ABSTRACT
Bioaugmentation has been frequently proposed in wastewater and soil treatment to remove toxic aromatic compounds. The performance of bioaugmentation is affected by a number of biological and environmental factors, including the interaction between the target pollutant and the augmented bacterial cells. In this study, using Comamonas testosteroni and 3-chloroaniline (3-CA) as the model organism and target pollutant, we explored the influence of toxic aromatic pollutants on the biofilm lifestyle of bacteria capable of degrading aromatic compounds toward a better understanding of cell-pollutant interaction in bioaugmentation. Our results showed that the exposure to 3-CA greatly reduced the retention of C. testosteroni cells in packed-bed bioreactors (from 22% to 15% after three pore volumes), which could be attributed to the altered bacterial motility and cell surface hydrophobicity. To further understand the molecular mechanisms, we employed an integrated genomic and transcriptomic analysis to examine the influence of 3-CA on the expression of genes important to the biofilm lifestyle of C. testosteroni. We found that exposure to 3-CA reduced the intracellular c-di-GMP level by downregulating the expression of genes encoding c-di-GMP synthases and induced massive cell dispersal from the biofilms. Our findings provide novel environmental implications on bioaugmentation, particularly in biofilm reactors, for the treatment of wastewater containing recalcitrant industrial pollutants.
IMPORTANCE Bioaugmentation is a bioremediation approach that often has been described in the literature but has almost never been successfully applied in practice. Many biological and environmental factors influence the overall performance of bioaugmentation. Among these, the interaction between the target pollutant and the augmented bacterial cells is one of the most important factors. In this study, we revealed the influence of toxic aromatic pollutants on the biofilm lifestyle of bacteria capable of degrading aromatic compounds toward a better understanding of cell-pollutant interaction in bioaugmentation. Our findings provide novel environmental implications on bioaugmentation for the treatment of wastewater containing recalcitrant industrial pollutants; in particular, the exposure to toxic pollutants may reduce the retention of augmented organisms in biofilm reactors by reducing the c-di-GMP level, and approaches to elevating or maintaining a high c-di-GMP level may be promising to establish and maintain sustainable bioaugmentation activity.
INTRODUCTION
Aromatic compounds are widely used in chemical and pharmaceutical industries (1). These organic compounds contain chemically stable benzene ring structures, hence their degradation by naturally occurring microbial communities in the contaminated environments is often slow (2, 3). This is because the indigenous microbial community usually lacks efficient catabolic capabilities for these anthropogenic compounds. To efficiently remove these toxic compounds from contaminated water or soil, bioaugmentation has been a frequently investigated bioremediation approach in which active bacteria capable of degrading recalcitrant organic compounds are introduced into the bioremediation system (4, 5). However, the performance of bioaugmentation remains difficult to predict, and in many cases bioaugmentation failed because of the complex interactions among the augmented organisms (6), indigenous microbial community (7), and environmental factors (8).
Previous studies have shown that bioavailability, concentration, and toxicity of the target pollutant are important characteristics influencing the performance of bioaugmentation (9). Further molecular-level mechanistic studies on responses of bacteria to the pollutants have improved our understanding of the interaction between augmented population and the pollutants and its potential effects on bioaugmentation. Most such studies used planktonic bacterial cultures and focused on the pollutant-induced changes in catabolic activity responsible for biodegradation of the pollutant and detoxification mechanisms employed by the bacteria at the gene, mRNA, or protein level (10–12). However, bacterial cells that are introduced into bioaugmentation systems often exist in the form of biofilms by attaching onto biotic and abiotic surfaces (13, 14). Bacterial cells in biofilms differ physiologically and genetically from their planktonic counterparts. Compared with cells in planktonic cultures, the biofilm lifestyle provides cells with a number of advantages under unfavorable conditions, including exposure to toxic chemicals (15–17), although toxic chemicals may compromise biofilm stability by inducing detachment of cells from the biofilms and result in a loss of the bioaugmented function (18, 19). Hence, the influence of toxic aromatic pollutants on the biofilm lifestyle of the introduced bacterial population is a critical aspect contributing to the overall performance of bioaugmentation, which remains largely unexplored.
The objective of this study was to reveal the influence of toxic aromatic pollutants on the biofilm lifestyle of bacteria capable of degrading aromatic compounds in order to better understand cell-pollutant interaction in bioaugmentation. Comamonas testosteroni and 3-chloroaniline (3-CA) were used as the model organism and the industrial pollutant, respectively, because biodegradation of 3-CA by C. testosteroni and bioaugmentation with C. testosteroni for the treatment of wastewater containing 3-CA have been well documented (13, 20, 21). By using a packed-bed biofilm reactor, we quantified the effect of 3-CA exposure on the retention of C. testosteroni cells in the bioreactor. To further understand the mechanisms, we employed an integrated genomic and transcriptomic analysis to examine the influence of 3-CA on the expression of genes important to the biofilm lifestyle of C. testosteroni.
MATERIALS AND METHODS
Bacterial strain and culture conditions.
C. testosteroni WDL7, a Gram-negative, aerobic organism capable of degrading 3-CA (18), was precultured in LB medium at 30°C. The overnight seed cultures then were inoculated in M9 minimal medium which was supplemented with 10 mM pyruvic acid sodium salt as the sole carbon source (18). Bacterial growth was monitored by measuring the optical density at 600 nm (OD600).
Retention of C. testosteroni cells in biofilm reactors.
Packed-bed column reactors were used to characterize bacterial retention under continuous-flow conditions. The column reactors were set up as previously described by using 10-ml syringes (Luer-Lok tip, latex free; BD) packed with glass beads (diameter, 2.0 mm) (solid glass beads; Sigma-Aldrich) (18). Each packed-bed reactor contains 1,520 ± 18 beads with a total surface area of 19,091 ± 305 mm2. The reactors were sterilized with 70% ethanol and then saturated with phosphate-buffered saline (PBS) buffer (pH 7.4) with or without 200 ppm 3-CA at an interstitial rate of 5.99 cm/h against gravity. For each condition, replicate experiments in three independent reactors were conducted. C. testosteroni cells were cultivated in M9 minimal medium with pyruvate as a carbon source. At the mid-exponential phase, one set of the cultures was treated with 3-CA at a final concentration of 200 ppm, while the other set of cultures with no 3-CA addition served as the control. After a 2-h treatment, bacterial cells were centrifuged and resuspended in PBS buffer with or without 200 ppm of 3-CA to obtain a cell density of 3.2 × 109 cells/ml. Approximately 0.4-ml aliquots of the cell suspensions were introduced into the bioreactors, and effluent samples were collected periodically. Cell density in the effluent samples was determined using a drop-plate method as previously described (18). Three technical replicates were performed to assess the average cell density for each biological replicate.
Mathematical analysis.
The breakthrough curves of the treated and control groups were obtained based on cell density in the effluents. The bacterial transport process was modeled using the following one-dimensional advection-dispersion equation (22, 23):
where R represents the retardation factor, b is the cell density in the aqueous phase (normalized by inoculum density), D is the longitudinal hydrodynamic dispersion coefficient, z is the longitudinal distance, and km represents the irreversible retention coefficient. R, D, and km were determined by fitting the advection-dispersion model to the experimental breakthrough curve data via optimizing the least-squares error by using CXTFIT/EXCEL code (24).
Motility assay.
The swimming motility of the C. testosteroni cells was determined on agar (0.3%) (18, 25) containing M9 minimal medium supplemented with 10 mM pyruvic acid sodium salt in the presence or absence of 200 ppm of 3-CA. Briefly, 1 μl of overnight culture (OD600 of ∼1.4) was inoculated into the agar plate using a sterile needle (25 gauge; Braun) by the half-penetration method.
Cell surface hydrophobicity assay.
Cell surface hydrophobicity was determined by bacterial adhesion to hydrocarbons (BATH) (26). Cells grown in M9 media with 10 mM pyruvate in the presence or absence of 3-CA (200 ppm) were harvested and resuspended in PBS buffer to a final OD600 of 0.20. Two milliliters of the cell suspensions was mixed with 2 ml of hexane. After 2 min of vortexing, the mixture was left for 15 min at room temperature, and then the OD600 of the aqueous phase was measured. The hydrophobicity was quantified by calculating the percentage of cells transferred to the hexane phase.
DNA and RNA extraction.
After overnight cultivation in LB medium, the culture was harvested and DNA was extracted by a Qiagen genomic kit (Qiagen K.K., Tokyo, Japan). The DNA concentration was monitored by using a NanoDrop spectrophotometer (Thermo Scientific, DE, USA), while the integrity and quality were examined by loading DNA on an agarose gel. For the total RNA extraction, cells grown in M9 media with or without 3-CA exposure (1 h of exposure at late exponential phase) were harvested by centrifugation, and RNA was extracted by using Qiagen mini-RNA prep kits. The concentration and quality of extracted total RNA were determined by a NanoDrop spectrophotometer (Thermo Scientific, DE, USA). For each experimental condition, there were three independent biological replicates.
Genome sequencing, draft genome assembly, and annotation.
Genomic DNA was sequenced on Illumina's MiSeq platform (Illumina, CA, USA) with 250-bp paired-end sequences (5,733,600 reads). Quality filtering and adapter removal were performed by using cutadapt 1.2.1 (27). Filtered reads were assembled using Velvet 1.2.08 (k = 153) (28). All contigs were annotated on the Integrated Microbial Genomes Expert Review (IMG ER) platform (29, 30). The genome sequence and annotation information were available from the IMG database (analysis project identifier [ID] Ga0022323). Prophage- and transposase-containing regions were analyzed by Phast and ISsaga (31, 32). The membrane-bound transporter genes were predicted in IMG using the Transport Classification Database (33, 34). Protein subcellular localization prediction was done by using PSORTb 3.0 (35).
RNA-seq analysis.
After RNA isolation, paired-end sequencing was performed on the Illumina Hi-Seq 2500 platform with a read length of 100 nucleotides. The sequence reads were assembled and analyzed in the RNA-seq (whole-transcriptome shotgun sequencing) and expression analysis application of CLC Genomics Workbench 6.0 (CLC Bio, Aarhus, Denmark). The sequences of the strain WDL7 genome and plasmid pWDL7::rfp (accession number GQ495894.1) were used as the references for assembly. Data were normalized by calculating the reads per kilobase per million mapped reads. Significant changes in gene expression were determined using selection criteria of a more than 2-fold change and P value of <0.05 (36).
Quantification of 3-CA.
The concentration of 3-CA in culture supernatant was determined by using reverse-phase high-performance liquid chromatography (HPLC) after filtering through 0.2-μm Acrodisc polyethersulfone membrane syringe filters (Pall Corporation, Singapore). In the HPLC system, an Ascentis C18 column (100 mm by 2.1 mm, 5 μm particle size), a mobile phase of methanol-water (50/50, vol/vol), a flow rate of 0.6 ml/min, and a UV detector set at 254 nm were used (18).
Extraction and quantification of c-di-GMP.
The extraction and quantification of bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) followed the methods described in a previous paper (18). Cells were grown at 30°C in M9 media using pyruvate as the sole carbon source. At the late exponential phase, 3-CA was added to the culture to a final concentration of 200 ppm. After the cells were harvested by centrifugation, c-di-GMP was extracted using organic solvent mixture (37). c-di-GMP concentration was analyzed by using a Thermo Accela 1250 series LC (liquid chromatography) system fitted with a Thermo Velos Pro Orbitrap mass spectrometer as described earlier (38). The c-di-GMP concentration was normalized to total protein.
Flow cell biofilms.
The response of C. testosteroni biofilms to 3-CA was examined using multichannel flow cells (BioCentrum-DTU, Denmark) (19, 36, 39). Each channel was inoculated using 1.5 ml diluted overnight culture in M9 medium (OD600 of ∼0.1). Air-saturated M9 medium containing 10 mM pyruvate was supplied at a flow rate of 3.0 ml/h for biofilm growth in each channel. 3-CA was introduced into the medium to a final concentration of 200 ppm after 4 days of growth. The experiments were conducted in triplicate. The cell detachment rate was determined by a drop-plate method as previously described (18).
Confocal laser scanning microscopy and image processing.
Biofilms were stained with Live/Dead BacLight bacterial viability kit L7012 (Molecular Probes, Inc.) (40). Diluted SYTO 9 (11.1 μM) and propidium iodide (66.7 μM) solution (∼0.7 ml) was injected into each flow channel. After staining for 15 min in the dark, the flow cell biofilms were imaged by using a confocal laser scanning microscope (CLSM) (LSM 780; Carl Zeiss Microscopy). Five images for each biofilm were collected by CSLM and analyzed with IMARIS software (version 8.0.0; Bitplane, Zurich, Switzerland).
Accession numbers.
The sequence assembled during the course of this work was deposited in the NCBI's assembly database under accession number ASM126683v1. The RNA-seq raw data were deposited in the NCBI GEO Sequence Read Archive under numbers SRX1124987 and SRX1124990.
RESULTS AND DISCUSSION
Exposure to 3-CA reduced retention of C. testosteroni cells in bioreactors.
Compared with the control, C. testosteroni cells exposed to 3-CA exhibited a higher migration rate through the packed-bed biofilm reactors (Fig. 1). Cells from the control group began to elute at a 0.71 pore volume, and the normalized cell density in the effluent reached a peak value of 0.085 ± 0.006 at a 1.18 pore volume. For the 3-CA-treated group, cells began to elute from the bioreactor after reaching a pore volume of 0.56, and the highest normalized cell density in the effluent (0.122 ± 0.009) was reached rapidly at a pore volume of 0.86.
FIG 1.
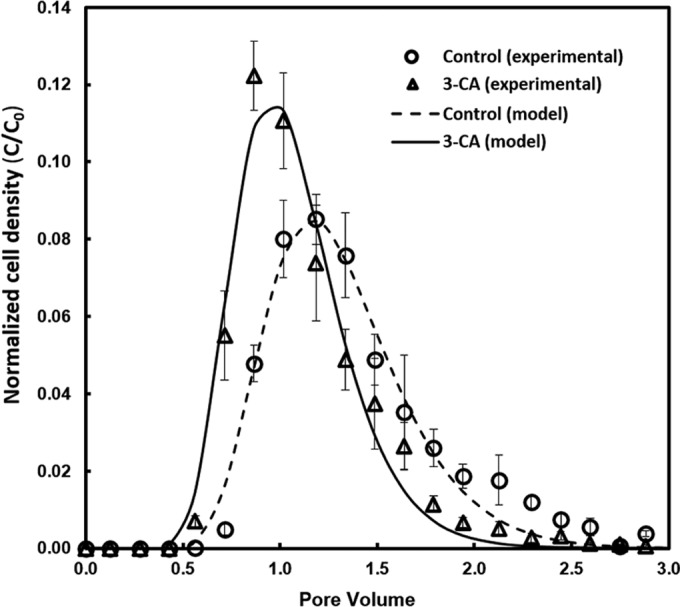
Breakthrough curves of C. testosteroni WDL7 with the absence or presence of 3-CA in packed-bed reactors. Experimental data for cell concentration in the effluent from the reactor with 3-CA (empty triangle) and without 3-CA (empty circle) were normalized by cell density of the influent cell suspension. Simulated data based on a one-dimensional advection-dispersion model is represented by the solid line for the group with 3-CA and a dashed line for the group without 3-CA.
After fitting the experimental data with the advection-dispersion model, we obtained the kinetic parameters for the breakthrough curves (Table 1). Compared with the control, the cells exposed to 3-CA had a lower dispersion coefficient (1.406 ± 0.033 cm2/h versus 1.487 ± 0.168 cm2/h), a lower retardation factor (R; 1.003 ± 0.023 versus 1.245 ± 0.017; P < 0.05), and lower irreversible retention (km; 0.151 ± 0.032 h−1 versus 0.237 ± 0.043 h−1; P < 0.05). Lower R and km values for cells exposed to 3-CA suggest that both the reversible and irreversible retention of C. testosteroni cells in the bioreactors were reduced (23). The 3-CA-treated cells also exhibited a shorter mean travel time, which suggested 3-CA would decrease the residence time of bacterial cells in the bioreactors. Compared with control cells, the treated cells retained in the reactors over three pore volumes decreased from 22.4% ± 3.2% to 15.4% ± 3.2%.
TABLE 1.
Fitted kinetics obtained from advection-dispersion model and parameters for simulated breakthrough curves
| Group | R | km (h−1) | D (cm2/h) | (C/C0)maxa | Mean travel time (pore vol) | Retention rateb (%) |
|---|---|---|---|---|---|---|
| Control | 1.245 ± 0.017 | 0.237 ± 0.043 | 1.487 ± 0.168 | 0.085 ± 0.006 | 1.25 ± 0.02 | 22.4 ± 3.2 |
| Treated | 1.003 ± 0.023 | 0.151 ± 0.032 | 1.406 ± 0.033 | 0.116 ± 0.005 | 1.02 ± 0.02 | 15.4 ± 3.2 |
C/C0 represents the normalized cell density in the effluent (normalized by the initial cell density in the influent).
Retention rate means the proportion of the cells retained in the bioreactors.
Oxygen availability in the medium could affect the transport and biofilm formation of the bacterial cells. However, the effect of oxygen availability can be ruled out in this study because the presence of 3-CA did not affect the dissolved oxygen concentration in the medium (6.89 ± 0.04 mg/liter [medium only] versus 6.96 ± 0.05 mg/liter [medium containing 3-CA]). The reduction in cell retention in the bioreactors could be caused by a decrease in bacterial motility that might result in a low effective motility coefficient and a low hydrodynamic dispersion coefficient (41). When growing in or on the agar containing 200 ppm of 3-CA, C. testosteroni cells formed much smaller halos or colonies, confirming a repressed swimming motility in the presence of 3-CA (see Fig. S1 in the supplemental material). In addition, the hydrophobicity assay showed that about 40% of the 3-CA-treated cells and only 28% of the control cells were transferred to the hexane phase, suggesting an increase in cell surface hydrophobicity upon exposure to 3-CA. An increased hydrophobicity of the cells after 3-CA treatment may loosen the interaction between bacteria and the hydrophilic surfaces of borosilicate glass beads, which also could contribute to the observed lower retention in the bioreactors.
Genome analysis suggested versatile environmental controls of biofilm lifestyle.
To explore molecular mechanisms of the influence of 3-CA on the retention of WDL7 cells in the bioreactors, we conducted an integrated genomic and transcriptomic analysis. The draft genome of C. testosteroni WDL7 was 5.49 Mbp in length and comprised 65 contigs with a GC content of 61.47%. A total of 5,068 genes with 4,972 protein-coding genes were predicted, whereas 4,130 (81.49% of total genes) protein-coding genes were associated with predicted functions. It also had genomic features that were similar to those of other Comamonas strains (see Table S1 in the supplemental material).
In previous genomic analysis, C. testosteroni strains were found to harbor a high number of putative c-di-GMP controlling enzymes, many of which contained a sensor domain (42). c-di-GMP is one of the most important signaling molecules in biofilm regulatory networks that coordinate biofilm formation and detachment (43). A previous study showed that c-di-GMP signaling not only plays an important role in biofilm formation but also influences catabolic activity of C. testosteroni (18). The c-di-GMP level is controlled by synthases (diguanylate cyclases) and hydrolases (phosphodiesterases). In the genome of C. testosteroni WDL7, 51 putative c-di-GMP controlling proteins were identified (see Fig. S2A and Table S2 in the supplemental material). Among these putative proteins, 23 contain a sensor domain (see Fig. S2B and Table S2). Remarkably, 38 of the 51 putative c-di-GMP-controlling proteins were predicted to locate on the cytoplasmic membranes, while 20 of them contain a sensor domain. The high abundance of sensor domain-containing c-di-GMP-controlling proteins on the inner membranes suggests versatile environmental controls of the intracellular c-di-GMP concentration.
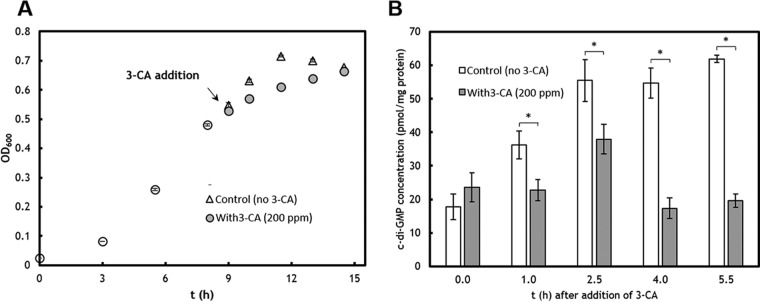
Transcriptomic analysis suggested an elevated catabolic activity but decreased c-di-GMP synthesis in C. testosteroni exposed to 3-CA.
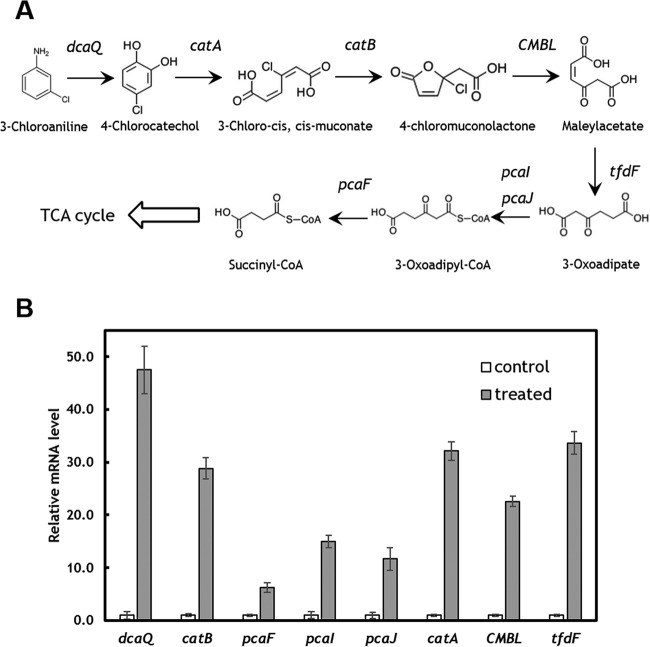
By using the draft genome as a reference, we further investigated the global transcriptomic changes after 3-CA treatment. Due to the toxicity, reduced growth was observed after 3-CA was added (see Fig. 3A). By using filtering criteria (P ≤ 0.05 and log2 fold change of ≥1), 1,696 genes were significantly downregulated and only 104 genes were upregulated. According to COG classification, various metabolic activities were repressed. More than 30% of genes in amino acid, carbohydrate, coenzyme, energy, inorganic, lipid, and nucleotide metabolisms were significantly downregulated (see Fig. S3 in the supplemental material). Other energy-intensive activities were also highly inhibited, especially motility. More than 50% of motility-related genes were downregulated (see Fig. S3), which is consistent with the bacterial motility assay (see Fig. S1). The oil/water partition coefficient of 3-CA is 1.88, suggesting its potential effects on cell membrane integrity and function (44, 45). Based on the transcriptomic data, 395 of 853 membrane-bound transporter genes exhibited a decreased expression level. The altered expression of membrane proteins may contribute partially to the changes in surface hydrophobicity. When exposed to 3-CA, one catabolic pathway for the degradation of 3-CA was induced (Fig. 2). C. testosteroni WDL7 originally was isolated from herbicide-treated soil and is capable of mineralizing 3-CA (46). Based on the induced expression of catabolic enzymes, WDL7 degraded 3-CA through the orthocleavage pathway, which is in contrast to the metacleavage pathway employed by other strains, such as C. testosteroni I2 (42). In addition, two exopolysaccharide biosynthesis genes and six exopolysaccharide export genes were found to be significantly downregulated in cells exposed to 3-CA (see Table S3). In Congo red assays, the colonies formed by C. testosteroni cells in the presence of 3-CA were of a lighter red color (see Fig. S4). As Congo red dye binds the extracellular polymeric substance (EPS) components cellulose and fimbriae, a lighter color suggested less EPS production by the cells exposed to 3-CA (47–49).
FIG 3.
Inhibited growth and decreased intracellular c-di-GMP concentration by exposure to 3-CA (200 ppm). (A) The growth of C. testosteroni WDL7 on pyruvate and inhibition by addition of 3-CA. (B) Decreased intracellular c-di-GMP after treatment with 3-CA for different lengths of time. An asterisk indicates significant difference (P < 0.05).
FIG 2.
Upregulation of 3-CA degradation pathway. (A) The orthocleavage degradation pathway. (B) The upregulated expression of catabolic genes. The transcript level was normalized to the corresponding genes in the control group. dcaQ, glutamine synthesis; catA, catechol 1,2-dioxygenase; catB, muconate cycloisomerase; CMBL, carboxymethylenebutenolidase; tfdF, maleylacetate reductase; pcaI, 3-oxoadipate coenzyme A (CoA)-transferase, alpha subunit; pcaJ, 3-oxoadipate CoA-transferase, beta subunit; pcaF, 3-oxoadipyl-CoA thiolase.
The above-described genomic analysis revealed that C. testosteroni has a sophisticated signaling system to regulate biofilm lifestyle. After being exposed to 3-CA, 28 c-di-GMP-controlling genes were downregulated (fold change > 1.5), and 24 of them are predicted to be c-di-GMP synthases (Table 2). The downregulation of this group of genes implies a decrease in the c-di-GMP level after exposure to 3-CA.
TABLE 2.
3-CA treatment decreased the expression of putative c-di-GMP-synthases
| Feature ID | Description | Log2 ratioa (3-CA/control) |
|---|---|---|
| WDL7contig_02402 | PAS domain S-box/diguanylate cyclase (GGDEF) domain | −2.24 |
| WDL7contig_01842 | Diguanylate cyclase (GGDEF) domain | −2.11 |
| WDL7contig_03518 | Diguanylate cyclase (GGDEF) domain | −1.81 |
| WDL7contig_00113 | PAS domain S-box/diguanylate cyclase (GGDEF) domain | −1.40 |
| WDL7contig_03907 | Diguanylate cyclase (GGDEF) domain | −1.36 |
| WDL7contig_02218 | Diguanylate cyclase (GGDEF) domain | −1.31 |
| WDL7contig_00230 | PAS domain S-box/diguanylate cyclase (GGDEF) domain | −0.99 |
| WDL7contig_00747 | Response regulator containing a CheY-like receiver domain and a GGDEF domain | −0.98 |
| WDL7contig_01803 | Diguanylate cyclase (GGDEF) domain | −0.98 |
| WDL7contig_02055 | Diguanylate cyclase (GGDEF) domain | −0.97 |
| WDL7contig_02791 | Diguanylate cyclase (GGDEF) domain | −0.87 |
| WDL7contig_00546 | Diguanylate cyclase (GGDEF) domain | −0.83 |
| WDL7contig_01961 | PAS domain S-box/diguanylate cyclase (GGDEF) domain | −0.78 |
| WDL7contig_02793 | PAS domain S-box/diguanylate cyclase (GGDEF) domain | −0.72 |
| WDL7contig_04387 | Diguanylate cyclase (GGDEF) domain | −0.67 |
| WDL7contig_02014 | Diguanylate cyclase (GGDEF) domain | −0.65 |
| WDL7contig_02488 | Diguanylate cyclase (GGDEF) domain | −0.65 |
| WDL7contig_00724 | Diguanylate cyclase (GGDEF) domain | −0.64 |
| WDL7contig_00769 | Diguanylate cyclase with PAS/PAC sensor | −0.64 |
| WDL7contig_03415 | Response regulator receiver modulated diguanylate cyclase | −0.62 |
Ratio indicates gene expression level in cells treated by 3-CA relative to those in the control group.
Exposure to 3-CA decreased intracellular c-di-GMP level in C. testosteroni and induced biofilm dispersal.
The intracellular c-di-GMP level was quantified before and after exposure to 3-CA (Fig. 3A). For the control cultures with no 3-CA exposure, the c-di-GMP concentration gradually increased when the cultures were growing from the exponential phase to the stationary phase (Fig. 3B).
When exposed to 3-CA, the intracellular c-di-GMP concentration was lower than that of the control cultures (Fig. 3B). At the stationary phase, the c-di-GMP level for the control cultures was about 2.5-fold of that of the 3-CA-treated cultures. Previous studies found the decreased c-di-GMP would downregulate the expression of adhesin and exopolysaccharide, which would negatively influence the irreversible attachment (49, 50). Therefore, the decreased c-di-GMP level could explain the reduced cell retention observed for the packed-bed bioreactors.
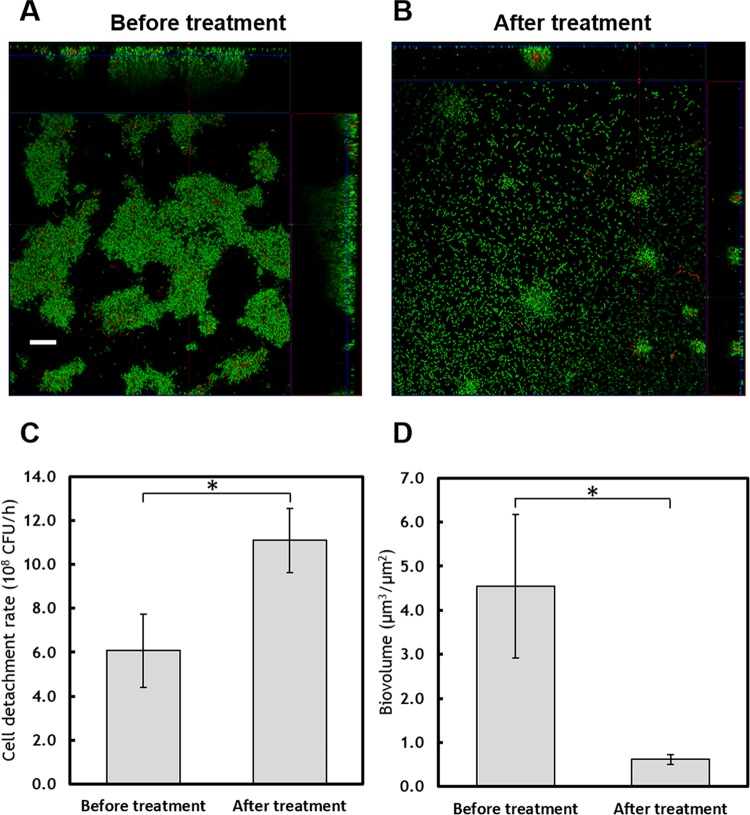
Flow cell biofilms were used to further investigate the consequences of reduced c-di-GMP upon 3-CA exposure. The WDL7 biofilms were grown using pyruvate as the sole carbon source. After 4 days of growth, the towering biofilm structure, a typical feature of mature biofilms in flow cells, was observed (Fig. 4A). Upon exposure to 3-CA, massive detachment of cells from the mature biofilms occurred (Fig. 4B). The cell detachment rate increased from 6.07 × 108 cells/h to 1.11 × 109 cells/h after 3-CA was introduced to the biofilms (Fig. 4C). Quantitative image analysis showed a loss of 86% of the biofilm biomass after 5 h of exposure to 3-CA (Fig. 4D). The massive detachment also resulted in a thinner and more even biofilm, which has a decreased mean thickness (from 13.3 ± 2.2 μm to 8.4 ± 2.0 μm; P < 0.05) and roughness (from 6.8 ± 1.3 to 4.9 ± 0.9; P < 0.05).
FIG 4.
(A) Biofilms developed in flow cells after 4 days of growth. (B) Biofilms after treatment by 3-CA (200 ppm) for 5 h. The scale bar represents 20 μm. (C) Cell detachment rate from biofilms before and after 5 h of exposure to 3-CA. (D) Biovolume of the biofilms before and after 3-CA treatment (n = 5). An asterisk indicates significant difference (P < 0.05).
Previous studies on Pseudomonas putida and Bacillus subtilis found that when exposed to aromatic hydrocarbons, cells minimize energy expenditure by repressing growth phase-dependent genes rather than pursuing a better environment (11, 12). Similar inhibition of 3-CA on cell motility and basic metabolisms of C. testosteroni were found in this study. Although in most Gram-negative bacteria a low level of c-di-GMP often leads to increased motility via the activation of the c-di-GMP-binding effectors (43), the influence of 3-CA on motility could not be predicted solely by the reduced level of c-di-GMP, because a lower-energy metabolism was also observed. Meanwhile, 3-CA treatment also results in biofilm dispersal. It suggested that, when exposed to toxic chemicals, bacterial cells prefer the free-suspended form to the biofilm lifestyle so that they can move passively with the aqueous phase.
On the community level, toxic contaminants such as 3-CA were found to have a negative impact on the whole wastewater treatment system (21, 51). In addition to inhibiting metabolic activities, exposure to 3-CA also decreased the ability of activated sludge to settle (21). A previous study showed that the granulation process was correlated with an increase in the intracellular c-di-GMP level (52). When the intracellular c-di-GMP level was reduced, a decrease in EPS content and a loss of granular stability were observed (53). Hence, the negative influence on the biofilm lifestyle and a decrease in the c-di-GMP concentration in C. testosteroni caused by the exposure to 3-CA would have important implications on bioaugmentation.
Environmental implications.
In bioaugmentation bioprocesses for the treatment of wastewater or soil containing toxic industrial pollutants, the organisms to be augmented usually are prepared in medium containing the targeted pollutants to induce desirable catabolic activities and to allow the cells to adapt well to the toxic conditions (13, 54). In this study, we demonstrate that although the catabolic activities can be induced, the preexposure to the pollutants has a great impact on the biofilm lifestyle of the bacteria, including decreases in bacterial motility and the intracellular c-di-GMP level. Consequently, when introducing the treated cultures into the bioaugmentation systems, cells would have a lower dispersion and retention coefficient, which leads to a rapid flow-out and less biomass retention. In addition, the reduced c-di-GMP level would destabilize biofilms and cause massive cell dispersal, which may eventually lead to bioaugmentation failure. For example, in real-world bioaugmentation cases, the augmented organism with weak biofilm formation ability resulted in a failed bioaugmentation (55), while enhanced biofilm formation capability improved the bioaugmentation performance (56). As a decreased c-di-GMP level leads to less initial attachment and biofilm dispersal, approaches to elevating or maintaining a high c-di-GMP level may be promising to establish and maintain sustainable bioaugmentation activity. In a previous study, an elevated intracellular c-di-GMP concentration for C. testosteroni WDL7 was achieved by constitutively expressing a c-di-GMP synthase (18). The high c-di-GMP level was found to alleviate inhibition by 3-CA at high concentration and enhance biofilm-based degradation. Genetic engineering may not be favored in environmental applications; alternative approaches may be explored to maintain a high c-di-GMP level to enhance overall biofilm robustness and reduce cell detachment under unfavorable conditions. For example, when nitrate was present in aqueous phase, C. testosteroni I2 cells residing at the bottom of the biofilm maintain metabolic activity by respiring on nitrate and also have a high c-di-GMP level to reduce biofilm dispersal (36).
In bioaugmentation, pure cultures of bacteria capable of degrading a targeted pollutant to be remediated are usually introduced into the bioremediation system. Therefore, in this study, we used pure cultures of a model organism (C. testosteroni) and one targeted pollutant (3-CA) to investigate the influence of the pollutant on the transport and biofilm lifestyle of the augmented organism. The findings from this model system provide an improved understanding of the transport and biofilm development of the augmented bacteria, which is essential for the development of novel strategies to achieve more robust bioaugmentation. When multiple pollutants are present in the bioremediation system, pollutants with similar chemical structures would likely display similar effects on the augmented bacteria; otherwise, a combined effect of different pollutants needs to be considered. When multiple organisms, for example, microbial consortia, are augmented, we would expect that both the organism-pollutant interaction and the organism-organism interaction contribute to the overall performance. The influence of the targeted pollutants on the transport and biofilm development of the augmented microbial consortia would be an intriguing research topic for future exploration.
Supplementary Material
ACKNOWLEDGMENTS
The c-di-GMP quantification was carried out with the help of Peter Imre Benke and Sanjay Swarup in the Metabolomics Laboratory of the Singapore Centre for Environmental Life Sciences Engineering (SCELSE). Sequencing of DNA and RNA was carried out with the help of Daniela Moses and Stephan Schuster using the sequencing facilities at SCELSE. We thank Hari Seshan for assistance in sample preparation, Krithika Arumugam for assistance in genome assembly, and Yingdan Zhang for assistance in oxygen measurement.
This research was supported by the National Research Foundation and Ministry of Education Singapore under its Research Centre of Excellence Programme, Singapore Centre for Environmental Life Sciences Engineering (SCELSE) (M4330005.C70), Nanyang Technological University, Singapore.
We have no competing financial interests to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00874-16.
REFERENCES
- 1.Key BD, Howell RD, Criddle CS. 1997. Fluorinated organics in the biosphere. Environ Sci Technol 31:2445–2454. doi: 10.1021/es961007c. [DOI] [Google Scholar]
- 2.Kulkarni M, Chaudhari A. 2007. Microbial remediation of nitro-aromatic compounds: an overview. J Environ Manag 85:496–512. doi: 10.1016/j.jenvman.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Spain JC. 1995. Biodegradation of nitroaromatic compounds. Annu Rev Microbiol 49:523–555. doi: 10.1146/annurev.mi.49.100195.002515. [DOI] [PubMed] [Google Scholar]
- 4.Mrozik A, Piotrowska-Seget Z. 2010. Bioaugmentation as a strategy for cleaning up of soils contaminated with aromatic compounds. Microbiol Res 165:363–375. doi: 10.1016/j.micres.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 5.El Fantroussi S, Agathos SN. 2005. Is bioaugmentation a feasible strategy for pollutant removal and site remediation? Curr Opin Biotechnol 8:268–275. [DOI] [PubMed] [Google Scholar]
- 6.Thompson IP, Van Der Gast CJ, Ciric L, Singer AC. 2005. Bioaugmentation for bioremediation: the challenge of strain selection. Environ Microbiol 7:909–915. doi: 10.1111/j.1462-2920.2005.00804.x. [DOI] [PubMed] [Google Scholar]
- 7.Bouchez T, Patureau D, Dabert P, Juretschko S, Dore J, Delgenes P, Moletta R, Wagner M. 2000. Ecological study of a bioaugmentation failure. Environ Microbiol 2:179–190. doi: 10.1046/j.1462-2920.2000.00091.x. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein R, Mallory L, Alexander M. 1985. Reasons for possible failure of inoculation to enhance biodegradation. Appl Environ Microbiol 50:977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogel TM. 1996. Bioaugmentation as a soil bioremediation approach. Curr Opin Biotechnol 7:311–316. doi: 10.1016/S0958-1669(96)80036-X. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Wu J-F, Zeyer J, Meng B, Liu L, Jiang C-Y, Liu S-Q, Liu S-J. 2009. Proteomic and molecular investigation on the physiological adaptation of Comamonas sp. strain CNB-1 growing on 4-chloronitrobenzene. Biodegradation 20:55–66. doi: 10.1007/s10532-008-9199-x. [DOI] [PubMed] [Google Scholar]
- 11.Tam LT, Eymann C, Albrecht D, Sietmann R, Schauer F, Hecker M, Antelmann H. 2006. Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis. Environ Microbiol 8:1408–1427. doi: 10.1111/j.1462-2920.2006.01034.x. [DOI] [PubMed] [Google Scholar]
- 12.Domínguez-Cuevas P, González-Pastor J-E, Marqués S, Ramos J-L, de Lorenzo V. 2006. Transcriptional tradeoff between metabolic and stress-response programs in Pseudomonas putida KT2440 cells exposed to toluene. J Biol Chem 281:11981–11991. doi: 10.1074/jbc.M509848200. [DOI] [PubMed] [Google Scholar]
- 13.Boon N, Goris J, De Vos P, Verstraete W, Top EM. 2000. Bioaugmentation of activated sludge by an indigenous 3-chloroaniline-degrading Comamonas testosteroni strain, I2gfp. Appl Environ Microbiol 66:2906–2913. doi: 10.1128/AEM.66.7.2906-2913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov V, Wang X-H, Tay ST-L, Tay J-H. 2006. Bioaugmentation and enhanced formation of microbial granules used in aerobic wastewater treatment. Appl Microbiol Biotechnol 70:374–381. doi: 10.1007/s00253-005-0088-5. [DOI] [PubMed] [Google Scholar]
- 15.Cao B, Majors PD, Ahmed B, Renslow RS, Silvia CP, Shi L, Kjelleberg S, Fredrickson JK, Beyenal H. 2012. Biofilm shows spatially stratified metabolic responses to contaminant exposure. Environ Microbiol 14:2901–2910. doi: 10.1111/j.1462-2920.2012.02850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh R, Paul D, Jain RK. 2006. Biofilms: implications in bioremediation. Trends Microbiol 14:389–397. doi: 10.1016/j.tim.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Ding Y, Cohen Y, Cao B. 2015. Elevated level of the second messenger c-di-GMP in Comamonas testosteroni enhances biofilm formation and biofilm-based biodegradation of 3-chloroaniline. Appl Microbiol Biotechnol 99:1967–1976. doi: 10.1007/s00253-014-6306-2. [DOI] [PubMed] [Google Scholar]
- 19.Ding Y, Peng N, Du Y, Ji L, Cao B. 2014. Disruption of putrescine biosynthesis in Shewanella oneidensis enhances biofilm cohesiveness and performance in Cr(VI) immobilization. Appl Environ Microbiol 80:1498–1506. doi: 10.1128/AEM.03461-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boon N, De Gelder L, Lievens H, Siciliano SD, Top EM, Verstraete W. 2002. Bioaugmenting bioreactors for the continuous removal of 3-chloroaniline by a slow release approach. Environ Sci Technol 36:4698–4704. doi: 10.1021/es020076q. [DOI] [PubMed] [Google Scholar]
- 21.Boon N, Top EM, Verstraete W, Siciliano SD. 2003. Bioaugmentation as a tool to protect the structure and function of an activated-sludge microbial community against a 3-chloroaniline shock load. Appl Environ Microbiol 69:1511–1520. doi: 10.1128/AEM.69.3.1511-1520.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M, Ford RM. 2009. Transverse bacterial migration induced by chemotaxis in a packed column with structured physical heterogeneity. Environ Sci Technol 43:5921–5927. doi: 10.1021/es901001t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adadevoh JS, Triolo S, Ramsburg CA, Ford RM. 2016. Chemotaxis increases the residence time of bacteria in granular media containing distributed contaminant sources. Environ Sci Technol 50:181–187. doi: 10.1021/acs.est.5b03956. [DOI] [PubMed] [Google Scholar]
- 24.Tang G, Mayes MA, Parker JC, Jardine PM. 2010. CXTFIT/Excel–a modular adaptable code for parameter estimation, sensitivity analysis and uncertainty analysis for laboratory or field tracer experiments. Comput Geosci 36:1200–1209. doi: 10.1016/j.cageo.2010.01.013. [DOI] [Google Scholar]
- 25.Murray TS, Kazmierczak BI. 2008. Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J Bacteriol 190:2700–2708. doi: 10.1128/JB.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prabhu Y, Phale P. 2003. Biodegradation of phenanthrene by Pseudomonas sp. strain PP2: novel metabolic pathway, role of biosurfactant and cell surface hydrophobicity in hydrocarbon assimilation. Appl Microbiol Biotechnol 61:342–351. doi: 10.1007/s00253-002-1218-y. [DOI] [PubMed] [Google Scholar]
- 27.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 28.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markowitz VM, Mavromatis K, Ivanova NN, Chen I-MA, Chu K, Kyrpides NC. 2009. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 30.Hyatt D, Chen G-L, LoCascio P, Land M, Larimer F, Hauser L. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M. 2011. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol 12:R30. doi: 10.1186/gb-2011-12-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markowitz VM, Chen I-MA, Chu K, Szeto E, Palaniappan K, Pillay M, Ratner A, Huang J, Pagani I, Tringe S. 2014. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res 42:D568–D573. doi: 10.1093/nar/gkt919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. 1999. Protein interaction maps for complete genomes based on gene fusion events. Nature 402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 35.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y, Shukal S, Mukherjee M, Cao B. 2015. Involvement in denitrification is beneficial to the biofilm lifestyle of Comamonas testosteroni: a mechanistic study and its environmental implications. Environ Sci Technol 49:11551–11559. doi: 10.1021/acs.est.5b03381. [DOI] [PubMed] [Google Scholar]
- 37.Kuchma S, Ballok A, Merritt J, Hammond J, Lu W, Rabinowitz J, O'Toole GA. 2010. Cyclic-di-GMP-mediated repression of swarming motility by Pseudomonas aeruginosa: the pilY1 gene and its impact on surface-associated behaviors. J Bacteriol 192:2950–2964. doi: 10.1128/JB.01642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spangler C, Böhm A, Jenal U, Seifert R, Kaever V. 2010. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J Microbiol Methods 81:226–231. doi: 10.1016/j.mimet.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 39.Mohanty A, Wei L, Lu L, Chen Y, Cao B. 2015. Impact of sublethal levels of single-wall carbon nanotubes on pyoverdine production in Pseudomonas aeruginosa and its environmental implications. Environ Sci Technol Lett 2:105–111. doi: 10.1021/acs.estlett.5b00057. [DOI] [Google Scholar]
- 40.Teal TK, Lies DP, Wold BJ, Newman DK. 2006. Spatiometabolic stratification of Shewanella oneidensis biofilms. Appl Environ Microbiol 72:7324–7330. doi: 10.1128/AEM.01163-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camper AK, Hayes JT, Sturman PJ, Jones WL, Cunningham AB. 1993. Effects of motility and adsorption rate coefficient on transport of bacteria through saturated porous media. Appl Environ Microbiol 59:3455–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Y, Arumugam K, Tay MQX, Seshan H, Mohanty A, Cao B. 2015. Comparative genome analysis reveals genetic adaptation to versatile environmental conditions and importance of biofilm lifestyle in Comamonas testosteroni. Appl Microbiol Biotechnol 99:3519–3532. doi: 10.1007/s00253-015-6519-z. [DOI] [PubMed] [Google Scholar]
- 43.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 44.Zok S, Görge G, Kalsch W, Nagel R. 1991. Bioconcentration, metabolism and toxicity of substituted anilines in the zebrafish (Brachydanio rerio). Sci Total Environ 109:411–421. [DOI] [PubMed] [Google Scholar]
- 45.Ramos JL, Duque E, Gallegos M-T, Godoy P, Ramos-González MI, Rojas A, Terán W, Segura A. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu Rev Microbiol 56:743–768. doi: 10.1146/annurev.micro.56.012302.161038. [DOI] [PubMed] [Google Scholar]
- 46.Dejonghe W, Berteloot E, Goris J, Boon N, Crul K, Maertens S, Höfte M, De Vos P, Verstraete W, Top EM. 2003. Synergistic degradation of linuron by a bacterial consortium and isolation of a single linuron-degrading Variovorax strain. Appl Environ Microbiol 69:1532–1541. doi: 10.1128/AEM.69.3.1532-1541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedman L, Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol 51:675–690. [DOI] [PubMed] [Google Scholar]
- 48.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ueda A, Wood TK. 2009. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog 5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. 2007. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falk MW, Wuertz S. 2010. Effects of the toxin 3-chloroaniline at low concentrations on microbial community dynamics and membrane bioreactor performance. Water Res 44:5109–5115. doi: 10.1016/j.watres.2010.08.043. [DOI] [PubMed] [Google Scholar]
- 52.Wan C, Yang X, Lee D-J, Wang X-Y, Yang Q, Pan X. 2014. Aerobic granulation of aggregating consortium X9 isolated from aerobic granules and role of cyclic di-GMP. Bioresour Technol 152:557–561. doi: 10.1016/j.biortech.2013.11.052. [DOI] [PubMed] [Google Scholar]
- 53.Wan C, Zhang P, Lee D-J, Yang X, Liu X, Sun S, Pan X. 2013. Disintegration of aerobic granules: role of second messenger cyclic di-GMP. Bioresour Technol 146:330–335. doi: 10.1016/j.biortech.2013.07.073. [DOI] [PubMed] [Google Scholar]
- 54.Gentry TJ, Newby DT, Josephson KL, Pepper IL. 2001. Soil microbial population dynamics following bioaugmentation with a 3-chlorobenzoate-degrading bacterial culture. Biodegradation 12:349–357. doi: 10.1023/A:1014394709703. [DOI] [PubMed] [Google Scholar]
- 55.Fu S, Fan H, Liu S, Liu Y, Liu Z. 2009. A bioaugmentation failure caused by phage infection and weak biofilm formation ability. J Environ Sci 21:1153–1161. doi: 10.1016/S1001-0742(08)62396-7. [DOI] [PubMed] [Google Scholar]
- 56.Wang M-Z, Zheng X, Zhang K, Ding Y-C, He H-Z, Shen D-S, Feng H-J. 2014. A new method for rapid construction of a Pseudomonas sp. HF-1 bioaugmented system: accelerating acylated homoserine lactones secretion by pH regulation. Bioresour Technol 169:229–235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.