ABSTRACT
Intestinal homeostasis mechanisms must protect the host intestinal tissue from endogenous lipopolysaccharides (LPSs) produced by the intestinal microbiota. In this report, we demonstrate that murine intestinal fecal lipids effectively block Toll-like receptor 4 (TLR4) responses to naturally occurring Bacteroidetes sp. LPS. Cardiolipin (CL) represents a significant proportion of the total intestinal and fecal lipids and, furthermore, potently antagonizes TLR4 activation by reducing LPS binding at the lipopolysaccharide binding protein (LBP), CD14, and MD-2 steps of the TLR4 signaling pathway. It is further demonstrated that intestinal lipids and CL are less effective at neutralizing more potent Enterobacteriaceae-type LPS, which is enriched in feces obtained from mice with dextran sodium sulfate (DSS)-treated inflammatory bowel disease. The selective inhibition of naturally occurring LPS structures by intestinal lipids may represent a novel homeostasis mechanism that blocks LPS activation in response to symbiotic but not dysbiotic microbial communities.
IMPORTANCE The guts of animals harbor a variety of Gram-negative bacteria associated with both states of intestinal health and states of disease. Environmental factors, such as dietary habits, can drive the microbial composition of the host animal's intestinal bacterial community toward a more pathogenic state. Both beneficial and harmful Gram-negative bacteria are capable of eliciting potentially damaging inflammatory responses from the host intestinal tissues via a lipopolysaccharide (LPS)-dependent pathway. Physical mucosal barriers and antibodies produced by the intestinal immune system protect against the undesired inflammatory effects of LPS, although it is unknown why some bacteria are more effective at overcoming the protective barriers than others. This report describes the discovery of a lipid-type protective barrier in the intestine that reduces the deleterious effects of LPSs from beneficial bacteria but is less effective in dampening the inflammatory effects of LPSs from harmful bacteria, providing a novel mechanistic insight into inflammatory intestinal disorders.
INTRODUCTION
The intestinal tract is constantly exposed to the potentially harmful effects of lipopolysaccharide (LPS), commonly referred to as endotoxin. Both internal sources, such as the intestinal microbiome, and external sources, such as food, provide a constant potential for endotoxin-mediated local or systemic injury. Several different innate and adaptive protective mechanisms present in the intestine facilitate a homeostatic relationship with the resident commensal microbiota and buffer food intake such that LPS does not impair normal intestinal tissue functions (1–5).
Studies have shown that a major mechanism by which the intestine limits exposure to the potentially harmful effects of LPS is by preventing intestinal LPS-epithelial cell interactions through a physiochemical barrier consisting of a family of glycoproteins referred to as mucins. Mucins provide an effective barricade restricting the ability of both commensal and pathogenic microbiota and their respective bacterial components to gain access to the intestinal epithelium, thereby limiting the magnitude of potential LPS interactions with intestinal epithelial cells (6). In addition, LPS-modifying enzymes produced in intestinal tissue can render the LPS less toxic, reducing the potential for inappropriate inflammation (7, 8). Furthermore, host adaptive immune responses include the production of IgA by plasma cells present in the intestinal mucosa, and TH1- and TH17-mediated immunity (9) provides an additional significant level of protection of the intestinal epithelium from the resident microbiota. Other studies have presented evidence that selective Toll-like receptor (TLR) expression and epithelial cell polarization may hinder intestinal epithelial cell exposure to LPS, preventing destructive inflammation (10).
However, the mechanisms by which the host protects intestinal tissues from LPS exposure are impaired in certain forms of inflammatory bowel disease (IBD). For example, anti-TLR4 antibodies have been shown to reduce intestinal damage in dextran sodium sulfate (DSS)-treated mice (11), consistent with a significant increase in water-soluble pattern recognition receptor ligands in the feces of DSS-treated mice, such as the LPS receptor TLR4 (12). Similarly, in humans, individuals with Crohn's disease have increased circulating levels of MD-2, a key LPS coreceptor for TLR4 which results in significantly increased TLR4-dependent intestinal epithelial cell responses in vitro (13). In addition, evidence suggesting that LPS exposure is involved in acute-phase responses in both Crohn's disease and experimental murine colitis has been presented (14). Furthermore, a high-fat diet also results in intestinal epithelial cell exposure to elevated levels of LPS, altering the permeability of intestinal epithelial cells by a TLR4-dependent mechanism, contributing to metabolic syndrome (15–17).
These findings of LPS-associated intestinal abnormalities stand in stark contrast to the findings of studies which have shown that orally administered LPS interacts with intestinal tissue and contributes to the maintenance of a healthy homeostatic relationship (18–20). One rationale for this apparent contradiction is the observation that intestinal dysbiotic communities alter normal intestinal homeostatic mechanisms. A common theme found in intestinal dysbiotic communities is a significant increase in the proportion of members of the family Enterobacteriaceae (15, 21–23). This change in microbial community composition significantly alters the amount and type of LPS to which intestinal tissue is exposed. In fact, it was recently demonstrated in a mouse model system that Rag1−/− mice harboring a high proportion of isolates of the family Enterobacteriaceae developed colitis (24). However, little is known concerning how the host may regulate LPS exposure to intestinal tissue in order to accomplish intestinal homeostasis and how LPS exposure is detrimental and associated with destructive inflammatory disease.
In this report, a novel mechanism by which the host selectively suppresses LPS activation in the intestinal tract is described. It was found that cardiolipin (CL), which is normally present in feces as a result of intestinal epithelial cell turnover, blocked TLR4 responses to LPS. Furthermore, LPS isolated from mouse feces containing normal intestinal microbiota or from Bacteroides thetaiotaomicron, a human intestinal symbiont, was more effectively neutralized than LPS obtained from the feces of DSS-treated mice or Escherichia coli, a bacterium found to be present in increased proportions in patients with IBD. These observations are consistent with the notion that alterations in the LPS composition arising from modulations in the Gram-negative microbial community can overwhelm the lipid antagonist barrier, contributing to the progression of intestinal dysbiosis-related diseases.
MATERIALS AND METHODS
Mouse strains and animal maintenance.
All animal experiments described in this report were performed after review and approval by both the Queen Mary University of London and the University of Washington Institutional Animal Care and Use Committees, which exceed the guidelines required by the NIH for funding. All animal experiments were conducted in accredited facilities in accordance with the United Kingdom Animals (Scientific Procedures) Act 1986 (Home Office license number 7006844). Germfree (GF) C3H/He mice were maintained in sterile isolators at the Royal Veterinary College, University of London. Germfree status was confirmed by aerobic and anaerobic culture of oral swabs and fecal pellets on nonselective media and by PCR using universal 16S rRNA-specific primers. Specific-pathogen-free (SPF) C3H/He mice were maintained in individually ventilated cages at the biological service unit of Queen Mary University of London.
Isolation of bacterial LPS and mouse fecal LPS.
Purified E. coli LPS (EcLPS) was purchased from Sigma Chemical Co. (St. Louis, MO). B. thetaiotaomicron LPS (BtLPS), SPF mouse fecal LPS, GF mouse fecal LPS, and DSS-treated mouse fecal LPS were isolated by the Tri-reagent extraction procedure as previously described (25, 26). The SPF mouse fecal LPS extract, which is based upon the starting dry weight of the fecal sample, was observed to contain a minimum of 50 ng LPS/mg total extract, translating to approximately 5 ng/ml in crude fecal LPS (100 μg/ml) aqueous extracts, as determined by a Limulus amebocyte lysate (LAL) assay using BtLPS standards (data not shown).
Total lipid extraction from GF and SPF mouse feces.
The chloroform-methanol-based extraction procedure used for isolation of the lipid fractions from both GF and SPF mouse feces was performed as described previously (27) with modifications which avoided the use of water as an extraction component. Briefly, GF mouse feces (3.4 g) were resuspended in 40 to 50 ml of deionized water, and the suspension was lyophilized. Freeze-dried GF mouse feces (600 mg) were resuspended in 35 to 40 ml of a mixture of chloroform-methanol (1:1, vol/vol), and extraction was carried out under vigorous stirring for 1 to 1.5 h at room temperature. Insoluble debris was removed from the resulting suspension by filtration through cotton wool. The excess of the organic solvent was removed in vacuo, and the dried residue was reconstituted by adding 2.5 to 3 ml of methanol. The resulting suspension was filtered using an Acrodisc LC polyvinylidene difluoride (PVDF) filter membrane (diameter, 25 mm; pore size, 0.45 μm; Gelman, United Kingdom). Excess methanol was removed under a stream of oxygen-free nitrogen to yield 46 mg of the crude lipid fraction, which was stored at −20°C for either further analysis or a second round of purification.
Total lipid extraction from SPF and GF mouse intestinal mucosa and intestinal contents.
Samples of intestinal mucosa and the intestinal (ileum) contents from both SPF and GF mice were collected, and each sample was suspended in 1.5 ml of deionized water, followed by freeze-drying. Each freeze-dried sample was weighed, transferred into a 4-ml glass screw-top vial, and resuspended in 1.5 ml of a mixture of chloroform-methanol (1:1, vol/vol). Extraction of the lipid components was carried out as described above for the total lipid extraction of the mouse feces. After extraction, insoluble debris was removed from each sample by passing the suspension through a cotton wool filter, and the samples were dried under a stream of oxygen-free nitrogen. The solid residue in each sample was resuspended in 0.5 ml of methanol (MeOH), the insoluble material was removed by final filtration using the Acrodisc LC PVDF filter membrane and dried as described above, and the dried samples were stored at −20°C for further analysis.
Quantitative analysis of CL in SPFfeccm, GFfeccm, GFimcm, SPFimcm, and DSSfecd1–d7 cm.
Quantitative analysis of the cardiolipin (CL) in chloroform-methanol lipid extracts of the feces of SPF mice (SPFfeccm) and GF mice (GFfeccm), chloroform-methanol lipid extracts of the intestinal mucosa (ileum) of SPF mice (SPFimcm) and GF mice (GFimcm), and chloroform-methanol lipid extracts of the feces of DSS-treated mice after 1 day of treatment (DSSfecd1 cm) and 7 days of treatment (DSSfecd7 cm) was carried out using a modified protocol for fluorometric determination of CL described previously (28, 29). For each fluorometric determination of the CL species in the fecal lipid extracts, duplicate dilutions of a stock solution of CL (bovine heart CL, sodium salt; Sigma-Aldrich) in methanol (3.3 mM) were prepared over a concentration range of from 0 to 2.5 mM on each 96-well microtiter plate (F96 Microwell black polystyrene microplate; Thermo Scientific, Denmark) to generate a calibration curve. To generate the calibration curves and determine the CL contents of the fecal and mucosal lipid extracts in the CL binding assay, 50 μl of CL standard solutions in methanol and 50 μl of duplicate solutions of extracts in methanol (50 μg to 200 μg) were added to 96-well microtiter plates, followed by evaporation of the organic solvent overnight at 4°C. Fifty microliters of a 4 mM solution of 10-N-nonyl-acridine orange (NAO; Biotium, Cambridge Biosciences, United Kingdom) in methanol-water (1:1, vol/vol) was added to each well, and the microtiter plate was incubated at room temperature in the dark for 30 min. Nonbound NAO was removed by washing the wells three times with a solution of phosphate-buffered saline (PBS) in methanol-water (1:1, vol/vol). Fluorescence was detected by use of a fluorescence model SIAFR Synergy HT microplate reader (Bio-Tek Instruments [USA]) using two filter sets, as follows: filter set 1 had an excitation λ of 485/20 nm and an emission λ of 528/20 nm, and filter set 2 had an excitation λ of 485/20 nm and an emission λ of 590/35 nm. The data were acquired and processed using KC4 Bio-Tek software (v.3.4). The concentrations of the CL species in the lipid extracts were determined from the bovine heart CL standard curves by linear interpolation of the fluorescence intensity as a function of the molar concentrations of CL and expressed as the amount of CL (in milligrams) per milligram of the lipid extract. Based upon determination of the CL concentrations from extracts derived from the dry weight of the intestinal contents (n = 6) and assuming that the dry weight was approximately 20% of the wet weight, we estimated that the mean concentration of CL in the intestinal lumen was as high as 5 mg/ml, which exceeded the amounts of CL employed in the in vitro assays of this study that were sufficient for BtLPS inhibition (1 to 100 μg/ml).
Model of DSS treatment and determination of bacterial contents in DSS-treated mice.
DSS (molecular weight, 40,000; Sigma-Aldrich) was prepared fresh daily by dissolving DSS in ultrapure water and adjusting the final pH to 8.5. Female mice (age, 8 weeks; weight, 20 to 23 g) received 5% (wt/vol) DSS ad libitum for 7 days. Healthy age-matched controls received ultrapure water only. A disease activity index was derived from the major clinical signs of weight loss, diarrhea, and rectal bleeding as described by Murthy et al. (30).
The mice were weighed daily, and feces from the DSS-treated and control groups were collected and pooled daily. At the end of the experiment, the animals were euthanized by carbon dioxide asphyxiation. Intestines and colons were opened longitudinally, and the contents were collected. Open intestines were rinsed in ice-cold PBS and placed on a clean glass surface, and the intestinal mucosa was scraped off with a clean glass slide.
Intestines and colon contents (10 mg) were suspended in sterile reduced transport medium for serial dilution. Blood agar plates containing 5% defibrinated horse blood were seeded and incubated for 3 days in an anaerobic atmosphere of 80% N2, 10% H2, and 10% CO2 (Don Whitely Scientific). Colonies were counted to determine the number of CFU per gram, pure cultures of bacteria were obtained, and the bacteria were identified by 16S rRNA sequencing.
MALDI-TOF MS analyses of fecal and intestinal lipids.
For matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (MS) analyses, total fecal lipid samples were dissolved in 10 μl of a mixture of 5-chloro-2-mercaptobenzothiazole (20 mg/ml) in chloroform-methanol (1:1, vol/vol), and 0.5 μl of each sample was analyzed in both positive and negative ion modes on an AutoFlex analyzer (Bruker Daltonics). Data were acquired with a 50-Hz repletion rate, and up to 3,000 shots were accumulated for each spectrum. Instrument calibration and all other tuning parameters were optimized using an HP Calmix (Sigma-Aldrich, St. Louis, MO). Data were acquired and processed using flexAnalysis software (Bruker Daltonics).
Gel permeation chromatography and HPLC.
The total chloroform-methanol lipid extract from either GF or SPF mouse feces was dissolved in 1.5 ml of MeOH, followed by gel permeation chromatography on a ToyoPearl HW-40 (S) column (35 cm by 2 cm [inside diameter {i.d.}]); Tosoh, Anachem Ltd., United Kingdom) equilibrated with the same solvent. The column effluent was monitored for changes in the refractive index (RI) with a Knauer Wellchrom K-2400 RI detector (Wissenschaftliche Geratebau, Dr. Ing. Herbert Knauer GmbH, Berlin, Germany). Peak fractions (1.5 ml) were pooled, and an excess of the solvent was removed by a stream of oxygen-free nitrogen. Each peak fraction was tested for biological activity, and those fractions showing TLR4-inhibitory properties were used for further purification. The high-molecular-weight lipid fraction, which was isolated by gel permeation chromatography and which showed potent inhibition of BtLPS-dependent TLR4 activation, was subjected to another round of fractionation on a Glyco-Pac N column (7.8 mm by 300 mm [i.d.]; Waters, USA) in MeOH using a Beckman Coulter high-performance liquid chromatography (HPLC) system (32 Karat software, v.7). The column effluent was monitored for changes in RI as described above. Fraction peaks were collected, the excess solvent was removed by a stream of nitrogen, and the high-molecular-weight fractions, which showed TLR4-inhibitory activity, were pooled. The high-molecular-weight fraction obtained from the fractionation of GFfeccm was referred to as fraction 1 and used for further analysis.
Acidic methanolysis of lipid fraction samples.
Fatty acids from the lipid fraction samples were liberated by acid-catalyzed methanolysis as described previously (31). Briefly, 3 N methanolic HCl (300 μl; Supelco, USA) was added to the dried sample (2 to 3 mg), which was placed in a glass screw-top vial (4 ml) with a perforated (open-top, silicone septum) screw cap, and the reaction mixture was heated at 85°C for 12 h. Samples for subsequent analysis by gas chromatography (GC)-MS were extracted into 0.5 ml hexane, excess solvent was removed by a stream of oxygen-free nitrogen, and the samples were kept dried.
Fatty acid composition.
Analysis of the composition of the fatty acids present in the lipid fractions was carried out by GC-MS using a 6890N Network GC system and a 5975 inert mass-selective detector (Agilent Technologies, United Kingdom) equipped with a Sepelcowax 10 capillary column (30 m by 0.25 mm [i.d.]; film thickness [df], 0.25 μm; Sepelcowax, United Kingdom). Analysis of the fatty acid methyl esters was carried out by acid methanolysis of the samples, followed by dissolution of the methanolysate in 250 ml of hexane; 1 ml of this solution was used for GC-MS analysis. The injector temperature was set to 60°C, and the following temperature gradient was used: 60°C for 3 min, followed by an increment of 10°C/min up to 140°C and 2°C/min up to 230°C, where the sample was maintained for 10 min. Helium was used as a carrier gas at a flow rate of 1.0 ml/min. The analytes were ionized in the positive ion mode by electron impact (EI) at 70 eV. The spectra were collected over the m/z 50 to m/z 500 range, and extracted ion chromatograms were analyzed by searching the spectral library NIST standard reference database 1A (NIST 05; NIST, Gaithersburg, MD, USA). The ratios of the fatty acid methyl esters in the analytes were calculated on the basis of the integration of the area under the peaks of interest using the diagnostic fragment ion species at m/z 74 and m/z 87. The molecular masses of the polyunsaturated fatty acids were estimated according to the values of characteristic fragment ions m/z [M-31]+.
Fatty acid analysis of the high-molecular-weight fraction (fraction 1), which was obtained after chromatography of GFfeccm on a ToyoPearl HW-40 (S) column, revealed the presence of four major fatty acids, which were found to be hexadecanoic acid (C16:0), 14-methylpentadecanoic acid (iso-C16:0), monounsaturated 9-octadecenoic acid (C18:1, oleic acid), and diunsaturated 9,12-octadecadienoic acid (C18:2, linoleic acid), which were present in a molar ratio of 3.8:1:3.8:3 (see Table S1 in the supplemental material). In addition, traces of octadecanoic acid (C18:0) were detected.
NMR spectroscopy.
Deuterium exchange of lipid fractions was carried out by repeated dissolution of the sample in CD3OD (99.90% deuterium; Sigma-Aldrich) and drying of the sample under a stream of oxygen-free nitrogen. Finally, the lipid fraction sample was dissolved in 0.60 ml of CD3OD (99.90% deuterium; Sigma-Aldrich). Acetone was used as an internal standard (δH 2.225). The one-dimensional 1H nuclear magnetic resonance (NMR) spectrum was recorded at 25°C on a Bruker AV400 spectrometer. Data were acquired and processed using Bruker TOPSPIN (v.2) software.
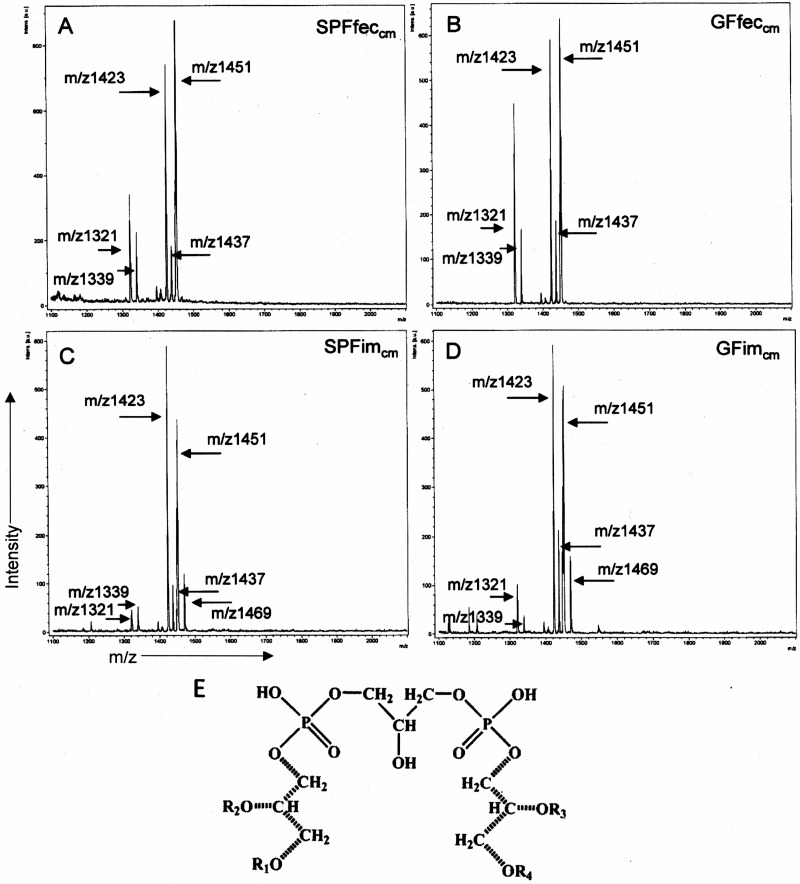
Analysis of the 1H NMR spectral data for fraction 1 (Table 1; see also Fig. S2B in the supplemental material) revealed the presence of signals for protons at 4.29 ppm and in the region from 5.0 to 5.10 ppm. The signal at 4.29 ppm corresponds to protons linked at C-1, and the signal in the region from 5.0 to 5.10 ppm corresponds to protons linked at glycerol C-2 side chain residues. These are the glycerol carbons linked to fatty acids R1 to R4 in Fig. 4E (32).
TABLE 1.
1H NMR spectral data for purified lipid extract of feces from GF micea
| Residue | δH (ppm) | Vicinal coupling (3JH,H) | Reference chemical shiftb |
|---|---|---|---|
| Side chain glycerol residues acylated with glycerol fatty acids R1 to R4c | |||
| CH2OR1(4) | 4.29 | JH1b,H2 = 6.7 Hz, JH1b,H1a = 12.1 Hz | 4.36 |
| CHOR2(3) | 5.0–5.10 | 5.15 | |
| CH2OP(O) | 4.00, 4.05 | JH3a,H2 = 6.2 Hz, JH3a,H3b = 10.4 Hz | 3.92 |
| Central residue POCH2CH(OH)CH2OP | 3.70–3.90 | 3.84–3.93 |
FIG 4.
Lipid fractions from SPF and GF mouse feces and intestinal mucosa display similar major mass ion species that correspond to cardiolipin isoforms. Negative ion MALDI-TOF MS was performed to assess the predominant mass ions present in chloroform-methanol lipid extracts of the feces and intestinal mucosa of SPF and GF mice. The atomic mass units assigned to the major peaks are indicated. (A to D) Major negative mass ions present in SPFfeccm (A), GFfeccm (B), SPFimcm (C), and GFimcm (D). (E) Structure of the major cardiolipin [1,3-bis(3-sn-phosphatidyl)glycerol] species corresponding to the negative mass ions present in GFfeccm, SPFfeccm, GFimcm and SPFimcm. R1 to R4, fatty acids residues attached to the side chain glycerol residues. The positions of the 1H NMR signals for protons of side chain glycerol residues CH2OR1(4), CHOR2(3), and CH2OP(O) and for protons of the central glycerol CH2OP(O) and CHOH are shown in Table 1. The side chain glycerol residues are indicated by dotted lines. intens., intensity; a.u., atomic mass units.
Signals in the 1H NMR spectrum at 4.00 ppm and 4.05 ppm were assigned to the protons at position C-3 of the side chain glycerol residues which are phosphorylated at position O-3 (Table 1; see also Fig. S2B in the supplemental material). The group of signals for protons with different intensities in the region from 3.70 to 3.90 ppm is indicative of H′1-H′3 of the central glycerol residue, therefore confirming the identity of this overall molecular species to be a CL entity (33).
1H NMR spectrum of fraction 1 also revealed the presence of signals for protons of both saturated and unsaturated fatty acid residues (see Fig. S2A and Table S2 in the supplemental material). The comparison of the intensities of the proton signals at 2.14 ppm and 2.68 ppm, which corresponded to α-methylene protons of all fatty acid residues and bisallylic protons of linoleic acid, respectively (34), gave a ratio value of 4:1. These data are in accordance with the ratio of fatty acids estimated by GC-MS-based quantitative analysis of fatty acids of fraction 1 (see Table S1 in the supplemental material).
TLR4 reporter assays for LPS activation and LPS antagonism.
HEK293 cell-based TLR4 reporter assays were performed essentially as described previously (35). Briefly, HEK293 cells were plated in 96-well plates at a density of 4 × 104 cells per well and on the following day were transfected with plasmids bearing firefly luciferase, Renilla luciferase, human TLR4, and MD-2 by standard calcium phosphate precipitation. The test wells were stimulated in triplicate for 4 h at 37°C with the doses of LPS indicated below that had been combined with fecal lipids, frozen at −20°C overnight, and subsequently thawed and suspended in Dulbecco modified Eagle medium containing 10% fetal bovine serum to achieve the final stimulatory concentrations. Following stimulation of the TLR4-transfected HEK293 cells, the cells were rinsed with phosphate-buffered saline and lysed with 50 μl of passive lysis buffer (Promega, Madison, WI). Luciferase activity was measured using a dual-luciferase assay reporter system (Promega, Madison, WI). Data are expressed as the fold increase of NF-κB activity, which represents the ratio of NF-κB-dependent firefly luciferase activity to β-actin promoter-dependent Renilla luciferase activity. Where indicated in the figure legends, data were analyzed by two-tailed unpaired Student t tests (GraphPad Prism).
LBP, CD14, and MD-2 binding assays.
The supernatants that were used in the binding assays were generated as follows: HEK293T cells were plated in 10-cm plates, grown to approximately 60% confluence, and transfected with 20 μg of plasmids carrying the soluble MD-2 (MD-2) with a 6×His tag, soluble lipopolysaccharide binding protein (LBP) with a 6×His tag, or soluble CD14 (CD14) with a 6×His tag by the calcium phosphate method for 2 to 4 h. Subsequently, the transfection complexes were removed, 10 ml of fresh growth medium was added per plate, and the cells were cultured for 2 days prior to harvesting of the supernatants. Binding assays were performed essentially according to the method of Visintin et al. (36), as follows: for each binding reaction, biotinylated EcLPS and fecal lipid antagonists were combined and frozen at −20°C overnight. The LPS-antagonist complexes were then resuspended in 4 ml of recombinant protein supernatant at the doses indicated below and in the figure legends. LPS-antagonist-protein interactions were allowed to form by incubation for 3 to 4 h at 21°C. The LPS-LBP-6×His, LPS-CD14-6×His, and LPS–MD-2–6×His complexes were all captured by addition of 40 μl (packed volume) streptavidin-agarose beads (Sigma-Aldrich, St. Louis, MO) to 4 ml supernatant, followed by agitation overnight on a rotator at 4°C. The agarose beads were pelleted by centrifugation for 30 s at 8,000 × g and 4°C. The beads were washed three times with PBS (LBP and CD14 complexes) or PBS containing 0.5% Tween 20 (MD-2 complexes). The washed beads were then resuspended in SDS sample buffer (Invitrogen, Carlsbad, CA), and the proteins in the sample were resolved in nonreducing, denaturing 4% to 20% Tris-glycine polyacrylamide gels (Invitrogen, Carlsbad, CA) and then transferred to a PVDF membrane (Invitrogen, Carlsbad, CA). The membranes were probed overnight at 4°C using a monoclonal anti-tetra-His antibody (Qiagen, Valencia, CA). The blot was washed and probed with goat anti-mouse IgG antibody-horseradish peroxidase conjugate (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 h at room temperature. Detection of the proteins was achieved by treating the blots with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL) followed by exposure to BioMax MS film (Kodak).
RESULTS
LPS activation is suppressed in the mouse gastrointestinal tract.
The gastrointestinal tract is exposed to LPS from both endogenous sources, such as the intestinal microbiota, and exogenous sources, such as LPS found in the diet (37). Therefore, we determined if LPS was able to elicit TLR4 activation during passage through the intestinal tract and incorporation in the feces. To accomplish this, intestinal and colon contents as well as feces were suspended in endotoxin-free water, and the ability of these aqueous suspensions to activate TLR4 in an HEK293 cell-based TLR4- and MD-2-mediated NF-κB activation assay was compared to that of crude LPS preparations obtained from the same samples. For these comparisons, crude LPS was extracted and resuspended in the same volume as the original intestinal sample, such that the same LPS concentration was examined in both extracted and nonextracted samples (Fig. 1). We observed that the fecal LPS aqueous extract (100 μg/ml) from SPF mice contained a minimum LPS content (5 ng/ml), as determined by an LAL assay (data not shown).
FIG 1.
The gastrointestinal contents of both SPF mice and GF mice contain activity inhibiting TLR4 activation by gastrointestinal lipopolysaccharides. Recombinant TLR4 activity was measured using a HEK293 cell-based luciferase reporter assay. (A) The relative abilities of SPF mouse feces (SPFfec) or LPS extracts from SPF mouse feces (SPFfecLPS) to activate TLR4 were compared. (B) The relative abilities of SPF mouse colon contents (SPFcc) or LPS extracts from SPF mouse colon contents (SPFccLPS) to activate TLR4 were compared. (C) The relative abilities of SPF mouse intestinal contents (SPFic) or LPS extracts from SPF mouse intestinal contents (SPFicLPS) to activate TLR4 were compared. (D) The relative abilities of GF mouse feces (GFfec) or LPS extracts from GF mouse feces (GFfecLPS) to activate TLR4 were compared. (E) The relative abilities of GF mouse colon contents (GFcc) or LPS extracts from GF mouse colon contents (GFccLPS) to activate TLR4 were compared. (F) The relative abilities of GF mouse intestinal contents (GFic) or LPS extracts from GF mouse intestinal contents (GFicLPS) to activate TLR4 were compared. The results are representative of those from at least two independent experiments and are plotted as the mean fold induction ± standard deviation from triplicate determinations relative to that for the unstimulated control. hTLR4, human TLR4; hMD-2, human MD-2. Asterisks represent significant differences in NF-κB activation elicited by the sample and derivative LPSs, as indicated by the dashed lines (*, P < 0.05; **, P < 0.001; ***, P < 0.0001; ns, not significant; unpaired Student t tests).
In SPF mice, where both the endogenous microbiota and dietary intake can contribute to LPS in the intestinal tract, comparison of aqueous extracts to the corresponding LPS extracts revealed that the fecal or colon contents displayed significantly lower levels of TLR4 activation, demonstrating that the colon and fecal contents suppress TLR4 activation (Fig. 1A and B). In contrast, there was no significant difference in the ability of the intestinal aqueous or crude LPS preparation to elicit a TLR4 response (Fig. 1C). The lack of a significant difference in TLR4 activation between intestinal aqueous and crude LPS preparations was further examined. First, it was determined that the number of bacteria found in the intestine was at least 2 log units lower than that found in the colon or feces (compare the results with those for the control in Fig. 7A and B [see below]). Second, aqueous suspensions of the SPF and GF mouse diets were tested in the TLR4 HEK293 cell-based assay, and the results revealed that the ability of the SPF mouse chow to activate TLR4 (see Fig. S1 in the supplemental material) was at least a 5-fold less than that of the GF mouse chow. Therefore, the lack of a sufficient number of bacteria combined with the low TLR4 activity found in SPF mouse chow but not GF mouse chow may have precluded the detection of LPS activation in the crude LPS preparations due to the sensitivity of our assay. In GF mice, where only the effect of dietary LPS was examined, all three components of the intestinal tract examined displayed a significantly lower ability to activate TLR4 than their corresponding LPS preparations (Fig. 1D to F). These data demonstrate that in GF mice the contents of all three components of the gastrointestinal tract examined demonstrate potent suppression of TLR4 activation.
FIG 7.
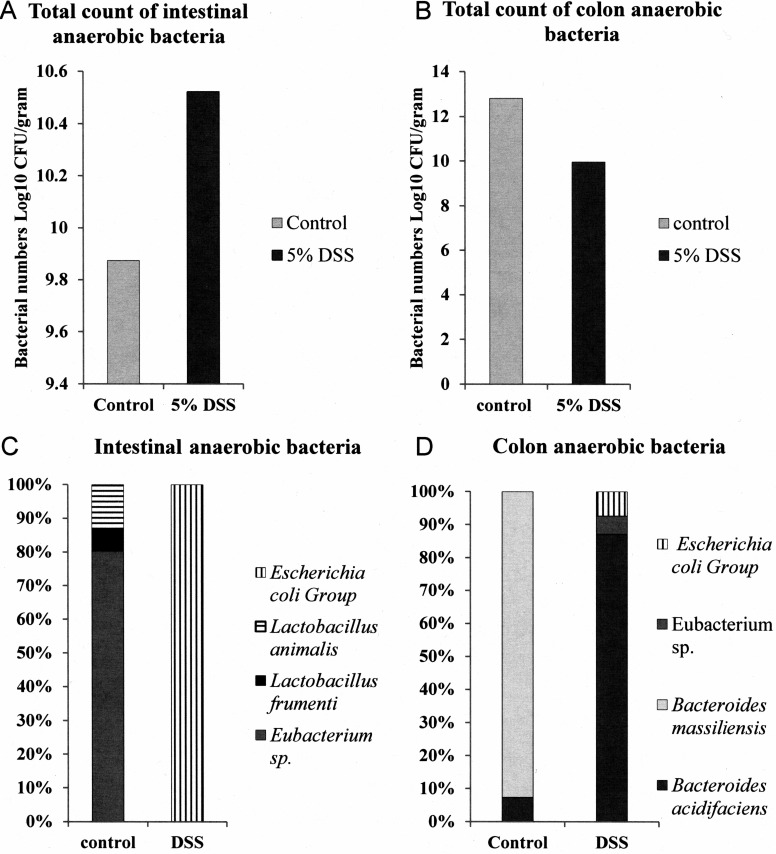
The intestinal anaerobic microbiota of DSS-fed mice displays a bacterial load and composition altered from those of the intestinal anaerobic microbiota of conventionally fed mice. DSS promotes a pronounced shift in the anaerobic Gram-negative microbial composition of mouse feces and intestinal contents. The intestinal and colon anaerobic bacterial loads and compositions of control SPF mice and DSS-fed mice (day 7) were determined. (A) Intestines from DSS-treated mice mouse obtained on day 7 of treatment exhibit an increased anaerobic bacterial load compared to that of control SPF mouse intestines. (B) Colons from DSS-treated mice obtained on day 7 of treatment exhibit a decreased anaerobic bacterial load compared to that of control SPF mouse colons. (C) Gram-negative Enterobacteriaceae are more prevalent in intestines from DSS-treated mice mouse obtained on day 7 of treatment than control SPF mouse intestines. (D) Gram-negative Enterobacteriaceae are more prevalent in colons from DSS-treated mice obtained on day 7 of treatment than control SPF mouse intestines.
Therefore, LPS activation of TLR4 from either endogenous sources, such as the intestinal microbiota, or exogenous sources, such as the GF mouse diet, is suppressed during transit through the gastrointestinal tract.
Total lipid extracts derived from mouse feces specifically inhibit LPS-dependent TLR4 proinflammatory responses.
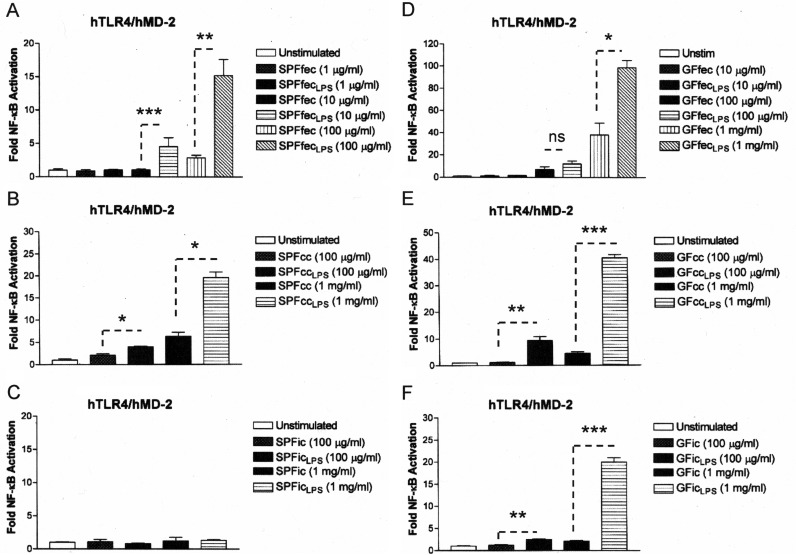
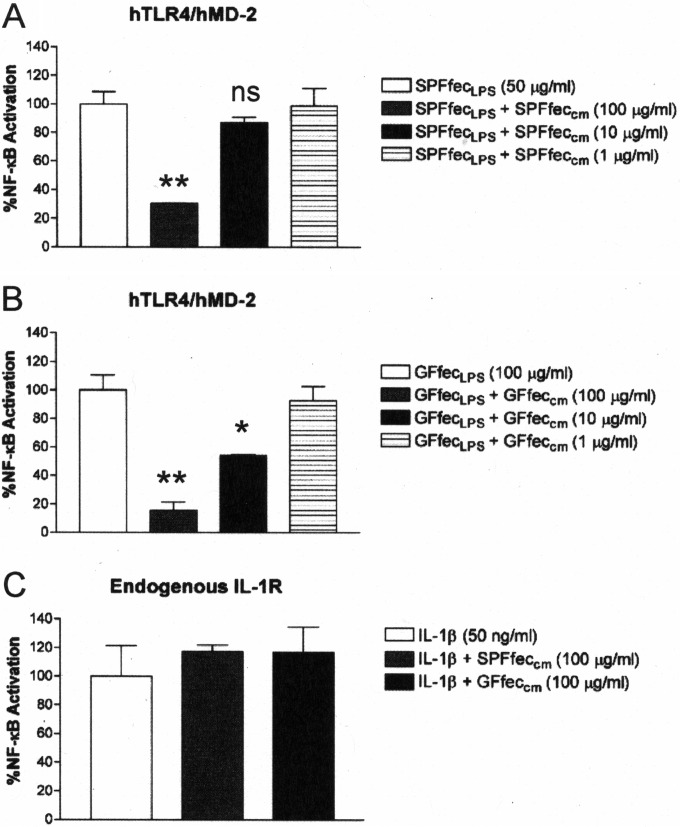
It has been reported that host phospholipids can inhibit LPS activation (38). Therefore, we tested the possibility that phospholipids present in mouse feces (39) contributed to the inhibition of TLR4. A chloroform-methanol-based extraction procedure was employed to obtain the total lipid fraction from both SPF and GF mouse feces (27). The chloroform-methanol lipid fractions were then examined for their ability to inhibit TLR4 activation in response to the crude fecal LPS preparations obtained from SPF and GF mouse feces described above (Fig. 2A and B). The chloroform-methanol preparations obtained from both SPF and GF mouse feces significantly antagonized the activation of TLR4 signaling by the fecal LPS preparations in a dose-dependent manner. Furthermore, the chloroform-methanol preparations did not inhibit the closely related interleukin-1 (IL-1) receptor (IL-1R)-dependent NF-κB activation pathway at the highest concentration of the fecal lipid extracts tested (100 μg/ml) (Fig. 2C), demonstrating that the inhibition of TLR4 activation was specific for LPS in the HEK293 cell-based assay.
FIG 2.
The lipid fractions from SPF and GF mouse feces contain a substance that specifically inhibits feces-derived LPS-dependent TLR4 activation. Recombinant TLR4 or endogenous IL-1R activity was measured using a HEK293 cell-based luciferase reporter assay. (A) Chloroform-methanol lipid extracts from SPF mouse feces (SPFfeccm) inhibit SPF feces-derived LPS-dependent TLR4 activation. (B) Chloroform-methanol lipid extracts from GF mouse feces (GFfeccm) inhibit GF mouse feces-derived LPS-dependent TLR4 activation. (C) Chloroform-methanol lipid extracts from either SPF mouse feces or GF mouse feces do not inhibit IL-1β-dependent IL-1R activation. The results obtained with an agonist (LPS or IL-1β) combined with SPFfeccm or GFfeccm are representative of those from at least two independent experiments and were plotted as percent activation minus the background ± standard deviation from triplicate determinations relative to that for the agonist alone (LPS or IL-1β) minus the background, which was assigned a value of 100% activation. Asterisks represent significant differences in NF-κB activation observed for LPS alone compared to that observed for LPS in combination with the lipid fraction (*, P < 0.05; **, P < 0.001; ns, not significant; unpaired Student t tests).
Lipids derived from feces and the host intestinal mucosa antagonize Bacteroides thetaiotaomicron LPS-dependent TLR4 activation.
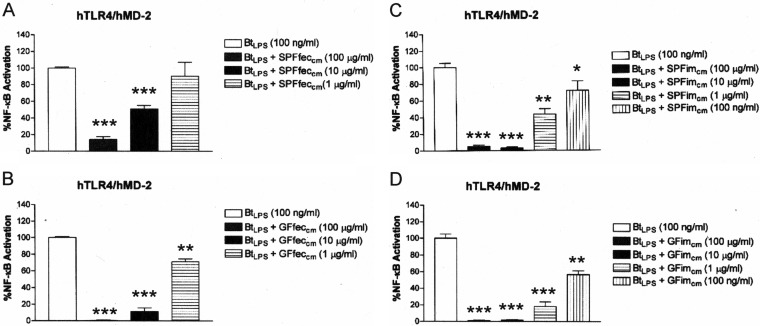
The ability of the fecal chloroform-methanol preparations described above as well as intestinal chloroform-methanol preparations to suppress TLR4 activation in response to a purified B. thetaiotaomicron LPS preparation was determined. LPS purified from B. thetaiotaomicron was employed as a representative LPS found in the intestines of healthy mice (35). Similar to the findings obtained with the crude fecal LPS preparations, TLR4 activation in response to B. thetaiotaomicron LPS was significantly suppressed in a dose-dependent manner by the fecal chloroform-methanol preparations obtained from either SPF mouse feces (Fig. 3A) or GF mouse feces (Fig. 3B). Next the ability of chloroform methanol extracts obtained from the intestinal mucosa (ileum) to suppress TLR4 responses to B. thetaiotaomicron LPS was examined (Fig. 3). Chloroform-methanol extracts of the GF and SPF mouse intestinal mucosa (colon plus intestine) were obtained by the same procedure used to isolate fecal lipids. It was found that lipid extracts obtained from both the SPF and GF mouse mucosa were potent inhibitors of TLR4 activation in response to B. thetaiotaomicron LPS (Fig. 3C and D, respectively). Collectively, these data demonstrate that the feces as well as the intestinal mucosa contain lipids that block the ability of B. thetaiotaomicron LPS to activate TLR4.
FIG 3.
The lipid fractions from SPF and GF mouse feces and intestinal mucosa contain a substance that potently inhibits BtLPS-dependent TLR4 activation. Recombinant TLR4 activity was measured using a HEK293 cell-based luciferase reporter assay. (A) Chloroform-methanol lipid extracts of feces from SPF mice (SPFfeccm) inhibit BtLPS-dependent TLR4 activation. (B) Chloroform-methanol lipid extracts of feces from GF mice (GFfeccm) inhibit BtLPS-dependent TLR4 activation. (C) Lipid extracts from SPF mouse intestinal mucosa (SPFimcm) inhibit BtLPS-dependent TLR4 activation. (D) Lipid extracts from GF mouse intestinal mucosa (GFimcm) inhibit BtLPS-dependent TLR4 activation. The results obtained with a combination of the agonist (BtLPS) and the lipid fractions (SPFfeccm, SPFimcm, GFfeccm, or GFimcm) are representative of those from at least two independent experiments and were plotted as percent activation minus the background ± standard deviation from triplicate determinations relative to that obtained with BtLPS alone minus the background, which was considered to be 100% activation. Asterisks represent significant differences in NF-κB activation observed for BtLPS alone compared to that observed for BtLPS in combination with the indicated lipid fraction (*, P < 0.05; **, P < 0.001; ***, P < 0.0001; unpaired Student t tests).
CL is a major fecal and intestinal lipid LPS antagonist.
MALDI-TOF MS was performed to identify the major mass ion structures present in the fecal and intestinal mucosal chloroform-methanol extracts of the GF and SPF mice (Fig. 4). A major cluster containing prominent ions at m/z 1,423 and m/z 1,451 was consistently identified to be the predominant structure found in chloroform-methanol extracts obtained from SPF or GF mouse feces as well as SPF or GF mouse intestinal mucosa (Fig. 4A to D). These ions correspond to CL species. Additionally, GC-MS analysis demonstrated the presence of a range of fatty acids which varied in chain length and degree of saturation, i.e., from C16:0 to C18:1 and C18:2 (see Table S1 in the supplemental material). 1H NMR characterization (see Fig. S2A and B in the supplemental material) of the chloroform-methanol extract obtained from GF mouse feces confirmed the presence of fatty acids (see Table S2 in the supplemental material). Moreover, this 1H NMR spectrum also contained signals for glycerol side chains and central glycerol moieties, the positions of which are specific for CL (Table 1). On the basis of these data, we propose that a major component of both fecal and intestinal lipid extracts corresponds to different CL isoform structures with various degrees of fatty acid substitutions (Fig. 4E). Consistent with CL representing the major mass ions found in the MALDI-TOF MS analysis, quantitative analysis of the CL content by a fluorometric assay using 10-N-nonyl-acridine orange (NAO) demonstrated that CL constituted between 9% and 28% of the total mass of lipid extracts of the feces and intestinal mucosa of both germfree and SPF mice, providing compelling evidence that this phospholipid is a major constituent of intestinal lipids (see Fig. S3 in the supplemental material). Assuming that the dry weight of the intestinal contents is approximately 20% of the wet weight, we estimate that the CL content in the contents of the intestinal lumen may approach 5 mg/ml.
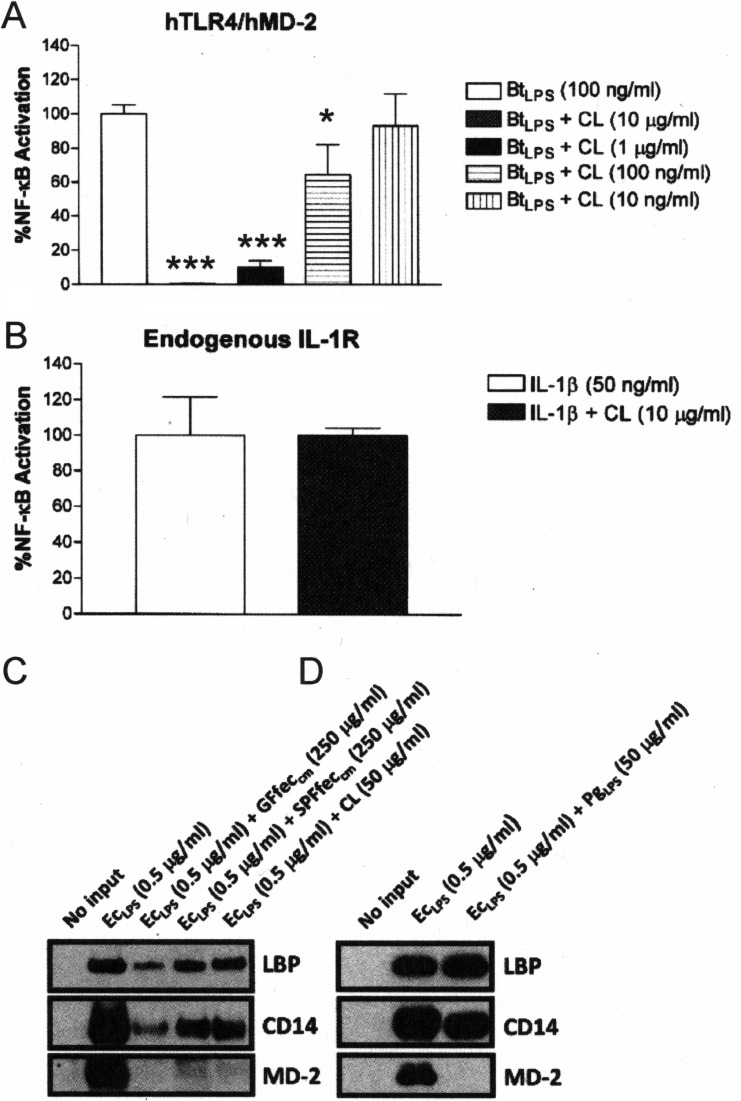
CL has previously been shown to inhibit tumor necrosis factor alpha production from mononuclear cells in response to LPS, most likely by inhibition of LPS interactions with lipopolysaccharide binding protein (LBP) (38), a protein that contributes to significantly increasing the sensitivity of TLR4 to LPS (40). Therefore, the ability of commercially obtained, purified CL to inhibit TLR4 activation in response to both LPS and IL-1β was determined in the HEK293 cell-based NF-κB assay (Fig. 5). CL specifically inhibited B. thetaiotaomicron LPS-mediated TLR4 activation (Fig. 5A), while it failed to inhibit IL-1β-dependent NF-κB activation (Fig. 5B). These findings are consistent with CL being a major TLR4 antagonist in the chloroform-methanol extracts.
FIG 5.
CL and mouse fecal lipids antagonize LPS-dependent TLR4 activation by a similar mechanism. The HEK293 cell-based luciferase assay was used to assess recombinant TLR4 activation or endogenous IL-1R activation. (A) CL inhibits BtLPS-dependent TLR4 activation. (B) CL does not inhibit IL-1β-dependent IL-1R activation. The results obtained with an agonist (LPS or IL-1β) combined with CL are representative of those from at least two independent experiments and were plotted as percent activation minus the background ± standard deviation from triplicate determinations relative to that obtained with the agonist alone (LPS or IL-1β) minus the background, which was assigned a value of 100% activation. Asterisks represent significant differences in NF-κB activation observed for BtLPS alone compared to that observed for BtLPS in combination with the indicated doses of CL (*, P < 0.05; ***, P < 0.0001; unpaired Student t tests). (C) CL, SPFfeccm, and GFfeccm inhibit the ability of LPS to interact with LBP, CD14, and MD-2. (D) P. gingivalis LPS (PgLPS) inhibits the ability of LPS to interact with MD-2 but not LBP or CD14. EcLPS with biotin alone or in combination with the indicated fractions was allowed to form complexes with recombinant LBP, CD14, or MD-2. Subsequently, the complexes were recovered by streptavidin-agarose-pulldown assays, and the proteins bound to EcLPS with biotin were visualized by Western blotting. The results are representative of those from at least two independent experiments.
Further evidence that CL represented the major component in the chloroform-methanol extracts that inhibits TLR4 responses to LPS was obtained by elucidating the molecular mechanism of fecal lipid LPS antagonism (Fig. 5C). A highly specific binding assay which utilizes recombinant sLBP, sCD14, and sMD-2 was employed. The basis for this assay depends upon the ability of the LPS antagonist to compete for binding at sLBP, sCD14, or sMD-2 with a biotinylated E. coli LPS preparation (41). A reduction in the signal intensity of autoradiogram images corresponding to LBP, CD14, or MD-2 detected by Western blotting indicates the specific step at which antagonism occurs. Figures 5C and D depict the results of the binding assay when chloroform-methanol extracts obtained from GF and SPF mouse feces were examined. It was found that purified CL was similar to chloroform-methanol extracts of SPF and GF mouse feces, in that antagonism against E. coli LPS was observed at each step in the activation pathway, LBP, CD14, and MD-2 (Fig. 5C). In contrast, the LPS antagonist derived from the oral pathogen Porphyromonas gingivalis, which was included as a comparative control for lipid A-type antagonism, blocked E. coli LPS binding exclusively at the TLR4 coreceptor, MD-2 (Fig. 5D), consistent with our previous results (25, 41). These data demonstrate that the intestinal lipids present in SPF and GF mouse feces mediate antagonism against E. coli LPS by a mechanism that is distinct from the mechanism displayed by the lipid A-type antagonist P. gingivalis LPS and is consistent with that observed with purified CL.
Feces from DSS-treated mice exhibit a pronounced increase in TLR4 activation.
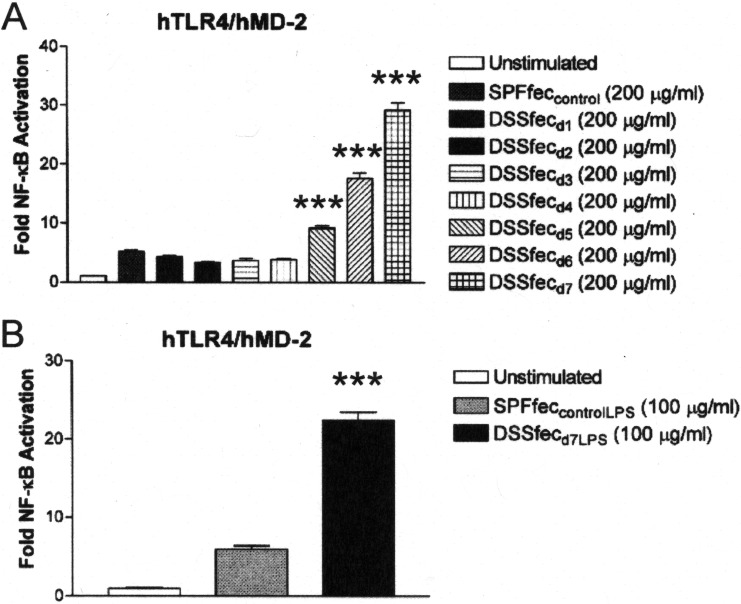
Inflammatory bowel disease has been associated with an increase in TLR4 activation (11), and therefore, we determined if feces from an IBD mouse model were altered in their capacity to activate TLR4 (Fig. 6A). IBD was induced in dextran sodium sulfate (DSS)-treated mice, an experimental model commonly utilized to explore the molecular and cellular mechanisms of IBD-related ulcerative colitis and colorectal cancer (42). Initially, TLR4 activation by feces obtained from conventionally fed mice was compared to that by feces obtained from mice that were administered DSS daily from 1 to 7 days of treatment. These data revealed that after 5 days of DSS administration, when the mice began to show signs of IBD by the disease activity index (see Fig. S4 and S5 in the supplemental material), the feces displayed significant TLR4 activation (Fig. 6A). In contrast, similar to what we described above, TLR4 activation was suppressed in feces obtained from control SPF mice not treated with DSS and presymptomatic DSS-treated mice (up to day 4).
FIG 6.
Feces and fecal LPS from DSS-fed mice fail to suppress TLR4 activation, whereas feces and fecal LPS from conventionally fed mice do suppress TLR4 activation. Recombinant TLR4 activity was measured using a HEK293 cell-based luciferase reporter assay. (A) The relative abilities of feces from mice treated with DSS and collected on from day 1 of treatment (DSSfecd1) to day 7 of treatment (DSSfecd7) and feces from SPF mouse controls (SPFfeccontrol) to activate TLR4 were compared. (B) The relative abilities of LPS derived from the feces of DSS-treated mice on day 7 of treatment (DSSfecd7LPS) and LPS derived from the feces of SPF mouse controls (SPFfeccontrolLPS) to activate TLR4 were compared. The results are representative of those from at least two independent experiments and were plotted as the mean fold induction ± standard deviation from triplicate determinations relative to that for the unstimulated control. Asterisks represent significant differences in NF-κB activation observed for feces from SPF mice compared to that observed for feces from DSS-treated mice or significant differences in NF-κB activation observed for LPS derived from the feces of SPF mouse controls compared to that observed for LPS derived from the feces of DSS-treated mice on day 7 of treatment (***, P < 0.0001; unpaired Student t tests).
The significant increase in the TLR4 activity of feces from DSS-treated mice obtained on day 7 of treatment (Fig. 6A) suggests that the potency of the endogenous LPS had been altered relative to that of the LPS in normal feces. To test this possibility, crude LPS preparations were prepared from extracts of feces obtained from DSS-treated mice on day 7 of treatment, and the TLR4 activity was compared to that of LPS preparations from control SPF mouse feces (Fig. 6B). These analyses revealed that the LPS preparation from the feces of DSS-treated mice obtained on day 7 of treatment resulted in significantly more potent TLR4 activity (approximately 4-fold) than control LPS preparations from the feces of SPF mice.
In addition, chloroform-methanol extracts were generated from feces collected from DSS-treated mice on day 1 to day 7 of treatment to determine if the onset of IBD also correlated with the loss of CL and the subsequent loss of TLR4-suppressing activity. However, it was found that the CL content of the feces obtained from DSS-treated mice on day 1 to day 7 of treatment did not significantly vary from that of the feces of control mice and that the extracts generated from these feces maintained their ability to suppress TLR4 activation in response to B. thetaiotaomicron LPS (see Fig. S6 and S7 in the supplemental material).
DSS promotes a marked shift in the intestinal microbial composition that correlates with increased TLR4 activity.
We suspected that the increase in TLR4 activity of the feces of DSS-treated mice obtained on day 7 of treatment (Fig. 6A) and the LPS obtained from these feces (Fig. 6B) may be due to a change in the microbial composition, which in turn might increase the abundance of more potent LPS species. It is known that DSS alters the intestinal microbial composition of mice (22) as well as increases soluble TLR4 ligand activity in mouse feces (12). Therefore, the microbial load of the intestinal and colonic contents derived from either control SPF mice or DSS-treated mice on day 7 of treatment was quantified by determining the cultivatable anaerobic bacteria (Fig. 7). These data demonstrated that the cultivatable microbial loads in both the intestine (Fig. 7A) and colon (Fig. 7B) differed significantly between DSS-treated mice and SPF mice. Furthermore, analyses of the microbial composition by 16S rRNA sequencing additionally confirmed an increase in the proportion of members of the family Enterobacteriaceae in the intestines (Fig. 7C) and colons (Fig. 7D) of DSS-treated mice, which is consistent with the findings of Lupp et al. (22). We note that the variability in the overall microbial composition observed in our study compared to that observed in other studies likely arose due to the method used to culture the isolates from the mouse microbiota and mouse-to-mouse batch variability.
LPS in feces from DSS-treated mice and E. coli LPS are less susceptible to TLR4 suppression by intestinal lipids.
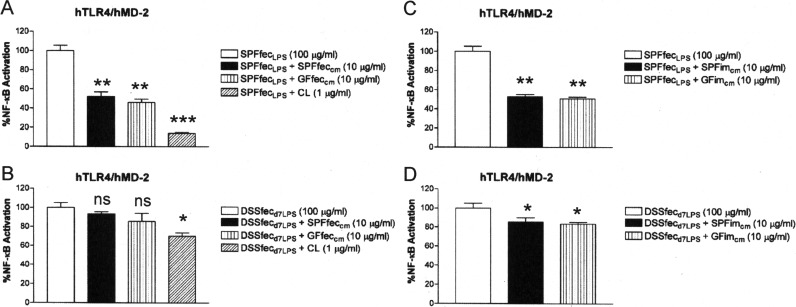
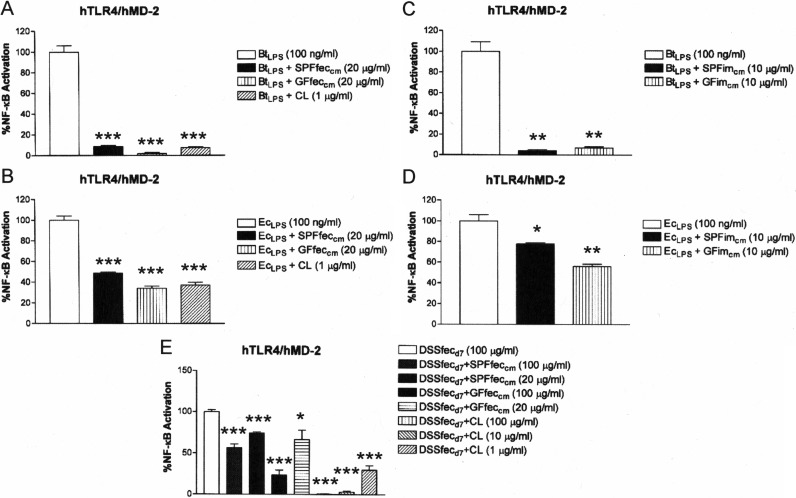
The observations that feces from DSS-treated mice showed increasing activation of TLR4 (Fig. 6), even though there was no change in the CL content of the feces of DSS-treated mice (see Fig. S6 in the supplemental material), and that extracts of feces from DSS-treated mice retained the ability to inhibit TLR4 activation by B. thetaiotaomicron LPS (see Fig. S7 in the supplemental material) indicated that the changed microbial composition of DSS-treated mice (Fig. 7) produces LPS that is less susceptible to intestinal lipid and CL inhibition. Consistent with this finding, it was found that fecal and intestinal chloroform-methanol extracts from either SPF or germfree mice as well as purified CL exhibited a significantly reduced ability to suppress TLR4 activation in response to LPS collected from the feces of DSS-treated mice on day 7 of treatment compared to their ability to suppress TLR4 activation in response to LPS from control SPF mice (compare Fig. 8A to B and compare Fig. 8C to D). To determine if the increase in the proportion of members of the family Enterobacteriaceae (at the expense of the Bacteroides sp.) could account for the TLR4 activation by feces collected from DSS-treated mice on day 7 of treatment and their LPS, we compared how effectively CL and lipid extracts suppressed TLR4 activation by B. thetaiotaomicron and E. coli LPSs. It was observed that both fecal and intestinal lipid extracts and CL were significantly more effective at neutralizing TLR4 activation in response to B. thetaiotaomicron LPS than at neutralizing TLR4 activation in response to E. coli LPS (compare Fig. 9A to B and compare Fig. 9C to D).
FIG 8.
Cardiolipin and mouse intestinal lipids inhibit SPFfecLPS-dependent TLR4 activation more potently than DSSfecd7LPS-dependent TLR4 activation. Recombinant TLR4 activity was measured using a HEK293 cell-based luciferase reporter assay. (A) Abilities of CL and mouse fecal lipids to inhibit SPFfecLPS-dependent TLR4 activation. (B) Abilities of CL and mouse fecal lipids to inhibit DSSfecd7LPS-dependent TLR4 activation. (C) Abilities of mouse intestinal mucosa lipids to inhibit SPFfecLPS-dependent TLR4 activation. (D) Abilities of mouse intestinal mucosa lipids to inhibit DSSfecd7LPS-dependent TLR4 activation. The results obtained with agonists (SPFfecLPS or DSSfecd7LPS) combined with antagonists (CL or chloroform-methanol lipid fractions of feces or intestinal mucosa) are representative of those from at least two independent experiments and were plotted as percent activation minus the background ± standard deviation from triplicate determinations relative to that for the agonists alone (SPFfecLPS or DSSfecd7LPS) minus the background, which was considered to be 100% activation. Asterisks represent significant differences in NF-κB activation observed for SPFfecLPS or DSSfecd7LPS alone compared to the results for SPFfecLPS or DSSfecd7LPS in combination with the indicated lipid fraction (*, P < 0.05; **, P < 0.001; ***, P < 0.0001; ns, not significant; unpaired Student t tests).
FIG 9.
Cardiolipin and mouse intestinal lipids inhibit BtLPS-dependent TLR4 activation more potently than EcLPS-dependent TLR4 activation. Recombinant TLR4 activity was measured using a HEK293 cell-based luciferase reporter assay. (A) Abilities of CL and mouse fecal lipids to inhibit BtLPS-dependent TLR4 activation. (B) Abilities of CL and mouse fecal lipids to inhibit EcLPS-dependent TLR4 activation. (C) Abilities of mouse intestinal mucosa lipids to inhibit BtLPS-dependent TLR4 activation. (D) Abilities of mouse intestinal mucosa lipids to inhibit EcLPS-dependent TLR4 activation. (E) The relative abilities of exogenous mouse fecal lipids and exogenous CL to inhibit DSSfecd7-dependent TLR4 activation were measured. The results obtained with agonists (BtLPS, EcLPS, or DSSfecd7) combined with antagonists (CL or chloroform-methanol lipid fractions of feces or intestinal mucosa) are representative of those from at least two independent experiments and were plotted as percent activation minus the background ± standard deviation from triplicate determinations relative to that for the agonists alone minus the background, which was considered to be 100% activation. Asterisks represent significant differences in NF-κB activation observed for BtLPS, EcLPS, or feces from DSS-treated mice alone compared to that for BtLPS, EcLPS, or feces from DSS-treated mice in combination with the indicated lipid fraction (*, P < 0.05; **, P < 0.001; ***, P < 0.0001; unpaired Student t tests).
These data provide novel evidence that feces from DSS-fed mice exhibit a progressively increased proinflammatory potential with respect to TLR4 signaling. The increase in TLR4 activity is not due to the loss of CL or the inability to suppress TLR4 activation in response to B. thetaiotaomicron LPS. Rather, the increase in the TLR4 activity of the crude LPS preparation obtained from DSS-treated mice on day 7 of treatment is consistent with a significant increase in the proportion of members of the family Enterobacteriaceae, which contain a significantly more potent LPS that is less susceptible to neutralization by the intestinal lipids found in mouse feces.
To gain formal evidence that an excess of exogenously added CL can suppress the LPS activity present in the feces of DSS-treated mice on day 7 of treatment, purified CL was added to the feces of DSS-treated mice on day 7 of treatment and tested for the ability to stimulate TLR4. It was found that increasing concentrations of exogenously added CL could effectively reduce the TLR4 response in the feces of DSS-treated mice on day 7 of treatment to near background levels (Fig. 9E). These data strongly suggest that CL, when present in excess, can reinforce the protective barrier established by the host to limit interactions between the endogenous intestinal Gram-negative microbiota and TLR4.
DISCUSSION
It has recently become clear that the lipid A structures of LPS to which intestinal tissue is exposed are significantly different in healthy and diseased individuals (15, 16, 20, 21, 23). Specifically, healthy individuals are exposed to significantly more LPS from the family Bacteroidaceae than individuals with dysbiotic microbial communities, where significant increases in the proportions of members of the family Enterobacteriaceae are found (15, 22). For example, obesity (15, 16), necrotizing enterocolitis in infants (43), as well as ulcerative colitis and Crohn's disease are all associated with intestinal dysbiotic communities, all of which result in significantly different lipid A structures in the intestinal lumen (9, 44). In this report, we describe a phospholipid-based barrier found in the intestine which has the ability to discriminate between these two different types of LPS lipid A structures. Previous reports have described both physical and spatial barriers which preclude interactions between the intestinal lumen components and the epithelial cell lining of intestinal tissue that are nonspecific, in that they prevent a wide variety of different bacterial and bacterial component interactions with intestinal tissue. For example, IgA produced by the gut-associated lymphoid tissue has been implicated in neutralizing the proinflammatory activity of the commensal microbiota via interaction with bacterial surface molecules, including the O antigen of LPS (45). However, the intestinal lipid barrier described here differs in two important aspects. First, the barrier does not rely upon a physiospatial separation of bacteria and their products from the luminal epithelium. Instead it is mediated via a neutralization mechanism that depends upon endogenous CL that is derived from the host intestinal epithelium and that directly inhibits TLR4 signaling. Second, the barrier is significantly more effective at inhibiting TLR4 activation in response to the LPS that is found in healthy intestinal microbiomes and that is typified by the LPS found in B. thetaiotaomicron rather than members of the family Enterobacteriaceae, which is typified by the LPS found in E. coli. This differential specificity could be viewed as an intestinal surveillance mechanism which is able to block LPS structures found in symbiotic microbial communities and is overcome during episodes of dysbiosis.
The competitive inhibition studies described here demonstrated CL inhibition of LPS binding at both LBP and CD14 in the LBP–CD14–MD-2/TLR4 activation pathway. CL was previously shown to inhibit LPS activation of mononuclear cells presumably by its structural similarity to compound 406, a lipid A structural antagonist (38). Furthermore, it was shown that although other host phospholipids were less potent than CL, they were able to inhibit mononuclear cell LPS responses by targeting LBP, although the mechanism was not elucidated (38). Since the chloroform-methanol extracts that inhibited LPS activation of TLR4 demonstrated similar levels of competitive inhibition of LBP and CD14 binding and CL was a major component in those extracts, it was concluded that CL and perhaps other phospholipids were responsible for TLR4 inhibition. Studies in germfree mice revealed that the source of the CL was the host and most likely arose in the intestinal lumen by the turnover of intestinal epithelial cells (39, 46). Indeed, studies have shown that intestinal commensal bacteria (19, 39, 46) and, specifically, LPS can regulate epithelial cell proliferation in a TLR4-dependent fashion in both health and disease (47, 48).
In the present study, we employed the LPS derived from the human gut symbiont B. thetaiotaomicron as a commensal symbiotic representative of the family Bacteroidaceae. To the best of our knowledge, the LPS structures of commensal symbiotic Bacteroides spp. have not been well characterized, although they are likely to have structures and, hence, endotoxic properties similar to those of B. thetaiotaomicron LPS. For example, the human symbionts Bacteroides fragilis and B. thetaiotaomicron display similar LPS structures as well as similar abilities to act as TLR4 agonists (49). In contrast, LPS structures derived from related commensals that are considered to be more pathogenic, such as the oral pathogen Porphyromonas gingivalis or the intestinal pathogen Bacteroides vulgatus, exhibit distinct structural properties and function as very weak agonists or even antagonists at TLR4 (50, 51). The selective inhibition of the family Bacteroidaceae-type LPS compared to that of the family Enterobacteriaceae-type LPS by CL and other phospholipids can be accounted for by lipid A structural features which differ between members of these two bacterial families. Family Enterobacteriaceae LPS lipid A structures demonstrate higher binding affinities for the LBP–CD14–MD-2/TLR4 activation pathway than family Bacteroidaceae lipid A structures (52). Lipid A components that significantly alter binding and subsequent activation in the TLR4 activation pathway are the number of phosphate groups and the number and length of fatty acids found in the lipid A (53–55). For example, members of the family Bacteroidaceae contain a lipid A that is monophosphorylated, whereas members of the family Enterobacteriaceae contain a diphosphorylated lipid A structure (56). The presence or absence of mono- or diphosphorylated lipid A structures appears to be highly conserved among members of the same bacterial family (57, 58). In addition, it has recently been demonstrated that 733 independent intestinal isolates of members of the family Bacteroidaceae demonstrate antimicrobial peptide resistance which has been correlated with a monophosphorylated lipid A structure (35, 57). In contrast, the lipid A in the family Enterobacteriaceae is diphosphorylated and consists mostly of shorter-chain-length fatty acids (58). Although other lipid A structural modifications have been described for members of both of these families (59), reflecting the well-known lipid A structural heterogeneity within a bacterial species, the monophosphorylated and altered fatty acid acylation patterns are two lipid A structural features that are fairly uniform among family members (56, 60). Structure-activity studies examining these lipid A characteristics have shown that they are associated with significantly less potent TLR4 activation (61). Direct comparisons of the B. thetaiotaomicron and E. coli LPSs have confirmed that the B. thetaiotaomicron LPS is 100 to 1,000 times less potent than the E. coli LPS in its ability to elicit TLR4 activation in the concentration range of fecal LPS concentrations tested in the present study (∼1 to 10 ng/ml) (49). Reasons for the decreased potency of B. thetaiotaomicron LPS compared to that of the Enterobacteriaceae LPS include significantly lower affinities for binding to both LBP and CD14 (52), two accessory proteins which greatly increase the potency of TLR4. In addition, the degree and amount of lipid A phosphorylation as well as the number and types of fatty acids can influence TLR4 potency after LPS has been delivered to the MD-2/TLR4 receptor complex (54). Therefore, dysbiotic shifts in the intestinal microbiome which result in significant increases in the proportions of members of the family Enterobacteriaceae are associated with more potent lipid A structures, which can render intestinal tissue more susceptible to TLR4 activation simply by reducing the amount of LPS necessary to potently activate TLR4. In fact, this has recently been demonstrated in a Rag1−/− mouse model of colitis (24). It was demonstrated that LPS with higher levels of TLR4 activity was a key determinant of whether mice develop colitis in the adaptive transfer model. A recent review predicted that significantly more potent LPS contributes to inflammation associated with inflammatory bowel disease in dysbiotic communities and discussed the ramifications of dysbiotic shifts in polymicrobial communities (56).
The maintenance of intestinal homeostasis can be accomplished in an LPS-rich intestinal lumen when the majority of the lipid A structures are less able to enter the LBP–CD14–MD-2/TLR4 activation pathway due to the presence of excess host CL and phospholipids. It has recently been shown that less potent monophosphorylated family Bacteroidaceae-type lipid A structures are maintained in the intestinal lumen, presumably as a mechanism to maintain intestinal homeostasis (57). In addition, it has been demonstrated that host intestinal alkaline phosphatase, which renders diphosphorylated family Enterobacteriaceae-type LPS, which is a potent activator of TLR4, similar to less potent monophosphorylated family Bacteroidaceae-type LPS, can prevent metabolic syndrome in mice through the detoxification of LPS (8). Shed CL as well as the intestinal phospholipid barrier may represent another mechanism by which monophosphorylated family Bacteroidaceae-type LPS in the intestinal lumen due to significantly lower binding to LBP and transfer to CD14 (52) is neutralized before engagement of TLR4. In addition, both LBP and CD14 are made by intestinal epithelial cells (62, 63), and evidence that they contribute to intestinal homeostasis has been reported (64, 65). Furthermore, modulation of TLR4 activity by LPB may have some clinical utility in treating necrotizing enterocolitis (66). Indeed, it might also be expected that both the bile salt (67) and phospholipid (68) barrier would display selective inhibition of the family Bacteroidaceae-type LPS rather than the family Enterobacteriaceae-type LPS for the same reasons that CL was shown here to be a more effective family Bacteroidaceae-type LPS blocker at LBP and CD14. Consistent with this, the intestinal phospholipid barrier has been shown to be altered in ulcerative colitis, and this may contribute to increased LPS activation (1). Furthermore, the crystal structure of the MD-2/TLR4 LPS receptor also predicts that the family Bacteroidaceae-type LPS could be more effectively blocked from active engagement due to the loss of a phosphate group (69). The relationship between the lipid A structure and TLR4 activation as it relates to the innate host response has previously been addressed by Munford and Varley (60); however, the less potent TLR4 activation displayed by the family Bacteroidaceae LPS, representing a significant factor contributing to the maintenance of intestinal homeostasis, was not considered at that time.
Supplementation of the inflammatory fecal extracts from DSS-treated mice with excess CL significantly reduced TLR4 activation, suggesting that the addition of CL to the diet may reduce the inflammation associated with dysbiotic communities. Indeed, it has been shown that dietary gangliosides can reduce proinflammatory signaling in individuals with IBD (70). The work presented here suggests that a novel mechanism by which host CL and the other phospholipids may modulate intestinal inflammation is by inhibition of the LPS entering the LBP–CD14–MD-2/TLR4 pathway.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the assistance of Margaret Collins during manuscript preparation.
This study was funded by a National Institute of Dental and Craniofacial Research grant awarded to R.P.D. (R01DE012768).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00463-16.
REFERENCES
- 1.Braun A, Treede I, Gotthardt D, Tietje A, Zahn A, Ruhwald R, Schoenfeld U, Welsch T, Kienle P, Erben G, Lehmann WD, Fuellekrug J, Stremmel W, Ehehalt R. 2009. Alterations of phospholipid concentration and species composition of the intestinal mucus barrier in ulcerative colitis: a clue to pathogenesis. Inflamm Bowel Dis 15:1705–1720. doi: 10.1002/ibd.20993. [DOI] [PubMed] [Google Scholar]
- 2.Dial EJ, Tran DM, Hyman A, Lichtenberger LM. 2013. Endotoxin-induced changes in phospholipid dynamics of the stomach. J Surg Res 180:140–146. doi: 10.1016/j.jss.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macpherson AJ, Slack E, Geuking MB, McCoy KD. 2009. The mucosal firewalls against commensal intestinal microbes. Semin Immunopathol 31:145–149. doi: 10.1007/s00281-009-0174-3. [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee S, Hooper LV. 2015. Antimicrobial defense of the intestine. Immunity 42:28–39. doi: 10.1016/j.immuni.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 5.Saleh M, Elson CO. 2011. Experimental inflammatory bowel disease: insights into the host-microbiota dialog. Immunity 34:293–302. doi: 10.1016/j.immuni.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lievin-Le Moal V, Servin AL. 2006. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin Microbiol Rev 19:315–337. doi: 10.1128/CMR.19.2.315-337.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen KT, Malo MS, Beasley-Topliffe LK, Poelstra K, Millan JL, Mostafa G, Alam SN, Ramasamy S, Warren HS, Hohmann EL, Hodin RA. 2011. A role for intestinal alkaline phosphatase in the maintenance of local gut immunity. Dig Dis Sci 56:1020–1027. doi: 10.1007/s10620-010-1396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaliannan K, Hamarneh SR, Economopoulos KP, Nasrin Alam S, Moaven O, Patel P, Malo NS, Ray M, Abtahi SM, Muhammad N, Raychowdhury A, Teshager A, Mohamed MM, Moss AK, Ahmed R, Hakimian S, Narisawa S, Millan JL, Hohmann E, Warren HS, Bhan AK, Malo MS, Hodin RA. 2013. Intestinal alkaline phosphatase prevents metabolic syndrome in mice. Proc Natl Acad Sci U S A 110:7003–7008. doi: 10.1073/pnas.1220180110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maloy KJ, Powrie F. 2011. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 10.Abreu MT, Fukata M, Arditi M. 2005. TLR signaling in the gut in health and disease. J Immunol 174:4453–4460. doi: 10.4049/jimmunol.174.8.4453. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Zhang Z, Wang L, Li J, Dong L, Yue W, Chen J, Sun X, Zhong L, Sun D. 2010. TLR4 monoclonal antibody blockade suppresses dextran-sulfate-sodium-induced colitis in mice. J Gastroenterol Hepatol 25:209–214. doi: 10.1111/j.1440-1746.2009.06046.x. [DOI] [PubMed] [Google Scholar]
- 12.Erridge C, Duncan SH, Bereswill S, Heimesaat MM. 2010. The induction of colitis and ileitis in mice is associated with marked increases in intestinal concentrations of stimulants of TLRs 2, 4, and 5. PLoS One 5:e9125. doi: 10.1371/journal.pone.0009125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seksik P, Sokol H, Grondin V, Adrie C, Duboc H, Pigneur B, Thomas G, Beaugerie L, Trugnan G, Masliah J, Bachelet M. 2010. Sera from patients with Crohn's disease break bacterial lipopolysaccharide tolerance of human intestinal epithelial cells via MD-2 activity. Innate Immun 16:381–390. doi: 10.1177/1753425909357076. [DOI] [PubMed] [Google Scholar]
- 14.Pasternak BA, D'Mello S, Jurickova II, Han X, Willson T, Flick L, Petiniot L, Uozumi N, Divanovic S, Traurnicht A, Bonkowski E, Kugathasan S, Karp CL, Denson LA. 2010. Lipopolysaccharide exposure is linked to activation of the acute phase response and growth failure in pediatric Crohn's disease and murine colitis. Inflamm Bowel Dis 16:856–869. doi: 10.1002/ibd.21132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim KA, Gu W, Lee IA, Joh EH, Kim DH. 2012. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS One 7:e47713. doi: 10.1371/journal.pone.0047713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luche E, Cousin B, Garidou L, Serino M, Waget A, Barreau C, Andre M, Valet P, Courtney M, Casteilla L, Burcelin R. 2013. Metabolic endotoxemia directly increases the proliferation of adipocyte precursors at the onset of metabolic diseases through a CD14-dependent mechanism. Mol Metab 2:281–291. doi: 10.1016/j.molmet.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherji A, Kobiita A, Ye T, Chambon P. 2013. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 19.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. 2008. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A 105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, Timmis KN, Schreiber S. 2004. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gronbach K, Flade I, Holst O, Lindner B, Ruscheweyh HJ, Wittmann A, Menz S, Schwiertz A, Adam P, Stecher B, Josenhans C, Suerbaum S, Gruber AD, Kulik A, Huson D, Autenrieth IB, Frick JS. 2014. Endotoxicity of lipopolysaccharide as a determinant of T-cell-mediated colitis induction in mice. Gastroenterology 146:765–775. doi: 10.1053/j.gastro.2013.11.033. [DOI] [PubMed] [Google Scholar]
- 25.Reife RA, Coats SR, Al-Qutub M, Dixon DM, Braham PA, Billharz RJ, Howald WN, Darveau RP. 2006. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cell Microbiol 8:857–868. doi: 10.1111/j.1462-5822.2005.00672.x. [DOI] [PubMed] [Google Scholar]
- 26.Yi EC, Hackett M. 2000. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 125:651–656. doi: 10.1039/b000368i. [DOI] [PubMed] [Google Scholar]
- 27.Gustafsson BE, Karlsson KA, Larson G, Midtvedt T, Stromberg N, Teneberg S, Thurin J. 1986. Glycosphingolipid patterns of the gastrointestinal tract and feces of germ-free and conventional rats. J Biol Chem 261:15294–15300. [PubMed] [Google Scholar]
- 28.Kaewsuya P, Danielson ND, Ekhterae D. 2007. Fluorescent determination of cardiolipin using 10-N-nonyl acridine orange. Anal Bioanal Chem 387:2775–2782. doi: 10.1007/s00216-007-1135-0. [DOI] [PubMed] [Google Scholar]
- 29.Nanda SK, Scholz WK. 2000. A distinctive surface for phospholipid assays. Clin Chem 46:1508–1510. [PubMed] [Google Scholar]
- 30.Murthy SN, Cooper HS, Shim H, Shah RS, Ibrahim SA, Sedergran DJ. 1993. Treatment of dextran sulfate sodium-induced murine colitis by intracolonic cyclosporin. Dig Dis Sci 38:1722–1734. doi: 10.1007/BF01303184. [DOI] [PubMed] [Google Scholar]
- 31.Rahman MM, Kolli VS, Kahler CM, Shih G, Stephens DS, Carlson RW. 2000. The membrane phospholipids of Neisseria meningitidis and Neisseria gonorrhoeae as characterized by fast atom bombardment mass spectrometry. Microbiology 146(Pt 8):1901–1911. doi: 10.1099/00221287-146-8-1901. [DOI] [PubMed] [Google Scholar]
- 32.Kim J, Hoppel CL. 2011. Monolysocardiolipin: improved preparation with high yield. J Lipid Res 52:389–392. doi: 10.1194/jlr.D010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mastronicolis SK, Arvanitis N, Karaliota A, Magiatis P, Heropoulos G, Litos C, Moustaka H, Tsakirakis A, Paramera E, Papastavrou P. 2008. Coordinated regulation of cold-induced changes in fatty acids with cardiolipin and phosphatidylglycerol composition among phospholipid species for the food pathogen Listeria monocytogenes. Appl Environ Microbiol 74:4543–4549. doi: 10.1128/AEM.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, Li H, Wu D, Lei X, Zhu X, Zhang A. 2010. Application of a hybrid variable selection method for the classification of rapeseed oils based on 1H NMR spectral analysis. Eur Food Res Technol 230:981–988. doi: 10.1007/s00217-010-1241-7. [DOI] [Google Scholar]
- 35.Coats SR, Berezow AB, To TT, Jain S, Bainbridge BW, Banani KP, Darveau RP. 2011. The lipid A phosphate position determines differential host Toll-like receptor 4 responses to phylogenetically related symbiotic and pathogenic bacteria. Infect Immun 79:203–210. doi: 10.1128/IAI.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Visintin A, Halmen KA, Latz E, Monks BG, Golenbock DT. 2005. Pharmacological inhibition of endotoxin responses is achieved by targeting the TLR4 coreceptor, MD-2. J Immunol 175:6465–6472. doi: 10.4049/jimmunol.175.10.6465. [DOI] [PubMed] [Google Scholar]
- 37.Hrncir T, Stepankova R, Kozakova H, Hudcovic T, Tlaskalova-Hogenova H. 2008. Gut microbiota and lipopolysaccharide content of the diet influence development of regulatory T cells: studies in germ-free mice. BMC Immunol 9:65. doi: 10.1186/1471-2172-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller M, Brandenburg K, Dedrick R, Schromm AB, Seydel U. 2005. Phospholipids inhibit lipopolysaccharide (LPS)-induced cell activation: a role for LPS-binding protein. J Immunol 174:1091–1096. doi: 10.4049/jimmunol.174.2.1091. [DOI] [PubMed] [Google Scholar]
- 39.Nair P, Lagerholm S, Dutta S, Shami S, Davis K, Ma S, Malayeri M. 2003. Coprocytobiology: on the nature of cellular elements from stools in the pathophysiology of colonic disease. J Clin Gastroenterol 36:S84–S93. doi: 10.1097/00004836-200305001-00015. [DOI] [PubMed] [Google Scholar]
- 40.Hailman E, Lichenstein HS, Wurfel MM, Miller DS, Johnson DA, Kelley M, Busse LA, Zukowski MM, Wright SD. 1994. Lipopolysaccharide (LPS)-binding protein accelerates the binding of LPS to CD14. J Exp Med 179:269–277. doi: 10.1084/jem.179.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coats SR, Do CT, Karimi-Naser LM, Braham PH, Darveau RP. 2007. Antagonistic lipopolysaccharides block E. coli lipopolysaccharide function at human TLR4 via interaction with the human MD-2 lipopolysaccharide binding site. Cell Microbiol 9:1191–1202. doi: 10.1111/j.1462-5822.2006.00859.x. [DOI] [PubMed] [Google Scholar]
- 42.Melgar S, Karlsson L, Rehnstrom E, Karlsson A, Utkovic H, Jansson L, Michaelsson E. 2008. Validation of murine dextran sulfate sodium-induced colitis using four therapeutic agents for human inflammatory bowel disease. Int Immunopharmacol 8:836–844. doi: 10.1016/j.intimp.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 43.Claud EC, Keegan KP, Brulc JM, Lu L, Bartels D, Glass E, Chang EB, Meyer F, Antonopoulos DA. 2013. Bacterial community structure and functional contributions to emergence of health or necrotizing enterocolitis in preterm infants. Microbiome 1:20. doi: 10.1186/2049-2618-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whelan K, Quigley EM. 2013. Probiotics in the management of irritable bowel syndrome and inflammatory bowel disease. Curr Opin Gastroenterol 29:184–189. doi: 10.1097/MOG.0b013e32835d7bba. [DOI] [PubMed] [Google Scholar]
- 45.Mantis NJ, Rol N, Corthesy B. 2011. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol 4:603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams JM, Duckworth CA, Burkitt MD, Watson AJ, Campbell BJ, Pritchard DM. 2015. Epithelial cell shedding and barrier function: a matter of life and death at the small intestinal villus tip. Vet Pathol 52:445–455. doi: 10.1177/0300985814559404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukata M, Michelsen KS, Eri R, Thomas LS, Hu B, Lukasek K, Nast CC, Lechago J, Xu R, Naiki Y, Soliman A, Arditi M, Abreu MT. 2005. Toll-like receptor-4 is required for intestinal response to epithelial injury and limiting bacterial translocation in a murine model of acute colitis. Am J Physiol Gastrointest Liver Physiol 288:G1055–G1065. doi: 10.1152/ajpgi.00328.2004. [DOI] [PubMed] [Google Scholar]
- 48.Ruemmele FM, Beaulieu JF, Dionne S, Levy E, Seidman EG, Cerf-Bensussan N, Lentze MJ. 2002. Lipopolysaccharide modulation of normal enterocyte turnover by Toll-like receptors is mediated by endogenously produced tumour necrosis factor alpha. Gut 51:842–848. doi: 10.1136/gut.51.6.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berezow AB, Ernst RK, Coats SR, Braham PH, Karimi-Naser LM, Darveau RP. 2009. The structurally similar, penta-acylated lipopolysaccharides of Porphyromonas gingivalis and Bacteroides elicit strikingly different innate immune responses. Microb Pathog 47:68–77. doi: 10.1016/j.micpath.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coats SR, Reife RA, Bainbridge BW, Pham TT, Darveau RP. 2003. Porphyromonas gingivalis lipopolysaccharide antagonizes Escherichia coli lipopolysaccharide at Toll-like receptor 4 in human endothelial cells. Infect Immun 71:6799–6807. doi: 10.1128/IAI.71.12.6799-6807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hashimoto M, Kirikae F, Dohi T, Adachi S, Kusumoto S, Suda Y, Fujita T, Naoki H, Kirikae T. 2002. Structural study on lipid A and the O-specific polysaccharide of the lipopolysaccharide from a clinical isolate of Bacteroides vulgatus from a patient with Crohn's disease. Eur J Biochem 269:3715–3721. doi: 10.1046/j.1432-1033.2002.03062.x. [DOI] [PubMed] [Google Scholar]
- 52.Cunningham MD, Seachord C, Ratcliffe K, Bainbridge B, Aruffo A, Darveau RP. 1996. Helicobacter pylori and Porphyromonas gingivalis lipopolysaccharides are poorly transferred to recombinant soluble CD14. Infect Immun 64:3601–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chilton PM, Embry CA, Mitchell TC. 2012. Effects of differences in lipid A structure on TLR4 pro-inflammatory signaling and inflammasome activation. Front Immunol 3:154. doi: 10.3389/fimmu.2012.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. 2007. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell 130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Maeshima N, Fernandez RC. 2013. Recognition of lipid A variants by the TLR4-MD-2 receptor complex. Front Cell Infect Microbiol 3:3. doi: 10.3389/fcimb.2013.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sansonetti PJ, Medzhitov R. 2009. Learning tolerance while fighting ignorance. Cell 138:416–420. doi: 10.1016/j.cell.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 57.Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, Goodman AL. 2015. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raetz CR, Reynolds CM, Trent MS, Bishop RE. 2007. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem 76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munford RS, Varley AW. 2006. Shield as signal: lipopolysaccharides and the evolution of immunity to gram-negative bacteria. PLoS Pathog 2:e67. doi: 10.1371/journal.ppat.0020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takada H, Kotani S. 1989. Structural requirements of lipid A for endotoxicity and other biological activities. Crit Rev Microbiol 16:477–523. doi: 10.3109/10408418909104475. [DOI] [PubMed] [Google Scholar]
- 62.Hansen GH, Rasmussen K, Niels-Christiansen LL, Danielsen EM. 2009. Lipopolysaccharide-binding protein: localization in secretory granules of Paneth cells in the mouse small intestine. Histochem Cell Biol 131:727–732. doi: 10.1007/s00418-009-0572-6. [DOI] [PubMed] [Google Scholar]
- 63.Huebner C, Petermann I, Lam WJ, Shelling AN, Ferguson LR. 2010. Characterization of single-nucleotide polymorphisms relevant to inflammatory bowel disease in commonly used gastrointestinal cell lines. Inflamm Bowel Dis 16:282–295. doi: 10.1002/ibd.21043. [DOI] [PubMed] [Google Scholar]
- 64.de Buhr MF, Hedrich HJ, Westendorf AM, Obermeier F, Hofmann C, Zschemisch NH, Buer J, Bumann D, Goyert SM, Bleich A. 2009. Analysis of Cd14 as a genetic modifier of experimental inflammatory bowel disease (IBD) in mice. Inflamm Bowel Dis 15:1824–1836. doi: 10.1002/ibd.21030. [DOI] [PubMed] [Google Scholar]
- 65.Lakatos PL, Kiss LS, Palatka K, Altorjay I, Antal-Szalmas P, Palyu E, Udvardy M, Molnar T, Farkas K, Veres G, Harsfalvi J, Papp J, Papp M. 2011. Serum lipopolysaccharide-binding protein and soluble CD14 are markers of disease activity in patients with Crohn's disease. Inflamm Bowel Dis 17:767–777. doi: 10.1002/ibd.21402. [DOI] [PubMed] [Google Scholar]
- 66.Richter JM, Schanbacher BL, Huang H, Xue J, Bauer JA, Giannone PJ. 2012. LPS-binding protein enables intestinal epithelial restitution despite LPS exposure. J Pediatr Gastroenterol Nutr 54:639–644. doi: 10.1097/MPG.0b013e31823a895a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kocsar LT, Bertok L, Varteresz V. 1969. Effect of bile acids on the intestinal absorption of endotoxin in rats. J Bacteriol 100:220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lichtenberger LM. 2013. Role of phospholipids in protection of the GI mucosa. Dig Dis Sci 58:891–893. doi: 10.1007/s10620-012-2530-8. [DOI] [PubMed] [Google Scholar]
- 69.Oblak A, Jerala R. 2015. The molecular mechanism of species-specific recognition of lipopolysaccharides by the MD-2/TLR4 receptor complex. Mol Immunol 63:134–142. doi: 10.1016/j.molimm.2014.06.034. [DOI] [PubMed] [Google Scholar]
- 70.Miklavcic JJ, Schnabl KL, Mazurak VC, Thomson AB, Clandinin MT. 2012. Dietary ganglioside reduces proinflammatory signaling in the intestine. J Nutr Metab 2012:280286. doi: 10.1155/2012/280286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.