ABSTRACT
Glycoside hydrolase family 7 (GH7) cellobiohydrolases (CBHs) are enzymes commonly employed in plant cell wall degradation across eukaryotic kingdoms of life, as they provide significant hydrolytic potential in cellulose turnover. To date, many fungal GH7 CBHs have been examined, yet many questions regarding structure-activity relationships in these important natural and commercial enzymes remain. Here, we present the crystal structures and a biochemical analysis of two GH7 CBHs from social amoeba: Dictyostelium discoideum Cel7A (DdiCel7A) and Dictyostelium purpureum Cel7A (DpuCel7A). DdiCel7A and DpuCel7A natively consist of a catalytic domain and do not exhibit a carbohydrate-binding module (CBM). The structures of DdiCel7A and DpuCel7A, resolved to 2.1 Å and 2.7 Å, respectively, are homologous to those of other GH7 CBHs with an enclosed active-site tunnel. Two primary differences between the Dictyostelium CBHs and the archetypal model GH7 CBH, Trichoderma reesei Cel7A (TreCel7A), occur near the hydrolytic active site and the product-binding sites. To compare the activities of these enzymes with the activity of TreCel7A, the family 1 TreCel7A CBM and linker were added to the C terminus of each of the Dictyostelium enzymes, creating DdiCel7ACBM and DpuCel7ACBM, which were recombinantly expressed in T. reesei. DdiCel7ACBM and DpuCel7ACBM hydrolyzed Avicel, pretreated corn stover, and phosphoric acid-swollen cellulose as efficiently as TreCel7A when hydrolysis was compared at their temperature optima. The Ki of cellobiose was significantly higher for DdiCel7ACBM and DpuCel7ACBM than for TreCel7A: 205, 130, and 29 μM, respectively. Taken together, the present study highlights the remarkable degree of conservation of the activity of these key natural and industrial enzymes across quite distant phylogenetic trees of life.
IMPORTANCE GH7 CBHs are among the most important cellulolytic enzymes both in nature and for emerging industrial applications for cellulose breakdown. Understanding the diversity of these key industrial enzymes is critical to engineering them for higher levels of activity and greater stability. The present work demonstrates that two GH7 CBHs from social amoeba are surprisingly quite similar in structure and activity to the canonical GH7 CBH from the model biomass-degrading fungus T. reesei when tested under equivalent conditions (with added CBM-linker domains) on an industrially relevant substrate.
INTRODUCTION
Dictyostelia are a class of social amoebae (historically known as slime molds) containing four different groups of terrestrial bacterivores from the kingdom Amoebozoa. During vegetative growth, dictyostelia exist as single-celled organisms; upon starvation, a lack of nutrients becomes preventive for vegetative growth and the cells aggregate into a multicellular slug. Slugs have a defined posterior and anterior, have the ability to migrate, are sensitive to light and temperature, and exhibit an innate immune system. When conditions are sufficiently severe, the slug can form a fruiting body, where cells differentiate into a spore and stalk. During the formation of the slug and fruiting body, proteins and cellulose are deposited as an extracellular matrix, providing the organism with environmental protection and structural rigidity (1–4). Cellulose is also found in the sheath that surrounds the cell aggregates and is deposited in the stalk, stalk cell walls, and spore coats (2, 5). Thus, the deposition and reorganization of cellulose upon morphogenesis into the fruiting body are crucial to the development and propagation of the organism.
Cellulose is the homopolymer of β-(1,4)-d-glucose, and individual cellulose chains pack into dense, recalcitrant crystalline microfibrils in plant cell walls and other biological tissues (6). In nature, the intrinsic crystallinity and recalcitrance of cellulose are of significant benefit to plants and other organisms that employ it as a structural material, such as those in the genus Dictyostelium. Besides providing a structural framework for biological tissues, cellulose also provides significant protection from physical, chemical, and biological damage. The genomes of Dictyostelium discoideum and Dictyostelium purpureum exhibit >40 genes related to cellulose synthesis and hydrolysis, including glycoside hydrolase (GH) family 3 (GH3) β-glucosidases, glycoside hydrolase family 5 (GH5) and 9 (GH9) endoglucanases, and a single gene with sequence homology to the gene for a GH family 7 (GH7) cellobiohydrolase (CBH) (7). GH7 CBHs are of particular interest, in that they exhibit significant hydrolytic potential and are commonly used by fungi and other biomass-degrading eukaryotes for cellulose hydrolysis (8). GH7 CBHs also form the basis of most industrial cellulase cocktails for industrial lignocellulosic biomass conversion (8–10). The D. discoideum GH7 CBH, Cel7A (DdiCel7A; previously CbhA) (7, 11), is a developmentally regulated enzyme, with the highest levels of expression occurring during formation of the fruiting body (11). The deletion of DdiCel7A results in normal phenotypes but delayed morphogenesis from slug to fruiting body (11), suggesting that DdiCel7A is required for the breakdown and rearrangement of cellulose during these phases of development. Given the similarities in organism development between D. discoideum and D. purpureum, it is likely that D. purpureum Cel7A (DpuCel7A) serves a similar function.
Structural and biochemical data for GH7s obtained over the last several decades have revealed that they have similar overall folds and catalytic arrangements (8). The structures of GH7 catalytic domains are known from 10 CBHs, Trichoderma reesei Cel7A (TreCel7A) (12–14), Heterobasidion irregulare Cel7A (HirCel7A) (15), Phanerochaete chrysosporium Cel7D (PchCel7D) (16), Talaromyces emersonii Cel7A (RemCel7A) (17), Trichoderma harzianum Cel7A (ThaCel7A) (18), Melanocarpus albomyces Cel7B (MalCel7B) (19), Aspergillus fumigatus Cel7A (AfuCel7A) (20), Humicola grisea var. thermoidea Cel7A (HgtCel7A) (21) Limnoria quadripunctata Cel7B (LquCel7B) (22), and Geotrichum candidum Cel7A (23), and three endoglucanases (EGs), T. reesei Cel7B (24), Humicola insolens Cel7B (25), and Fusarium oxysporum Cel7B (26). All GH7s share a β-jelly roll fold, with two antiparallel β-sheets packing face to face to form a curved β-sandwich. Long loops extend the edges of the β-sandwich and form an ∼45-Å-long binding groove along the entire length of the enzyme. In CBHs, loops A1 to A4 and B1 to B4 (the nomenclature is defined in reference 8) are further elongated and enclose the cellulose chain in a tunnel using a threading-like mechanism, whereas EGs exhibit a more open cleft. Many GH7 enzymes are bimodular in nature, having a family 1 carbohydrate-binding module (CBM) connected to the catalytic domain (CD) by a glycosylated, flexible linker comprised of about 30 amino acids (27–29).
Deconstruction of cellulose by GH7 CBHs is a multistep process that includes substrate binding, formation of the catalytically active complex, hydrolysis, product release, and processive translation along the substrate chain (8, 9, 30, 31). These cellulases act from the reducing end of cellulose chains and perform multiple, processive hydrolytic events before disassociating from a cellulose chain (32). For cellulases, such as the model GH7 CBH from T. reesei (TreCel7A), this process continues until the enzyme either runs into an obstruction where the enzyme is stalled and eventually releases the substrate or the end of the cellulose chain is reached (33–35). TreCel7A exhibits the most extensively enclosed tunnel among known GH7 CBH structures, while PchCel7D displays the most open active site due to several loop deletions and residue size reductions on the tips of tunnel-enclosing loops (36). It has been suggested that this more open active site can have increased activity on microcrystalline cellulose by increasing the rate of endoinitiation while decreasing the effect of product inhibition (34, 36).
While the posthydrolysis release of cellobiose is important for processivity, it also has an undesired consequence of becoming a strong inhibitor of hydrolysis. Product inhibition slows the overall conversion rate of cellulose to glucose, particularly when substrate concentrations are high (>10%) (37, 38). In commercial cellulase cocktails and natural organismal secretomes, this product inhibition is relieved by the action of β-glucosidases, glycoside hydrolases that cleave cellobiose into two glucose molecules. As GH7 CBHs comprise the majority of industrial cellulase mixtures and are significantly inhibited by cellobiose, overcoming product inhibition in GH7 CBHs is of paramount importance for achieving high product yields in the enzymatic hydrolysis of biomass.
Although the study of GH7 CBHs has now spanned several decades, our collective understanding of their action is still being revealed (8). The development of accurate structure-activity relationships of these enzymes, especially given their significant societal importance in the renewable fuels industry as well as their natural importance in the turnover of the most abundant biological material on Earth, warrants concerted efforts to elucidate, understand, and improve their function. With the wealth of new genomics and metagenomics data, new GH7 CBHs have recently been revealed, as described above, outside the fungal kingdom (7, 39, 40), thus presenting new opportunities to study these key enzymes in branches of life phylogenetically distant from fungi (41). To that end, we present here the structures of two GH7 CBHs from the social amoebas D. discoideum and D. purpureum from the Amoebozoa kingdom and a corresponding biochemical characterization of these CBHs. We also constructed and expressed chimeras of these GH7 CBHs in an industrially relevant fungal host, T. reesei, with the family 1 CBM and linker from TreCel7A such that performance characterizations could be conducted on the basis of equivalence to the full-length model GH7 CBH TreCel7A. Approaches such as this, wherein full-length enzymes are produced in the same expression host to ensure similar glycosylation patterns (42), will be essential to conduct comparisons of cellulase activities on a consistent basis (43). Using this approach, we demonstrate that both Dictyostelium GH7 CBHs are equally as efficient as TreCel7A in the hydrolysis of crystalline cellulose at their relative temperature and pH optima and also exhibit increased tolerance to product inhibition.
MATERIALS AND METHODS
Cloning and protein expression.
The genomes of D. discoideum and D. purpureum have been fully sequenced (7, 40), and each has been shown to contain one GH7 CBH (Cel7A, previously CbhA) (7, 11), herein referred to as DdiCel7A and DpuCel7A, respectively, to conform with current nomenclature (8). The DdiCel7A and DpuCel7A sequences were obtained from the Joint Genome Institute genome database (44, 45). Dictyostelium GH7 CBH genes were codon optimized and synthesized (GeneArt) without introns and cloned into and expressed in the linearized pTrEno plasmid, and plasmid pTrEno was transformed into T. reesei strain QM6a from which cbh1 was deleted as described previously (46). In nature, DdiCel7A and DpuCel7A do not exhibit a CBM; thus, the TreCel7A linker, which connects the CD and family 1 CBM, was added to the 3′ end to generate a chimeric Cel7A, referred to here as DdiCel7ACBM and DpuCel7ACBM, respectively. The linker sequence starts at the GNPPGGNPP motif from the TreCel7A linker.
Expression medium was inoculated with spores from transformed spore stocks, and the production volume was scaled up stepwise from 0.1 liter to a final volume of 8 liters in a 14-liter bioreactor (BioFlo3000; New Brunswick) as previously described (46).
Protein purification.
Prior to protein purification, expression of the Dictyostelium GH7 CBHs with and without the TreCel7A CBM-linker domain was verified by determination of their activity on p-nitrophenyl-β-d-lactopyranoside (pNPL; Sigma-Aldrich, St. Louis, MO) and visualization on SDS-polyacrylamide gels (see Fig. S1 in the supplemental material) and Western blots with rabbit anti-Cel7A polyclonal IgG.
For protein purification, fermentation broths were filtered through a 0.45-μm-pore-size filter, concentrated ∼40 times, and exchanged into 20 mM bis-Tris buffer, pH 6.5, by ultrafiltration through a hollow fiber with a 10,000-Da-molecular-mass cutoff. The filtrate was adjusted to 1.5 M (NH4)2SO4 by the addition of 4.0 M (NH4)2SO4 in 20 mM bis-Tris buffer, pH 6.5, and filtered through a 0.2-μm-pore-size polyethersulfone membrane prior to hydrophobic interaction chromatography on a 26/10 phenyl Sepharose fast-flow column eluted with a 1.5 M to 0.0 M (NH4)2SO4 gradient in 20 mM bis-Tris buffer, pH 6.5, over 8 column volumes. Fractions containing Cel7s, identified by activity on pNPL, were pooled, desalted, and separated on a Source 15Q 10/100 Tricorn anion-exchange column with a gradient of 0 to 500 mM NaCl in 20 mM bis-Tris buffer, pH 6.5, for 30 column volumes. Purity was assessed by SDS-PAGE and Western blotting. The anion-exchange chromatography was completed twice for the native recombinant DdiCel7A and DpuCel7A enzymes only. Finally, the enzymes were size separated and buffer exchanged over a 26/60 Superdex 75 size exclusion column in 20 mM acetate buffer, pH 5.0, 100 mM NaCl containing 0.02% NaN3. Purity was determined by SDS-PAGE. Trypsin digestion, followed by liquid chromatography-mass spectrometry (MS) and peptide mapping, confirmed the protein identities.
DSC.
Thermal stability was evaluated by differential scanning microcalorimetry (DSC) using a MicroCal model VP-DSC calorimeter (MicroCal, Inc., Northampton, MA). Data were collected by Origin for DSC software (MicroCal). Samples contained 50 μg/ml protein at pH 5.0 in 20 mM acetate buffer, 100 mM NaCl. The calorimeter scan rate was 60°C/h over a temperature range of from 30°C to 110°C.
pH and temperature optima.
pH profiles for enzymatic activity were generated in duplicate by incubating 1.6 mM pNPL with 0.66 μM the indicated enzyme in 150 μl of MacIlvaine buffer (citrate, phosphate, NaCl) with a constant conductivity of between 19 and 21 mS/cm in a 96-well plate at pH 2.9, 3.9, 5.0, 6.1, 7.1, and 8.3 at 30°C for 0, 1, 5, 10, 20, and 30 min. The reaction was stopped by the addition of 25 μl of 1.0 M sodium carbonate, and the A405 was determined by use of a colorimetric microtiter plate reader. The absorbance values were correlated to the p-nitrophenol (pNP) concentration by the use of pNP standards, and the pH profile graphs were generated by plotting the linear rate of pNP production as a function of pH.
Temperature profiles were generated in duplicate by incubating 1.6 mM pNPL with 0.66 μM enzyme in 2.0 ml of 20 mM sodium acetate buffer, pH 5.0, 100 mM NaCl, and 0.02% NaN3. Reaction mixtures were incubated at 30, 35, 40, 45, 50, 55, 60, and 65°C in a water bath. At 0, 1, 5, 10, 20, and 30 min, 150 μl was removed from the reaction vial and placed in a 96-well plate containing 25 μl of 1.0 M sodium carbonate. The absorbance values were determined as described above for pH, and temperature profile graphs were generated by plotting the linear rate of pNPL turnover as a function of temperature.
Enzyme kinetics.
Vmax and apparent Km (Kmapp) values were determined in pNPL solutions of 0.0, 0.8, 1.6, 2.5, 4.2, 5.8 6.6, and 8.3 mM. pNPL was incubated with 0.66 μM CBH at 40°C or 45°C in 20 mM acetate buffer, pH 5.0, 100 mM NaCl in 96-well microtiter plates. The reactions were quenched by the addition of 25 μl of 1.0 M sodium carbonate at 0, 1, 5, 10, 15, 20, and 30 min. The absorbance values were converted to molar product concentrations using pNP standard curves. The data were fit to the Michaelis-Menten expression. Fitting and plots were generated using Kaleidagraph software (Synergy Software).
Inhibition experiments were completed with 0, 0.16.6, 41.6, 75.0, 91.6, and 108.3 μM cellobiose and 0.0, 0.8, 1.7, 2.5, and 4.2 mM pNPL incubated with 0.6 μM Cel7A for 0, 5, 10, 15, and 20 min. The reactions were quenched by the addition of 25 μl of 1.0 M sodium carbonate, and the A405 was recorded. To fit kinetic parameters, theoretical rates were calculated from starting input values of Vmax, Kmapp, and Ki using the Michaelis-Menten expression for competitive enzyme inhibition. Data were also evaluated for noncompetitive, uncompetitive, and mixed inhibition.
Nonlinear regression fitting was accomplished using the Excel Solver add-in (Microsoft, Redmond WA). Weighted squared residuals were calculated for each data point using three different weighting schemes—simple, (νobs − νcalc)2; proportional, [(νobs − νcalc)/νcalc]2; and statistical, [(νobs − νcalc)2/νcalc)], where νobs is the observed value of ν and νcalc is the calculated value of ν—and the sum of residuals was minimized. The best fit to the experimental data obtained was used for the reported kinetic parameters derived.
Hydrolysis of crystalline and amorphous cellulose.
Hydrolysis reaction mixtures contained 5.0 mg/ml of the following cellulose substrates: Avicel (Sigma-Aldrich PH-101), pretreated corn stover (PCS; NREL dilute acid-pretreated corn stover [P050921]), or phosphoric acid-swollen cellulose (PASC; from cotton linter). The substrates were incubated with 20 mg CBH per gram of glucan, 1.0 mg β-glucosidase (Aspergillus niger; Novozymes 188) per gram of glucan, and 1.0 mg Cel5A CD with a Y245G substitution (Acidothermus cellulolyticus) per gram of glucan in 20 mM sodium acetate buffer, pH 5.0, 100 mM NaCl, and 0.02% NaN3. Reactions were conducted in triplicate in 1.8-ml sealed cryogenic vials with continuous mixing by rotational inversion at ∼10 rpm and incubation at 50°C or 40°C, as specified below. At the time points indicated below, 100-μl samples were removed from the reaction vials and diluted in water, and the mixture was boiled for 5 min to quench the reaction. Boiled samples were filtered through a 0.22-μm-pore-size syringe filter into glass high-performance liquid chromatography (HPLC) vials. Glucose and cellobiose were determined by HPLC analysis on an Aminex HPX-87P column operated at 65°C with water as the mobile phase.
Crystallization and X-ray data collection.
The purified DdiCel7A and DpuCel7A proteins (without CBM) were desalted to 20 mM sodium acetate buffer, pH 5.0, prior to crystallization. Initial screens for crystallization conditions were performed by sitting-drop vapor diffusion in 96-well plates and were set up using a Mosquito crystallization robot (TTP Labtech, Cambridge, United Kingdom). Equal amounts of protein (12 mg/ml) and well solution were mixed in 0.3-μl crystallization drops. The most promising crystallization hits for DdiCel7A and DpuCel7A were obtained with the Morpheus (Molecular Dimensions) and PEG Ion II (Hampton Research) screens, respectively.
Crystals used for data collection were grown at 20°C by the hanging-drop vapor diffusion method (47) by mixing protein (7 mg/ml in 20 mM sodium acetate, pH 5.0) and precipitant solutions at a 1:1 ratio, as follows: DdiCel7A with 12.5% (wt/vol) polyethylene glycol (PEG) 1000, 12.5% (wt/vol) PEG 3350, 12.5% (vol/vol) MPD (2-methyl-2,4-pentanediol), 0.1 M sodium HEPES-MOPS (morpholinepropanesulfonic acid), pH 7.5, 0.03 M MgCl2, and 0.03 M CaCl2 and DpuCel7A with 0.1 M sodium malonate, pH 7.0, and 12% (wt/vol) PEG 3350.
Data collection and processing and structure solution.
X-ray diffraction data were collected at 100 K from single crystals of DdiCel7A and DpuCel7A at the MAX-Lab I911-3 (Lund, Sweden) and the European Synchrotron Radiation Facility (ESRF) ID23-1 (Grenoble, France) beam lines, respectively. The data were indexed and integrated using the IMOSFLM2 program and scaled with the SCALA3 tool in the CCP4 program suite (48). Five percent of the reflections were set aside for calculation of Rfree factors. The crystal structures were solved by molecular replacement using the Phaser program (49) in the CCP4 package. A structure of TreCel7A (Protein Data Bank [PDB] accession number 4D5J) (50) was used as the search model for DdiCel7A, which was then used as the model for DpuCel7A. The DdiCel7A structure was solved in space group P212121 with a single molecule in the asymmetric unit, and the DpuCel7A structure was solved in space group P212121 with two molecules in the asymmetric unit. Both structures were refined by alternating cycles of maximum likelihood refinement with the REFMAC5 program (51) and manual rebuilding using the Coot program (52). Statistics from diffraction data processing and structure refinement are summarized in Results.
Sequence alignments and analyses.
A structure-based sequence alignment was first generated using the Swiss-PdbViewer (v. 4.1) application (53; http://www.expasy.org/spdbv/) by superimposing the structures of DdiCel7A (PDB accession number 4ZZQ), DpuCel7A (PDB accession number 4ZZP), and GH7 CBHs from PDB (TreCel7A, PDB accession number 4C4C; ThaCel7A, PDB accession number 2YOK; AfuCel7A, PDB accession number 4V20; RemCel7A, PDB accession number 3PL3; HgtCel7A, PDB accession number 4CSI; MalCel7B, PDB accession number 2RFZ; HirCel7A, PDB accession number 2XSP; PchCel7D, PDB accession number 1Z3V; LquCel7B, PDB accession number 4IPM). This alignment was opened in the Jalview (v. 2.9.0b2) program (54), and full-length sequences corresponding to the PDB entries and a collection of 38 nonfungal GH7 sequences were added, followed by multiple-sequence alignment using the MUSCLE web service with default settings (55). Regions flanking the GH7 domain (e.g., the signal peptide, CBM, linker) were trimmed off. The sequences selected for use in Fig. 7 and Table 4 were realigned with the MUSCLE web service, and the program Indonesia (D. Madsen, P. Johansson, N. Johansson, S. Arent, M. Harris, and G. Kleywegt, unpublished) was used to calculate pairwise sequence identities and similarities using the Gonnet substitution matrix (56). Phylogenetic analyses of an extended set of 113 GH7 protein sequences were conducted with the program MEGA (v. 7) (57). Sequences were collected by a pBLAST search of the sequence of each nonfungal GH7 against the sequences in the NCBI and UniProt databases, and a selection of taxonomically diverse hits among the many fungal entries was retrieved. The sequences were aligned by use of MUSCLE (55), and regions flanking the GH7 domain were trimmed off. A 90% identity threshold was applied to remove redundancy, and endoglucanases lacking the B4 loop (marked in Fig. 1 and Fig. 4A) were excluded. The evolutionary history was inferred using the minimum evolution method (58) and bootstrap phylogeny testing with 700 replicates (59). The optimal tree with the sum of the branch length of 19.8 is shown below. Branch lengths are drawn to scale, with evolutionary distances, computed using the Dayhoff matrix-based method (60), being in units of number of amino acid substitutions per site. The minimum evolution tree was searched using the close-neighbor-interchange (CNI) algorithm (61) at a search level of 1. The neighbor-joining algorithm (62) was used to generate the initial tree. All ambiguous positions were removed for each sequence pair. There were a total of 520 positions in the final data set.
FIG 7.
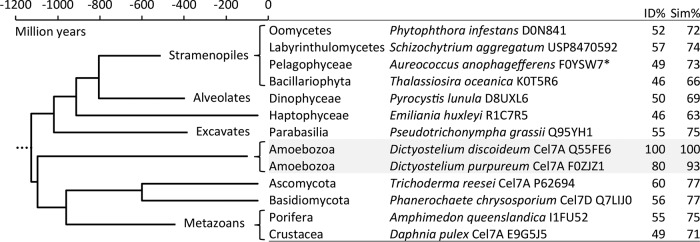
Eukaryote evolution timeline of branches where GH7 genes have been found and the percent identities (ID%) and similarities (Sim%) of the protein sequences within the GH7 domain to the D. discoideum Cel7A sequence. Branch names and time points of divergence are from reference 72. Only points of early divergence (<600 million years ago) are included; the later divergence among stramenopiles, amoebozoa, and metazoans is not resolved here. The multiple-sequence alignment of selected GH7 sequences was done with the MUSCLE web service, and flanking regions (e.g., the signal peptide, CBM) were trimmed off before calculation of pairwise sequence identities and similarities using the Gonnet substitution matrix. UniProt accession numbers are provided, except in the case of Schizochytrium aggregatum, for which the U.S. patent number is indicated (76). *, the Aureococcus anophagefferens GH7 sequence (UniProt accession number F0YSW7) appears to be a fragment containing only 135 residues of the C-terminal part of the GH7 domain in the alignment.
TABLE 4.
Pairwise protein sequence identities and similarities within the GH7 CDa
Pairwise protein sequence identities are to the upper right and similarities are to the lower left of the diagonal of boldface numbers. The boldface numbers are the number of residues in the alignment for each sequence. The shading highlights values for the Dictyostelium sequences.
Designations in parentheses are UniProt accession numbers for the GH7 sequences except in the case of Schizochytrium aggregatum, which is a U.S. patent number from reference 76.
FIG 1.
Sequence alignment of the GH7 CBHs that have been structurally characterized to date. The regions highlighted in blue denote N-glycosylation motifs, the residues in red (EXDXXE motif) denote the catalytic residues, and the residues enclosed in boxes represent the A1 to A4 and B1 to B4 loops enclosing the active-site tunnel.
FIG 4.
Superposition of the structures of DpuCel7A (A) and DdiCel7A (B) with the structure of the cellononaose Michaelis complex of TreCel7A (PDB accession number 4C4C). The key substrate-binding loops are labeled in panel A, and the substrate-binding sites are labeled in panel B.
Protein structure accession numbers.
The coordinates and structure factors for the DdiCel7A and DpuCel7A structures have been deposited in PDB under accession numbers 4ZZQ and 4ZZP, respectively.
RESULTS
Expression and purification of enzymes.
DdiCel7A, DdiCel7ACBM, DpuCel7A, DpuCel7ACBM, and TreCel7A were recombinantly expressed in a newly developed T. reesei eno expression system (46). The constitutive eno promoter drives expression of Cel7A to allow growth on glucose, which represses the native cellulase system. Coupling of this system with the Δcbh strain of T. reesei ensures that only the recombinant Cel7A protein is expressed, as previously described (46). For evaluation of cellulose digestion on various solid substrates, we added the family 1 CBM from TreCel7A to the CD of native DdiCel7A and DpuCel7A (dubbed DdiCel7CBM and DpuCel7ACBM, respectively). Fed-batch fermentations typically yielded 5 to 25 mg/liter recombinant protein after processing and purification. All proteins were purified to electrophoretic homogeneity, with the exception of DpuCel7A, which exhibited a doublet band by SDS-PAGE (see Fig. S1 in the supplemental material). Sequencing mass spectrometry identified both bands to be native DpuCelA, and it is thus unclear why there were two populations. However, as we were not able to separate the two populations, the mixed population was provided for crystallization.
Temperature, pH, and thermostability profiles.
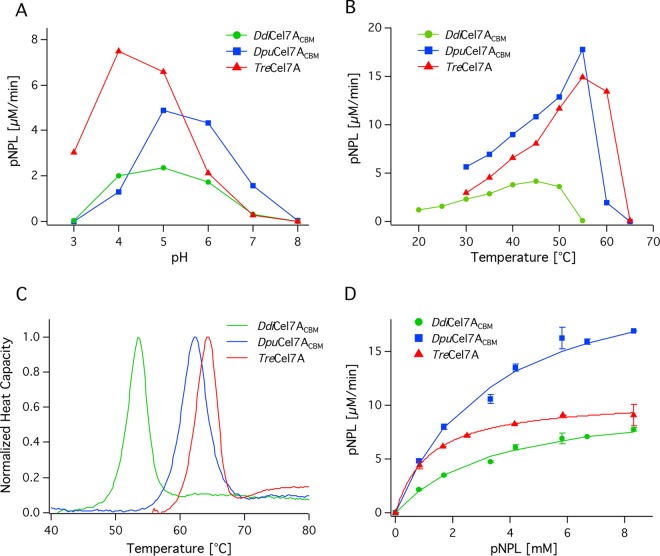
The dependence of activity on temperature and pH for DdiCel7ACBM, DpuCel7ACBM, and, for comparison, TreCel7A was determined using pNPL as the substrate. Activities were measured at eight temperatures at pH 5.0, and the data point with the highest activity was recorded as the temperature optimum in Table 1. DdiCel7ACBM exhibited a lower temperature optimum, 45°C, than DpuCel7ACBM and TreCel7A, which exhibited optimal activity at about 55°C, as shown in Fig. 2. For consistency, the optimal pH was determined in MacIlvaine buffer with a constant ionic strength. Activity was evaluated at 40°C for DdiCel7ACBM and 45°C for DpuCel7ACBM and TreCel7A to account for the difference in temperature optima. The pH of maximum pNPL turnover for DpuCel7A and DdiCel7A, pH 5, was higher than that of TreCel7A, pH 4, as shown in Fig. 2.
TABLE 1.
Biochemical and kinetic characterization on pNPL
| Enzyme | Optimum temp (°C) | Optimum pH | Tm (°C) | kcat (min−1) | Kmapp (mM) | Ki (μM) |
|---|---|---|---|---|---|---|
| DdiCel7ACBM | 45 | 5 | 53 | 16.1 ± 2.8 | 3.4 ± 0.1 | 205 |
| DpuCel7ACBM | 55 | 5 | 63 | 36.0 ± 6.7 | 3.4 ± 0.3 | 130 |
| TreCel7A | 55 | 4 | 64 | 16.1 ± 3.0 | 1.2 ± 0.1 | 29 |
FIG 2.
Biochemical and kinetic characterization of DdiCel7ACBM, DpuCel7ACBM, and TreCel7A. (A) Comparison of activity on 2 mM pNPL at 40°C and at various pH values; (B) comparison of activity on 2 mM pNPL at pH 5 and various temperatures; (C) DSC traces comparing the denaturation temperature (Tms) of all three enzymes; (D) Michaelis-Menten plot for comparison of activities with various concentrations of pNPL at 45°C and pH 5.
The denaturation temperature was measured by DSC in the absence of substrate. The melting temperatures (Tms) of DdiCel7ACBM, DpuCel7ACBM, and TreCel7A were 53°C, 62°C, and 64°C, respectively, as shown in Table 1 and Fig. 2.
Enzyme kinetics on pNPL.
The kinetic properties of DdiCel7ACBM, DpuCel7ACBM, and TreCel7A on pNPL were compared (Table 1). Vmax and Kmapp were determined by plotting the turnover rate on pNPL for increasing substrate concentrations and evaluated by use of the Michaelis-Menten model, as shown in Fig. 2. The catalytic rate constant (kcat) of DpuCel7ACBM was significantly higher at 36.2 min−1 than the kcats of DdiCel7ACBM and TreCel7A, which were nearly equal at 16.2 and 15.9 min−1, respectively. However, the Kmapp values of both DpuCel7ACBM and DdiCel7ACBM were 3.4 mM, which was higher than the Kmapp value of TreCel7A, 1.2 mM, indicating a lower binding affinity for pNPL.
Cellulose hydrolysis.
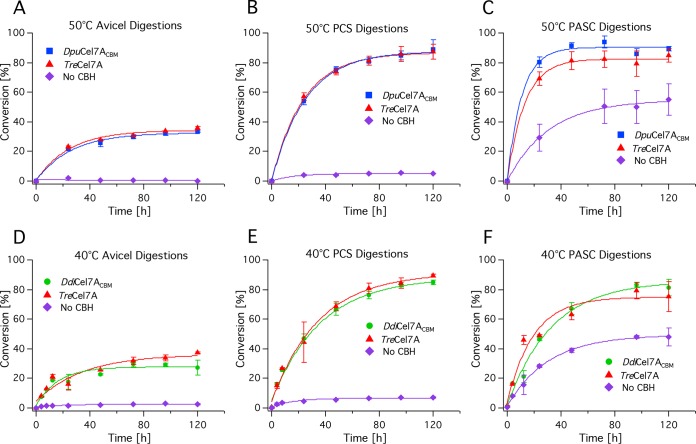
Enzymatic activity comparisons were conducted on 0.5% Avicel, pretreated corn stover (PCS), and phosphoric acid-swollen cellulose (PASC) substrates. The enzyme cocktails comprised β-glucosidase from Novozymes 188, the E1 endoglucanase from A. cellulolyticus, and DpuCel7ACBM, DdiCel7ACBM, or TreCel7A (63). Assays were completed near the previously determined pH and temperature optima for DpuCel7ACBM and DdiCel7ACBM, at pH 5.0 and 50°C and 40°C, respectively. Glucose and cellobiose concentrations were determined by HPLC and plotted as a function of time. Curves were fit to a single exponential; the time constant (τ) for cellulose hydrolysis is reported in Table 2 and indicates that cocktails with both DpuCel7ACBM and DdiCel7ACBM convert cellulose to glucose in a manner equivalent to that for TreCel7A on all three cellulose substrates. All hydrolysis profiles were strikingly similar (Fig. 3), with only one minor difference in the hydrolysis of Avicel by DpuCel7ACBM being seen (Fig. 3), where the enzyme reached a lower maximum conversion than TreCel7A but did so faster than TreCel7A.
TABLE 2.
Cellulose hydrolysis kinetic parameters determined by use of a single exponential curvea
| Enzyme | Temp (°C) | τ (h) |
||
|---|---|---|---|---|
| Avicel | PCS | PASC | ||
| DdiCel7ACBM | 40 | 23 ± 7 | 36 ± 5 | 34 ± 6 |
| TreCel7A | 40 | 35 ± 13 | 35 ± 5 | 33 ± 6 |
| DpuCel7ACBM | 50 | 26 ± 5 | 25 ± 2 | 11 ± 2 |
| TreCel7A | 50 | 25 ± 5 | 24 ± 2 | 13 ± 1 |
The equation for the exponential curve was A · (1 − exp−t/τ) + C, where A is the amplitude, t is time, τ is the time constant for cellulose hydrolysis, and C is the baseline offset.
FIG 3.
Cellulose hydrolysis on Avicel (A, D), PCS (B, E), and PASC (C, F) in a three-enzyme cocktail consisting of the experimental CBH, endoglucanase (E1), and β-glucosidase. (A to C) Comparison of DpuCel7ACBM and TreCel7A at 40°C and pH 5.0; (D to F) comparison of DdiCel7ACBM and TreCel7A at 50°C and pH 5.0. All digestions were evaluated against a no-CBH control.
DdiCel7A and DpuCel7A structures.
After extensive screening, crystals of the T. reesei-expressed DdiCel7A and DpuCel7A proteins that could be used for synchrotron radiation diffraction data collection were obtained. However, in both cases the crystals were small and the diffraction data were notoriously weak and suffered from twinning and high degrees of mosaicity. This is reflected by high R and Rfree factors of 0.25 and 0.35, respectively, for the DdiCel7A structure and 0.23 and 0.31, respectively, for the DpuCel7A structure, which were refined at resolutions of 2.1 and 2.7 Å, respectively, and a rather high average temperature factor (B factor) of 28.5 Å2 for DdiCel7A. Nevertheless, almost the entire backbone could be traced with confidence. All amino acid side chains were placed and refined, apart from Lys-438 of DdiCel7A and Asn-438 and Asn-439 of DpuCel7A, which were not visible in the electron density and thus excluded from the final models. The DdiCel7A structure was solved in space group P212121 with a single protein chain in the asymmetric unit, and DpuCel7A was solved in space group P212121 with two chains, A and B, in the asymmetric unit. In both structures, the N-terminal glutamine residue is cyclized to pyroglutamate (PCA1), Pro-388 is cis-proline, all 18 cysteines form disulfide bonds, N-glycosylation is evident at Asn-269 with density for one N-acetylglucosamine unit, and Lys437 is the last visible residue. In DpuCel7A, an additional GlcNAc is attached to Asn-112. Statistics for diffraction data processing and structure refinement are summarized in Table 3.
TABLE 3.
Data collection and refinement statistics for DdiCel7A and DpuCel7A
| Parameter | Value(s) fora: |
|
|---|---|---|
| DdiCel7A | DpuCel7A | |
| Data collection statistics | ||
| PDB accession no. | 4ZZQ | 4ZZP |
| Beam line | I911-3, MAX-Lab | ID23-1, ESRF |
| Space group | P212121 | P212121 |
| a, b, c (Å) unit cell dimensions | 44.2, 62.9, 132.9 | 55.6, 85.2, 168.5 |
| Wavelength (Å) | 1.0 | 0.87 |
| Resolution (Å) | 45.7–2.10 (2.21–2.10) | 46.9–2.70 (2.85–2.70) |
| No. of unique reflections | 22,434 | 22,787 |
| Multiplicity | 8.3 (8.3) | 6.6 (6.3) |
| Completeness (%) | 100 (100) | 99.9 (99.9) |
| I/σI | 5.5 (1.3) | 5.3 (3.2) |
| Rmergeb | 0.33 (1.6) | 0.25 (0.5) |
| Refinement statistics | ||
| Rwork/Rfree (%) | 25.6/35.1 | 23.5/31.4 |
| Mean B factor (Å2) | 28.5 | 15.16 |
| No. of amino acids | 437 | 437 |
| Completeness (%) | 99.9 | 99.8 |
| RMSD | ||
| Bond angle (°) | 1.26 | 1.27 |
| Bond length (Å) | 0.0073 | 0.0058 |
Numbers in parentheses correspond to the highest-resolution bins.
Rmerge = ∑h∑i|I(h)i − 〈I(h)〉|/∑h∑i I(h)i, where I(h)i is the intensity (I) of reflection h and 〈I(h)〉 is the average value over multiple measurements.
Structural alignments and analysis.
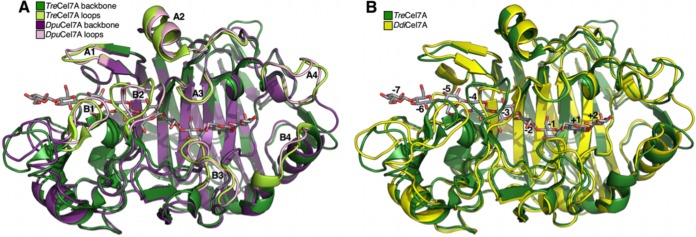
As expected from the high degree of amino acid sequence identity (Fig. 1), the folds of DdiCel7A and DpuCel7A were very similar to one another (80% identity; root mean square deviation [RMSD], 0.45 Å) as well as to the fold of the catalytic module of TreCel7A (59% identity; RMSD, 0.66 and 0.62 Å, respectively) (Fig. 4). The catalytic residues for the Dictyostelium Cel7A structures (Glu211, Asp213, and Glu216, corresponding to TreCel7A Glu212, Asp214, and Glu217, respectively) and aromatic residues that stack on the binding sites at positions −7, −4, −2, and +1 (W40, W38, W373, and W382, respectively) overlap the positions of the analogous residues in TreCel7A (PDB accession number 4C4C). Figures 4A and B show the superposition of DdiCel7A and DpuCel7A with the cellononaose Michaelis complex of TreCel7A (PDB accession number 4C4C), where surface loops of interest and glucoside-binding subsites are indicated (14). The corresponding loop regions are also highlighted in the structure-based sequence alignment of the catalytic modules of the GH7 CBH structures available in PDB (Fig. 1).
Alignments of the structures of DdiCel7A and DpuCel7A to the known structures of other GH7 CBHs showed that the eight major binding tunnel loops (A1 to A4 and B1 to B4) of DdiCel7A and DpuCel7A are exceptionally similar to each other and to those of TreCel7A, revealing a highly enclosed active site (Fig. 5). Loops A3 and B3 of DdiCel7A and DpuCel7A lack at their tips the Tyr residues that are present in TreCel7A. However, the Dictyostelium Cel7A enzymes have a tyrosine (Tyr202) that protrudes from the base of the B2 loop whose side chain resides in the same general space as the B3 tyrosine in the other GH7 structures (Tyr247 in TreCel7A) (Fig. 6). The opposing residue on loop A3 (Tyr371 in TreCel7A) is Gln377 in DdiCel7A and Leu377 in DpuCel7A. Glutamine at this position is a rare motif among GH7 CBHs and is found only in another protist, Pseudotrichonympha grassii, a parabasilian symbiont in the gut of termites. A few distantly related basidiomycetes have glutamate at this position; e.g., in HirCel7A, molecular dynamics simulations showed that the glutamate side chain can interact with a bound cellulose chain (15). DpuCel7A and DdiCel7A also possess on the A1 loop two glutamine residues (Q97 and Q99) that would be in direct contact with the cellulose surface when the enzyme is adsorbed (64); TreCel7A possesses only the former of these. Near the entrance to the tunnel, an aromatic residue in DpuCel7A (Phe6) may potentially provide an additional subsite at position −8 and help to guide a cellulose chain into the tunnel. Leucine is found at the corresponding position in DdiCel7A and TreCel7A.
FIG 5.
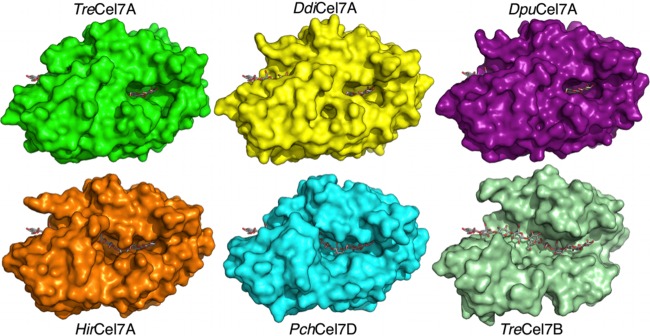
Space-fill GH7 structures comparing the substrate tunnel enclosures of CBHs from T. reesei (PDB accession number 4C4C), D. discoideum, D. purpureum, H. irregulare (PDB accession number 2YG1), P. chrysosporium (PDB accession number 1GPI), and the endoglucanase (EG) Cel7B from T. reesei (PDB accession number 1EG1). In all frames, the cellononaose ligand from the TreCel7A Michaelis complex is shown as gray sticks.
FIG 6.
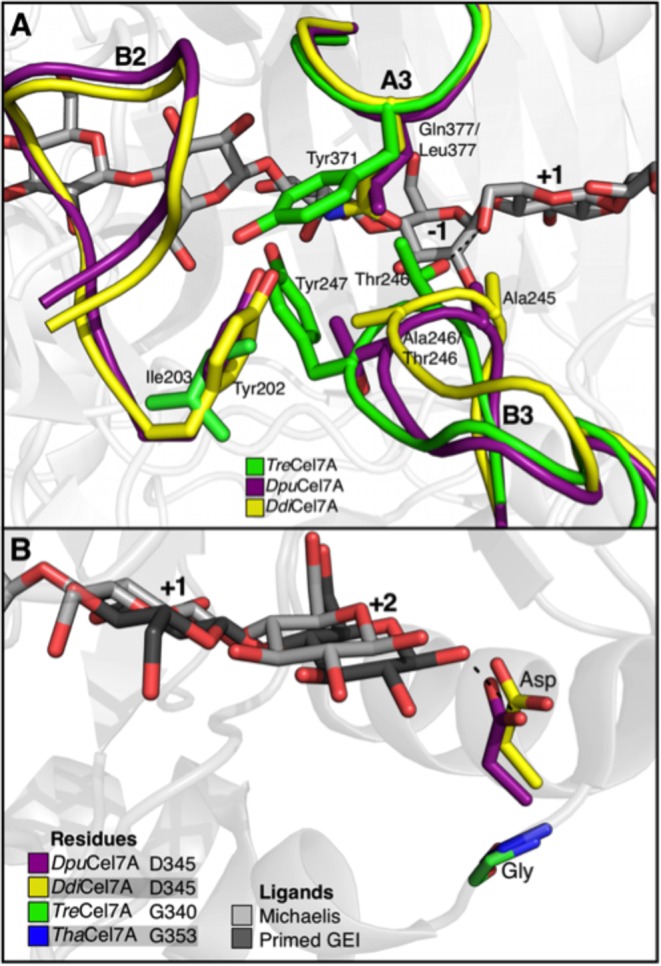
Alignments of the structures of DdiCel7A and DpuCel7A with the known structures of other GH7 CBHs. (A) Alignment around the active site illustrating the interactions of the B2, B3, and A3 loops discussed in the text. (B) Product-binding sites illustrating the relative positioning of the commonly found aspartate residue that interacts with the glucosyl unit at position +2 but which is replaced by glycine in TreCel7A and ThaCel7A. Both the Michaelis (PDB accession number 4C4C) and primed glycosyl-enzyme intermediate (GEI; PDB accession number 3CEL) ligands are from the TreCel7A structure. The aspartic acid side chains of PchCel7D, HirCel7A, RemCel7A, AfuCel7A, and LquCel7B essentially overlay those of DpuCel7A and DdiCel7A and are not shown for clarity.
TreCel7A has four attachment sites on the CD for N-glycosylation: N45, N64, N270, and N384. Only the N270 glycan site is maintained (at N269) in DpuCel7A and DdiCel7A, where one attached N-acetylglucosamine is visible in both structures. At the position analogous to TreCel7A N45, both DdiCel7A and DpuCel7A have a 1-residue deletion, and TreCel7A N384 is replaced by a threonine. Instead, the Dictyostelium sequences exhibit an N-glycosylation site at N112, which is also found in ThaCel7A and where GlcNAc is seen in the DpuCel7A structure, and at an additional site in DpuCel7A, N433, although there is not clear electron density to observe glycan attachment. We also note that TreCel7A has 10 disulfide bridges, whereas the Dictyostelium Cel7A enzymes have only 9. The more common scenario in GH7 cellulases is for them to lack this extra disulfide bridge between Cys4 and Cys72 (TreCel7A numbering). In fact, TreCel7A and T. harzianum Cel7A are the only GH7 members with a solved structure that possess the 10th disulfide bridge.
Lastly, the product-binding sites of DdiCel7A and DpuCel7A are quite similar to those of TreCel7A, with the exception of a few notable differences among residues that have been extensively studied and discussed in past GH7 studies (8). Thr246 at the tip of loop B3 in TreCel7A is replaced by Ala245 in DdiCel7A and DpuCel7A, resulting in the loss of a hydrogen bond to the substrate at the subsite at position +1 (Fig. 6A). At the same time, the side chain of Asp345 in loop B4 of DdiCel7A and DpuCel7A is suitably positioned for H bonding to OH1 of the glucosyl unit at position +2 of the reducing end. Actually, most GH7 CBHs have aspartate at the corresponding position, but TreCel7A and other Trichoderma species lack this interaction due to a 1-residue deletion in loop B4 (15, 50, 65).
Cellobiose inhibition.
In light of the structural differences found in the product-binding site, Ki studies for product inhibition were carried out using pNPL as the substrate. Simultaneous nonlinear regression was used to evaluate the events for competitive, uncompetitive, noncompetitive, and mixed inhibition. The data collected suggest that these enzymes are competitively inhibited by cellobiose, and the Kis for competitive inhibition are reported in Table 1. Cellobiose is 5 to 7 times less inhibitory to DdiCel7ACBM and DpuCel7ACBM than to TreCel7A, with Ki values of 205, 130, and 29 μM, respectively. This is consistent with the findings of previous studies investigating similar structural changes in the GH7 CBH active site (36).
DISCUSSION
GH7 CBHs are complex, important enzymes in both nature and the growing biofuels industry. Continued structural and biochemical studies of these enzymes are essential to developing detailed structure-function relationships, given their importance. The GH7 CBHs from D. discoideum and D. purpureum are now the second and third known nonfungal GH7 enzyme structures, with the first one being from a salt water-inhabiting, wood-boring isopod, L. quadripunctata (22).
Although many GH7 CBHs are bimodular, exhibiting a catalytic domain (CD) connected to a carbohydrate-binding module (CBM) by an ∼30-amino-acid flexible linker, the native sequences of DdiCel7A and DpuCel7A do not exhibit a CBM-linker domain. It has long been known that the CBM aids the enzyme by increasing the binding to its crystalline cellulose substrate (66); however, it is likely that there are evolutionary reasons for the lack of a CBM, such as environments of high cellulose density where the substrate concentration greatly surpasses the Km of the CBM or CD. Indeed, a recent study by Varnai et al. demonstrated that TreCel7A without the CBM-linker is able to achieve the same extent of conversion on Avicel and pretreated wheat straw at a high level (20%) of solids loadings, while at a lower level of solids loading, such as the 0.5% used here, the CBM-linker aids in conversion by binding crystalline cellulose and increasing the local concentration (67). Consequently, for biochemical studies, we have added the family 1 CBM and linker from TreCel7A to the CD of native DdiCel7A and DpuCel7A (DdiCel7CBM and DpuCel7ACBM, respectively) to be able to compare more directly GH7 CBH conversion at low levels of solids loadings. Going forward, self-consistent comparisons of GH7 CBHs with (or without) CBM-linkers, enabled by the development of a T. reesei expression host from which Cel7A is deleted (46), will be important to the continued development of structure-activity relationships and the development of the baseline activity of GH7 CBHs from natural diversity in the context of a background enzyme cocktail on process-relevant substrates.
Since T. reesei is known to be one of the most cellulolytic organisms, it is surprising that both Dictyostelium GH7s were equally active on biomass and crystalline cellulose at their respective temperature optima (Table 1). However, previous investigations of the cellulose synthase in D. discoideum have shown that the cellulose deposited in the sheath is highly crystalline cellulose I (68) and that the high crystallinity is positively correlated with an increased strength and rigidity of the stalk (69). Thus, it is not unreasonable to surmise that the Dictyostelium Cel7s have evolved a highly efficient mechanism for rearrangement of crystalline cellulose I to resourcefully modify the cell wall and sheath structure and potentially recycle glucose during formation of the stalk and fruiting body. Additionally, Eichinger et al. speculate that dictyostelia may also utilize the cellulose- and hemicellulose-degrading enzymes in their genomes to degrade plant tissue for food (40). Immunolabeling of GH7 CBHs is a well-known technique to visualize these enzymes (70) and could potentially be used to examine the distribution of these enzymes when they are expressed in the presence and absence of either self-generated cellulose or exogenous sources to determine the function of GH7 CBHs in the life cycle of dictyostelia.
In a previous report, following the in vivo expression of GH7 CBH in D. discoideum, the enzyme was found to be highly controlled by the developmental phase of the organism (11). While the actual quantity of GH7 CBH at each stage was not reported, it may be that this organism expresses only a small amount of Cel7 at critical developmental phases and that the expression of large quantities, like those secreted by T. reesei during biomass degradation, could perhaps be detrimental to the organism. It is also possible that the hydrolytic ability of these enzymes is advantageous, in that it enables the organism to progress through morphogenesis quickly, therefore allowing it to rapidly escape ill-fated environments. Given this tight regulation of expression, it would also be interesting to investigate the regulation of the GH7 CBH promoter during morphogenesis to better understand the process of morphogenesis or the mechanism of transcription regulation, as well as for its potential use in future GH7 expression systems for industrial applications.
The crystal structures of native DdiCel7A and DpuCel7A are exceptionally similar to each other and to the crystal structure of TreCel7A (Fig. 4). Indeed, the degree of binding tunnel closure of native DdiCel7A and DpuCel7A is the closest to that of TreCel7A to have been observed among GH7 structures to date (Fig. 5). Functionally, this suggests increased processivity and a reduced dissociation constant that are perhaps similar to those of TreCel7A. Of particular significance is the lack of tyrosine in the B3 and A3 loops, whose side chains maintain some degree of flexibility (exemplified by their different positions in the TreCel7A structures with PDB accession numbers 1CEL and 4C4C) and partially enclose the binding tunnel at the catalytic center (8). To compensate, the Dictyostelium Cel7A enzymes have a unique tyrosine (Y202) that protrudes from the base of the B2 loop, whose side chain resides in the same general space as the side chains of the A3 and B3 tyrosines in the other GH7 structures (Fig. 6A). The result is that the degree of binding tunnel enclosure in DpuCel7A and DdiCel7A in this region is comparable to or greater than that in the other GH7 CBHs. In addition to the aforementioned implications for processivity and the dissociation constant, this may suggest that the Dictyostelium Cel7A enzymes may have elevated probabilities of endoinitiation compared with the probability for TreCel7A (34).
The product-binding sites of DdiCel7A and DpuCel7A are similar to those of TreCel7A with a few notable, potentially important differences, namely, T246 (TreCel7A numbering) and D345 (DpuCel7A and DdiCel7A numbering) (Fig. 6A). Ståhlberg et al. (71) speculated on the flexibility of the T246 side chain, as evidenced by elevated temperature factors and weaker electron density when the binding site at position +1 was vacant. However, when the binding site at position +1 was filled, the position of T246 was as stable as the rest of the protein, thus providing further evidence for the importance of this hydrogen bond. von Ossowski et al. (36) discussed the impact of this residue's absence in PchCel7D, as did Ubhayasekera et al. (65). Ubhayasekera et al. (65) concluded that the two most significant differences in ligand binding between TreCel7A and PchCel7D are the direct hydrogen bond with T246 (which is absent in PchCel7D) and the D336 interaction at the binding tunnel exit, in which the interaction is present in PchCel7D but in which D336 is replaced by glycine in TreCel7A. Like PchCel7D, both Dictyostelium structures possess an aspartate at this position (D345 in DdiCel7A and DpuCel7A), as illustrated in Fig. 6B. The conservation of this interaction in GH7 cellulases has been discussed (65); as mentioned in Results, the aspartate residue here is rather well conserved in all GH7 CBHs except those in the Trichoderma genus and a few others, where the B4 loop is 1 residue shorter. This aspartate is additionally absent in GH7 EGs due to deletion of the B4 loop, implying that this residue may play a role in cellulose chain processivity in some GH7 CBHs.
To verify the functional significance of the unique product site motif, we investigated the biochemical characteristics and activities of DdiCel7ACBM and DpuCel7ACBM on soluble (Table 1) and insoluble (Table 2) substrates. We found that on the soluble substrate pNPL, DpuCel7ACBM has a higher specific activity than either DdiCel7ACBM or TreCel7A. Interestingly, we could find no structural difference between the CDs of DdiCel7A and DpuCel7A to fully explain the differences in activities. Evolutionarily, it is unclear why the Dictyostelium Cel7s are more cellobiose tolerant. Perhaps the high viscosity of the extracellular matrix amplifies the effective cellobiose concentration due to limited diffusion. Both Dictyostelium organisms studied here contain multiple genes predicted to have β-glucosidase activity (seven for D. discoideum and eight for D. purpureum) (7); however, to our knowledge, it has not been reported if the organism utilizes the glucose for energy or for some other purpose, such as feeding a bacterial symbiote, as has been suggested previously (40). The cosecretion of β-glucosidases alongside the GH7 CBHs has also not been examined, to our knowledge.
GH7 genes occur in branches of the eukaryotic tree of life that diverged over 1 billion years ago (Fig. 7) (72), suggesting either an ancient ancestral GH7 gene or gene uptake via horizontal gene transfer (HGT). These genes have been well characterized and identified in many cellulolytic fungi and, more recently, in animals, such as marine wood borers (Crustacea, e.g., the gribble [Limnoria quadripunctata]) (39). Moreover, they have been found in additional eukaryote branches, such as amoeba (40), oomycetes (stramenopiles, e.g., the potato blight pathogen Phytophthora infestans), haptophytes (e.g., the phytoplankton Emiliania huxleyi), and parabasilids (Excavata, e.g., the termite hindgut symbiont Pseudotrichonympha grassii) (39, 73). Compared to other GH families, GH7 CBHs are noteworthy in how conserved they are in terms of sequence identity (>40%; Fig. 7; Table 4). If GH7 cellulases were derived from a single ancestor before the earliest point of divergence in the eukaryotic tree of life, the sequence conservation is remarkable, given the early event of divergence. If this is indeed the case, these findings suggest that GH7 CBHs cannot accommodate a broad sequence space for primary function. However, we cannot rule out the possibility that HGT occurred on a more recent time scale, thus limiting the extent of sequence divergence. We performed a phylogenetic analysis of taxonomically diverse GH7 protein sequences to construct an evolutionary tree on the basis of the sequence of the GH7 domain alone. A simplified tree is shown in Fig. 8, and the complete tree with all sequence names is shown in Fig. S2 in the supplemental material. The amoebae are placed among ascomycete sequences, indeed suggesting HGT from an early ascomycete to a dictyostelial ancestor. The event should then have occurred after the separation of the Ascomycota and Basidiomycota (∼600 million years ago) but before the divergence of D. discoideum and D. purpureum, which is estimated to have been about 400 million years ago (7), and supposedly much earlier, as indicated by additional nodes further down in the Amoebozoa subtree and the high degree of similarity between DdiCel7A and DpuCel7A (Fig. 8). HGT has been hypothesized to be a primary means of genome evolution in D. discoideum (40). Given the lifestyle of dictyostelia, which inhabit forest soil, it seems plausible that HGT of the genes for GH7 CBHs from fungi may have been the primary mechanism for the presence of these enzymes present in slime mold genomes (or vice versa). Similar arguments have been made for HGT into wood-boring crustaceans, such as L. quadripunctata (39). We note, however, that all the genes in dictyostelia that are strongly supported to have been acquired by HGT are genes of bacterial origin (7) and prokaryote-eukaryote HGT is generally easier to detect than HGT of nuclear genes between eukaryotes (74). For oomycetes, extensive whole-genome, gene-to-gene phylogenetic analysis has been applied to detect putative HGT between fungi and plant-pathogenic oomycetes, and the results indicate that HGT has been fundamental to the evolution of plant-parasitic traits within the oomycetes (75). Interestingly, though, the GH7 gene was not present among the 34 candidate genes acquired by HGT identified in the analysis, suggesting that it does not show strong signs of being acquired by HGT in oomycetes [while a GH10 gene annotated as a β(1,4)-xylanase is], which is also consistent with the findings of our phylogenetic analysis (Fig. 8). Regardless of the gene transfer mechanism or ancestry, we find it remarkable that, despite a distant phylogenetic and temporal relationship between dictyostelial GH7 CBHs and their more well-studied fungal counterparts, such high degrees of homology and similarity in activity and structure are preserved.
FIG 8.
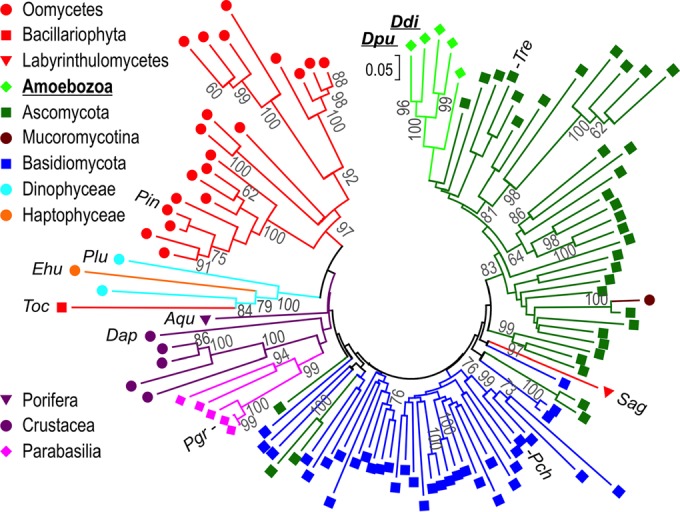
Phylogenetic tree of 113 GH7 CBH protein sequences. The evolutionary history was inferred using the minimum evolution method (58) with 700 bootstrap replicates in MEGA (v. 7) software (57) from a multiple-sequence alignment of the GH7 domain by MUSCLE, as described in Materials and Methods. Branch lengths are drawn to scale, with evolutionary distances, which were computed using the Dayhoff matrix-based method, being given in units of number of amino acid substitutions per site. Gray numbers are bootstrap values (in percent). A key to the colors and symbols is provided. Sequences listed in Figure 7 and Table 4 are indicated with the following abbreviations (UniProt accession numbers within parentheses): Ddi, Dictyostelium discoideum Cel7A (Q55FE6); Dpu, Dictyostelium purpureum Cel7A (F0ZJZ1); Tre, Trichoderma reesei Cel7A (P62694); Sag, Schizochytrium aggregatum (sequence from reference 76); Pch, Phanerochaete chrysosporium Cel7D (Q7LIJ0); Pgr, Pseudotrichonympha grassii (Q95YH1); Dap, Daphnia pulex Cel7A (E9G5J5); Aqu, Amphimedon queenslandica (I1FU52); Toc, Thalassiosira oceanica (K0T5R6); Ehu, Emiliania huxleyi (R1C7R5); Plu, Pyrocystis lunula (D8UXL6); Pin, Phytophthora infestans (D0N841). The complete tree with all sequence names is provided in Fig. S2 in the supplemental material.
In conclusion, this study presents the crystal structures of the GH7 CBHs of two social amoebae and shows that they are structurally and functionally very similar to the crystal structure of the well-studied TreCel7A, revealing a remarkable degree of conservation in these important cellulose-degrading enzymes across distant phylogenetic branches on the eukaryotic tree of life (39). Several key differences in the makeup of the binding tunnel loops and the product-binding sites likely give rise to the observed differences in kinetics on the small-molecule substrate pNPL and the reduction in product inhibition by cellobiose compared to the results for TreCel7A. Lastly, this study presents an approach to append a CBM-linker on GH7 CBHs to produce chimeric cellulases for direct comparison on insoluble cellulose-rich substrates. This approach will be essential to building a large collection of self-consistent results in the study of GH7 CBH activities and for determining accurate structure-function relationships for these important enzymes (8). The functional relevance of the GH7 CBHs in dictyostelia, likely intimately related to the reshaping of the extracellular matrix important to the slime mold life cycle in its morphogenesis from slug to fruiting body (11) or in its ability to digest cellulose-containing organisms in the forest soil (40), remains a question of particular interest to understand the evolutionary pressures on the activities of slime mold GH7 CBHs.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to Nils Högberg, Swedish University of Agricultural Sciences, for advice on phylogenetic analysis.
Funding Statement
S.E.H., B.C.K., L.E.T., K.K.P., T.A.V., M.E.H., S.R.D., and G.T.B. acknowledge the U.S. Department of Energy BioEnergy Technologies Office for funding. M.H.M., A.S.B., and J.S. acknowledge the Swedish Research Council Formas and the Faculty for Natural Resources and Agriculture at the Swedish University of Agricultural Sciences through the research program MicroDrivE. B.C.K. also thanks the National Renewable Energy Laboratory's Director's Fellowship Program for funding.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00163-16.
REFERENCES
- 1.Wang Y, Slade MB, Gooley AA, Atwell BJ, Williams KL. 2001. Cellulose-binding modules from extracellular matrix proteins of Dictyostelium discoideum stalk and sheath. Eur J Biochem 268:4334–4345. doi: 10.1046/j.1432-1327.2001.02354.x. [DOI] [PubMed] [Google Scholar]
- 2.Zhang P, McGlynn AC, Loomis WF, Blanton RL, West CM. 2001. Spore coat formation and timely sporulation depend on cellulose in Dictyostelium. Differentiation 67:72–79. doi: 10.1046/j.1432-0436.2001.067003072.x. [DOI] [PubMed] [Google Scholar]
- 3.Huber RJ, O'Day DH. 2015. Proteomic profiling of the extracellular matrix (slime sheath) of Dictyostelium discoideum. Proteomics 15:3315–3319. doi: 10.1002/pmic.201500143. [DOI] [PubMed] [Google Scholar]
- 4.Freeze H, Loomis WF. 1977. Isolation and characterization of a component of the surface sheath of Dictyostelium discoideum. J Biol Chem 252:820–824. [PubMed] [Google Scholar]
- 5.Freeze H, Loomis WF. 1978. Chemical analysis of stalk components of Dictyostelium discoideum. Biochim Biophys Acta 539:529–537. doi: 10.1016/0304-4165(78)90086-7. [DOI] [PubMed] [Google Scholar]
- 6.Nishiyama Y, Langan P, Chanzy H. 2002. Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc 124:9074–9082. doi: 10.1021/ja0257319. [DOI] [PubMed] [Google Scholar]
- 7.Sucgang R, Kuo A, Tian X, Salerno W, Parikh A, Feasley CL, Dalin E, Tu H, Huang E, Barry K, Lindquist E, Shapiro H, Bruce D, Schmutz J, Salamov A, Fey P, Gaudet P, Anjard C, Babu MM, Basu S, Bushmanova Y, van der Wel H, Katoh-Kurasawa M, Dinh C, Coutinho PM, Saito T, Elias M, Schaap P, Kay RR, Henrissat B, Eichinger L, Rivero F, Putnam NH, West CM, Loomis WF, Chisholm RL, Shaulsky G, Strassmann JE, Queller DC, Kuspa A, Grigoriev IV. 2011. Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum. Genome Biol 12:R20. doi: 10.1186/gb-2011-12-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Payne CM, Knott BC, Mayes HB, Hansson H, Himmel ME, Sandgren M, Ståhlberg J, Beckham GT. 2015. Fungal cellulases. Chem Rev 115:1308–1448. doi: 10.1021/cr500351c. [DOI] [PubMed] [Google Scholar]
- 9.Chundawat SP, Beckham GT, Himmel ME, Dale BE. 2011. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu Rev Chem Biomol Eng 2:121–145. doi: 10.1146/annurev-chembioeng-061010-114205. [DOI] [PubMed] [Google Scholar]
- 10.Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 11.Kunii M, Yasuno M, Shindo Y, Kawata T. 2014. A Dictyostelium cellobiohydrolase orthologue that affects developmental timing. Dev Genes Evol 224:25–35. doi: 10.1007/s00427-013-0460-x. [DOI] [PubMed] [Google Scholar]
- 12.Divne C, Stahlberg J, Reinikainen T, Ruohonen L, Pettersson G, Knowles JK, Teeri TT, Jones TA. 1994. The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science 265:524–528. doi: 10.1126/science.8036495. [DOI] [PubMed] [Google Scholar]
- 13.Divne C, Ståhlberg J, Teeri TT, Jones TA. 1998. High-resolution crystal structures reveal how a cellulose chain is bound in the 50 Å long tunnel of cellobiohydrolase I from Trichoderma reesei. J Mol Biol 275:309–325. doi: 10.1006/jmbi.1997.1437. [DOI] [PubMed] [Google Scholar]
- 14.Knott BC, Haddad Momeni M, Crowley MF, Mackenzie LF, Götz AW, Sandgren M, Withers SG, Ståhlberg J, Beckham GT. 2013. The mechanism of cellulose hydrolysis by a two-step, retaining cellobiohydrolase elucidated by structural and transition path sampling studies. J Am Chem Soc 136:321–329. doi: 10.1021/ja410291u. [DOI] [PubMed] [Google Scholar]
- 15.Haddad Momeni M, Payne CM, Hansson H, Mikkelsen NE, Svedberg J, Engstrom A, Sandgren M, Beckham GT, Ståhlberg J. 2013. Structural, biochemical, and computational characterization of the glycoside hydrolase family 7 cellobiohydrolase of the tree-killing fungus Heterobasidion irregulare. J Biol Chem 288:5861–5872. doi: 10.1074/jbc.M112.440891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muñoz IG, Ubhayasekera W, Henriksson H, Szabó I, Pettersson G, Johansson G, Mowbray SL, Ståhlberg J. 2001. Family 7 cellobiohydrolases from Phanerochaete chrysosporium: crystal structure of the catalytic module of Cel7D (CBH58) at 1.32 Å resolution and homology models of the isozymes. J Mol Biol 314:1097–1111. doi: 10.1006/jmbi.2000.5180. [DOI] [PubMed] [Google Scholar]
- 17.Grassick A, Murray PG, Thompson R, Collins CM, Byrnes L, Birrane G, Higgins TM, Tuohy MG. 2004. Three-dimensional structure of a thermostable native cellobiohydrolase, CBH IB, and molecular characterization of the cel7 gene from the filamentous fungus, Talaromyces emersonii. Eur J Biochem 271:4495–4506. doi: 10.1111/j.1432-1033.2004.04409.x. [DOI] [PubMed] [Google Scholar]
- 18.Textor LC, Colussi F, Silveira RL, Serpa V, Mello BL, Muniz JRC, Squina FM, Pereira N, Skaf MS, Polikarpov I. 2013. Joint X-ray crystallographic and molecular dynamics study of cellobiohydrolase I from Trichoderma harzianum: deciphering the structural features of cellobiohydrolase catalytic activity. FEBS J 280:56–69. doi: 10.1111/febs.12049. [DOI] [PubMed] [Google Scholar]
- 19.Parkkinen T, Koivula A, Vehmaanpera J, Rouvinen J. 2008. Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding. Protein Sci 17:1383–1394. doi: 10.1110/ps.034488.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moroz O, Maranta M, Shaghasi T, Harris P, Wilson K, Davies G. 2015. The three-dimensional structure of the cellobiohydrolase Cel7A from Aspergillus fumigatus at 1.5 Å resolution. Acta Crystallogr F Struct Biol Commun 71(Pt 1):114–120. doi: 10.1107/S2053230X14027307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haddad Momeni M, Goedegebuur F, Hansson H, Karkehabadi S, Askarieh G, Mitchinson C, Larenas EA, Ståhlberg J, Sandgren M. 2014. Expression, crystal structure and cellulase activity of the thermostable cellobiohydrolase Cel7A from the fungus Humicola grisea var. thermoidea. Acta Crystallogr D Biol Crystallogr 70:2356–2366. doi: 10.1107/S1399004714013844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kern M, McGeehan JE, Streeter SD, Martin RN, Besser K, Elias L, Eborall W, Malyon GP, Payne CM, Himmel ME. 2013. Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance. Proc Natl Acad Sci U S A 110:10189–10194. doi: 10.1073/pnas.1301502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borisova AS, Eneyskaya EV, Bobrov KS, Jana S, Logachev A, Polev DE, Lapidus AL, Ibatullin FM, Saleem U, Sandgren M. 2015. Sequencing, biochemical characterization, crystal structure and molecular dynamics of cellobiohydrolase Cel7A from Geotrichum candidum 3C. FEBS J 282:4515–4537. doi: 10.1111/febs.13509. [DOI] [PubMed] [Google Scholar]
- 24.Kleywegt GJ, Zou JY, Divne C, Davies GJ, Sinning I, Ståhlberg J, Reinikainen T, Srisodsuk M, Teeri TT, Jones TA. 1997. The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 Å resolution, and a comparison with related enzymes. J Mol Biol 272:383–397. doi: 10.1006/jmbi.1997.1243. [DOI] [PubMed] [Google Scholar]
- 25.MacKenzie L, Sulzenbacher G, Divne C, Jones T, Woldike H, Schulein M, Withers S, Davies G. 1998. Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 Å; resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate. Biochem J 335:409–416. doi: 10.1042/bj3350409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sulzenbacher G, Driguez H, Henrissat B, Schulein M, Davies GJ. 1996. Structure of the Fusarium oxysporum endoglucanase I with a nonhydrolyzable substrate analogue: substrate distortion gives rise to the preferred axial orientation for the leaving group. Biochemistry 35:15280–15287. doi: 10.1021/bi961946h. [DOI] [PubMed] [Google Scholar]
- 27.Stals I, Sandra K, Geysens S, Contreras R, Van Beeumen J, Claeyssens M. 2004. Factors influencing glycosylation of Trichoderma reesei cellulases. I. Postsecretorial changes of the O- and N-glycosylation pattern of Cel7A. Glycobiology 14:713–724. [DOI] [PubMed] [Google Scholar]
- 28.Beckham GT, Bomble YJ, Matthews JF, Taylor CB, Resch MG, Yarbrough JM, Decker SR, Bu L, Zhao X, McCabe C, Wohlert J, Bergenstråhle M, Brady JW, Adney WS, Himmel ME, Crowley MF. 2010. The O-glycosylated linker from the Trichoderma reesei family 7 cellulase is a flexible, disordered protein. Biophys J 99:3773–3781. doi: 10.1016/j.bpj.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sammond DW, Payne CM, Brunecky R, Himmel ME, Crowley MF, Beckham GT. 2012. Cellulase linkers are optimized based on domain type and function: insights from sequence analysis, biophysical measurements, and molecular simulation. PLoS One 7:e48615. doi: 10.1371/journal.pone.0048615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beckham GT, Bomble YJ, Bayer EA, Himmel ME, Crowley MF. 2011. Applications of computational science for understanding enzymatic deconstruction of cellulose. Curr Opin Biotechnol 22:231–238. doi: 10.1016/j.copbio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Bu L, Beckham GT, Shirts MR, Nimlos MR, Adney WS, Himmel ME, Crowley MF. 2011. Probing carbohydrate product expulsion from a processive cellulase with multiple absolute binding free energy methods. J Biol Chem 286:18161–18169. doi: 10.1074/jbc.M110.212076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vrsanska M, Biely P. 1992. The cellobiohydrolase-I from Trichoderma reesei Qm-9414—action on cello-oligosaccharides. Carbohydr Res 227:19–27. doi: 10.1016/0008-6215(92)85058-8. [DOI] [Google Scholar]
- 33.Igarashi K, Uchihashi T, Koivula A, Wada M, Kimura S, Okamoto T, Penttila M, Ando T, Samejima M. 2011. Traffic jams reduce hydrolytic efficiency of cellulase on cellulose surface. Science 333:1279–1282. doi: 10.1126/science.1208386. [DOI] [PubMed] [Google Scholar]
- 34.Kurasin M, Valjamae P. 2011. Processivity of cellobiohydrolases is limited by the substrate. J Biol Chem 286:169–177. doi: 10.1074/jbc.M110.161059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jalak J, Valjamae P. 2014. Multi-mode binding of cellobiohydrolase Cel7A from Trichoderma reesei to cellulose. PLoS One 9:e108181. doi: 10.1371/journal.pone.0108181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Ossowski I, Ståhlberg J, Koivula A, Piens K, Becker D, Boer H, Harle R, Harris M, Divne C, Mahdi S, Zhao Y, Driguez H, Claeyssens M, Sinnott ML, Teeri TT. 2003. Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A. A comparison with Phanerochaete chrysosporium Cel7D. J Mol Biol 333:817–829. doi: 10.1016/S0022-2836(03)00881-7. [DOI] [PubMed] [Google Scholar]
- 37.Andric P, Meyer AS, Jensen PA, Dam-Johansen K. 2010. Reactor design for minimizing product inhibition during enzymatic lignocellulose hydrolysis. II. Quantification of inhibition and suitability of membrane reactors. Biotechnol Adv 28:407–425. doi: 10.1016/j.biotechadv.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Gan Q, Allen SJ, Taylor G. 2002. Design and operation of an integrated membrane reactor for enzymatic cellulose hydrolysis. Biochem Eng J 12:223–229. doi: 10.1016/S1369-703X(02)00072-4. [DOI] [Google Scholar]
- 39.King AJ, Cragg SM, Li Y, Dymond J, Guille MJ, Bowles DJ, Bruce NC, Graham IA, McQueen-Mason SJ. 2010. Molecular insight into lignocellulose digestion by a marine isopod in the absence of gut microbes. Proc Natl Acad Sci U S A 107:5345–5350. doi: 10.1073/pnas.0914228107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, Tunggal B, Kummerfeld S, Madera M, Konfortov BA, Rivero F, Bankier AT, Lehmann R, Hamlin N, Davies R, Gaudet P, Fey P, Pilcher K, Chen G, Saunders D, Sodergren E, Davis P, Kerhornou A, Nie X, Hall N, Anjard C, Hemphill L, Bason N, Farbrother P, Desany B, Just E, Morio T, Rost R, Churcher C, Cooper J, Haydock S, van Driessche N, Cronin A, Goodhead I, Muzny D, Mourier T, Pain A, Lu M, Harper D, Lindsay R, Hauser H, et al. . 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cragg SM, Beckham GT, Bruce NC, Bugg TD, Distel DL, Dupree P, Etxabe AG, Goodell BS, Jellison J, McGeehan JE. 2015. Lignocellulose degradation mechanisms across the tree of life. Curr Opin Chem Biol 29:108–119. doi: 10.1016/j.cbpa.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beckham GT, Dai Z, Matthews JF, Momany M, Payne CM, Adney WS, Baker SE, Himmel ME. 2012. Harnessing glycosylation to improve cellulase activity. Curr Opin Biotechnol 23:338–345. doi: 10.1016/j.copbio.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 43.Singh A, Taylor LE, Vander Wall TA, Linger J, Himmel ME, Podkaminer K, Adney WS, Decker SR. 2014. Heterologous protein expression in Hypocrea jecorina: a historical perspective and new developments. Biotechnol Adv 33:142–154. doi: 10.1016/j.biotechadv.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 44.Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA. 2012. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res 40(Database issue):D26–D32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, Smirnova T, Grigoriev IV, Dubchak I. 2014. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res 42(Database issue):D26–D31. doi: 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linger J, Taylor L, Baker J, Vander Wall T, Hobdey S, Podkaminer K, Himmel M, Decker S. 2015. A constitutive expression system for glycosyl hydrolase family 7 cellobiohydrolases in Hypocrea jecorina. Biotechnol Biofuels 8:45. doi: 10.1186/s13068-015-0230-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McPherson A. 1982. Preparation and analysis of protein crystals. John Wiley & Sons, New York, NY. [Google Scholar]
- 48.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haddad Momeni M, Ubhayasekera W, Sandgren M, Ståhlberg J, Hansson H. 2015. Structural insights into the inhibition of cellobiohydrolase Cel7A by xylooligosaccharides. FEBS J 282:2167–2177. doi: 10.1111/febs.13265. [DOI] [PubMed] [Google Scholar]
- 51.Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PDB Viewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 54.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonnet GH, Cohen MA, Benner SA. 1992. Exhaustive matching of the entire protein sequence database. Science 256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- 57.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rzhetsky A, Nei M. 1992. A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol 9:945–967. [Google Scholar]
- 59.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 60.Schwartz R, Dayhoff M. 1978. Matrices for detecting distant relationships. Atlas Protein Sequence Struct 5:353–358. [Google Scholar]
- 61.Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 62.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 63.Baker JO, Ehrman CI, Adney WS, Thomas SR, Himmel ME. 1998. Hydrolysis of cellulose using ternary mixtures of purified celluloses. Appl Biochem Biotechnol 70–72:395–403. [DOI] [PubMed] [Google Scholar]
- 64.Payne CM, Resch MG, Chen L, Crowley MF, Himmel ME, Taylor LE, Sandgren M, Ståhlberg J, Stals I, Tan Z. 2013. Glycosylated linkers in multimodular lignocellulose-degrading enzymes dynamically bind to cellulose. Proc Natl Acad Sci U S A 110:14646–14651. doi: 10.1073/pnas.1309106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ubhayasekera W, Munoz IG, Vasella A, Ståhlberg J, Mowbray SL. 2005. Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors. FEBS J 272:1952–1964. doi: 10.1111/j.1742-4658.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- 66.Vantilbeurgh H, Tomme P, Claeyssens M, Bhikhabhai R, Pettersson G. 1986. Limited proteolysis of the cellobiohydrolase I from Trichoderma reesei—separation of functional domains. FEBS Lett 204:223–227. doi: 10.1016/0014-5793(86)80816-X. [DOI] [Google Scholar]
- 67.Varnai A, Siika-Aho M, Viikari L. 2013. Carbohydrate-binding modules (CBMs) revisited: reduced amount of water counterbalances the need for CBMs. Biotechnol Biofuels 6:30. doi: 10.1186/1754-6834-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blanton RL, Northcote DH. 1990. A 1,4-beta-d-glucan-synthase system from Dictyostelium discoideum. Planta 180:324–332. doi: 10.1007/BF01160387. [DOI] [PubMed] [Google Scholar]
- 69.Wakeman H. 1954. Mechanical properties of cellulose and its derivatives, p 1247–1355. In Ott E, Spurlin H (ed), Cellulose and cellulose derivatives. Wiley Interscience, New York, NY. [Google Scholar]
- 70.Resch MG, Donohoe BS, Baker JO, Decker SR, Bayer EA, Beckham GT, Himmel ME. 2013. Fungal cellulases and complexed cellulosomal enzymes exhibit synergistic mechanisms in cellulose deconstruction. Energy Environ Sci 6:1858–1867. doi: 10.1039/c3ee00019b. [DOI] [Google Scholar]
- 71.Ståhlberg J, Henriksson H, Divne C, Isaksson R, Pettersson G, Johansson G, Jones TA. 2001. Structural basis for enantiomer binding and separation of a common β-blocker: crystal structure of cellobiohydrolase Cel7A with bound (S)-propranolol at 1.9 Å resolution. J Mol Biol 305:79–93. doi: 10.1006/jmbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- 72.Berney C, Pawlowski J. 2006. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc Biol Sci 273:1867–1872. doi: 10.1098/rspb.2006.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parfrey LW, Grant J, Tekle YI, Lasek-Nesselquist E, Morrison HG, Sogin ML, Patterson DJ, Katz LA. 2010. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol 59:518–533. doi: 10.1093/sysbio/syq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keeling PJ, Palmer JD. 2008. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet 9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 75.Richards TA, Soanes DM, Jones MD, Vasieva O, Leonard G, Paszkiewicz K, Foster PG, Hall N, Talbot NJ. 2011. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Natl Acad Sci U S A 108:15258–15263. doi: 10.1073/pnas.1105100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brevnova EE, Flatt J, Gandhi C, Rajgarhia V, McBride J, Warner A. June 2013. Isolation and characterization of Schizochytrium aggregatum cellobiohydrolase I (Cbh 1). US patent 8,470,592 B2.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.