ABSTRACT
Animals are important reservoirs of zoonotic enteropathogens, and transmission to humans occurs more frequently in low- and middle-income countries (LMICs), where small-scale livestock production is common. In this study, we investigated the presence of zoonotic enteropathogens in stool samples from 64 asymptomatic children and 203 domestic animals of 62 households in a semirural community in Ecuador between June and August 2014. Multilocus sequence typing (MLST) was used to assess zoonotic transmission of Campylobacter jejuni and atypical enteropathogenic Escherichia coli (aEPEC), which were the most prevalent bacterial pathogens in children and domestic animals (30.7% and 10.5%, respectively). Four sequence types (STs) of C. jejuni and four STs of aEPEC were identical between children and domestic animals. The apparent sources of human infection were chickens, dogs, guinea pigs, and rabbits for C. jejuni and pigs, dogs, and chickens for aEPEC. Other pathogens detected in children and domestic animals were Giardia lamblia (13.1%), Cryptosporidium parvum (1.1%), and Shiga toxin-producing E. coli (STEC) (2.6%). Salmonella enterica was detected in 5 dogs and Yersinia enterocolitica was identified in 1 pig. Even though we identified 7 enteric pathogens in children, we encountered evidence of active transmission between domestic animals and humans only for C. jejuni and aEPEC. We also found evidence that C. jejuni strains from chickens were more likely to be transmitted to humans than those coming from other domestic animals. Our findings demonstrate the complex nature of enteropathogen transmission between domestic animals and humans and stress the need for further studies.
IMPORTANCE We found evidence that Campylobacter jejuni, Giardia, and aEPEC organisms were the most common zoonotic enteropathogens in children and domestic animals in a region close to Quito, the capital of Ecuador. Genetic analysis of the isolates suggests transmission of some genotypes of C. jejuni and aEPEC from domestic animals to humans in this region. We also found that the genotypes associated with C. jejuni from chickens were present more often in children than were those from other domestic animals. The potential environmental factors associated with transmission of these pathogens to humans then are discussed.
INTRODUCTION
Diarrheal diseases are a major cause of illness and death in low- and middle-income countries (LMICs), where there are over 1.5 billion diarrhea cases that occur annually among children less than 5 years old, resulting in nearly 700,000 deaths (1). Although the contribution of zoonotic pathogens to human diarrheal disease is significant (2), these pathogens are often overlooked, and their detection may be hindered by patterns of seasonality (3). Zoonotic enteropathogens comprise a large and diverse range of microorganisms that could be transmitted to humans by consumption of meat or dairy products, by direct contact with animals (or their feces) in the environment, or by consumption of food or water contaminated with animal feces (2, 4). In the United States, researchers estimated that 14% of enteric infections with 7 groups of zoonotic enteropathogens were attributable to direct contact with animals (5).
Most of the studies of zoonotic enteropathogens have taken place in high-income countries, and their results may not be applicable to LMICs with less developed sanitary infrastructure and different animal husbandry practices (4). LMICs report many zoonotic pathogens that are rare in industrialized nations and vice versa (4). For instance, Campylobacter jejuni, non-Typhi Salmonella, and enterohemorrhagic Escherichia coli (EHEC) are zoonotic pathogens associated with high morbidity in high-income countries (6, 7), but they are unusual causes of human disease in LMICs (8–10).
Although some zoonotic pathogens, such as Campylobacter jejuni, are recognized as the most frequent gastrointestinal bacterial pathogen in humans in industrialized countries (11, 12), the contribution of other zoonotic pathogens (such as enteropathogenic E. coli or many strains of Shiga-toxigenic E. coli) to human diarrhea is less understood (13–15).
Most zoonotic enteropathogens were thought to be generalists (able to infect a wide variety of animals, including humans); however, recent evidence suggests that some strains of Cryptosporidium parvum, Campylobacter jejuni, and Giardia lamblia are host adapted with low levels of transmission to humans (12, 16, 17). Furthermore, some domestic animals may harbor generalist strains while others carry more host-adapted strains (9).
The present study aimed to investigate the prevalence of 7 zoonotic enteropathogens (bacteria and protozoa) in children and domestic animals in a semirural community of Ecuador.
MATERIALS AND METHODS
Study location.
The study was conducted in Otón de Vélez-Yaruquí, a low-income semirural community east of Quito, at an altitude of 2,527 m above sea level. The main economic activities are agriculture and animal husbandry, particularly intensive poultry production (four chicken industrial operations were present in the community). Of the sampled households, 68% had chickens, 64.5% had guinea pigs (raised for food), 64.5% had dogs, 58% had pigs, 32.3% had rabbits, and 11.3% had cattle, and cats, ducks, quails, sheep, geese, and horses were present in 10%, 8%, 5%, 3%, 1.6% and 1.6% of the households, respectively (see Fig. S1 in the supplemental material).
Ethical considerations.
The study protocol was approved by the Institutional Animal Care and Use Committee at the George Washington University (IACUC number A296), as well as the Bioethics Committee at the Universidad San Francisco de Quito (2014-135M) and the George Washington University Committee on Human Research Institutional Review Board (IRB number 101355).
Sample collection.
Sixty-five households were recruited randomly between June and August 2014, during the dry season. Fifty-nine of the households had domestic animals (1 to 8 animal species), 3 did not have animals, and 3 households did not provide samples. Eleven households reported having someone in the home who worked in the poultry industry. We collected 64 stool samples from asymptomatic children (47% female and 53% male; ages were 3 to 12 months old [n = 11], 1 to 3 years old [n = 31], 3 to 5 years old [n = 18], and 6 years old [n = 4]) (see Table S1 in the supplemental material) and 203 samples from 12 animal species (Table 1; also see Fig. S1). Animal fecal samples were obtained either directly from the rectus (dogs, cats, sheep, and quail) or from pooled fecal matter when animals were maintained in enclosures (pigs, chickens, and cows) or cages (guinea pigs, rabbits, and quails). The stool samples were placed in a cooler on ice for transportation to the laboratory. All bacterial culturing and sample preservation began less than 8 h after collection.
TABLE 1.
Frequency of zoonotic enteropathogens identified in both children and domestic animals
| Source | No. of samples | No. (%) positive for: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C. jejuni | C. coli | aEPEC | Campylobacter spp.a | STEC | Salmonella spp. | Yersinia | Giardia lamblia | Cryptosporidium parvum | ||
| Children | 64 | 7 (10.9) | 3 (4.7) | 11 (17.2) | 1 (1.6) | 1 (1.6) | 0 | 0 | 22 (34.4) | 2 (3.1) |
| Chickens | 42 | 25 (59.5) | 7 (16.7) | 3 (7.1) | 0 | 1 (2.4) | 0 | 0 | 0 | 0 |
| Guinea pigs | 40 | 29 (72.5) | 2 (5.0) | 2 (5.0) | 0 | 1 (2.5) | 0 | 0 | 1 (2.5) | 0 |
| Dogs | 40 | 10 (25.0) | 1 (2.5) | 4 (10.0) | 0 | 0 | 5 (12.5) | 0 | 5 (12.5) | 0 |
| Pigs | 36 | 3 (8.3) | 14 (38.9) | 4 (11.1) | 10 (27.8) | 0 | 0 | 1 (2.8) | 2 (5.6) | 0 |
| Rabbits | 20 | 2 (10.0) | 0 | 0 | 0 | 0 | 0 | 0 | 4 (20.0) | 0 |
| Cattle | 7 | 1 (14.3) | 1 (14.3) | 2 | 0 | 4 (57.1) | 0 | 0 | 0 | 0 |
| Cats | 6 | 2 (33.3) | 1 (16.7) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ducks | 5 | 1 (20.0) | 1 (20.0) | 1 | 2 (40.0) | 0 | 0 | 0 | 0 | 0 |
| Quail | 3 | 2 (66.7) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Sheep | 2 | 0 | 1 (50.0) | 1 | 0 | 0 | 0 | 0 | 1 (50.0) | 1 (50.0) |
| Geese | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Horses | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 267 | 82 (30.7) | 31 (11.6) | 28 | 13 (4.9) | 7 (2.6) | 5 (1.9) | 1 (0.4) | 35 (13.1) | 3 (1.1) |
Campylobacter non-jejuni/coli species included C. hyointestinalis (pigs and child), C. lanienae (pig), and C. canadensis (ducks).
Identification of zoonotic enteropathogens.
Fecal samples were analyzed for seven zoonotic enteropathogens: Campylobacter spp., atypical enteropathogenic E. coli (aEPEC), Shiga toxin-producing E. coli (STEC), Salmonella spp., Yersinia spp., Cryptosporidium parvum, and Giardia lamblia.
Pathotypes of E. coli were obtained by culturing samples on MacConkey lactose agar (Difco, Sparks, MD) (at 37°C for 18 h), and lactose-fermenting colonies were plated in Chromocult coliform agar (Merck KGaA, Darmstadt, Germany) to identify the ß-d-glucuronidase activity. Five lactose-positive isolates were pooled as a random sample, suspended in 300 μl of sterile distilled water, and boiled for 10 min to release the DNA. The resulting supernatant was used for PCR to identify eae (18), bfpA (19), stx1, and stx2 as previously described (20). Isolates from positive pools for any loci were individually analyzed by PCR and cryopreserved in brain and hearth infusion medium (Difco, Sparks, MD) with 20% glycerol at −80°C for further analyses. aEPEC isolates were positive for eae (LEE gene) and negative for bfpA and stx genes (13); STEC isolates were positive for any of the stx genes by PCR but lacked the eae gene.
To isolate Yersinia spp., the samples were preenriched in 1× phosphate-buffered saline (PBS) for 21 days at 4°C and cultured in cefsulodin irgasan novobiocin agar (at 28°C for 24 and 48 h) (Oxoid Ltd., Basingstoke, Hampshire, England). Suspected colonies were confirmed with the following tests: oxidase test (Bactident oxidase; Merck), RapiD-20E (bioMérieux, Marcy l'Etoile, France) with identification percentages greater than 85%, lack of lactose fermentation on MacConkey lactose agar (Difco, Sparks, MD), and urease activity in Christenson urea agar (Difco, Sparks, MD).
To recover Salmonella spp., samples were preenriched in selenite broth (Merck KGaA, Darmstadt, Germany) (at 37°C for 18 h) and cultured in xylose-lysine-deoxycholate agar (Difco, Sparks, MD) (at 37°C for 18 h). Suspected colonies were subjected to RapiD-20E tests (bioMérieux, Marcy l'Etoile, France), with acceptable identification set at greater than 95%. The identification of serovars was performed by amplifying 10 pairs of primers by multiplex PCR in two separate reactions (assays STM and STY) as previously described (21). The STM amplification was performed in a 10-μl reaction mix with 1.4× PCR buffer, 2 mM MgCl2, 0.2 mm deoxynucleoside triphosphates (dNTPs), 0.3 μM each primer (STM1, STM2, STM3, STM4, and STM5), 0.75 U GoTaq polymerase, and 1 μl of DNA (∼10 ng/μl). Furthermore, the STY amplification reaction was performed in a final volume of 10 μl with 1.6× reaction buffer; 2 mM MgCl2; 0.2 mm dNTPs; 0.08 μM primers STY1, STY2, and STM6; 0.3 μM primer STY3; 0.1 μM primer STY4; 0.75 U GoTaq polymerase; and 1 μl of DNA. Both reactions used the same amplification program: initial denaturation at 94°C for 5 min, followed by 40 cycles of 94°C for 30 s, 62°C for 30 s, and 72°C for 1 min, and ending with a final extension at 72°C for 5 min. Electrophoresis conditions for displaying the results of STM and STY included a 2.5% agarose gel run for 2 h at 80 V.
To investigate thermophilic Campylobacter spp., samples were cultured on Campylobacter agar with 5% lysed horse blood and modified Preston Campylobacter selective supplement (Oxoid Ltd., Basingstoke, Hampshire, England) and incubated at 42°C during 48 h under microaerobic conditions using CampyGen CO2 (Oxoid Ltd., Basingstoke, Hampshire, England). The colonies were Gram stained and tested for oxidase (Bactident oxidase; Merck). Campylobacter jejuni/Campylobacter coli were confirmed by PCR of hippuricase and aspartokinase genes according to the protocol developed by Persson and Olsen in 2005 (22). Campylobacter species not belonging to C. jejuni/coli were identified through 16S rRNA gene sequencing by Functional Biosciences (Madison, WI), and sequences were uploaded to GenBank.
Giardia lamblia and Cryptosporidium parvum were detected using an enzyme-linked immunosorbent assay (Ridascreen Giardia and Ridascreen Cryptosporidium; r-Biopharm, Darmstadt, Germany).
Transmission between humans and domestic animals.
Transmission of bacterial pathogens (C. jejuni and aEPEC) between domestic animals and humans and between domestic animals was assessed by characterizing the clonal relationships of bacterial isolates using multilocus sequence typing (MLST). To detect clonally related bacteria, we screened C. jejuni isolates using nucleotide sequences of pgm genes and aEPEC isolates using nucleotide sequences of fumC genes; all isolates showing an identical DNA sequence were assumed to be clonally related and were subjected to a full MLST profiling to confirm this status.
MLST.
DNA extraction of C. jejuni and aEPEC isolates was performed using DNAzol reagent (Invitrogen, Carlsbad, CA, USA) by following the manufacturer's protocol. We screened 72 C. jejuni isolates by amplifying and sequencing the pgm allele (23); 55 isolates with identical pgm DNA sequences were subjected to additional analysis of genes glyA and tkt, and only the isolates with identical DNA sequences for pgm, glyA, and tkt alleles (48 out of 72 isolates) were subjected to full MLST analysis (23). Similarly, aEPEC isolates with identical fumC DNA sequences (14 out of 28 isolates) were subjected to full MLST analysis (24). The PCR products were sequenced by Functional Biosciences (Madison, WI), and sequences were uploaded to the PubMLST website for Campylobacter spp. (http://pubmlst.org/campylobacter/) and E. coli (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli/) to assign the allelic profiles.
Data analyses.
Evolutionary relationships of sequence types were inferred using eBURST V3 (http://eburst.mlst.net/). To visualize the microevolutionary processes of the STs, minimum-spanning trees were constructed with Prim's algorithm in the BioNumerics software according to species source (version 7.5; Applied-Maths, Sint Martens-Latem, Belgium). Statistical analyses were performed using Microsoft Office Excel 2013. Geographic distribution maps were developed using BaseCamp software version 4.4.7. and GPSvisualizer (http://www.gpsvisualizer.com/).
Nucleotide sequence accession numbers.
Sequences determined during the course of this work were deposited in GenBank under accession numbers KU362553 to KU362565.
RESULTS
Prevalence and geographic distribution of zoonotic enteropathogens.
Campylobacter jejuni was the most prevalent pathogen in all samples (30.7%; n = 82), followed by Giardia lamblia (13.1%; n = 35), C. coli (11.6%; n = 31), aEPEC (10.5%; n = 28), Campylobacter non-jejuni/coli (5.2%; n = 13), STEC (2.6%; n = 7), Salmonella spp. (1.9%; n = 5), Cryptosporidium parvum (1.1%; n = 3), and Yersinia enterocolitica (0.4%; n = 1) (Table 1). The geographic distribution of isolates is shown in Fig. S2 in the supplemental material.
Campylobacter spp. were found in 17.2% of children (7 samples were C. jejuni, 3 were C. coli, and 1 was C. hyointestinalis), and 57.1% of samples (n = 112) were positive in domestic animals. High percentages of guinea pigs (77.5%) and chickens (76%) were positive for Campylobacter spp. (mostly C. jejuni) (chickens, 59.5%; guinea pigs, 72.5%). Also, 75% of pigs were positive for Campylobacter spp., including C. coli (38.9%) and C. hyointestinalis (27.8%). Dogs carried Campylobacter spp. (30%; n = 12), mainly C. jejuni (25%). In addition, Campylobacter also was present in rabbits, cows, cats, ducks, and quails but at a lower prevalence (Table 1). Other Campylobacter species identified were C. canadensis in ducks (2 out of 5 samples) and C. lanienae in a pig (1 out of 36 samples).
Atypical EPEC was present in a wide range of hosts, including children (17.2%; n = 11), dogs (10.0%; n = 4), pigs (11.1%; n = 4), chickens (7.1%; n = 3), guinea pigs (5.0%; n = 2), cattle (28.6%; n = 2), ducks (20.0%; n = 1), and sheep (50.0%; n = 1) (Table 1).
Seven STECs were isolated from a child (n = 1), cattle (n = 4), a guinea pig (n = 1), and a chicken (n = 1). Giardia lamblia was present mainly in children (n = 22) and was detected in 13 animal fecal samples from guinea pigs, dogs, pigs, rabbits, and a sheep (Table 1). Salmonella spp. were detected in 5 samples from dogs (S. enterica serovar Infantis), 2 children and 1 sheep were positive for Cryptosporidium parvum, and 1 pig was positive for Yersinia enterocolitica (Table 1).
The copresence of pathogens was found in 9 children, 7 pigs, 4 guinea pigs, 5 dogs, 2 cattle, 3 chickens, 1 sheep, 1 cat, and 1 duck (see Table S4 in the supplemental material). The most common one was Campylobacter-aEPEC, found in 11 (4.1%) samples, followed by Campylobacter-Giardia in 8 (3%) samples and aEPEC-Giardia in 7 (2.6%) samples.
Zoonotic enteropathogens were present in all age groups of children; however, most of the pathogens were detected in children aged 1 to 3 years old (56.4% of 47 positive samples; 27 of 31 children of this age group), followed by children aged 3 to 5 years old (31.9% of 47 positive samples; 15 of 18 children of this category) (see Table S1 in the supplemental material). The youngest children infected were one 3-month-old baby with aEPEC, a 3-month-old baby with G. lamblia, a 10-month-old infant with C. coli, a 13-month-old infant with coinfection of C. parvum and C. jejuni, a 4-year-old child with C. hyointestinalis, and a 5-year-old child with STEC (see Table S1 and Fig. S3). Interestingly, none of the subjects showed evidence of acute diarrhea.
Campylobacter jejuni MLST.
We identified 21 C. jejuni STs in the 48 analyzed isolates, of which 14 were novel: ST-7643, ST-7759, ST-7760, ST-7662, ST-7669, ST-7671, ST-7672, ST-7775, ST-7777, ST-7778, ST-7779, ST-7780, ST-7781, and ST-7789 (Table 2). Four of the STs were detected in both children and domestic animals: ST-137, ST-1233, ST-3515, and ST-7671 (Table 2 and Fig. 1). Ten STs belonged to 5 clonal complexes (CC). The most common was CC-353, comprised of 8 isolates (2 from children and 6 from domestic animals), followed by CC-607, comprised of 7 isolates from different domestic animal species but no humans, and CC-354, with 4 isolates from avian species (3 chickens and 1 quail). In three households we found animals that shared isolates with the same ST (1 guinea pig and 1 chicken, 1 rabbit and 1 chicken, and 1 dog and 1 quail). The rest of the STs and CCs were randomly distributed in the community (Table 2; also see Table S2 in the supplemental material). Further, guinea pigs were the major reservoir of C. jejuni; 21 isolates of this animal species were analyzed by MLST, and nine novel STs were identified solely in guinea pigs (25).
TABLE 2.
Number of isolates of C. jejuni, by sequence type, recovered from children 0 to 3 years of age and from each animal source
| CCa | STb | No. of isolates from: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Children | Chickens | Guinea pigs | Dogs | Pigs | Rabbits | Cattle | Cats | Quails | ||
| CC-607 | 607 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| 1212 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | |
| CC-353 | 1233 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3515 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 7643 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | |
| CC-354 | 354 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7662 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| 7669 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| CC-464 | 464 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| CC-45 | 137 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Unassigned CC | 7671 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 7672 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 7759 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 7760 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 7775 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 7777 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 7778 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 7779 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 7780 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 7781 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 7789 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
CC, clonal complex.
The STs marked in boldface correspond to new STs.
FIG 1.
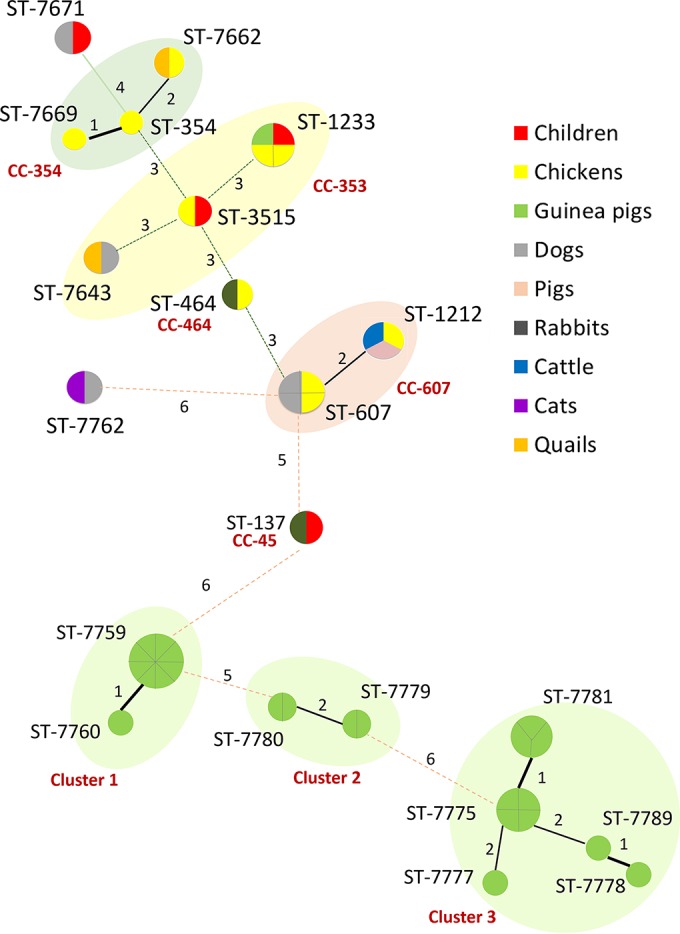
Minimum spanning tree analysis of 48 C. jejuni isolates based on MLST profile and according to source (the color of the circle indicates the species). Each circle represents the ST, and the size of the circle and circle divisions indicate the number of isolates within any given ST. Lines indicate strength length (distance) symbolized with numbers (number of nonshared alleles) and thickness. Clonal complexes and genetic clusters are labeled and represented by the surrounding shading.
Atypical EPEC MLST.
Fourteen aEPEC isolates belonged to 9 STs; 4 STs (ST-20, ST-137, ST-517, and ST-4550) were present in both children and domestic animals. Isolates from a sheep and a duck belonged to ST-317 (Fig. 2). There were no predominant clonal complexes: 5 STs belonged to 5 different CCs, while 4 STs were not assigned to any CC. None of the STs shared by isolates from humans and domestic animals belonged to the same household; indeed, identical STs were found in households distantly located (Table 3; also see Table S3 in the supplemental material). ST-4550 was present in a child and a chicken living in close proximity and was genetically related to ST-29 identified in a child (identical at 6 of 7 loci).
FIG 2.
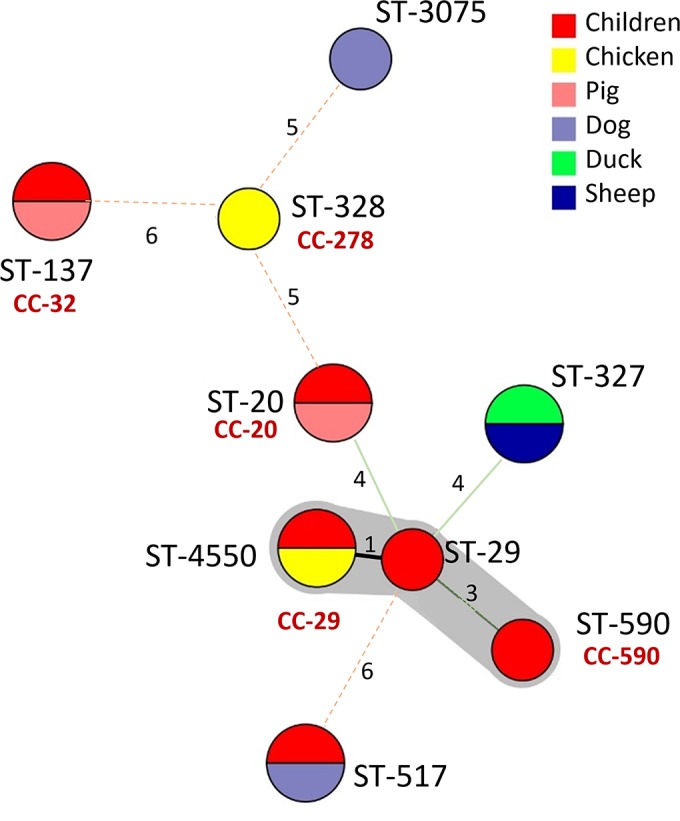
Minimum spanning tree analysis of 14 aEPEC isolates based on MLST profile and according to source (color of the circle indicates the species). Each circle represents the ST, and the size of the circle and circle divisions indicate the number of isolates within any given ST. Lines indicate strength length (distance) symbolized with numbers (number of nonshared alleles) and thickness. The genetic cluster is labeled and represented by the surrounding shading.
TABLE 3.
Number of isolates of aEPEC, by sequence type, recovered from children 0 to 5 years of age and from each animal source
| CC | ST | No. of isolates from: |
|||||
|---|---|---|---|---|---|---|---|
| Children | Chickens | Dogs | Pigs | Ducks | Sheep | ||
| CC-20 | 20 | 1 | 0 | 0 | 1 | 0 | 0 |
| CC-29 | 29 | 1 | 0 | 0 | 0 | 0 | 0 |
| CC-32 | 137 | 1 | 0 | 0 | 1 | 0 | 0 |
| CC-278 | 328 | 0 | 1 | 0 | 0 | 0 | 0 |
| CC-590 | 590 | 1 | 0 | 0 | 0 | 0 | 0 |
| Unassigned | 327 | 0 | 0 | 0 | 0 | 1 | 1 |
| 517 | 1 | 0 | 1 | 0 | 0 | 0 | |
| 3075 | 0 | 0 | 1 | 0 | 0 | 0 | |
| 4550 | 1 | 1 | 0 | 0 | 0 | 0 | |
DISCUSSION
Consistent with previous studies (26–29), our MLST analysis indicated that chickens were an important source of C. jejuni for both humans and domestic animals; this animal species commonly shared the same STs with other species (2 STs in 2 isolates from humans and 5 STs from 7 isolates of domestic animals) (Table 2 and Fig. 1). On the other hand, STs of C. jejuni from guinea pigs (the animal with the highest prevalence of C. jejuni) seemed to infect only this animal species (Table 2) (25). Previous MLST analyses have found that some C. jejuni strains may be more adapted to one animal species and less likely to infect humans or other hosts (25, 29, 30). The association of C. jejuni's STs and CCs with specific animal species seemed to concur with findings previously reported (28, 30, 31).
Similarly, MLST analysis of aEPEC revealed that pigs, chickens, and dogs shared genotypes of aEPEC found in children; hence, these animals could be involved in the transmission of aEPEC to humans (Table 3 and Fig. 2). Additionally, several animal species, like guinea pigs, cattle, ducks, and sheep, carried aEPEC (Table 3) as reported in previous studies (32, 33). Most STs were not found in animal species previously reported except for ST-4550 and ST-327, which were described previously in chickens and ruminants, respectively (http://mlst.warwick.ac.uk/mlst/).
Although our findings suggest zoonotic transmission of C. jejuni and aEPEC, they do not provide conclusive evidence for transmission from domestic animals to humans. This is especially critical for aEPEC, a pathogen of uncertain zoonotic potential, and it is possible that domestic animals became colonized by aEPEC from humans (14, 15, 34). It is important to note, however, that most households in this community have improved water and sanitation (i.e., piped drinking water and flush toilets inside the home), which prompt us to suggest that the main route of infection of these zoonotic pathogens for humans was contact with animals or a contaminated environment. In fact, we detected by PCR C. jejuni and C. coli in water from an irrigation channel in the community (data not shown).
The dissemination of zoonotic enteric pathogens could be influenced by the use of animal fecal matter to fertilize soils and the presence of four large poultry facilities (total capacity of ∼200,000 chickens) within the community, which corresponds to the second largest conglomerate of poultry farms in Ecuador (http://www.conave.org/informacionlistall.php?pagina=2). Transmission could occur by workers in the poultry industry who subsequently expose their households to enteropathogens.
Additionally, E. coli (and probably pathogenic members of Enterobacteriaceae) grows massively in fresh fecal matter when exposed to oxygen in the environment (35, 36). A high prevalence of enteric pathogens in the environment may increase the possibility of crop contamination or the high presence of these pathogens in animal products. This area possibly represents a hot spot for zoonotic pathogens, especially for Campylobacter species, and their food products can represent a health risk to urban areas and may be associated with traveler's diarrhea when susceptible individuals visit this region, which is in close proximity to the Quito International airport.
Giardia lamblia was the most prevalent enteric pathogen among children (34.4%), and its level was higher than that in previous studies in Ecuador (ranging from 11 to 24%) (8, 37). Dogs, rabbits, pigs, guinea pigs, and sheep also carried G. lamblia in this location, which is an indication of transmission among animal species. However, we were not able to analyze genetic markers of these protozoa. Of the seven genotypes of Giardia (A to G), humans are susceptible to genotypes A and B, and its zoonotic transmission is mainly related to companion animals, such as dogs and cats, while livestock and contaminated water appear to be uncommon sources (16).
We also detected Cryptosporidium parvum in 2 samples from children (3.1%) and STEC in 1 sample from a child (1.6%). Both pathogens were detected in ruminants, and STEC was also detected in chickens and guinea pigs, which concurs with previous studies (38). Cryptosporidium spp. are associated with morbidity and mortality in young children in developing countries (39) and may be an important cause of diarrhea in Ecuadorian rural villages (8). Although Cryptosporidium spp. are highly prevalent in livestock (40), several studies in developing countries suggest that zoonotic transmission of Cryptosporidium spp. is uncommon (17). Meanwhile, STEC is considered a food-borne disease with ruminants as the main reservoir (7); however, symptomatic disease in humans seems to be uncommon in Ecuador (8, 10).
Despite the proximity of study households to poultry industrial operations, Salmonella was not detected in children, although it was isolated from five dogs (S. enterica serovar Infantis). Yersinia enterocolitica was present only in one pig fecal sample (pigs are known as the main reservoir for Y. enterocolitica) (7, 41).
The high prevalence of pathogens analyzed in this study of domestic animals (77% of the birds and 59% of the mammals) may contribute to environmental contamination and subsequent human infection. In fact, the distribution of certain species of animals in the community appeared to be related to the geographic presence of particular pathogens (see Fig. S1 and S2 in the supplemental material); for instance, the geographic location of guinea pigs and chickens corresponded to C. jejuni, pigs to C. coli, and ruminants to STEC (see Fig. S1 and S2).
Most children under 5 years of age (59.4%) carried intestinal pathogens but were asymptomatic (nondiarrheic stool), a phenomenon also observed with nonzoonotic human enteric pathogens in LMICs (42–46), and this may be due to herd immunity resulting from regular exposure to these pathogens (47); immune mothers may transfer immunoglobulins to their offspring through the placenta and breast milk. Another factor protecting people from symptomatic infection may be the microbiota composition (48). Despite the absence of diarrhea, asymptomatic infections, such as campylobacteriosis and cryptosporidiosis, may reduce growth in children (43, 44). The present work had some limitations; the first one was the small number of samples analyzed. The second limitation was the scheme used to assess clonality, as the initial selection of bacteria with one identical allele may have prevented us from detecting additional related clones (members of the same clonal complex). Finally, we were unable to carry out genotyping of Giardia lamblia, which was one of the most abundant enteric pathogens in the study.
The control of the dissemination of these pathogens calls for a comprehensive and multidisciplinary approach (49). It is necessary to have a complete analysis of the spatial, ecological, evolutionary, social, economic, and epidemiological aspects in order to reduce pathogen transmission. This report suggests that the area we investigated is heavily contaminated with zoonotic enteropathogens, which calls for additional research to detect pathogens in the environment (water, soil, and possibly crops).
Supplementary Material
ACKNOWLEDGMENTS
We greatly appreciate the assistance of Valeria Garzon, the Yaruquí community, and Gabriela Vasco, as well as our colleagues in the Microbiology Institute at the Universidad San Francisco de Quito, in conducting this research.
This research was supported by the Fogarty International Center of the National Institutes of Health under award number K01 TW 009484.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00795-16.
REFERENCES
- 1.Walker CLF, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, O'Brien KL, Campbell H, Black RE. 2013. Global burden of childhood pneumonia and diarrhoea. Lancet 381:1405–1416. doi: 10.1016/S0140-6736(13)60222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zambrano LD, Levy K, Menezes NP, Freeman MC. 2014. Human diarrhea infections associated with domestic animal husbandry: a systematic review and meta-analysis. Trans R Soc Trop Med Hyg 108:313–325. doi: 10.1093/trstmh/tru056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lal A, Hales S, French N, Baker MG. 2012. Seasonality in human zoonotic enteric diseases: a systematic review. PLoS One 7:e31883. doi: 10.1371/journal.pone.0031883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Croll NA, Cross JH. 2013. Human ecology and infectious diseases. Academic Press, New York, NY. [Google Scholar]
- 5.Hale CR, Scallan E, Cronquist AB, Dunn J, Smith K, Robinson T, Lathrop S, Tobin-D'Angelo M, Clogher P. 2012. Estimates of enteric illness attributable to contact with animals and their environments in the United States. Clin Infect Dis 54(Suppl 5):S472–S479. doi: 10.1093/cid/cis051. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2014. Estimates of foodborne illness in the United States. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/foodborneburden/ Accessed 3 September 2015. [Google Scholar]
- 7.European Food Safety Authority. 2015. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J 13:3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasco G, Trueba G, Atherton R, Calvopina M, Cevallos W, Andrade T, Eguiguren M, Eisenberg JN. 2014. Identifying etiological agents causing diarrhea in low income Ecuadorian communities. Am J Trop Med Hyg 91:563–569. doi: 10.4269/ajtmh.13-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Todd E. 1997. Epidemiology of foodborne diseases: a worldwide review. World Health Stat Q 50:30–50. [PubMed] [Google Scholar]
- 10.Trueba G, Garcés V, Colman RE, Seymour M, Vogler AJ, Keim P. 2013. Escherichia coli O157: H7 in Ecuador: animal reservoirs, yet no human disease. Vector Borne Zoonotic Dis 13:295–298. doi: 10.1089/vbz.2012.1128. [DOI] [PubMed] [Google Scholar]
- 11.Wagenaar JA, Newell DG, Kalupahana RS, Mughini-Gras L. 2015. Campylobacter: animal reservoirs, human infections, and options for control, p 159–177. In Sing A. (ed), Zoonoses–infections affecting humans and animals. Springer, Dordrecht, Netherlands. [Google Scholar]
- 12.Kaakoush NO, Castano-Rodriguez N, Mitchell HM, Man SM. 2015. Global epidemiology of Campylobacter infection. Clin Microbiol Rev 28:687–720. doi: 10.1128/CMR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernandes RT, Elias WP, Vieira MA, Gomes TA. 2009. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol Lett 297:137–149. doi: 10.1111/j.1574-6968.2009.01664.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Wakushima M, Aota T, Yoshida Y, Kita T, Maehara T, Ogasawara J, Choi C, Kamata Y, Hara-Kudo Y, Nishikawa Y. 2013. Specific properties of enteropathogenic Escherichia coli isolates from diarrheal patients and comparison to strains from foods and fecal specimens from cattle, swine, and healthy carriers in Osaka City, Japan. Appl Environ Microbiol 79:1232–1240. doi: 10.1128/AEM.03380-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moura RA, Sircili MP, Leomil L, Matte MH, Trabulsi LR, Elias WP, Irino K, Pestana de Castro AF. 2009. Clonal relationship among atypical enteropathogenic Escherichia coli strains isolated from different animal species and humans. Appl Environ Microbiol 75:7399–7408. doi: 10.1128/AEM.00636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson RC. 2004. The zoonotic significance and molecular epidemiology of Giardia and giardiasis. Vet Parasitol 126:15–35. doi: 10.1016/j.vetpar.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Xiao L, Feng Y. 2008. Zoonotic cryptosporidiosis. FEMS Immunol Med Microbiol 52:309–323. doi: 10.1111/j.1574-695X.2008.00377.x. [DOI] [PubMed] [Google Scholar]
- 18.Karns J, Van Kessel J, McClusky B, Perdue M. 2007. Incidence of Escherichia coli O157: H7 and E. coli virulence factors in US bulk tank milk as determined by polymerase chain reaction. J Dairy Sci 90:3212–3219. doi: 10.3168/jds.2006-009. [DOI] [PubMed] [Google Scholar]
- 19.Tornieporth NG, John J, Salgado K, de Jesus P, Latham E, Melo MC, Gunzburg ST, Riley LW. 1995. Differentiation of pathogenic Escherichia coli strains in Brazilian children by PCR. J Clin Microbiol 33:1371–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollard D, Johnson W, Lior H, Tyler S, Rozee K. 1990. Rapid and specific detection of verotoxin genes in Escherichia coli by the polymerase chain reaction. J Clin Microbiol 28:540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Frye JG, Hu J, Fedorka-Cray PJ, Gautom R, Boyle DS. 2006. Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp. enterica. J Clin Microbiol 44:3608–3615. doi: 10.1128/JCM.00701-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Persson S, Olsen KE. 2005. Multiplex PCR for identification of Campylobacter coli and Campylobacter jejuni from pure cultures and directly on stool samples. J Med Microbiol 54:1043–1047. doi: 10.1099/jmm.0.46203-0. [DOI] [PubMed] [Google Scholar]
- 23.Dingle K, Colles F, Wareing D, Ure R, Fox A, Bolton F, Bootsma H, Willems R, Urwin R, Maiden M. 2001. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol 39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham JP, Vasco K, Trueba G. 11 May 2016. Hyperendemic Campylobacter jejuni in guinea pigs (Cavia porcellus) raised for food in a semi-rural community of Quito, Ecuador. Environ Microbiol Rep. doi: 10.1111/1758-2229.12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullner P, Shadbolt T, Collins-Emerson JM, Midwinter AC, Spencer SE, Marshall J, Carter PE, Campbell DM, Wilson DJ, Hathaway S, Pirie R, French NP. 2010. Molecular and spatial epidemiology of human campylobacteriosis: source association and genotype-related risk factors. Epidemiol Infect 138:1372–1383. doi: 10.1017/S0950268809991579. [DOI] [PubMed] [Google Scholar]
- 27.Levesque S, Fournier E, Carrier N, Frost E, Arbeit RD, Michaud S. 2013. Campylobacteriosis in urban versus rural areas: a case-case study integrated with molecular typing to validate risk factors and to attribute sources of infection. PLoS One 8:e83731. doi: 10.1371/journal.pone.0083731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strachan NJ, Gormley FJ, Rotariu O, Ogden ID, Miller G, Dunn GM, Sheppard SK, Dallas JF, Reid TM, Howie H, Maiden MC, Forbes KJ. 2009. Attribution of Campylobacter infections in northeast Scotland to specific sources by use of multilocus sequence typing. J Infect Dis 199:1205–1208. doi: 10.1086/597417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson DJ, Gabriel E, Leatherbarrow AJ, Cheesbrough J, Gee S, Bolton E, Fox A, Fearnhead P, Hart CA, Diggle PJ. 2008. Tracing the source of campylobacteriosis. PLoS Genet 4:e1000203. doi: 10.1371/journal.pgen.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheppard SK, Didelot X, Jolley KA, Darling AE, Pascoe B, Meric G, Kelly DJ, Cody A, Colles FM, Strachan NJ, Ogden ID, Forbes K, French NP, Carter P, Miller WG, McCarthy ND, Owen R, Litrup E, Egholm M, Affourtit JP, Bentley SD, Parkhill J, Maiden MC, Falush D. 2013. Progressive genome-wide introgression in agricultural Campylobacter coli. Mol Ecol 22:1051–1064. doi: 10.1111/mec.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheppard SK, Maiden MC. 2015. The evolution of Campylobacter jejuni and Campylobacter coli. Cold Spring Harb Perspect Biol 7:a018119. doi: 10.1101/cshperspect.a018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hornitzky MA, Mercieca K, Bettelheim KA, Djordjevic SP. 2005. Bovine feces from animals with gastrointestinal infections are a source of serologically diverse atypical enteropathogenic Escherichia coli and Shiga toxin-producing E. coli strains that commonly possess intimin. Appl Environ Microbiol 71:3405–3412. doi: 10.1128/AEM.71.7.3405-3412.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanco M, Schumacher S, Tasara T, Zweifel C, Blanco JE, Dahbi G, Blanco J, Stephan R. 2005. Serotypes, intimin variants and other virulence factors of eae positive Escherichia coli strains isolated from healthy cattle in Switzerland. Identification of a new intimin variant gene (eae-η2). BMC Microbiol 5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krause G, Zimmermann S, Beutin L. 2005. Investigation of domestic animals and pets as a reservoir for intimin (eae) gene positive Escherichia coli types. Vet Microbiol 106:87–95. doi: 10.1016/j.vetmic.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Russell JB, Jarvis GN. 2001. Practical mechanisms for interrupting the oral-fecal lifecycle of Escherichia coli. J Mol Microbiol Biotechnol 3:265–272. [PubMed] [Google Scholar]
- 36.Vasco G, Spindel T, Carrera S, Grigg A, Trueba G. 2015. The role of aerobic respiration in the life cycle of Escherichia coli: public health implications. Av Cienc Ingenierías 7:B7–B9. [Google Scholar]
- 37.Jacobsen KH, Ribeiro PS, Quist BK, Rydbeck BV. 2007. Prevalence of intestinal parasites in young Quichua children in the highlands of rural Ecuador. J Health Popul Nutr 25:399–405. [PMC free article] [PubMed] [Google Scholar]
- 38.Beutin L, Geier D, Steinrück H, Zimmermann S, Scheutz F. 1993. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J Clin Microbiol 31:2483–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. [DOI] [PubMed] [Google Scholar]
- 40.Palacios-Ordóñez TE. 2014. Prevalencia de Cryptosporidium spp. y Giardia spp. en terneros, como contaminante de los recursos hídricos y su efecto en la población infantil en el cantón San Fernando. M.S. thesis. Universidad Estatal de Cuenca, Cuenca, Ecuador. [Google Scholar]
- 41.Berger S. 2016. Yersiniosis: global status. GIDEON Informatics Inc, Los Angeles, CA. [Google Scholar]
- 42.Alikhani MY, Mirsalehian A, Aslani MM. 2006. Detection of typical and atypical enteropathogenic Escherichia coli (EPEC) in Iranian children with and without diarrhoea. J Med Microbiol 55:1159–1163. [DOI] [PubMed] [Google Scholar]
- 43.Checkley W, Gilman RH, Epstein LD, Suarez M, Diaz JF, Cabrera L, Black RE, Sterling CR. 1997. Asymptomatic and symptomatic cryptosporidiosis: their acute effect on weight gain in peruvian children. Am J Epidemiol 145:156–163. [DOI] [PubMed] [Google Scholar]
- 44.Lee G, Pan W, Penataro Yori P, Paredes Olortegui M, Tilley D, Gregory M, Oberhelman R, Burga R, Chavez CB, Kosek M. 2013. Symptomatic and asymptomatic Campylobacter infections associated with reduced growth in Peruvian children. PLoS Negl Trop Dis 7:e2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quiroga M, Oviedo P, Chinen I, Pegels E, Husulak E, Binztein N, Rivas M, Schiavoni L, Vergara M. 2000. Asymptomatic infections by diarrheagenic Escherichia coli in children from Misiones, Argentina, during the first twenty months of their lives. Rev Inst Med Trop Sao Paulo 42:9–15. [DOI] [PubMed] [Google Scholar]
- 46.Ramirez JD, Heredia RD, Hernandez C, Leon CM, Moncada LI, Reyes P, Pinilla AE, Lopez MC. 2015. Molecular diagnosis and genotype analysis of Giardia duodenalis in asymptomatic children from a rural area in central Colombia. Infect Genet Evol 32:208–213. [DOI] [PubMed] [Google Scholar]
- 47.Belongia EA, Chyou PH, Greenlee RT, Perez-Perez G, Bibb WF, DeVries EO. 2003. Diarrhea incidence and farm-related risk factors for Escherichia coli O157:H7 and Campylobacter jejuni antibodies among rural children. J Infect Dis 187:1460–1468. [DOI] [PubMed] [Google Scholar]
- 48.Singh Y, Ahmad J, Musarrat J, Ehtesham NZ, Hasnain SE. 2013. Emerging importance of holobionts in evolution and in probiotics. Gut Pathog 5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rabinowitz P, Conti L. 2013. One health and emerging infectious diseases: clinical perspectives, p 17–28. In Mackenzie JS, Jeggo M, Daszak P, Richt JA (ed), One health: the human-animal-environment interfaces in emerging infectious diseases. The concept and examples of a one health approach. Springer, Berlin, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


