ABSTRACT
Antimicrobial peptides offer potential as novel therapeutics to combat food spoilage and poisoning caused by pathogenic and nonpathogenic bacteria. Our previous studies identified the peptide human beta-defensin 3 (HBD3) as a potent antimicrobial agent against a wide range of beer-spoiling bacteria. Thus, HBD3 is an excellent candidate for development as an additive to prevent food and beverage spoilage. To expand the repertoire of peptides with antimicrobial activity against bacteria associated with food spoilage and/or food poisoning, we carried out an in silico discovery pipeline to identify peptides with structure and activity similar to those of HBD3, focusing on peptides of plant origin. Using a standardized assay, we compared the antimicrobial activities of nine defensin-like plant peptides to the activity of HBD3. Only two of the peptides, fabatin-2 and Cp-thionin-2, displayed antimicrobial activity; however, the peptides differed from HBD3 in being sensitive to salt and were thermostable. We also compared the activities of several ultrashort peptides to that of HBD3. One of the peptides, the synthetic tetrapeptide O3TR, displayed biphasic antimicrobial activity but had a narrower host range than HBD3. Finally, to determine if the peptides might act in concert to improve antimicrobial activity, we compared the activities of the peptides in pairwise combinations. The plant defensin-like peptides fabatin-2 and Cp-thionin-2 displayed a synergistic effect with HBD3, while O3TR was antagonistic. Thus, some plant defensin-like peptides are effective antimicrobials and may act in concert with HBD3 to control bacteria associated with food spoilage and food poisoning.
IMPORTANCE Food spoilage and food poisoning caused by bacteria can have major health and economic implications for human society. With the rise in resistance to conventional antibiotics, there is a need to identify new antimicrobials to combat these outbreaks in our food supply. Here we screened plant peptide databases to identify peptides that share structural similarity with the human defensin peptide HBD3, which has known antimicrobial activity against food-spoiling bacteria. We show that two of the plant peptides display antimicrobial activity against bacteria associated with food spoilage. When combined with HBD3, the peptides are highly effective. We also analyzed the activity of an easily made ultrashort synthetic peptide, O3TR. We show that this small peptide also displays antimicrobial activity against food-spoiling bacteria but is not as effective as HBD3 or the plant peptides. The plant peptides identified are good candidates for development as natural additives to prevent food spoilage.
INTRODUCTION
Antimicrobial peptides (AMPs) are short oligopeptides ranging from 4 to 100 amino acids that display antimicrobial activity against a broad range of pathogens, including Gram-positive and -negative bacteria, fungi, viruses, protists, parasites, and even insects (1). AMPs form part of the innate immune response, acting as a first line of defense and assisting in the rapid elimination of invading pathogens (2). The peptides can also invoke adaptive immune responses as a secondary response (3). The host range and specificity of antimicrobial activity are unique to each peptide.
AMPs can be linear or cyclic and are generally grouped into specific classes based on their biological and physical properties. The major classes of peptides are (i) anionic, (ii) cationic, (iii) cationic and amphipathic, and (iv) α-helical (4, 5). More than 5,000 AMPs have been identified from a wide range of organisms, including bacteria, plants, invertebrates, and vertebrates, through biochemical characterization of purified peptides or through in silico analyses of peptides based on gene sequence or on structural and/or physicochemical similarities to known AMPs (6–11). Rational combinatorial peptide design, coupled with high-throughput screening, has also been employed to design novel synthetic AMPs (12).
The majority of AMPs elicit antimicrobial activity through interaction with cellular membranes, and several models have been proposed to describe their mode of action (13). The barrel-stave model proposes that positively charged peptides interact with the cellular membranes through interaction with polar phospholipid head groups. Once a critical concentration is reached, the peptides self-aggregate and are inserted into the membrane perpendicularly to create transmembrane pores lined with peptides, resulting in the disruption of ionic and proton gradients (14). A related model, the toroidal model, hypothesizes that at a critical concentration of peptides, the strain induces membranes to curve inward, creating donut-like pores lined with peptides and phospholipid head groups (15). An alternative model, the carpet model, proposes that small areas of membrane become coated with peptides, creating hydrophilic pockets, which disrupt the lipid bilayer (16).
Besides interacting with the membrane, some peptides exert antimicrobial activity through interacting with other cellular targets, inhibiting cellular processes such as nucleic acid and protein synthesis and disrupting multienzyme complexes such as the electron transport chain and cell wall synthesis complexes (17–19).
One of the most important categories of AMPs is the defensins, a class of small cationic, amphipathic peptides that have been identified in both vertebrates and invertebrates (20). Like other cationic peptides, defensins rapidly and efficiently permeabilize membrane bilayers (21, 22) and also appear to inhibit bacterial cell wall biosynthesis (19, 23). In mammals, the peptides are produced in cells and tissues such as salivary glands and Paneth cells and are secreted into tissues that come into contact with invading microbes, for example, the buccal cavity and the intestinal lining (24, 25). Defensins are also produced in abundance in plants, mainly in seeds, where they are constitutively expressed in storage organs or are induced either systemically or locally in response to attack by pathogenic microorganisms (26, 27).
Defensins generally range from 45 to 54 amino acids, and although the peptides differ greatly at the amino acid sequence level, they possess many conserved structural and biochemical features, such as twisted antiparallel beta-sheets, a short amphipathic α-helix, conserved cysteine residues, an overall positive net charge, and a net negative hydrophobicity score (28). Based on the pattern of intramolecular disulfide bonds formed by the cysteine residues, defensins are classified as alpha- or beta-defensins (29). A third type of cyclic defensins, named theta, has so far been found only in Old World rhesus macaque monkeys (30). While beta-defensins are found in vertebrates, invertebrates, and plants, alpha-defensins are confined to mammals and generally lack the short α-helix (31). Defensins can have broad or narrow antimicrobial activity and are effective against Gram-positive and -negative bacteria, fungi, and some viruses.
The prototypical defensin is human beta-defensin-3 (HBD3; encoded by the DEFB103A gene), a broad-spectrum antimicrobial with activity against Gram-positive and -negative bacteria. The peptide is either expressed constitutively or induced upon bacterial challenge (32). Due to its broad specificity, HBD3 is a good candidate for development as an antimicrobial agent, particularly in light of the increasing instances of bacterial resistance to conventional antibiotics.
We recently demonstrated that HBD3 displays antimicrobial activity against beverage-spoiling bacteria, such as Lactobacillus, Pectinatus, and Pediococcus spp., and that when expressed in situ in yeasts, HBD3 can provide protection against beer-spoiling bacteria during industrial fermentation (33). To expand the repertoire of peptides with antimicrobial activity against bacteria associated with food spoilage and food poisoning, we carried out an in silico screen to search for peptides with structures and activities similar to those of HBD3. Since plants produce and store in seeds a wide and diverse group of antimicrobial peptides, we initially focused our analysis on defensin-like peptides of plant origin. We also included in our study several nondefensin AMPs native to plants and/or of a synthetic origin. The antimicrobial activities of the peptides against a range of Gram-positive and -negative food-spoiling bacteria were examined in parallel and were compared to that of the prototype, HBD3.
MATERIALS AND METHODS
Peptide discovery and bioinformatic analysis.
Potential candidate antimicrobial peptides from plants were mined from the PhytAMP database (http://phytamp.pfba-lab-tun.org/main.php), which contains information on 273 plant antimicrobials. The database was queried for defensin and defensin-like molecules that had been shown previously to have antimicrobial activity. Peptide sequences for the candidate peptides identified were retrieved from the UniProt database (http://www.uniprot.org/), and any containing the “knottin” peptide motif and scorpion toxin-like motifs were rejected. Where structures were available, they were retrieved from the Protein Data Bank server (http://www.rcsb.org/pdb/home/home.do). Candidate peptides with no experimentally determined structure were modeled using the protein structure prediction program Robetta (http://robetta.bakerlab.org/). All modeling was conducted using default protocols and parameters (34, 35). Template structures against which modeling was conducted were identified using the Ginzu PDB template identification and domain prediction protocol. Templates for modeling were chosen using an all-atom energy-based selection process. Structures were modeled using the Structure Prediction Tool (robetta.bakerlab.org; Domain Parsing & 3-D Modeling service), set to default parameters. The template and alignment most enriched in the lowest-energy population of all-atom refined models was then selected.
Candidate peptide structures were aligned to, and compared with, the empirically determined structure of human beta-defensin 3 (HBD3; PDB identifier [ID] 1KJ6). Alignment was carried out separately against the helix and beta-sheet regions of the peptides. Alignment root mean square deviation (RMSD) scores of 9.0 for the beta-sheet region and 4.5 for the helix region were used as cutoffs. The peptides were visualized using the PyMOL software suite (PyMOL Molecular Graphics System, version 1.7.4; Schrödinger, LLC). The distribution of charged and hydrophobic residues of the candidate peptides was visually compared to that of HBD3. Peptide net charge, hydrophilicity, and isoelectric point were calculated using Bachem's and Innovagen's peptide property calculation tools. The average hydrophobicity was calculated using the online GRAVY (grand average of hydropathy) tool (http://www.gravy-calculator.de/index.php) or was determined manually (36).
Peptide synthesis.
Synthetic antimicrobial peptides were chemically synthesized at a purity of ≥70% (GL Biochem Ltd., Shanghai, People's Republic of China). All peptides contain a C-terminal amide group. For some peptides, an acetyl group (CH3CO) was added to the N terminus. The peptides were dissolved in 100 mM acetic acid at 20 mg ml−1 and were stored frozen at −70°C. The stocks were diluted in phosphate-buffered saline (PBS, comprising 10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, and 2.6 mM KCl [pH 7.4]; Oxoid, Ltd., Basingstoke, United Kingdom) or the appropriate buffer immediately before use.
Strains and growth conditions.
Lactobacillus brevis L1055 and Pediococcus (Lactobacillus) dextrinicus DSM20293 (from the Deutsche Sammlung von Mikroorganismen and Zellkulturen, Leibniz Institute, Braunschweig, Germany) were grown in MRS broth (Lab M Ltd., Lancashire, United Kingdom) at 30°C under aerobic and microaerobic conditions, respectively. Staphylococcus aureus strain Newman, Salmonella enterica subsp. enterica serovar Typhimurium SL1344, Escherichia coli MG1655, and Pseudomonas aeruginosa NCIMB 950, sourced from the Moyne Institute Culture Collection, Trinity College Dublin, Dublin, Ireland, were grown under aerobic conditions at 37°C in tryptic soy broth (TSB; Sigma Chemical Co., Poole, United Kingdom). Bacillus cereus NCIMB 1526 (Moyne Institute Culture Collection) was grown under aerobic conditions at 30°C in L broth (Thermo Fisher Scientific, MA, USA). Saccharomyces cerevisiae strain S-1502B was cultured under aerobic conditions in yeast extract-peptone-dextrose (YEPD) broth at 30°C (37).
Antimicrobial activity assay.
The antimicrobial activities of selected peptides were assessed using a standard assay for defensin (33, 38, 39). Unless otherwise stated, all media and solutions were prewarmed to the permissive temperature for strain growth. Bacterial cultures were diluted to an optical density at 600 nm (OD600) of ∼0.1 to 0.2 in 10 ml of medium and were grown to mid-exponential phase (OD600, ∼0.5). The cultures were diluted in PBS to a final concentration of 2.0 × 103 to 4.5 × 103 cells ml−1. An initial high-throughput spectrophotometry-based screening assay was conducted as follows. AMPs, at various concentrations, were added to 96-well plates in 50 μl of PBS. To this 50 μl, bacteria were added. As a control, the bacterial cultures were added to PBS with no added peptides. The microtiter plate was incubated for 1.5 h at either 30°C or 37°C depending on the strain growth conditions. Postchallenge, 100 μl of 2× medium was added to each well. The plates were incubated at the strain permissive temperature, and the absorbance at a λ of 540 nm was measured immediately (0 h) and following a 24-h incubation for Staphylococcus aureus Newman, Salmonella enterica, E. coli, Pseudomonas aeruginosa, and Bacillus cereus 1526, after 48 h for Lactobacillus brevis, and after 72 h for Pediococcus (Lactobacillus) dextrinicus DSM20293.
For a more quantitative assessment of AMP activity, a spread plate assay was performed as follows. Briefly, 100 μl of AMPs, at various concentrations in prewarmed PBS, were mixed in a 96-well microtiter plate with 100 μl of L. brevis, diluted to a concentration of 4.5 × 103 cells ml−1 in prewarmed PBS. As a control, the diluted bacterial culture was added to prewarmed PBS with no added peptides. Following incubation for 1.5 h at 30°C, 100 μl from each well was spread onto an MRS agar plate. Surviving colonies were counted after 48 h of incubation at 30°C. To exclude any loss of viability or bacterial growth during the in vitro assay, bacterial CFU were measured immediately after dilution in PBS (without added peptides) and after incubation for 1.5 h. No observed growth or loss of viability was observed during the 1.5 h of incubation in PBS in the absence of added peptides (data not shown). Bacterial survival following AMP treatment was assessed relative to the survival of untreated control samples and was expressed as a percentage of control survival, taken as 100%. The concentration of a peptide required to inhibit 50% or 95% of bacteria (IC50 or IC95, respectively) under the experimental conditions was determined.
To examine the thermostability of AMPs, the peptides, dissolved in PBS to the IC95 in a volume of 150 μl, were first incubated at 100°C for 10 min and then allowed to cool for 30 min at a controlled ambient temperature (25°C) prior to use in the AMP assay. Alternatively, the peptide solution was rapidly cooled by plunging it into an ice bath at 4°C. Likewise, the influence of the salt concentration on antimicrobial activity was assessed by conducting the AMP assay in phosphate buffer (PB, comprising10 mM Na2HPO4, 1.8 mM KH2PO4, and 2.6 mM KCl [pH 7.4]) containing different NaCl concentrations (0 to 150 mM). Bacterial survival was enumerated following incubation on MRS agar plates for 48 h.
Kinetics of action and synergism of peptides.
The kinetics of action of the antimicrobial peptides were determined as follows. Briefly, 100 μl of AMPs at the IC95 was incubated with 100 μl of L. brevis at a concentration of 4.5 × 103 cells ml−1 for increasing time intervals. Bacterial survival was enumerated following incubation on MRS agar plates for 48 h.
To examine the effects of combinations of peptides on bacterial survival, the assay was repeated using AMPs in different combinations at ≤IC50 or IC95. Bacterial survival was enumerated following incubation on MRS agar plates for 48 h. A combination index (CI) score was calculated for each combination of peptides, using the time required to achieve IC50 to IC90 as the measured effect. The CI score was calculated using the formula [(time to reach ICn with peptide 1+ peptide 2)/(time to reach ICn with peptide 1)] + [(time to reach ICn with peptide 1+ peptide 2)/(time to reach ICn with peptide 2)]. A CI score of <1.0 indicates synergistic activity of the pair of peptides, and a CI score of 1.0 indicates additive effects, while a CI score of >1.0 indicates antagonistic effects (40).
Statistical analysis.
Experiments were conducted using three technical replicates and were repeated on at least two to three independent occasions unless otherwise stated. The data were analyzed using Microsoft Excel and GraphPad Prism (version 5.0) software. P values were determined using a Student t test or one-way ANOVA (analysis of variance) statistical tests. P values of <0.05 were considered to be statistically significant.
RESULTS
More than 350 defensins and defensin-like peptides have been identified to date and have been designated defensins based on structural characteristics, antimicrobial activities, and biochemical characterization (41). Most of these peptides have been characterized individually, but rarely have they been directly compared to each other or their combined effects assessed in a defined assay. The reported antimicrobial specificities and levels of effectiveness of the peptides often differ from study to study, largely due to the variety of methods used to determine inhibitory concentrations (IC) or lethal concentrations (LC) of the peptides. Furthermore, the lack of conservation at the primary sequence level hinders the discovery of new defensin peptides.
To expand the repertoire of peptides with antimicrobial activity against bacteria associated with food spoilage and/or food poisoning, we undertook an initial selection screen to identify peptides with structures and activities similar to those of HBD3. The screening criteria were as follows: (i) the peptides were of plant origin; (ii) peptides containing undesirable motifs associated with cytotoxic and toxin activities were excluded; (iii) the predicted structures of the peptides were compared to that of HBD3; and (iv) antimicrobial activity and host range, as determined in previous studies, were scored. Initially, 52 plant defensin-like peptides were selected. The list of peptides was further reduced to 33 when peptides with undesirable motifs with possible toxic effects were excluded. A further seven peptides were excluded due to lack of structural alignment to HBD3. Of the remaining peptides, structural modeling revealed that Cp-thionin-2 (KT43C), fabatin (LL44C), and mung bean defensin PDF-1 (KT46C) were most closely related structurally to HBD3 and displayed similar placement of hydrophobic and cationic residues (Fig. 1). Three other peptides, hevein-like AMP (QQ42G), antimicrobial seed protein (AC34H), and TK-AMP-D1 (R46C) (see Fig. S1 in the supplemental material), share some structural features with HBD3, such as the stacked antiparallel beta-sheets, and were carried forward for further analysis based on reported antibacterial activity. Added to the list for analysis was the cyclic peptide circulin (C30P) (see Fig. S1 in the supplemental material), which we define here as defensin-like because it contains six cysteine residues that putatively form three disulfide bonds in the molecule. Two defensin-like peptides, gamma-2-purothionin (K47C) from wheat grain and Rs-AFP-1 (Q23N) from radishes, were initially excluded based on the selection criteria, because they possess knottin motifs. However, both peptides are structurally very similar to HBD3 (see Fig. S1) (42) and were thus included for analysis. The antibacterial activities of the latter two peptides have not been characterized previously. The peptides are referred to below by their IDs (i.e., identifier names generated from the amino acid sequence [e.g., in “LL44C,” “LL” refers to the first two amino acids, “44” to the length of the peptide, and “C” to the last amino acid]) (Table 1).
FIG 1.
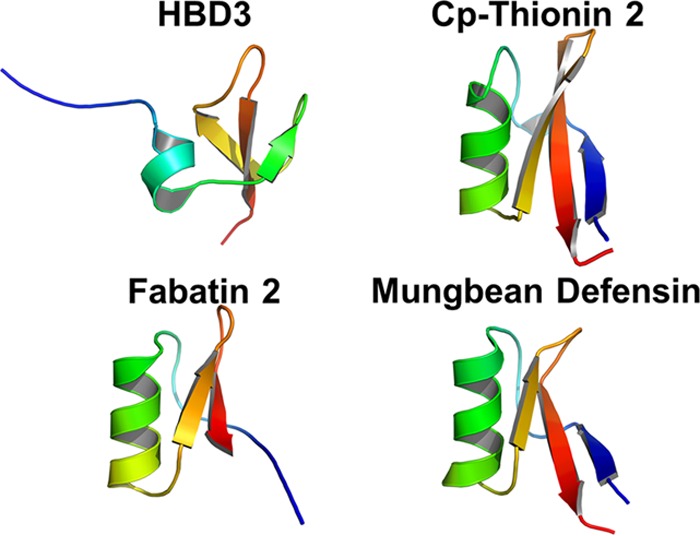
Structures of defensin peptides. Defensin-like peptides were modeled using the Robetta protein structure prediction tool. Candidate peptide structures were aligned to, and compared with, the empirically determined structure of human beta-defensin 3 (HBD3; PDB ID 1KJ6) using the PyMOL Molecular Graphics System. Peptides displaying structures most similar to HBD3 are shown. The peptides are colored blue to red from the N to the C terminus.
TABLE 1.
List of peptides
| Peptide | ID | Source organism | Amino acid sequence | Source or reference |
|---|---|---|---|---|
| Human beta-defensin 3 | HBD3 | Human | GIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK | 33 |
| Cp-Thionin-2 | KT43C | Vigna unguiculata | KTCMTKKEGWGRCLIDTTCAHSCRKYGYMGGKCQGITRRCYCLLNC | 50 |
| Fabatin-2 | LL44C | Vicia faba | LLGRCKVKSNRFNGPCLTDTHCSTVCRGEGYKGGDCHGLRRRCMCLC | 51 |
| Mung bean defensin PDF-1 | KT46C | Vigna radiata | KTCENLANTYRGPCFTTGSCDDHCKNKEHLRSGRCRDDFRCWCTRNC | 52 |
| TK-AMP-D1.1 | R46C | Triticum kiharae | RDCESDSHKFHGACFSDTNCANVCQTEGFAGKCVGVQRHCHCTKDC | 53 |
| Hevein-like AMP | QQ42G | Euonymus europaeus | QQCGRQAGNRRCANNLCCSQYGYCGRTNEYCCTSQGCQSQCRRCG | 54 |
| Antimicrobial seed protein | AC34H | Phytolacca americana | ACIKNGGRCVASGGPPYCCSNYCLQIAGQSYGVCKKH | 55 |
| Circulin-A | C30P | Chassalia parviflora | GIPCGESCVWIPCISAALGCSCKNKVCYRN | 56 |
| Gamma-2-purothionin | K47C | Triticum aestivum | KVCRQRSAGFKGPCVSDKNCAQVCLQEGWGGGNCDGPFRRCKCIRQC | 57 |
| Rs-AFP-1 | Q23N | Raphanus sativus | QKLCERPSGTWSGVCGNNNACKN | 58 |
| JC-Pep-7 | KV5K | Jatropha curcas | KVFLGLK | 59 |
| Acetylated JC-Pep-7 | Ac-KV5K | Jatropha curcas | Acetyl-KVFLGLK | This study |
| Synthetic tetrapeptide | O3TR | Synthetic | OOWW | 44 |
| Acetylated synthetic tetrapeptide | Ac-O3TR | Synthetic | Acetyl-OOWW | This study |
In addition to including defensin-like peptides, we included in the analysis a small linear heptapeptide (JC-Pep-7) from Jatropha curcas and a synthetic tetrapeptide (O3TR) recently reported as displaying antimicrobial activity (43, 44) (Table 1). To modify the net charges of the latter two peptides, an acetyl group was added to the NH2 terminus during peptide synthesis. The peptides were chemically synthesized (Table 1), and their physicochemical properties were compared (Table 2). The net charges of the peptides ranged from −0.2 to 11.6; HBD3 was the most positively charged peptide. Isoelectric points ranged from 6.9 to 14, with most in the range of 8 to 10.7. The majority of peptides displayed negative hydropathy scores (Table 2). The defensin-like peptides contain 3 to 8 cysteine residues, while the small linear peptides contain none.
TABLE 2.
Physicochemical properties of peptides
| Peptide | No. of cysteines | Net charge | Isoelectric point | Avg hydropathy score |
|---|---|---|---|---|
| Human beta-defensin 3 | 6 | 11.6 | 10.72 | −0.7 |
| Cp-thionin-2 | 8 | 7.5 | 9.07 | −0.439 |
| Fabatin-2 | 8 | 6.7 | 8.87 | −0.423 |
| Gamma-2-purothionin | 8 | 6.5 | 8.82 | −0.596 |
| Hevein-like AMP | 10 | 5.3 | 8.3 | −1.038 |
| Antimicrobial seed protein | 6 | 4.7 | 8.72 | −0.1 |
| Mung bean defensin PDF-1 | 8 | 3.7 | 8.12 | −1.174 |
| JC-Pep-7 | 0 | 3 | 14 | 0.914 |
| Rs-AFP-1 | 3 | 2.8 | 8.88 | −0.597 |
| Circulin-A | 6 | 2.6 | 8.13 | 0.416 |
| Acetylated JC-Pep-7 | 0 | 2 | 14 | 0.914 |
| Synthetic tetrapeptide | 0 | 1 | 14 | 0.19 |
| Acetylated synthetic tetrapeptide | 0 | 0 | 7 | 0.19 |
| TK-AMP-D1.1 | 8 | −0.2 | 6.92 | −0.613 |
Kinetics of action of AMPs.
To identify AMPs showing antimicrobial activity in actively growing bacterial cultures, such as might be encountered in food or beverage preparations, peptides in the concentration range of 40 to 50 μg ml−1 were initially incubated with L. brevis in vitro for 1.5 h and were then further incubated in growth medium until control cultures, in the absence of AMPs, reached stationary phase. Of the 14 AMPs tested, only HBD3, LL44C, KT43C, and O3TR showed antimicrobial activity against L. brevis under the chosen experimental conditions and concentration range (Fig. 2) (see Materials and Methods). The peptides were subsequently tested for antimicrobial activity against a range of Gram-positive and -negative bacteria, chosen for their importance as contaminants of food and beverages, as well as several pathogenic bacterial species often associated with food poisoning (Table 3). S. aureus was included as a positive control, because this bacterium is regularly used for the assessment of AMP activity. By far, HBD3 showed the broadest host range under the chosen experimental conditions, displaying activity against the Gram-positive bacteria L. brevis, P. dextrinicus, S. aureus, and B. cereus and against the Gram-negative bacterium P. aeruginosa but not against E. coli or Salmonella enterica. Peptides LL44C and KT43C displayed activity against L. brevis, P. dextrinicus, and P. aeruginosa but not against S. aureus, B. cereus, E. coli, or Salmonella enterica. The synthetic tetrapeptide O3TR displayed activity only against L. brevis and P. dextrinicus, while an acetylated form of the same peptide had no activity. The rest of the peptides were ineffectual against the range of bacteria tested under the chosen experimental conditions. None of the peptides showed antimicrobial activity against the yeast Saccharomyces cerevisiae (data not shown).
FIG 2.
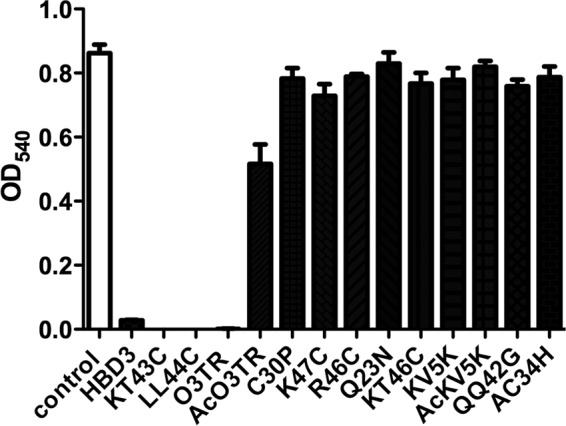
Antimicrobial activities of AMPs in actively growing bacterial cultures. L. brevis (2 × 103 to 4.5 × 103 cells ml−1) in 50 μl PBS was incubated with 50 μl of peptide in the concentration range of 40 to 50 μg ml−1 for 1.5 h in a 96-well microtiter plate. One hundred microliters of 2× medium was then added, and cultures were incubated at 30°C for 48 h. Bacterial survival was assessed by measuring the absorbance at a λ of 540 nm. L. brevis cultures incubated in the absence of peptides served as the control (open bar). Error bars represent the standard errors of the means from three independent experiments. Each experiment was conducted with triplicate samples.
TABLE 3.
IC95 of peptides
| Bacterium | Gram stain | IC95 (μg ml−1)a |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HBD3 | LL44C | KT43C | O3TR | AcO3TR | C30P | KT46C | K47C | R46C | Q23N | KV5K | AcKV5K | QQ42G | AC34H | ||
| L. brevis | Positive | 1.3 | 10.3 | 12.7 | 14.4 | >40 | >50 | >40 | >50 | >50 | >50 | >50 | >50 | >40 | >45 |
| P. dextrinicus | Positive | 2.1 | 11.2 | 10.7 | 17.3 | >40 | >50 | >40 | >50 | >50 | >50 | >50 | >50 | >40 | >45 |
| S. aureus | Positive | 12.5 | >45 | >40 | >40 | >40 | >50 | >40 | >50 | >50 | >50 | >50 | >50 | >40 | >45 |
| B. cereus | Positive | 5.8 | >45 | >40 | >40 | >40 | ND | >40 | ND | ND | ND | >50 | >50 | >40 | >45 |
| S. enterica | Negative | >50 | >45 | >40 | >40 | >40 | >50 | >40 | >50 | >50 | >50 | >50 | >50 | >40 | >45 |
| E. coli | Negative | >50 | >45 | >40 | >40 | >40 | ND | >40 | ND | ND | ND | >50 | >50 | >40 | >45 |
| P. aeruginosa | Negative | 3.5 | 10.3 | 21.4 | >40 | >40 | ND | >40 | ND | ND | ND | >50 | >50 | >40 | >45 |
ND, not determined.
Peptides displaying antimicrobial activity were further tested at lower concentrations to determine the concentration required for 95% growth inhibition after 48 h of incubation (IC95:48 h). For L. brevis grown in continuous culture for 48 h in the presence of peptides, the IC95:48 h of HBD3, LL44C, KT43C, or O3TR was 1.3, 10.3, 12.7, or 14.4 μg ml−1, respectively, and the IC95:48 h against P. dextrinicus was in the same range (Table 3).
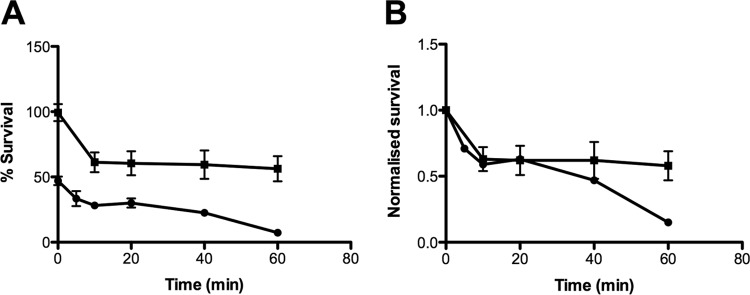
To examine the kinetics of inhibition in greater detail, the peptides, in various concentrations, were incubated in vitro for 1.5 h with L. brevis as a model bacterium. Following the incubation, the bacteria were immediately plated onto MRS agar without the added extended growth period (48 h) in growth medium. The concentration of peptide required to obtain 50% or 95% inhibition of growth (IC50:1.5 h or IC95:1.5 h, respectively) was determined (Fig. 3). Following the 1.5-h in vitro incubation, the IC95:1.5 h for HBD3, LL44C, KT43C, or O3TR was determined to be 2.5, 13.7, 21.4, or 60 μg ml−1, respectively, while the IC50:1.5 h was 2.2, 7.3, 10.5, or 19.6 μg ml−1, respectively. Comparison of the IC95:1.5 h (Fig. 3) to the IC95:48 h (Table 3) suggests that inhibition is immediate for HBD3, LL44C, and KT43C, while for O3TR, prolonged incubation is required to achieve the same effect as the other peptides. We explored this apparent discrepancy between IC95:1.5-h and IC95:48-h values for O3TR by examining the survival kinetics of L. brevis following incubation with O3TR at the two peptide concentrations (Fig. 4A). At the higher concentration of peptide (IC95:1.5 h; 60 μg ml−1), inhibition was almost instantaneous, with the percentage of survival decreasing to 50% in <1 min. Survival dropped to 28% after 10 min and to 7% by 60 min. At the lower concentration of peptide (IC95:48 h, 14.4 μg ml−1), survival dropped to 60% after 10 min and remained at this level when the incubation period was extended to 60 min. Thus, at both peptide concentrations, the survival kinetics appear to be biphasic, with an initial rapid decrease in cell survival to approximately 60% by 10 min, followed by a lower rate of killing thereafter (Fig. 4B).
FIG 3.
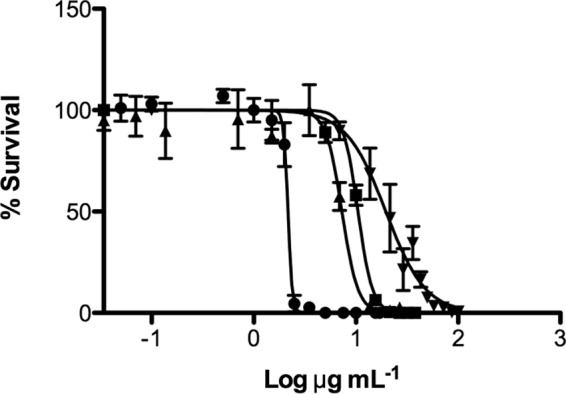
Inhibitory activities of HBD3, KT43C, LL44C, and O3TR. Cultures of L. brevis (4.5 × 103 cells ml−1; 100 μl) were mixed with increasing concentrations of peptides (100 μl). Following incubation for 1.5 h at 30°C, 100 μl of each sample was added to MRS agar and was incubated for 48 h at 30°C. Bacterial survival was expressed as a percentage of the survival of untreated control cultures, taken as 100%. Error bars represent the standard errors of the means from two independent experiments. Each experiment was conducted with triplicate samples. Symbols: ●, HBD3; ■, KT43C; ▲, LL44C; ▼, O3TR. Lines were generated by nonlinear regression analysis of the data. R2 values were 0.9781 (●), 0.9836 (■), 0.9026 (▲), and 0.9172 (▼).
FIG 4.
Comparison of antimicrobial activities of two concentrations of O3TR. (A) O3TR at the IC95:1.5 h (60 μg ml−1) (●) or at the IC95:48 h (14.4 μg ml−1) (■) in 100 μl was incubated with 100 μl of L. brevis (4.5 × 103 cells ml−1) for increasing time intervals (0 to 60 min). Following incubation for 1.5 h at 30°C, 100 μl of each sample was added to MRS agar and was incubated for 48 h at 30°C. Bacterial survival was expressed as a percentage of the survival of untreated control cultures, taken as 100%. Error bars represent the standard errors of the means from 3 to 5 independent experiments. Each experiment was conducted with triplicate samples. (B) The data presented in panel A were normalized relative to the percentage of bacterial survival at 0 min (1.0). Error bars represent the standard errors of the means.
With these facts in mind, the antimicrobial activities of the four peptides were directly compared at the IC95:1.5 h (Fig. 5). As observed in Fig. 4A, O3TR at the IC95:1.5 h resulted in an instantaneous 53% reduction in survival, while 50% reduction was achieved after 7 min with HBD3. Both KT43C and LL44C displayed similar kinetics of inhibition, achieving 50% inhibition by 40 and 44 min, respectively.
FIG 5.
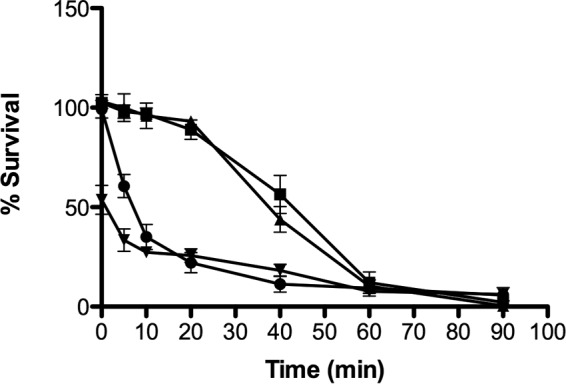
Kinetics of action of the antimicrobial peptides. One hundred microliters of a peptide (HBD3 [●], KT43C [▲], LL44C [■], or O3TR [▼]) at the IC95:1.5 h was incubated with 100 μl of L. brevis (4.5 × 103 cells ml−1) for increasing times (0 to 90 min). Following incubation, 100 μl of each sample was added to MRS agar and was incubated for 48 h at 30°C. Bacterial survival was calculated as a percentage of the survival of the untreated bacterial sample. Error bars represent the standard errors of the means for three independent experiments, with each experiment conducted with triplicate samples.
Thermostability and salt tolerance of peptides.
To be effective as antimicrobials in food and beverages, it may be advantageous in some cases (for example, bread making) for the peptides to be thermostable. To examine this, peptides at the IC95:1.5 h were incubated at 100°C for 10 min and were then cooled at 25°C prior to addition to L. brevis cultures (Fig. 6A). The peptide-bacterium mixture was then incubated for 1.5 h prior to being plated for enumeration. The LL44C and O3TR peptides were thermostable, while KT43C was slightly inhibited (approximately 18%) by the pretreatment. However, the antimicrobial activity of HBD3 was severely affected by the pretreatment, showing a 70% reduction. The method of cooling after the heat treatment did not alter the activities of the peptide, as evidenced by the fact that rapid cooling by plunging the mixture into an ice bath generated similar results (data not shown).
FIG 6.
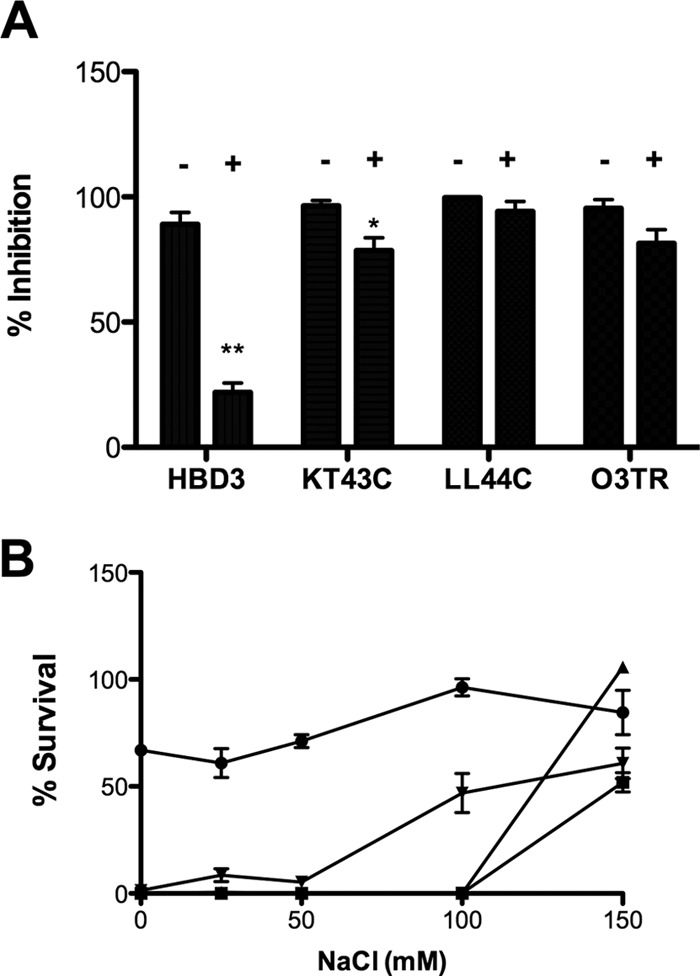
Thermostability and salt sensitivity of antimicrobial peptides. (A) The antimicrobial peptide HBD3, KT43C, LL44C, or O3TR (100 μl) at the IC95:1.5 h was heated to 100°C for 10 min (+) and then cooled to room temperature before addition to 100 μl of L. brevis (4.5 × 103 cells ml−1). (B) The peptide HBD3 (1.5 μg ml−1) (●), KT43C (10.5 μg ml−1) (■), LL44C (5.5 μg ml−1) (▲), or O3TR (14.4 μg ml−1) (▼) and L. brevis (4.5 × 103 cells ml−1) were diluted in PB containing different NaCl concentrations. Following incubation for 1.5 h at 30°C, 100 μl of each sample was added to MRS agar and was incubated for 48 h at 30°C. The percentage of inhibition (A) or of survival (B) was calculated relative to the level for untreated bacterial cultures, taken as 100%. Error bars represent the standard errors of the means for three independent experiments, with each experiment conducted with triplicate samples. **, P < 0.001; *, P < 0.05. For the percentage of survival (B), peptides showed values significantly different from the HBD3 values (for LL44C and O3TR, P < 0.001; for KT43C, P < 0.05) at ≤100 mM NaCl.
To determine if the peptide activities are influenced by the salt concentration, the AMP assay was repeated in PB containing different concentrations of NaCl and using the IC50:1.5 h or lower for the peptides (Fig. 6B). The antimicrobial activities of KT43C, LL44C, and O3TR were extremely sensitive to salt concentrations. Complete inhibition was achieved by KT43C and LL44C at NaCl concentrations of ≤100 mM and by O3TR at ≤50 mM. On the other hand, the activity of HBD3 was less affected by decreasing the salt concentration: there was a slight decrease (approximately 20%) in activity at NaCl concentrations of ≤50 mM. The same trends in salt sensitivity for all four peptides were observed when the AMP concentrations were varied (data not shown).
Antimicrobial activity using peptide combinations.
To determine if the peptides interact with each other to exert additive or reductive effects on their individual antimicrobial activities, L. brevis was incubated with each peptide individually or in combination for increasing time intervals. Based on the kinetics of action of each peptide (Fig. 5), a suboptimal concentration of each peptide, sufficient to assay its activity, was chosen in order to allow the quantification of combined effects. Accordingly, combinatorial experiments were carried out with HBD3 and O3TR at concentrations lower than the IC50:1.5 h (1.5 and 14.4 μg ml−1, respectively) due to their rapid kinetics, while LL44C and KT43C were maintained at the IC95:1.5 h (13.6 and 21.4 μg ml−1, respectively), due to their slow kinetics of action (Fig. 5). HBD3 in combination with LL44C or KT43C resulted in a more rapid reduction in the survival of L. brevis than that achieved by either peptide alone (Fig. 7A and B). The increased rate of inhibition from the combination of the two peptides was statistically significant between 10 and 50 min for HBD3 and LL44C and between 5 and 50 min for HBD3 and KT43C. Combining LL44C and KT43C also resulted in an increased rate of inhibition (Fig. 7C). Combining the tetrapeptide O3TR with any of the three defensin peptides did not appear to increase the rate of inhibition (Fig. 7D to F), except for the combination of O3TR with KT43C, where an immediate reduction in survival at <1 min was observed (Fig. 7E). Thereafter, the rate of inhibition for the combined peptides was not statistically significant from that for either peptide alone.
FIG 7.
Interactions between antimicrobial peptides. Cultures of L. brevis (4.5 × 103 cells ml−1; 100 μl) were incubated for increasing times with the antimicrobial peptides (100 μl), either singly or in pairwise combinations (▲), at concentrations of 1.5 μg ml−1 for HBD3 (●), 13.7 μg ml−1 for LL44C (■), 21.4 μg ml−1 for KT43C (◆), and 14.4 μg ml−1 for O3TR (○). The combination of peptides in each experiment is shown in the key on the right of each panel. Following incubation for 1.5 h at 30°C, 100 μl of each sample was added to MRS agar and was incubated for 48 h at 30°C. Bacterial survival was calculated as a percentage of the survival of the control without AMP treatment (taken as 100%). Error bars represent the standard deviations from one to five independent experiments. Each experiment was conducted with triplicate samples.
To determine if the increased rate of inhibition observed with some of the combined peptides resulted from the peptides acting in synergy or in an additive manner, a combination index (CI) score was calculated using IC50:1.5 h to IC90:1.5 h as the measured effect (Table 4). Synergistic effects were observed using the peptide combinations HBD3 plus LL44C, HBD3 plus KT43C, and O3TR plus KT43C, although in the last case, synergism was evident only in the first 10 min of the reaction (Fig. 7E). Peptide LL44C in combination with either KT43C or O3TR produced an additive effect, while the combination of HBD3 and O3TR was antagonistic. Taking the findings together, the most potent effect on bacterial inhibition was observed for the combination of HBD3 with either of the plant defensin-like peptides KT43C and LL44C.
TABLE 4.
Combination index scores of peptides
| IC | CI scorea |
|||||
|---|---|---|---|---|---|---|
| HBD3 + LL44C | HBD3 + KT43C | KT43C + LL44C | HBD3 + O3TR | KT43C + O3TR | LL44C + O3TR | |
| 90% | 0.47 | 0.34 | ND | 11.36 | 0.00 | 0.99 |
| 80% | 0.46 | 0.30 | 0.99 | 5.64 | 0.00 | 1.12 |
| 70% | 0.45 | 0.38 | 1.09 | 4.27 | 0.00 | 1.20 |
| 60% | 0.45 | 0.40 | 1.12 | 3.57 | 0.39 | 1.25 |
| 50% | 0.43 | 0.44 | 1.19 | ND | ND | ND |
ND, no data.
DISCUSSION
The discovery and characterization of a vast array of small peptides displaying antimicrobial activity offer great potential for the development of novel therapeutics to combat food poisoning and spoilage caused by pathogenic and nonpathogenic bacteria, especially in light of the growing upsurge in resistance to conventional antibiotics. Our previous studies had identified the peptide human beta-defensin 3 (HBD3) as a potent inhibitor of a wide range of beer-spoiling bacteria, and expression and secretion of HBD3 by lager yeast provided prophylaxis against bacterial infection in yeast fermentations (33). Since the bacterial species implicated in beer spoilage are also associated with the spoilage of dairy-based food and other yeast-based fermented products, HBD3 is an excellent candidate for development as a food additive to prevent spoilage.
Here we set out to identify HBD3-like peptides and focused on peptides of plant origin due to the diverse array of AMPs previously identified in plants (27). Antimicrobial peptides in plants are generally stored in seeds as endospermal reserves, and humans and animals consume substantial quantities of such seeds without any known ill effects. While more than 350 defensin-like peptides have been described (41), to date the antimicrobial activity and host range of each peptide have mainly been studied individually using differing experimental conditions. This has led a variety of reported IC and often-contradictory reporting on host ranges.
We developed a screening pipeline to select candidate peptides for a comparative analysis with HBD3, employing criteria such as source of peptides, elimination of peptides containing potentially toxic motifs, structural similarity to HBD3, and prior reported antimicrobial activity. Peptides containing knottin motifs were excluded because of their similarity to scorpion toxin peptides, which have been shown to be cytotoxic to mammalian cells and therefore would not be likely candidates for development as potential food additives. Two exceptions were made to the latter criterion: peptides gamma-2-purothionin and RS-AFP-1 were included in the analysis due to their structural similarity to HBD3. By use of defined experimental conditions, the peptides were tested in parallel for antimicrobial activity against representative Gram-positive and Gram-negative food-spoiling bacteria or pathogenic bacteria associated with food poisoning.
Of the defensin-like peptides analyzed, only two, fabatin-2 (LL44C) from the broad bean, Vicia faba, and Cp-thionin (KT43C) from the cowpea, Vigna unguiculata, displayed antimicrobial activity; however, the IC95 was higher than that of HBD3. Both peptides were effective against L. brevis and Pediococcus dextrinicus (recently renamed Lactobacillus dextrinicus) but not against S. aureus or B. cereus, which were inhibited by HBD3. HBD3, KT43C, and LL44C were ineffective against the Gram-negative species E. coli and Salmonella enterica; however, all three displayed antimicrobial activity against P. aeruginosa.
The structural analysis of the peptides identified fabatin-2, Cp-thionin-2, and mung bean PDF-1 as most structurally similar to HBD3; however, we found that mung bean PDF-1 did not show antimicrobial activity under the experimental conditions chosen here. Likewise, the other peptides identified through structural similarity to HBD3 did not display antibacterial activity. A comparison of physicochemical properties of the peptides indicated that the only parameter that appeared to correlate with antimicrobial activity was overall net charge, since HBD3, Cp-thionin-2, and fabatin-2 were the top three ranking peptides based on net positive charge, while mung bean PDF-1 had a much lower net positive charge. Thus, including net charge as a selection criterion may be informative in identifying the most active peptides.
An analysis of the kinetics of the antimicrobial activity of the three defensin-like peptides indicated that HBD3 has a much higher rate of activity than either KT43C or LL44C. However, the antimicrobial activities of KT43C and LL44C were greatly influenced by salt concentrations: at NaCl concentrations of <100 mM, both peptides were more effective than HBD3. Previous studies have shown that the antimicrobial activity of HBD3 was not influenced by salt concentrations (19, 45, 46); however, the salt sensitivities of fabatin-2 and Cp-thionin-2 had not been analyzed previously. Additionally, both peptides were resistant to heat treatment, while HBD3 was sensitive, a property that might be useful if peptides are to be used in thermally processed foods. Thus, the plant peptides fabatin-2 and Cp-thionin-2 are good candidates for the prevention of food spoilage (47). Given the observed salt sensitivities of fabatin-2 and Cp-thionin-2, it is possible that antimicrobial activity might be detected for the other nonactive peptides (Table 3) by reducing the salt concentrations in the assay. Likewise it will be interesting to determine whether the host range can be expanded at lower salt concentrations.
We also included in the analysis one nondefensin plant antimicrobial peptide, JC-Pep-7, from Jatropha curcas (Barbados nut), and a synthetic peptide, named O3TR. Both peptides are small and easy to synthesize and would be more cost-effective than the longer defensin-like peptides, which range from 45 to 54 amino acids. Of these nondefensin peptides, only O3TR displayed antimicrobial activity. This peptide was originally identified as part of a rational design study to generate small cationic peptides with antimicrobial potential (43). The tetrapeptide contains two ornithine residues, which are nonnatural charged amino acids with high resistance to proteases, and two tryptophans, which have a strong preference for the membrane interface. Subsequent modifications of the peptide by modification of the N terminus with p-hydroxyl-cinnamic acid or acetic anhydride improved the antimicrobial activity of the tetrapeptide. Further modifications, such as the addition of a fatty acyl tail to the N terminus, improved the antimicrobial activity, although the reported MICs and minimum bactericidal concentrations were higher than that observed for unmodified O3TR in this study (44). In the present study, the acetylated form of the peptide was ineffectual against the range of bacteria tested. The concentration of O3TR required to achieve a 95% reduction in the bacterial load in 1.5 h of incubation was approximately 40-fold higher than the corresponding concentration of HBD3 and approximately 3- to 5-fold higher than those of LL44C and KT43C. Interestingly, O3TR displayed biphasic kinetics of antimicrobial activity. Upon addition to L. brevis, the peptide caused a rapid reduction in bacterial survival: there was an almost instantaneous 50% reduction of the bacterial load. Thereafter, the rate of bacterial killing was reduced. The initial high rate of killing was independent of the peptide concentration.
We examined the question of whether the peptides might act in concert to improve antimicrobial activity by determining the rate of bacterial killing achieved by peptides individually or in pairwise combinations. Our analysis revealed that the defensin-like plant peptides KT43C and LL44C act in synergy with HBD3 to improve the rate of bacterial killing; on the other hand, when combined with each other, the two plant peptides produced an additive effect. In combination with HBD3, the tetrapeptide O3TR was antagonistic, while it was additive with LL44C. There appear to be some synergistic effects for O3TR in combination with KT43C; however, synergism is apparent only immediately upon incubation of the peptides with L. brevis (0 to 10 min) and not thereafter.
Defensins are purported to exert antimicrobial activity through insertion into the plasma membranes of bacterial cells, leading to depolarization of the membrane and cell lysis (13, 48). Membrane insertion requires a critical minimum concentration. The very narrow concentration range required to reduce bacterial survival from 100% to <5% supports this mode of action. The interaction of defensins with the cell membrane is influenced by physiological conditions, such as localized pH (46). There is also evidence indicating that HBD3 treatment leads to disorganization of membrane-bound multienzyme machines, such as the cell wall biosynthesis complex and the electron transport chain, even in the absence of cell depolarization (19). The synergistic effects of defensin-like peptides may result from facilitation of the insertion of one defensin-like peptide into the cell membrane by another, through the formation of membrane pores with mixed types of peptides or by physiological modulation of the pH or ionic strength of the local environment. Further research will be required to tease out the synergistic mechanism.
The mode of action of the ultrasmall cationic peptides, such as O3TR, is not well understood; however, recent evidence suggests that small cationic peptides can be inserted into phospholipid bilayers, leading to disruption of essential processes, such as respiration and cell wall biosynthesis (49). Thus, similar cellular processes are disrupted by defensin and nondefensin cationic peptides. The reason for the antagonistic effect of O3TR on HBD3 activity is not understood, but this effect may point to an initial competition for binding of the peptides to the bacterial membrane.
Taken together, the data presented indicate that the plant defensin-like peptides fabatin-2 and Cp-thionin-2 are effective antimicrobials and may act in concert with HBD3 to control food-spoiling and food-poisoning bacteria. Comparative analysis of the tertiary structures of the plant peptides and HBD3 may aid in the design of synthetic peptides with enhanced antimicrobial activity.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by a grant (13/F/462) from the Department of Agriculture, Food and the Marine to U.B. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00558-16.
REFERENCES
- 1.Reddy KVR, Yedery RD, Aranha C. 2004. Antimicrobial peptides: premises and promises. Int J Antimicrob Agents 24:536–547. doi: 10.1016/j.ijantimicag.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Pasupuleti M, Schmidtchen A, Malmsten M. 2012. Antimicrobial peptides: key components of the innate immune system. Crit Rev Biotechnol 32:143–171. doi: 10.3109/07388551.2011.594423. [DOI] [PubMed] [Google Scholar]
- 3.Arnett E, Seveau S. 2011. The multifaceted activities of mammalian defensins. Curr Pharm Des 17:4254–4269. doi: 10.2174/138161211798999348. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharjya S, Ramamoorthy A. 2009. Multifunctional host defense peptides: functional and mechanistic insights from NMR structures of potent antimicrobial peptides. FEBS J 276:6465–6473. doi: 10.1111/j.1742-4658.2009.07357.x. [DOI] [PubMed] [Google Scholar]
- 5.Harris F, Dennison SR, Phoenix DA. 2009. Anionic antimicrobial peptides from eukaryotic organisms. Curr Protein Pept Sci 10:585–606. doi: 10.2174/138920309789630589. [DOI] [PubMed] [Google Scholar]
- 6.Blondelle SE, Lohner K. 2010. Optimization and high-throughput screening of antimicrobial peptides. Curr Pharm Des 16:3204–3211. doi: 10.2174/138161210793292438. [DOI] [PubMed] [Google Scholar]
- 7.Hammami R, Ben Hamida J, Vergoten G, Fliss I. 2009. PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res 37:D963–D968. doi: 10.1093/nar/gkn655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Luca M, Maccari G, Maisetta G, Batoni G. 2015. BaAMPs: the database of biofilm-active antimicrobial peptides. Biofouling 31:193–199. doi: 10.1080/08927014.2015.1021340. [DOI] [PubMed] [Google Scholar]
- 9.Pirtskhalava M, Gabrielian A, Cruz P, Griggs HL, Squires RB, Hurt DE, Grigolava M, Chubinidze M, Gogoladze G, Vishnepolsky B, Alekseev V, Rosenthal A, Tartakovsky M. 2016. DBAASP v.2: an enhanced database of structure and antimicrobial/cytotoxic activity of natural and synthetic peptides. Nucleic Acids Res 44:D1104–D1112. doi: 10.1093/nar/gkv1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waghu FH, Gopi L, Barai RS, Ramteke P, Nizami B, Idicula-Thomas S. 2014. CAMP: collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res 42:D1154–D1158. doi: 10.1093/nar/gkt1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang G, Mishra B, Lau K, Lushnikova T, Golla R, Wang X. 2015. Antimicrobial peptides in 2014. Pharmaceuticals (Basel) 8:123–150. doi: 10.3390/ph8010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rathinakumar R, Wimley WC. 2010. High-throughput discovery of broad-spectrum peptide antibiotics. FASEB J 24:3232–3238. doi: 10.1096/fj.10-157040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiesner J, Vilcinskas A. 2010. Antimicrobial peptides: the ancient arm of the human immune system. Virulence 1:440–464. doi: 10.4161/viru.1.5.12983. [DOI] [PubMed] [Google Scholar]
- 14.Bessin Y, Saint N, Marri L, Marchini D, Molle G. 2004. Antibacterial activity and pore-forming properties of ceratotoxins: a mechanism of action based on the barrel stave model. Biochim Biophys Acta 1667:148–156. doi: 10.1016/j.bbamem.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Yoneyama F, Imura Y, Ohno K, Zendo T, Nakayama J, Matsuzaki K, Sonomoto K. 2009. Peptide-lipid huge toroidal pore, a new antimicrobial mechanism mediated by a lactococcal bacteriocin, lacticin Q. Antimicrob Agents Chemother 53:3211–3217. doi: 10.1128/AAC.00209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez DI, Le Brun AP, Whitwell TC, Sani MA, James M, Separovic F. 2012. The antimicrobial peptide aurein 1.2 disrupts model membranes via the carpet mechanism. Phys Chem Chem Phys 14:15739–15751. doi: 10.1039/c2cp43099a. [DOI] [PubMed] [Google Scholar]
- 17.Brogden KA. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 18.Abedinzadeh M, Gaeini M, Sardari S. 2015. Natural antimicrobial peptides against Mycobacterium tuberculosis. J Antimicrob Chemother 70:1285–1289. doi: 10.1093/jac/dku570. [DOI] [PubMed] [Google Scholar]
- 19.Sass V, Pag U, Tossi A, Bierbaum G, Sahl HG. 2008. Mode of action of human beta-defensin 3 against Staphylococcus aureus and transcriptional analysis of responses to defensin challenge. Int J Med Microbiol 298:619–633. doi: 10.1016/j.ijmm.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Silva PM, Goncalves S, Santos NC. 2014. Defensins: antifungal lessons from eukaryotes. Front Microbiol 5:97. doi: 10.3389/fmicb.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kagan BL, Ganz T, Lehrer RI. 1994. Defensins: a family of antimicrobial and cytotoxic peptides. Toxicology 87:131–149. doi: 10.1016/0300-483X(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 22.Morgera F, Antcheva N, Pacor S, Quaroni L, Berti F, Vaccari L, Tossi A. 2008. Structuring and interactions of human beta-defensins 2 and 3 with model membranes. J Pept Sci 14:518–523. doi: 10.1002/psc.981. [DOI] [PubMed] [Google Scholar]
- 23.Sass V, Schneider T, Wilmes M, Korner C, Tossi A, Novikova N, Shamova O, Sahl HG. 2010. Human beta-defensin 3 inhibits cell wall biosynthesis in staphylococci. Infect Immun 78:2793–2800. doi: 10.1128/IAI.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salzman NH. 2010. Paneth cell defensins and the regulation of the microbiome: detente at mucosal surfaces. Gut Microbes 1:401–406. doi: 10.4161/gmic.1.6.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abiko Y, Nishimura M, Kaku T. 2003. Defensins in saliva and the salivary glands. Med Electron Microsc 36:247–252. doi: 10.1007/s00795-003-0225-0. [DOI] [PubMed] [Google Scholar]
- 26.Carvalho Ade O, Gomes VM. 2011. Plant defensins and defensin-like peptides—biological activities and biotechnological applications. Curr Pharm Des 17:4270–4293. doi: 10.2174/138161211798999447. [DOI] [PubMed] [Google Scholar]
- 27.Tam JP, Wang S, Wong KH, Tan WL. 2015. Antimicrobial peptides from plants. Pharmaceuticals (Basel) 8:711–757. doi: 10.3390/ph8040711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pazgier M, Li X, Lu W, Lubkowski J. 2007. Human defensins: synthesis and structural properties. Curr Pharm Des 13:3096–3118. doi: 10.2174/138161207782110381. [DOI] [PubMed] [Google Scholar]
- 29.Clarke DJ, Campopiano DJ. 2006. Structural and functional studies of defensin-inspired peptides. Biochem Soc Trans 34(Pt 2):251–256. doi: 10.1042/BST0340251. [DOI] [PubMed] [Google Scholar]
- 30.Conibear AC, Craik DJ. 2014. The chemistry and biology of theta defensins. Angew Chem Int Ed Engl 53:10612–10623. doi: 10.1002/anie.201402167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehrer RI, Lu W. 2012. α-Defensins in human innate immunity. Immunol Rev 245:84–112. doi: 10.1111/j.1600-065X.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 32.Bauer B, Wex T, Kuester D, Meyer T, Malfertheiner P. 2013. Differential expression of human beta defensin 2 and 3 in gastric mucosa of Helicobacter pylori-infected individuals. Helicobacter 18:6–12. doi: 10.1111/hel.12000. [DOI] [PubMed] [Google Scholar]
- 33.James TC, Gallagher L, Titze J, Bourke P, Kavanagh J, Arendt E, Bond U. 2014. In situ production of human beta defensin-3 in lager yeasts provides bactericidal activity against beer-spoiling bacteria under fermentation conditions. J Appl Microbiol 116:368–379. doi: 10.1111/jam.12382. [DOI] [PubMed] [Google Scholar]
- 34.Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D. 2013. High-resolution comparative modeling with RosettaCM. Structure 21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei J, Kim D, Kellogg E, DiMaio F, Lange O, Kinch L, Sheffler W, Kim BH, Das R, Grishin NV, Baker D. 2009. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins 77(Suppl 9):S89–S99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolboaca SD, Jäntschi L. 2008. Modelling analysis of amino acids hydrophobicity. Match Commun Math Comput Chem 60:1021–1032. [Google Scholar]
- 37.Canavan R, Bond U. 2007. Deletion of the nuclear exosome component RRP6 leads to continued accumulation of the histone mRNA HTB1 in S-phase of the cell cycle in Saccharomyces cerevisiae. Nucleic Acids Res 35:6268–6279. doi: 10.1093/nar/gkm691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higgs R, Lynn DJ, Cahalane S, Alaña I, Hewage CM, James T, Lloyd AT, O'Farrelly C. 2007. Modification of chicken avian beta-defensin-8 at positively selected amino acid sites enhances specific antimicrobial activity. Immunogenetics 59:573–580. doi: 10.1007/s00251-007-0219-5. [DOI] [PubMed] [Google Scholar]
- 39.Higgs R, Lynn DJ, Gaines S, McMahon J, Tierney J, James T, Lloyd AT, Mulcahy G, O'Farrelly C. 2005. The synthetic form of a novel chicken beta-defensin identified in silico is predominantly active against intestinal pathogens. Immunogenetics 57:90–98. doi: 10.1007/s00251-005-0777-3. [DOI] [PubMed] [Google Scholar]
- 40.Chou TC. 2010. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 41.Seebah S, Suresh A, Zhuo S, Choong YH, Chua H, Chuon D, Beuerman R, Verma C. 2007. Defensins knowledgebase: a manually curated database and information source focused on the defensins family of antimicrobial peptides. Nucleic Acids Res 35:D265–D268. doi: 10.1093/nar/gkl866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fant F, Vranken W, Broekaert W, Borremans F. 1998. Determination of the three-dimensional solution structure of Raphanus sativus antifungal protein 1 by 1H NMR. J Mol Biol 279:257–270. doi: 10.1006/jmbi.1998.1767. [DOI] [PubMed] [Google Scholar]
- 43.Bisht GS, Rawat DS, Kumar A, Kumar R, Pasha S. 2007. Antimicrobial activity of rationally designed amino terminal modified peptides. Bioorg Med Chem Lett 17:4343–4346. doi: 10.1016/j.bmcl.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 44.Laverty G, McLaughlin M, Shaw C, Gorman SP, Gilmore BF. 2010. Antimicrobial activity of short, synthetic cationic lipopeptides. Chem Biol Drug Des 75:563–569. doi: 10.1111/j.1747-0285.2010.00973.x. [DOI] [PubMed] [Google Scholar]
- 45.Harder J, Bartels J, Christophers E, Schroder JM. 2001. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J Biol Chem 276:5707–5713. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 46.Abou Alaiwa MH, Reznikov LR, Gansemer ND, Sheets KA, Horswill AR, Stoltz DA, Zabner J, Welsh MJ. 2014. pH modulates the activity and synergism of the airway surface liquid antimicrobials beta-defensin-3 and LL-37. Proc Natl Acad Sci U S A 111:18703–18708. doi: 10.1073/pnas.1422091112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Souza Cândido E, e Silva Cardoso MH, Sousa DA, Viana JC, de Oliveira-Junior NG, Miranda V, Franco OL. 2014. The use of versatile plant antimicrobial peptides in agribusiness and human health. Peptides 55:65–78. doi: 10.1016/j.peptides.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 48.Bechinger B, Salnikov ES. 2012. The membrane interactions of antimicrobial peptides revealed by solid-state NMR spectroscopy. Chem Phys Lipids 165:282–301. doi: 10.1016/j.chemphyslip.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 49.Wenzel M, Chiriac AI, Otto A, Zweytick D, May C, Schumacher C, Gust R, Albada HB, Penkova M, Kramer U, Erdmann R, Metzler-Nolte N, Straus SK, Bremer E, Becher D, Brotz-Oesterhelt H, Sahl HG, Bandow JE. 2014. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc Natl Acad Sci U S A 111:E1409–E1418. doi: 10.1073/pnas.1319900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franco OL, Murad AM, Leite JR, Mendes PA, Prates MV, Bloch C Jr. 2006. Identification of a cowpea gamma-thionin with bactericidal activity. FEBS J 273:3489–3497. doi: 10.1111/j.1742-4658.2006.05349.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Lewis K. 1997. Fabatins: new antimicrobial plant peptides. FEMS Microbiol Lett 149:59–64. doi: 10.1111/j.1574-6968.1997.tb10308.x. [DOI] [PubMed] [Google Scholar]
- 52.Lin KF, Lee TR, Tsai PH, Hsu MP, Chen CS, Lyu PC. 2007. Structure-based protein engineering for alpha-amylase inhibitory activity of plant defensin. Proteins 68:530–540. doi: 10.1002/prot.21378. [DOI] [PubMed] [Google Scholar]
- 53.Odintsova TI, Egorov TA, Musolyamov A, Odintsova MS, Pukhalsky VA, Grishin EV. 2007. Seed defensins from T. kiharae and related species: genome localization of defensin-encoding genes. Biochimie 89:605–612. doi: 10.1016/j.biochi.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 54.Van den Bergh KP, Van Damme EJ, Peumans WJ, Coosemans J. 2002. Ee-CBP, a hevein-type antimicrobial peptide from bark of the spindle tree (Euonymus europaeus L.). Meded Rijksuniv Gent Fak Landbouwkd Toegep Biol Wet 67:327–331. [PubMed] [Google Scholar]
- 55.Liu Y, Luo J, Xu C, Ren F, Peng C, Wu G, Zhao J. 2000. Purification, characterization, and molecular cloning of the gene of a seed-specific antimicrobial protein from pokeweed. Plant Physiol 122:1015–1024. doi: 10.1104/pp.122.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tam JP, Lu YA, Yang JL, Chiu KW. 1999. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc Natl Acad Sci U S A 96:8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colilla FJ, Rocher A, Mendez E. 1990. γ-Purothionins: amino acid sequence of two polypeptides of a new family of thionins from wheat endosperm. FEBS Lett 270:191–194. doi: 10.1016/0014-5793(90)81265-P. [DOI] [PubMed] [Google Scholar]
- 58.Terras FR, Schoofs HM, De Bolle MF, Van Leuven F, Rees SB, Vanderleyden J, Cammue BP, Broekaert WF. 1992. Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J Biol Chem 267:15301–15309. [PubMed] [Google Scholar]
- 59.Xiao J, Zhang H, Niu L, Wang X. 2011. Efficient screening of a novel antimicrobial peptide from Jatropha curcas by cell membrane affinity chromatography. J Agric Food Chem 59:1145–1151. doi: 10.1021/jf103876b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.