ABSTRACT
Mycoplasmas are minimal, wall-less bacteria but have retained the ability to secrete complex carbohydrate polymers that constitute a glycocalyx. In members of the Mycoplasma mycoides cluster, which are important ruminant pathogens, the glycocalyx includes both cell-attached and cell-free polysaccharides. This report explores the potential secretion of polysaccharides by M. agalactiae, another ruminant pathogen that belongs to a distant phylogenetic group. Comparative genomic analyses showed that M. agalactiae possesses all the genes required for polysaccharide secretion. Notably, a putative synthase gene (gsmA) was identified, by in silico reconstruction of the biosynthetic pathway, that could be involved in both polymerization and export of the carbohydrate polymers. M. agalactiae polysaccharides were then purified in vitro and found to be mainly cell attached, with a linear β-(1→6)-glucopyranose structure [β-(1→6)-glucan]. Secretion of β-(1→6)-glucan was further shown to rely on the presence of a functional gsmA gene, whose expression is subjected to high-frequency phase variation. This event is governed by the spontaneous intraclonal variation in length of a poly(G) tract located in the gsmA coding sequence and was shown to occur in most of the M. agalactiae clinical isolates tested in this study. M. agalactiae susceptibility to serum-killing activity appeared to be dictated by ON/OFF switching of β-(1→6)-glucan secretion, suggesting a role of this phenomenon in survival of the pathogen when it invades the host bloodstream. Finally, β-(1→6)-glucan secretion was not restricted to M. agalactiae but was detected also in M. mycoides subsp. capri PG3T, another pathogen of small ruminants.
IMPORTANCE Many if not all bacteria are able to secrete polysaccharides, either attached to the cell surface or exported unbound into the extracellular environment. Both types of polysaccharides can play a role in bacterium-host interactions. Mycoplasmas are no exception despite their poor overall metabolic capacity. We showed here that M. agalactiae secretes a capsular β-(1→6)-glucopyranose thanks to a specific glycosyltransferase with synthase activity. This secretion is governed by high-frequency ON/OFF phase variation that might be crucial in mycoplasma host dissemination, as cell-attached β-(1→6)-glucopyranose increases serum-killing susceptibility. Our results provide functional genetic data about mycoplasmal glycosyltransferases with dual functions, i.e., assembly and export of the sugar polymers across the cell membrane. Furthermore, we demonstrated that nonprotein epitopes can be subjected to surface antigenic variation in mycoplasmas. Finally, the present report contributes to unravel the role of secreted polysaccharides in the virulence and pathogenicity of these peculiar bacteria.
INTRODUCTION
Many bacterial species are able to synthesize and secrete carbohydrate polymers that either remain attached to the cell surface (capsular polysaccharides [CPS]) or are exported unbound into the extracellular environment, sometimes forming a loose slime layer (exopolysaccharides [EPS]) (1). The term “CPS” refers here to cell-attached glycoconjugates whether or not they form a true capsule that can be visualized by electron microscopy. There is growing awareness of the role played by CPS and EPS in cross talk among bacteria and between the bacterium and its host cell or the overall environment (2, 3). In various bacterial models, secreted polysaccharides were shown to modulate the intrinsic properties of the producing bacteria (surface adhesion, biofilm formation, resistance to host immune defenses such as phagocytosis and serum killing) as well as the host cell response through immunomodulation. Hence, bacterial exopolysaccharides have been studied both as true virulence factors in several infectious diseases (4) and as important components of probiotics due to their positive impact on the host (5). The interplay between the host and the polysaccharide-secreting bacteria is complex and commonly involves phase variation of the surface-exposed polysaccharides to modulate their level of expression during colonization or infection processes (6, 7).
Mycoplasmas constitute a peculiar group of wall-less bacteria with small genomes. Despite the paucity of their metabolic pathways, these minimal cells have been shown to produce complex macromolecules with carbohydrate moieties, the so-called glycocalyx (8). Polysaccharide secretion in mycoplasmas was first evidenced in 1958 (9), and since then the number of species reported to have a capsule has been constantly increasing (8). The current list of mycoplasmas producing CPS or EPS is not exhaustive, however, as purification of polysaccharides has been hampered for many years by the use of complex culture media. A detailed description of the polysaccharides secreted by nonruminant mycoplasmas is available only for Mycoplasma pulmonis and M. pneumoniae (10, 11).
Our group recently contributed to identification of the polysaccharides secreted by several species that are pathogenic for ruminants and classified in the so-called M. mycoides cluster (12, 13). These are (i) galactan, a β-(1→6)-galactofuranose polymer, secreted by M. mycoides subsp. mycoides and M. mycoides subsp. capri serovar LC; and (ii) β-(1→2)-glucan, a linear β-(1→2)-glucopyranose polysaccharide, produced by mycoplasmas in the M. capricolum and M. leachii species (see Table 1 for a summary). The role of β-(1→2)-glucan has not yet been studied, but the galactan in M. mycoides subsp. mycoides strain Afadé has been found both to exhibit anti-inflammatory properties in EPS form (14) and to protect the cells from the bactericidal activity of the serum when in CPS form (15).
TABLE 1.
Polysaccharide secretion among members of the M. mycoides cluster and M. agalactiae
| Phylogenetic group | Mycoplasma species | Main host(s) | Disease(s)a | Polysaccharide |
||
|---|---|---|---|---|---|---|
| Structure | Secreted form(s)b | Reference(s) or source | ||||
| Spiroplasma | M. mycoides subsp. mycoides | Bovine | CBPP | Polymer of β(1→6)-galactofuranose | CPS, EPS | 12, 13 |
| M. mycoides subsp. capri serovar LC | Caprine | CA | Polymer of β(1→6)-galactofuranose | CPS | 12 | |
| M. mycoides subsp. capri serovar capri | Caprine | CA | Polymer of β(1→6)-glucopyranose | CPS, EPS | This study | |
| M. leachii | Bovine | Mastitis, arthritis | Polymer of β(1→2)-glucopyranose | CPS, EPS | 12 | |
| M. capricolum subsp. capripneumoniae | Caprine | CCPP | Polymer of β(1→2)-glucopyranose | CPS, EPS | 12 | |
| M. capricolum subsp. capricolum | Caprine | CA | Polymer of β(1→2)-glucopyranose | CPS | 12 | |
| Hominis | M. agalactiae | Caprine, ovine | CA | Polymer of β(1→6)-glucopyranose | CPS | This study |
CBPP, contagious bovine pleuropneumonia; CCPP, contagious caprine pleuropneumonia; CA, contagious agalactia (CA is a syndrome that includes several clinical signs such as mastitis, arthritis, keratoconjunctivitis, and pneumonia).
CPS, capsular polysaccharide; EPS, exopolysaccharide.
Polysaccharide biosynthesis in members of the M. mycoides cluster is predicted to occur as a two-step process (12, 13) in which sugars are first activated by the addition of a nucleoside-diphosphate group and then linked to an appropriate acceptor molecule by a variety of glycosyltransferases, resulting in the synthesis of different glycoconjugates (16). Among the glycosyltransferases involved in EPS/CPS secretion, one specific subfamily of membrane-embedded synthases has been described in several Gram-negative bacteria whose members are able both to assemble and to export the sugar polymers across the cell membrane (17). In M. mycoides subsp. mycoides, only one of the three predicted glycosyltransferases, the MSC_0108 product, exhibits structural features indicative of a synthase function, namely, the presence of multiple transmembrane domains (TMD) and a large cytoplasmic loop bearing two specific motifs, DXD and R/QXXRW (13) (Table 2; see also Fig. S1 in the supplemental material). The homology of the MSC_0108 product with the BcsA subunit of Rhodobacter sphaeroides cellulose synthase, an enzyme responsible for cellulose synthesis and translocation across the cytoplasmic membrane, prompted us to propose that the MSC_0108 product is the galactan synthase catalyzing both UDP-galactofuranose polymerization and galactan secretion across the mycoplasma cytoplasmic membrane (13).
TABLE 2.
Enzymes involved in the biosynthesis of polysaccharides in the M. mycoides cluster and putative homologs in M. agalactiae
| Enzyme name or category (reaction)e |
Mycoplasma mycoides cluster gene(s)a |
Mycoplasma agalactiae gene(s)b |
||||
|---|---|---|---|---|---|---|
| M. mycoides subsp. mycoides strain PG1T | M. capricolum subsp. capripneumoniae strain Ambosa | M. mycoides subsp. capri strain PG3T | Strain PG2T | Strain 5632 | Strain 14628 | |
| Pgm (Glc-6P→Glc-1P) | MSC_0829 | MCCP01_0854 | MMC_0730 | MAG4650 | MAGa5120 | MAGb_1060 |
| GalU (Glc-1P→UDP-Glc) | MSC_0110, MSC_0990 | MCCP01_0082 | MMC_6550 | MAG4580 | MAGa4810 | MAGb_0980 |
| GalE (UDP-Glc↔UDP-Galp) | MSC_0971, MSC_0978 | ND | ND | ND | ND | ND |
| Glf (UDP-Galp→UDP-Galf) | MSC_0984 | ND | ND | ND | ND | ND |
| Glycosyltransferases predicted as synthasesc (polymerization and secretion of polysaccharides) | MSC_0108 (449 aa/4 TMDs) | MCCP01_0081 (524 aa/7 TMDs) | MMC_0120 (514 aa/7 TMDs), MMC_6560 (523 aa/7 TMDs) | MAG4860d (244 aa/2 TMDs) | MAGa5320d (96) (351 aa/2 TMDs) | MAGb_1260 (96) (533 aa/7 TMDs) |
| Other glycosyltransferases | MSC_0109, MSC_0771 | MCCP01_0815 | MMC_1050 | MAG0570, MAG3010 | MAGa0600 (98), MAGa3400 (97) | MAGb_4980 (90), MAGb_7470 (97) |
Biosynthetic pathways were predicted by Bertin et al. (12, 13). Genes are designated by their locus tags.
Glycosyltransferases in PG2 were predicted using the CAZy database, and homologs in strains 5632 and 14628 were determined by BLASTP analysis using PG2T sequences (identity percentages are indicated in parentheses). aa, amino acids. Some enzymes were not detected (ND) using BLASTP searches.
Polysaccharide synthases were identified in this study by analyzing their secondary structure predicted by TMHMM 2.0 and their similarity with other known synthases using Phyre. The length of the predicted synthases and the number of transmembrane domains (TMDs) are indicated.
Truncated protein.
Pgm, phosphoglucomutase; GalU, glucose-1-phosphate uridylyltransferase; GalE, UDP-glucose 4-epimerase; Glf, UDP-galactofuranose mutase; Glc, glucose; Galp, galactopyranose; Galf, galactofuranose.
Interestingly, comparative genomic data have shown that several genes involved in polysaccharide biosynthesis have been horizontally transferred between members of the M. mycoides cluster and other mycoplasma species that are also ruminant pathogens but belong to a distant and distinct phylogenetic group (12, 18, 19). These include M. agalactiae, a major pathogen responsible for contagious agalactia in small ruminants, which is listed by the World Organization of Animal Health (OIE) due to its economic impact in countries with important dairy industries. The M. agalactiae model is thus of particular interest because it shares common hosts and ecological niches with some members of the M. mycoides cluster, and this situation could be favorable to gene transfers.
Although CPS and/or EPS may have an important role in M. agalactiae infection, polysaccharide secretion has not yet been investigated in this pathogen. The present study was undertaken to fill this gap. By combining comparative genomics with biochemistry and classical molecular biology, we demonstrated that M. agalactiae can secrete a linear β-(1→6)-glucopyranose which is attached to the cell surface. We then reconstructed the corresponding biosynthetic pathway in silico and showed that this secretion was governed by high-frequency phase variation, which could play a role in M. agalactiae-host interactions.
MATERIALS AND METHODS
Mycoplasma strains and culture conditions.
The genome sequences of M. agalactiae strains PG2T, 5632, and 14628 and M. mycoides subsp. capri serovar capri strain PG3T are available in databases (GenBank accession numbers CU179680, FP671138, and AJPR00000000.1 and JFAE00000000.1, respectively). Other M. agalactiae clinical isolates came from a collection maintained in our laboratory. Most isolates were collected within the framework of the Vigimyc national surveillance network (20) or during specific epidemiological studies (19). Strains were grown in PPLO medium supplemented as previously described (21). Tetracycline (2 μg/ml) was added to the medium for the propagation of mycoplasma transformants. Mycoplasma titers (quantified in CFU per milliliter) were determined by plating serial dilutions of broth cultures onto solid medium and counting the colonies after incubation for 4 days at 37°C. Each CFU titer determination was done in triplicate.
DNA extractions and PCR amplifications.
Mycoplasma genomic DNA was extracted from logarithmic-phase cultures (2 ml) using a Blood & Cell Culture DNA kit from Qiagen. Specific oligonucleotide primers were used to amplify the gsmA region surrounding the poly(G) tract (5′-CTATGAAACAGTCATTTTGG-3′ and 5′-GGCGCATATAGTGATTGTC-3′) or the full gsmA gene (5′-GCATCACAATTGAAAACG-3′ and 5′-CAGTTTATATTCCTAAAACTGG-3′). PCR amplifications (25-μl volume) were performed according to the recommendations of the Taq DNA polymerase supplier (Promega). All PCR amplifications were conducted in a Bio-Rad thermocycler with an initial denaturation step of 5 min at 95°C, followed by 35 cycles, including a denaturation step at 95°C (30 s), primer annealing at 47°C (30 s), and elongation at 72°C [40 s or 2 min for poly(G) or gsmA amplification, respectively], and finally an elongation step at 72°C (2 min). PCR products were sequenced using an external facility at Beckman Coulter Genomics.
Purification and quantification of capsular and cell-free polysaccharides.
Mycoplasma cells from 50-ml stationary-phase cultures were counted (time zero [T0] in Table 3), harvested by centrifugation at 12,000 × g for 30 min at 18°C, washed with phosphate-buffered saline (PBS), and further incubated in 12 ml of CMRL-1066 medium (Life Technologies) for 72 h at 37°C. After counting the cells (h 72 [T72] in Table 3), the cultures were centrifuged at 14,000 × g for 1 h at 4°C to separate the cells from the supernatant. EPS were extracted from the cell-free supernatant as described previously (13). CPS were purified from the cell pellet recovered after centrifugation using the method described by Shi et al. (22) with some modifications. Briefly, cells were suspended in 1 ml of sterile PBS and lysed with proteinase K (0.1 μg/ml) for 4 h at 37°C. The lysate was incubated for an additional 12 h at 37°C after adding SDS (1% [wt/vol]), DNase (100 U/ml), and RNase (500 μg/ml). Residual proteins were removed by trichloroacetic acid precipitation (10% [wt/vol], 1 h, 4°C) followed by centrifugation (14,000 × g, 1 h at 4°C). Polysaccharides were precipitated from the resulting supernatant with 10 volumes of cold acetone (−20°C) for 72 h and centrifuged at 14,000 × g for 1 h at 4°C. The polysaccharide pellet was allowed to dry at room temperature and was resuspended in sterile ultrapure water. Absence of protein contamination was assessed by SDS-polyacrylamide gel electrophoresis and silver staining. The sugar concentration was estimated by the phenol/sulfuric acid method using glucose as the standard (23). For each sample, the determination was done three times and the results were expressed as means ± standard deviations (SD). The limit of quantification (LOQ) was calculated using the following equation: LOQ = Mbs + 10 × SDbs, where Mbs is the mean of 10 blank samples (without sugar) and SDbs is the standard deviation of the blank samples. The estimated LOQ of the phenol/sulfuric assay in our experimental conditions was 0.8 μg of glucose/ml.
TABLE 3.
Quantification of the production of exopolysaccharides and capsular polysaccharides by M. agalactiae and by M. mycoides subsp. capri
| Mycoplasma | Titer (CFU/ml)a |
Polysaccharide production (μg/ml)b |
||
|---|---|---|---|---|
| T0 | T72 | EPS | CPS | |
| M. agalactiae 14628 | 2 × 109 | 3 × 106 | 1.1 ± 0.1 | 9.7 ± 0.1 |
| M. agalactiae PG2T | 3 × 1010 | 1 × 104 | <0.8c | 1.3 ± 0.1 |
| M. agalactiae 5632 | 7 × 1010 | 7 × 104 | <0.8c | 1.6 ± 0.1 |
| M. agalactiae 5632pO/T | 7 × 1010 | 1 × 106 | <0.8c | 1.2 ± 0.1 |
| M. agalactiae 5632pO/T1260 | 6 × 1010 | 3 × 104 | 1.5 ± 0.1 | 10.0 ± 0.3 |
| M. agalactiae 5632pOT/1260mod | 6 × 1010 | 8 × 103 | 1.9 ± 0.1 | 11.1 ± 0.5 |
| M. mycoides subsp. capri PG3T | 2 × 1011 | 6 × 108 | 5.1 ± 0.2 | 10.9 ± 0.1 |
Data represent mycoplasma titers at h 0 (T0) and after 72 h of incubation (T72) in CMRL-1066 medium.
Data represent averages of results of three biological repeats ± standard deviations. Results are expressed as micrograms of glucose per milliliter of CMRL.
Concentrations were below the detection limit (0.8 μg/ml).
Composition and structure of the capsular polysaccharides.
CPS were purified as described above except that the CMRL phase was omitted and the cell pellets were collected directly from PPLO broth cultures and washed twice with PBS. After resuspension in pure water, the CPS extract was further dialyzed against regularly renewed ultrapure sterile water for 48 h at room temperature using 3.5-kDa-cutoff dialysis tubing (Spectrum Laboratories) to eliminate potential contaminants. The monosaccharide components were determined after hydrolysis of 1 mg of CPS with 4 M CF3CO2H (100°C, 4 h) and dried under nitrogen. After dilution in pure water, samples were analyzed by high-performance anion-exchange chromatography (HPAEC) using a pulsed amperometric detector (Dionex ICS 3000 system). Samples were then passed through a Propac PA1 precolumn (Dionex) (4 by 50 mm) followed by a CarboPak PA 1 column at 30°C. Gradient elution was performed at a flow rate of 1 ml/min with a multistep gradient as follows: 0 to 25 min, 90% H2O and 10% NaOH (160 mM); 25 to 34 min, 100% NaOH (200 mM); 35 to 50 min, 90% H2O and 10% NaOH (160 mM). Peak analysis was performed using Chromeleon software, version 7.0.
For nuclear magnetic resonance (NMR) spectroscopy, CPS were exchanged twice with 99.9% D2O (Euriso-top), dried under vacuum, and dissolved in 99.96% D2O (3 mg/0.5 ml). 1H NMR spectra were recorded, at 80°C, on a Bruker Avance 500 spectrometer equipped with a BBI probe (5-mm sample diameter) and Topspin 1.3 software. 1H NMR spectra were accumulated using a 30° pulse angle, a recycle time of 1 s, and an acquisition time of 2 s for a spectral width of 3,000 Hz for 32-K data points after presaturation of the HOD signal using an experimental sequence provided by Bruker. 13C NMR experiments were conducted using the same spectrometer operating at 125.48 MHz with 2 s as the relaxation delay. The two-dimensional (2D) 1H/1H correlation spectroscopy (COSY), 1H/1H total correlation spectroscopy (TOCSY), 1H/1H nuclear Overhauser effect spectroscopy (NOESY), 1H/13C heteronuclear single quantum coherence (HSQC), and 1H/13C heteronuclear multiple bond correlation (HMBC) spectra were acquired with standard pulse sequences delivered by Bruker.
Detection of polysaccharide secretion by colony immunostaining.
Colony immunostaining experiments were performed as described previously (24). The mmc4 serum prepared against the type strain of M. mycoides subsp. capri (PG3T) was kindly provided by the CIRAD laboratory in Montpellier, France. The anti-β-(1→6)-glucan polyclonal serum was prepared as described by Montijn et al. (25). Briefly, purified β-(1→6)-glucans were coupled with bovine serum albumin (BSA) and used to inoculate rabbits in a 28-day immunization protocol (Proteomics Services of Eurogentec). Sera collected at the end of the immunization period were tested by dot blotting (13). No reaction was observed with galactan purified from M. mycoides subsp. mycoides strain Afadé or with β-(1→2)-glucan from M. capricolum subsp. capricolum strain 14232, while a positive signal was observed with β-(1→6)-glucan purified from M. mycoides subsp. capri serovar capri strain PG3T and from M. agalactiae strain 14628.
Plasmids, DNA constructions, and transformation of M. agalactiae.
Plasmid p20-1miniO/T (here termed “pO/T”), which contains parts of the M. agalactiae origin of replication (oriC), was used as a stable vector for protein expression in this species (26). The gsmA gene from strain 14628 (MAGb_1260) was cloned downstream of the P40 gene promoter region, as previously described (26). Briefly, the promoter region was amplified first by using oligonucleotide primers p40RF-CC (5′-ACGGGGCTAAAGAAGCTGAT-3′) and p40/MAGb1260 (5′-ACACATTTTGTTTCTTTTTCATAATTATTTATATCCTTTTC-3′) to generate a 200-bp DNA fragment overlapping MAGb_1260 at the ATG codon. The MAGb_1260 locus was then amplified by using the overlapping DNA fragment and the primer MAGb1260_R (5′-TTATTTTTGTTGTGCACCTG-3′). The resulting PCR product was cloned into pGEM-T Easy (Promega) before subcloning at the NotI site of the pO/T plasmid was performed to generate pO/T1260. Plasmid pO/T1260mod is a modified version of pO/T1260 in which the poly(G) (9G) tract in MAGb_1260 has been replaced by a synonymous, nonhomopolymeric nucleotide sequence (GGAGGTGGC). Site-directed mutagenesis was performed by PCR amplification using the overlapping oligonucleotide primers MAGb1260mut_F (5′-GTAAATGGAGGTGGCAACTTATTCCAATACTTGCTCT-3′) and MAGb1260mut_R (5′-TAAGTTGCCACCTCCATTTACTTTTGTTTTATTTAT-3′). Briefly, two overlapping PCR products were generated from pO/T1260 by using the primer sets p40RF-CC/MAGb1260mut_R and MAGb1260mut_F/MAGb1260_R. Overlapping PCR products were then assembled by using the primer set p40RF-CC/MAGb1260_R. The final PCR product was used to generate pO/T1260mod, as described above.
Cloned sequences were verified by DNA sequencing at the GeT-Purpan sequencing facility (Toulouse, France). PCRs were performed using Phusion High-Fidelity DNA polymerase (New England BioLabs). Transformation of M. agalactiae with DNA constructions was obtained with polyethylene glycol 8000 (27).
Mycoplasma susceptibility to serum-killing and complement-killing activity.
Mycoplasma cells from 1-ml cultures (∼109 CFU) were centrifuged at 12,000 × g (room temperature) for 15 min and resuspended in 1 ml of CMRL-1066 medium. For the complement-killing assay, 100 μl of mycoplasma cells in CMRL medium was incubated for 1 h at 37°C with 100 μl of fresh or heat-inactivated (1 h at 56°C) guinea pig serum (Sigma-Aldrich) diluted in CMRL-1066 (1:3 [vol/vol]). The mix was then spread on an agar plate, and mycoplasma colonies were counted after incubation for 4 days at 37°C. Mycoplasma survival after treatment with complement was determined from the percentage of CFU recovered after the treatment with fresh guinea pig serum in relation to the CFU recovered after treatment with heat-inactivated serum. For the goat serum sensitivity assay, 100 μl of M. agalactiae cells in CMRL was incubated for 90 min at 37°C with 100 μl of (i) a pool of 4 serum samples collected from mycoplasma-free goats (preimmune; diluted 1:1 in CMRL), (ii) a pool of sera from the same animals collected 23 days postinfection (23DPI) with strain 5632 (diluted 1:1 in CMRL), and (iii) CMRL, as a serum-negative control. Experimental infections with strain 5632 were conducted as previously described (28). Percentages of survival were calculated from the fraction of CFU recovered after incubation with goat serum divided by the CFU recovered from the incubation without serum. For each experiment, the β-(1→6)-glucan secreted by the culture was determined by colony immunostaining using mmc4 serum or anti-β-(1→6)-glucan serum.
In silico analysis of potential polysaccharide biosynthetic pathways.
BLASTP analyses were conducted in the Molligen database (29) to search for enzymes potentially involved in glucose uptake and phosphorylation and for the synthesis of UDP-sugar in M. agalactiae genomes, using the proteins described for members of the M. mycoides cluster (12, 13). Homologs of enzymes described in Fig. 1 for M. mycoides subsp. mycoides PG1 and M. capricolum subsp. capripneumoniae strain Ambosa were first searched using BLASTP. Glycosyltransferases from M. agalactiae strain PG2T were retrieved from the CAZy databases (30), and the homologous enzymes in strains 14628 and 5632 were identified using BLASTP analyses and the Molligen database (29). Glycosyltransferases showing (i) several TMDs predicted by TMHMM2 (31) with a large cytoplasmic loop bearing DXD and R/QXXRW-like motifs (identified by alignment with the galactan synthase MSC_0108 with Clustal Omega [32]) and (ii) homology with the cellulose synthase of Rhodobacter sphaeroides cellulose synthase subunit a (Uniprot accession number Q3J125) as shown using Phyre (33) were classified as synthases. Transmembrane proteins (see Fig. S1 in the supplemental material) were represented by using the TOPO2 software (http://www.sacs.ucsf.edu/TOPO2/).
FIG 1.
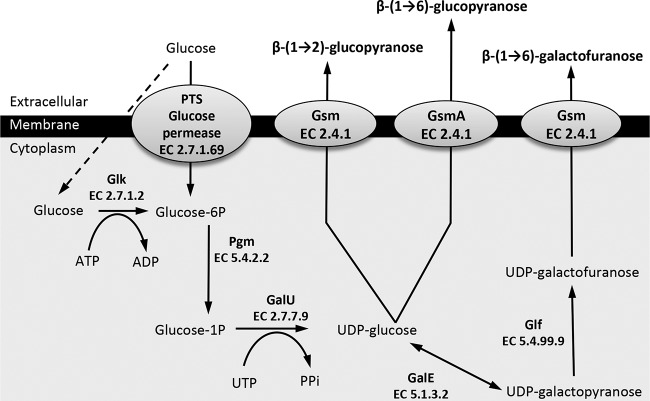
Predicted biosynthetic pathways of β-(1→2)-glucopyranose, β-(1→6)-glucopyranose, and β-(1→6)-galactofuranose polysaccharides in mycoplasma species of the M. mycoides cluster and M. agalactiae. Glucose uptake and activation into glucose-6-phosphate are mediated by the phosphotransferase system (PTS)-glucose permease or by a two-step process that includes nonspecific permeases (dashed arrow) and a glucokinase (Glk) for phosphorylation. The glucose-6-phosphate is then isomerized by a phosphoglucomutase (Pgm) into glucose-1-phosphate, which in turn is transformed in UDP-glucose by a glucose-1-phosphate uridylyltransferase (GalU). The UDP-glucose is used either directly by a glycosyltransferase with synthase activity (Gsm; glycan synthase of mollicutes) to build and export a β-(1→2)-glucopyranose or a β-(1→6)-glucopyranose polymer [common name, β-(1→2)-glucan or β-(1→6)-glucan, respectively] or further transformed into UDP-galactofuranose by the successive action of an UDP-glucose 4-epimerase (GalE) and an UDP-galactofuranose mutase (Glf). The last-named UDP-sugar monomer is then used by another specific synthase and polymerized into galactan, the common name for the β-(1→6)-galactofuranose polymer. Each enzyme is also designated by its EC number.
Phylogenetic analyses of synthases among mycoplasmas.
Glycosyltransferase sequences of diverse mycoplasma species were obtained from the CAZy database or from the nonredundant protein database of the National Center for Biotechnology Information by carrying out BLASTP searches against the synthase sequences identified in this study or previously described (13). Among the putative glycosyltransferases, synthases were specifically identified as described above and selected to construct a phylogenetic tree using the www.phylogeny.fr website (34, 35) and the methodology of Ulvskov et al. (36). Briefly, the amino acid sequences were aligned with Muscle v3.8.31 using the default parameters. Gaps were removed, and the resulting sequences were used to build a maximum likelihood phylogenetic tree using the default settings and the WAG substitution matrix.
RESULTS
A complete polysaccharide biosynthetic pathway is encoded by M. agalactiae.
Metabolic pathways involved in the biosynthesis of β-(1→2)-glucopyranose and β-(1→6)-galactofuranose polysaccharides have already been predicted for several mycoplasma species within the M. mycoides cluster (Fig. 1). These data served as a basis to identify putative polysaccharide biosynthetic pathways in M. agalactiae. BLASTP analyses were conducted using whole-genome sequences of M. agalactiae available in databases to detect sequence similarity with enzymes of the M. mycoides cluster putatively involved in glucose uptake, transformation, and activation as UDP monomers. Homologs of Pgm and GalU were predicted in each M. agalactiae genome tested (similarity of >55% with MSC_0829 and MSC_0110, respectively), but no sequence similarity was evidenced for phosphotransferase system (PTS) permeases, Glk, GalE, or Glf (Table 2). Putative glycosyltransferase genes were further explored using the CAZy database (30). For strain PG2T, the only M. agalactiae strain available in the CAZy database, three genes were predicted to encode glycosyltransferases, namely, MAG4860, MAG0570, and MAG3010 (Table 2). Homologs of the corresponding glycosyltransferases in strains 14628 and 5632 were retrieved by conducting BLASTP searches (Table 2). Remarkably, only the MAGb_1260 product (strain 14628) was found to display structural similarity to the previously described galactan synthase encoded by M. mycoides subsp. mycoides PG1T MSC_0108 (13). Both are transmembrane proteins carrying a cytoplasmic domain with DXD and R/QXXRW motifs of the GT-A glycosyltransferase family (37) (see Fig. S1 in the supplemental material). However, the MAGb_1260 product is composed of 7 TMDs (see Fig. S1) whereas the MSC_0108 product has only 4 (13). This structural difference might be associated with the nature of the exported polymers. Indeed, all the glycosyltransferases predicted in glucan-producing strains of the species M. capricolum (12) were analyzed by TMHMM2 and shown to possess 7 TMDs like MAGb_1260 (see, for instance, MCCP01_0081) (Table 2; see also Fig. S1), whereas the M. mycoides species shown to produce galactan (13) displayed 4 TMD synthases (see, for instance, MSC_0108 in Table S1 and Fig. S1).
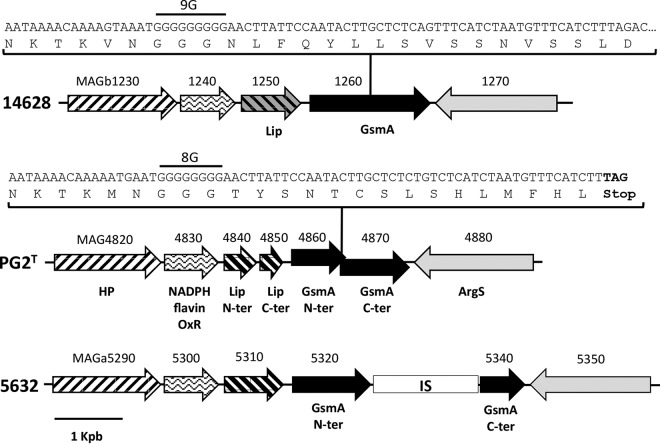
In contrast to the MAGb_1260 product in strain 14628, both of the glycosyltransferase homologs in strains PG2 and 5632 (MAG4860 and MAGa5320, respectively) were truncated and had only two TMDs in their N-terminal portion (Table 2; see also Fig. S1 in the supplemental material). Their respective genes were disrupted by an insertion sequence in strain 5632 (MAGa5330) and a premature STOP codon in PG2T resulting from a nucleotide deletion in a poly(G) stretch (Fig. 2). Apart from this difference, the overall synteny of the genomic region surrounding the synthase gene in the three strains was well conserved (Fig. 2).
FIG 2.
Schematic representation of the genome locus containing the gsmA gene in M. agalactiae strains 14628, PG2T, and 5632. The coding sequences (CDSs) are indicated by arrows. Identical motifs indicate similar predicted functions. CDS numbers refer to the locus tag, and CDS-encoded products are indicated by the following abbreviations: HP, hypothetical protein; NADPH flavin OxR, NADPH flavin oxidoreductase; Lip, lipase/esterase; ArgS, arginyl-tRNA synthetase; N-ter, N-terminal part of the protein; C-ter, C-terminal part of the protein; IS, insertion sequence ISmag1. gsmA sequences with a focus on the poly(G) stretch region for PG2T and 14628 are detailed in boxes. The number of nucleotides inside the poly(G) region is indicated upstream of the poly(G) sequence. The TAG stop codon in the PG2T sequence is shown in bold.
Thus, in silico analysis revealed that a complete polysaccharide biosynthetic pathway is encoded by the M. agalactiae strain 14628 genome. However, the apparent lack of GalE and Glf, two enzymes involved in β-(1→6)-galactofuranose synthesis, suggests that the polysaccharides produced by M. agalactiae might be restricted to glucopyranose polymers. This is consistent with the 7 TMDs in its putative membrane-associated glycosyltransferase, proposed as the synthase and here named “gsmA” (for “glycan synthase of mollicutes in M. agalactiae”).
M. agalactiae secretes a cell-attached β-(1→6)-glucopyranose polymer.
To further investigate the in vitro production of polysaccharides by M. agalactiae, capsular polysaccharides (CPS) and exopolysaccharides (EPS) were purified and quantified as described in Materials and Methods. As shown in Table 3, CPS production in strain 14628 was 6-fold higher than in PG2T and 5632, attaining up to 9 μg per ml of culture. The polysaccharides secreted in strain 14628 were preferentially attached to the cell, since the production of CPS was 8-fold higher than that of EPS. Interestingly, no marked difference was observed between the amounts of EPS produced by the different M. agalactiae strains, which were close to the limit of quantification.
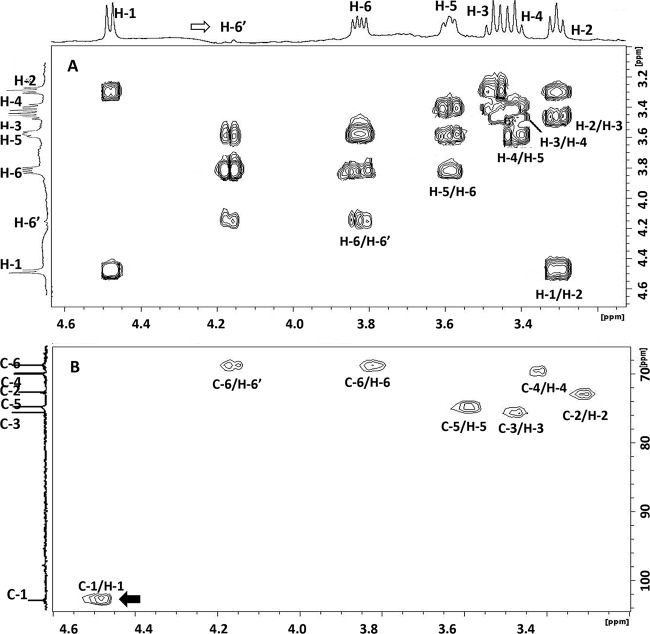
Antibodies raised against the β-(1→2)-glucopyranose and β-(1→6)-galactofuranose polysaccharides produced by Bertin et al. (12) failed to react with purified CPS, suggesting a different structure for the polysaccharide secreted by M. agalactiae 14628 (data not shown). The chemical composition and structure of the CPS in M. agalactiae were therefore elucidated by HPAEC analysis and NMR spectra, as described in Materials and Methods. Both analyses revealed that the CPS produced by strain 14628 was composed solely of glucose. Indeed, the correlations in the 2D NMR COSY (Fig. 3A) and TOCSY spectra made it possible to attribute various chemical shifts to protons of the glucosyl residues (4.481 [H-1], 3.308 [H-2], 3.473 [H-3], 3.418 [H-4], 3.589 [H-5], 3.825 [H-6], and 4.168 [H-6′] ppm). These results were further confirmed by the 2D NMR HSQC spectrum (Fig. 3B), in which the observed connectivities between H-1 and C-1 (102.63 ppm), H-2 and C-2 (72.92 ppm), H-3 and C-3 (75.57 ppm), H-4 and C-4 (69.65 ppm), H-5 and C-5 (74.79 ppm), and H-6 or H-6′ and C-6 (68.52 ppm) were characteristic of a glucose polymer. The H-1 signal at 4.48 ppm (1,2J = 7.83 Hz) and C-1 at 102.63 ppm are typical of d-glucose residues with a β-pyranoside configuration. The H-6′ chemical values indicate the presence of a linkage on the C-6 in glucose residues (38, 39). This linkage was confirmed by the presence of connectivities between the H-1 of glucose (δ 4.481 ppm) and the signal at δ 4.168 ppm, which corresponded to the H-6′ of glucose in the NOESY spectrum. Similar connectivities were also observed in the 1H/13C HMBC spectrum.
FIG 3.
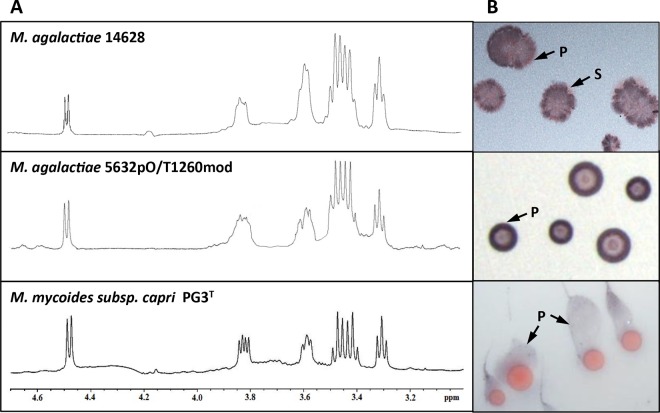
2D 1H/1H COSY NMR spectrum (A) and 2D 1H/13C HSQC NMR spectrum (B) of CPS purified from M. agalactiae strain 14628. The H-1 signal at 4.48 ppm and that of C-1 at 102.63 ppm are typical of d-glucose residues with a β-pyranoside configuration (black arrow). The H-6/C-6′ chemical values indicate the presence of a linkage on the C-6 glucose residue (white arrow).
These results demonstrate unambiguously that the CPS in M. agalactiae is a β-(1→6)-glucopyranose polymer, commonly known as β-(1→6)-glucan. This polysaccharide is unusual in prokaryotes but a common component of fungal cell walls (39).
GsmA is involved in β-(1→6)-glucan synthesis.
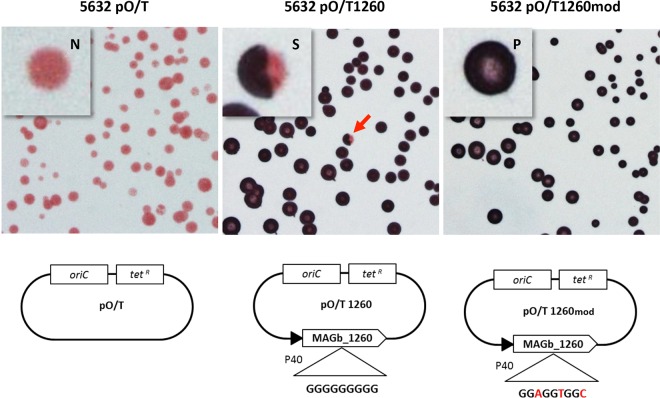
To confirm the role of GsmA in the production of β-(1→6)-glucans in M. agalactiae, strain 5632 was transformed with plasmid pO/T1260 carrying the gsmA gene from strain 14628 (MAGb_1260) under the control of the constitutive P40 promoter (Fig. 4). Strain 5632 was selected for transformation because of the nonreversible disruption of its endogenous gsmA gene by an insertion sequence (Fig. 2). Transformation of strain 5632 with pO/T1260 restored CPS production to a level equivalent to that of strain 14628 compared to the negative control using pO/T (Table 3). The nature of the CPS produced by 5632pO/T1260 was confirmed by colony immunostaining using a specific anti-β-(1→6)-glucan polyclonal serum produced in rabbits (see Materials and Methods for details) (Fig. 4). Interestingly, colony immunostaining of 5632pO/T1260 with anti-β-(1→6)-glucan antibodies revealed positive, negative, and sectored colonies (Fig. 4), a pattern characteristic of high-frequency phase variation in expression. Indeed, phase variation in β-(1→6)-glucan expression could be mediated by the poly(G) tract detected in the gsmA coding sequence, since this homopolymeric stretch was shown to be the cause of the frameshift responsible for gsmA truncation in PG2T (Fig. 2). This was investigated by using plasmid pO/T1260mod, in which the poly(G) tract was replaced by a synonymous, nonhomopolymeric nucleotide sequence (Fig. 4). Strain 5632 transformed with plasmid pO/T1260mod produced a quantity of CPS similar to the quantities produced by strains 14628 and 5632pO/T1260 (Table 3). As expected, colony immunostaining of 5632pO/T1260mod failed to reveal negative or sectored colonies, thereby confirming the constitutive expression of β-(1→6)-glucan in this transformant (Fig. 4). The nature of the CPS produced by 5632 transformants was corroborated by the NMR proton spectra (Fig. 5). These results confirmed that gsmA is essential for β-(1→6)-glucan production in M. agalactiae and that the poly(G) tract found in the 5′ region of gsmA is responsible for phase variations in β-(1→6)-glucan secretion. Based on the colony immunostaining data, the switch in expression was estimated to occur at a frequency of 10−3 in 5632pO/T1260. This very high frequency is usual in other types of phase variation spontaneously occurring in mycoplasmas, with 10−2 to 10−5 events/cell/generation classically reported in vitro (40).
FIG 4.
Colony immunostaining with anti-β-(1→6)-glucan serum of different M. agalactiae 5632 transformants. Different OriC/Tetr-encoding plasmids, containing (i) no gene (pO/T), (ii) the native MAGb_1260 gene (pO/T1260), or (iii) a modified MAGb_1260 gene with no poly(G) tract (pO/T1260mod) under the control of the P40 promoter, were used for transformation. The population consisted of colonies that were positive (P; dark blue), negative (N; pink), or sectored (S; red arrow).
FIG 5.
(A) NMR proton spectra of CPS purified from M. agalactiae strain 14628, transformant 5632pO/T1260mod, and M. mycoides subsp. capri strain PG3T. (B) Corresponding colony immunostaining using the anti-β-(1→6)-glucan serum (P, positive staining; S, sectored colony).
Synthesis of β-(1→6)-glucan is not restricted to M. agalactiae and is shared by phylogenetically distant mycoplasma species.
Although gsmA (MAGb_1260) had been predicted for horizontal gene transfer (HGT) between members of the M. mycoides cluster and M. agalactiae (19), no β-(1→6)-glucan had been found in the CPS or EPS detected for several strains in this cluster (12). However, the best BLAST hit analyses identified the MMC_0120 and MMC_6560 products in M. mycoides subsp. capri strain PG3T as orthologs of GsmA. The putative synthase activity of these two proteins was further supported by their predicted secondary structure, which was very similar to that predicted for GsmA (see Fig. S1 in the supplemental material). The existence of a complete pathway for the secretion of glucan-type polysaccharides in M. mycoides subsp. capri strain PG3T was also corroborated by the presence of genes encoding Pgm and GalU, which are involved in UDP-glucose synthesis (12) (Table 2). Finally, both EPS and CPS were detected in cultures of M. mycoides subsp. capri strain PG3T (Table 3) and gave a positive reaction in dot blot experiments with anti-β-(1→6)-glucan (data not shown). NMR analysis showed that the CPS of M. mycoides subsp. capri PG3T and M. agalactiae 14628 were identical (Fig. 5). Detailed examination of the CPS structure in M. mycoides subsp. capri further confirmed a β-(1→6)-glucopyranose homopolymer (data not shown) identical to that of M. agalactiae 14628. Likewise, colony immunostaining of M. mycoides subsp. capri strain PG3T with anti-β-(1→6)-glucan antibodies resulted in a positive signal (Fig. 5). However, the contrast and quality of the immunostaining were very different from those of 5632pO/T1260mod or 14628. This could have resulted from differences in the levels of β-(1→6)-glucan production (Table 3) and/or a difference between M. mycoides subsp. capri and M. agalactiae in cell membrane anchorage.
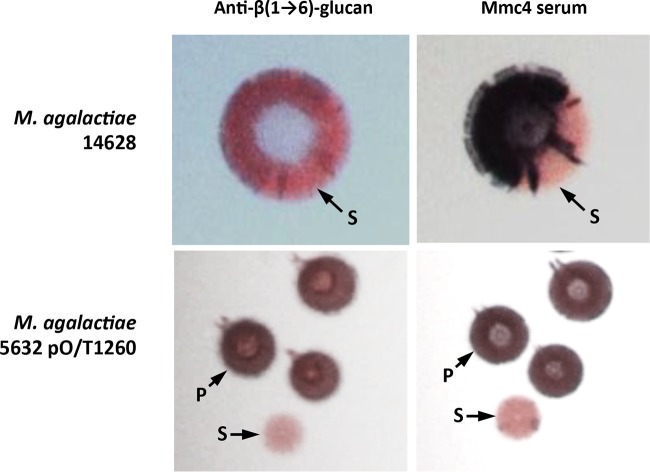
Interestingly, recent studies have identified an antigenic structure shared by M. agalactiae strain 14628 and M. mycoides subsp. capri strain PG3T that is recognized by a polyclonal antiserum raised against PG3T, the so-called mmc4 antibody (19). A panel of strains, with various expression statuses, was subjected to colony immunostaining to see if the antigenic target of the mmc4 antibody was β-(1→6)-glucan (see Fig. S2 in the supplemental material). As expected, strain 14628 displayed a majority of positive colonies whereas the results for strain 5632 remained negative unless it was transformed with plasmid constructs expressing GsmA. Sectored colonies, consistent with the phase-variable expression of β-(1→6)-glucan, were observed with strains 14628 and 5632pO/T1260. To further confirm the recognition of β-(1→6)-glucan by mmc4 antibodies, double-colony immunostaining of 5632pO/T1260 and strain 14628 was carried out with mmc4 and anti-β-(1→6)-glucan serum (Fig. 6). The results showed that (i) individual colonies which gave a positive reaction with anti-β-(1→6)-glucan serum were also detected with the mmc4 serum and (ii) mmc4 and anti-β-(1→6)-glucan sera generated identical patterns on single-sectored colonies of strain 14628 (Fig. 6). Furthermore, 17 and 16 single 14628 colonies, respectively, that gave positive and negative results with mmc4 were picked and analyzed to determine the length of the gsmA poly(G) sequence. All positive colonies had a poly(G) tract of 9 nucleotides (nt), preserving the gsmA coding frame. The 16 negative colonies had two poly(G) configurations composed of 8 or 10 G's, leading both to a frameshift and to inactivation of the gsmA gene. These results clearly demonstrate that the mmc4 detection pattern is consistent with β-(1→6)-glucan production and that the structure of β-(1→6)-glucan is a common antigenic structure shared by M. agalactiae strain 14628 and M. mycoides subsp. capri strain PG3T.
FIG 6.
Double-colony immunostaining of M. agalactiae clone 5632pO/T1260 and strain 14628 using anti-β-(1→6)-glucan serum (1st column) and mmc4 serum (2nd column). The population consists of colonies that are positive (P; dark blue) or sectored (S).
Phase-variable secretion of β-(1→6)-glucan occurs in most M. agalactiae field isolates.
To find out whether β-(1→6)-glucan production by strain 14628 was representative of M. agalactiae strains circulating in the field, 12 clinical isolates collected from different animal hosts, countries, and tissues were tested for their capacity to react with mmc4 serum in colony immunostaining experiments (Table 4; see also Fig. S2 in the supplemental material). The mmc4 serum was preferred to anti-β-(1→6)-glucan serum for this experiment, because of its better sensitivity and the better contrasting images that it generates (Fig. 6). Strains PG2T, 5632, and 14628 were used as controls. For most of the isolates, both mmc4-positive and mmc4-negative colonies were identified on a single agar plate, together with a variable number of sectored colonies (see Fig. S2). This strongly suggested that most of the clinical strains were able to produce β-(1→6)-glucan, whatever their clinical history, and that secretion of this polysaccharide was controlled by phase variation. The only exception was isolate 4867, for which no positive or sectored colonies were detected among the 2,000 examined.
TABLE 4.
Length of the poly(G), 3′ sequence tract in gsmA and β-(1→6)-glucan secretion in M. agalactiae field isolates
| Strain | Origin | Host | Clinical sign | Yr | Poly(G) and 3′ sequencea | Length of the poly(G) sequence (nt) | gsmA in frame | β-(1→6)-Glucan secretionb |
|---|---|---|---|---|---|---|---|---|
| 15291 | France | Goat | Arthritis | 1987 | GGGGGGGGGGAC TTA TTC | 10 | Yes | Yes |
| 14628 | France | Ibex | Pneumonia | 2006 | GGGGGGGGG AAC TTA TTC | 9 | Yes | Yes |
| 15341 | France | Chamois | Pneumonia | 2009 | GGGGGGGGG AAA CTA ATC | 9 | Yes | Yes |
| 13387 | France | Ibex | Pneumonia | 2003 | GGGGGGGGG AAC TTA TTC | 9 | Yes | Yes |
| 15310 | France | Ibex | Pneumonia | 2009 | GGGGGGGGG AAC TTA TTC | 9 | Yes | Yes |
| 7323 | Greece | Goat | Mastitis | 1993 | GGGGGGGGG AAC TTA TTC | 9 | Yes | Yes |
| 7320 | Greece | Sheep | Mastitis | 1993 | GGGGGGGGG AAC TTA TTC | 9 | Yes | Yes |
| 7337 | Greece | Goat | Mastitis | 1993 | GGGGGGGGG AAC TTA TTC | 9 | Yes | Yes |
| 13378 | France | Goat | None | 2003 | GGGGGGGGG AAC TTA TTC | 9 | Yes | Yes |
| PG2T | Spain | Goat | Unknown | 1952 | GGGGGGGG- AAC TTA TTC | 8 | No | No |
| 4908 | France | Goat | Mastitis | 1990 | GGGGGGGAC --- TTA TTC | 7 | Yes | Yes |
| 6355 | Spain | Sheep | Arthritis | 1992 | GGGGGGGAC --- TTA TTC | 7 | Yes | Yes |
| 5632 | Spain | Goat | Arthritis | <1991 | GGGGGC --- AAC TTG TTC | 5 | Noc | No |
| 14668 | France | Goat | Pneumonia | 2006 | GGGGGC --- AAC TTG TTC | 5 | Yes | Yes |
| 4867 | Spain | Goat | Mastitis | 1990 | GGGGGC --- AAC TTG TTC | 5 | Noc | No |
Sequence details of the gsmA locus containing the poly(G) stretch (indicated with underlined boldface characters). Nucleotides are grouped in codons according to the open reading frame. Base deletions are indicated by hyphens.
As estimated by the majority of colonies positively stained by mmc4 antiserum.
gsmA is disrupted by an insertion sequence (ISmag1) or by a single base deletion upstream of the poly(G) sequence for strain 5632 or strain 4867, respectively.
The gsmA locus of 12 clinical isolates was PCR amplified and the corresponding poly(G) tract profile defined by sequencing (Table 4). Seven isolates were found with a 9-nt-long poly(G) tract identical to that of strain 14628 and were shown to produce β-(1→6)-glucan. The remaining 5 isolates had a poly(G) tract composed of 5, 7, or 10 nt (Table 4). Careful examination of the nucleotide sequence located immediately downstream revealed compensatory deletions or modifications that preserved the gsmA coding frame but that most likely introduced the deletion and/or change of one amino acid compared to all the other strains (Table 4). Overall, these findings were coherent with the detection of β-(1→6)-glucan production in all isolates except isolate 4867, in which the gsmA coding frame is disrupted by a single base deletion upstream of the poly(G) (data not shown). All isolates except 4867 were also shown to undergo phase variation in their β-(1→6)-glucan production, as evidenced by colony immunostaining (see Fig. S2 in the supplemental material).
Phase variation in β-(1→6)-glucan secretion protects M. agalactiae from serum-killing activity.
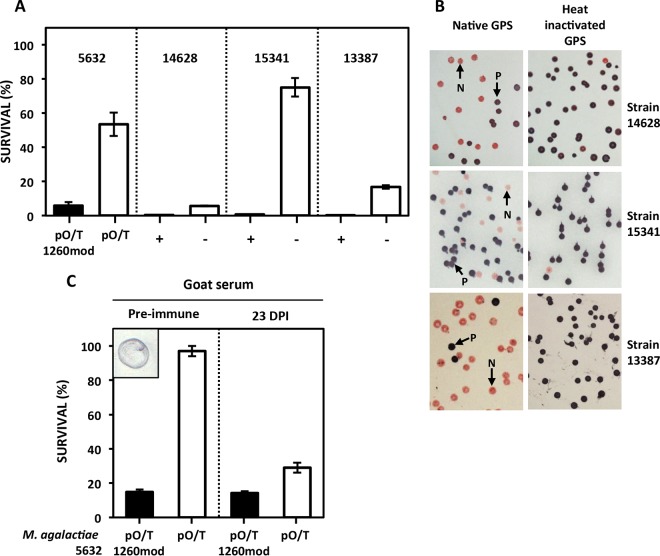
Recent studies with M. mycoides subsp. mycoides have shown that galactan-CPS can protect this mycoplasma species from the killing activity of serum complement (15). This prompted us to examine the role of β-(1→6)-glucan CPS in preventing complement-mediated killing of M. agalactiae. Mycoplasma survival assays were carried by using fresh guinea pig serum (GPS) as a source of complement and heat-inactivated GPS as a control (Fig. 7A). Survival of 5632pO/T1260mod, which constitutively produces β-(1→6)-glucan as CPS, was drastically reduced upon incubation with fresh GPS, whereas 5632pO/T, which does not produce β-(1→6)-glucan, was more resistant to complement-killing activity. This result was corroborated by taking several clinical isolates and selecting mmc4-positive and mmc4-negative clones. Despite considerable differences in the survival rates of mmc4-negative clones (6% to 75%), β-(1→6)-glucan-producing clones (mmc4+) displayed higher sensitivity to complement-killing activity and a survival rate of less than 1%. These results suggest that β-(1→6)-glucan CPS not only failed to protect M. agalactiae from complement-killing activity but in fact increased its susceptibility. To further test this hypothesis, strains 14628, 15341, and 13387 were incubated for 1 h with fresh or inactivated GPS, plated, grown for 4 days at 37°C, and subjected to immunostaining with mmc4. Results showed that the proportions of negative-mmc4 colonies were higher after incubation with native GPS (50%, 32%, and 90%, respectively) than after incubation with inactivated GPS (2.0%, 0.4%, and 0.4%, respectively) (Fig. 7B). Hence, mycoplasmas that do not produce β-(1→6)-glucans are clearly favored in the presence of GPS complement. Besides this innate complement pathway, the classical pathway triggered by antibody-antigen complexes might also play a role in goats, as anti-β-(1→6)-glucan antibodies are commonly found in their bloodstream due to its frequent colonization by fungi (41). We showed that sera from goats with no prior contact with mycoplasmas and known to have been free of mycoplasma for years (28) indeed reacted with purified β-(1→6)-glucans (Fig. 7C). Survival assays further confirmed that incubation with these sera (designated “Pre-immune” in Fig. 7C) resulted in a loss of viable cells only in the β-(1→6)-glucan-producing 5632pO/T1260mod clone and not in 5632 pO/T. In contrast, sera collected at 23 days postinfection with strain 5632 (28) displayed an adaptive specific immune response that affected the two clones equally (Fig. 7C).
FIG 7.
Effect of guinea pig serum (GPS) complement (A and B) and goat sera (C) on the survival of M. agalactiae. (A) M. agalactiae transformants (5632pO/T and 5632pOT/1260mod) or clones (14628, 15341, and 13387), selected for the presence (black bars; “+” for mmc4-positive clones) or absence (white bars, “−” for mmc4-negative clones) of the capacity to produce β-(1→6)-glucan, were incubated with native GPS used as a source of complement proteins. Data represent averages of results of three biological repeats. Error bars indicate standard deviations. (B) Colony immunostaining with mmc4 serum of M. agalactiae strains incubated with native or heated GPS. Positive colonies (P) are dark blue and negative ones (N) pink. (C) Survival of M. agalactiae transformants after incubation with preimmune goat sera or sera recovered 23 days postinfection (DPI) with strain 5632. As shown in the small insert on the left, preimmune sera gave a positive reaction to β-(1→6)-glucan purified from M. agalactiae 14628 CPS and spotted (1 μg) on a nitrocellulose membrane.
DISCUSSION
The present report shows that M. agalactiae secretes a β-(1→6)-glucopyranose homopolymer as a CPS. This particular polysaccharide is commonly known as β-(1→6)-glucan and has never before been described in mycoplasmas. Remarkably, we also detected its secretion in two other ruminant (sub)species: M. mycoides subsp. capri (strain PG3T), where it is present as both CPS and EPS; and M. bovis (data not shown). Interestingly, this carbohydrate polymer is usually present in the cell walls of many, if not all, fungal species but is very rare in prokaryotes (39). So far it has been reported only in the Actinobacillus genus and, more specifically, in the swine pathogen Actinobacillus suis serotype O1/K1, where it forms part of the lipopolysaccharide-O chain in the cell wall and capsule (39). Whether β-(1→6)-glucan secretion is restricted to ruminant mycoplasma species or is widespread among mycoplasmas has still to be explored.
In M. agalactiae, the secretion of β-(1→6)-glucans is governed by the GsmA synthase. In silico analysis of ruminant mycoplasma genomes revealed two categories of putative synthases based on the number of predicted TMDs and the nature of the carbohydrates secreted (see Table S1 in the supplemental material). Indeed, all glucan-producing (sub)species, including M. agalactiae, M. capricolum subsp. capricolum, M. capricolum subsp. capripneumoniae, and M. leachii, harbor synthases with 7 TMDs, while galactan-producing (sub)species, including M. mycoides subsp. mycoides, harbor synthases with 4 TMDs (see Table S1 and Fig. S1). This is also true at the serovar level as shown by glucan-producing M. mycoides subsp. capri serovar capri (7 TMDs) and galactan-producing M. mycoides subsp. capri serovar LC (4 TMDs). Orthologous synthases have been identified in several other species of mycoplasmas isolated from a wide variety of animal hosts, seven of which are detailed in Table S1 in the supplemental material. Most of them exhibit features indicative of a synthase function such as multiple TMDs and a large cytoplasmic loop bearing specific DXD and R/QXXRW-like motifs (see Table S1). Interestingly, the number of predicted TMDs in putative mycoplasma synthases seems to be limited to 4 or 7 domains, suggesting the exclusive production of either galactan or glucan. Only one species, namely, M. bovigenitalium, was shown to harbor both categories of synthases, suggesting the secretion of 2 different polysaccharides. A phylogenetic tree was constructed using 18 sequences of mycoplasma synthases (see Fig. S3). As expected, this phylogenetic construction split into two main clusters, one containing the 4-TMD synthases, including all known galactan-producing species, and a second cluster containing the 7-TMD synthases and encompassing the glucan-producing mycoplasma species. More interestingly, the synthases involved in β-(1→6)-glucan and β-(1→2)-glucan secretion are divided into two branches within the second cluster.
It is tempting to speculate that most of the species listed in Table S1 in the supplemental material are able to produce EPS or CPS, since they have a synthase and most of the enzymes involved in sugar activation. This conserved pathway for the production of polysaccharides in minimal bacteria is highly suggestive of an important biological function. Among the systems described for polysaccharide synthesis and secretion in bacteria, the synthase pathway is the simplest, with a single enzyme catalyzing both polymerization and translocation of the newly synthesized polysaccharide across the cytoplasmic membrane (42). The presence of this multifunctional enzyme corresponds well with the need of mycoplasmas for optimization of their poor overall metabolic capacity. However, the synthase-dependent pathway might not be the only polysaccharide secretion pathway in mycoplasmas, as M. pulmonis was shown to produce two polysaccharides in the absence of predicted synthase genes (10).
In M. agalactiae, β-(1→6)-glucan is essentially cell attached, whereas it is secreted as both CPS and EPS in M. mycoides subsp. capri PG3 (Table 3). This difference in polysaccharide production could explain the appearance of M. mycoides subsp. capri PG3T colonies after immunostaining with anti-β-(1→6)-glucan, which resulted in a substantial slime layer (Fig. 5), and is certainly due to the different membrane anchoring systems and acceptor molecules in the two species. The anchoring moiety is as yet unknown; however, a gene encoding a lipoprotein (MMC_1040) was predicted to be part of the membrane anchoring system in M. mycoides subsp. capri PG3T but is absent from M. agalactiae strains (12). Unlike M. agalactiae, the M. mycoides subsp. capri strain PG3T genome harbors two synthase genes, namely, MMC_0120 and MMC_6560 (Table 2), located in different genetic environments. These two synthases share 65% identity and are located in the same branch of the mycoplasma synthase phylogenetic tree (see Fig. S3 in the supplemental material) corresponding to β-(1→6)-glucan production. Whether the different specificities of the two synthases, and the subsequent secretion of either β-(1→6)-glucan EPS or CPS, could be due to gene duplication in different loci and their sequence divergence has yet to be elucidated. Although M. mycoides subsp. capri PG3T and M. agalactiae produce the same carbohydrate, they use different pathways for glucose uptake and phosphorylation. M. mycoides subsp. capri PG3T encodes two enzymes involved in the formation of glucose-6-phosphate, a glucose PTS permease involved in glucose transport and phosphorylation and a glucokinase (Glk) that catalyzes the phosphorylation of intracellular glucose (12). Remarkably, both proteins are lacking in M. agalactiae. Because M. agalactiae uses glucose as a carbon source, it might harbor unidentified glucose transporters and kinases.
Unraveling the role of secreted polysaccharides in the virulence and pathogenicity of mycoplasmas has only just started. Galactan was shown to protect M. mycoides subsp. mycoides from serum killing when cell attached (15) and to possess anti-inflammatory properties when secreted as an exopolysaccharide (14). We have demonstrated here that M. agalactiae strains producing β-(1→6)-glucan in contact with goat sera or the complement fraction of guinea pig sera were rapidly killed, suggesting that they might be rapidly eliminated from the bloodstream of infected animals, prior to the acquisition of antimycoplasma immunity. Whether this results from the capacity of complement proteins to bind directly to β-(1→6)-glucans (43) or from antibody-mediated complement lysis has yet to be elucidated. Indeed, anti-β-(1→6)-glucan antibodies are commonly found in the bloodstream of human and animal hosts as a consequence of their frequent colonization by fungi (41, 44). Hence, bacteria producing β-(1→6)-glucans can be expected to be poorly virulent in a host previously infected by fungi. This is the case for the bacterium A. suis serotype O1/K1, which infects its swine host less efficiently than other serotypes expressing non-β-(1→6)-glucan polysaccharides (45).
Interestingly, the production of β-(1→6)-glucan was not a constitutive phenotypic trait in M. agalactiae, as we demonstrated phase variation in the expression of GsmA associated with changes in length of a homopolymeric tract in its coding sequence. This phenomenon was responsible for highly heterogeneous mycoplasma populations containing both β-(1→6)-glucan producers and nonproducers. The high frequency of the ON/OFF switch is consistent with the high-frequency phase variation usually observed in other mycoplasmas (40). A similar mechanism of polysaccharide phase variation involving homopolymeric tracts has been described for glycosyltransferases of Helicobacter pylori (46), Neisseria gonorrhoeae (47), and Campylobacter jejuni (48). It provides a rapid and simple way for bacteria to generate clonal subpopulations without using complex transcriptional-regulation mechanisms. Interestingly, in the M. agalactiae model, variant selection in vitro was shown to differ considerably between different isolates even when they shared the same poly(G) tract structure (see Fig. S2 in the supplemental material). Phenotypic switching, as observed for β-(1→6)-glucan phase variation, can be a real asset when the environment imposes strong selection (49). During the course of infection, M. agalactiae is disseminated from the primary site of infection (respiratory tract mucosa, small intestine, or the mammary gland alveoli, depending on the routes of transmission) to different vital organs by the bloodstream (50). Switching off β-(1→6)-glucan allows the generation of a M. agalactiae subpopulation that is less susceptible to the bactericidal activity of goat serum and to the lytic action of the complement (Fig. 7). We therefore propose that the switching off of β-(1→6)-glucan would enhance M. agalactiae dissemination through the blood to deep organs and secondary infection sites. A similar adaptation pattern during different colonization steps was suggested for M. mycoides subsp. mycoides through phase variation leading to a switch from CPS to EPS galactan secretion (15). Once in the deep organs, M. agalactiae β-(1→6)-glucan would promote the proliferation of immune cells in infected tissues, this proinflammatory effect being an important feature in development of mycoplasmoses (51). Such a role has been suggested in diverse species of pathogenic fungi. For instance, β-(1→6)-glucans present in the cell wall of Pneumocystis jirovecii were shown to promote the activation of inflammatory cells in the lung (52) and those present in Candida albicans induced neutrophil migration to the infected host tissues (53). Although the exact part played by β-(1→6)-glucans in the interplay between mycoplasmas and their hosts has yet to be elucidated, the conserved capacity to produce β-(1→6)-glucan in most M. agalactiae field isolates and in M. mycoides subsp. capri strain PG3T is a strong argument in favor of a significant biological role.
Conclusion.
Genomic and biochemical data led to the discovery of β-(1→6)-glucopyranose polymer production and secretion by M. agalactiae and M. mycoides subsp. capri, two mycoplasma species involved in contagious agalactia of small ruminants. Prediction of the biosynthetic pathway combined with genetic experiments revealed the essential role of membrane-embedded glycosyltransferases (named “Gsm” for glycan synthases of mollicutes). These synthases are widespread in different mycoplasma species, and their structure is correlated with the type of polysaccharide secreted. M. agalactiae gsmA presents the unique feature of having a poly(G) tract within its coding frame, for which spontaneous variation in length can modulate expression of the gene and result in ON/OFF antigenic phase variation. Cell-attached M. agalactiae β-(1→6)-glucopyranose increased serum-killing susceptibility, suggesting that phase variation might play a crucial role in mycoplasma host dissemination.
Supplementary Material
ACKNOWLEDGMENTS
We thank Monika Gjorgjieva, Marie Guillot, Patrice Cuchet, and Véronique Lefriand for their excellent technical assistance. We are grateful to François Poumarat, who first observed mmc4 antigenic phase variation in M. agalactiae isolates many years ago. Special thanks to all members of the VIGIMYC network for supplying several M. agalactiae isolates with documented clinical histories.
P.G., F.T., E.B., and C.C. were responsible for the conception and design of the experiments; C.P.-R. was responsible for the monomer composition and NMR analysis; P.G., E.B., and E.S. performed all of the other experiments; P.G., F.T., C.P.-R., E.B., and C.C. performed the data analysis; F.T., P.G., C.P.-R., E.B., and C.C. were responsible for the draft of the manuscript; all of us were responsible for critical review of the manuscript and approval of the final version.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00274-16.
REFERENCES
- 1.Tytgat HL, Lebeer S. 2014. The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol Mol Biol Rev 78:372–417. doi: 10.1128/MMBR.00007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nwodo UU, Green E, Okoh AI. 2012. Bacterial exopolysaccharides: functionality and prospects. Int J Mol Sci 13:14002–14015. doi: 10.3390/ijms131114002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willis LM, Whitfield C. 2013. Structure, biosynthesis, and function of bacterial capsular polysaccharides synthesized by ABC transporter-dependent pathways. Carbohydr Res 378:35–44. doi: 10.1016/j.carres.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Cullen L, McClean S. 2015. Bacterial adaptation during chronic respiratory infections. Pathogens 4:66–89. doi: 10.3390/pathogens4010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hidalgo-Cantabrana C, Nikolic M, Lopez P, Suarez A, Miljkovic M, Kojic M, Margolles A, Golic N, Ruas-Madiedo P. 2014. Exopolysaccharide-producing Bifidobacterium animalis subsp. lactis strains and their polymers elicit different responses on immune cells from blood and gut associated lymphoid tissue. Anaerobe 26:24–30. doi: 10.1016/j.anaerobe.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Lukácová M, Barák I, Kazár J. 2008. Role of structural variations of polysaccharide antigens in the pathogenicity of Gram-negative bacteria. Clin Microbiol Infect 14:200–206. doi: 10.1111/j.1469-0691.2007.01876.x. [DOI] [PubMed] [Google Scholar]
- 7.Reid CW, Fulton KM, Twine SM. 2010. Never take candy from a stranger: the role of the bacterial glycome in host-pathogen interactions. Future Microbiol 5:267–288. doi: 10.2217/fmb.09.103. [DOI] [PubMed] [Google Scholar]
- 8.Daubenspeck JA, Jordan DS, Dybvig K. 2014. The glycocalyx of mollicutes, p 131–147. In Browning GF, Citti C (ed), Mollicutes: molecular biology and pathogenesis. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 9.Plackett P, Buttery SH. 1958. A galactan from Mycoplasma mycoides. Nature 182:1236–1237. doi: 10.1038/1821236a0. [DOI] [PubMed] [Google Scholar]
- 10.Daubenspeck JM, Bolland JR, Luo W, Simmons WL, Dybvig K. 2009. Identification of exopolysaccharide-deficient mutants of Mycoplasma pulmonis. Mol Microbiol 72:1235–1245. doi: 10.1111/j.1365-2958.2009.06720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmons WL, Daubenspeck JM, Osborne JD, Balish MF, Waites KB, Dybvig K. 2013. Type 1 and type 2 strains of Mycoplasma pneumoniae form different biofilms. Microbiology 159:737–747. doi: 10.1099/mic.0.064782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertin C, Pau-Roblot C, Courtois J, Manso-Silvan L, Tardy F, Poumarat F, Citti C, Sirand-Pugnet P, Gaurivaud P, Thiaucourt F. 2015. Highly dynamic genomic loci drive the synthesis of two types of capsular or secreted polysaccharides within the “Mycoplasma mycoides” cluster. Appl Environ Microbiol 81:676–687. doi: 10.1128/AEM.02892-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertin C, Pau-Roblot C, Courtois J, Manso-Silvan L, Thiaucourt F, Tardy F, Le Grand D, Poumarat F, Gaurivaud P. 2013. Characterization of free exopolysaccharides secreted by Mycoplasma mycoides subsp. mycoides. PLoS One 8:e68373. doi: 10.1371/journal.pone.0068373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Totté P, Puech C, Rodrigues V, Bertin C, Manso-Silvan L, Thiaucourt F. 2015. Free exopolysaccharide from Mycoplasma mycoides subsp. mycoides possesses anti-inflammatory properties. Vet Res 46:122. doi: 10.1186/s13567-015-0252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaurivaud P, Lakhdar L, Le Grand D, Poumarat F, Tardy F. 2014. Comparison of in vivo and in vitro properties of capsulated and noncapsulated variants of Mycoplasma mycoides subsp. mycoides strain Afade: a potential new insight into the biology of contagious bovine pleuropneumonia. FEMS Microbiol Lett 359:42–49. [DOI] [PubMed] [Google Scholar]
- 16.Gloster TM. 2014. Advances in understanding glycosyltransferases from a structural perspective. Curr Opin Struct Biol 28:131–141. doi: 10.1016/j.sbi.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitney JC, Howell PL. 2013. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol 21:63–72. doi: 10.1016/j.tim.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sirand-Pugnet P, Lartigue C, Marenda M, Jacob D, Barre A, Barbe V, Schenowitz C, Mangenot S, Couloux A, Segurens B, de Daruvar A, Blanchard A, Citti C. 2007. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet 3:e75. doi: 10.1371/journal.pgen.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tardy F, Baranowski E, Nouvel LX, Mick V, Manso-Silvan L, Thiaucourt F, Thebault P, Breton M, Sirand-Pugnet P, Blanchard A, Garnier A, Gibert P, Game Y, Poumarat F, Citti C. 2012. Emergence of atypical Mycoplasma agalactiae strains harboring a new prophage and associated with an alpine wild ungulate mortality episode. Appl Environ Microbiol 78:4659–4668. doi: 10.1128/AEM.00332-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chazel M, Tardy F, Le Grand D, Calavas D, Poumarat F. 2010. Mycoplasmoses of ruminants in France: recent data from the national surveillance network. BMC Vet Res 6:32. doi: 10.1186/1746-6148-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poumarat F, Perrin B, Longchambon D. 1991. Identification of ruminant mycoplasmas by dot immunobinding on membrane filtration (MF dot). Vet Microbiol 29:329–338. doi: 10.1016/0378-1135(91)90140-B. [DOI] [PubMed] [Google Scholar]
- 22.Shi F, Harada T, Ogawa Y, Ono H, Ohnishi-Kameyama M, Miyamoto T, Eguchi M, Shimoji Y. 2012. Capsular polysaccharide of Erysipelothrix rhusiopathiae, the causative agent of swine erysipelas, and its modification with phosphorylcholine. Infect Immun 80:3993–4003. doi: 10.1128/IAI.00635-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. 1956. Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- 24.Gaurivaud P, Persson A, Grand DL, Westberg J, Solsona M, Johansson KE, Poumarat F. 2004. Variability of a glucose phosphotransferase system permease in Mycoplasma mycoides subsp. mycoides small colony. Microbiology 150:4009–4022. [DOI] [PubMed] [Google Scholar]
- 25.Montijn RC, van Rinsum J, van Schagen FA, Klis FM. 1994. Glucomannoproteins in the cell wall of Saccharomyces cerevisiae contain a novel type of carbohydrate side chain. J Biol Chem 269:19338–19342. [PubMed] [Google Scholar]
- 26.Baranowski E, Guiral S, Sagne E, Skapski A, Citti C. 2010. Critical role of dispensable genes in Mycoplasma agalactiae interaction with mammalian cells. Infect Immun 78:1542–1551. doi: 10.1128/IAI.01195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dordet-Frisoni E, Sagné E, Baranowski E, Breton M, Nouvel LX, Blanchard A, Marenda MS, Tardy F, Sirand-Pugnet P, Citti C. 2014. Chromosomal transfers in mycoplasmas: when minimal genomes go mobile. mBio 5(6):e01958-14. doi: 10.1128/mBio.01958-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tardy F, Maigre L, Tricot A, Poumarat F, Nguyen L, Le Grand D. 2011. Comparison of isolates of Mycoplasma mycoides subspecies capri from asymptomatic and septicaemic goats. J Comp Pathol 144:70–77. doi: 10.1016/j.jcpa.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Barré A, de Daruvar A, Blanchard A. 2004. MolliGen, a database dedicated to the comparative genomics of mollicutes. Nucleic Acids Res 32:D307–D310. doi: 10.1093/nar/gkh114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Möller S, Croning MD, Apweiler R. 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 32.Sievers F, Higgins DG. 2014. Clustal Omega. Curr Protoc Bioinformatics 48:1.25.1–1.25.33. doi: 10.1002/0471250953.bi0313s48. [DOI] [PubMed] [Google Scholar]
- 33.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 34.Dereeper A, Audic S, Claverie JM, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulvskov P, Paiva DS, Domozych D, Harholt J. 2013. Classification, naming and evolutionary history of glycosyltransferases from sequenced green and red algal genomes. PLoS One 8:e76511. doi: 10.1371/journal.pone.0076511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breton C, Snajdrová L, Jeanneau C, Koca J, Imberty A. 2006. Structures and mechanisms of glycosyltransferases. Glycobiology 16:29R–37R. [DOI] [PubMed] [Google Scholar]
- 38.Kruppa MD, Lowman DW, Chen YH, Selander C, Scheynius A, Monteiro MA, Williams DL. 2009. Identification of (1→6)-beta-d-glucan as the major carbohydrate component of the Malassezia sympodialis cell wall. Carbohydr Res 344:2474–2479. doi: 10.1016/j.carres.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monteiro MA, Slavic D, St Michael F, Brisson JR, MacInnes JI, Perry MB. 2000. The first description of a (1→6)-beta-d-glucan in prokaryotes: (1→6)-beta-d-glucan is a common component of Actinobacillus suis and is the basis for a serotyping system. Carbohydr Res 329:121–130. doi: 10.1016/S0008-6215(00)00148-8. [DOI] [PubMed] [Google Scholar]
- 40.Citti C, Nouvel LX, Baranowski E. 2010. Phase and antigenic variation in mycoplasmas. Future Microbiol 5:1073–1085. doi: 10.2217/fmb.10.71. [DOI] [PubMed] [Google Scholar]
- 41.Ishibashi K, Dogasaki C, Motoi M, Miura N, Adachi Y, Ohno N. 2010. Anti-fungal cell wall beta-glucan antibody in animal sera. Nihon Ishinkin Gakkai Zasshi 51:99–107. doi: 10.3314/jjmm.51.99. [DOI] [PubMed] [Google Scholar]
- 42.Schmid J, Sieber V, Rehm B. 2015. Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front Microbiol 6:496. doi: 10.3389/fmicb.2015.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubin-Bejerano I, Abeijon C, Magnelli P, Grisafi P, Fink GR. 2007. Phagocytosis by human neutrophils is stimulated by a unique fungal cell wall component. Cell Host Microbe 2:55–67. doi: 10.1016/j.chom.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noss I, Wouters IM, Smit LA, Meijer E, Pronk A, Heederik DJ, Doekes G. 2012. IgG to various beta-glucans in a human adult population. Int Arch Allergy Immunol 157:98–108. doi: 10.1159/000324674. [DOI] [PubMed] [Google Scholar]
- 45.Deutschmann R, Boncheff AG, MacInnes JI, Monteiro MA. 2011. Discovery and characterization of a fructosylated capsule polysaccharide and sialylated lipopolysaccharide in a virulent strain of Actinobacillus suis. Biochem Cell Biol 89:325–331. doi: 10.1139/o11-001. [DOI] [PubMed] [Google Scholar]
- 46.Sanabria-Valentín E, Colbert MT, Blaser MJ. 2007. Role of futC slipped strand mispairing in Helicobacter pylori Lewisy phase variation. Microbes Infect 9:1553–1560. doi: 10.1016/j.micinf.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banerjee A, Wang R, Supernavage SL, Ghosh SK, Parker J, Ganesh NF, Wang PG, Gulati S, Rice PA. 2002. Implications of phase variation of a gene (pgtA) encoding a pilin galactosyl transferase in gonococcal pathogenesis. J Exp Med 196:147–162. doi: 10.1084/jem.20012022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parker CT, Gilbert M, Yuki N, Endtz HP, Mandrell RE. 2008. Characterization of lipooligosaccharide-biosynthetic loci of Campylobacter jejuni reveals new lipooligosaccharide classes: evidence of mosaic organizations. J Bacteriol 190:5681–5689. doi: 10.1128/JB.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kussell E, Leibler S. 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 50.Kumar A, Rahal A, Chakraborty S, Verma AK, Dhama K. 2014. Mycoplasma agalactiae, an etiological agent of contagious agalactia in small ruminants: a review. Vet Med Int 2014:286752. doi: 10.1155/2014/286752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Browning GF, Noormohammadi AH, Markham PF. 2014. Identification and characterization of virulence genes in mycoplasmas, p 77–90. In Browning GF, Citti C (ed), Mollicutes: molecular biology and pathogenesis. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 52.Kottom TJ, Hebrink DM, Jenson PE, Gudmundsson G, Limper AH. 2015. Evidence for proinflammatory beta-1,6 glucans in the Pneumocystis carinii cell wall. Infect Immun 83:2816–2826. doi: 10.1128/IAI.00196-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato T, Iwabuchi K, Nagaoka I, Adachi Y, Ohno N, Tamura H, Seyama K, Fukuchi Y, Nakayama H, Yoshizaki F, Takamori K, Ogawa H. 2006. Induction of human neutrophil chemotaxis by Candida albicans-derived beta-1,6-long glycoside side-chain-branched beta-glucan. J Leukoc Biol 80:204–211. doi: 10.1189/jlb.0106069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.