ABSTRACT
The remarkable metal resistance of many microorganisms is related to the presence of multiple metal resistance operons. Pseudomonas putida KT2440 can be considered a model for these microorganisms since its arsenic resistance is due to the action of proteins encoded by the two paralogous arsenic resistance operons ARS1 and ARS2. Both operons contain the genes encoding the transcriptional regulators ArsR1 and ArsR2 that control operon expression. We show here that purified ArsR1 and ArsR2 bind the trivalent salt of arsenic (arsenite) with similar affinities (~30 μM), whereas no binding is observed for the pentavalent salt (arsenate). Furthermore, trivalent salts of bismuth and antimony showed binding to both paralogues. The positions of cysteines, found to bind arsenic in other homologues, indicate that ArsR1 and ArsR2 employ different modes of arsenite recognition. Both paralogues are dimeric and possess significant thermal stability. Both proteins were used to construct whole-cell, lacZ-based biosensors. Whereas responses to bismuth were negligible, significant responses were observed for arsenite, arsenate, and antimony. Biosensors based on the P. putida arsB1 arsB2 arsenic efflux pump double mutant were significantly more sensitive than biosensors based on the wild-type strain. This sensitivity enhancement by pump mutation may be a convenient strategy for the construction of other biosensors. A frequent limitation found for other arsenic biosensors was their elevated background signal and interference by inorganic phosphate. The constructed biosensors show no interference by inorganic phosphate, are characterized by a very low background signal, and were found to be suitable to analyze environmental samples.
IMPORTANCE Arsenic is at the top of the priority list of hazardous compounds issued by the U.S. Agency for Toxic Substances and Disease. The reason for the stunning arsenic resistance of many microorganisms is the existence of paralogous arsenic resistance operons. Pseudomonas putida KT2440 is a model organism for such bacteria, and their duplicated ars operons and in particular their ArsR transcription regulators have been studied in depth by in vivo approaches. Here we present an analysis of both purified ArsR paralogues by different biophysical techniques, and data obtained provide valuable insight into their structure and function. Particularly insightful was the comparison of ArsR effector profiles determined by in vitro and in vivo experimentation. We also report the use of both paralogues to construct robust and highly sensitive arsenic biosensors. Our finding that the deletion of both arsenic efflux pumps significantly increases biosensor sensitivity is of general relevance in the biosensor field.
INTRODUCTION
Arsenic is omnipresent in nature and found in many environmental biotopes such as seawater, freshwater, soils, and rocks. It is either released through various natural processes such as weathering or hydrothermal emissions or generated by a number of anthropogenic activities such as the use of arsenic-containing pesticides, mining activities, and combustion of fossil fuels (1, 2).
Arsenic is very toxic and was found to be associated with an increased risk of a wide range of health issues, including cancers of the skin, lung, bladder, liver, and kidney as well as neurological and cardiovascular diseases (3). The toxicity of arsenic, combined with its omnipresence in nature, represents a worldwide health concern (4). In fact, it was placed first on the priority list of hazardous compounds issued by the U.S. Agency for Toxic Substances and Disease Registry (see http://www.atsdr.cdc.gov/SPL/index.html). Of particular relevance is groundwater contamination with arsenic in Asiatic countries (5, 6), and environmental arsenic monitoring is thus of primary importance (7, 8).
At neutral pH, arsenic is found mostly as trivalent arsenite or pentavalent arsenate. The principles of toxicity differ for both forms. Whereas arsenite toxicity can be associated with its ability to react with sulfhydryl groups in proteins, the harmful effects caused by arsenate are associated with its capacity to mimic phosphate groups in a wide number of cellular reactions (9, 10). Bacteria have developed many strategies in response to the environmental presence of arsenic (1). One of these strategies consists of the extrusion of arsenic, and the corresponding proteins are encoded by ars operons. The minimal set of ars operon genes, consisting of the arsR regulator (11), the transmembrane arsB arsenite efflux pump, and the arsenate reductase arsC, has been observed in a number of different bacteria (12). Interestingly, several strains, such as Pseudomonas putida KT2440 (13–15), Corynebacterium glutamicum (16), and Ochrobactrum tritici (17), possess two functional copies of the ars operon, providing high arsenic resistance.
The present study was conducted by using P. putida KT2440, which is a metabolically versatile soil bacterium that serves as a model not only to study metal resistance but also in the fields of bioremediation, plant-microbe interactions, and bacterial signaling (18–20). Strain KT2440 is highly resistant to arsenic but is also resistant to other metalloids and heavy metals, and genome analyses indicated that several resistance systems are duplicated (21). The two ars operon copies of P. putida KT2440, termed operons ARS1 and ARS2, show a high degree of sequence identity and are extended by the arsH gene, encoding an organoarsenical oxidase (22) (Fig. 1). Different aspects of the two ars operons in P. putida KT2440 were recently reported. The ARS2 operon was found to be present in the core genome of P. putida strains, whereas the existence of operon ARS1 is specific to KT2440 and may have been acquired by horizontal gene transfer (14). The gene products of both operons work synergistically, which may account for the high arsenite tolerance of this strain (14). Another study reported that the effects of operons ARS1 and ARS2 are not additive at the optimal growth temperature of 30°C, whereas at 15°C, the contribution of operon ARS2 to arsenite resistance was largely superior to that mediated by the operon ARS1 system (15).
FIG 1.
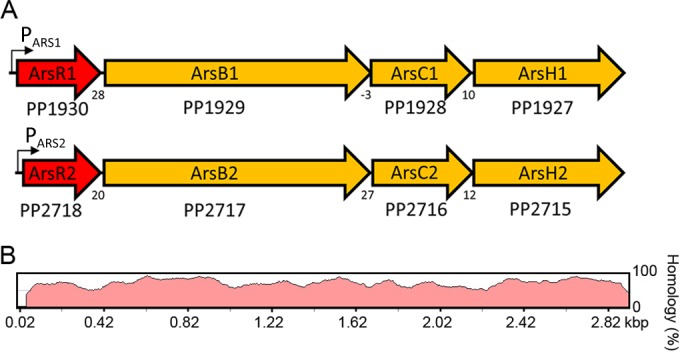
Two paralogous ars operons in P. putida KT2440. (A) Genetic organization. The ArsR transcriptional regulators are shown in red, arsB encodes an arsenite efflux pump, arsH encodes an organoarsenical oxidase, and arsC encodes an arsenate reductase. Numbers below the arrows represent the intergenic distance between contiguous genes, with negative numbers indicating overlapping genes. (B) DNA sequence homology between both operons. The alignments were performed by using the wgVISTA algorithm (56).
ArsR together with the SmtB regulator give a name to a family of regulators that can be associated with heavy metal resistance (23). Members of the ArsR/SmtB protein family operate by a derepression mechanism, where aporepressors are DNA bound, and the binding of different heavy metals causes protein dissociation and transcriptional activation. The ArsR protein of Escherichia coli has been shown in vitro to bind arsenite as well as the trivalent salts of bismuth and antimony (24, 25). In vivo studies show that arsenate, in addition to these three compounds, causes transcriptional activation (24). The in vivo effect of arsenate is caused by its cellular conversion into arsenite by the arsenate reductase ArsC (24). Members of the ArsR family share the same overall structure (26). Arsenite binds to ArsR proteins via three cysteine residues. However, experiments conducted with regulators from E. coli (27), Corynebacterium glutamicum (28), and Acidithiobacillus ferrooxidans (29) have resulted in the identification of three different arsenite binding modes, characterized by distinct cysteine binding motifs that are located in different parts of the structures (see Fig. S1 in the supplemental material). The ArsR1 and ArsR2 paralogues of P. putida KT2440 share 56% sequence identity among each other (see Fig. S1 in the supplemental material). As indicated in Fig. S1 in the supplemental material, the positions of cysteine residues differ in ArsR1 and ArsR2 and, in addition, are different from those of the studied ArsR proteins of E. coli, C. glutamicum, and A. ferrooxidans. This indicates that arsenite recognition at ArsR1 and ArsR2 occurs but by another cysteine motif (see Fig. S1 in the supplemental material). It has been proposed that the evolution of spatially unrelated arsenite binding sites is the result of convergent evolution (28).
The two ArsR paralogues of P. putida KT2440 are a model system to understand the concerted action of paralogous regulators on metal resistance (14, 15). We report here the generation of purified, recombinantly produced ArsR1 and ArsR2 and their analysis by an array of biochemical and biophysical approaches. As mentioned above, environmental arsenic monitoring is of central importance, and several ArsR-based biosensors have been described (7, 30). In the second part of this work, we describe the construction of ArsR1- and ArsR2-based biosensors. We demonstrate that biosensors based on the mutation of both arsenite efflux pumps, ArsB1 and ArsB2, possess higher sensitivity than wild-type (wt)-based sensors. The finding that efflux pump mutation translates into a significant increase in the sensitivity of the corresponding sensor is of general relevance for biosensor construction.
MATERIALS AND METHODS
Materials.
NaAsO2, Na2HAsO4, SbCl3, Bi(NO3)3, and NaH2PO4 (referred to throughout this article as arsenite, arsenate, antimony, bismuth, and inorganic phosphate, respectively) were purchased from Sigma-Aldrich.
Strains and culture conditions.
Strains and plasmids used in this study are listed in Table 1. P. putida KT2440 and its isogenic arsB1 arsB2 double mutant (14) were grown in M9 medium with 5 mM glucose as a carbon source or LB medium at 30°C with shaking, unless otherwise stated. Escherichia coli DH5α, the host for gene cloning, was cultured in LB medium at 37°C. When required, antibiotics were added at the following final concentrations: chloramphenicol (Cm) at 30 μg/ml, kanamycin (Km) at 25 μg/ml, gentamicin (Gm) at 30 μg/ml, and tetracycline (Tc) at 10 μg/ml.
TABLE 1.
Strains and plasmids used in this studya
| Strain, plasmid, or biosensor | Characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| P. putida KT2440 | Considered wild type in this study; spontaneous restriction-deficient derivative of strain mt-2 cured of TOL plasmid pWW0 | 57 |
| P. putida KT2440 arsB1 arsB2 | Double mutation of arsB1 arsB2 pCHESI::ARS1/mini-Tn5::ARS2 (Kmr Gmr) | 14 |
| E. coli DH5α | supE44 lacU169(ϕ80lacZΔM15) hsdR17(rK− mK−) recA1 endA1 gyrA96 thi-1 relA1 | 58 |
| E. coli BL21(DE3) | F− ompI hsdSB(rB− mB−) gal dam met | 59 |
| Plasmids | ||
| pET28a | Protein expression vector; Kmr | Novagen |
| pET_arsR1 | pET28a containing arsR1; Kmr | This work |
| pET_arsR2 | pET28a containing arsR2; Kmr | This work |
| pMP220 | Transcriptional fusion vector; lacZ; Tcr | 60 |
| pBiosen_arsR1 | Transcriptional fusion PARS1_ARS1_lacZ in pMP220; Tcr | This work |
| pBiosen_arsR2 | Transcriptional fusion PARS2_ARS2_lacZ in pMP220; Tcr | This work |
| Biosensors | ||
| Bs_WtR1 | P. putida KT2440 harboring pBiosen_arsR1 | This work |
| Bs_WtR2 | P. putida KT2440 harboring pBiosen_arsR2 | This work |
| Bs_MutR1 | P. putida KT2440 arsB1 arsB2 strain harboring pBiosen_arsR1 | This work |
| Bs_MutR2 | P. putida KT2440 arsB1 arsB2 strain harboring pBiosen_arsR2 | This work |
Km, kanamycin; Gm, gentamicin; Tc, tetracycline.
Construction of expression plasmids for ArsR1 and ArsR2, cell culture, and protein purification.
The arsR1 and arsR2 sequences were amplified by PCR using specific primers containing NdeI and XhoI restriction sites (Table 2). The PCR products were digested with the corresponding enzymes and cloned into pET28a (Novagen) previously cleaved with the same enzymes. Ligation mixtures were used to electroporate E. coli DH5α, and colonies were selected on LB plates containing Km. The resulting plasmids (pET_arsR1 and pET_arsR2) were verified by sequencing of the insert and flanking regions. E. coli BL21(DE3) was transformed with either pET_arsR1 or pET_arsR2. Cultures were grown in 2-liter Erlenmeyer flasks containing 400 ml LB medium supplemented with 25 μg ml−1 kanamycin at 37°C until the culture reached an optical density at 660 nm (OD660) of 0.6. At this point, protein expression was induced by the addition of 0.1 mM isopropyl-β-d-1-thiogalactopyranoside. Growth was continued at 18°C overnight prior to cell harvest by centrifugation (15 min at 4,400 × g). All manipulations were carried out at 4°C. The pellet was resuspended in buffer A (20 mM Tris-HCl, 500 mM NaCl, 10 mM imidazole, 10% [vol/vol] glycerol, 10 mM β-mercaptoethanol [pH 8.5]), sonicated for cell rupture, and centrifuged at 20,000 × g for 1 h. The supernatant was filtered and loaded onto a 5-ml HisTrap HP column (Amersham GE Healthcare) previously equilibrated with buffer A. Proteins were eluted by using an imidazole gradient (0 to 1 M) in buffer A. Protein purity was assessed by SDS-PAGE, and pooled fractions were dialyzed into analysis buffer [20 mM HEPES, 150 mM NaCl, 2 mM tris(2-carboxyethyl)phosphine (TCEP) (pH 7.2)] unless otherwise stated. Sample concentrations were determined by a Bradford assay (31) and by measurement of the absorbance at 280 nm using the theoretical extinction coefficient of 19,730 M−1 cm−1 determined for both proteins by the Expasy ProtParam algorithm (32).
TABLE 2.
Oligonucleotides used in this study
| Procedure and oligonucleotide | Sequence (5′–3′) (restriction site)a |
|---|---|
| Protein expression | |
| arsR1His_for | AACATATGATGCGAGAAATACTGACTCC (NdeI) |
| arsR1His_rev | TTCTCGAGTCAAGCACAAGCAACAGGGCTC (XhoI) |
| arsR2His_for | AACATATGATGATCACACCGCCCGATGT (NdeI) |
| arsR2His_rev | TTCTCGAGTCAGCAGCAGACGGAATCAC (XhoI) |
| Biosensor construction | |
| ars1biosen_for | AAGAATTCGACCGCTTCCTTCAATGCCCCC (EcoRI) |
| ars1biosen_rev | AACTGCAGGCGAAGGGAAGTCTCAAGCACA (PstI) |
| ars2biosen_for | AAGAATTCGCGTAGCCGGTGACACCGATCG (EcoRI) |
| ars2biosen_rev | AATCTAGAGGTGATGATCAGCAGCAGACGG (XbaI) |
| EMSA | |
| Pars1_for | GACCCTTGCGGCGATCCATCA |
| Pars1_rev | AAGGTCTTGGGTCGGCAAGCC |
| Pars2_for | TGGCGACTGATCTTGGGTTGGC |
| Pars2_rev | GGAGACACCTTCAGCGTAGCCG |
| Primers for nonspecific fragments | AGAGGTAGCTCACATGATb |
| CATGCCGCTTCCTTGGTTTb |
Restriction sites are underlined.
From Serratia plymuthica strain A153.
Analytical size exclusion chromatography.
Analytical size exclusion chromatography experiments were conducted by using an Åkta fast protein liquid chromatography (FPLC) system (Amersham GE Healthcare). Purified protein (at 25 μM) was loaded onto a Superdex-200 10/300GL column (Amersham GE Healthcare) previously equilibrated with analysis buffer. Elution was performed at a constant flow rate of 0.4 ml/min, and the absorbance of the eluate was monitored at 280 nm. The molecular mass of the ArsR paralogues was estimated from a calibration curve constructed from analysis of the following standards: cytochrome c (12.4 kDa), carbonic anhydrase (29 kDa), bovine serum albumin (monomer, 66 kDa, and dimer, 132 kDa), and β-amylase (200 kDa).
Circular dichroism spectroscopy.
Circular dichroism (CD) measurements were performed with a Jasco J-715 spectropolarimeter (Tokyo, Japan) equipped with a temperature-controlled cell holder. Measurements of the far-UV CD spectra (260 to 200 nm) were made with a 1-mm-path-length quartz cuvette at a protein concentration of 10 μM in analysis buffer. Spectra were recorded at a scan rate of 100 nm/min, with a 1-nm step resolution, a 1-s response, and a 1-nm bandwidth. Spectra shown are averages of results of 5 scans. Spectra were corrected by baseline subtraction using the buffer spectrum, and the CD signal was normalized to the mean residue molar ellipticity (θ) (in degrees per decimole per square centimeter). The spectra were deconvoluted to determine the relative abundance of secondary structure elements by using the CDNN software package (33).
Differential scanning calorimetry.
Differential scanning calorimetry (DSC) experiments were carried out on a VP capillary cell microcalorimeter (MicroCal, Northampton, MA). A temperature range from 5°C to 85°C was scanned at a rate of 180°C/h. Calorimetric cells (operating volume, 0.134 ml) were kept under an excess pressure of 60 lb/in2 to prevent degassing. Several buffer-buffer baselines were obtained before each protein run to ascertain proper instrument equilibration. Protein samples were exhaustively dialyzed at 4°C against analysis buffer. The protein concentration was 20 μM, and arsenite and arsenate were added at a concentration of 250 μM. Thermograms were normalized by using the concentration of protein monomers.
Intrinsic tryptophan fluorescence spectroscopy.
Measurements were made on a PTI QM-2003 fluorimeter with a xenon lamp as a light source (Photon Technology International, Lawrenceville, NJ) at 25°C, using an excitation wavelength of 290 nm and slit widths of 5 nm. Tryptophan emission spectra were recorded from 300 to 400 nm. Spectra were taken in 1-nm increments with 1 s of integration per increment. Protein samples of 5 μM were prepared in analysis buffer. Titrations were conducted with freshly prepared solutions of arsenite (1 mM stock solution), antimony, and bismuth. Due to the low solubility of antimony and bismuth, these stock solutions were prepared in methanol at concentrations of 20 mM and 2 mM, respectively. Quenching of the tryptophan emission was performed by successive additions of the ligands to the cuvette, followed by gentle sample mixing, incubation for 2 min, and spectrum recording. The fluorescence intensities were corrected for absorption of the exciting light and reabsorption of the emitted light to decrease the inner filter effect by using the equation Fcorr = Fobs × 10(Aexc + Aem)/2, where Fcorr is the corrected fluorescence value, Fobs is the measured fluorescence value, Aexc is the absorption value at the excitation wavelength, and Aem is the absorption value at the emission wavelength (34). Spectra were then normalized by the total protein concentration after each titration, and the starting fluorescence signal was set to 100% fluorescence. Fluorescence titration curves were fitted in Origin by using the equation for one binding site to determine the dissociation constants.
Electrophoretic mobility shift assays (EMSAs).
DNA fragments of ∼500 bp containing the PARS1 and PARS2 promoter regions were amplified by PCR using the primer pairs Pars1_for/Pars1_rev and Pars2_for/Pars2_rev, respectively (Table 2). Amplified DNA was isolated from agarose gels and end labeled with [γ-32P]dATP using T4 polynucleotide kinase. A 10-μl sample containing 2 nM labeled DNA (1.5 × 104 cpm) was incubated at 30°C with various concentrations of purified ArsR for 15 min in binding buffer (10 mM Tris-HCl, 20 mM NaCl, 100 mM Mg acetate, 0.02 mM EDTA, 0.2 mM dithiothreitol [DTT], 1% [vol/vol] glycerol [pH 7.5]) containing 20 μg/ml of poly(dI-dC) and 200 of μg/ml bovine serum albumin. The DNA-protein complexes were resolved by electrophoresis in 4% (wt/vol) nondenaturing polyacrylamide gels at 50 V for 3 h in a solution containing 50 mM Tris-HCl and 20 mM glycine (pH 7.8). The gels were dried and analyzed with a GS525 molecular imager (Bio-Rad).
Construction of arsenic biosensors.
The genes of operons ARS1 and ARS2, besides their respective promoter regions, were amplified by PCR using primers ars1biosen_for (EcoRI) and ars1biosen_rev (PstI) (for operon ARS1) and primers ars2biosen_for (EcoRI) and ars2biosen_rev (XbaI) (for operon ARS2). The Expand high-fidelity PCR system (Roche) was used for all PCR amplifications. PCR products were cleaned by using a QIAquick PCR purification kit (Qiagen) and then cloned into pMP220 digested with the corresponding enzymes. Ligation mixtures were electroporated into E. coli, and clones were selected on Tc-containing LB plates. Clones were verified by plasmid restriction analysis and DNA sequencing. The resulting plasmids, pBiosen_arsR1 and pBiosen_arsR2, were transferred into P. putida KT2240 and its arsB1 arsB2 double mutant.
β-Galactosidase assays.
Cultures grown overnight were diluted 50-fold in LB medium supplemented with different ArsR ligand concentrations and incubated with shaking until an OD600 of 0.8 was reached. Assays were carried out on permeabilized cells as previously described (35). Data shown are means and standard deviations from three independent experiments, each conducted in duplicate.
Inductively coupled plasma-optical emission spectroscopy.
Inductively coupled plasma-optical emission spectroscopy (ICP-OES) analyses were carried out by the scientific instrumentation service of the Estación Experimental del Zaidín, Granada, Spain (see http://www.eez.csic.es/?q=es/node/1187).
RESULTS
Generation of recombinant ArsR1 and ArsR2.
ArsR1 and ArsR2 of P. putida KT2440 were overexpressed in E. coli and purified from the soluble fraction of the cell lysate by affinity chromatography. Relatively modest average yields of 2 and 4 mg of pure protein per liter of E. coli culture were obtained for ArsR1 and ArsR2, respectively. Far-UV CD experiments with ArsR1 and ArsR2 were carried out to assess their secondary structure (Fig. 2A). The spectra of both proteins were very similar, indicating that ArsR1 and ArsR2 possess similar secondary structure contents. The curves showed minima at 208 and ∼220 nm, characteristic of α-helical elements. The deconvolution of spectra resulted in 42 and 41% α-helices for ArsR1 and ArsR2, respectively, as well as 28 and 29% random coils, respectively. Attempts to make homology models were successful only in the case of ArsR1 (see Fig. S2 in the supplemental material), but the high degree of sequence similarity suggests that both proteins share similar secondary structure contents. This model shows 41% α-helix, assuming a nonhelical conformation of the His tag (not modeled). The agreement of the experimentally determined α-helical contents with those derived from the homology model strongly suggests that the purified proteins are in their folded and active state.
FIG 2.
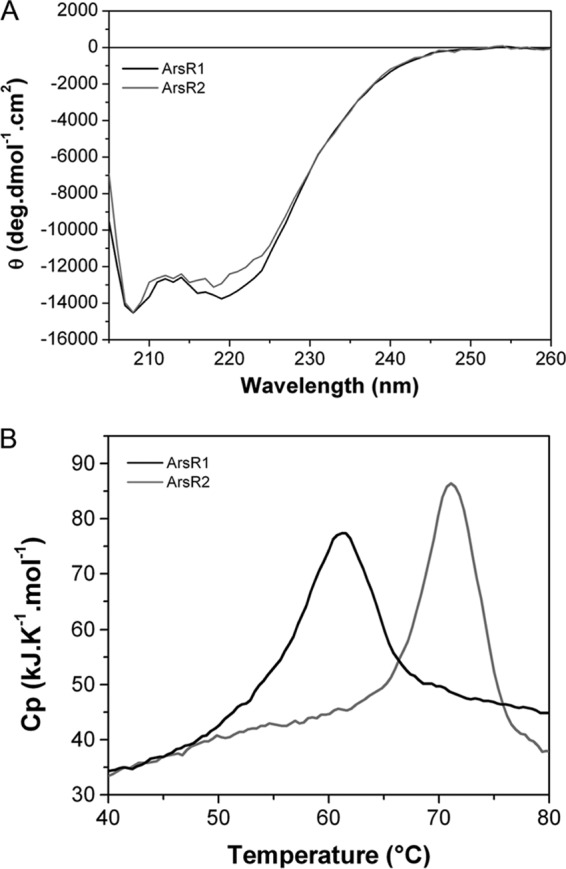
Comparison of the secondary structures and thermal stabilities of the ArsR paralogues. (A) Analysis of ArsR1 and ArsR2 by far-UV circular dichroism spectroscopy. (B) Analysis of thermal unfolding by differential scanning calorimetry. Cp, heat capacity.
ArsR1 and ArsR2 are dimeric in solution.
To determine the oligomeric state of both proteins, analytical size exclusion chromatography experiments were carried out, showing that both proteins eluted as a single symmetric peak (Fig. 3). Eluted proteins were submitted to SDS-PAGE, which resulted in the confirmation of the molecular integrity of both proteins (see Fig. S3 in the supplemental material). With the help of standard curves (Fig. 3), apparent molecular masses of 28 kDa and 26 kDa were determined for ArsR1 and ArsR2, respectively. Considering the sequence-derived masses of ArsR1 and ArsR2 monomers (15,757 Da and 15,407 Da, respectively), we conclude that both proteins are dimeric in solution. To assess the effect of arsenite and arsenate on the oligomeric state of both proteins, experiments were repeated in the presence of 1 mM arsenite or arsenate, which was added to the protein sample and the gel filtration buffer. The obtained chromatograms are highly similar to those recorded in the absence of the ligand (Fig. 3), indicating that ligand binding does not alter the protein oligomeric state.
FIG 3.
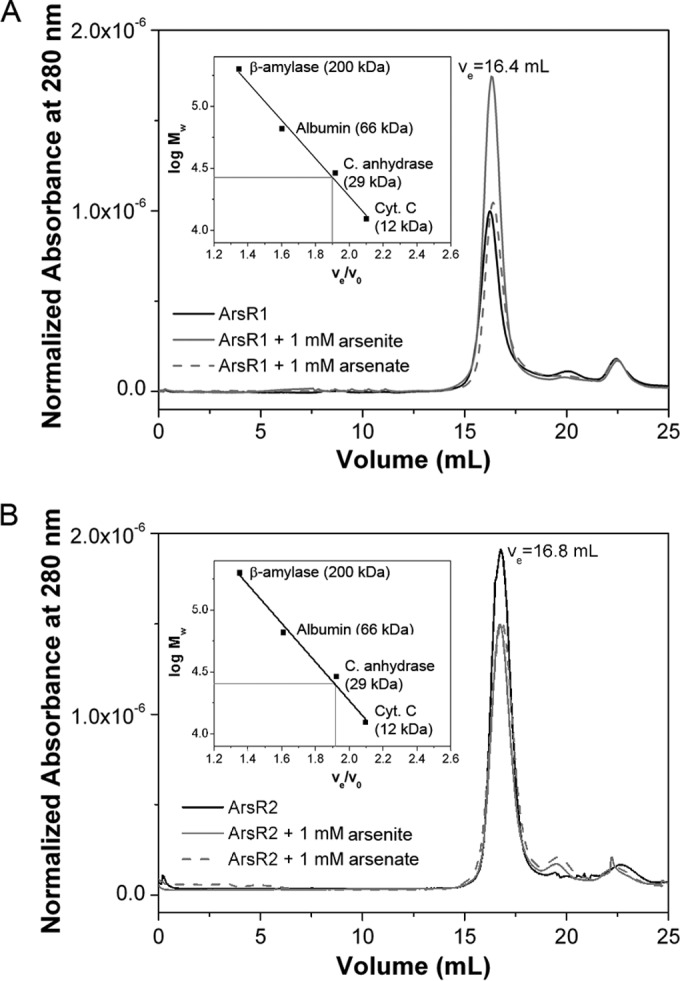
Determination of the oligomeric states of both ArsR paralogues. Shown are data from size exclusion chromatography for ArsR1 (A) and ArsR2 (B) in the absence and presence of 1 mM arsenite or 1 mM arsenate. The gray lines in the insets show the experimentally determined elution volume (ve)/void volume (vo) ratios as well as the corresponding extrapolation of the molecular weight (Mw). C. anhydrase, carbonic anhydrase; Cyt. C, cytochrome c.
The ArsR paralogues unfold in a single event at temperatures beyond 60°C.
The protein stability of both paralogues was analyzed by DSC. This technique permits the determination of the midpoint temperature (Tm) value, which corresponds to the temperature at which half the protein is unfolded and the remaining part is in its native state, as well as the enthalpy change upon unfolding. The DSC thermograms of both proteins (Fig. 2B) show that both proteins possess significant thermal stabilities, and Tm values of 61.5°C and 71.1°C were determined for ArsR1 and ArsR2, respectively (Table 3). At the end of the temperature up-scan, samples were cooled to 5°C, and no unfolding events were observed in the subsequent up-scan, indicating that protein unfolding is irreversible.
TABLE 3.
Thermodynamic parameters of protein unfolding monitored by DSC in the absence and presence of different ligandsd
| Condition | ArsR1 |
ArsR2 |
||
|---|---|---|---|---|
| Tm (°C)a | Mean ΔHm (kJ · mol−1) ± SD | Tm (°C)a | Mean ΔHm (kJ · mol−1) ± SD | |
| Ligand-free | 61.5 | 280 ± 2 | 71.1 | 265 ± 10 |
| 0.2 mM arsenite | 67.5/73.4b | 239 ± 10c | 77.8/87.4b | 274 ± 6c |
| 0.2 mM arsenate | 61.2 | 282 ± 9 | 71.5 | 266 ± 6 |
| 1.0 mM inorganic phosphate | 61.6 | 277 ± 13 | 71.0 | 251 ± 5 |
Internal calibration of the DSC equipment gives an error of ±0.10°C in Tm values.
The protein unfolded in two events, and both Tm values are provided.
Provided is the sum of both events.
The corresponding data are shown in Fig. 4. ΔHm, molar enthalpy change.
ArsR paralogues bind arsenite with similar affinities.
In vivo, the ArsR transcriptional regulators mediate responses to arsenate and arsenite (12, 36). Since both compounds can be interconverted in the cell, it is unclear whether both compounds or only one of them is recognized by the regulators. Ligand binding typically causes increases in the thermal stability of proteins and, consequently, the Tm value (37). To study ligand binding at ArsR1 and ArsR2, DSC studies were repeated in the presence of saturating concentrations of arsenate and arsenite. Due to the structural similarities between inorganic phosphate and arsenite/arsenate, it was observed previously that these ligands are recognized by the same receptor protein (38–40). To assess this issue, DSC analyses were also carried out in the presence of inorganic phosphate. The corresponding thermograms (Fig. 4) showed an increase in the Tm exclusively in the presence of arsenite. Apart from the increase in the Tm (Table 3), protein folding occurred in two sequential events. Importantly, the Tm values of the major unfolding event in the presence of arsenite were ∼12°C and 16°C higher than that of the ligand-free protein. In marked contrast, the thermograms (Fig. 4) and the derived unfolding parameters in the presence of arsenate and inorganic phosphate (Table 3) are almost identical to those of the apoproteins. These data show that arsenite but not arsenate or inorganic phosphate binds to ArsR1 and ArsR2.
FIG 4.
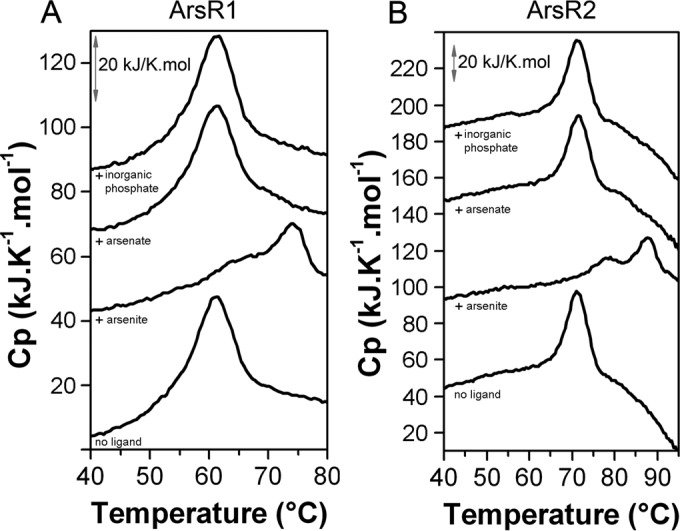
Studies of the stability of both ArsR paralogues. Shown are data from differential scanning calorimetry experiments with ArsR1 (A) and ArsR2 (B) in the absence and presence of arsenite, arsenate, and inorganic phosphate. Ligands were added at a concentration of 200 μM, and the protein was added at a concentration of 20 μM. The derived thermodynamic parameters are given in Table 3. For clarity, traces were moved arbitrarily on the y axis.
To determine the affinity of both regulators for arsenite, we conducted isothermal titration calorimetry experiments (41). However, titrations of both paralogues with arsenite, conducted at several analysis temperatures, revealed only minor binding heat changes, which did not permit the calculation of binding parameters. We therefore resorted to intrinsic tryptophan fluorescence spectroscopy measurements (42). The inspection of the ArsR2 homology model revealed the presence of a Trp residue (W79) in the immediate vicinity of cysteines 43 and 45, which are involved in ligand binding (see Fig. S1 in the supplemental material). Following excitation at 290 nm, the emission spectra showed maxima at 336 nm (Fig. 5). The addition of different concentrations of arsenite caused fluorescence quenching. The plot of corrected fluorescence at the maximum of emission as a function of the arsenite concentration is shown in Fig. 5C. Data fitting resulted in KD (equilibrium dissociation constant) values of ∼30 μM for both paralogues (Table 4).
FIG 5.
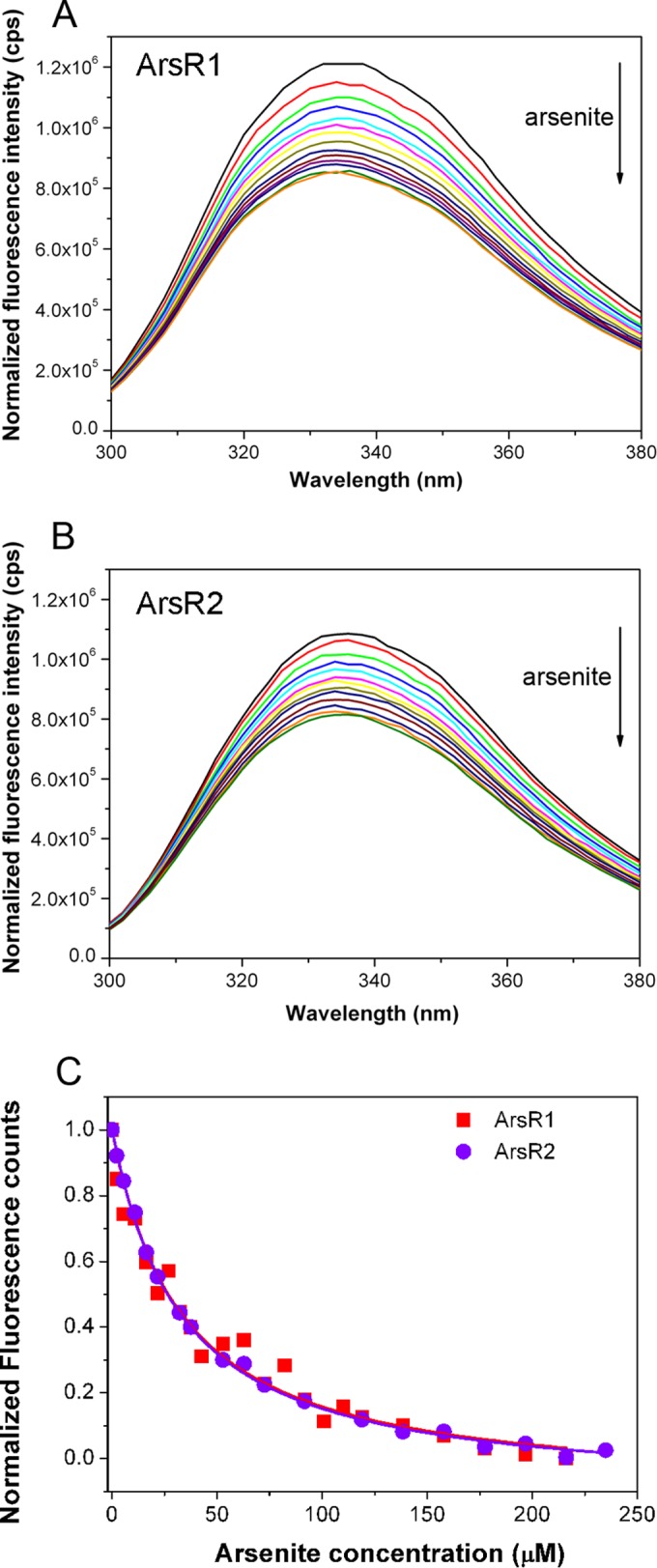
Studies of binding of arsenite to ArsR1 and ArsR2. (A and B) Intrinsic tryptophan fluorescence spectra with 5 μM ArsR1 (A) and ArsR2 (B) in the absence (black line) and presence (colored lines) of increasing concentrations of arsenite (up to 230 μM). (C) Decrease in maximum fluorescence intensity as a function of the arsenite concentration.
TABLE 4.
Interactions of different metal salts with ArsR1 and ArsR2a
| Ligand | Mean KD (μM) ± SD |
|
|---|---|---|
| ArsR1 | ArsR2 | |
| Arsenate | No binding | No binding |
| Arsenite | 30.8 ± 3 | 29.7 ± 1 |
| Antimony | 28.1 ± 1 | 21.8 ± 2 |
| Bismuth | 9.2 ± 1 | 6.3 ± 1 |
Shown are dissociation constants derived from fluorescence titrations of proteins with different ligands (see Materials and Methods for further information). Data are means and standard deviations from three independent experiments.
Evidence for ArsR1 and ArsR2 cross talk.
The DNA segments harboring the ARS1 and ARS2 operons are highly homologous. However, the sequences of both target promoters, PARS1 and PARS2, show significant sequence divergence (see Fig. S4 in the supplemental material). To assess whether ArsR1 and ArsR2 bind to both promoters or exclusively to their cognate promoter, we conducted EMSAs using purified ArsR1 and ArsR2 as well as DNA fragments covering both promoter regions. Although a wide variety of experimental conditions were tested, satisfactory EMSA results could be obtained only for the promoter PARS1 (Fig. 6). Interestingly, both proteins bound to the PARS1 fragment. A single protein-DNA complex with an estimated KD in the submicromolar range could be detected for ArsR2. In contrast, ArsR1 appears to form multiple DNA complexes. No binding to control DNA was observed. Unfortunately, when experiments were repeated in the presence of arsenite and arsenate, a smear and no distinct bands appeared, which made interpretation impossible. The fact that both paralogues bind to PARS1 suggests the existence of cross talk between both systems.
FIG 6.
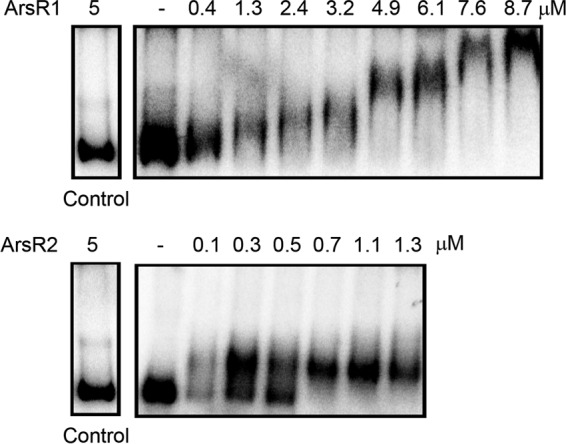
Binding of ArsR1 and ArsR2 to the promoter PARS1. Shown are data from electrophoretic mobility shift assays with both ArsR paralogues using a 476-bp DNA fragment spanning the PARS1 promoter. The control is a DNA fragment amplified from the genome of Serratia plymuthica. The corresponding primers for the amplification of DNA probes are given in Table 2.
Construction and validation of KT2440-based arsenic biosensors.
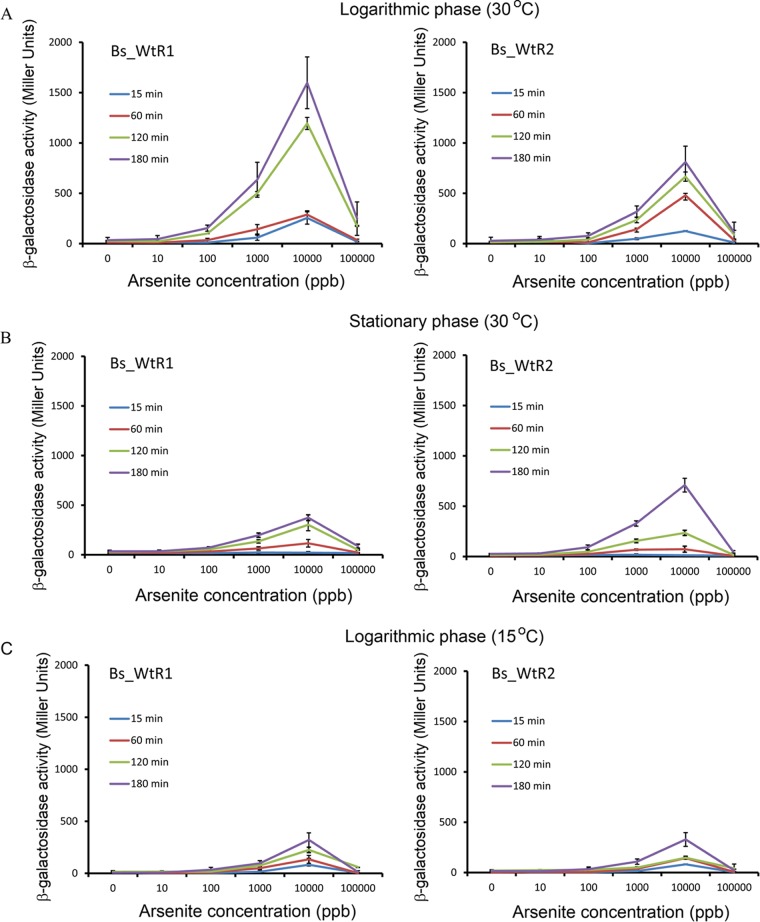
To generate whole-cell biosensors, the arsR1 and arsR2 sequences and their respective promoter regions were cloned into pMP220 to generate the transcriptional fusions PARS1_ARS1_lacZ and PARS2_ARS2_lacZ, respectively. The resulting plasmids were inserted into P. putida KT2440 to give rise to the biosensors Bs_WtR1 and Bs_WtR2, respectively. Beta-galactosidase measurements were made to assess biosensor function. Initial experiments were conducted to determine the influence of bacterial growth phase, temperature, and time of response on the performance of the different biosensors. As shown in Fig. 7A and B, cells induced in the logarithmic phase gave stronger responses than those induced in the stationary phase. Paez-Espino et al. reported that at 15°C, products of the ARS2 operon afforded much higher resistance to arsenite than did those of the ARS1 operon (15). To assess the influence of temperature, cultures were grown at 30°C to early logarithmic phase, arsenite was added, and cultures were then continued at either 30°C or 15°C. Figure 7 shows that the reduction in the temperature translated into a reduced response for both sensors. Experiments at 30°C showed that the biosensor Bs_WtR1 gave stronger responses than did the Bs_WtR2 counterpart (Fig. 7A). Responses were detected as early as 15 min after arsenite addition, whereas maximal readings were obtained following a 3-h incubation. The onset of the response was at ∼10 ppb, and the sensitivity of the biosensors is described in further detail below. Maximal responses were obtained at a concentration of 10,000 ppb, whereas at a concentration of 100,000 ppb, a very significant drop in activity was measured, which was due to toxicity.
FIG 7.
Assessment of the influences of growth phase, growth temperature, and time of arsenite exposure on biosensor performance. Cells were pregrown at 30°C, and arsenite was added to cells in either the logarithmic phase (OD660 of ∼0.3 to 0.4) (A and C) or stationary phase (cultures grown overnight) (B). Cultures were then incubated at either 30°C (A and B) or 15°C (C). In all cases, measurements were made 15, 60, 120, or 180 min after arsenite addition. Shown are means and standard deviations from three independent experiments.
ArsB mutant-based biosensors show higher sensitivity and higher magnitudes of response than wt-based sensors.
It has been shown that a mutant of the arsenic efflux pumps ArsB1 and ArsB2 (Fig. 1) is more sensitive to arsenic than the wt (14), which is due to cellular arsenite accumulation. To verify whether this accumulation translates into higher biosensor sensitivity, two additional biosensors were constructed, in which both transcriptional fusions were inserted into the ArsB1/ArsB2 double mutant to give rise to the biosensors Bs_MutR1 and Bs_MutR2. The dose-response curves for all four biosensors with arsenate and arsenite are shown in Fig. 8A and B. We noted that mutant-based biosensors caused a higher magnitude of response than the biosensors using the wt strain. Response patterns for arsenate and arsenite differed significantly. Whereas a strong reduction in the response was noted at 100,000 ppb arsenite (due to toxicity), maximal responses for arsenate were observed at an analyte concentration of 100,000 ppb.
FIG 8.
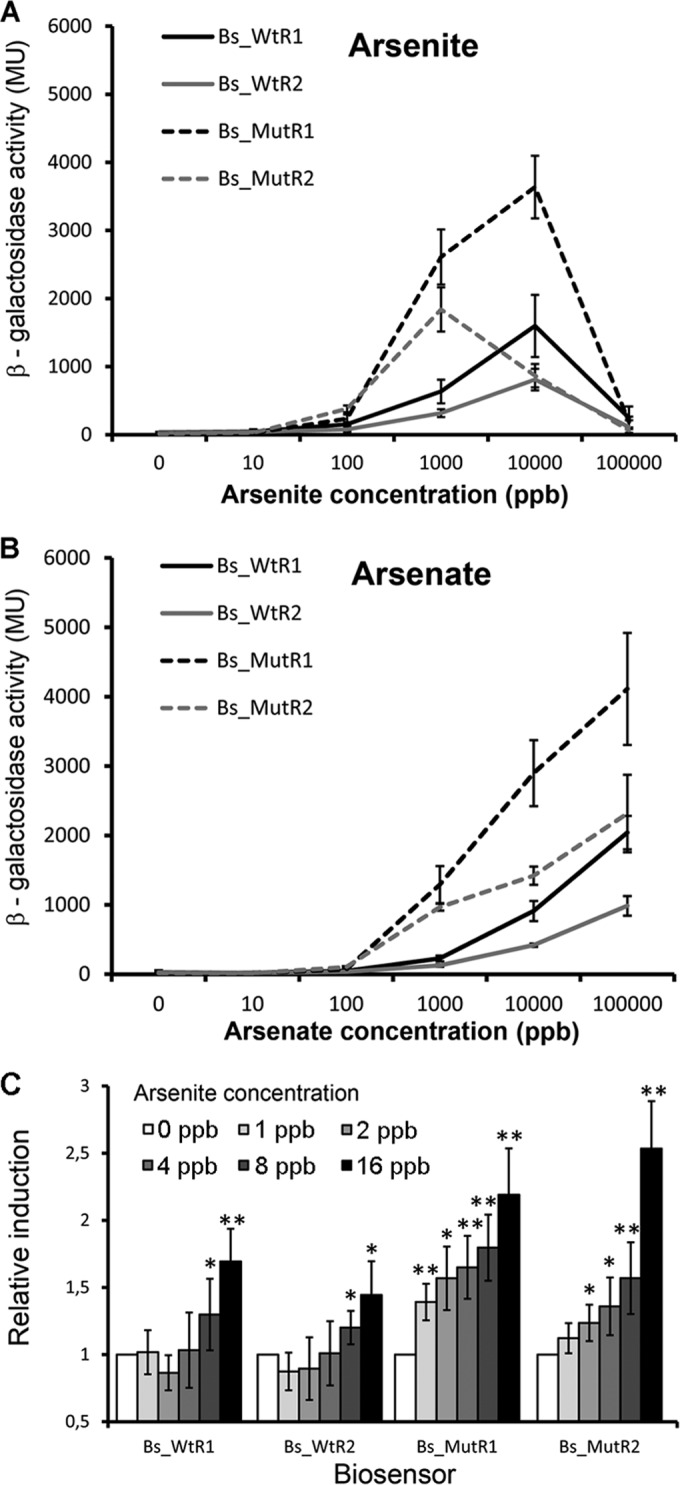
Biosensor responses to different arsenite and arsenate concentrations. Cells were grown at 30°C until the OD660 reached 0.3 to 0.4, prior to the addition of either arsenite or arsenate. Measurements were made 3 h after induction. (A and B) Dose-response curves for arsenite and arsenate. (C) Determination of the onset of the response. Shown are means and standard deviations from three independent experiments. *, P value of <0.05; **, P value of <0.01 (with respect to arsenite-free samples, as determined by Student's t test). MU, Miller units.
A characteristic feature of any analytical tool is its sensitivity, or the lowest analyte concentration at which significant readings are obtained. To assess the sensitivity of the four biosensors, their responses in the concentration window between 1 and 16 ppb arsenite were determined. Analysis of data (Fig. 8C) revealed that the first significant measurements were obtained at 8 ppb for the wt-based sensors Bs_WtR1 and Bs_WtR2. In contrast, the first statistically significant measurements were obtained at 1 and 2 ppb for both ArsB mutant-based biosensors BS_MutR1 and Bs_MutR2, respectively. These data thus demonstrate that the mutation of both arsenite efflux pumps translated into not only higher magnitudes of response but also significant increases in biosensor sensitivity. These results differ from those from analogous experiments using E. coli, where mutation of the arsenite efflux pump did not cause an increase in biosensor sensitivity (43).
Biosensors also respond to trivalent salts of antimony.
In vivo gene expression studies have shown that next to arsenate and arsenite, E. coli ArsR also responds to trivalent salts of antimony and bismuth (24). To verify whether this is also the case for the P. putida paralogues, we conducted a series of gene expression studies. Data show that ArsR1 and ArsR2 mediate significant responses to antimony (see Fig. S5 in the supplemental material), which were particularly pronounced for ArsR1. Interestingly, the responses of wt- and ArsB mutant-based biosensors were similar, strongly suggesting that antimony is not an ArsB pump substrate. Responses observed for bismuth (see Fig. S5 in the supplemental material) were negligible compared to those measured for arsenate, arsenite, and antimony. To verify whether antimony and bismuth bind to the purified ArsR paralogues, we conducted fluorescence titration assays similar to those with arsenite, as shown in Fig. 5. Antimony bound to both paralogues with affinities comparable to those for arsenite (Table 4). Surprisingly, the tightest-binding ligand was bismuth (KD values of 9 and 6 μM for ArsR1 and ArsR2, respectively), which contrasted with the extremely weak response in vivo. This may be due to limitations in bismuth uptake.
Phosphate does not interfere with the performance of arsenite biosensors.
A major limitation of other arsenic biosensors is interference by inorganic phosphate (38–40). To assess any possible interference of inorganic phosphate in the functioning of the arsenite biosensors, the responses of Bs-WtR1 and Bs-WtR2 to 1,000 ppb arsenite in the absence and presence of up to 100 mM inorganic phosphate were monitored. As shown in Fig. S5 in the supplemental material, the presence of inorganic phosphate did not cause any significant interference with the biosensors. These data agree with the above-reported absence of binding of inorganic phosphate to ArsR1 and ArsR2 in vitro (Fig. 4).
Use of the arsenic biosensors for analysis of environmental samples.
To assess the suitability of the developed biosensors to analyze environmental samples, we carried out experiments with water recovered from a mine. This sample was submitted to inductively coupled plasma-optical emission spectroscopy to precisely quantify the arsenic content. These measurements resulted in an arsenic concentration of 596 ± 32 ppb. Data from the analysis of 1:10-diluted samples using the four different biosensors are shown in Fig. S6 in the supplemental material, demonstrating that all biosensors were able to detect arsenic. The strongest signals were obtained with the two ArsB mutant-based sensors, confirming the above-mentioned conclusions that these sensors possess higher sensitivities. These measurements illustrate the usefulness of the developed biosensors to detect low concentrations of arsenic in environmental samples.
DISCUSSION
As mentioned in the introduction, the duplication of ars operons appears to be a more general strategy of bacteria to enhance arsenic resistance, and the double ars operons in P. putida are a model system to study the coexisting ars resistance systems. A number of in vivo studies of this model have been reported (14, 15), which contrasts with the absence of any biochemical or biophysical data on these proteins.
An unusual feature of ArsR regulators is that there are at least three different modes by which arsenite is recognized. In all three cases, a cysteine triad binds arsenite, but the locations of these residues in the structure differ for proteins of E. coli (27), Corynebacterium glutamicum (28), and Acidithiobacillus ferrooxidans (29) (see Fig. S1 in the supplemental material). The sequence alignment shows that the P. putida proteins employ yet another binding mechanism, since none of the identified cysteine triads are conserved. The ArsR2 sequence motif C-X-C-(X)3-C may correspond to a version of the E. coli binding motif of C-X-C-(X)2-C. ArsR1 has a C-X-C motif, but the positions of the two other cysteine residues are not conserved. We show here that despite this difference, both paralogues bind arsenite with similar affinities of ∼30 μM. These similarities in affinities may at least partially account for the similarities in the sensitivities of ArsR1- and ArsR2-based biosensors (Fig. 8C).
To our knowledge, these are the first direct binding studies in vitro, although others previously reported the binding of arsenite to the regulators based on the capacity of arsenite to trigger protein release from DNA. Using such an approach, an apparent KD of 12 μM was reported previously for arsenite binding to ArsR of Acidithiobacillus ferrooxidans (29), and a value of 150 μM was reported previously for ArsR of Corynebacterium glutamicum (28).
E. coli ArsR, the primary model protein, shows only modest sequence similarity to ArsR1 (40% identity) and ArsR2 (42% identity). However, there are clear parallels in their ligand susceptibilities, suggesting that these are features common to the family of ArsR proteins. (i) Data from in vivo experiments with P. putida and E. coli ArsR proteins show strong comparable responses to arsenite, arsenate, and antimony (Fig. 8) (24). In both cases, very minor responses were observed for bismuth. (ii) In vitro and in vivo data show that inorganic phosphate does not act on ArsR (Fig. 4; see also Fig. S5 in the supplemental material) (24). (iii) Data show that arsenite, antimony, and bismuth, but not arsenate, bind to the ArsR proteins (Fig. 4 and 5) (24). An unexpected finding was that bismuth, which caused only weak responses in vivo, bound to ArsR1 and ArsR2 with significant affinities (Table 4). A similar discrepancy between in vitro binding and in vivo efficiency has been observed for binding of antimony to ArsR of C. glutamicum, for which a KD of 10 μM was determined, a value lower than the corresponding one for arsenite (KD = 150 μM). As suggested by the authors of that study, these differences may be caused by a relatively poor uptake of antimony or bismuth compared with that of arsenite.
Our demonstration that ArsR1 and ArsR2 are dimeric coincides with similar findings for homologous proteins in E. coli and Acidithiobacillus ferrooxidans (29, 44), suggesting that the dimeric state may also be a feature common to the ArsR protein family. We have shown here that effector binding did not cause any alterations to the oligomeric state of ArsR, which is an issue that has so far not been investigated.
The construction of a significant number of whole-cell arsenic biosensors has been reported in the literature (7, 30), which hence raises the question of why further sensors were constructed. However, most of the biosensors use E. coli as a host, whereas no biosensors using P. putida strains have been reported. The choice of the latter organism was based on its stunning resistance to a wide range of natural and anthropogenic stresses such as metals, organic solvents, and pollutants and its ease in adapting to changing physicochemical parameters of the environment such as water and oxidative stress or pH changes (21, 45–48). Considering that in situ arsenite detection will involve the analysis of complex samples of different natures, the use of a robust bacterium is likely to confer robustness to the analysis.
The World Health Organization (WHO) has fixed the maximum level of arsenic in drinking water to 10 ppb, and the Food and Agriculture Organization (FAO) has set a maximum arsenic level of 100 ppb in irrigation water (49). As shown in Fig. 8C, the AsR1- and ArsR2-based biosensors can be used to assess the quality of drinking and irrigation water. Both biosensors based on the native P. putida strain had a sensitivity of 8 ppb. In contrast, the biosensors based on the double mutation of the arsenic efflux pumps were effective in detecting 1 and 2 ppb. These data indicate that the intracellular arsenic level is higher in the double mutant than in the wt strain, which is due to the perturbation of arsenite efflux.
A biosensor based on a mutation of the ars operon in E. coli has also been reported (43). However, in contrast to our findings, the elimination of the arsenic efflux pump did not translate into an improvement of biosensor performance. The sensitivity of our biosensors is superior to those of the majority of whole-cell arsenic sensors reported previously (30).
A major hurdle for the successful construction of arsenic biosensors is interference with inorganic phosphate (30), which has been observed in a number of cases (50–52). We show here that both ArsR paralogues do not bind inorganic phosphate (Fig. 4) and that inorganic phosphate concentrations as high as 100 mM do not interfere with arsenite sensing (see Fig. S5 in the supplemental material). Another undesirable property of biosensors is their elevated background signal (43, 51, 53–55). The biosensors described here are characterized by very low background signals, and β-galactosidase activity of ∼10 Miller units was observed in the absence of a ligand, translating into very favorable signal-to-noise ratios for arsenic measurements. The measurements conducted were based on an analyte incubation time of 3 h, which is comparable to those for most other sensors described previously (30).
These analytical parameters combined with the robustness of the host strain make the constructed biosensors convenient and robust analytical tools for arsenite detection in diverse and complex samples. The enhancement of biosensor sensitivity and magnitude of response by deleting the efflux pump of the cognate analyte may be an interesting strategy for the construction of biosensors for other analytes.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Abdelali Daddaoua for valuable advice in setting up the EMSAs. We thank the Biophysics and Molecular Biotechnology group from the Department of Physical Chemistry (University of Granada) for permitting access to the DSC and CD instruments.
We acknowledge financial support from FEDER funds and the Fondo Social Europeo through grants from the Spanish Ministry for Economy and Competitiveness (RECUPERA grant 20134R057 to T.K. and grant BIO2013-42297 to T.K.) from the Junta de Andalucía (grant CVI-7335 to T.K.).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00606-16.
REFERENCES
- 1.Slyemi D, Bonnefoy V. 2012. How prokaryotes deal with arsenic. Environ Microbiol Rep 4:571–586. doi: 10.1111/j.1758-2229.2011.00300.x. [DOI] [PubMed] [Google Scholar]
- 2.McCarty KM, Hanh HT, Kim KW. 2011. Arsenic geochemistry and human health in South East Asia. Rev Environ Health 26:71–78. doi: 10.1515/reveh.2011.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argos M, Ahsan H, Graziano JH. 2012. Arsenic and human health: epidemiologic progress and public health implications. Rev Environ Health 27:191–195. doi: 10.1515/reveh-2012-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh R, Singh S, Parihar P, Singh VP, Prasad SM. 2015. Arsenic contamination, consequences and remediation techniques: a review. Ecotoxicol Environ Saf 112:247–270. doi: 10.1016/j.ecoenv.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed A, Edward A, Burnham G. 2004. Health indicators for mothers and children in rural Herat Province, Afghanistan. Prehosp Disaster Med 19:221–225. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee A, Sengupta MK, Hossain MA, Ahamed S, Das B, Nayak B, Lodh D, Rahman MM, Chakraborti D. 2006. Arsenic contamination in groundwater: a global perspective with emphasis on the Asian scenario. J Health Popul Nutr 24:142–163. [PubMed] [Google Scholar]
- 7.Merulla D, Buffi N, Beggah S, Truffer F, Geiser M, Renaud P, van der Meer JR. 2013. Bioreporters and biosensors for arsenic detection. Biotechnological solutions for a world-wide pollution problem. Curr Opin Biotechnol 24:534–541. doi: 10.1016/j.copbio.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Ron EZ. 2007. Biosensing environmental pollution. Curr Opin Biotechnol 18:252–256. doi: 10.1016/j.copbio.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Dopson M, Baker-Austin C, Koppineedi PR, Bond PL. 2003. Growth in sulfidic mineral environments: metal resistance mechanisms in acidophilic micro-organisms. Microbiology 149:1959–1970. doi: 10.1099/mic.0.26296-0. [DOI] [PubMed] [Google Scholar]
- 10.Baker-Austin C, Dopson M, Wexler M, Sawers RG, Stemmler A, Rosen BP, Bond PL. 2007. Extreme arsenic resistance by the acidophilic archaeon ‘Ferroplasma acidarmanus’ Fer1. Extremophiles 11:425–434. doi: 10.1007/s00792-006-0052-z. [DOI] [PubMed] [Google Scholar]
- 11.San Francisco MJ, Hope CL, Owolabi JB, Tisa LS, Rosen BP. 1990. Identification of the metalloregulatory element of the plasmid-encoded arsenical resistance operon. Nucleic Acids Res 18:619–624. doi: 10.1093/nar/18.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paez-Espino D, Tamames J, de Lorenzo V, Canovas D. 2009. Microbial responses to environmental arsenic. Biometals 22:117–130. doi: 10.1007/s10534-008-9195-y. [DOI] [PubMed] [Google Scholar]
- 13.Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VA, Fouts DE, Gill SR, Pop M, Holmes M, Brinkac L, Beanan M, DeBoy RT, Daugherty S, Kolonay J, Madupu R, Nelson W, White O, Peterson J, Khouri H, Hance I, Lee PC, Holtzapple E, Scanlan D, Tran K, Moazzez A, Utterback T, Rizzo M, Lee K, Kosack D, Moestl D, Wedler H, Lauber J, Stjepandic D, Hoheisel J, Straetz M, Heim S, Kiewitz C, Eisen JA, Timmis KN, Dusterhoft A, Tummler B, Fraser CM. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol 4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez M, Udaondo Z, Niqui JL, Duque E, Ramos JL. 2014. Synergic role of the two ars operons in arsenic tolerance in Pseudomonas putida KT2440. Environ Microbiol Rep 6:483–489. doi: 10.1111/1758-2229.12167. [DOI] [PubMed] [Google Scholar]
- 15.Paez-Espino AD, Durante-Rodriguez G, de Lorenzo V. 2015. Functional coexistence of twin arsenic resistance systems in Pseudomonas putida KT2440. Environ Microbiol 17:229–238. doi: 10.1111/1462-2920.12464. [DOI] [PubMed] [Google Scholar]
- 16.Ordonez E, Letek M, Valbuena N, Gil JA, Mateos LM. 2005. Analysis of genes involved in arsenic resistance in Corynebacterium glutamicum ATCC 13032. Appl Environ Microbiol 71:6206–6215. doi: 10.1128/AEM.71.10.6206-6215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Branco R, Chung AP, Morais PV. 2008. Sequencing and expression of two arsenic resistance operons with different functions in the highly arsenic-resistant strain Ochrobactrum tritici SCII24T. BMC Microbiol 8:95. doi: 10.1186/1471-2180-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regenhardt D, Heuer H, Heim S, Fernandez DU, Strompl C, Moore ER, Timmis KN. 2002. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ Microbiol 4:912–915. doi: 10.1046/j.1462-2920.2002.00368.x. [DOI] [PubMed] [Google Scholar]
- 19.Espinosa-Urgel M, Kolter R, Ramos JL. 2002. Root colonization by Pseudomonas putida: love at first sight. Microbiology 148:341–343. doi: 10.1099/00221287-148-2-341. [DOI] [PubMed] [Google Scholar]
- 20.Jimenez JI, Canales A, Jimenez-Barbero J, Ginalski K, Rychlewski L, Garcia JL, Diaz E. 2008. Deciphering the genetic determinants for aerobic nicotinic acid degradation: the nic cluster from Pseudomonas putida KT2440. Proc Natl Acad Sci U S A 105:11329–11334. doi: 10.1073/pnas.0802273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canovas D, Cases I, de Lorenzo V. 2003. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ Microbiol 5:1242–1256. doi: 10.1111/j.1462-2920.2003.00463.x. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Bhattacharjee H, Rosen BP. 2015. ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol Microbiol 96:1042–1052. doi: 10.1111/mmi.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osman D, Cavet JS. 2010. Bacterial metal-sensing proteins exemplified by ArsR-SmtB family repressors. Nat Prod Rep 27:668–680. doi: 10.1039/b906682a. [DOI] [PubMed] [Google Scholar]
- 24.Wu J, Rosen BP. 1993. Metalloregulated expression of the ars operon. J Biol Chem 268:52–58. [PubMed] [Google Scholar]
- 25.Oden KL, Gladysheva TB, Rosen BP. 1994. Arsenate reduction mediated by the plasmid-encoded ArsC protein is coupled to glutathione. Mol Microbiol 12:301–306. doi: 10.1111/j.1365-2958.1994.tb01018.x. [DOI] [PubMed] [Google Scholar]
- 26.Busenlehner LS, Pennella MA, Giedroc DP. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol Rev 27:131–143. doi: 10.1016/S0168-6445(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 27.Shi W, Wu J, Rosen BP. 1994. Identification of a putative metal binding site in a new family of metalloregulatory proteins. J Biol Chem 269:19826–19829. [PubMed] [Google Scholar]
- 28.Ordonez E, Thiyagarajan S, Cook JD, Stemmler TL, Gil JA, Mateos LM, Rosen BP. 2008. Evolution of metal(loid) binding sites in transcriptional regulators. J Biol Chem 283:25706–25714. doi: 10.1074/jbc.M803209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin J, Fu HL, Ye J, Bencze KZ, Stemmler TL, Rawlings DE, Rosen BP. 2007. Convergent evolution of a new arsenic binding site in the ArsR/SmtB family of metalloregulators. J Biol Chem 282:34346–34355. doi: 10.1074/jbc.M706565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaur H, Kumar R, Babu JN, Mittal S. 2015. Advances in arsenic biosensor development—a comprehensive review. Biosens Bioelectron 63:533–545. doi: 10.1016/j.bios.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 32.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607. In Walker JM. (ed), The proteomics protocols handbook. Humana Press, Totowa, NJ. [Google Scholar]
- 33.Bohm G, Muhr R, Jaenicke R. 1992. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng 5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- 34.van de Weert M. 2010. Fluorescence quenching to study protein-ligand binding: common errors. J Fluoresc 20:625–629. doi: 10.1007/s10895-009-0572-x. [DOI] [PubMed] [Google Scholar]
- 35.Miller JE. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 36.Xu C, Shi W, Rosen BP. 1996. The chromosomal arsR gene of Escherichia coli encodes a trans-acting metalloregulatory protein. J Biol Chem 271:2427–2432. doi: 10.1074/jbc.271.5.2427. [DOI] [PubMed] [Google Scholar]
- 37.Chiu MH, Prenner EJ. 2011. Differential scanning calorimetry: an invaluable tool for a detailed thermodynamic characterization of macromolecules and their interactions. J Pharm Bioallied Sci 3:39–59. doi: 10.4103/0975-7406.76463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hughes MF. 2002. Arsenic toxicity and potential mechanisms of action. Toxicol Lett 133:1–16. doi: 10.1016/S0378-4274(02)00084-X. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez D, Richez M, Bergonzi C, Chabriere E, Elias M. 2014. Crystal structure of the phosphate-binding protein (PBP-1) of an ABC-type phosphate transporter from Clostridium perfringens. Sci Rep 4:6636. doi: 10.1038/srep06636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beene LC, Halluer J, Yoshinaga M, Hamdi M, Liu Z. 2011. Pentavalent arsenate transport by zebrafish phosphate transporter NaPi-IIb1. Zebrafish 8:125–131. doi: 10.1089/zeb.2011.0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krell T. 2008. Microcalorimetry: a response to challenges in modern biotechnology. Microb Biotechnol 1:126–136. doi: 10.1111/j.1751-7915.2007.00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eftink MR. 1997. Fluorescence methods for studying equilibrium macromolecule-ligand interactions. Methods Enzymol 278:221–257. doi: 10.1016/S0076-6879(97)78013-3. [DOI] [PubMed] [Google Scholar]
- 43.Tauriainen S, Virta M, Chang W, Karp M. 1999. Measurement of firefly luciferase reporter gene activity from cells and lysates using Escherichia coli arsenite and mercury sensors. Anal Biochem 272:191–198. doi: 10.1006/abio.1999.4193. [DOI] [PubMed] [Google Scholar]
- 44.Xu C, Rosen BP. 1997. Dimerization is essential for DNA binding and repression by the ArsR metalloregulatory protein of Escherichia coli. J Biol Chem 272:15734–15738. doi: 10.1074/jbc.272.25.15734. [DOI] [PubMed] [Google Scholar]
- 45.Nikel PI, de Lorenzo V. 2014. Robustness of Pseudomonas putida KT2440 as a host for ethanol biosynthesis. N Biotechnol 31:562–571. doi: 10.1016/j.nbt.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Dominguez-Cuevas P, Gonzalez-Pastor JE, Marques S, Ramos JL, de Lorenzo V. 2006. Transcriptional tradeoff between metabolic and stress-response programs in Pseudomonas putida KT2440 cells exposed to toluene. J Biol Chem 281:11981–11991. doi: 10.1074/jbc.M509848200. [DOI] [PubMed] [Google Scholar]
- 47.Chavarria M, Nikel PI, Perez-Pantoja D, de Lorenzo V. 2013. The Entner-Doudoroff pathway empowers Pseudomonas putida KT2440 with a high tolerance to oxidative stress. Environ Microbiol 15:1772–1785. doi: 10.1111/1462-2920.12069. [DOI] [PubMed] [Google Scholar]
- 48.Timmis KN. 2002. Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ Microbiol 4:779–781. doi: 10.1046/j.1462-2920.2002.00365.x. [DOI] [PubMed] [Google Scholar]
- 49.Rahaman S, Sinha AC, Pati R, Mukhopadhyay D. 2013. Arsenic contamination: a potential hazard to the affected areas of West Bengal, India. Environ Geochem Health 35:119–132. doi: 10.1007/s10653-012-9460-4. [DOI] [PubMed] [Google Scholar]
- 50.Cai J, DuBow MS. 1997. Use of a luminescent bacterial biosensor for biomonitoring and characterization of arsenic toxicity of chromated copper arsenate (CCA). Biodegradation 8:105–111. doi: 10.1023/A:1008281028594. [DOI] [PubMed] [Google Scholar]
- 51.Liao VH, Ou KL. 2005. Development and testing of a green fluorescent protein-based bacterial biosensor for measuring bioavailable arsenic in contaminated groundwater samples. Environ Toxicol Chem 24:1624–1631. doi: 10.1897/04-500R.1. [DOI] [PubMed] [Google Scholar]
- 52.Ivanina AV, Shuvaeva OV. 2009. Use of a bacterial biosensor system for determining arsenic in natural waters. J Anal Chem 64:310–315. doi: 10.1134/S1061934809030186. [DOI] [Google Scholar]
- 53.Scott DL, Ramanathan S, Shi W, Rosen BP, Daunert S. 1997. Genetically engineered bacteria: electrochemical sensing systems for antimonite and arsenite. Anal Chem 69:16–20. doi: 10.1021/ac960788x. [DOI] [PubMed] [Google Scholar]
- 54.Roberto FF, Barnes JM, Bruhn DF. 2002. Evaluation of a GFP reporter gene construct for environmental arsenic detection. Talanta 58:181–188. doi: 10.1016/S0039-9140(02)00266-7. [DOI] [PubMed] [Google Scholar]
- 55.Tani C, Inoue K, Tani Y, Harun-ur-Rashid M, Azuma N, Ueda S, Yoshida K, Maeda I. 2009. Sensitive fluorescent microplate bioassay using recombinant Escherichia coli with multiple promoter-reporter units in tandem for detection of arsenic. J Biosci Bioeng 108:414–420. doi: 10.1016/j.jbiosc.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 56.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res 32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bagdasarian M, Lurz R, Ruckert B, Franklin FC, Bagdasarian MM, Frey J, Timmis KN. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237–247. [DOI] [PubMed] [Google Scholar]
- 58.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res 17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi SH, Couloux A, Lee SW, Yoon SH, Cattolico L, Hur CG, Park HS, Segurens B, Kim SC, Oh TK, Lenski RE, Studier FW, Daegelen P, Kim JF. 2009. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J Mol Biol 394:644–652. doi: 10.1016/j.jmb.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 60.Spaink HP, Okker RJ, Wijffelman CA, Pees E, Lugtenberg BJ. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol 9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.