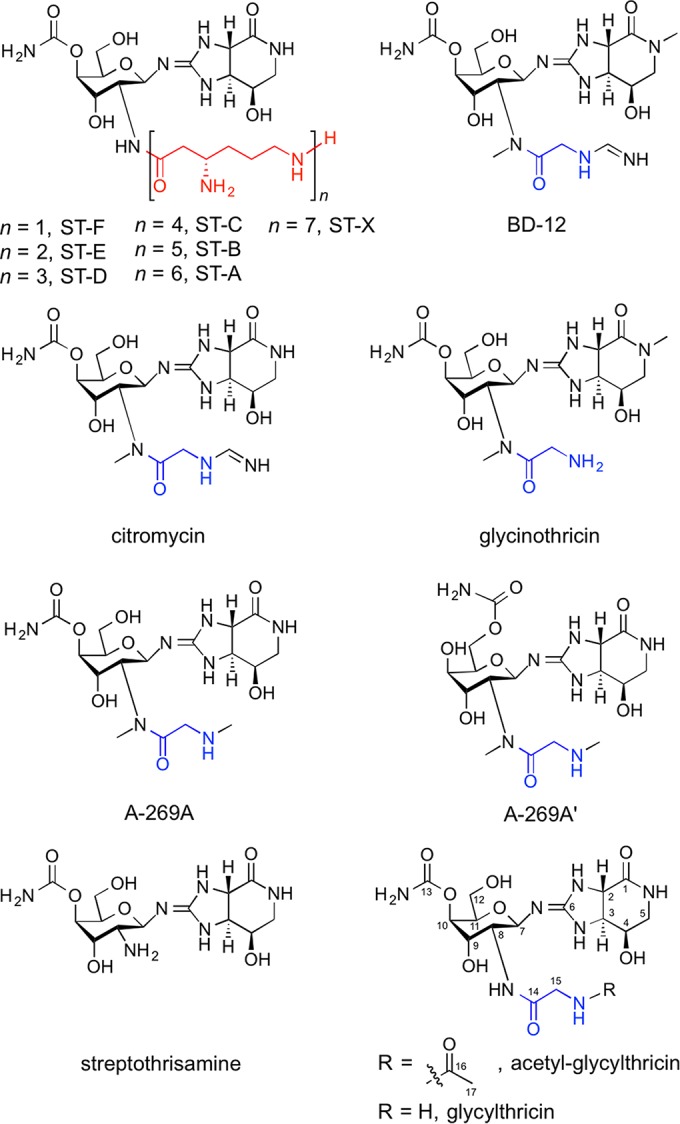
Chemical structures of STs and the ST-related antibiotics. The β-Lys and glycine residues are shown in red and blue, respectively.
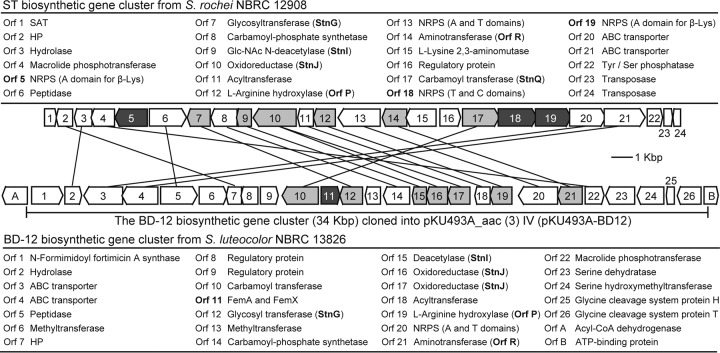
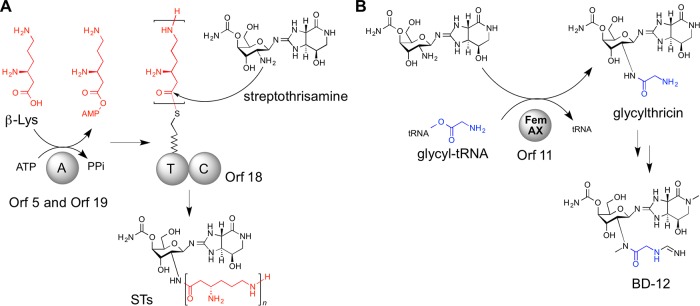
We previously identified the ST-biosynthetic gene cluster (accession no. AB684619) in Streptomyces rochei NBRC 12908 (Fig. 2) and elucidated the biosynthetic mechanisms of the oligo(β-Lys) side chains (20). Nonribosomal peptide synthetases (NRPSs) are known to catalyze the assembly of a myriad of structurally complex peptide natural products (21). However, in the previous study, we identified three unique stand-alone NRPSs among the ST-biosynthetic enzymes and showed that they assembled the structurally simple peptide oligo(β-Lys) (Fig. 3A). The biosynthesis is initiated by adenylation of β-Lys in Orf5 (stand-alone adenylation [A] domain), and the resulting l-β-lysyl-O-AMP is loaded onto the thiolation (T) domain of Orf18. β-Lys molecules adenylated by Orf19 (a second stand-alone A domain) are not directly loaded onto the T domain but are used as extending units for elongation of the oligo(β-Lys) chain on Orf18. Surprisingly, peptide bond formations for the elongation are iteratively catalyzed by Orf19 itself. The condensation (C) domain of Orf18 catalyzes the subsequent condensation reaction between the covalently bound oligo(β-Lys) (or β-Lys) and a freely diffusible amino sugar intermediate, streptothrisamine, ultimately releasing STs (20). Thus, the peptide bond between streptothrisamine and the amino acid side chain is formed by the NRPS machinery during ST biosynthesis.
FIG 2.
Gene organization of the ST- and BD-12-biosynthetic gene clusters. The ST-biosynthetic gene cluster (accession no. AB684619) from S. rochei NBRC 12908 (top) and the BD-12-biosynthetic gene cluster (accession no. LC122485) from S. luteocolor NBRC 13826 (bottom) are shown. The dark-gray-shaded genes are the genes responsible for the peptide bond formation between streptothrisamine and the amino acid residues. The light-gray-shaded genes are the genes responsible for streptothrisamine biosynthesis. The lines connect homologuous genes in the two biosynthetic gene clusters.
FIG 3.
Peptide bond formation between amino sugars and amino acids in ST (A) and BD-12 (B) biosyntheses. The β-Lys and glycine residues are shown in red and blue, respectively.
In consideration of this, we hypothesized a similar NRPS pathway for glycine attachment in the biosynthesis of the ST analogues. Among the ST analogues, we focus here on BD-12 (Fig. 1), produced by Streptomyces luteocolor NBRC 13826, and describe the identification of its biosynthetic gene cluster. We demonstrate that, unexpectedly, the formation of a peptide bond between the streptothrisamine and the glycine residue is catalyzed, not by an NRPS, but by a Fem-like enzyme (Orf11) in a tRNA-dependent manner. Thus, the structural diversity of the side chains of the ST-type antibiotics occurring in nature is generated by two distinct peptide bond-forming mechanisms.
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. luteocolor NBRC 13826 was used as a BD-12 producer. Streptomyces lividans TK23 and Streptomyces avermitilis SUKA17 (22) were used as heterologous host strains for the gene expression experiments (Table 1). Streptomyces integrating vectors, pKU493A_aac(3)IV and pKU1016 (22), were used for the gene expression experiments (Table 1).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference(s) |
|---|---|---|
| Streptomyces strains | ||
| S. luteocolor NBRC 13826 | BD-12 producer | 14, 15 |
| S. lividans TK23 | Host strain for gene expression experiments | 25 |
| S. avermitilis SUKA17 | Host strain for gene expression experiments | 22 |
| Plasmids | ||
| pKU493A_aac(3)IV | Apmr, oriT of RK2, pMB1 ori; ϕK38-1-integrating vector derived from pKU460 | 22 |
| pKU1016 | Neor (aphII), oriT of RK2, pMB1 ori; ϕC31-integrating vector derived from pKU460 | 22 |
| pKU518 | Ampr Neor (aphII); ϕBT1-integrating BAC vector | 24 |
| pRED-sacB-aph | Neor (aphII) sacB Cmr; cloning vector for E. coli | 22 |
| pKU518BD12P2-C23 | pKU518 carrying the genomic DNA fragment (200,999 bp) that contains the BD-12-biosynthetic gene cluster (34 kbp) from S. luteocolor NBRC 13826 | This study |
| pRED/BD-12cluster | pRED-sacB-aph carrying the BD-12-biosynthetic gene cluster (34 kbp) from S. luteocolor NBRC 13826 | This study |
| pKU493A-BD12 | pKU493A_aac(3)IV carrying the BD-12-biosynthetic gene cluster (34 kbp) from S. luteocolor NBRC 13826 | This study |
| pKU493A-STΔorf18 | pKU493A_aac(3)IV carrying the ST-biosynthetic gene cluster (30 kbp) with in-frame deletion of the orf18 gene (STΔorf18) from S. rochei NBRC12908 | This study |
| pKU1016-orf11 | pKU1016 carrying the orf11 gene from the BD-12-biosynthetic gene cluster | This study |
Apmr, apramycin resistant; Neor, neomycin resistant; Cmr, chloramphenicol resistant; Ampr, ampicillin resistant.
Draft genome sequencing of S. luteocolor NBRC 13826.
Draft genome sequences of S. luteocolor NBRC 13826 were determined by using a MiSeq desktop sequencer (Illumina, San Diego, CA, USA). A genomic library with an insert size of 500 bp was constructed by using a Nextera XT DNA Sample Prep kit (Illumina). The sequencing run yielded 19,156,126 reads (accession no. DRA003165), with 300-bp paired-end sequences, providing over 600-fold genome coverage. The genome was assembled using a GS de novo Assembler (Newbler) version 2.7 (454 Life Sciences, Branford, CT, USA). The final assembly consisted of 262 scaffolds containing 9,298,666 bp, with a G+C content of 71.5% and an N50 scaffold size of 56,991 bp. Gene prediction was carried out with MetaGeneAnnotator (23) to identify the BD-12-biosynthetic gene cluster (accession no. LC122485).
Cloning of the BD-12-biosynthetic gene cluster.
To clone the putative BD-12-biosynthetic gene cluster, the bacterial artificial chromosome (BAC) genomic library of S. luteocolor NBRC 13826 was prepared according to a method previously reported (24). Clones carrying the entire gene cluster for BD-12 biosynthesis were screened by PCR using two pairs of primers corresponding to the upstream and downstream regions of the gene cluster, ORF1_F plus ORF1_R and ORF26-ORF27_F plus ORF26-ORF27_R, respectively (Table 2). In the PCR screening, we found one of the BAC clones, pKU518BD12P2-C23 (inset size, 200,999 bp), which contained the putative BD-12-biosynthetic gene cluster (34 kbp) (Table 1 and Fig. 2).
TABLE 2.
Oligonucleotides used in this study
The 34-kbp DNA fragment was further subcloned from pKU518BD12P2-C23 by an in vivo gene replacement mediated by λ-Red recombinase. Approximately 500-bp homologous regions upstream and downstream of the gene cluster were amplified by PCR with pKU518BD12P2-C23 as a template using two primer pairs, BD-12cluster_up_SpeI_F plus BD-12cluster_up_BsrGI_R and BD-12cluster_down_BsrGI_F plus BD-12cluster_down_NheI_R, respectively (Table 2). The two amplified fragments were ligated with pRED-sacB-aph (Table 1) (22). The resulting plasmid, BD12cluster-up-down/pRED, was linearized by PCR using the primer pair BD-12cluster_down_BsrGI_F and BD-12cluster_up_BsrGI_R (Table 2). The amplified fragment was mixed with the BAC clone, pKU518BD12P2-C23, that had been linearized by digestion with BsrGI. The mixture was introduced into Escherichia coli BW25113 carrying pKD46 (http://cgsc.biology.yale.edu/), in which λ-Red recombinase had been expressed, to generate a circular plasmid (pRED/BD-12cluster) (Table 1) that carried the 34-kbp fragment of the putative BD-12-biosynthetic gene cluster. After the digestion of pRED/BD-12cluster with SpeI and NheI, a 34-kbp DNA fragment was obtained and cloned into a Streptomyces integrating vector, pKU493A_aac(3)IV, to generate pKU493A-BD12 (Table 1).
Heterologous expression of the BD-12-biosynthetic gene cluster in S. lividans TK23 and S. avermitilis SUKA17.
The constructed integration vector, pKU493-BD12, was introduced into two heterologous host strains, S. lividans TK23 and S. avermitilis SUKA17, by standard procedures (24, 25). The S. lividans TK23 transformant harboring pKU493-BD12 was cultured in S10.3 medium (20) for 2 days at 28°C. The S. avermitilis SUKA17 transformant harboring pKU493-BD12 was cultured in AVM medium containing 6% (wt/vol) glucose, 0.2% (wt/vol) yeast extract (Difco Laboratories, Franklin Lakes, NJ, USA), 0.2% (wt/vol) (NH4)SO4, 0.5% (wt/vol) CaCO3, 0.2% (wt/vol) NaCl, 0.05% (wt/vol) K2HPO4, 0.01% (wt/vol) MgSO4 · 7H2O, 0.005% (wt/vol) FeSO4 · 7H2O, 0.005% (wt/vol) ZnSO4 · 7H2O, and 0.005% (wt/vol) MnSO4 · 4H2O (pH 7.0) for 5 days at 28°C. To examine BD-12 productivity, the supernatants of the culture broth were analyzed by high-performance liquid chromatography and high-resolution electrospray ionization mass spectrometry analysis (HPLC-HR–ESI-MS) (LTQ/Orbitrap; Thermo Scientific, Waltham, MA, USA) or HPLC–ESI-MS (Esquire 4000; Bruker, Billerica, MA, USA).
Heterologous coexpression of the ST-biosynthetic gene cluster possessing in-frame deletions of the orf18 gene and the orf11 gene from the BD-12-biosynthetic gene cluster.
The ST-biosynthetic gene cluster (30 kbp) possessing an in-frame deletion of the orf18 gene (STΔorf18) of S. rochei NBRC12908, which was constructed in a previous study (20), was cloned into the pRED-sacB-aph vector using the same methods used for the construction of pRED/BD-12cluster. The STΔorf18 fragment was then cloned into the integrating vector, pKU493A_aac(3)IV, to generate pKU493A-STΔorf18 (Table 1). pKU493A-STΔorf18 was introduced into S. avermitilis SUKA17, and the resulting transformant was used for further experiments.
The orf11 gene from the BD-12-biosynthetic gene cluster was amplified by PCR using one set of primers (orf 11_BamHI_F and orf 11_HindIII_R) (Table 2). The amplified DNA fragment was ligated into a Streptomyces integrating vector, pKU1016, to yield pKU1016-orf11 (Table 1). The pKU1016-orf11 vector was then introduced into the S. avermitilis SUKA17 transformant harboring pKU493A-STΔorf18, and the resulting transformant was cultured in AVM medium for 5 days at 28°C. The supernatant of the culture broth was analyzed by HPLC–ESI-MS.
Purification of acetyl-glycylthricin.
The culture broth (1,000 ml) of the S. avermitilis SUKA17 transformant harboring pKU493A-STΔorf18 and pKU1016-orf11 was centrifuged, and the supernatant obtained was mixed with 1,000 ml of chloroform. After vigorous shaking, the aqueous layer from the centrifugation was mixed with 200 ml acetonitrile and the mixture was filtered. The filtrate was loaded onto a Dowex 50W × 4 column (100 to 200 mesh; H+ form; 4.0 by 10 cm; Dow Chemical, Midland, MI, USA), and the column was washed with 200 ml of 20% (vol/vol) acetonitrile in water. The sample was eluted in a stepwise fashion with 200 ml of 0.1, 0.15, 0.3, 0.45, and 0.6 M NaCl in 20% (vol/vol) acetonitrile. The acetyl-glycylthricin fractions eluted with 0.15, 0.3, and 0.45 M NaCl were combined. After removal of the organic solvent, the aqueous layer was lyophilized to give a white powder. This sample was dissolved in 100 ml of methanol, the insoluble materials were removed by centrifugation, and the methanol-soluble fraction was evaporated. The dried sample was dissolved in a small volume of water and fractionated by preparative HPLC using a reversed-phase column (Sunniest RP-AQUA; 5 μm; 250 by 10 mm; ChromaNik Technologies) at 40°C at a flow rate of 7 ml min−1 and with a mobile phase composed of 2% (vol/vol) acetonitrile and 0.1% (vol/vol) n-heptafluorobutyric acid (HFBA). Fractions were collected and monitored with a UV detector at 210 nm. The fraction containing acetyl-glycylthricin was lyophilized, dissolved in a small volume of 50% (vol/vol) acetonitrile in water, and fractionated by preparative HPLC using a hydrophilic-interaction liquid chromatography (HILIC) column (ZIC-HILIC; 5 μm; 250 by 10 mm; Merck, Kenilworth, NJ, USA) at 55°C at a flow rate of 8 ml min−1 and with a mobile phase composed of 77% (vol/vol) acetonitrile, 5 mM HCOONH4, and 0.1% (vol/vol) formic acid (FA). Fractions were collected and monitored with a UV detector at 210 nm. The fraction containing acetyl-glycylthricin was lyophilized to give a white powder. This sample was dissolved in a small volume of water and fractionated by preparative HPLC using a reversed-phase column under the conditions described above. The fraction containing acetyl-glycylthricin was lyophilized and dissolved in a small volume of 50% (vol/vol) acetonitrile in water. To remove the residual HFBA from the sample, the solution was passed through a Varipure IPE column (Agilent Technologies, Santa Clara, CA, USA). The sample was lyophilized to obtain the purified acetyl-glycylthricin (approximately 1.5 mg), whose chemical structure was then determined by nuclear magnetic resonance (NMR) analysis.
Overexpression and purification of rOrf11 and rOrf1-SAT.
The following two sets of PCR primers were designed and used to amplify the orf11 gene from the BD-12-biosynthetic gene cluster and the orf1 gene (named orf1-SAT in this study) (ST acetyltransferase [SAT]) from the ST-biosynthetic gene cluster: rORF11_BamHI_F and rORF11_HindIII_R for construction of N-terminally His6-tagged Orf11 (rOrf11) and S.rochei_SAT_F and S.rochei_SAT_R for N-terminally His6-tagged Orf1-SAT (rOrf1-SAT) (Table 1). The amplified fragments (orf11, 1.0 kbp; orf1-SAT, 0.7 kbp) were then ligated into pQE30 (Qiagen, Hilden, Germany). After confirmation of their sequences, the resulting plasmids, pQE30-ORF 11 and pQE30-ORF 1-SAT, were introduced into E. coli M15(pREP4) for expression as N-terminally His6-tagged fusion proteins. The recombinant enzymes were purified by standard protocols with nickel-nitriloacetic acid (Ni-NTA) Sepharose (Qiagen).
Enzyme reactions with rOrf11.
In the enzyme reaction, an E. coli T7 S30 Extract System for Circular DNA kit (Promega, Madison, WI, USA) was employed to supply aminoacyl-tRNA. A reaction mixture (200 μl) consisting of 50 mM Tris-HCl (pH 8.0), 1 mM streptothrisamine, 1 mM glycine (or 0.5 mM each proteinogenic amino acid to investigate the substrate specificity for aminoacyl-tRNAs), 20 μl S30 premix, 15 μl S30 extract, and 100 μg ml−1 rOrf11 was incubated at 37°C for 3 h. In addition, the enzyme reaction was carried out with or without RNase (10 μg ml−1). The reaction mixture was mixed with 200 μl chloroform. After vigorous shaking, the aqueous layer from centrifugation was analyzed by HPLC–ESI-MS.
Enzyme reactions with rOrf1-SAT.
A reaction mixture (200 μl) consisting of 100 mM Tris-HCl (pH 8.0), 1 mM ST-F, 1 mM acetyl-coenzyme A (CoA), and 100 μg ml−1 rOrf1-SAT was incubated at 30°C for 2 h. The reaction mixture was mixed with 200 μl chloroform. After vigorous shaking, the aqueous layer from centrifugation was analyzed by HPLC–ESI-MS. To confirm the acetylation of glycylthricin, 1 mM acetyl-CoA and 100 μg ml−1 rOrf1-SAT were added to the reaction mixture with rOrf11. After the incubation at 30°C for 2 h, the reaction mixture was analyzed by HPLC–ESI-MS.
Nucleotide sequence accession numbers.
The sequence data for the draft genome and the BD-12- biosynthetic gene cluster of S. luteocolor NBRC 13826 were deposited in GenBank/DDBJ under Sequence Read Archive (SRA) accession number DRA003165 and GenBank accession number LC122485, respectively.
RESULTS
Identification of the BD-12-biosynthetic gene cluster from S. luteocolor NBRC13826.
Recently, two research groups reported on genes responsible for biosynthesis of the streptolidine lactam and amino sugar moieties in ST (26, 27). The orfP and orfR genes from the ST producer Streptomyces lavendulae BCRC 12163 were found to participate in the early steps of the streptolidine lactam biosynthesis (26). The stnG, stnI, stnJ, and stnQ genes from the ST producer Streptomyces sp. strain TP-A0356 were involved in the amino sugar moiety of the ST biosynthesis (27). Their homologues also exist in the ST-biosynthetic gene cluster from S. rochei NBRC12908 (Fig. 2) (20). In the draft genome sequence of the BD-12 producer S. luteocolor NBRC13826 (accession no. DRA003165), we identified an ST-biosynthetic homologous gene cluster (34 kbp; accession no. LC122485) (Fig. 2). The gene cluster possessed the orfP, orfR, stnG, stnI, stnJ, and stnQ homologue genes. In addition to these genes, two methyltransferase genes (orf6 and orf13), which could be responsible for the two N-methyl groups of BD-12, were found. This suggested that the ST-biosynthetic homologous gene cluster may encode the biosynthesis of BD-12. Surprisingly, NRPS genes homologous to orf5 and orf18, which are present in the ST-biosynthetic gene cluster, were not found in this putative BD-12-biosynthetic gene cluster. This suggested that an NRPS-independent mechanism was responsible for the glycine attachment in the biosynthesis of BD-12.
Functional analysis of the BD-12-biosynthetic gene cluster by heterologous expression.
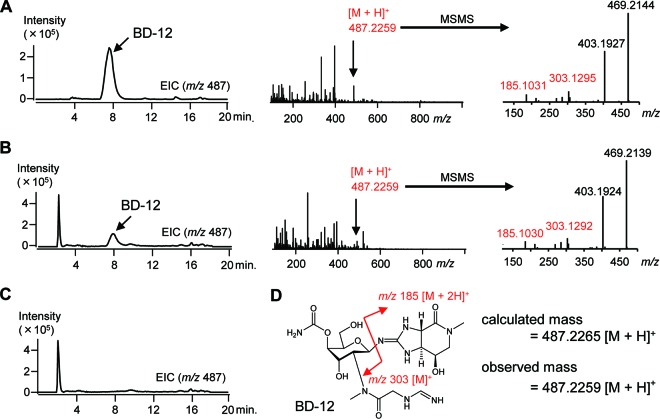
To investigate the involvement of the gene cluster (34 kbp) from NBRC13826 in BD-12 biosynthesis, we introduced a Streptomyces integrating vector carrying the 34-kbp DNA fragment (pKU493A-BD12) (Fig. 2) into the BD-12 nonproducer S. lividans TK23. The resulting transformant was cultivated, and analysis of the culture broth by HPLC-HR–ESI-MS revealed that the transformant produced BD-12 (Fig. 4), while production of BD-12 was not observed in a transformant harboring an empty vector, pKU493A_aac(3)IV (Fig. 4). Introducing pKU493A-BD12 into the versatile genome-minimized host strain S. avermitilis SUKA17 (22) also revealed BD-12, although the productivity was significantly lower (see Fig. S1 in the supplemental material). These findings clearly demonstrate that the 34-kbp DNA fragment carries a complete set of genes responsible for BD-12 biosynthesis, including a certain gene(s) involved in the peptide bond formation between the glycine (or glycine derivative) and streptothrisamine.
FIG 4.
HPLC-HR–ESI-MS analysis of BD-12 produced by the Streptomyces strains. (A to C) S. luteocolor NBRC 13826 (BD-12 producer) (A), S. lividans TK23 harboring pKU493A-BD12 (B), and S. lividans TK23 harboring the empty vector [pKU493A_aac(3)IV] (C) were cultivated, and the culture broths were analyzed by HPLC-HR–ESI-MS using a reversed-phase column (Sunniest RP-AQUA; 3 μm; 100 by 2.0 mm; ChromaNik Technologies) at 30°C at a flow rate of 0.3 ml min−1 and with an initial gradient of 8% (vol/vol) acetonitrile in water to 16% (vol/vol) acetonitrile over 11 min, which was then ramped to 40% (vol/vol) acetonitrile over 5 min. Both acetonitrile and water contained 0.05% (vol/vol) HFBA and 0.05% (vol/vol) FA. Extracted ion chromatograms (EICs) for BD-12 (m/z = 487) are shown. (D) BD-12 chemical structure with the fragmentation pattern shown in red.
Using HHpred, an online program for detecting the structural homology of hypothetical proteins (28), we searched for a candidate gene encoding a peptide-forming enzyme on the BD-12-biosynthetic gene cluster. We detected the structural homologues of the orf11 gene product (Orf11), FemA (Staphylococcus aureus) (29) and FemX (Weissella viridescens) (30), although the amino acid sequence of Orf11 showed no significant similarity to those of FemA and FemX. Fem enzymes catalyze the aminoacyl-tRNA-dependent peptide bond formation between the peptidoglycan precursor and amino acids; FemA and FemX utilize Gly-tRNAGly and Ala-tRNAAla, respectively. We thus expected that Orf11 could similarly utilize Gly-tRNAGly as a substrate for the addition of glycine to streptothrisamine via a peptide bond (Fig. 3B).
Functional analysis of the orf11 gene encoding the Fem-like enzyme.
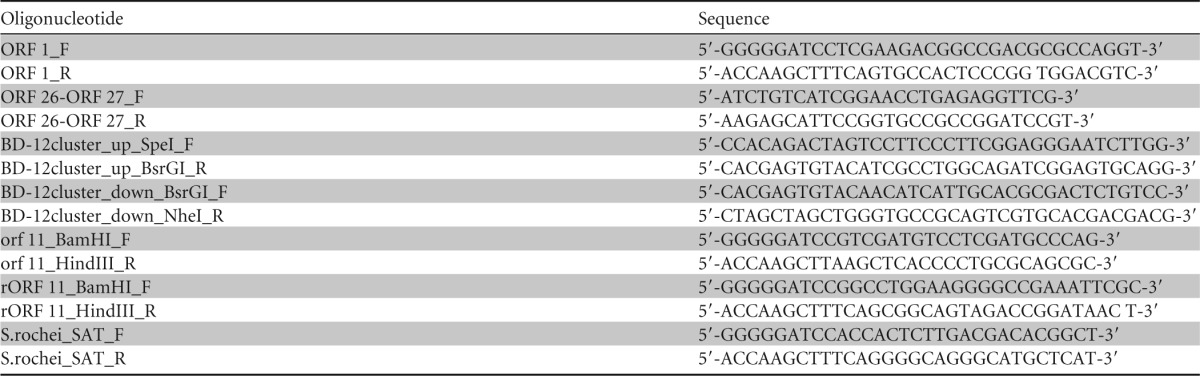
In order to directly assess the function of the orf11 gene in the BD-12-biosynthetic gene cluster, it was coexpressed with a mutated ST-biosynthetic gene cluster carrying an in-frame deletion of the orf18 gene (STΔorf18) because STΔorf18 produces streptothrisamine as the ST intermediate (20) and can provide Orf11 with streptothrisamine as a substrate. For this experiment, we constructed two integration vectors, pKU493A-STΔorf18 and pKU1016-orf11, which carried STΔorf18 and the orf11 gene under the control of the constitutive ermE promoter, respectively. We introduced these two vectors into S. avermitilis SUKA17 and analyzed its culture broth by HPLC–ESI-MS. We anticipated that the resulting transformant would produce a new compound, glycylthricin (Fig. 1), with a molecular mass of 431.3 Da (m/z = 432.3 [M + H]+). To our surprise, the transformant provided a compound with a molecular mass of 473.3 Da (m/z = 474.3 [M + H]+) (Fig. 5A) rather than the expected compound, glycylthricin. This was considered to be an orf11 gene-dependent compound, because a transformant harboring pKU493A-STΔorf18 and a pKU1016 empty vector produced no such compound (data not shown). The mass difference (42 Da) between the detected compound and our expected compound (glycylthricin) suggested that the detected compound was an acetylated derivative of glycylthricin. NMR analysis of the detected compound purified from the culture broth showed that it was indeed the acetylated form of glycylthricin (Table 3), and the compound was therefore designated acetyl-glycylthricin (Fig. 1).
FIG 5.
Functional analysis of Orf11 in vivo and in vitro. EICs and tandem mass spectrometry of the compounds are shown on the left and right, respectively. (A) The compound produced by the S. avermitilis SUKA17 strain harboring pKU493A-STΔorf18 and pKU1016-orf11 was analyzed by HPLC–ESI-MS. (B) rOrf11 was incubated with Gly-tRNAGly and streptothrisamine. (C) rOrf11 was incubated with RNase, Gly-tRNAGly, and streptothrisamine. (D) rOrf11 was incubated with Ala-tRNAAla and streptothrisamine. (E) The rOrf11 product (glycylthricin) was incubated with rOrf1-SAT and acetyl-CoA. These samples were analyzed by HPLC–ESI-MS using a reversed-phase column (Sunniest RP-AQUA; 3 μm; 100 by 2.0 mm) at 30°C at a flow rate of 0.3 ml min−1 and with a gradient of acetonitrile-water in 0.05% (vol/vol) HFBA and 0.05% (vol/vol) FA run over 15 min (3% [vol/vol] acetonitrile for 3 min, 3 to 15% [vol/vol] acetonitrile for 2 min, and 15% [vol/vol] for 10 min). EICs for acetyl-glycylthricin (m/z = 474.3) and glycylthricin (m/z = 432.3) are shown.
TABLE 3.
Assignments of 13C and 1H NMR data for acetyl-glycylthricina
| Position | δC | δH (multiplicity; J in Hz) | 1H-1H COSY | 1H-13C HMBC |
|---|---|---|---|---|
| 1 | 173.0 | |||
| 2 | 56.3 | 4.31 (d, 14.1) | 3 | 1, 4 |
| 3 | 52.2 | 4.35 (dd, 14.1, 2.2) | 2 | |
| 4 | 62.1 | 4.62 (ddd, 5.6, 2.2, 1.2) | 5 | 2 |
| 5 | 49.3 | 3.78 (dd, 14.4, 5.6) | 4 | 1 |
| 3.36 (dd, 14.4, 1.2) | 1 | |||
| 6 | 164.2 | |||
| 7 | 79.8 | 5.12 (d, 9.6) | 8 | 6, 11 |
| 8 | 50.1 | 4.13 (dd, 9.6, 2.9) | 7 | 10, 14 |
| 9 | 66.9 | 4.12 (t, 2.9) | 10 | 8, 11 |
| 10 | 70.4 | 4.75 (dd, 3.5, 2.9) | 9 | 8, 13 |
| 11 | 73.4 | 4.26 (dt, 6.5, 3.5) | 12 | 7 |
| 12 | 60.5 | 3.70 (d, 6.5) | 11 | |
| 13 | 158.2 | |||
| 14 | 171.6 | |||
| 15 | 42.6 | 3.92 (s) | 14, 16 | |
| 16 | 175.0 | |||
| 17 | 21.8 | 2.06 (s) | 16 |
NMR assignment of acetyl-glycylthricin. 1H and 13C NMR spectra were recorded at 600 and 150 MHz, respectively, using the Varian NMR system 600 NB CL (Varian, Palo Alto, CA, USA). One- and two-dimensional experiments (double-quantum-filtered correlation spectroscopy [DQF-COSY] and constant-time heteronuclear multiple-bond connectivity [CT-HMBC]) were performed at ambient temperature, and the correlation positions are shown. The samples were dissolved in D2O, and the solvent peak was used as an internal standard (δH, 4.80 ppm).
In vitro analysis of Orf11.
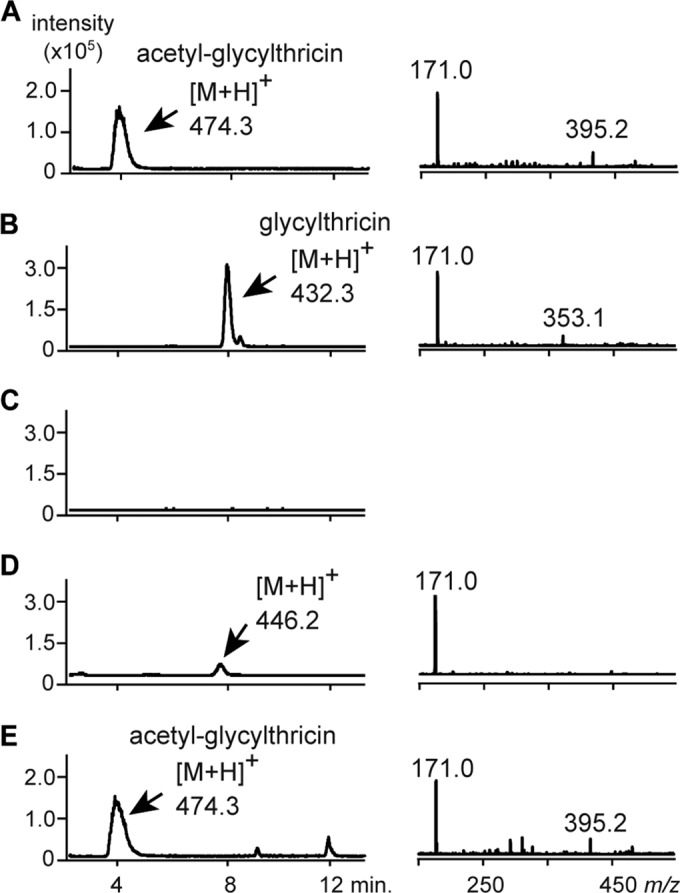
To gain a better understanding of the enzymatic function and substrate of Orf11, we performed an enzyme reaction using N-terminally His6-tagged Orf11 (rOrf11) produced as a homodimer in E. coli (Fig. 6). Streptothrisamine was incubated with rOrf11 and Gly-tRNAGly, and the reaction product was shown to be the expected compound, glycylthricin, based on HPLC–ESI-MS analysis (Fig. 5B). On the other hand, when RNase was added to the enzyme reaction, glycylthricin was not produced (Fig. 5C), demonstrating that Orf11 catalyzes the aminoacyl transfer reaction in a tRNA-dependent manner. Moreover, the investigation of substrate specificity suggested that rOrf11 can accept Ala-tRNAAla as a substrate in vitro, although the productivity of the reaction product with a molecular mass of 445.2 Da (m/z = 446.2 [M + H]+) was much lower than that of the reaction using Gly-tRNAGly (Fig. 5D).
FIG 6.
Enzymatic characterization of rOrf11. Purified rOrf11 was subjected to SDS-PAGE. Proteins were stained with CBB R-250. The relative molecular mass of rOrf11 was estimated by gel filtration chromatography (SunSec Diol; 4 mm; 300 by 4.6 mm; ChromaNik Technologies, Osaka, Japan). Glutamate dehydrogenase (290 kDa), lactate dehydrogenase (142 kDa), enolase (67 kDa), myokinase (32 kDa), and cytochrome c (12.4 kDa) were used as the standard molecular masses.
Identification of the acetyltransferase gene responsible for acetyl-glycylthricin production.
Considering that Orf11 accepts Gly-tRNAGly as a substrate, glycylthricin should be converted into acetyl-glycylthricin by an acetyltransferase in the coexpression experiment described above. In the ST-biosynthetic gene cluster from S. rochei NBRC12908, we identified the orf1 gene as a putative ST self-resistance gene that encodes an ST acetyltransferase (SAT) (Fig. 2) (20). SATs in different ST-producing strains are well known to catalyze the acetyl-CoA-dependent acetylation at the β-amino group of the β-Lys moiety in ST-F, conferring ST resistance (31, 32). Although no reports about the substrate specificity of SATs have been published, the orf1 gene product (Orf1-SAT) was considered to be a candidate for the enzyme that acetylates glycylthricin. To confirm the function of Orf1-SAT as an acetyltransferase, we first examined the acetylation of ST-F using N-terminally His6-tagged Orf1-SAT (rOrf1-SAT) produced as a homotetramer in E. coli (see Fig. S2 in the supplemental material). HPLC–ESI-MS analysis of the reaction mixture confirmed the acetyl-CoA-dependent acetylation of ST-F (see Fig. S3 in the supplemental material). In addition, the E. coli strains expressing rOrf1-SAT showed resistance to ST-F (MIC, 250 μM), whereas a control strain not producing rOrf1-SAT was sensitive to ST-F (MIC, 8 μM), implying that the β-amino group of the β-Lys moiety in ST-F was acetylated by rOrf1-SAT. Next, glycylthricin was used as a substrate for rOrf1-SAT. Predictably, the enzyme reaction yielded a compound with a retention time corresponding to that of acetyl-glycylthricin (Fig. 5E). The tandem-MS (MS-MS) spectrum was also identical to that of acetyl-glycylthricin, confirming that rOrf1-SAT converted glycylthricin into acetyl-glycylthricin in vitro. These findings also demonstrated that glycylthricin produced in the in vivo coexpression experiment was indeed converted into acetyl-glycylthricin by the ST self-resistance gene in the ST-biosynthetic gene cluster.
Antibacterial activities of the new compounds generated in this study.
As in the case of acetylated ST-F, we detected no antibiotic activity of acetyl-glycylthricin against E. coli W3110. However, we reliably detected the antibiotic activity of glycylthricin (Fig. 7).
FIG 7.
Antibacterial activities of glycylthricin and acetyl-glycylthricin. (A) rOrf11 was incubated with Gly-tRNAGly and streptothrisamine, producing glycylthricin. (B) rOrf11 was incubated with RNase, Gly-tRNAGly, and streptothrisamine, producing no compound (control). (C) The rOrf11 product (glycylthricin) was incubated with rOrf1-SAT and acetyl-CoA, producing acetyl-glycylthricin. The antibiotic activities of the reaction mixtures (50 μl) were investigated by paper disk assay using E. coli W3110.
DISCUSSION
Through these experiments, it was demonstrated that Orf11, the FemXA-like protein, transfers glycine from Gly-tRNAGly to the amino group of streptothrisamine to produce glycylthricin (Fig. 3B). Glycylthricin is a possible intermediate in BD-12 biosynthesis, and three sequential reactions by two methyltransferases encoded by the orf6 and orf13 genes and an N-formimidoyl fortimicin A synthase homologue encoded by the orf1 gene would produce BD-12. Glycylthricin, which was enzymatically synthesized by rOrf11, was a new ST-related compound. We were unable to confirm the chemical structure of glycylthricin by NMR analysis due to the small amount produced in vitro. However, the extensive analysis with rOrf1-SAT in vitro (Fig. 5B and E) clarified the chemical structure. Glycylthricin is closely related to glycinothricin (Fig. 1), which possesses two methyl groups and exhibits antibacterial activity. Because glycylthricin also showed antibacterial activity, the methyl groups of glycinothricin are unlikely to be essential for its antibacterial activity. More importantly, this also reveals that the ε-amino group and alkyl chain of the β-Lys residue in ST-F do not play an important role in the antibacterial activity of ST-F. Thus, the identification and functional analysis of Orf11 in the BD-12 biosynthesis study not only clarified the unique biosynthetic mechanism with aminoacyl-tRNA, but also provided important information regarding the structure-activity relationship of the ST-related antibiotics.
In the substrate specificity study of rOrf11, we demonstrated that the enzyme can accept Ala-tRNAAla in addition to Gly-tRNAGly (Fig. 5D). However, we did not detect Ala-tRNAAla-derived compounds in the coexpression experiment with the orf11 gene and STΔorf18, probably due to the low productivities.
Although aminoacyl-tRNAs are normally dedicated to protein biosynthesis, a few microbial FemXA-like proteins have been reported to use aminoacyl-tRNAs and to catalyze peptide bond formation in natural-product biosynthesis. Ser-tRNASer is used for the biosynthesis of valanimycin (33), while PacB transfers l-alanine from l-Ala-tRNAAla to the N terminus of a tetrapeptide intermediate during biosynthesis of pacidamycin (34). In the biosynthesis of dehydrophos, DphH and DphK both possess FemX domains at their C termini and utilize Leu-tRNALeu and Gly-tRNAGly, respectively, to form the peptide structure of dehydrophos (35). It was very recently demonstrated that, in the human pathogen Pseudomonas aeruginosa, tRNA-dependent aminoacylation of the polar head group of phosphatidylglycerol with alanine or lysine is catalyzed by FemX-like proteins to confer resistance to antibiotics (36). In addition, tRNA-dependent cyclodipeptide synthase (37) and tRNA-dependent lantibiotic dehydratase (NisB) (38) were reported, although they lack FemXA-like features. Although reports of amide bond-forming tRNA-dependent enzymes have been forthcoming, to our knowledge, Orf11 is the first characterized tRNA-dependent aminoacylating enzyme that can accept an amino sugar as a substrate.
In conclusion, we clarified that the tRNA-dependent peptide bond-forming enzyme Orf11 mediates the condensation reaction between streptothrisamine and glycine in the biosynthesis of BD-12, whereas the biosynthesis of ST-F employs NRPS machinery. Thus, the structural diversity of the amino acid side chains of the ST-type compounds occurring in nature is generated by two distinct peptide bond-forming mechanisms. STs and ST-related antibiotics are not currently used therapeutically, because they are toxic to both prokaryotes and eukaryotes. In order to engineer new ST-related compounds displaying bacterium-specific activities, genetic modification of these peptide bond-forming machineries would be advantageous.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00725-16.
REFERENCES
- 1.Waksman SA. 1943. Production and activity of streptothricin. J Bacteriol 46:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haupt I, Huebener R, Thrum H. 1978. Streptothricin F, an inhibitor of protein synthesis with miscoding activity. J Antibiot 31:1137–1142. doi: 10.7164/antibiotics.31.1137. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein AL, McCusker JH. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553. [DOI] [PubMed] [Google Scholar]
- 4.Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM. 2005. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 22:1013–1019. doi: 10.1002/yea.1291. [DOI] [PubMed] [Google Scholar]
- 5.Shen J, Guo W, Kohler JR. 2005. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect Immun 73:1239–1242. doi: 10.1128/IAI.73.2.1239-1242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Idnurm A, Reedy JL, Nussbaum JC, Heitman J. 2004. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot Cell 3:420–429. doi: 10.1128/EC.3.2.420-429.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi PB, Webb JR, Davies JE, McMaster WR. 1995. The gene encoding streptothricin acetyltransferase (sat) as a selectable marker for Leishmania expression vectors. Gene 156:145–149. doi: 10.1016/0378-1119(95)00042-5. [DOI] [PubMed] [Google Scholar]
- 8.Takemoto T, Inamori Y, Kato Y, Kubo M, Morimoto K, Morisaka K, Sakai M, Sawada Y, Taniyama H. 1980. Physiological activity of streptothricin antibiotics. Chem Pharm Bull 28:2884–2891. doi: 10.1248/cpb.28.2884. [DOI] [PubMed] [Google Scholar]
- 9.Chamberlain DA, Brettell RIS, Last DI, Witrzens B, McElroy D, Dolferus R, Dennis ES. 1994. The use of the Emu promoter with antibiotic and herbicide resistance genes for the selection of transgenic wheat callus and rice plants. Aust J Plant Physiol 21:95–112. doi: 10.1071/PP9940095. [DOI] [Google Scholar]
- 10.Inamori Y, Sunagawa S, Tsuruga M, Sawada Y, Taniyama H, Saito G, Daigo K. 1978. Toxicological approaches to streptothricin antibiotics. I. Implications of delayed toxicity in mice. Chem Pharm Bull 26:1147–1152. [DOI] [PubMed] [Google Scholar]
- 11.Inamori Y, Morimoto K, Morisaka K, Saito G, Sawada Y, Taniyama H, Matsuda H. 1979. Toxicological approaches to streptothricin antibiotics. II. The developmental mechanism of delayed toxicity in mice and rats. Chem Pharm Bull 27:230–234. [DOI] [PubMed] [Google Scholar]
- 12.Inamori Y, Kato Y, Morimoto K, Morisaka K, Saito G, Sawada Y, Taniyama H. 1979. Toxicological approaches to streptothricin antibiotics. III. Biological studies on delayed toxicity of streptothricin antibiotics in rats. Chem Pharm Bull 27:2570–2576. [DOI] [PubMed] [Google Scholar]
- 13.Kato Y, Kubo M, Morimoto K, Saito G, Morisaka K, Inamori Y, Hama I, Maekawa T, Mazaki H, Ishimasa T, Sawada Y, Taniyama H. 1981. Toxicological approaches to streptothricin antibiotics. IV. Toxicity of streptothricin antibiotics to the blood. Chem Pharm Bull 29:580–584. [DOI] [PubMed] [Google Scholar]
- 14.Furumai T, Kaneko K, Matsuzawa N, Sato M, Okuda T. 1968. New basic water-soluble antibiotics BD-12 and BY-81. I. Taxonomy of the producing organisms and antibiotic production. J Antibiot 21:283–289. [DOI] [PubMed] [Google Scholar]
- 15.Ito Y, Ohashi Y, Sakurai Y, Sakurazawa M, Yoshida H, Awataguchi S, Okuda T. 1968. New basic water-soluble antibiotics BD-12 and BY-81. II. Isolation, purification and properties. J Antibiot 21:307–312. [DOI] [PubMed] [Google Scholar]
- 16.Kusakabe Y, Yamauchi Y, Nagatsu C, Abe H, Akasaki K. 1969. Citromycin, a new antibiotic. I. Isolation and characterization. J Antibiot 22:112–118. [DOI] [PubMed] [Google Scholar]
- 17.Taniyama H, Sawada Y. 1971. The identity of citromycin with LL-AC541, E-749-C, and BY-81. J Antibiot 24:708–710. doi: 10.7164/antibiotics.24.708. [DOI] [PubMed] [Google Scholar]
- 18.Sawada Y, Kawakami S, Taniyama H. 1977. Glycinothricin, a new streptothricin-class antibiotic from Streptomyces griseus. J Antibiot 30:460–467. doi: 10.7164/antibiotics.30.460. [DOI] [PubMed] [Google Scholar]
- 19.Kido Y, Furuie T, Suzuki K, Sakamoto K, Yokoyama Y, Uyeda M, Kinjyo J, Yahara S, Nohara T, Shibata M. 1987. A streptothricin-like antibiotic mixture, A-269A (and A-269A′). J Antibiot 40:1698–1706. doi: 10.7164/antibiotics.40.1698. [DOI] [PubMed] [Google Scholar]
- 20.Maruyama C, Toyoda J, Kato Y, Izumikawa M, Takagi M, Shin-ya K, Katano H, Utagawa T, Hamano Y. 2012. A stand-alone adenylation domain forms amide bonds in streptothricin biosynthesis. Nat Chem Biol 8:791–797. doi: 10.1038/nchembio.1040. [DOI] [PubMed] [Google Scholar]
- 21.Marahiel MA, Stachelhaus T, Mootz HD. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev 97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 22.Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. 2010. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci U S A 107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noguchi H, Taniguchi T, Itoh T. 2008. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res 15:387–396. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komatsu M, Komatsu K, Koiwai H, Yamada Y, Kozone I, Izumikawa M, Hashimoto J, Takagi M, Omura S, Shin-ya K, Cane DE, Ikeda H. 2013. Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Synth Biol 2:384–396. doi: 10.1021/sb3001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical streptomyces genetics. John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 26.Chang CY, Lyu SY, Liu YC, Hsu NS, Wu CC, Tang CF, Lin KH, Ho JY, Wu CJ, Tsai MD, Li TL. 2014. Biosynthesis of streptolidine involved two unexpected intermediates produced by a dihydroxylase and a cyclase through unusual mechanisms. Angew Chem Int Ed Engl 53:1943–1948. doi: 10.1002/anie.201307989. [DOI] [PubMed] [Google Scholar]
- 27.Guo Z, Li J, Qin H, Wang M, Lv X, Li X, Chen Y. 2015. Biosynthesis of the carbamoylated d-gulosamine moiety of streptothricins: involvement of a guanidino-N-glycosyltransferase and an N-acetyl-d-gulosamine deacetylase. Angew Chem Int Ed Engl 54:5175–5178. doi: 10.1002/anie.201412190. [DOI] [PubMed] [Google Scholar]
- 28.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benson TE, Prince DB, Mutchler VT, Curry KA, Ho AM, Sarver RW, Hagadorn JC, Choi GH, Garlick RL. 2002. X-ray crystal structure of Staphylococcus aureus FemA. Structure 10:1107–1115. doi: 10.1016/S0969-2126(02)00807-9. [DOI] [PubMed] [Google Scholar]
- 30.Biarrotte-Sorin S, Maillard AP, Delettre J, Sougakoff W, Arthur M, Mayer C. 2004. Crystal structures of Weissella viridescens FemX and its complex with UDP-MurNAc-pentapeptide: insights into FemABX family substrates recognition. Structure 12:257–267. [DOI] [PubMed] [Google Scholar]
- 31.Horinouchi S, Furuya K, Nishiyama M, Suzuki H, Beppu T. 1987. Nucleotide sequence of the streptothricin acetyltransferase gene from Streptomyces lavendulae and its expression in heterologous hosts. J Bacteriol 169:1929–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krugel H, Fiedler G, Haupt I, Sarfert E, Simon H. 1988. Analysis of the nourseothricin-resistance gene (nat) of Streptomyces noursei. Gene 62:209–217. doi: 10.1016/0378-1119(88)90559-8. [DOI] [PubMed] [Google Scholar]
- 33.Garg RP, Qian XL, Alemany LB, Moran S, Parry RJ. 2008. Investigations of valanimycin biosynthesis: elucidation of the role of seryl-tRNA. Proc Natl Acad Sci U S A 105:6543–6547. doi: 10.1073/pnas.0708957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Ntai I, Kelleher NL, Walsh CT. 2011. tRNA-dependent peptide bond formation by the transferase PacB in biosynthesis of the pacidamycin group of pentapeptidyl nucleoside antibiotics. Proc Natl Acad Sci U S A 108:12249–12253. doi: 10.1073/pnas.1109539108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bougioukou DJ, Mukherjee S, van der Donk WA. 2013. Revisiting the biosynthesis of dehydrophos reveals a tRNA-dependent pathway. Proc Natl Acad Sci U S A 110:10952–10957. doi: 10.1073/pnas.1303568110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hebecker S, Krausze J, Hasenkampf T, Schneider J, Groenewold M, Reichelt J, Jahn D, Heinz DW, Moser J. 2015. Structures of two bacterial resistance factors mediating tRNA-dependent aminoacylation of phosphatidylglycerol with lysine or alanine. Proc Natl Acad Sci U S A 112:10691–10696. doi: 10.1073/pnas.1511167112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belin P, Moutiez M, Lautru S, Seguin J, Pernodet JL, Gondry M. 2012. The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways. Nat Prod Rep 29:961–979. doi: 10.1039/c2np20010d. [DOI] [PubMed] [Google Scholar]
- 38.Ortega MA, Hao Y, Zhang Q, Walker MC, van der Donk WA, Nair SK. 2015. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517:509–512. doi: 10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.