ABSTRACT
Hybrid isolates of Shiga toxin-producing Escherichia coli (STEC) and enterotoxigenic E. coli (ETEC) encoding heat-stable enterotoxin (ST) are being reported with increasing frequency from a variety of sources. However, information regarding the plasmids that these strains harbor is scarce. In this study, we sequence and characterize a plasmid, p7v, from the STEC/ETEC hybrid strain 7v. Whole-genome phylogenetic analyses of STEC/ETEC hybrid strains and prototype E. coli isolates of other pathotypes placed 7v in the Escherichia sp. cryptic lineage 1 (CL1) clade. The complete plasmid, p7v, was determined to be 229,275 bp and encodes putative virulence factors that are typically carried on STEC plasmids as well as those often carried on ETEC plasmids, indicating that the hybrid nature of the strain extends beyond merely encoding the two toxins. Plasmid p7v carries two copies of sta with identical sequences, which were discovered to be divergent from the sta sequences found in the prototype human ETEC strains. Using a nomenclature scheme based on a phylogeny constructed from sta and stb sequences, the sta encoded on p7v is designated STa4. In silico analysis determined that p7v also encodes the K88 fimbria, a colonization factor usually associated with porcine ETEC plasmids. The p7v sequence and the presence of plasmid-encoded virulence factors are compared to those of other STEC/ETEC CL1 hybrid genomes and reveal gene acquisition/loss at the strain level. In addition, the interrogation of 24 STEC/ETEC hybrid genomes for identification of plasmid replicons, colonization factors, Stx and ST subtypes, and other plasmid-encoded virulence genes highlights the diversity of these hybrid strains.
IMPORTANCE Hybrid Shiga toxin-producing Escherichia coli/enterotoxigenic Escherichia coli (STEC/ETEC) strains, which have been isolated from environmental, animal, and human clinical samples, may represent an emerging threat as food-borne pathogens. Characterization of these strains is important for assessing virulence potential, aiding in the development of pathogen detection methods, and understanding how the hybrid strains evolve to potentially have a greater impact on public health. This study represents, to our knowledge, both the first characterization of a closed plasmid sequence from a STEC/ETEC hybrid strain and the most comprehensive phylogenetic analysis of available STEC/ETEC hybrid genomes to date. The results demonstrate how the mobility of plasmid-associated virulence genes has resulted in the creation of a diverse plasmid repertoire within the STEC/ETEC hybrid strains.
INTRODUCTION
Pathogenic Escherichia coli strains are categorized into pathotypes or pathovars (pathogenic variants) based on virulence-associated genes whose expression contributes to particular disease symptoms in the human host (1). The virulence-associated genes that are unique to a pathotype have been used as molecular markers to define the pathotype of a given E. coli isolate. Two E. coli pathotypes that cause a significant burden of illness throughout the world are Shiga toxin-producing E. coli (STEC) and enterotoxigenic E. coli (ETEC) (1, 2). STEC genomes possess at least one allele variant of either the stx1 or stx2 genes encoding Shiga toxin. The Shiga toxin alleles have been classified into 10 subtypes by sequence similarity: stx1a, stx1c, stx1d, stx2a, stx2b, stx2c, stx2d, stx2e, stx2f, and stx2g (3, 4). Colonization of the human large intestine with STEC can result in severe hemorrhagic colitis and, in some cases, a potentially fatal complication, hemolytic-uremic syndrome (HUS), resulting from elaboration of the Shiga toxin. ETEC strains colonize primarily in the small intestine and cause watery diarrhea in the human host by expressing heat-stable enterotoxin (ST) and/or heat-labile enterotoxin (LT) (1). There are two classes of ST, STa (encoded by sta) and STb (encoded by stb), which differ in sequence and mechanism of action (1). STa is associated with human disease, while STb is typically associated with ETEC infection in pigs, although the stb gene has been found in human ETEC isolates (1, 4–6). Some ETEC strains express fimbrial intestinal colonization factors (CFs) that are specific to animals and are an important cause of diarrheal disease in pigs (1, 7).
Many of the virulence-associated genes that define a pathotype are carried on mobile genetic elements and, thus, allow potential genetic combinations that result in hybrid pathotype strains. Highly virulent hybrid pathotype strains that possess molecular markers associated with more than one E. coli pathotype have been reported and include, for example, the STEC/enteroaggregative E. coli O104:H4 hybrid that caused an outbreak with a high percentage of HUS in Germany in 2011 (8). A recent report describes an STEC/extraintestinal pathogenic E. coli O80:H2 hybrid that caused HUS and bacteremia (9). STEC/uropathogenic E. coli O2:H6 hybrid strains have also been reported (10). An additional recent report of an E. coli strain isolated from a child with acute diarrhea describes the strain as an enteropathogenic E. coli/ETEC hybrid since the genome contains the locus of enterocyte effacement (LEE) pathogenicity island and a gene encoding LT (11). STEC/ETEC hybrid pathotype strains, carrying a gene expressing Shiga toxin and a gene expressing ST, have been isolated from many sources, including humans, animals, food, and water (4, 12–18). Some STEC/ETEC hybrid strains have been associated with diarrheal disease and HUS in humans (14, 17, 19), whereas others have been associated with diarrheal disease in pigs (13, 20).
In human isolates, LEE-positive STEC (also known as enterohemorrhagic E. coli [EHEC]) strains predominantly form two phylogenetic clusters based on core genome sequences (21); however, LEE-negative STEC strains have more diverse genetic backbones (4, 19, 21). Although discrete lineages of ETEC have been identified in high-resolution genomic analysis, ETEC strains are considerably diverse genetically (22, 23). Various genome comparison studies using core genome multilocus sequence typing (MLST) (19), MLST based on six genes (7), or pulsed-field gel electrophoresis (16, 17) have revealed that hybrid STEC/ETEC genomes are also quite diverse. In addition to strains belonging to the Escherichia coli species, hybrid Shiga toxin-producing Escherichia spp./ETEC strains have been identified that are classified phylogenetically as Escherichia sp. cryptic lineage 1 (Escherichia sp. CL1) (4, 19, 24, 25). Escherichia sp. CL1 strains are phenotypically indistinguishable from E. coli species but form a distinct clade by genomic analysis (4, 25–27). One such strain, 7v, possesses the genes that encode Stx2g and STa (4, 17). For simplicity, the hybrid 7v strain and other Shiga toxin-producing Escherichia sp. CL1 strains carrying genes that encode ST will be referred to as STEC/ETEC hybrids throughout this work.
While the Shiga toxin gene is carried in a prophage integrated in the chromosome, the sta or stb gene is usually carried by a plasmid in ETEC (2). ETEC strains frequently contain more than one plasmid, one of which, not necessarily encoding ST, often encodes a fimbrial adhesin referred to as a colonization factor (CF) (2, 28). ETEC strains isolated from humans elaborate human-specific CFs, while porcine ETEC isolates primarily harbor plasmids encoding the CF K88 or F18 (1, 2, 7). Not all ETEC strains carry known CFs, and these strains may represent CF-negative strains or possess CFs that are not yet identified (22). In addition to STs and CFs, a virulence factor often encoded on ETEC plasmids is the serine protease EatA (2, 28, 29). STEC also commonly carries a gene encoding a serine protease, in this case espP, on a plasmid (2). Another virulence factor carried by most LEE-positive and some LEE-negative STEC plasmids is enterohemolysin, which is encoded by the ehxCABD operon (2, 30–32). The catalase-peroxidase gene, katP, is frequently found on LEE-positive STEC plasmids but is not normally encountered on LEE-negative STEC plasmids (4, 30–32).
In general, ETEC plasmids contain an extensive number of insertion sequences and possess a mosaic structure leading to a highly diverse plasmid repertoire (33). While LEE-positive STEC plasmids are not as highly diverse as ETEC plasmids, LEE-negative STEC plasmids appear to exhibit great variation (4) but remain largely uncharacterized with the exception of pO113 (31). Additionally, the sequences of Escherichia sp. CL1 plasmids have not been previously characterized; however, STEC/ETEC strain 7v is reported to harbor a large plasmid encoding STa (17). It has been noted that, although there is heterogeneity between virulence plasmids carried by a particular E. coli pathotype, the plasmids display a greater level of similarity within a pathotype than between pathotypes (2). In this study, the virulence plasmid p7v harbored in hybrid STEC/ETEC strain 7v is sequenced and characterized, and we address the question of whether virulence plasmids harbored in STEC/ETEC hybrid strains bear more resemblance to those typical of STEC or ETEC strains and whether this is strain dependent. Plasmid-encoded virulence factors that are carried in the genomes of 24 hybrid STEC/ETEC strains are identified. As p7v carries a previously undescribed STa-encoding allele, a phylogeny of ST-encoding gene sequences from ETEC and hybrid STEC/ETEC strains is constructed, and we determine the extent to which this allele is found in other STEC/ETEC hybrid strains. In addition, this work presents the first description, to our knowledge, of a plasmid harbored in a CL1 strain, for which information is scarce, thus aiding in elucidating the extent to which plasmids in CL1 strains potentially carry plasmid-encoded virulence factors found in E. coli species.
MATERIALS AND METHODS
STEC/ETEC hybrid strains included in this study.
STEC/ETEC hybrid strains included in this study are listed in Table 1. The novel STEC/ETEC strains that were sequenced and characterized in this study are MDP04-01392, MDP04-02111, MI02-35, 857226, and 628591-2 and were kindly provided by Peter Feng, while strain E149 was obtained from the STEC Center (www.shigatox.net/new). The draft genomes of the remaining 18 STEC/ETEC hybrid strains were obtained from GenBank (Table 1).
TABLE 1.
Characteristics of the STEC/ETEC hybrid strains included in this study
| Straina | Serotype | Phylogroup | Location/year | Source | Shiga toxin(s) | Heat-stable enterotoxin(s) | Colonization factor | Plasmid replicon(s) | Accession no. (reference)b |
|---|---|---|---|---|---|---|---|---|---|
| 7v | O2:H25 | CL1 | Hong Kong | Feces of healthy cattle | Stx2g | STa4c | K88 | FIB(AP001918), FII(pSE11) | AEXD02000000 (50) |
| 628591-2 | O2:H25 | CL1 | USA/2010 | Coriander | Stx2gc | STa4 | K88 | FIB(AP001918), FII(pSE11) | JZDN01000000 (this study) |
| 1.2741 | O2:H25 | CL1 | Unknown | Bovine | Stx1a, Stx2g | STa4, STa5 | K88 | FIB(AP001918), FII(pSE11) | AEZI02000000 |
| FE95160 | O2:H25 | CL1 | Burkina Faso/2008 | Bovine intestine | Stx1a | STa5 | FIB(AP001918), FII(pSE11) | LFZI01000000 (15) | |
| E807 | O2:H45 | CL1 | Australia | Freshwater sediment | Stx1a, Stx2a | STa5 | K88 | FIB(AP001918), FII(pSE11), Col156 | AEJX01000000 (24) |
| IH53473 | O101:H33 | A | Finland/2001 | Infant with HUS | Stx2a | STa4 | FII(29), X1 | LFZH01000000 (16) | |
| CFSAN026836 | O109:H48 | A | USA/1999 | Pond water | Stx1a | STa4 | K88 | FIB(AP001918), FII(pSE11), FIC | LDCY01000000 |
| CFSAN026835 | O109:H48 | A | USA/1999 | Bovine | Stx1a | STa4 | K88 | FIB(AP001918), FII(pSE11), FIC | LDCZ01000000 |
| CVMN33742PS | O136:H16 | A | USA/2011 | Farm environment | Stx2g | STa4 | K88 | FIB(AP001918), FII(pSE11), Col156, Col(MG828), ColRNAI, Col(MGD2) | JUDF01000000 |
| CVMN33429PS | O136:H16 | A | USA/2011 | Farm environment | Stx1a | STa4, STa5 | K88 | FIB(AP001918), FII(pSE11), FIC, ColRNAI | JWZR01000000 |
| CFSAN026844 | O136:H16 | A | USA/1999 | Bovine | Stx1a | STa4, STa5 | K88 | FIB(AP001918), FII(pSE11), A/C2, ColRNAI | LDCW01000000 |
| CFSAN026843 | O136:H16 | A | USA/1999 | Deer | Stx1a | STa4, STa5 | K88 | FIB(AP001918), FII(pSE11), A/C2, ColRNAI | LGZN01000000 |
| S1191 | O141:H4 | A | Unknown | Pig with edema disease | Stx2e | STb1 | F18 | FII(pCoo), FII, X1, X4, I1, Y | AFEA02000000 (51) |
| K88 | O141:H4 | A | China | Pig | Stx2e | STb1 | K88 | FIB(AP001918), FII(pSE11), FII(pCoo), FIC, X4, I1, pO111, ColRNAI | LBBN01000000 |
| UMNF18 | O147:H4 | A | Iowa/2007 | Pig with diarrhea | Stx2e | STa1 | F18 | FII(pCoo), X1, X4, I1, ColRNAI | AGTD01000000 (7) |
| 2.3916 | O147:H4 | A | Unknown | Pig | Stx2e | STa1, STb1 | F18 | FII(pCoo), X1, I1, P, Y | AFAB02000000 |
| IH57218 | O2:H27 | A | Finland/1997 | Child with diarrhea | Stx2a | STa4 | FII | LFZJ01000000 (16) | |
| E149 | O138:H14 | D | England | Pig | Stx2e | STa1, STb1 | F18 | FII(pCoo), X1, I1, P, ColRNAI | JZXA01000000 (12) |
| C165-02 | O73:H18 | D | Denmark | Patient with bloody diarrhea | Stx2d | STb2 | Tcfd | FIB(AP001918), FII(29), FIA, I1, pO111 | AFDR02000000 (52) |
| 3020-98 | O187:H52 | B1 | Virginia/1998 | Infant with diarrhea | Stx1c | STa4 | FIB(AP001918), FII | AVRH01000000 (12) | |
| MDP04-01392 | O187:H52 | B1 | Maryland/2004 | Cantaloupe | Stx1c | STa4 | FIB(AP001918), FII | JZDJ01000000 (12) | |
| MDP04-02111 | O187:H52 | B1 | Michigan/2004 | Cilantro | Stx1c | STa4 | FIB(AP001918), FII | JZDK01000000 (12) | |
| MI02-35 | O187:H52 | B1 | Michigan/2002 | Patient with bloody diarrhea | Stx1c | STa4 | FIB(AP001918), FII, L/M(pMU407) | JZDL01000000 (12) | |
| 857226 | O187:H52 | B1 | Unknown | Raw milk Havarti cheese | Stx1c | STa4 | FIB(AP001918), FII | JZDM01000000 (this study) |
Strains in bold font were sequenced as part of this study.
The accession number is unpublished if no reference is given.
The genome contains two copies.
Tcf, Typhi colonization factor.
DNA isolation and whole-genome sequencing.
Genomic DNA was extracted from overnight cultures grown in Luria broth at 37°C using the DNeasy blood and tissue kit (Qiagen, Germantown, MD, USA). Sequencing libraries were prepared from genomic DNA with the Nextera DNA sample preparation kit (Illumina, San Diego, CA, USA) and were sequenced on the Illumina MiSeq platform, generating paired-end 250-bp reads in sufficient quantity to provide between 104- and 219-fold coverage for each genome. Raw reads were trimmed and draft genome sequences were assembled de novo with CLC Genomics Workbench v7.5 or v8.0 (CLC bio, Boston, MA, USA).
Plasmid isolation, sequencing, and characterization.
Plasmid DNA was extracted from STEC/ETEC strain 7v using the Qiagen Large-Construct kit following the protocol included for plasmid extraction without removal of contaminating genomic DNA (Qiagen). The enriched plasmid DNA was sheared using a Covaris M220 Focused-ultrasonicator (Covaris, Woburn, MA, USA), a sequencing library was prepared with the TruSeq DNA sample preparation kit (Illumina), and the library was sequenced on the Illumina MiSeq platform, generating paired-end 300-bp reads. A mate pair sequencing library was also prepared from the purified plasmid DNA using the Nextera mate pair library preparation kit (Illumina). The mate pair library was sequenced on the Illumina MiSeq platform, generating paired-end 250-bp reads. Raw reads were trimmed, and contigs were assembled de novo with CLC Genomics Workbench v6.5.1 (CLC bio) including reads from both sequencing libraries. To obtain a closed plasmid sequence, suspected plasmid contigs were joined bioinformatically by overlapping sequence, and one physical gap was closed by manual PCR and sequencing. This was accomplished using primer pair 5′-TGGTTACTGGCAGAGATGAGC-3′ and 5′-CTGGAATGATAAAGCAGTCTAGG-3′, resulting in a PCR product that was sequenced using Sanger technology with a GeXP8800 instrument (Beckman Coulter Inc., Fullerton, CA). Samples were analyzed using the GeXP8800 software, and the resulting sequence trace files were exported for further analysis with the SeqMan Pro module of the Lasergene software package (DNASTAR Inc., Madison, WI). Joined sequences were then verified by mapping the reads onto the joined sequence using CLC Genomics Workbench (CLC bio). The sequence of the completely closed plasmid p7v was annotated using RAST (34). Manual annotation using BLASTp (http://blast.ncbi.nlm.nih.gov/Blast.cgi) for selected predicted coding sequences was also performed.
Whole-genome phylogeny.
Using the E. coli K-12 MG1655 genome as a reference, BLASTn analyses of 42 representative closed E. coli/Shigella genomes identified 2,542 conserved chromosomal genes present in at least 41 of the genomes with at least 95% sequence similarity to K-12 MG1655. Strain relatedness was determined through phylogenetic analysis of sequence variation of these 2,542 conserved chromosomal genes. Sequences for these genes were extracted from the draft genomes of the 24 STEC/ETEC hybrid strains via BLASTn searches using the sequences from strain K-12 MG1655 as queries, and for each genome, the matching bases were aligned in one file for each core gene. Each core gene alignment was then scanned for nucleotide variation at each position. Draft genomes of five CL1 strains and closed genomes of 26 reference E. coli prototype strains from other E. coli pathotypes were included in the phylogenetic analysis for comparative purposes (GenBank accession numbers are given in Fig. 1). A total of 235,269 polymorphic sites were identified among the 55 genomes, which were then concatenated and imported into MEGA 3.1 (35) for neighbor-joining analysis using a p distance matrix and 500 bootstrap replications, with gaps in the data handled by the pairwise deletion option. E. coli phylogroups were assigned to the STEC/ETEC hybrid strains on the basis of their location within the resulting phylogenetic tree.
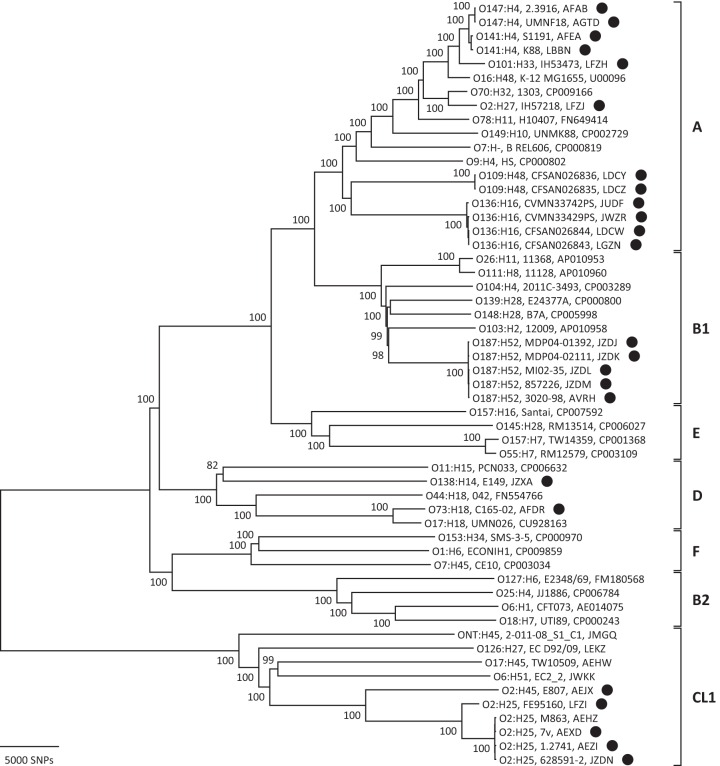
FIG 1.
Phylogenetic relationships of STEC/ETEC hybrid strains to other E. coli strains. This neighbor-joining tree was constructed using a p distance matrix. Molecular serotypes, strain designations, and GenBank accession numbers (or WGS prefixes) are given at the branch tips. STEC/ETEC hybrid strains are indicated by the black circles. E. coli (A, B1, B2, D, E, and F) and Escherichia sp. cryptic lineage 1 (CL1) phylogroups are marked by the square brackets. SNPs, single nucleotide polymorphisms.
Molecular serotyping and plasmid sequence comparisons.
The molecular serotypes of the STEC/ETEC hybrid strains were determined from the draft genomes by BLASTn analysis utilizing an in-house custom database, including the wzx, wzy, wzm, wzt, fliC, flkA, fllA, flmA, and flnA loci (36–42). BLASTn analysis that utilized an in-house custom database was also used to identify the allele subtypes of the Shiga toxin genes and the presence of heat-stable enterotoxin-encoding genes. The STEC/ETEC draft genomes were interrogated in silico using BLASTn analysis with primer sequences obtained from published work for ETEC colonization factors CS1, CS2, CS3, CS4, CS5, CS6, CS7, CS8, CS12, CS13, CS14, CS15, CS17, CS18, CS19, CS20, CS21, CS22, and CFA/I (43, 44). In addition, the genomes were queried by tBLASTn using sequences for the animal versions of the colonization factors, including all eight K88 fimbrial proteins, FaeC, FaeD, FaeE, FaeF, FaeG, FaeH, FaeI, and FaeJ (plasmid pUMNK88_K88, GenBank accession no. CP002730.1), and the four F18 fimbrial proteins, FedA, FedB, FedC, and FedE (E. coli strain UMNF18, GenBank accession no. AGTD01000000). Additional tBLASTn searches using sequences for Typhi colonization factor (Yersinia kristensenii TcfC, GenBank accession no. CNL86260, and Salmonella enterica subsp. enterica serovar Senftenberg strain ATCC 43845 TcfC, GenBank accession no. ESF46116.1) were performed. Plasmid replicons were identified in the STEC/ETEC genomes using PlasmidFinder 1.3 (45), with a threshold identity of 85%. To determine whether CL1 strains other than 7v harbor a plasmid with a similar sequence to p7v, BLASTn analysis was performed on CL1 draft genomes using the sequence of p7v as a query. For hybrid STEC/ETEC CL1 strains, as well as for ETEC CL1 strain M863, the BLAST Ring Image Generator (BRIG) v0.95 was utilized for sequence comparison (46). STEC/ETEC strains were also interrogated by tBLASTn analysis for the presence of the STEC virulence genes katP, espP, and ehxA, which are carried on the virulence plasmid pO157 harbored in EHEC strain EDL933 (GenBank accession no. CP008958.1), eatA (GenBank accession no. FN649418.1), and astA (GenBank accession no. YP_009077498). For all tBLASTn analyses, a gene was considered present with an amino acid sequence identity of ≥80%.
Heat-stable enterotoxin phylogeny.
Gene sequences for STa and STb were downloaded from GenBank or extracted from genomes sequenced in this work. A total of 228 sta and 22 stb sequences were included, of which 21 unique sta and 5 unique stb sequences were identified. The 26 unique sequences were aligned using the ClustalW algorithm as implemented in the MegAlign module of the Lasergene software package (DNASTAR Inc.). The resulting alignment was imported into MEGA for neighbor-joining analysis using a p distance matrix and 500 bootstrap replications.
Nucleotide sequence accession numbers.
The sequence of plasmid p7v was deposited in GenBank under accession no. KT992796. The draft genome sequences of the STEC/ETEC strains MDP04-01392, MDP04-02111, MI02-35, 857226, 628591-2, and E149 were deposited in GenBank under the accession numbers given in Table 1.
RESULTS
Whole-genome phylogeny.
Similar to strain 7v, the hybrid STEC/ETEC strain FE95160 was determined by Nyholm et al. to be an Escherichia sp. CL1 strain (19). In order to discover whether there are other hybrid STEC/ETEC CL1 strains and how these hybrid strains compare phylogenetically to other CL1 strains as well as to other STEC/ETEC hybrid strains in general, a core genome phylogeny was determined. The 24 STEC/ETEC hybrid strains included were obtained from a variety of sources. Molecular serotyping based on the genome sequences revealed an assortment of serotypes (Table 1). Phylogenetic analysis revealed that five STEC/ETEC hybrid strains cluster in the CL1 phylogroup, inclusive of strain 7v (Fig. 1). In fact, two strains, 628591-2 and 1.2741, share 99.96% core nucleotide sequence similarity with strain 7v. The remainder of the STEC/ETEC hybrid strains were distributed throughout E. coli phylogroups A (12 strains), B1 (5 strains), and D (2 strains). No STEC/ETEC hybrid strains clustered with phylogroup B2, E, or F. Although the results determined that 12 strains are members of phylogroup A, they are not found in a single cluster; instead, some strains are found on more distant branches within the phylogroup. In contrast, the CL1 STEC/ETEC hybrid strains appear to cluster more closely within the CL1 lineage. Visual inspection of the amount of backbone variation (i.e., branch lengths) that is present among the CL1 hybrid strains appears comparable to that observed within each of the E. coli phylogroups as opposed to the species-wide variation across the phylogroups, suggesting that the CL1 hybrid strains belong to a single phylogroup within CL1 (Fig. 1).
Characterization of plasmid p7v.
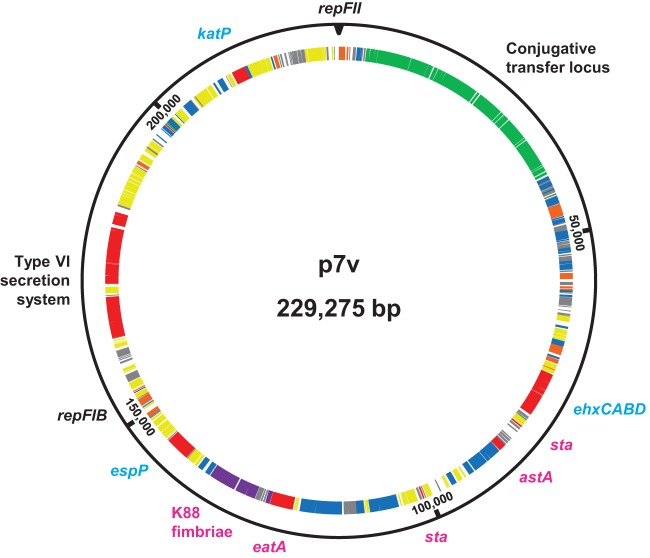
The closed sequence of the large virulence plasmid harbored in STEC/ETEC strain 7v was determined to be 229,275 bp in size. Annotation of the plasmid sequence using RAST (34) revealed 340 coding sequences. Interestingly, p7v is a hybrid plasmid that encodes virulence factors associated with STEC plasmids as well as those associated with ETEC plasmids (Fig. 2). STEC plasmid-associated genes carried by p7v include enterohemolysin ehxCABD (2, 30–32), serine protease espP (2, 31), and catalase katP (4, 30–32). ETEC plasmid-associated genes carried by p7v include heat-stable enterotoxin sta and serine protease eatA (2, 28, 29). The astA gene that encodes the EAST1 toxin, associated with several E. coli pathotypes including ETEC and STEC (47), is also carried on p7v. Plasmid p7v was found to contain a conjugative transfer locus, frequently found on E. coli plasmids, regardless of pathotype. ETEC strains often carry a plasmid-encoded CF locus, so we sought to determine whether p7v carries a CF locus. tBLASTn analysis and annotation of p7v revealed that p7v possesses all eight genes constituting an operon similar to the K88 fimbria CF, which is typically found on plasmids harbored in porcine ETEC strains (2). The FaeC, FaeD, FaeE, FaeF, FaeH, FaeI, and FaeJ proteins encoded on p7v share between 82% and 96% amino acid sequence identity with those encoded on the reference plasmid pUMNK88_K88. However, FaeG, the major subunit, shares only 29% amino acid sequence identity with the FaeG protein encoded on pUMNK88_K88. Although p7v encodes the K88 fimbria that is typically found on porcine ETEC plasmids, in silico analysis revealed that p7v does not encode the STb variant form of ST, which is encoded on the plasmid harbored in the reference porcine ETEC strain UMNK88 (7). Instead, p7v encodes two identical copies of STa. The virulence factors typical of ETEC plasmids are adjacent to each other in the p7v sequence, and those typical of STEC plasmids, although spread over a greater length of the sequence, are also adjacent to each other (Fig. 2). Characteristic of many large E. coli virulence plasmids, especially ETEC plasmids (33), p7v contains many insertion sequences that are distributed throughout the plasmid sequence. Replicons from two different incompatibility groups were identified on p7v, namely, RepFIB and RepFII. In silico PCR and PlasmidFinder (45) were used to verify the presence of these two replicons and the absence of others in the STEC/ETEC strain 7v genome. The two replicons are found within the segment of p7v that contains the STEC virulence factors. Along with the typical STEC virulence factors mentioned above, the STEC-like segment of p7v carries additional genes that may encode factors aiding in virulence or survival, such as a type VI secretion system, a serine-threonine protein kinase, a microcin, the insecticidal toxin SepC, and the PhoP/Q-regulated protein PqaA. Sequence analysis determined that two enzymes related to metabolic processes, namely, formate dehydrogenase and glycosyltransferase, are encoded on p7v within the ETEC-like sequence segment. Many open reading frames that encode hypothetical proteins are distributed throughout the plasmid sequence (Fig. 2).
FIG 2.
Plasmid p7v. The closed plasmid sequence was determined and annotated, revealing a hybrid plasmid. Virulence genes typically associated with STEC are labeled in turquoise, and those typically associated with ETEC are labeled in magenta. Open reading frames are categorized as virulence (red), adherence (purple), plasmid maintenance (orange), conjugative transfer (green), coding sequence (blue), insertion sequence (yellow), and hypothetical (gray).
Comparison of p7v to plasmids harbored in other STEC/ETEC hybrid strains.
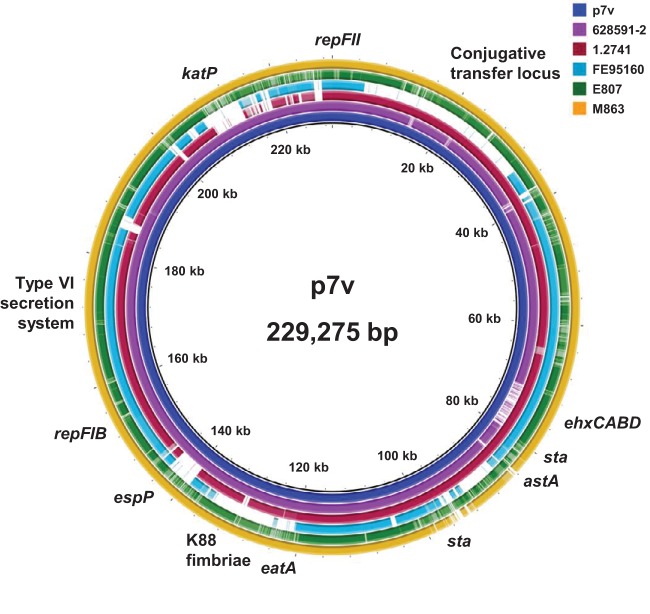
Having characterized the large virulence plasmid harbored in strain 7v and having determined that four additional STEC/ETEC hybrid strains are phylogenetically classified as CL1, we inquired as to whether they harbored a plasmid similar to p7v. In silico analysis identified the RepFIB and RepFII plasmid replicons on p7v; thus, the replicons contained in the genomes of all of the STEC/ETEC hybrid strains were examined utilizing PlasmidFinder (45). We discovered that the genomes of all five (inclusive of 7v) CL1 hybrid strains contain the RepFIB(AP001918) and RepFII(pSE11) replicons (Table 1). In fact, the genomes of seven of the STEC/ETEC hybrid strains clustering in phylogroup A carry these two plasmid replicons along with other replicons. In particular, the RepFIB(AP001918) plasmid replicon was found in 18 of the 24 STEC/ETEC hybrid strains. The genomes of many strains possess several replicons, suggesting that additional plasmids may be present in these strains (Table 1). Since the genomes of the STEC/ETEC CL1 hybrid strains were determined to contain the same plasmid replicons as p7v, a comparison of the nucleotide sequence of p7v to the genomes of the other STEC/ETEC CL1 strains was conducted using the BLAST analysis-based software BRIG (46) (Fig. 3). The ETEC pathotype CL1 strain M863 that clusters with strain 7v by whole-genome single nucleotide polymorphism (SNP) analysis (Fig. 1) was also included. The results reveal that strains 628591-2 and M863 most likely harbor large virulence plasmids similar to p7v (Fig. 3). These two strains each exhibit ≥90% nucleotide identity to the p7v sequence over 89% and 95% of the length of p7v, respectively. Strain 1.2741 also exhibits high sequence identity (≥90% nucleotide identity over 85% of the length of p7v), and although the 1.2741 plasmid appears to be missing several genes, strain 1.2741 most likely also carries a plasmid similar to p7v. STEC/ETEC strains FE95160 and E807 demonstrate much less sequence similarity (<55% of the length of p7v has ≥90% nucleotide identity to the p7v sequence) and most likely do not carry a complete plasmid resembling p7v. It was determined by BLASTn analyses of the remaining CL1 strains included in Fig. 1 (nonhybrid strains) that these strains do not harbor a plasmid similar to p7v, as <36% of the length of p7v has ≥90% sequence identity to the p7v sequence, and they do not contain either of the two plasmid replicons found on p7v. Inspection of the plasmid comparison results revealed the possible absence of several virulence genes in several of the STEC/ETEC CL1 genomes (Fig. 3). Utilizing tBLASTn analysis, the STEC/ETEC genomes were interrogated for the presence of the virulence genes katP, espP, ehxA, eatA, and astA specifically. Strain 7v is the only strain that carries all five genes (Table 2). ETEC CL1 strain M863, however, is lacking only the astA gene. The results of the gene-specific analysis are in agreement with the results of the BRIG analysis (Table 2; Fig. 3). The katP and espP genes are not present in any of the non-CL1 STEC/ETEC hybrid strains, with the exception of espP in strain IH53473 (Table 2).
FIG 3.
Comparison of the p7v sequence to CL1 STEC/ETEC strains. Sequence comparison was performed using BRIG (46). CL1 ETEC strain M863 was included. Lighter colored areas denote less than 75% sequence identity to p7v, and white areas denote less than 50% sequence identity. Gene labels identify the locations of genes on p7v.
TABLE 2.
Presence of selected virulence genes carried on p7v in the STEC/ETEC genomes
| Strain | Phylogroup | Presence of genea: |
||||
|---|---|---|---|---|---|---|
| katP | espP | ehxA | eatA | astA | ||
| 7v | CL1 | + | + | + | + | + |
| 628591-2 | CL1 | + | + | − | + | − |
| 1.2741 | CL1 | − | − | + | + | + |
| FE95160 | CL1 | − | − | + | − | + |
| E807 | CL1 | − | − | + | − | − |
| IH53473 | A | − | + | + | − | − |
| CFSAN026836 | A | − | − | + | + | − |
| CFSAN026835 | A | − | − | + | + | − |
| CVMN33742PS | A | − | − | + | + | + |
| CVMN33429PS | A | − | − | + | − | + |
| CFSAN026844 | A | − | − | + | − | − |
| CFSAN026843 | A | − | − | + | − | − |
| S1191 | A | − | − | − | − | − |
| K88 | A | − | − | − | − | + |
| UMNF18 | A | − | − | − | − | − |
| 2.3916 | A | − | − | − | − | − |
| IH57218 | A | − | − | + | − | − |
| E149 | D | − | − | − | − | − |
| C165-02 | D | − | − | − | − | − |
| 3020-98 | B1 | − | − | − | − | + |
| MDP04-01392 | B1 | − | − | − | − | + |
| MDP04-02111 | B1 | − | − | − | − | + |
| MI02-35 | B1 | − | − | − | − | + |
| 857226 | B1 | − | − | − | − | + |
+, gene was present in the strain; −, gene was not present in the strain.
Characterization of p7v revealed that along with STEC and ETEC virulence genes, p7v encodes an adherence factor, the K88 fimbria. BLASTn results of the other four STEC/ETEC CL1 hybrids confirm the result obtained by BRIG analysis that shows the presence of homologs of the entire K88-encoding operon in all but strain FE95160 (Table 1; Fig. 3). While STEC/ETEC strain 628591-2 possesses an faeG homolog similar to that found on p7v, STEC/ETEC strains 1.2741 and E807 encode an FaeG protein with similarity neither to that encoded on p7v nor to that on UMNK88_pK88. The K88 CF was also present in seven STEC/ETEC hybrid strains that cluster in E. coli phylogroup A. However, it was determined that none of these strains encode the homolog of FaeG encoded on p7v, and only STEC/ETEC strain K88 encodes the homolog of FaeG encoded on UMNK88_pK88. Another CF that was associated with porcine ETEC strains, F18, was determined to be harbored by four STEC/ETEC hybrid strains, all isolated from pigs. These strains contain the four fimbrial proteins, FedA, FedB, FedC, and FedE, that are encoded on the reference plasmid UMNF18_87. The amino acid sequence identities range from 96% to 100% compared to those encoded on plasmid UMNF18_87 for the four fimbrial proteins in the STEC/ETEC hybrid strains. Interestingly, none of the CFs typically found on plasmids harbored in human ETEC strains were present in the STEC/ETEC hybrid strains, even those isolated from humans. However, the STEC/ETEC hybrid strain C165-02, isolated from a human, harbors a plasmid carrying genes encoding the Typhi colonization factor (Tcf), which is present in the genomes of some strictly human-adapted pathogens (48).
Heat-stable enterotoxin phylogeny.
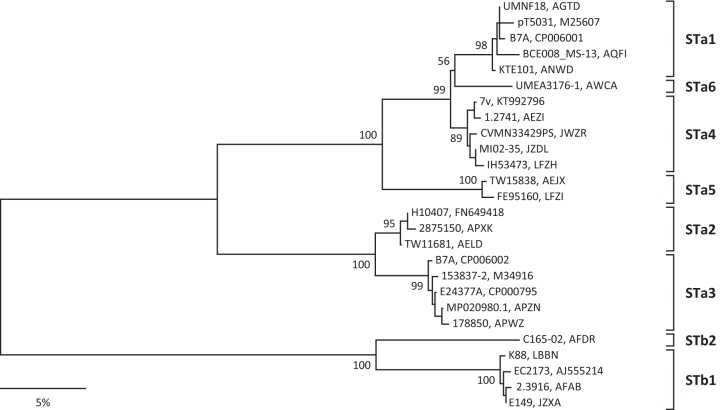
As noted in the plasmid description, p7v carries two copies of sta. Furthermore, the sta gene sequence on p7v is divergent from that of the sta gene sequences carried on plasmids in the reference human ETEC strains H10407 (sta1 and sta2) and E24377A (sta3) (28, 33). To further investigate the ST-encoding gene sequence differences, a phylogeny was constructed that included the sta and stb genes found in the 24 STEC/ETEC hybrid strains along with representative sta and stb sequences from ETEC strains submitted to GenBank (Fig. 4). The sta1 genes and the sta gene carried on p7v exhibit 96.4% nucleotide sequence similarity, which is only slightly greater than the 95.4% sequence similarity found in a comparison of sta2 and sta3 sequences. Examination of the distance matrix from the sta and stb phylogenetic analysis and defining an sta and stb sequence similarity of less than 97% between different clusters in the phylogeny lead to the designation of six sta allele subtypes and two stb allele subtypes (Fig. 4; see also Fig. S1 in the supplemental material). In this scheme, the STa-encoding genes carried on p7v would be considered a new allele subtype that we have designated sta4. Utilizing the sta and stb phylogeny to assign allele subtypes to the ST-encoding genes carried in the 24 STEC/ETEC hybrid strains demonstrates that 16 of the hybrid genomes also contain the sta4 allele type (Table 1). Another allele that is newly identified here, sta5, is carried by six STEC/ETEC hybrid strains, including three of the CL1 strains. Four additional strains carry both sta4 and sta5 (Table 1). Five of the STEC/ETEC hybrid strains possess an STb-encoding allele, either alone or in combination with an STa-encoding gene. In addition, the results reveal a novel stb gene that is carried by the STEC/ETEC hybrid strain C165-02 and designated stb2 (Table 1; Fig. 4).
FIG 4.
Phylogenetic relationships of ST alleles. This neighbor-joining tree was constructed using a p distance matrix. Strain designations and GenBank accession numbers (or WGS prefixes) are given at the branch tips. Bootstrap values based on 500 replications are given at the internal nodes. Allele subtypes are given next to the square brackets.
Shiga toxin allele subtypes present in STEC/ETEC hybrid strains.
Shiga toxin gene allele subtypes found in the STEC/ETEC genomes were assigned based on BLASTn analyses, and the results demonstrate that a variety of stx alleles are carried in the STEC/EHEC hybrid strains (Table 1). Among only the CL1 strains, three different stx subtypes are represented. The two CL1 strains that exhibit the greatest core genome sequence similarity to p7v, namely, 628591-2 and 1.2741 (Fig. 1), carry the stx2g allele, which is found in the 7v genome. Thirteen strains possess an Stx1-encoding gene, and 13 possess an Stx2-encoding gene (two CL1 strains carry both stx1 and stx2 alleles). Despite the assortment of stx subtypes found in the 24 STEC/ETEC hybrid strains, two trends emerge. First, the four strains that harbor a plasmid encoding STb1 also carry the stx2e gene in their genomes, and second, all five STEC/ETEC hybrid strains that cluster in E. coli phylogroup B1 possess the stx1c allele subtype.
DISCUSSION
The acquisition of additional virulence-associated genes may render an E. coli strain that is already considered pathogenic capable of causing more severe illness. Many virulence genes are plasmid encoded (1, 2); thus, it is important to characterize plasmids harbored in hybrid E. coli strains, as these plasmids may represent mobile genetic elements that can create new combinations of virulence genes that may impact/affect the pathogenicity of existing E. coli strains. The work presented here is the first report, to our knowledge, of the sequencing and characterization of a complete plasmid isolated from a STEC/ETEC hybrid strain. Plasmid p7v was determined to carry the RepFIB and RepFII replicons as were plasmids from 17 other STEC/ETEC hybrid strains included in this study (Table 1) despite some of the plasmids having very low sequence similarity with p7v. This is not unexpected considering that this replicon combination is fairly commonly found on large virulence plasmids harbored in E. coli (2). Sequence analysis revealed many insertion (IS) elements scattered throughout p7v (Fig. 2), and it has been suggested that the ancestral RepFIB/FIIA plasmid backbone has acquired IS elements that have led to the introduction of virulence genes (2). Therefore, it would not be surprising to find many variations in RepFIB/FII plasmid gene repertoires among STEC/ETEC hybrid strains considering that ST-encoding genes are often found adjacent to IS elements. Even the plasmids determined to be most similar in sequence to p7v, namely, those harbored in STEC/ETEC CL1 strains 62859-2 and 1.2741 as well as the plasmid harbored in ETEC CL1 strain M863, demonstrate variation in virulence gene repertoire (Tables 1 and 2; Fig. 3). Excluding STs and CFs, of the five virulence genes specifically queried, strain 62859-2 and 1.2741 each possess three but only carry one gene, eatA, in common. Potentially, multiple parallel genomic insertion/deletion events occurred in individual STEC/ETEC hybrid CL1 strain plasmids subsequent to acquiring an ancestral plasmid backbone. Alternatively, the combination of plasmid factors from ETEC and STEC is advantageous, and the recombination of these plasmids has likely occurred multiple times in the context of the CL1 background.
A recent comprehensive study of ETEC phylogeny and evolution determined that, although ETEC strains can be found in most E. coli phylogroups, the majority are clustered in phylogroups A and B1 (22). Additionally, ETEC strains clustering in the CL1 clade have been reported (25). LEE-negative STEC strains also possess diverse core genome sequences (4); thus, it is not surprising to find that many of the STEC/ETEC hybrids are distributed in several phylogroups (Fig. 1). The STEC/ETEC hybrid strains in phylogroup B1 exhibit similarity in that they share core genome sequence similarity, identical serotype, ST allele subtype, Shiga toxin allele subtype, and even similar plasmid replicons (Table 1; Fig. 1). This is despite having a variety of isolation sources, locations, and dates (Table 1). Interestingly, the two STEC/ETEC hybrid strains clustering in phylogroup D, E149 and C165-02, do not share toxin or plasmid replicon profiles; rather, strain E149 shares identical ST, Stx, and CF profiles with strain 2.3916, which clusters in phylogroup A (Table 1; Fig. 1). Furthermore, strains E149 and 2.3916 share four out of five plasmid replicon sequences. While STEC/ETEC hybrid strain CVMN33742PS does not harbor a plasmid similar to strain 7v, the two strains carry an stx2g-encoding phage, the sta4 gene, and the K88 CF. These examples demonstrate that acquisition of a particular ST-encoding plasmid is not necessarily linked to a particular backbone sequence type, and similar toxin and CF profiles are not necessarily indicators of similar backbone sequences.
Although ETEC strains clustering with CL1 have been isolated from sick children (25), none of the STEC/ETEC hybrid CL1 strains included in our study were isolated from human clinical samples. However, multiple STEC/ETEC hybrid strains possessing stx2g and sta have been isolated from patients with diarrhea, fever, and abdominal pain (17). Two of the strains included in that report have serotype O136:H16, the serotype of four of the strains clustering in phylogroup A included in the present study. Interestingly, our O136:H16 strains (CVMN33742PS, CVMN33429PS, CFSAN026844, and CFSAN026843) carry the K88 CF, which is considered to be specific for porcine infection (2, 7). In fact, none of the strains in the present study that possess either the K88 or F18 CF were isolated from human clinical samples (Tables 1 and 2). Sequence analysis determined that none of the five clinical isolates harbor a known ETEC CF, but strain C165-02 does possess Tcf, which is thought to be a human-specific colonization factor (48). In addition, the HUS-causing strain IH53473 carries the LEE pathogenicity island, which encodes the intimin adhesin. ETEC strains that are negative for known CFs have been isolated from clinical samples and may possess unidentified new human-specific CFs (22), which may also be the case for STEC/ETEC human isolates. Virulence of a strain may be partially determined by the ability of the strain to adhere to the human intestine. Considering that several of the hybrid STEC/ETEC CL1 strains carry known virulence genes in addition to Stx and ST (Table 2), they may have the potential to cause disease in humans if they are able to, or acquire the ability to, adhere to the human intestine.
Another factor that is often considered to affect virulence is the Shiga toxin allele subtype encoded in the genome. The subtypes linked with serious human disease are stx1a, stx2a, stx2c, and stx2d (49). Two of the STEC/ETEC hybrid strains isolated from humans exhibiting illness, IH53473 and IH57218, express Stx2a, and another, C165-02, expresses Stx2d (Table 1). Interestingly, the remaining two clinical isolates, 3020-98 and MI02-35, one of which was isolated from a patient exhibiting bloody diarrhea, carry the stx1c subtype, which is rarely isolated from humans and is typically linked to mild disease when it is isolated from humans (49). These two strains reportedly express Stx1 but not ST (12). The clinical isolate IH53473 was also shown to express Stx but not ST, while strain IH57218 is reported to produce Stx and STa (19). Prager et al. also identified two STEC/ETEC hybrid clinical isolates expressing Stx and ST (17). In contrast to the STEC/ETEC hybrid strains isolated from humans included in the present study, where ST may not play much of a role in the pathogenicity of the bacteria, human clinical STEC/ETEC hybrids expressing STa, but not Stx, have been reported (17). Like STEC/ETEC hybrid strain 7v, these strains express Stx2g; however, strain 7v does express Stx2g albeit at low levels (4, 17). Strain 7v is reported to express ST (17), and our sequence analysis of p7v combined with sta and stb phylogeny results demonstrates that p7v carries two STa4-encoding genes. ETEC and STEC/ETEC hybrid strains that carry more than one gene encoding ST have been reported (13, 18, 20, 28), and our results demonstrate that 7 of the 24 STEC/ETEC hybrid strains included in the present study possess two ST-encoding genes. Interestingly, strains 2.3916 and E149 possess an STb-encoding allele variant, stb1, as well as an STa-encoding allele variant, sta1. While the Stx2e that they encode is not linked to serious human disease (49), STa1 is encoded on a plasmid harbored in the human ETEC reference strain H10407 (28). ETEC strains that possess stb have been primarily isolated from animals, particularly pigs, but have been infrequently isolated from human clinical samples (5, 6). However, it is not clear that STb is a contributing factor to the illness caused by the strains isolated from humans, such as strain C165-02 in this study, which also expresses stx2d at a low level (4).
In conclusion, sequence analysis of the large virulence plasmid, p7v, that is harbored in hybrid STEC/ETEC CL1 strain 7v revealed that p7v is itself hybrid in nature, as it encodes both putative STEC-associated and ETEC-associated virulence factors. Core genome phylogenetic analysis of a set of 24 STEC/ETEC hybrid strains, in conjunction with a plasmid-encoded virulence gene, CF, and plasmid replicon analysis, highlights the diversity of these strains, and this study demonstrates how the mobility of plasmid-associated virulence genes, irrespective of core genome sequence, has resulted in the creation of STEC/ETEC hybrid strains in multiple parallel events. STEC/ETEC hybrid strains have been isolated from human clinical samples and from a variety of other sources, including food; thus, they may represent an emerging threat as a food-borne pathogen. Walk has suggested that the recent emergence in humans of ETEC strains belonging to a subset of CL1 may be due to their ability to cause disease (25). This reasoning may be extended to the STEC/ETEC hybrid strains, yet it may also be possible that, in some cases, multiple virulence factors that define the E. coli pathotypes were not queried for in a single isolate. Potentially, as more clinical isolates are sequenced and thus allow differentiation between the E. coli species and Escherichia CL1, STEC/ETEC hybrid CL1 strains that cause disease in humans may be identified. The results of this study, which demonstrates the lack of both the LEE pathogenicity island and typical human-specific ETEC CFs in the majority of the clinical STEC/ETEC hybrid strains, emphasize the need to further explore the adherence of these strains to the human intestine.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter Feng (U.S. FDA) and Shannon Manning (STEC Center, Michigan State University) for providing the STEC/ETEC hybrid strains sequenced in this work.
S.R.L. was supported by a fellowship appointment administered by Oak Ridge Institute for Science and Education.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01129-16.
REFERENCES
- 1.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Johnson TJ, Nolan LK. 2009. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev 73:750–774. doi: 10.1128/MMBR.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheutz F, Teel LD, Beutin L, Pierard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol 50:2951–2963. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steyert SR, Sahl JW, Fraser CM, Teel LD, Scheutz F, Rasko DA. 2012. Comparative genomics and stx phage characterization of LEE-negative Shiga toxin-producing Escherichia coli. Front Cell Infect Microbiol 2:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lortie LA, Dubreuil JD, Harel J. 1991. Characterization of Escherichia coli strains producing heat-stable enterotoxin b (STb) isolated from humans with diarrhea. J Clin Microbiol 29:656–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamoto K, Fujii Y, Akashi N, Hitotsubashi S, Kurazono H, Karasawa T, Takeda Y. 1993. Identification and characterization of heat-stable enterotoxin II-producing Escherichia coli from patients with diarrhea. Microbiol Immunol 37:411–414. doi: 10.1111/j.1348-0421.1993.tb03230.x. [DOI] [PubMed] [Google Scholar]
- 7.Shepard SM, Danzeisen JL, Isaacson RE, Seemann T, Achtman M, Johnson TJ. 2012. Genome sequences and phylogenetic analysis of K88- and F18-positive porcine enterotoxigenic Escherichia coli. J Bacteriol 194:395–405. doi: 10.1128/JB.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Moller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK. 2011. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med 365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mariani-Kurkdjian P, Lemaitre C, Bidet P, Perez D, Boggini L, Kwon T, Bonacorsi S. 2014. Haemolytic-uraemic syndrome with bacteraemia caused by a new hybrid Escherichia coli pathotype. New Microbes New Infect 2:127–131. doi: 10.1002/nmi2.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bielaszewska M, Schiller R, Lammers L, Bauwens A, Fruth A, Middendorf B, Schmidt MA, Tarr PI, Dobrindt U, Karch H, Mellmann A. 2014. Heteropathogenic virulence and phylogeny reveal phased pathogenic metamorphosis in Escherichia coli O2:H6. EMBO Mol Med 6:347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutta S, Pazhani GP, Nataro JP, Ramamurthy T. 2015. Heterogenic virulence in a diarrheagenic Escherichia coli: evidence for an EPEC expressing heat-labile toxin of ETEC. Int J Med Microbiol 305:47–54. doi: 10.1016/j.ijmm.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Monday SR, Keys C, Hanson P, Shen Y, Whittam TS, Feng P. 2006. Produce isolates of the Escherichia coli Ont:H52 serotype that carry both Shiga toxin 1 and stable toxin genes. Appl Environ Microbiol 72:3062–3065. doi: 10.1128/AEM.72.4.3062-3065.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beutin L, Kruger U, Krause G, Miko A, Martin A, Strauch E. 2008. Evaluation of major types of Shiga toxin 2e-producing Escherichia coli bacteria present in food, pigs, and the environment as potential pathogens for humans. Appl Environ Microbiol 74:4806–4816. doi: 10.1128/AEM.00623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller D, Greune L, Heusipp G, Karch H, Fruth A, Tschape H, Schmidt MA. 2007. Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl Environ Microbiol 73:3380–3390. doi: 10.1128/AEM.02855-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martikainen O, Kagambega A, Bonkoungou IJ, Barro N, Siitonen A, Haukka K. 2012. Characterization of Shigatoxigenic Escherichia coli strains from Burkina Faso. Foodborne Pathog Dis 9:1015–1021. doi: 10.1089/fpd.2012.1228. [DOI] [PubMed] [Google Scholar]
- 16.Nyholm O, Heinikainen S, Pelkonen S, Hallanvuo S, Haukka K, Siitonen A. 2015. Hybrids of Shigatoxigenic and enterotoxigenic Escherichia coli (STEC/ETEC) among human and animal isolates in Finland. Zoonoses Public Health 62:518–524. doi: 10.1111/zph.12177. [DOI] [PubMed] [Google Scholar]
- 17.Prager R, Fruth A, Busch U, Tietze E. 2011. Comparative analysis of virulence genes, genetic diversity, and phylogeny of Shiga toxin 2g and heat-stable enterotoxin STIa encoding Escherichia coli isolates from humans, animals, and environmental sources. Int J Med Microbiol 301:181–191. doi: 10.1016/j.ijmm.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Mohlatlole RP, Madoroba E, Muchadeyi FC, Chimonyo M, Kanengoni AT, Dzomba EF. 2013. Virulence profiles of enterotoxigenic, Shiga toxin and enteroaggregative Escherichia coli in South African pigs. Trop Anim Health Prod 45:1399–1405. doi: 10.1007/s11250-013-0377-4. [DOI] [PubMed] [Google Scholar]
- 19.Nyholm O, Halkilahti J, Wiklund G, Okeke U, Paulin L, Auvinen P, Haukka K, Siitonen A. 2015. Comparative genomics and characterization of hybrid Shigatoxigenic and enterotoxigenic Escherichia coli (STEC/ETEC) strains. PLoS One 10:e0135936. doi: 10.1371/journal.pone.0135936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byun JW, Jung BY, Kim HY, Fairbrother JM, Lee MH, Lee WK. 2013. Real-time PCR for differentiation of F18 variants among enterotoxigenic and Shiga toxin-producing Escherichia coli from piglets with diarrhoea and oedema disease. Vet J 198:538–540. doi: 10.1016/j.tvjl.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 21.Hazen TH, Sahl JW, Fraser CM, Donnenberg MS, Scheutz F, Rasko DA. 2013. Refining the pathovar paradigm via phylogenomics of the attaching and effacing Escherichia coli. Proc Natl Acad Sci U S A 110:12810–12815. doi: 10.1073/pnas.1306836110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Mentzer A, Connor TR, Wieler LH, Semmler T, Iguchi A, Thomson NR, Rasko DA, Joffre E, Corander J, Pickard D, Wiklund G, Svennerholm AM, Sjoling A, Dougan G. 2014. Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution. Nat Genet 46:1321–1326. doi: 10.1038/ng.3145. [DOI] [PubMed] [Google Scholar]
- 23.Sahl JW, Steinsland H, Redman JC, Angiuoli SV, Nataro JP, Sommerfelt H, Rasko DA. 2011. A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect Immun 79:950–960. doi: 10.1128/IAI.00932-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT. 2011. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci U S A 108:7200–7205. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walk ST. 30 July 2015. The “cryptic” Escherichia. EcoSal Plus 2015. doi: 10.1128/ecosalplus.ESP-0002-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walk ST, Alm EW, Gordon DM, Ram JL, Toranzos GA, Tiedje JM, Whittam TS. 2009. Cryptic lineages of the genus Escherichia. Appl Environ Microbiol 75:6534–6544. doi: 10.1128/AEM.01262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clermont O, Gordon DM, Brisse S, Walk ST, Denamur E. 2011. Characterization of the cryptic Escherichia lineages: rapid identification and prevalence. Environ Microbiol 13:2468–2477. doi: 10.1111/j.1462-2920.2011.02519.x. [DOI] [PubMed] [Google Scholar]
- 28.Crossman LC, Chaudhuri RR, Beatson SA, Wells TJ, Desvaux M, Cunningham AF, Petty NK, Mahon V, Brinkley C, Hobman JL, Savarino SJ, Turner SM, Pallen MJ, Penn CW, Parkhill J, Turner AK, Johnson TJ, Thomson NR, Smith SG, Henderson IR. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J Bacteriol 192:5822–5831. doi: 10.1128/JB.00710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wajima T, Sabui S, Kano S, Ramamurthy T, Chatterjee NS, Hamabata T. 2013. Entire sequence of the colonization factor coli surface antigen 6-encoding plasmid pCss165 from an enterotoxigenic Escherichia coli clinical isolate. Plasmid 70:343–352. doi: 10.1016/j.plasmid.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K, Tobe T, Hattori M, Hayashi T. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A 106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newton HJ, Sloan J, Bulach DM, Seemann T, Allison CC, Tauschek M, Robins-Browne RM, Paton JC, Whittam TS, Paton AW, Hartland EL. 2009. Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement. Emerg Infect Dis 15:372–380. doi: 10.3201/eid1503.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan X, Fratamico PM, Needleman DS, Bayles DO. 2012. DNA sequence and analysis of a 90.1-kb plasmid in Shiga toxin-producing Escherichia coli (STEC) O145:NM 83-75. Plasmid 68:25–32. doi: 10.1016/j.plasmid.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Rasko DA, Rosovitz MJ, Myers GS, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 36.Iguchi A, Iyoda S, Kikuchi T, Ogura Y, Katsura K, Ohnishi M, Hayashi T, Thomson NR. 2015. A complete view of the genetic diversity of the Escherichia coli O-antigen biosynthesis gene cluster. DNA Res 22:101–107. doi: 10.1093/dnares/dsu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Rothemund D, Curd H, Reeves PR. 2003. Species-wide variation in the Escherichia coli flagellin (H-antigen) gene. J Bacteriol 185:2936–2943. doi: 10.1128/JB.185.9.2936-2943.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tominaga A. 2004. Characterization of six flagellin genes in the H3, H53 and H54 standard strains of Escherichia coli. Genes Genet Syst 79:1–8. doi: 10.1266/ggs.79.1. [DOI] [PubMed] [Google Scholar]
- 39.Tominaga A, Kutsukake K. 2007. Expressed and cryptic flagellin genes in the H44 and H55 type strains of Escherichia coli. Genes Genet Syst 82:1–8. doi: 10.1266/ggs.82.1. [DOI] [PubMed] [Google Scholar]
- 40.Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Wang Q, Reeves PR, Wang L. 2008. Structure and genetics of Shigella O antigens. FEMS Microbiol Rev 32:627–653. doi: 10.1111/j.1574-6976.2008.00114.x. [DOI] [PubMed] [Google Scholar]
- 41.Feng L, Liu B, Liu Y, Ratiner YA, Hu B, Li D, Zong X, Xiong W, Wang L. 2008. A genomic islet mediates flagellar phase variation in Escherichia coli strains carrying the flagellin-specifying locus flk. J Bacteriol 190:4470–4477. doi: 10.1128/JB.01937-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratiner YA, Sihvonen LM, Liu Y, Wang L, Siitonen A. 2010. Alteration of flagellar phenotype of Escherichia coli strain P12b, the standard type strain for flagellar antigen H17, possessing a new non-fliC flagellin gene flnA, and possible loss of original flagellar phenotype and genotype in the course of subculturing through semisolid media. Arch Microbiol 192:267–278. doi: 10.1007/s00203-010-0556-x. [DOI] [PubMed] [Google Scholar]
- 43.Rodas C, Iniguez V, Qadri F, Wiklund G, Svennerholm AM, Sjoling A. 2009. Development of multiplex PCR assays for detection of enterotoxigenic Escherichia coli colonization factors and toxins. J Clin Microbiol 47:1218–1220. doi: 10.1128/JCM.00316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nada RA, Shaheen HI, Touni I, Fahmy D, Armstrong AW, Weiner M, Klena JD. 2010. Design and validation of a multiplex polymerase chain reaction for the identification of enterotoxigenic Escherichia coli and associated colonization factor antigens. Diagn Microbiol Infect Dis 67:134–142. doi: 10.1016/j.diagmicrobio.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 45.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veilleux S, Dubreuil JD. 2006. Presence of Escherichia coli carrying the EAST1 toxin gene in farm animals. Vet Res 37:3–13. doi: 10.1051/vetres:2005045. [DOI] [PubMed] [Google Scholar]
- 48.Nuccio SP, Baumler AJ. 2007. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev 71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melton-Celsa AR. 2014. Shiga toxin (Stx) classification, structure, and function. Microbiol Spectr 2:EHEC-0024-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leung PH, Peiris JS, Ng WW, Robins-Browne RM, Bettelheim KA, Yam WC. 2003. A newly discovered verotoxin variant, VT2g, produced by bovine verocytotoxigenic Escherichia coli. Appl Environ Microbiol 69:7549–7553. doi: 10.1128/AEM.69.12.7549-7553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinstein DL, Jackson MP, Samuel JE, Holmes RK, O'Brien AD. 1988. Cloning and sequencing of a Shiga-like toxin type II variant from Escherichia coli strain responsible for edema disease of swine. J Bacteriol 170:4223–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Persson S, Olsen KE, Ethelberg S, Scheutz F. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J Clin Microbiol 45:2020–2024. doi: 10.1128/JCM.02591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.