ABSTRACT
Ascochyta blight, caused by the necrotrophic ascomycete Didymella pinodes, is responsible for severe losses in winter and spring pea crops. Despite different climatic conditions, epidemics on winter and spring crops are due to a single population of D. pinodes, suggesting gene flow either between the two crops or from reservoir sources during the cropping season. This should lead to similar pathogenicity characteristics in isolates sampled from the two crops. However, these hypotheses have never been formally tested. We therefore sampled a total of 520 D. pinodes strains throughout a growing season from winter and spring pea plots (WP and SP, respectively) and from winter and spring trap plants (TWP and TSP). Amplified fragment length polymorphism (AFLP) markers revealed high genetic diversity within subpopulations, whereas pathogenicity tests showed that mean aggressiveness increases over the course of an epidemic. These results support the idea that alloinoculum contributes to the carryover of epidemics between winter and spring crops and that the most aggressive isolates are selected as an epidemic progresses.
IMPORTANCE Ascochyta blight, caused by Didymella pinodes, is responsible for severe losses in pea crops. While previous studies have shown that ascochyta blight epidemics on winter and spring crops are due to a single population of D. pinodes, suggesting that isolates from the two crops present similar pathogenicity characteristics, that hypothesis have never been tested. Genetic analysis of subpopulations sampled throughout a growing season from winter and spring pea plots revealed high genetic diversity within subpopulations, whereas pathogenicity tests showed that mean aggressiveness increases over the course of an epidemic.
INTRODUCTION
The spatiotemporal dynamics of crop diseases are simultaneously impacted by pathogens, host plants, the environment, and human activities (1, 2). Indeed, whether or not hosts and pathogens interact is determined largely by spatial and temporal components of host and pathogen life history traits (3, 4). These interactions can thus be conceptualized as a continuous sequence of biological cycles, including dormancy, growth, reproduction, dispersal, and pathogenesis (5).
Gene flow, resulting from pathogen reproduction and dispersal, can drastically increase the extent to which pathogen epidemics spread across a landscape (6). As such, it is a main factor in the transmission of disease to previously uninfected areas and drives the spatial structure of pathogen populations in fragmented landscapes by influencing the long-term survival and genetic composition of populations (7–9). Individual dispersal events, occurring over periods of days or weeks during both the cropping and intercropping seasons (2), originate from a large number of potential inoculum sources: resting structures in soil (mycelium, oospores, chlamydospores, or sclerotia), infested stubble left on the soil surface, infested seed, and alternative hosts (wild or cultivated plants, including volunteers). The degree of connectivity among host populations is thus likely to influence spatial patterns of disease occurrence and persistence (10–14). Dispersal events also shape the structure and changes of population genetic variability. The role of gene flow within and among plant pathogen populations is still insufficiently characterized but is crucial to understanding of the distribution of alleles conferring virulence or fungicide resistance within populations. Disease dispersal can favor contact between wild and cultivated areas, or between different cultivated areas, during the cropping and the intercropping season, and thus, gene flow between these compartments can influence the genetic structure of populations. By mixing populations initially subjected to different selection pressures, gene flow can also negate the effect of local selection on adaptation and can result in local or general maladaptation (15).
Reproduction is the other important factor that impacts the genetic structure of plant pathogens (16, 17). The life cycles of many fungal plant pathogens alternate between asexual multiplication and episodes of sexual reproduction, but their relative importance differs both within and among species (18). Asexual multiplication rapidly increases the size of populations. Strains with the best reproductive success are amplified and tend to decrease the overall population genotypic diversity in time (9, 19). For instance, in the case of grapevine powdery mildew, a positive relationship between spatial and genetic distances shows that epidemics result from the spread of clones within a crop (20). Besides, genotypic diversity may be increased with genetic mixing by sexual reproduction (18, 21). For the potato late blight pathogen Phytophthora infestans, clonal reproduction has been reported in populations with restricted levels of genotypic diversity from the United Kingdom (22) and France (15, 23), whereas sexual reproduction has been reported in populations with high genotypic diversity from Nordic countries (21, 24).
Despite recent advances, the role of gene flow (and the roles of its two main triggers, reproduction and dispersal) in the local evolution of pathogen populations is still poorly assessed. This is especially the case for epidemics occurring on both winter and spring genotypes of the same host. This situation raises specific questions, particularly regarding the respective roles of autoinoculum and alloinoculum (the inoculum fraction produced by a source exogenous to the plot [25]) in the epidemic process and in pathogenicity changes within populations. To address these questions, we analyzed the phenotypic and genotypic diversity in populations of an aggressive foliar pathogen of peas, Didymella pinodes, along the course of an epidemic. D. pinodes is a necrotrophic, polyphagous, polycyclic, homothallic fungus that causes ascochyta blight on winter and spring pea fields worldwide (26, 27). This pathogen is known to present individuals differing widely in their abilities to cause disease on peas (28–32). In contrast to many necrotrophic fungi, D. pinodes can simultaneously develop its anamorph (asexual) and teleomorph (sexual) forms on the same plant during the growing season (32, 33). Moreover, due to the indeterminate growth of the pea crop, both pycnidia (asexual) and pseudothecia (sexual) can be observed simultaneously on the same plant organs. Whereas pycnidia are produced on both green and senescent plant organs, pseudothecia appear only on the senescent parts (33). This fungal pathogen has various ways of persisting in the environment and dispersing over more or less long distances (splashing, wind) (27, 34, 35). These features are more important for D. pinodes than for pathogens whose modes of reproduction are dissociated, since they help to increase population diversity and have an impact on pathogenicity. A recent study (32) showed that epidemics on winter and spring pea crops are due to a single D. pinodes population. Despite the long intercropping season, D. pinodes populations from winter and spring pea crops display high, but similar, genetic variability. In contrast to other natural plant-pathogen associations, a high rate of annual extinction is not observed (36–40). Population turnover is thus likely to be driven by both local selection and demographic stochasticity due to seasonal population growth and decline. During the spring, abundant spore production (41) leads to high colonization rates. The fact that winter and spring pea crops are attacked by the same populations of D. pinodes suggests that, as observed by Laine and Hanski (42) for another ascomycete plant pathogen, regional persistence of D. pinodes populations could occur at the scale of metapopulations, consisting of many coupled populations.
The origin and availability of inoculum sources during the cropping season are still questionable. Do D. pinodes populations colonizing winter pea crops constitute the only inoculum sources for the spring pea crops? Does their apparent genetic similarity translate into similar pathogenicity levels? This study, designed to address these questions, is based on a collection of D. pinodes isolates sampled throughout a single cropping season (January to June) on both winter pea (WP) and spring pea (SP) crops, but also on winter and spring trap plants (TWP and TSP, respectively) catching the alloinoculum. The specific objectives of this work were (i) to determine if and how D. pinodes aggressiveness evolves during the cropping season, (ii) to determine if D. pinodes populations that developed on winter pea crops constitute the main inoculum source for spring pea crops, and (iii) to analyze the relationships between crop populations and alloinoculum caught on trap plants.
MATERIALS AND METHODS
Field experiments.
Two field experiments were carried out at the INRA experimental station of Le Rheu, western France (48°06.00′N, 1°48.00′W; 30 m above mean sea level) in 2004–2005. The two fields, separated by >3 km, shared similar pedoclimatic environments and were free from soilborne inoculum, due to a rotation without a pea crop during the five previous years. These fields were, respectively, sown with a winter (Cheyenne; GAE Recherche, France) and a spring (Baccara; Florimond-Desprez, France) pea cultivar. The winter cultivar was sown on 25 October 2004, and the spring crop was sown on 24 February 2005. For both crops, the experimental design consisted of a 30-m2 (3-m-wide by 10-m-long) plot divided into 30 microplots (1 m2 each).
Trap plants.
Alloinoculum, defined as the inoculum fraction produced by a source exogenous to the plot (25), was assessed through trap plants. Each week from mid-January to the end of June 2005, one tray containing 20 trap plants (Cheyenne or Baccara seedlings at the 5-leaf stage for the winter or spring trial, respectively) was placed at each of the four corners of the plot. After 7 days of exposure in the field, trap trays were brought back and were incubated in a dew chamber (12-h photoperiod; 20°C; 100% relative humidity) for 4 days. Deposition of viable ascospores on trap plants was assessed by isolating strains from the small, purple-black, irregular flecks on the five lower stipules of the plants after incubation (41).
Spatiotemporal disease development.
Disease started in each crop without an artificial supply of inoculum. Disease severity was assessed visually once a month in the different microplots from January to June. To determine disease development at the plot level, one plant was randomly collected from each microplot on different sampling dates (five sampling dates for the winter pea crop and three sampling dates for the spring pea crop). Disease was scored on each stipule of the sampled plants using the adapted disease scale of Tivoli (41, 43), and a mean disease score (Di) was calculated. Disease distribution maps were created for each sampling date using ArcMap software and a deterministic method based on estimation of the weighted inverse distance (44).
Fungal strains.
A total of 520 strains of D. pinodes collected from infected field pea plants and trap plants were used in this study. Each field was divided into 30 microplots where one plant was randomly sampled on each sampling date. One hundred fifty strains were sampled from winter pea (WP) crops during the winter cropping season (February to May). Ninety strains were sampled from spring pea (SP) crops during the spring cropping season (May and June). Two hundred fifteen strains were sampled from trap plants placed at the corners of the winter pea plots (TWP), and 66 strains were sampled from trap plants placed at the corners of the spring pea plots (TSP). For each population (WP, SP, TWP, and TSP), the different subpopulations correspond to the different sampling dates (Table 1). For isolation, approximately 5 mm2 of diseased leaf tissue was surface-sterilized for 1 min in 70% ethanol, rinsed three times in sterile water, placed on sterile filter paper to remove excess water, and plated for 14 days on V8 medium (99 ml V8 vegetable juice [Campbell, France], 35 g agar, and 801 ml distilled water, autoclaved at 105°C for 30 min) distributed in petri dishes. Pycnidiospores from the resulting cultures were spread on 2% malt agar and were incubated for 12 h as described by Onfroy et al. (45). Single germinating pycnidiospores were transferred to fresh V8 plates under a dissecting microscope, and cultures were incubated at 20°C with a 12-h photoperiod under cool white fluorescent lamps. Single-spore cultures were then maintained on malt slants and were stored in the dark at 4°C.
TABLE 1.
Genetic characterization of Didymella pinodes strains sampled on winter or spring pea plots and on winter or spring trap plants
| Sourcea | Subpopulation | Sample date (day/mo) | n | Nab | Ic | uhd | H′ |
|---|---|---|---|---|---|---|---|
| WP | WP7 | 17 February | 30 | 1.97 | 0.51 | 0.35 | 0.83 |
| WP8 | 28 February | 30 | 2.00 | 0.58 | 0.41 | ||
| WP10 | 29 March | 30 | 1.96 | 0.49 | 0.34 | ||
| WP12 | 25 April | 30 | 1.99 | 0.61 | 0.43 | ||
| WP14 | 23 May | 30 | 1.99 | 0.56 | 0.39 | ||
| SP | SP1 | 30 May | 30 | 1.98 | 0.59 | 0.42 | 0.86 |
| SP2 | 13 June | 30 | 1.97 | 0.57 | 0.40 | ||
| SP3 | 27 June | 30 | 1.99 | 0.58 | 0.41 | ||
| TWP | TWP19 | 28 January | 13 | 1.83 | 0.50 | 0.37 | 0.70 |
| TWP20 | 4 February | 14 | 1.77 | 0.43 | 0.31 | ||
| TWP21 | 11 February | 5 | 1.06 | 0.18 | 0.15 | ||
| TWP22 | 18 February | 15 | 1.94 | 0.55 | 0.40 | ||
| TWP23 | 25 February | 13 | 1.81 | 0.44 | 0.31 | ||
| TWP24 | 4 March | 12 | 1.75 | 0.46 | 0.34 | ||
| TWP28 | 1 April | 15 | 1.82 | 0.48 | 0.35 | ||
| TWP29 | 8 April | 15 | 1.88 | 0.53 | 0.39 | ||
| TWP30 | 15 April | 14 | 1.82 | 0.48 | 0.35 | ||
| TWP31 | 22 April | 15 | 1.79 | 0.39 | 0.28 | ||
| TWP32 | 29 April | 15 | 1.48 | 0.28 | 0.20 | ||
| TWP33 | 5 May | 13 | 1.86 | 0.48 | 0.35 | ||
| TWP34 | 16 May | 12 | 1.49 | 0.31 | 0.23 | ||
| TWP35 | 20 May | 13 | 1.85 | 0.52 | 0.38 | ||
| TWP36 | 27 May | 15 | 1.41 | 0.28 | 0.20 | ||
| TWP37 | 3 June | 15 | 1.86 | 0.47 | 0.34 | ||
| TSP | TSP2 | 20 May | 4 | 1.06 | 0.25 | 0.23 | 0.76 |
| TSP3 | 27 May | 9 | 1.86 | 0.51 | 0.39 | ||
| TSP4 | 3 June | 10 | 1.84 | 0.49 | 0.37 | ||
| TSP5 | 10 June | 14 | 1.84 | 0.48 | 0.35 | ||
| TSP6 | 20 June | 14 | 1.83 | 0.46 | 0.33 | ||
| TSP7 | 27 June | 15 | 1.78 | 0.45 | 0.33 |
WP, winter pea plots; SP, spring pea plots; TWP, winter trap plants; TSP, spring trap plants.
Na, number of different alleles.
I, Shannon's information index, calculated as −1∑{[p × ln(p)] + [q × ln(q)]}, where p is the band frequency and q is 1 − p (GenAlEx, version 6.5).
h is diversity, calculated as 1 − (p2 + q2); uh is unbiased diversity, calculated as [N/(N − 1)] × h, where N is the total number of samples.
DNA extraction and AFLP typing.
The single strains were grown in 75 ml of LP liquid medium (10 g tryptone, 5 g extract of yeast powder, 5 g NaCl, 1 liter distilled water; autoclaved at 115°C for 20 min) supplemented with streptomycin (1.5 g) and penicillin (0.75 g). Inoculated vials were incubated, under agitation, for 14 days at 20°C with a 12-h photoperiod under cool white fluorescent lamps. Mycelia were harvested by vacuum filtration through two layers of sterilized Miracloth (Calbiochem, CN Biosciences, Inc., La Jolla, CA), rinsed twice in sterile water, and stored at −80°C until lyophilized. DNA was extracted from lyophilized mycelium as described by Lodhi et al. (46), quantified by measuring the optical densities of extracts at 260 and 280 nm with a NanoDrop 1000 spectrophotometer (Thermo Scientific), and adjusted to a final concentration of 100 ng · liter−1 for amplified fragment length polymorphism (AFLP) analysis. AFLP analysis was carried out as described by Vos et al. (47) with modifications used by Le May et al. (32). AFLP reactions were performed independently three times, using the same set of primers with reference strains and a random sample of 10 isolates from the collection, as well as independent DNA preparations of the same strains, to estimate the repeatability of fragment scoring.
AFLP analysis.
The raw data were analyzed with GeneMapper (version 3.5; Applied Biosystems). The presence and absence of all fragments between 100 and 400 bp were scored in each of the 520 strains. Bands with molecular sizes exceeding 400 bp were not scored because of insufficient resolution. The data set obtained was based on the assumption that bands of the same molecular weight were identical.
(i) Genotypic diversity.
The AFLP data were used to define multilocus genotypes (MLGs) and to check for repeated MLGs, i.e., the strains sharing the same alleles at all loci, using the Microsoft Excel add-in GenAlEx, version 6.5 (48). Genotypic diversity was calculated for each population using the Shannon-Wiener index H′ (49, 50). The number of different alleles (Na), Shannon's information index (I), and unbiased diversity (uh) were computed using GenAlEx for each population and subpopulation. Clonality was assessed using the index of association (IA), the traditional measure of multilocus linkage disequilibrium, calculated with Multilocus software, version 3.1b (51). The “distance” (number of loci at which isolates differ) between all pairs of individuals is calculated, and the variance of these distances compared to that expected if there is no linkage disequilibrium is determined. IA is calculated as [(VD)/(∑varj)] − 1, where VD is the variance of the distances between two isolates over all loci (i.e., the number of loci at which the isolates differ), and varj corresponds to the variance of the mean distance (either 0 or 1) between all possible pairs of isolates [with all possible pairs calculated as n(n − 1)/2]. The higher the IA, the more clonal the population. Departure from the null hypothesis, i.e., complete panmixia, was checked by permuting alleles between individuals independently for each locus (500 permutations).
(ii) Population differentiation.
Analysis of molecular variance (AMOVA) (calculated with Arlequin software, version 3.1, hosted online by the Department of Anthropology, University of Geneva, Switzerland [52]) was used to partition molecular variance between populations, between subpopulations within populations, and within subpopulations. Pairwise FST values (variance of allele frequencies between populations) were calculated with Arlequin for each pair of subpopulations of D. pinodes and were compared within plots (SP-SP and WP-WP) or trap plants (TSP-TSP and TWP-TWP), between plots (SP-WP), and between plots and related trap plants (SP-TSP and WP-TWP).
(iii) Genetic structure.
Principal-component analysis (PCA) was performed using the procedure available in the adegenet package (53) for the statistical freeware R, version 3.1.1 (2014; The R Foundation for Statistical Computing). PCA has an important advantage over other methods, such as the Bayesian clustering algorithm implemented in STRUCTURE (54), because it does not require strong assumptions about an underlying genetic model, such as the Hardy-Weinberg equilibrium or the absence of linkage disequilibrium between loci (53).
Aggressiveness of D. pinodes strains.
Aggressiveness levels were evaluated for a random sample of D. pinodes strains sampled on winter (WP7, WP10, WP12, WP14) and spring (SP1, SP3) pea plots and of D. pinodes strains sampled on winter (TWP20, TWP24, TWP30, TWP34) and spring (TSP2, TSP3, TSP6) trap plants (for sampling dates, see Table 1). Five D. pinodes strains were randomly chosen within each subpopulation, and their aggressiveness was evaluated on three pea genotypes: the winter cultivar Enduro (Florimond-Desprez, France), the spring cultivar Lumina (Nickerson, France), and the spring breeding line DP (55). Enduro and Lumina were chosen to replace Cheyenne and Baccara, respectively, for which seeds were no longer available at the time of the pathogenicity tests. They presented similar and high levels of susceptibility to the disease, while DP had a higher level of quantitative resistance. Plants were grown at 18 to 20°C for 3 weeks, until they reached the 5- to 6-leaf stage, before inoculation. Plant preparation and experimental design have been described in reference 45. The inoculation method was based on that proposed by Onfroy et al. (56). Briefly, strains were grown for 10 days on V8 medium under white light with a 12-h photoperiod at 20°C before pycnidiospore suspensions were prepared by flooding the surfaces of cultures with sterile distilled water, gently scraping with a glass rod, and filtering the suspension through two layers of sterile cheesecloth. The spore concentration was adjusted to 5 × 104 spores · ml−1, and Tween 20 (VWR International SAS, Strasbourg, France) was added as a wetting agent (2 drops per 500 ml of the spore suspension). Inoculation consisted of depositing a 10-μl drop of the spore suspension on the upper surfaces of freshly detached stipules floated, lower surface down, on tap water in a compartmentalized square petri dish (12-cm side; Gosselin, France). Drops were deposited away from the main veins. To avoid drop evaporation, petri dishes were placed in large, transparent plastic boxes immediately after inoculum deposition and were incubated in a climate chamber for 7 days with a continuous cycle of 14 h of light at 20°C.
Symptom development was assessed 2, 4, and 7 days after inoculation, as described by Onfroy et al. (56). A semiquantitative scale with scores from 0 to 3 (0, symptom free; 1, flecks appearing; 2, flecks covering half of the area of drop deposition; 3, coalescence of the flecks within the area of drop deposition) was used to score symptoms not extending past the inoculation droplet. For stipules with necrosis extending beyond the borders of inoculum drops, lesion diameter (in millimeters) was measured with a graduated ruler. Visual assessment using a scale with scores from 0 to 7 (0, symptom free; 1, flecks appearing; 2, flecks covering half of the drop deposit; 3, coalescence of the flecks in the area of the drop deposit; 4, lesion with a diameter of 3 to 6 mm; 5, lesion with a diameter of 6 to 9 mm; 6, lesion with a diameter of 9 to 12 mm; 7, lesion with a diameter of >12 mm), adapted from the work of Wroth (57), was also performed.
Two stipules from each of four different plants per genotype were inoculated. For each genotype, the area under the disease progress curve (AUDPC) was calculated as follows:
where Di,j and Di,j + 1 correspond to disease scores on two consecutive dates, tj and tj + 1 (58). Statistical analysis of the data was performed with R statistical software (version 3.1.1, 2014; The R Foundation for Statistical Computing). The normality and homogeneity of variances were checked by the Shapiro-Wilk and Levene tests, respectively. The effects of population (WP, SP, TWP, and TSP), pea genotype (DP, Lumina, or Enduro), and their interactions on AUDPC values were tested through multiway analysis of variance (ANOVA). The effects of subpopulations, pea genotypes, and their interactions were tested on AUDPC values using a second ANOVA model. When significant effects were detected, mean values were compared with Tukey tests (α = 0.05).
RESULTS
D. pinodes displayed homogeneous spatial development during the cropping season.
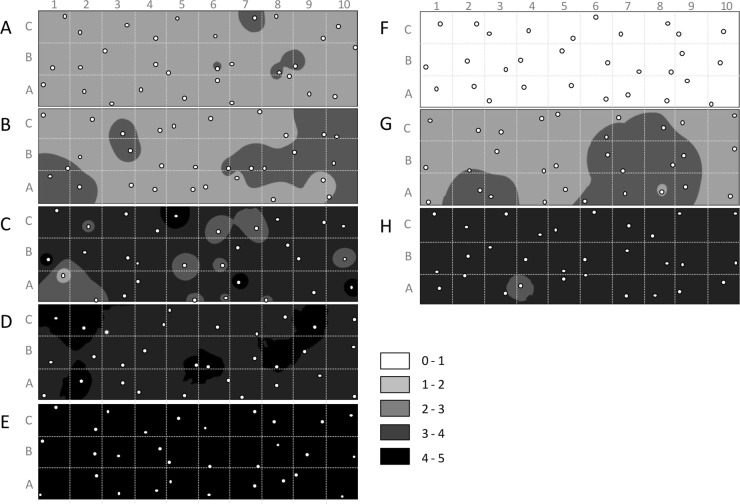
Disease layout maps for the winter crop (created with ArcMap software) were spatially similar to those for the spring crops (Fig. 1). Disease severity increased with time but was homogeneously distributed spatially through the plots, with no strong foci. The mean disease scores were higher for the winter pea crop than for the spring pea crop, essentially because of the longer growing season and the more-conducive climatic conditions in the late winter and early spring.
FIG 1.
Maps representing the evolution of disease severity due to ascochyta blight within winter and spring pea plots on different sampling dates. Winter sampling dates were 17 February (A), 28 February (B), 29 March (C), 25 April (D), and 23 May (E). Spring sampling dates were 30 May (F), 13 June (G), and 27 June (H). Each plot was divided into 1-m2 microplots (A1 to C10), where one plant was randomly sampled on each sampling date. Disease was scored on each stipule of the sampled plants using the adapted disease scale of Tivoli (40, 43), and a mean disease score (Di) was calculated. Disease distribution maps were created for each sampling date using ArcMap software.
Genotypic flow acted on the genetic structure of the D. pinodes population during the cropping season.
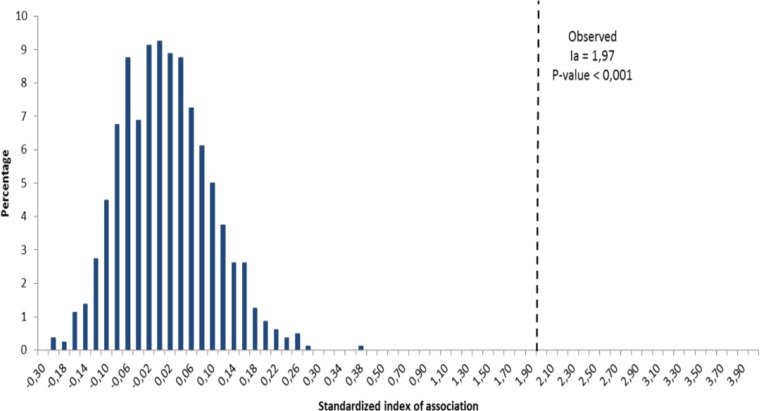
The three AFLP primer sets allowed the detection of 646 loci, 388 of which were polymorphic over all populations tested. The percentage of polymorphic markers in each subpopulation ranged from 29.90% in TWP21 to 100% in WP8. No repeated MLG was found among the 520 strains, indicating a very low incidence of clonality. However, the overall IA value differed significantly from zero (IA = 1.97), thus rejecting the null hypothesis of recombination (P = 0.001) (Fig. 2). The number of alleles (Na), Shannon's information index (I), and unbiased diversity (uh) showed the highest genetic diversity within subpopulations for WP12 (I = 0.61; uh = 0.43) and the lowest for TWP21 (I = 0.176; uh = 0.151). The low genetic diversity observed for subpopulation TWP21 may be due to the low number of isolates within this subpopulation. At the scale of populations (WP, SP, TWP, and TSP), the number of alleles, Shannon's information index, and unbiased diversity showed that genetic diversity was higher for populations sampled in the plots (SP and WP) than for populations sampled on trap plants (TWP and TSP) (Table 1).
FIG 2.
Standardized index of association (IA) calculated from the clone-corrected data set, rejecting the hypothesis of the absence of linkage disequilibrium among microsatellite loci (P < 0.001).
AMOVA revealed that 80.35% of the total genetic variance was partitioned within subpopulations (Table 2). A relatively low proportion of genetic variability was attributable to differences between populations (9.62%) and between subpopulations within populations (10.03%). High variances of FST values, which ranged from −0.055 to 0.586, were detected. Pairwise FST values were higher between plots and trap plants (WP-TWP and SP-TSP) than within plots (WP-WP or SP-SP) (Fig. 3) and quite low between plots (SP-WP) and within spring trap plants (TSP-TSP) but quite high within winter trap plants (TWP-TWP).
TABLE 2.
AMOVA for D. pinodes strains sampled from different plots and trap plantsa based on AFLP markers
| Source of variation | df | Sum of squares | Variance component | Variation (%) | Fixation index | P value |
|---|---|---|---|---|---|---|
| Among populations | 3 | 3,781.378 | 8.26273 | 9.62 | FCT, 0.09616 | <0.000001 |
| Among subpopulations within populations | 26 | 5,535.163 | 8.62056 | 10.03 | FSC, 0.11100 | <0.000001 |
| Within subpopulations | 490 | 33,830.767 | 69.04238 | 80.35 | FST, 0.19649 | <0.000001 |
Strains were sampled from winter and spring pea plots and from winter and spring trap plants.
FIG 3.
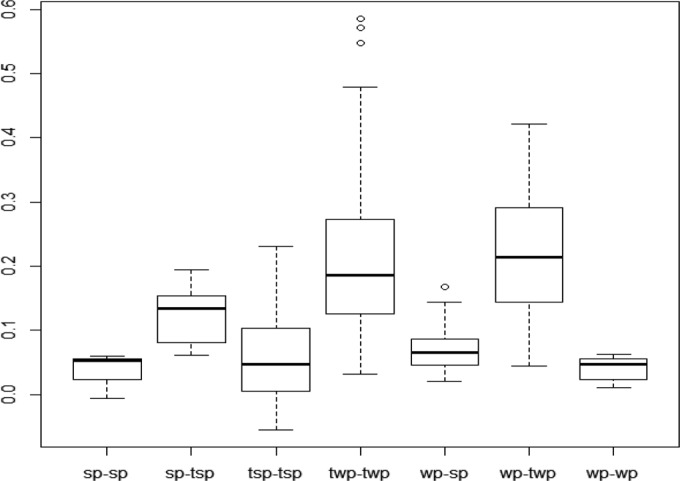
Box plots representing the distribution of pairwise FST values within each subpopulation (sp-sp, wp-wp, twp-twp, tsp-tsp) and between subpopulations (wp-sp, wp-twp, sp-tsp).
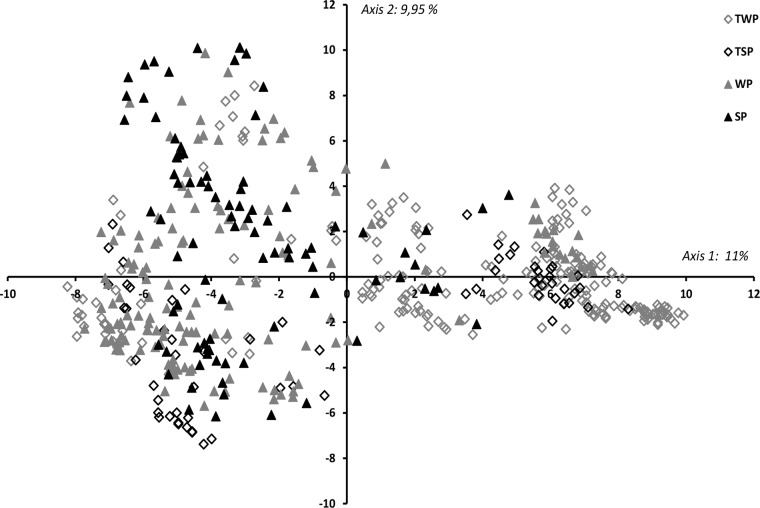
Principal-component analysis (PCA) failed to separate the strains of the subpopulations sampled on WP, SP, TWP, and TSP into different groups (Fig. 4). Moreover, the low percentage of genetic diversity explained by the two principal axes of the PCA (11% and 9.95%, respectively) suggested that these different subpopulations did not constitute distinct genetic groups and that all the strains belonged to a single population.
FIG 4.
PCA conducted on the D. pinodes subpopulations sampled in winter pea plots (WP) (150 strains), spring pea plots (SP) (90 strains), winter trap plants (TWP) (214 strains), and spring trap plants (TSP) (66 strains).
Aggressiveness changes during the cropping season differed between winter and spring pea populations.
The level of aggressiveness differed according to the test host genotype (Table 3, pea genotype): as expected, AUDPC values were significantly lower for the resistant pea genotype DP than for the pea genotype Enduro and were intermediate (and not significantly different from those for the other two genotypes) for the pea genotype Lumina. However, the lack of significant interaction between populations or subpopulations and pea genotypes (Table 3) showed that all populations shared the same ranking across hosts.
TABLE 3.
ANOVA of AUDPC values at the population and subpopulation levels
| Source of variationa | df | F value | P>Fb |
|---|---|---|---|
| At the population level | |||
| Subpopulation | 3 | 28.41 | <0.0001* |
| Pea genotype | 2 | 7.71 | 0.0006* |
| Subpopulation × pea genotype | 6 | 0.11 | 0.9555 |
| Error | 180 | ||
| At the subpopulation level | |||
| Subpopulation | 12 | 11.22 | <0.0001* |
| Pea genotype | 2 | 8.50 | 0.0003* |
| Subpopulation × pea genotype | 24 | 0.23 | 0.9999 |
| Error | 153 |
Sources of variation are the D. pinodes population (or subpopulation), the pea genotype, and the corresponding two-way interaction between these variables.
P>F, significance of a statistical analysis based on a Fisher test. Asterisks indicate statistically significant effects (P < 0.001).
Overall, strains sampled in winter plots and on trap plants were significantly more aggressive than strains sampled on spring plots (Fig. 5A). Moreover, at the subpopulation level, the aggressiveness of SP strains increased strongly during the growing season, whereas aggressiveness remained stable over time in the other three populations (Fig. 5B).
FIG 5.
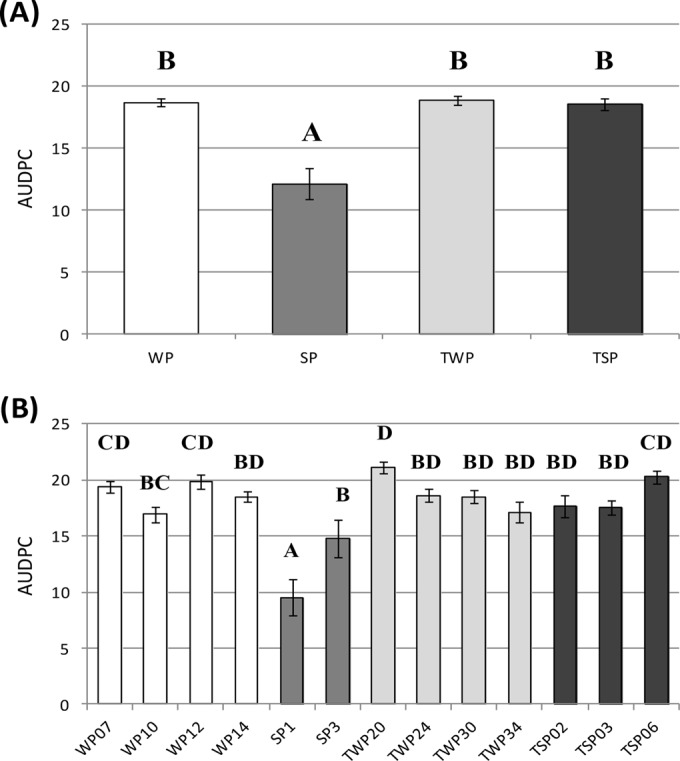
Mean aggressiveness level (AUDPC) for each D. pinodes population (A) and subpopulation (B). AUDPC was measured on detached stipules of three pea genotypes (Lumina, Enduro, and DP) for D. pinodes strains sampled on different dates from winter (WP) and spring (SP) pea plots and for D. pinodes strains sampled on winter (TWP) and spring (TSP) trap plants. AUDPC was calculated as described by Shaner and Finney (58) by estimating the integral of the disease progress curve, including assessment of the disease at 2, 4 and 7 days after inoculation. Vertical bars correspond to standard deviation, and different letters indicate significant differences between treatments.
Alloinoculum sampled on TWP or TSP showed an aggressiveness level similar to that for strains sampled from the winter plot (Fig. 5A and B). Taken together, these results tended to show that trap plants were always infected from the winter populations and that the spring plots initially select against aggressive isolates.
DISCUSSION
One of the objectives of this study was to determine how D. pinodes population structures change during a growing season. The amounts of genetic variation were similar within D. pinodes subpopulations sampled from winter or spring pea plots (H′, 0.83 and 0.86, respectively). The genetic variability observed at the beginning of the season remained roughly the same throughout the growing season. The main part of this genetic variability was observed within subpopulations (80%), confirming the PCA result of an absence of a clear genetic structure over time and suggesting that significant gene flow occurs between subpopulations. The low pairwise FST values observed within (SP-SP and WP-WP) and between (SP-WP) plots suggest also that subpopulations were not genetically differentiated. Previous work released on D. pinodes–pea pathosystems showed that D. pinodes populations from Canada, France, and Algeria displayed a high degree of genetic variability (32, 59). This variation, estimated on a different scale than in our study, is illustrated by the high value of Shannon's information index and Nei's gene diversity and by the high number of distinct haplotypes. Our study showed that population diversity estimated at a field level, and during the growing season, is changing, but slightly higher than the diversity estimated among countries or among locations within a country. Indeed, the H′ value in our study ranged from 0.70 to 0.83, whereas it ranged from 0.39 to 0.47 in D. pinodes populations collected from different locations in Algeria (59). Moreover, the H′ value was closely similar to those obtained for fungi showing great genetic diversity, such as Ascochyta rabiei (H′ = 0.58 [60]), Mycosphaerella fijiensis (H′ = 0.58 [61]), or Botrytis cinerea (H′ values ranging from 0.401 to 0.518 [62]). This change in the diversity level observed during the growing season reinforced the fact that sexual reproduction was important in the life cycle of D. pinodes, and the predominance of this mode of reproduction seemed to affect the previous population structure.
On trap plants, pairwise FST values were higher within winter subpopulations than within spring subpopulations, suggesting that the same alloinoculum sources must be active during the whole spring growing season, whereas other alloinoculum sources must be active during the winter cropping season. This result suggests that epidemics on winter crops are initiated from a large number of sources producing pseudothecia and releasing ascospores, whereas the release of ascospores, probably from infected winter crops, fuels spring crop epidemics with steadier and genetically similar sources of inoculum. It also suggests that asexual inoculum is not an important component of epidemic extension, although it might play a major role in local disease intensification. The spatially homogeneous disease severity observed throughout the winter and spring plots sampled here also point to a homogeneous distribution of initial inoculum consistent with ascospore showers from external sources rather than with short-distance splashes of asexual pycnidiospores. If ascospore release indeed constituted the main mechanism fueling epidemic development and spread, it would also provide a good explanation for the connected genetic structures of pathogen populations sampled in different plots. As indicated in Materials and Methods, our experiment was conducted only once, but we have assessed disease development and genetic diversity evolution on two different crops. Past studies showed that D. pinodes disease that developed on winter and spring pea crops was initiated by a single population, whose pathogenicity is a plastic trait modulated by the physiological status of the host plant (32). The fact that only a single year was studied here suggests different limitations. A second year of experimentation will help us to see if a pool of alloinoculum similar to that from the first year would be mobilized during the cropping season. Indeed, a second year of field experiments will impose different constraints: (i) climatic change, which will modify plant development, plant receptivity, and the availability of different sources of inoculum, (ii) change in the diversity of the agrosystem (change in the prevalence of legume species, diversity of cultivated species in a neighboring environment). As reported by Plantegenest et al. (63), the composition of the landscape determines the local abundance of potential reservoirs of inoculum that may obviously influence the global propagule pressure and hence the risk of infection of a plant. Those reservoirs may, in particular, consist of diseased individuals of the same host species or of alternative hosts, either cultivated or wild. Moreover, changes in landscape composition and diversity will change the pathogen dynamics.
The initial, natural hypothesis is that each type of pea crop would serve as the main inoculum source for the other, via exchanges between plots. However, since ascopores are most likely the main inoculum form starting and fueling the epidemics, and given the large dispersal capabilities of wind-borne ascospores, sources other than plants growing in adjacent or neighboring plots need to be considered inoculum reservoirs. Indeed, our results suggest that various parts of the metapopulation are mobilized during the growing season, particularly during the winter growing season. Two further sources of inoculum could be involved: alternative hosts and chlamydospores (35, 64).
Compared to infected pea debris, alternative hosts are generally considered of minor importance in the epidemiology of ascochyta blight (65). Recently, Trapero-Casas and Kaiser (66) showed that A. rabiei is capable of infecting different plant species, suggesting that these naturally infected alternative hosts could serve as significant sources of inoculum to initiate disease epidemics on cultivated chick peas. In recent work, Le May et al. (64) showed that the asexual stage of D. pinodes could be observed on pea, common vetch, and clover plants, suggesting that other legume species may also act as inoculum sources for epidemics on peas (65, 67–69). Legume species such as vetch or clover are generally sympatric to pea crops during the growing season but also persist during the intercropping season. As shown by Savage et al. (6), long-distance dispersal generally results in rapid transmission of disease to previously uninfected areas and can facilitate genetic interaction between spatially separated populations, resulting in the introduction of new virulent alleles into existing populations (70). Thus, more-extensive knowledge of the host range of D. pinodes and of the relatedness between populations from peas and other hosts could help us to estimate the risk of ascochyta blight epidemics of peas arising from alternate hosts, as in other pathosystems where cultivated and wild hosts grow sympatrically (71, 72).
D. pinodes can also survive in soil as mycelium or chlamydospores (73, 74). Davidson et al. (35) investigated the survival of ascochyta blight pathogens in soils of commercial pea-cropping paddocks and showed that the level of pathogen populations in the soil was related to the severity of the epidemic in the last pea crop grown. Although airborne ascospores of D. pinodes appear to be the primary inoculum during the establishment of field pea crops, soilborne inoculum has also been associated with disease (35). To evaluate the impact of this inoculum source on the genetic structure of D. pinodes populations, it would be interesting to develop a genetic approach to characterizing the genetic population variability of D. pinodes before and after harvest. It would be particularly interesting to define the level of diversity that is maintained between two growing seasons.
Understanding how this gene flow influences the aggressiveness of pathogen populations during the season is crucial for the development of sustainable strategies. In our study, two scenarios for the evolution of the aggressiveness of D. pinodes strains inoculated into the crop were observed: (i) the maintenance of an almost constant level of aggressiveness throughout the season in the winter pea crops and (ii) an increase in the average aggressiveness of D. pinodes strains inoculated into spring peas, following initially low aggressiveness levels. The “spring pea” scenario was quite surprising, since the inoculum released by the winter pea crop was rather aggressive. The low initial aggressiveness in the spring pea plots might therefore signify other infection sources, such as alternate hosts and/or soil, as discussed above. Moreover, the fact that the D. pinodes population sampled at the beginning of the cropping season displayed a lower aggressiveness level suggests an alternative hypothesis, that spring crops initially select the least aggressive components in the alloinoculum. However, there is no strong evidence or rationale to support this hypothesis. Interestingly, the fact that aggressiveness in spring crops later increased to levels similar to those observed in trap plants and in winter plots suggests a later influx of aggressive isolates from nearby winter pea fields and/or active selection of more-aggressive isolates by spring plants over the course of the epidemic—which would, in turn, imply polycyclic epidemic development within plots. The uniform genetic structure of D. pinodes populations, however, does not allow us to distinguish between the two hypotheses, since ascospores generating the late infections could come either from within or from outside the canopy.
This study shows that monitoring over time allows one to identify inoculum sources (autoinoculum or alloinoculum). Studying the temporal changes in gene and genotypic diversity between the beginning and the end of an epidemic would thus provide information about the different epidemiological processes involved (75). Many fungal plant pathogens alternate rounds of asexual multiplication with a single annual episode of sexual reproduction (9). The number of rounds of asexual multiplication not only has a demographic impact, since it corresponds mostly to the epidemic phase of the disease, but also has drastic consequences for the genetic characteristics of pathogen populations. As a result, one would expect both the genetic structure and the gene and genotypic diversity of plant pathogens to change during the asexual phase of the life cycle (9). Founder effects resulting from colonization by a few sexually derived spores and subsequent asexual reproduction may result in reduced genotypic diversity at the beginning of an epidemic. A few population genetic studies of airborne plant pathogens have used such a nested hierarchical sampling strategy. Gobbin et al. (76) explained the arrival of new genotypes and the erosion of clonal structure that they observed in Plasmopara viticola populations by the continual input of sexual spores. Examining the changes in clonal structure during the epidemic season can provide insights into the balance between auto- and alloinfection processes, thus ideally complementing direct epidemiological observations (i.e., disease monitoring) dedicated to the quantification of the autoinfection process only (19). In contrast to many necrotrophic fungi, D. pinodes can simultaneously develop its anamorph and teleomorph forms on the same plant during the growing season (32, 33). Due to the indeterminate growth of pea plants, during crop growth, both pycnidia and pseudothecia can be observed on the same plant organs (33). Thus, beyond these qualitative predictions, estimating the population genetic consequences of intermediate rates of asexual or sexual reproduction remains a challenging task, especially when the organism's life cycle consists of alternate phases of sexual and asexual reproduction. In this study, we show particularly that the alloinoculum, essentially constituted by ascospores, is a driving force in the epidemic dynamics of ascochyta blight of peas. As reported by Savage et al. (6), by connecting wild and cultivated compartments, this alloinoculum allows the pathogen to maintain a high level of genetic variability and to modulate pest pressure during the growing season. Our study also shows that origin of this alloinoculum is still a central question. Indeed, in French agriculture, which is intensive, in contrast to more-extensive cropping, such as that in Australia or Canada (35, 74), the main sources of inoculum are well managed by the growers. Hypotheses explaining the availability of ascospores throughout the cropping seasons of both winter and spring peas thus need to include ascospores released from stubble or volunteers before burying, which persist in the atmosphere for a long time (34), or potential reservoirs of inoculum, especially in the case of winter peas. Efficient control strategies should reduce the production of alloinoculum and, for that purpose, should encompass all possible alternative sources, probably dispersed through the agricultural landscape. Thus, it is important to consider landscape composition and characteristics, since they may influence pathogen ecology and especially the range of hosts simultaneously or successively exploited (63). However, additional knowledge of the possible role, dynamics, and characteristics of inoculum sources other than peas (alternate hosts, soil) is also required.
ACKNOWLEDGMENTS
The financial support for this work from the ARIMNET project “Medileg” (2012–2015; proposal 396, Breeding, agronomic and biotechnological approaches for reintegration and revalorization of legumes in Mediterranean agriculture), INRA, and Agrocampus Ouest is gratefully acknowledged.
REFERENCES
- 1.Zadoks JC, Schein RD. 1979. Epidemiology and plant disease management. Oxford University Press Inc., New York, NY. [Google Scholar]
- 2.Cook RJ, Veseth RJ. 2001. Wheat health management. APS Press, St. Paul, MN. [Google Scholar]
- 3.Burdon JJ, Thrall PH. 2008. Pathogen evolution across the agro-ecological interface: implications for disease management. Evol Appl 1:57–65. doi: 10.1111/j.1752-4571.2007.00005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao J, Wang L, Wang Z, Chen X, Zhang H, Yao J, Zhan G, Chen W, Huang L, Kang Z. 2013. Identification of eighteen Berberis species as alternate hosts of Puccinia striiformis f. sp tritici and virulence variation in the pathogen isolates from natural infection of barberry plants in China. Phytopathology 103:927–934. doi: 10.1094/PHYTO-09-12-0249-R. [DOI] [PubMed] [Google Scholar]
- 5.De Wolf ED, Isard SA. 2007. Disease cycle approach to plant disease prediction. Annu Rev Phytopathol 45:203–220. doi: 10.1146/annurev.phyto.44.070505.143329. [DOI] [PubMed] [Google Scholar]
- 6.Savage D, Barbetti MJ, MacLeod WJ, Salam MU, Renton M. 2012. Seasonal and diurnal patterns of spore release can significantly affect the proportion of spores expected to undergo long-distance dispersal. Microb Ecol 63:578–585. doi: 10.1007/s00248-011-9949-x. [DOI] [PubMed] [Google Scholar]
- 7.Brown JKM, Hovmøller MS. 2002. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297:537–541. doi: 10.1126/science.1072678. [DOI] [PubMed] [Google Scholar]
- 8.Nathan R, Sapir N, Trakhtenbrot A, Katul GG. 2005. Long-distance biological transport processes through the air: can nature's complexity be unfolded in silico? Divers Distrib 11:131–137. doi: 10.1111/j.1366-9516.2005.00146.x. [DOI] [Google Scholar]
- 9.Barrès B, Dutech C, Andrieux A, Halkett F, Frey P. 2012. Exploring the role of asexual multiplication in poplar rust epidemics: impact on diversity and genetic structure. Mol Ecol 21:4996–5008. doi: 10.1111/mec.12008. [DOI] [PubMed] [Google Scholar]
- 10.Thrall PH, Burdon JJ. 1997. Host-pathogen dynamics in a metapopulation context: the ecological and evolutionary consequences of being spatial. J Ecol 85:743–753. doi: 10.2307/2960598. [DOI] [Google Scholar]
- 11.Aylor DE. 2003. Spread of plant disease on a continental scale: role of aerial dispersal of pathogens. Ecology 84:1989–1997. doi: 10.1890/01-0619. [DOI] [Google Scholar]
- 12.Agrios GN. 2004. Plant pathology, 5th ed Elsevier Academic Press, London, United Kingdom. [Google Scholar]
- 13.Burie JB, Langlais M, Calonnec A. 2011. Switching from a mechanistic model to a continuous model to study at different scales the effect of vine growth on the dynamic of a powdery mildew epidemic. Ann Bot 107:885–895. doi: 10.1093/aob/mcq233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakraborty S, Newton AC. 2011. Climate change, plant diseases and food security: an overview. Plant Pathol 60:2–14. doi: 10.1111/j.1365-3059.2010.02411.x. [DOI] [Google Scholar]
- 15.Glais I, Montarry J, Corbière R, Pasco C, Marquer B, Magalon H, Andrivon D. 2014. Long-distance gene flow outweighs a century of local selection and prevents local adaptation in the Irish famine pathogen Phytophthora infestans. Evol Appl 7:442–452. doi: 10.1111/eva.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen RS, McDonald BA. 1996. Sexual reproduction plays a major role in the genetic structure of populations of the fungus Mycosphaerella graminicola. Genetics 142:1119–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald BA, Linde C. 2002. Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40:349–379. doi: 10.1146/annurev.phyto.40.120501.101443. [DOI] [PubMed] [Google Scholar]
- 18.Burdon JJ, Silk J. 1997. Sources and patterns of diversity in plant-pathogenic fungi. Phytopathology 87:664–669. doi: 10.1094/PHYTO.1997.87.7.664. [DOI] [PubMed] [Google Scholar]
- 19.Lannou C, Soubeyrand S, Frezal L, Chadoeuf J. 2008. Autoinfection in wheat leaf rust epidemics. New Phytol 177:1001–1011. doi: 10.1111/j.1469-8137.2007.02337.x. [DOI] [PubMed] [Google Scholar]
- 20.Brewer MT, Frenkel O, Milgroom MG. 2012. Linkage disequilibrium and spatial aggregation of genotypes in sexually reproducing populations of Erysiphe necator. Phytopathology 102:997–1005. doi: 10.1094/PHYTO-11-11-0321. [DOI] [PubMed] [Google Scholar]
- 21.Brurberg MB, Elameen A, Le VH, Naerstad R, Hermansen A, Lehtinen A, Hannukkala A, Nielsen B, Hansen J, Andersson B. 2011. Genetic analysis of Phytophthora infestans populations in the Nordic European countries reveals high genetic variability. Fungal Biol 115:335–342. doi: 10.1016/j.funbio.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Cooke BM, Jones DG, Kaye B. 2006. The epidemiology of plant diseases, 2nd ed Chapter 2, Disease assessment and yield loss. Springer, Dordrecht, Netherlands. [Google Scholar]
- 23.Montarry J, Andrivon D, Glais I, Corbière R, Mialdea G, Delmotte F. 2010. Microsatellite markers reveal two admixed genetic groups and an ongoing displacement within the French population of the invasive plant pathogen Phytophthora infestans. Mol Ecol 19:1965–1977. doi: 10.1111/j.1365-294X.2010.04619.x. [DOI] [PubMed] [Google Scholar]
- 24.Sjöholm L, Anderson B, Hogberg N, Widmark AK, Yuen J. 2013. Genotypic diversity and migration patterns of Phytophthora infestans in the Nordic countries. Fungal Biol 117:722–730. doi: 10.1016/j.funbio.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Lepoivre P. 2005. Phytopathologie. Bases moléculaires et biologiques des pathosystèmes et fondements des stratégies de lutte, p 193–209. De Boeck Université/Presses agronomiques de Gembloux, Gembloux, Belgium. [Google Scholar]
- 26.Bretag TW, Ramsey M. 2001. Foliar diseases caused by fungi: Ascochyta spp., p 24–28. In Kraft JM, Pfleger FL (ed), Compendium of pea diseases and pests, 2nd ed APS Press, St. Paul, MN. [Google Scholar]
- 27.Tivoli B, Banniza S. 2007. Comparison of the epidemiology of ascochyta blights on grain legumes. Eur J Plant Pathol 119:59–76. doi: 10.1007/s10658-007-9117-9. [DOI] [Google Scholar]
- 28.Ali MS, Paterson J, Crosby J. 1982. A standard technique for detecting seed-borne pathogens in peas, chemical control, and testing commercial seed in South Australia. Aust J Exp Agric 32:1121–1125. [Google Scholar]
- 29.Nasir M, Hoppe HH, Ebrahim-Nesbat F. 1992. The development of different pathotype groups of Mycosphaerella pinodes in susceptible and partially resistant pea leaves. Plant Pathol 41:187–194. doi: 10.1111/j.1365-3059.1992.tb02337.x. [DOI] [Google Scholar]
- 30.Xue AG, Warkentin TD. 2001. Partial resistance to Mycosphaerella pinodes in field pea. Can J Plant Sci 81:535–540. doi: 10.4141/P00-103. [DOI] [Google Scholar]
- 31.Zhang JX, Fernando WGD, Xue AG. 2004. Temporal and spatial dynamics of mycosphaerella blight [Mycosphaerella pinodes] in field pea. Can J Plant Pathol 26:522–532. doi: 10.1080/07060660409507173. [DOI] [Google Scholar]
- 32.Le May C, Guibert M, Leclerc A, Andrivon D, Tivoli B. 2012. A single, plastic population of Mycosphaerella pinodes causes ascochyta blight on winter and spring peas (Pisum sativum) in France. Appl Environ Microbiol 78:8431–8440. doi: 10.1128/AEM.01543-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roger C, Tivoli B. 1996. Spatio-temporal development of pycnidia and perithecia and dissemination of spores of Mycosphaerella pinodes on pea (Pisum sativum). Plant Pathol 45:518–528. doi: 10.1046/j.1365-3059.1996.d01-139.x. [DOI] [Google Scholar]
- 34.Salam MU, Galloway J, Diggle AJ, MacLeod WJ, Maling T. 2011. Predicting regional-scale spread of ascospores of Didymella pinodes causing ascochyta blight disease on field pea. Australas Plant Pathol 40:640–647. doi: 10.1007/s13313-011-0064-8. [DOI] [Google Scholar]
- 35.Davidson JA, Krysinska-Kaczmarek M, Wilmshurst CJ, McKay A, Scott ES. 2011. Distribution and survival of ascochyta blight pathogens in field pea cropping soils of Australia. Plant Dis 95:1217–1223. doi: 10.1094/PDIS-01-11-0077. [DOI] [PubMed] [Google Scholar]
- 36.Jarosz AM, Burdon JJ. 1991. Host-pathogen interactions in natural populations of Linum marginale and Melampsora lini. II. Local and regional variation in patterns of resistance and racial structure. Evolution 45:1618–1627. [DOI] [PubMed] [Google Scholar]
- 37.Antonovics J, Iwasa Y, Hassell MP. 1995. A generalized model of parasitoid, venereal, and vector-based transmission processes. Am Nat 145:661–675. doi: 10.1086/285761. [DOI] [Google Scholar]
- 38.Burdon JJ, Thompson JN. 1995. Changed patterns of resistance in a population of Linum marginale attacked by the rust pathogen Melampsora lini. J Ecol 83:199–206. doi: 10.2307/2261558. [DOI] [Google Scholar]
- 39.Ericson L, Burdon JJ, Müller WJ. 1999. Spatial and temporal dynamics of epidemics of the rust fungus Uromyces valerianae on populations of its host Valeriana salina. J Ecol 87:649–658. doi: 10.1046/j.1365-2745.1999.00384.x. [DOI] [Google Scholar]
- 40.Smith DL, Ericson L, Burdon JJ. 2003. Epidemiological patterns at multiple spatial scales: an 11-year study of a Triphragmium ulmariae–Filipendula ulmaria metapopulation. J Ecol 91:890–903. doi: 10.1046/j.1365-2745.2003.00811.x. [DOI] [Google Scholar]
- 41.Schoeny A, Jumel S, Rouault F, Le May C, Tivoli B. 2007. Assessment of airborne primary inoculum availability and modelling of disease onset of ascochyta blight in field peas. Eur J Plant Pathol 119:87–97. doi: 10.1007/s10658-007-9163-3. [DOI] [Google Scholar]
- 42.Laine AL, Hanski I. 2006. Large-scale spatial dynamics of a specialist plant pathogen in a fragmented landscape. J Ecol 94:217–226. doi: 10.1111/j.1365-2745.2005.01075.x. [DOI] [Google Scholar]
- 43.Tivoli B. 1994. Conséquences des attaques parasitaires foliaires sur l'eìlaboration du rendement des plantes à croissance indeìtermineìe, p 199–219. In UNIP-ITCF-INRA (ed), Agrophysiologie du pois proteìagineux. ITCF, Paris, France. [Google Scholar]
- 44.Fisher NI, Lewis T, Embleton BJJ. 1987. Statistical analysis of spherical data. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 45.Onfroy C, Tivoli B, Corbière R, Bouznad Z. 1999. Cultural, molecular and pathogenic variability of Mycosphaerella pinodes and Phoma medicaginis var. pinodella isolates from dried pea (Pisum sativum) in France. Plant Pathol 48:218–229. doi: 10.1046/j.1365-3059.1999.00323.x. [DOI] [Google Scholar]
- 46.Lodhi MA, Ye GN, Weeden NF, Reisch BI. 1994. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol Biol Rep 12:6–13. doi: 10.1007/BF02668658. [DOI] [Google Scholar]
- 47.Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Res 6:288–295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon CE, Weaver W. 1949. The mathematical theory of communication. University of Illinois Press, Urbana, IL. [Google Scholar]
- 50.Grünwald NJ, Goodwin SB, Milgroom MG, Fry WE. 2003. Analysis of genotypic diversity data for populations of microorganisms. Phytopathology 93:738–746. doi: 10.1094/PHYTO.2003.93.6.738. [DOI] [PubMed] [Google Scholar]
- 51.Agapow PM, Burst A. 2001. Indices of multilocus linage disequilibrium. Mol Ecol Notes 1:101–102. doi: 10.1046/j.1471-8278.2000.00014.x. [DOI] [Google Scholar]
- 52.Excoffier L, Laval G, Schneider S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 53.Jombart T, Devillard S, Dufour AB, Pontier D. 2008. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101:92–103. doi: 10.1038/hdy.2008.34. [DOI] [PubMed] [Google Scholar]
- 54.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baranger A, Aubert G, Arnau G, Lainé AL, Deniot G, Potier J, Weinachter C, Lejeune I, Lallemand J, Burstin J. 2004. Genetic diversity within Pisum sativum using protein and PCR-based markers. Theor Appl Genet 108:1309–1321. doi: 10.1007/s00122-003-1540-5. [DOI] [PubMed] [Google Scholar]
- 56.Onfroy C, Baranger A, Tivoli B. 2007. Biotic factors affecting the expression of partial resistance in pea to ascochyta blight in a detached stipule assay. Eur J Plant Pathol 119:13–27. doi: 10.1007/s10658-007-9153-5. [DOI] [Google Scholar]
- 57.Wroth JM. 1998. Possible role for wild genotypes of Pisum spp. to enhance ascochyta blight resistance in pea. Aust J Exp Agric 38:469–479. doi: 10.1071/EA98024. [DOI] [Google Scholar]
- 58.Shaner G, Finney RE. 1977. The effect of nitrogen fertilization on the expression of slow-mildewing resistance in Knox wheat. Phytopathology 67:1051–1056. [Google Scholar]
- 59.Setti B, Bencheikh M, Henni DE, Neema C. 2012. Genetic variability and population structure of Mycosphaerella pinodes in western Algeria using AFLP fingerprinting. J Plant Pathol 94:127–133. doi: 10.4454/jpp.fa.2012.014. [DOI] [Google Scholar]
- 60.Rhaiem A, Cherif M, Peever TL, Dyer PS. 2008. Population structure and mating system of Ascochyta rabiei in Tunisia: evidence for the recent introduction of mating type 2. Plant Pathol 57:540–551. doi: 10.1111/j.1365-3059.2007.01779.x. [DOI] [Google Scholar]
- 61.Carlier J, Lebrun MH, Zapater MF, Dubois C, Mourichon X. 1996. Genetic structure of the global population of banana black leaf streak fungus, Mycosphaerella fijiensis. Mol Ecol 5:499–510. [Google Scholar]
- 62.Alfonso C, Rapso R, Malgarejo P. 2000. Genetic diversity in Botrytis cinerea populations on vegetable crops in greenhouses in south-eastern Spain. Plant Pathol 49:243–251. doi: 10.1046/j.1365-3059.2000.00452.x. [DOI] [Google Scholar]
- 63.Plantegenest M, Le May C, Fabre F. 2007. Landscape epidemiology of plant diseases. J R Soc Interface 4:963–972. doi: 10.1098/rsif.2007.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Le May C, Guibert M, Baranger A, Tivoli B. 2014. A wide range of cultivated legume species act as alternative hosts for the pea ascochyta blight fungus, Didymella pinodes. Plant Pathol 63:877–887. doi: 10.1111/ppa.12154. [DOI] [Google Scholar]
- 65.Lawyer AS. 1984. Foliar diseases caused by fungi. Diseases caused by Ascochyta spp., p 11–15. In Hagedorn DJ. (ed), Compendium of pea diseases. APS Press, St Paul, MN. [Google Scholar]
- 66.Trapero-Casas A, Kaiser WJ. 2009. Alternative hosts and plant tissues for the survival, sporulation and spread of the Ascochyta blight pathogen of chickpea. Eur J Plant Pathol 125:573–587. doi: 10.1007/s10658-009-9507-2. [DOI] [Google Scholar]
- 67.Weimer JL. 1947. Resistance of Lathyrus spp. and Pisum spp. to Ascochyta pinodella and Mycosphaerella pinodes. J Agric Res 75:181–190. [Google Scholar]
- 68.Gurung AM, Pang ECK, Taylor PWJ. 2002. Examination of Pisum and Lathyrus species as sources of ascochyta blight resistance for field pea (Pisum sativum). Australas Plant Pathol 31:41–45. doi: 10.1071/AP01069. [DOI] [Google Scholar]
- 69.Bretag TW. 2004. Ascochyta blight in field peas. Victorian Department of Primary Industries, Horsham, Victoria, Australia. [Google Scholar]
- 70.Singh AB, Babu CR. 1980. Diurnal periodicity of common allergenic pollen. Allergy 35:311–317. doi: 10.1111/j.1398-9995.1980.tb01772.x. [DOI] [PubMed] [Google Scholar]
- 71.Abbo S, Frenkel O, Sherman A, Shtienberg D. 2007. The sympatric Ascochyta pathosystems of Near Eastern legumes, a key for better understanding of pathogen biology. Eur J Plant Pathol 119:111–118. doi: 10.1007/s10658-007-9116-x. [DOI] [Google Scholar]
- 72.Akinsanmi OA, Chakraboty S, Backhouse D, Simpfendorfer S. 2007. Passage through alternative hosts changes the fitness of Fusarium graminearum and Fusarium pseudograminearum. Environ Microbiol 9:512–520. doi: 10.1111/j.1462-2920.2006.01168.x. [DOI] [PubMed] [Google Scholar]
- 73.Hare WW, Walker JC. 1944. Ascochyta diseases of canning pea. Wisc Res Bull 150:1–31. [Google Scholar]
- 74.Wallen VR, Jeun J. 1968. Factors limiting the survival of Ascochyta spp. of peas in soil. Can J Bot 46:1279–1286. doi: 10.1139/b68-170. [DOI] [Google Scholar]
- 75.Prugnolle F, De Meeûs T. 2010. Apparent high recombination rates in clonal parasitic organisms due to inappropriate sampling design. Heredity 104:135–140. doi: 10.1038/hdy.2009.128. [DOI] [PubMed] [Google Scholar]
- 76.Gobbin D, Jermini M, Loskill B, Pertot I, Raynal M, Gessler C. 2005. Importance of secondary inoculum of Plasmopara viticola to epidemics of grapevine downy mildew. Plant Pathol 54:522–534. doi: 10.1111/j.1365-3059.2005.01208.x. [DOI] [Google Scholar]