ABSTRACT
Polycyclic aromatic hydrocarbons (PAHs) are widespread environmental contaminants that are hazardous to human health. It has been demonstrated that members of the Mycobacterium genus are among the most effective degraders of PAHs, but few studies have focused on the degradation of PAH mixtures. In this study, single and mixed PAH metabolism was investigated in four phylogenetically distinct Mycobacterium species with respect to (i) parent compound degradation, (ii) bacterial growth, (iii) catabolic gene expression, and (iv) metabolite production. Synergistic and antagonistic effects on four model PAH compounds (benzo[a]pyrene, pyrene, fluoranthene, and phenanthrene) characterized degradation of mixtures in a strain- and mixture-dependent manner. The mixture of pyrene and phenanthrene, in particular, resulted in antagonized degradation by three out of four bacterial species, and further studies were narrowed to investigate the degradation of this mixture. Antagonistic effects persisted over time and were correlated with reduced bacterial growth. Antagonized degradation of PAH was not caused by preferential degradation of secondary PAHs, nor were mixture compounds or concentrations toxic to cells growing on sugars. Reverse transcription-PCR (RT-PCR) studies of the characterized catabolic pathway of phenanthrene showed that in one organism, antagonism of mixture degradation was associated with downregulated gene expression. Metabolite profiling revealed that antagonism in mixture degradation was associated with the shunting of substrate through alternative pathways not used during the degradation of single PAHs. The results of this study demonstrate metabolic differences between single and mixed PAH degradation with consequences for risk assessment and bioremediation of PAH-contaminated sites.
IMPORTANCE Mycobacterium species are promising organisms for environmental bioremediation because of their ubiquitous presence in soils and their ability to catabolize aromatic compounds. PAHs can be degraded effectively as single compounds, but mixed substrates often are subject to degradative inhibition, which may explain the persistence of these pollutants in soils. Single and mixed PAH degradation by diverse Mycobacterium species was compared, with associated bacterial growth, gene expression, and metabolite production. The results demonstrate that antagonism characterized degradation in a strain- and mixture-dependent manner. One strain that was versatile in its pathway use of single chemicals also efficiently degraded the mixture, whereas antagonism in other the strains was associated with altered metabolic profiles, indicating unusual pathway use. The impacts of this work on risk assessment and bioremediation modeling studies indicate the need to account for mixture-generated intermediates and to recognize mixture degradation as a property distinct from that of PAH substrate range.
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs) are a class of chemical pollutants that are widespread and persistent in the environment (1–3). Because of the carcinogenic and toxic effects of bioactivated forms of PAHs, especially those with high molecular weight (HMW), these compounds are priority pollutants that must be removed from soils to provide for the safety of humans and wildlife (4–7). Bioremediation is considered an attractive and viable cleanup strategy, such that much scientific work has focused on understanding the bacterial catabolism of PAHs (2). In particular, Mycobacterium species have been shown to be successful degraders of a wide variety of PAHs, including recalcitrant HMW PAHs (3, 8–15).
Studies of the degradation of PAH mixtures have been relatively few, but it has been demonstrated that mixture degradation is characterized by synergistic and antagonistic effects in diverse bacteria, including Pseudomonas, Sphingomonas, Stenotrophomonas, Brevibacterium, Arthrobacter, Rhodococcus, and Mycobacterium species (16–24). Synergy and antagonism refer to the enhanced and inhibited degradation, respectively, of one or more PAHs as mixture components compared to their removal as sole substrates. These effects are relevant to bioremediation studies because contaminated environments harbor complex mixtures of chemicals (2, 18, 23). If antagonistic effects on mixture degradation persist over time, they may reduce the ability of strains to remove target pollutants in the environment (18) and contribute to the long-term persistence of PAHs in the soil (12).
Underlying causes of synergistic or antagonistic degradation of multiple substrates have not been investigated, but some studies have offered possible explanations for the phenomena (12, 16–20, 23, 25, 26). Synergy in mixed PAH degradation might be caused by (i) additional enzyme induction by the secondary PAH enhancing degradation of the primary, (ii) the use of common enzymes to degrade both PAHs, or (iii) fortuitous growth of bacteria on one PAH that increases the degradative potential of the second PAH. Similarly, causes of antagonism in mixture degradation have been proposed. (i) Cells might preferentially utilize a more easily degradable or more bioavailable PAH, resulting in the apparent inhibited degradation of the other mixture component. Inhibition of degradation in mixtures may also be caused by (ii) repression of catabolic genes at the pathway level or global level (catabolite repression), (iii) competition of PAHs at the enzyme active sites, or (iv) toxicity of antagonizing PAHs or their metabolites. The causes of synergy and antagonism may be multifactorial or may vary by strain and PAH combination.
Bacterial metabolic pathways have been elucidated for phenanthrene (22, 27–32) and pyrene (14, 15, 33–37). The degradation of phenanthrene is initiated by dioxygenation at the C-1 and C-2, C-3 and C-4, or C-9 and C-10 positions. Some bacteria produce metabolites of two (27, 30, 38) or all three (32, 39, 40) pathways. The best-characterized pathway for phenanthrene degradation proceeds initially through dioxygenation at the C-3 and C-4 positions, and enzymes involved in breakdown through phthalic acid have been elucidated in Mycobacterium vanbaalenii PYR-1 (41). As a HMW PAH, pyrene is a more recalcitrant chemical for which metabolites and pathways have been best characterized in two related bacterial genera, Mycobacterium and Rhodococcus. The main mineralization pathway begins with dioxygenase attack at the C-4 and C-5 positions to form the cis-dihydrodiol. In subsequent steps, the pathway splits, with one branch preparing the molecule to enter the phenanthrene pathway and the other catabolizing through substituted biphenyl intermediates, both of which can lead to mineralization (39). Several enzymes involved in phenanthrene degradation are also induced during the catabolism of pyrene (11, 12), but important recent studies have shown that paralogs of initial ring-hydroxylating dioxygenases in Mycobacterium species act distinctly both in induction and regiospecific product formation with exposure to pyrene or phenanthrene (11, 42, 43).
Because of the widespread reports of antagonism and its relevance to bioremediation of contaminated sites, the mechanics of PAH mixture degradation must be studied further. In order to investigate the broader ability of Mycobacterium species to transform PAH mixtures, we selected four phylogenetically distinct and substrate-versatile species and studied the catabolism of four model PAHs as single compounds and in mixtures. Antagonistic mixture degradation was studied over time. The possibility of PAH mixture toxicity was investigated with growth experiments using sugars as primary carbon sources. Finally, gene expression studies and metabolite profiling were employed to compare differences in pathway use during single- and mixed-PAH degradation by the four strains.
MATERIALS AND METHODS
Bacterial strains.
Four Mycobacterium strains were used for this study. Three organisms were characterized recently as the novel species M. aromaticivorans JS19b1 (ATCC BAA-1378T), M. crocinum czh-3 (ATCC BAA-1371), and M. rutilum czh-117 (ATCC BAA-1375T) (10). The fourth organism, strain czh-101, was isolated in the same screen for PAH-degrading organisms, and it was identified to the species level using characterization methods described previously (10), including Gram stain, colony morphology, motility, substrate oxidation and utilization, biochemical tests, and sequencing of the 16S rRNA gene and rpoB.
Chemicals.
Phenanthrene (PHE), fluoranthene (FLA), pyrene (PYR), benzo[a]pyrene (BAP), diphenic acid, 1-hydroxy-2-naphthoic acid, 2-hydroxy-1-naphthoic acid, 2-carboxycinnamic acid, phthalic acid, 2-formylbenzoic acid, and protocatechuic acid were purchased from Sigma-Aldrich (Milwaukee, WI). Naphthalene-1,2-dicarboxylic acid anhydride, 1-hydroxy-2-naphthaldehyde, 2-hydroxy-1-naphthaldehyde, and naphthalene-1,2-diol were purchased from TCI America (Portland, OR). Chemicals that were not commercially available were synthesized previously in our laboratory, including 5,6-benzocoumarin, 7,8-benzocoumarin, 1-(2-carboxyvinyl)-2-naphthoic acid, 2-(2-carboxyvinyl)-1-naphthoic acid, 4-(2-hydroxynaphth-1-yl)-2-oxobut-3-enoic acid, 4-(1-hydroxynaphth-2-yl)-2-oxo-but-3-enoic acid (44), cis-9,10-dihydrophenanthrene-9,10-diol, naphthalene-1,2-dicarboxylic acid (32), 4-oxapyren-5-one, and phenanthrene-4,5-dicarboxylic acid (40). Solvents, including ethyl acetate, acetonitrile, acetone, ethanol, and methyl-tert-butyl ether (MTBE), were purchased from Fisher Scientific (Morris Plains, NJ). Metabolite standards were derivatized to corresponding methyl esters and methyl ethers, as previously described (32). Model PAH solubility levels in water (in milligrams per liter) are as follows: BAP, 0.003; PYR, 0.140; FLA, 0.260; and PHE, 1.300 (5).
PAH degradation by bacteria in liquid culture.
Quantitative degradation by Mycobacterium strains of four PAHs, BAP, PYR, FLA, and PHE, was tested in liquid culture. Large test tubes (25 mm) were sterilized and dried, and 100 μl of PAH stock solution (2.5 mg ml−1 BAP and 5 mg ml−1 PYR, FLA, and PHE in acetone) was delivered with a glass syringe to tubes. Acetone was evaporated in a laminar flow hood. Five milliliters of standard mineral base (SMB) medium (45) was added to tubes, with a final concentration of 50 ppm (BAP) or 100 ppm (other PAHs). Bacterial inocula (grown in 25 ml of tryptic soy broth [TSB] for 4 to 6 days at 29°C with a rotation speed of 180 rpm) were pelleted in sterile centrifuge tubes at 3,600 relative centrifugal force (RCF), washed with 10 ml of SMB, and pelleted again. Cells were resuspended in SMB to an optical density at 600 nm (OD600) of 0.3, and 100 μl of cell suspension was inoculated into growth tubes with PAH medium. Abiotic controls were prepared in the same way with sterile SMB added instead of bacteria. Samples and controls were prepared in triplicate. Tubes were capped and incubated with rotation in the dark at 29°C for 7 days.
To test the degradation of PAH mixtures, bacterial strains were incubated with binary combinations of the four PAHs. The combination of PYR and FLA was not tested due to the difficulty of chromatographic separation of the two. Triplicate samples and controls were prepared and incubated as described above, but with two PAH stock solutions in each sample.
After incubation, cultures were extracted by adding 5 ml of ethyl acetate and vortexing for 15 s on high. One milliliter was removed from the organic layer with a volumetric glass pipette into a clean vial, and the solvent was evaporated under a gentle nitrogen stream. Acetonitrile (2 ml) was used to reconstitute residues. Extracts were filtered (0.22-μm pore size) into amber high-performance liquid chromatography (HPLC) vials and capped. PAH analysis was done on a Dionex BioLC HPLC system with GP50 pump, PDA100 detector, and AS50 autosampler. Separation with a C18 column (Hypersil, 5-μm particle size, 4-mm diameter, and 250-mm length; Hewlett Packard) by gradient elution began with 5 min of 40% acetonitrile and 60% water (with 1% isopropanol), increasing to 100% acetonitrile over the next 30 min, with a final 5-min hold at 100% acetonitrile. The percent PAH degradation was determined by a comparison of the PAH recovered in experimental samples with that in abiotic controls.
Bacterial growth and PAH degradation over time.
Two organisms exhibiting antagonized PYR/PHE degradation were chosen for time course studies of PAH use over a 14-day incubation. Four samples per time point of strains czh-101 and czh-3 and three abiotic controls were prepared as described above with single or mixed PYR and PHE. The samples were incubated for 0, 2, 4, 7, 10, and 14 days. At each time point, one experimental sample was used to determine the bacterial count by serial dilution and colony count on tryptic soy agar (TSA) medium, and triplicate cultures were extracted and analyzed as described above for PAH quantification. PAH removal was calculated as the percent recovery compared to the time-zero sample. Strain JS19b1 was not tested in this section or the next because this strain grows in biofilm only (not in single colonies) and could not be plate counted.
Effects of PAHs on bacteria growing with sugar substrates.
Test tubes were supplied as described above with PHE, PYR, the mixture, or acetone alone (sugar-only samples), and acetone was evaporated sterilely. Five milliliters of sugar medium (filter-sterilized 1% [wt/vol] mannitol or glucose in SMB) and a 100-μl inoculum adjusted to approximately an OD600 of 0.1 were added to each tube. Another 100 μl was used for initial cell count. Samples were incubated as described above for 5 and 8 days, pelleted, washed, and plated in serial dilutions. The average plate counts from two replicate samples were used to compare the growth on sugars alone to that on sugar combined with PAH(s).
Culture preparation for metabolite and gene expression studies.
Cultures of four Mycobacterium species were grown in 300 ml of TSB and pelleted as described above, and the washed cells were frozen in SMB-glycerol in 1-ml aliquots for use as inocula. The initial CFU per 1 ml of frozen cells was determined by plate count. Six milliliters of one or both (single or mixture degradation, respectively) PAH stock solutions prepared in acetone (25 mg ml−1) was delivered to 3-liter sterile, dry Erlenmeyer flasks, and acetone was evaporated sterilely. SMB medium (1.5 liters) and 1 ml of bacterial inoculum were added to each flask. The flasks were incubated in the dark at 29°C with rotation (180 rpm) for 3 or 7 days.
After incubation, bacterial cultures were immediately placed at 4°C. To prepare for extraction, cultures were filtered through glass wool to remove unreacted PAH, divided into 250-ml centrifuge tubes, and pelleted (5,500 RCF) at 4°C for 15 min. The supernatant was reserved for metabolite analysis, and the pelleted cells were combined and centrifuged again. Cells were resuspended in 5 ml of supernatant with an equal volume of RNAprotect bacteria reagent (Qiagen, Valencia, CA) and stored at −20°C to preserve RNA integrity.
RNA extraction.
RNA extraction used a modified sonication protocol in combination with the RNeasy midikit (Qiagen). The bacteria in RNAprotect were divided into microcentrifuge tubes and pelleted at 14,000 rpm. The tubes were transferred to ice, the supernatant was removed, and the pellet was resuspended in 1 ml of lysis buffer with β-mercaptoethanol (Qiagen). The tubes were ultrasonicated (3 times for 10 s using a Misonix XL-2000 ultrasonicator on medium-low speed with 10-s rests on ice in between). Pure ethanol (750 μl) was added and an RNA isolation procedure performed according to the manufacturer's protocol. RNA was eluted in RNase-free water, and quality was verified with the 2100 Bioanalyzer (Agilent Technologies, Santa Rosa, CA) at the Greenwood Facility for Biotechnology, University of Hawaii at Manoa, Honolulu, HI.
DNase digestion and cDNA synthesis.
DNase digestion was adjusted from that recommended by the manufacturer (Qiagen) because of the difficult removal of residual contaminating DNA. The reaction mixtures contained 2 μg of RNA template, 13.6 U of DNase I enzyme, and 10 μl of RDD buffer in a 100-μl total volume, and they were incubated for 1 h at room temperature. cDNA was prepared using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) according to the manufacturer's protocol. Control PCR amplification was performed (i) on DNase-digested RNA to confirm the removal of contaminating genomic DNA and (ii) on cDNA to confirm synthesis. The control reactions were performed with 10 pmol of each sigA primer (Table 1), 1 μl of template, and 27 μl of Platinum PCR SuperMix high fidelity (Invitrogen, Carlsbad, CA). PCR began with initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturation for 30 s at 94°C, annealing for 45 s at 58°C, and extension for 25 s at 72°C, and a final extension step (7 min at 72°C).
TABLE 1.
Primer sequences and results of gene amplification from genomic DNA of Mycobacterium strainsa
| Gene targeted | Primer name | Primer sequence (5′ to 3′) | EPS (bp)b | AT (°C)c |
|---|---|---|---|---|
| Mycobacterium housekeeping sigma factor (sigA) | sigA 536F | GCGCCTACCTCAAGCAGATC | 89 | 59 |
| sigA 625R | CGTACAGGCCTGCCTCGAT | |||
| Initial dioxygenase alpha-subunit (nidA) | nidA 919F | GGCACCGTGTTCCCGAATTTG | 103 | 55 |
| nidA 1022R | GAGAGCGGATGCCATAC | |||
| Initial dioxygenase beta-subunit (nidB) | nidB 35F | AGGCTGTCGAGGCGTTCAT | 79 | 59 |
| nidB 114R | GTCGAGCAGACCCAACCATT | |||
| Extradiol dioxygenase (phdF) | phdF 500F | AGAAGAGCGATGAGATYT | 124 | 51 |
| phdF 624R | CACCTCGACCATCAC | |||
| 1-Hydroxy-2-naphthoate dioxygenase (phdI) | phdI 307F | CACCGMCACACSAT | 707 | 51 |
| phdI 1014R | CTCGGGCTCYTCGTA | |||
| 2-Carboxybenzaldehyde hydratase aldolase (phdJ) | phdJ 173F | TGACCAMWGGCACCTT | 136 | 52 |
| phdJ 309R | CGTRTCGCGGGTATTG | |||
| Phthalate dioxygenase alpha-subunit (phtAa) | phtAa 655F | CACACSTCGGTGGTSGAGAT | 344 | 58 |
| phtAa 999R | CTCGCTGATYGGCTGCCAC |
All genes were amplified successfully from every strain, with the exception of phdJ from JS19b1.
EPS, expected product size.
AT, annealing temperature used for PCR amplification.
Primer design and reverse transcription-PCR.
Primers were designed to target the housekeeping gene sigA and six catabolic genes involved in the conversion of PHE through the 3,4-dioxygenation pathway. An available Mycobacterium vanbaalenii sequence was subjected to a BLAST search, and then sequences from rapidly growing Mycobacterium species or other related organisms (Nocardia and Rhodococcus spp.) were aligned to find conserved regions of the genes. Primers were optimized using the DOS-based Primer Designer version 1.01 (Scientific and Educational Software) and Primer Express 3.0 (Applied Biosystems, Foster City, CA) programs to determine secondary structure or primer-dimer formation. Primer sequence specificity was verified with the BLAST program (NCBI). PCR was performed as described above, with annealing temperatures listed in Table 1 and using genomic DNA (10 to 50 ng) as a template, and the resulting PCR products were sequenced to determine the specificity of amplification. Primer pairs that resulted in amplification from genomic DNA were applied to amplification of cDNA samples from cell cultures of all four Mycobacterium species that had been incubated with single or mixed PAHs for 3 and 7 days. Negative results in amplification were repeated three times to check for consistency.
Metabolite extraction and analysis.
For the extraction of PAH metabolites, the culture supernatant was acidified to pH 2.3 using 6 N hydrochloric acid and then extracted with an equal volume of ethyl acetate. The organic layer was reserved and extracted with an equal volume of 10 mM aqueous sodium hydroxide. The remaining organic layer was dried over anhydrous sodium sulfate and concentrated to 10 ml (neutral fraction). The aqueous layer (NaOH) was acidified to pH 2.3 and extracted with an equal volume of ethyl acetate. The resulting organic layer was dried over anhydrous sodium sulfate and concentrated to 10 ml (acidic fraction). The acidic fraction was derivatized with diazomethane, and the neutral fraction was derivatized with n-butylboronic acid (27). The derivatized extracts were concentrated to 1 ml for analysis.
Gas chromatography-mass spectrometry (GC-MS) analysis was performed on an HP 6890 gas chromatograph with an HP 5973 mass spectrometry system. The column was a ZB-1 (55 m, 250-μm diameter, 0.25-μm film thickness; Phenomenex, Inc.). The carrier gas was helium at a constant flow of 2 ml min−1. The column temperature was held at 120°C for 6.5 min, increased at a rate of 4°C/min to 270°C, and held for 3.5 min. To remove any remaining compounds from the column, the analysis was finished with a ramp of 20°C/min to 320°C held for 20 min. The solvent cutoff was 0 to 6.5 min. The mass spectrometer was operated in electron impact (EI) mode at 70 eV in the full scan mode from 85 to 450 m/z over 6.5 to 85 min. The injection volume (splitless mode) was 2 μl (HP 7683 Autosampler). The injector and analyzer temperatures were 270°C and 280°C, respectively. An Agilent G1701DA ChemStation was used as the system and data controller.
RESULTS
Identification of strain czh-101 as Mycobacterium gilvum.
Strain czh-101 was identified through phenotypic and genotypic tests as Mycobacterium gilvum. The phenotypic properties of czh-101 were consistent with those of M. gilvum (46). Briefly, this organism was Gram positive, scotochromogenic, and nonmotile and had a yellow colony color, and it reduced nitrate. The substrates oxidized by strain czh-101 were similar to those utilized by M. gilvum (d-fructose, d-mannitol, d-mannose, and sucrose). This strain did not grow at 45°C, nor did it utilize citrate. One phenotypic characteristic was inconsistent with reported data on M. gilvum: it was catalase positive, whereas M. gilvum has been reported as catalase negative (47). The sequence of the 16S rRNA gene over 1,433 bp was 100% identical to that of M. gilvum strain PYR-GCK. The sequence for rpoB (389 bp) was 100% identical to that in M. gilvum strains DSM 44503 and PYR-GCK. Nucleotide sequences were deposited in GenBank under the accession numbers DQ370009 (16S rRNA gene) and DQ534003 (rpoB).
PAH degradation as single or mixed substrates.
Four Mycobacterium species were tested for degradation in liquid culture of BAP, PYR, FLA, and PHE. Compared to abiotic controls, as sole substrates for the 7-day incubation period, M. gilvum czh-101 could degrade PYR (56%) and FLA (26%); M. aromaticivorans JS19b1 could degrade PHE (68%); M. crocinum czh-3 could degrade PYR (75%), FLA (13%), and PHE (100%); and M. rutilum czh-117 could degrade PYR (48%) and PHE (100%). Where <10% of the substrate was removed over the incubation, degradation is not reported.
The degradation of binary mixtures of the four PAHs was tested to compare the removal of PAH sole substrates. The ability to degrade PAH mixtures varied with the strain and chemical combination. Table 2 shows the percent degradation of compounds that were degraded differently as mixture compounds than they were as sole substrates. There was only one synergistic interaction observed: FLA was degraded better (29%) by strain czh-3 in the mixture with PHE than it was as a sole carbon source (13%). On the other hand, many strains exhibited antagonism in mixture degradation (see Table 2), some of which had extreme effects. For example, whereas 75% and 100% of PYR and PHE, respectively, were degraded as sole substrates by strain czh-3, only 5% and 20% were degraded in a mixture.
TABLE 2.
Antagonistic and synergistic interactions observed in degradation of mixed PAHs by four Mycobacterium strains
| Strain | PAH 1a | % degradation ± SD of PAH 1: |
PAH 2a | % degradation ± SD of PAH 2: |
||
|---|---|---|---|---|---|---|
| As sole substrate | In mixture | As sole substrate | In mixture | |||
| czh-101 | PYR | 56 ± 6 | 0 ± 2b | PHE | 12 ± 2 | 5 ± 1 |
| FLA | 26 ± 5 | 15 ± 2b | PHE | 12 ± 2 | 10 ± 1 | |
| JS19b1 | PHE | 68 ± 10 | 30 ± 18b | BAP | 0 ± 1 | 0 ± 6 |
| PHE | 68 ± 10 | 2 ± 0b | PYR | 7 ± 2 | 0 ± 1 | |
| PHE | 68 ± 10 | 15 ± 5b | FLA | 5 ± 6 | 0 ± 4 | |
| czh-3 | PYR | 75 ± 4 | 10 ± 2b | BAP | 0 ± 15 | 0 ± 2 |
| PYR | 75 ± 4 | 5 ± 0b | PHE | 100 ± 1 | 20 ± 0b | |
| FLA | 13 ± 2 | 6 ± 2b | BAP | 0 ± 15 | 2 ± 3 | |
| FLA | 13 ± 2 | 29 ± 2c | PHE | 100 ± 1 | 32 ± 12b | |
| PHE | 100 ± 1 | 70 ± 7b | BAP | 0 ± 15 | 7 ± 7 | |
| czh-117 | PHE | 100 ± 1 | 41 ± 13b | FLA | 0 ± 1 | 9 ± 10 |
PAHs were supplied at 100 ppm, except for BAP (50 ppm).
Antagonistic interaction in mixture degradation.
Synergistic interaction in mixture degradation.
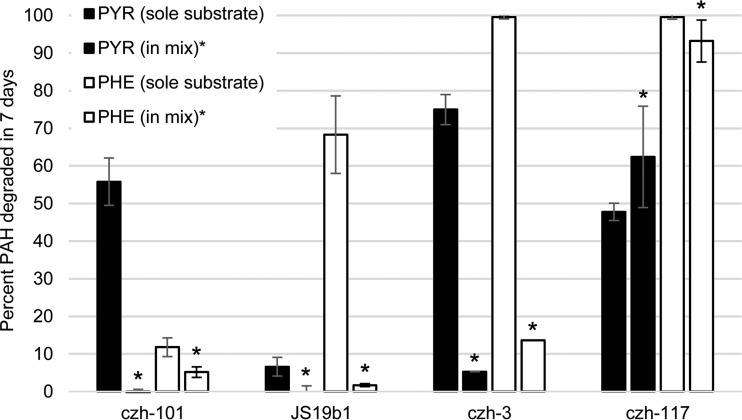
Indeed, the most striking changes in mixture degradation occurred with the combination of PYR and PHE. Each strain had a distinct pattern with regard to these two PAHs (Fig. 1). Whereas strain czh-101 could degrade PYR as a sole substrate, JS19b1 could degrade PHE, and strain czh-3 could degrade either chemical, none of these three strains could degrade the chemicals efficiently as mixture components. In contrast, strain czh-117 could degrade either chemical as a sole substrate and both in the mixture. This set of results provided a basis for the future comparison of pathway use via gene expression and metabolite production in closely related organisms with highly varied abilities to degrade the single chemicals and the mixture.
FIG 1.
Degradation of PYR and PHE as sole substrates and in a binary mixture by four Mycobacterium strains. The initial PYR or PHE concentration was 100 ppm (200 ppm total PAH in mixture). The error bars are the standard deviations. Antagonistic interactions were observed for three (czh-101, JS19b1, and czh-3) of the four strains.
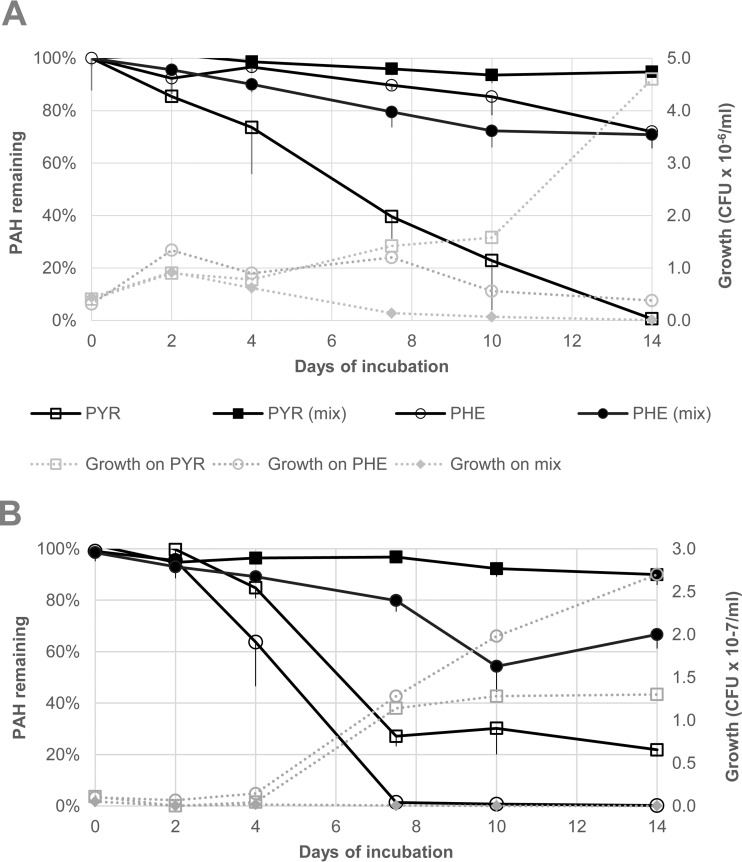
Antagonism of mixture degradation over 14-day incubation.
To determine whether antagonistic interactions observed were dependent on the sampling time point, strains were incubated with single and mixed PAHs over a 14-day time period. Two strains exhibiting antagonistic degradation of PYR and PHE were chosen for the time course study. The disappearance of PAH parent compounds (compared to time-zero samples) and bacterial growth were assessed at time points of 0, 2, 4, 7, 10, and 14 days. Figure 2 shows PAH utilization and associated growth of bacteria over an incubation period of 14 days. The results of these experiments demonstrated that the level of antagonism observed at day 7 was not mitigated by further incubation, and that bacterial growth was inhibited in mixed-substrate cultures.
FIG 2.
Time course study on the growth of Mycobacterium strains czh-101 (A) and czh-3 (B) during the degradation of PYR and PHE as sole substrates and as a binary PAH mixture. The initial PYR or PHE concentration was 100 ppm (200 ppm total PAH in mixture). The error bars are the standard deviations. Abiotic controls of PYR and PHE contained, respectively, 97% ± 2% and 83% ± 1% PAH remaining after 14 days of incubation.
Effects of PAHs on bacteria growing with sugar substrates.
Lack of cell growth was correlated with antagonism; however, it was not clear whether suppressed growth was the result or the cause of reduced substrate consumption. If antagonizing PAHs caused toxicity, their presence in a culture might result in growth inhibition and the consequential inability to utilize the substrate PAH. To test whether antagonizing PAHs (or increased PAH concentration in the mixture) were responsible for a lack of cell growth, strains czh-101 and czh-3 were supplied with substrates of sugar alone (mannitol or glucose), sugar with the antagonizing PAH, or sugar with the antagonized PAH combination. Strain czh-117, which showed no antagonism in PYR/PHE degradation, was also tested similarly for comparison with the antagonized strains. Growth was monitored by plate count at 5 and 8 days of incubation and compared for substrate treatments. The results (not shown) were similar among all the strains: the presence of the PAH combination or level did not result in the growth inhibition seen in cultures supplied with the antagonized PAH mixtures described in the previous section.
Catabolic pathway analysis.
Primers were designed (Table 1) to target conserved regions of the genes in the described 3,4-dioxygenation catabolic pathway of PHE (also involved in the conversion of later-stage PYR metabolites). The results from this study confirmed the presence of the full catabolic pathway in the genomic DNA of all four organisms, with the exception of phdJ (2-carboxybenzaldehyde hydratase aldolase gene) from strain JS19b1.
Aspects of gene expression during degradation differed by strain and by pathway (upper versus lower). Regarding genes from the upper pathway (nidAB and phdFIJ), in all treatments of three of the strains, czh-101, czh-3, and czh-117, no qualitative differences in gene expression were observed in a comparison of cells fed with sole substrates versus mixtures (Table 3). However, although amplified successfully from the genome of JS19b1, nidAB (initial dioxygenase, alpha- and beta-subunit genes) were not expressed under any experimental condition in this study. In this strain, we also observed qualitative differences in the expression of catabolic genes in cells grown on PHE as a sole substrate and those grown on the PAH mixture: phdF and phdI were expressed differentially at one time point between cells grown on PHE and those growing with the mixture (see Table 3).
TABLE 3.
RT-PCR gene expression in four Mycobacterium strains grown with single (PYR and PHE) and mixed (MIX) substrates at 3/7 days of incubation
| Gene | Substrate treatment by bacterial straina |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| czh-101 |
JS19b1 |
czh-3 |
czh-117 |
|||||||||
| PYR | MIX | PHE | PYR | MIX | PHE | PYR | MIX | PHE | PYR | MIX | PHE | |
| nidA | +/+ | +/+ | +/+ | −/− | −/− | −/− | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ |
| nidB | +/+ | +/+ | +/+ | −/− | −/− | −/− | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ |
| phdF | +/+ | +/+ | +/+ | −/− | +/− | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ |
| phdI | +/+ | +/+ | +/+ | −/− | −/− | −/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ |
| phdJ | +/+ | +/+ | +/+ | NA | NA | NA | +/+ | +/+ | +/+ | −/+ | +/+ | +/+ |
| phtAa | +/+ | +/+ | +/− | −/− | −/− | +/+ | +/+ | +/+ | +/+ | −/− | −/− | −/− |
+, gene amplified; −, gene not amplified; NA, gene not found in the genome of the strain. For all strains and treatments, the main housekeeping sigma factor (sigA) was amplified from cDNA as a positive control for amplification.
On the other hand, the expression of phtAa (phthalate dioxygenase) from a later stage in degradation varied more with the strain and treatment. For example, although it was present in the genome, phtAa was not detected in RNA samples of strain czh-117 under any condition in this study. In addition, phtAa was not detected in the RNA of other strains in which PAH degradation was minimal, like that of czh-101 with PHE at day 7, or all JS19b1 samples grown with PYR or the mixture.
Identification of metabolites in single and mixed PAH degradation.
In this study, four Mycobacterium species were characterized with respect to metabolites formed during the degradation of PHE, PYR, and a mixture of the two at 3 and 7 days of incubation. The purpose of this part of the study was to compare the metabolic profiles of (i) strains degrading single compounds versus mixtures and (ii) strains with antagonized versus nonantagonized mixture degradation. We specifically looked for metabolites or pathways that were present or absent in the extracts of cells grown on the PAH mixture versus those grown on a single substrate. Table 4 shows the metabolites surveyed by GC-MS in this study with their identifying characteristics of retention time and mass fragmentation pattern. Table 5 shows the metabolites that were identified in the extracts of cultures incubated with single and mixed PAHs.
TABLE 4.
Metabolites surveyed: chemical identification, GC retention times, and MS identifying ions
| IDa | Chemical name | Fractionb | GC RT (min)c | Mass spectra m/z (% relative abundance) |
|---|---|---|---|---|
| A-II | Phenanthrene-4,5-dicarboxylic acid (diME) | A | 62.37 | 294 [M+] (1), 263 (2), 235 (100), 220 (49) |
| A-III | Phenanthrene-4-carboxylic acid (ME) | A | 51.88 | 236 [M+] (100), 205 (92), 177 (77), 176 (61) |
| B-II | 4-Oxapyren-5-one | A/N | 57.16 | 220 [M+] (100), 192 (16), 163 (33) |
| A-IX | 7,8-Benzocoumarin | A/N | 44.80 | 196 [M+] (83), 168 (100), 139 (59) |
| A-X | 1-Hydroxy-2-naphthoic acid (ME) | A | 33.20 | 202 [M+] (31), 170 (100), 142 (5), 114 (51) |
| A-XI | 2-Carboxycinnamic acid (ME) | A | 31.81 | 206 [M+] (13), 146 (100), 133 (97), 105 (42) |
| A-XII | Phthalic acid (diME) | A | 18.10 | 194 [M+] (6), 163 (100), 133 (6) |
| C-I | 2-(2-Carboxyvinyl)-1-naphthoic acid (diME) | A | 50.71 | 211 (100), 197 (53), 181 (45), 152 (40) |
| C-II | Naphthalene-1,2-dicarboxylic acid (diME) | A | 44.43 | 244 [M+] (35), 213 (100) |
| C-III | Naphthalene-1,2-dicarboxylic acid anhydride | A/N | 36.92 | 198 [M+] (58), 154 (56), 126 (100) |
| E-IV | 5,6-Benzocoumarin | A/N | 47.15 | 196 [M+] (60), 168 (100), 139 (70) |
| F-II | cis-9,10-Dihydrophenanthrene-9,10-diol (diME) | A | 51.55 | 238 [M+] (100), 180 (55), 151 (90) |
| F-III | Diphenic acid (diME) | A | 41.35 | 211 [M+] (100), 196 (17), 180 (13), 152 (13) |
Metabolite identification (ID) corresponding with structures in Fig. 3. Other metabolites surveyed that were not found in any culture extract included: 4-(1-hydroxynaphth-2-yl)-2-oxobut-3-enoic acid (A-VIII), 1-hydroxy-2-naphthaldehyde, 4-(2-hydroxynaphth-1-yl)-2-oxobut-3-enoic acid (E-III), naphthalene-1,2-diol (C-IV), 2-hydroxy-1-naphthoic acid, 2-hydroxy-1-naphthaldehyde, and 1-(2-carboxyvinyl)-2-naphthoic acid (D-I).
Fractions, acidic (A) or neutral (N), from which metabolites were detected.
RT, retention time.
TABLE 5.
Metabolites detected in extracts from four Mycobacterium strains incubated for 3 and 7 days with pyrene, phenanthrene, or a mixture of the two chemicals
| Metabolite IDa | Chemical nameb | czh-101 |
JS19b1 |
czh-3 |
czh-117 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PYR | Mix | PHE | PYR | Mix | PHE | PYR | Mix | PHE | PYR | Mix | PHE | ||
| A-II | Phenanthrene-4,5-dicarboxylic acid (diME) | +/+ | +/+ | +/− | −/+ | +/+ | +/+ | +/d | |||||
| A-III | Phenanthrene-4-carboxylic acid (ME) | +/+ | +/− | −/+ | −/+ | +/+ | +/− | ||||||
| B-II | 4-Oxapyren-5-one | +/+ | +/+ | +/d | +/+ | +/+ | +/+ | d/+ | |||||
| A-IX | 7,8-Benzocoumarin | +/+ | +/+ | −/+ | d/+ | +/+ | +/+ | +/+ | +/d | +/+ | +/+ | +/+ | |
| A-X | 1-Hydroxy-2-naphthoic acid (ME) | −/d | d/+ | +/+ | −/+ | −/+ | +/+ | +/+ | +/+ | +/+ | +/+ | ||
| A-XI | 2-Carboxy-cinnamic acid (diME) | +/+ | −/+ | +/− | +/d | ||||||||
| A-XII | Phthalic acid (diME) | −/+ | −/+ | −/+ | +/− | +/+ | +/+ | +/+ | −/+ | +/+ | +/+ | +/+ | |
| C-I | 2-(2-Carboxy-vinyl)-1-naphthoic acid (diME) | −/+ | −/+ | −/d | +/+ | +/+ | +/+ | +/+ | |||||
| C-II | Naphthalene-1,2-dicarboxylic acid (diME) | −/+ | −/+ | ||||||||||
| C-III | Naphthalene-1,2-dicarboxylic acid anhydride | +/+ | +/+ | −/+ | −/+ | +/+ | −/d | +/+ | +/+ | ||||
| E-IV | 5,6-Benzocoumarin | +/+ | +/+ | +/+ | +/+ | ||||||||
| F-II | cis-9,10-Dihydrophenanthrene-9,10-diol (diME) | +/+ | −/+ | +/+ | |||||||||
| F-III | Diphenic acid (diME) | +/+ | +/+ | +/+ | +/− | −/+ | −/+ | −/+ | +/+ | +/+ | +/+ | +/+ | +/+ |
Metabolite letter-numeral combinations correspond with metabolite IDs in Fig. 3.
diME, dimethyl ester; ME, methyl ester; d, the identifying mass spectrum was detected in the sample, but the peak was not above the background.
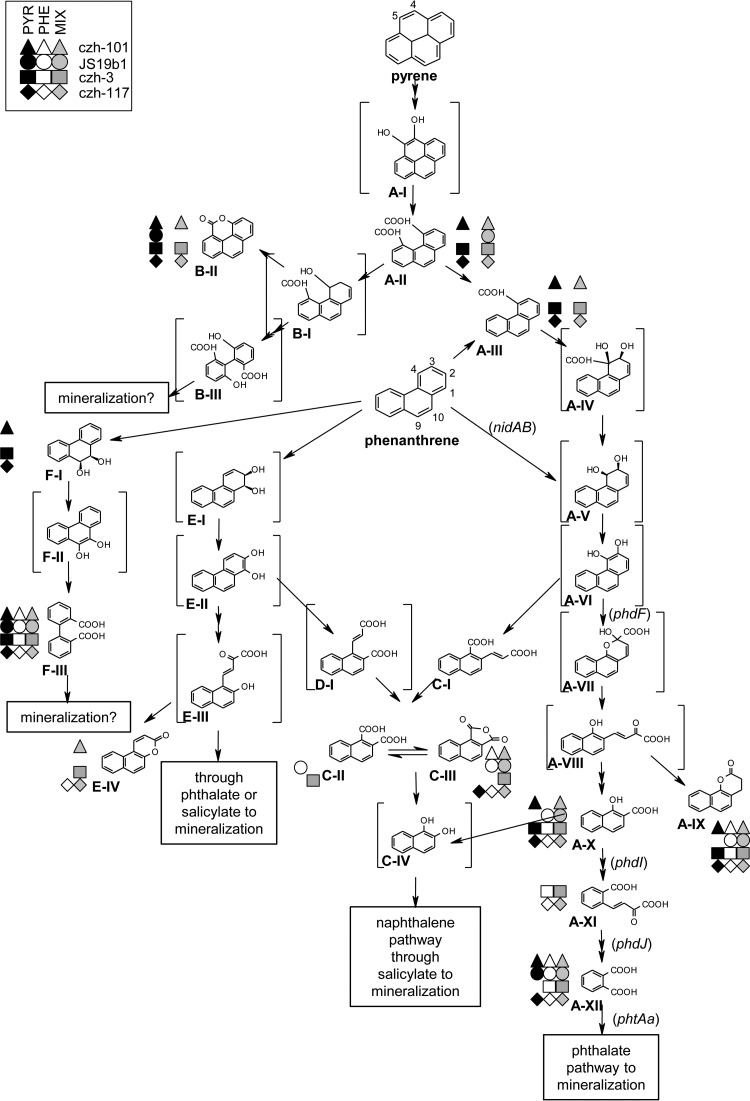
Metabolites and pathways of PHE, PYR, and mixture degradation by strain czh-101.
Strain czh-101 degraded about 12% of the available PHE in 7 days but could not use it as a sole carbon source for growth. In this study, metabolites detected from cultures of czh-101 incubated with PHE were identified as intermediates of the 3,4- and 9,10-dioxygenase pathways (Fig. 3, metabolite series A, C, and F; see this figure also for the letter-number combinations in parentheses). Detected metabolites of the 3,4-pathway included 7,8-benzocoumarin (A-IX), 2-(2-carboxyvinyl)-1-naphthoic acid (C-I), naphthalene-1,2-dicarboxylic acid anhydride (C-III), and phthalic acid (A-XII). The detection of later-stage metabolites (C-I/C-III and A-IX/A-XII) indicated that cis-3,4-dihydrophenanthrene-3,4-diol underwent both ortho- and meta-cleavage (27, 32, 40). The presence of diphenic acid (F-III) demonstrated a degradation route through the 9,10-dioxygenation pathway. In spite of the ability to transform PHE to metabolites similar to those produced by other Mycobacterium strains, czh-101 cannot grow with PHE as a sole substrate.
FIG 3.
Map of proposed degradation of pyrene (PYR), phenanthrene (PHE), and a mixture of the two chemicals (MIX) by four Mycobacterium species (czh-101, JS19b1, czh-3, and czh-117). Metabolites detected in each strain-chemical treatment combination are indicated by the symbols next to the metabolites. The metabolites in brackets are proposed intermediates. The initial PYR or PHE concentration was 100 ppm (200 ppm total PAH in mixture). Genes monitored by RT-PCR are indicated in italics next to arrows, showing catalytic conversion of metabolites. This map allows for a comparison of the metabolites detected in different strains and treatments.
PYR was degraded well by czh-101 (56% in 7 days at 100 μg ml−1 level), and metabolism proceeded as previously described (5, 14, 33, 39) through 4,5-phenanthrenedicarboxylic acid (A-II) and phenanthrene-4-carboxylic acid (A-III). A branching route described by Vila et al. (36) was also observed in strain czh-101 by the recovery of 4-oxapyrene-5-one (B-II). Metabolites of the PHE 3,4-dioxygenation pathway were detected, and it is probable that PYR is mineralized through this route. In the catabolism of PYR (unlike PHE, see above), evidence was found only for meta-cleavage of cis-3,4-dihydrophenanthrene-3,4-diol. Finally, diphenic acid (F-III) was detected during PYR degradation, possibly arising from the loss of the carboxyl group of 4-phenanthroic acid (48), with subsequent attack at the C-9 and C-10 positions of PHE.
Strain czh-101 did not degrade PYR or PHE efficiently when the two chemicals were mixture components. Metabolites of PYR (phenanthrene-4,5-dicarboxylic acid [A-II], phenanthrene-4-carboxylic acid [A-III], and 4-oxapyren-5-one [B-II]) were detected during mixture degradation. Whereas the use of the PHE 3,4- and 9,10-dioxygenation pathways was demonstrated in extracts from both single-PAH and mixture-grown cultures, interestingly, in mixture degradation, metabolites of a new PHE pathway came to the forefront. The detection of 5,6-benzocoumarin (E-IV) demonstrated degradation through the 1,2-dioxygenation pathway (28, 30, 32), occurring only during the metabolism of the mixture. Although degradation through a new pathway was observed in cultures exposed to the mixture, PHE degradation was not enhanced by the opening of this pathway.
Metabolites and pathways of PHE, PYR, and mixture degradation by strain JS19b1.
JS19b1 degraded PHE well (68% in 7 days at 100 μg ml−1 level) and could use it as a sole carbon source. Metabolites from the 3,4-dioxygenation pathway were detected; later-stage metabolites demonstrated that cis-3,4-dihydrophenanthrene-3,4-diol (A-V) undergoes meta-cleavage to form 1-hydroxy-2-naphthoic acid (A-X). Together with the detection of diphenic acid (F-III), these data corroborate (40) the use of both the 3,4- and 9,10-pathways for the degradation of PHE in strain JS19b1.
JS19b1 did not degrade PYR well and could not use it as a sole source of carbon under the study conditions. The PYR metabolite 4-oxapyren-5-one (B-II) was detected in culture extracts of JS19b1 grown on PYR. In addition, two later-stage metabolites, phthalic acid (A-XII) and diphenic acid (F-III), were detected. The scarcity of metabolites was consistent with the strain's inability to use PYR as a sole carbon source.
Mixture metabolites of strain JS19b1, 7,8-benzocoumarin (A-IX), 1-hydroxy-2-napthoic acid (A-X), and diphenic acid (F-III), were similar to those in extracts of cells grown on PHE. Metabolites of alternative PHE pathways were not detected in mixture extracts of this strain, as they were in those of the other strains that exhibited antagonism in degradation of the mixture (see czh-101, above, and czh-3, below). However, a new PYR metabolite (phenanthrene-4,5-dicarboxylic acid [A-II]) from a major mineralization pathway was detected in mixture extracts. As with other strains, the emergence of the new PYR metabolite was not indicative of enhanced PYR degradation by cells growing on the mixture.
Metabolites and pathways of PHE, PYR, and mixture degradation by strain czh-3.
Strain czh-3 also degraded PHE well (100% in 7 days at 100 μg ml−1 level) and could use it as a sole carbon source. The metabolism of PHE proceeded through the 3,4- and 9,10-dioxygenation pathways (A, C, and F series of metabolites in Fig. 3). As with strains czh-101 and czh-117 (see below), czh-3 evidently uses both meta- and ortho-cleavage mechanisms to break down cis-3,4-dihydrophenanthrene-3,4-diol (A-V).
PYR was degraded well by czh-3 (75% in 7 days at 100 μg ml−1 level), and metabolism proceeded through 4,5-phenanthrenedicarboxylic acid (A-II) and phenanthrene-4-carboxylic acid (A-III). Similar to czh-101, strain czh-3 metabolizes PYR through the PHE 3,4-dioxygenation pathway, and evidence was found only for meta-cleavage of cis-3,4-dihydrophenanthrene-3,4-diol (A-V). Other similarities in the degradation of PYR by czh-3 and czh-101 included the utilization of the second mineralization route via 4-oxapyrene-5-one (B-II) and the degradation of PYR through diphenic acid (see above section on czh-101 for more detail).
Degradation of the PAH mixture by strain czh-3 was antagonized such that neither substrate was degraded much. Strain czh-3 exhibited trends similar to those of strain czh-101 in the metabolic profiles of cells grown on mixed PAHs. In mixture degradation by czh-3, metabolites of the A (both PYR and PHE), B, C, E, and F pathways were found. Again, the presence of 5,6-benzocoumarin (E-IV) demonstrated the novel use of the 1,2-dioxygenation pathway in mixture degradation. As with strains czh-101 and JS19b1, the emergence of minor and novel pathways in mixture degradation did not correspond to increased degradation of either substrate.
Metabolites and pathways of PHE, PYR, and mixture degradation by strain czh-117.
Strain czh-117 degraded PHE well in 7 days (100% at the 100 μg ml−1 level), and it was the only organism in this study for which metabolites of all three initial dioxygenation pathways were detected in single-PAH degradation (A, E, and F series of metabolites in Fig. 3). As in other strains, both meta- and ortho-cleavage of cis-3,4-dihydrophenanthrene-3,4-diol (A-V) were demonstrated by the detection of later-stage metabolites, like 7,8-benzocoumarin (A-IX) and 2-(2-carboxyvinyl)-1-naphthoic acid (C-I), respectively. In contrast to other strains, 5,6-benzocoumarin (pathway E) was detected in single-substrate PHE degradation. Overall, czh-117 was more metabolically versatile than the other three strains with respect to PHE catabolism. Ring fission of phenanthrene-1,2-diol does not occur in Mycobacterium vanbaalenii PYR-1 (48).
PYR degradation by czh-117 was similar to that of other strains which degraded it well, with metabolism proceeding through the A-series metabolites (4,5-phenanthrenedicarboxylic acid [A-II] and phenanthrene-4-carboxylic acid [A-III]) and a B-series metabolite, 4-oxapyrene-5-one (B-II). Evidence was found only for meta-cleavage of cis-3,4-dihydrophenanthrene-3,4-diol (A-V), and diphenic acid (F-III) was detected. The detected products and pathways were similar in all three PYR-degrading strains, and this study demonstrates the likeness in PYR degradation across diverse Mycobacterium species, including the meta-cleavage (and not ortho-cleavage) of cis-3,4-dihydrophenanthrene-3,4-diol (A-V) and metabolism through diphenic acid (F-III).
In stark contrast to other strains, strain czh-117 showed little change between mixed- and single-PAH metabolic profiles. Most importantly, all three routes of PHE degradation were represented in single and mixed degradation. In contrast to the strains in which antagonized mixture degradation and shunting through alternate routes were observed, strain czh-117 showed pathway flexibility and an unambiguous ability to degrade mixed PYR/PHE.
DISCUSSION
As Mycobacterium is one of the few bacterial genera able to degrade HMW PAHs, and one that is being used in developing remediation strategies for real-world spill mixtures (49, 50), understanding the intricacies of PAH mixture degradation by Mycobacterium species is an area of importance in bioremediation research. The aim of this study was to investigate the degradation of PAH substrates by Mycobacterium species and characterize the metabolic changes associated with mixed-substrate antagonism. Using four species that were distinct in both taxonomy and PAH substrate range, we expected to be able to interpret the phenomenon of antagonism more broadly. Potential causes of antagonism in PAH mixture degradation were investigated, including the possibilities of preferential substrate degradation, toxicity of parent PAH or of PAH level, repression of catabolic genes, and metabolite formation. A major finding of this work demonstrates that the metabolism of PAH mixtures is characterized by the use of alternate degradation pathways.
Antagonistic interactions in mixture degradation with the bacterial species and PAHs tested (BAP, PYR, PHE, and FLA) were very common, and in fact, more common than degradation being unaffected by the addition of a second PAH. The most common antagonistic effect in this study and in other reports (16, 17, 23) involved decreased degradation of a substrate when a second nondegradable substrate was added. In other words, preferential substrate degradation of the second PAH cannot explain the numerous cases of antagonism reported. In 8 of 11 cases in this study, inhibition was caused by the addition of a more-complex and less-water-soluble substrate, which was not degraded in the mixture at all. This contradicts the previous suggestion (17) that water solubility or the simplicity of additional carbon sources affects antagonism. Because the most significant level of antagonism occurred with the combination of PYR and PHE, strains fed with those PAHs were further studied with respect to persistence of antagonism, toxicity of parent compounds or concentrations, catabolic gene expression, and metabolite profiles.
Time course studies demonstrated that the degradation of both PYR and PHE, regardless of molecular weight or aqueous solubility, was antagonized and persisted over a long incubation period. In addition, combined utilization of the two PAHs did not reach the level of a single well-utilized carbon source in percentage, weight, or number of moles. For example, whereas 2.47 μmol (497 μg) PYR was consumed by strain czh-101 over 14 days, the combined consumption of PYR and PHE in a mixture only reached and 0.95 μmol (172 μg). Furthermore, antagonism in mixture degradation was accompanied by substantially suppressed cell growth. The persistence over time of antagonistic effects in this study and others (18, 21, 23) is important to the field of bioremediation because it may explain the recalcitrance of PAHs in soil where bacteria that have the ability to degrade them are present (12, 18). Often, bioremediation potential has been measured by monitoring the presence of relevant microbes or catabolic genes (51, 52), but if in situ organisms experience antagonistic interference with degradation, the predictive power of these techniques may depend on which organisms and PAH mixtures are present.
Even with lack of growth or substrate transformation, cells fed with PAH mixtures were minimally alive: they could be plated, and housekeeping and catabolic genes were amplified from cDNA after incubation. Causes for suppressed growth included possible toxicity or the operational lack of carbon source (stemming from, for example, obstruction of PAH transport into cells, inhibition of catabolic enzymes, or repression of catabolic genes). Parent PAH species or concentrations were not toxic to cells in growth tests with mannitol and glucose. However, because cells grown with sugar generally use catabolite repression to suppress the degradation of energetically inferior substrates, like aromatics (31, 53, 54), this test could not resolve whether cells were subject to the toxicity of the metabolites produced during PAH mixture degradation.
Reverse transcription-PCR (RT-PCR) has been used in previous studies of aromatic degradation (42, 53, 55, 56) to monitor the expression of catabolic genes, which are generally tightly regulated and induced in the presence of relevant substrates (11, 35, 57–60). Induction in response to different PAHs is a strain-specific characteristic (12), such that the various abilities of the Mycobacterium species in this study and others to handle diverse PAHs and mixtures depend on not only the presence of catabolic operons but also their induction and regulation. Repression mediated at the transcriptional level has been identified as a cause for inhibited degradation of styrene in Pseudomonas putida (53). However, the expression data in this study did not suggest a strong role for repression in antagonism: qualitative differences in gene expression were not observed in two of the strains exhibiting antagonized mixture degradation. The application of a more discriminating technique, such as quantitative PCR, may be required to reveal subtler changes in the level of expression.
RT-PCR, however, did provide insight into the pathways used by the organisms in the study. Genes from the upper pathway (nidAB and phdFIJ) were expressed in all treatments in three of the strains, czh-101, czh-3, and czh-117. On the other hand, expression of phtAa (phthalate dioxygenase gene) from a later stage in degradation was not detected in the RNA of samples in which PAH degradation was minimal, like that of czh-101 with PHE at day 7 or all JS19b1 samples grown with PYR or the mixture. If regulated separately, as in other Mycobacterium species (61), this gene may be expressed only when there is sufficient substrate for the enzyme. Although it was present in the genome, phtAa was not expressed by strain czh-117 during the degradation of either sole substrate or the mixture. Strain czh-117 may not utilize the phthalate pathway and instead may catabolize naphthalene-1,2-diol further through the naphthalene pathway, as has been shown in other species of Mycobacterium (36), as well as other genera (28, 30). Whether circumvention of the phthalate pathway is related to the strain's ability to degrade mixed substrates without antagonism is not yet known.
Several data supported a previous reference (62) to a divergent PHE catabolic pathway in strain JS19b1. phdJ (2-carboxybenzaldehyde hydratase aldolase) could not be amplified from genomic DNA with the consensus primers designed for this study. In addition, although amplified successfully from the genome, nidAB (initial dioxygenase, alpha- and beta-subunit genes) transcripts were not detected with exposure to either PYR or PHE. Earlier proteomic profiling of this organism using PHE as a sole substrate resulted in the detection of a protein similar to phenanthrene dioxygenase PdoA2 but not NidAB (62). Although NidAB targets PYR (11, 43, 58), it has notably been induced with exposure to PHE (11, 55, 58), including in the three other strains of this study. A mutation in nidA in M. vanbaalenii PYR-1 has been shown to disrupt pyrene degradation (43), but with strain JS19b1, the presence in genomic DNA but the absence of transcripts under PYR, PHE, or mixture (MIX) growth in this study argues that the induction process may be affected. Indeed, the inability of strain JS19b1 to use PYR as a growth substrate under the conditions tested, unlike many PAH-utilizing Mycobacterium species with nid orthologs, might be due to a loss of induction of this initial dioxygenase. Also interesting with this strain is that the qualitative differences in expression between PHE and mixture treatments (phdF, phdI, and phtAa) were observed in genes encoding enzymes that have been associated with tight control in response to PAH (59). The presence of the PAH mixture in this case did correlate with a lack of expression of some catabolic genes, a suspected cause of antagonism characterizing the degradation of PAH mixtures. However, some enzymes of upper pathways can be induced by downstream metabolites (11), so a lack of induction of these genes in PYR or the mixture may simply reflect the low transformation of substrate.
Important results of the study came from the profiling of metabolites during single and mixed PAH degradation. Information on pathway usage for four taxonomically distinct Mycobacterium species furthers the knowledge of PYR and PHE degradation; in particular, PYR was degraded not only through the A series lower-pathway metabolites (see Fig. 3) but also through the F series, and strain czh-117 routinely produced ring fission products of the 1,2-dioxygenation pathway of PHE. A key novel aspect of this work comprises the demonstrated use of alternate pathways in mixture degradation. In two of three strains that exhibited antagonized mixture degradation, metabolites were identified that belonged to a pathway not normally used by the organism in the degradation of single PAHs. The occurrence of this in two organisms and the replication of these data at two time points lend consistency and weight to the result. In the third organism, JS19b1, an additional metabolite was found, but the use of a wholly new pathway was not established.
Recently, detailed work on initial ring-hydroxylating dioxygenases in Mycobacterium species has led to a more accurate understanding of the diversity of enzymes present, their induction by and transformation rates of PAH substrates, and regioselective product formation (11, 42, 43, 48, 60, 63, 64). nid homologs are subject to induction by different subsets of PAH substrates (11) and seem optimized to be well induced by the PAHs that they can efficiently convert into usable cis-dihydrodiol isomers (i.e., intermediates of mineralization pathways) (42, 60). On the other hand, heterologous expression (not requiring induction by PAH) studies revealed that when PAH transformation is catalyzed by a nonoptimal nid paralog, products with low regiospecificity (multiple cis-dihydrodiol isomers) result (11, 60). Though PYR and PHE were earlier thought to be degraded by similar enzymes, it has now been established that they are targeted by distinct nid paralogs (43), a point that is critical to understanding the antagonism in PYR-PHE mixtures. For example, in experiments of heterologous expression, NidAB, the PYR dioxygenase of Mycobacterium vanbaalenii PYR-1, converted 100% of the supplied PYR to cis-4,5-dihydrodiol, whereas NidA3B3 (FLA dioxygenase that also acts on PHE) converted only about 65% of the PYR, forming a mixed product that included dead-end metabolites (60). These studies together suggest that induction of a dioxygenase system by one PAH may have incidental or unintentional consequences on the degradation of another. In our study, the appearance of intermediates of new pathways in antagonized mixture degradation supports the idea of cross talk between enzymes and substrates: the initial dioxygenase induced by PYR may act on PHE to produce the metabolites of a pathway not normally used in single-PAH degradation. For example, the appearance of the 1,2-dioxygenation pathway intermediates in mixture degradation by two strains may result from the action of a PYR-induced enzyme on PHE to produce cis-1,2-dihydrodiol. Because subsequent pathway steps are thought to be catalyzed by enzymes with broad substrate ranges (48), ring fission products (5,6-benzocoumarin) were produced. Another piece of evidence pointing to enzyme cross talk was the high recovery in antagonized mixture samples of diphenic acid (60% of combined peak area of czh-3 MIX metabolites at day 7 compared to 32% of PHE sample or 2% of PYR sample), which is produced by K-region attack and ring fission analogous to that on the C-4 and C-5 positions of pyrene involved in typical pyrene pathways (29). NidA enzymes induced by PYR in the mixture may work on the K region of PHE to produce C-9 and C-10 dioxygenation pathway products in excess. In contrast, strain czh-117, which readily degraded the PAH mixture, utilized the same pathways and produced similar intermediates for single and mixed PAH degradation.
Novel pathway use during mixture degradation suggests two possible mechanisms for antagonism which will direct our future work. The shunting of substrate through a new pathway, in combination with poor growth and suppressed degradation, suggest mechanisms of antagonism involving either toxicity or enzyme-level inhibition, both with precedents in published work on aromatic catabolism (19, 25, 57, 65). For example, in Mycobacterium species and other bacteria, it is known that dihydrodiols are converted to diols, which autooxidize into toxic o-quinones (25, 35, 58, 66). Detoxification involves retroconversion to a diol by quinone reductase enzymes, which can have narrow substrate ranges (66). Either ring fission and further degradation in the case of mineralization pathways or inactivation by methyl conjugation in dead-end pathways can resolve the reactive cycling between diol and quinone (67). An organism like czh-101, which does not normally use the 1,2-dioxygenation pathway to degrade PHE, may lack the enzymatic means to detoxify phenanthrene-1,2-dione. Conversely, czh-117, which tolerated PYR-PHE mixtures and produced ring fission products of the 1,2-dioxygenation pathway in both single and mixed PAH degradation, uses endogenous mechanisms to detoxify quinones of that origin. Chemical analyses of suspected quinones in cell extracts, analysis of quinone reductase genes, and toxicity assays with suspected interfering chemicals are tests that can resolve this question. On the other hand, enzyme-level inhibition affects the conversion of some PAH substrates via disparate mechanisms. Deveryshetty and Phale (57) demonstrated competitive inhibition of 1-hydroxy-2-naphthoic acid dioxygenase by a stereoisomeric compound, 3-hydroxy-2-naphthoic acid. An alternative mechanism, oxidative (suicide) inactivation of extradiol dioxygenases, has been demonstrated with suboptimal substrates, and the level of inhibition depended on the sequence variation of catabolic enzymes (68). Similar mechanisms may cause enzyme inhibition and concomitant diminished growth/degradation if the alternative pathway intermediates of antagonized mixture catabolism interfere at enzyme active sites. Because enzyme inhibition and metabolite toxicity are known in aromatic degradation, they comprise reasonable hypotheses for future work about the mechanisms of antagonism through enzyme cross talk and the appearance of new disruptive pathways. Indeed, similar mechanisms may affect organisms like czh-101 and JS19b1 in their inability to degrade PHE and PYR, respectively. For example, if PHE enzymes in JS19b1 are aversely induced by PYR, the resulting metabolites may induce toxicity or simply fail to feed the organism by unproductive obstruction of active sites.
Knowledge about antagonistic interactions in PAH mixture degradation must have a large impact on studies of bioremediation strains and risk assessment of contaminated sites. First, support in this work for the occurrence of antagonistic phenomena in taxonomically diverse Mycobacterium species indicates that modeling studies must consider this property to be distinct from that of PAH substrate range. The presence of catabolic genes or the ability to degrade single chemicals may not predict bioremediation potential. In addition, risk assessment may be improved by accounting for the toxicity of mixture-produced intermediates in addition to those produced by the degradation of single PAHs. Further work on the underlying causes of antagonism may provide the means to improve risk assessment and promote in situ degradation.
ACKNOWLEDGMENTS
We thank Young Soo Keum and Jong-Su Seo for technical advice and Karl and Joan Yanagihara for analytical assistance. Very special thanks go to Wendy Kaneshiro-Sueno for thorough and conscientious work on the sugar/PAH experiments.
Funding Statement
The study design, data collection and interpretation, and the decision to submit for publication were independent of the funding agency.
REFERENCES
- 1.Mumtaz M, George J. 1995. Toxicological profile for polycyclic aromatic hydrocarbons. Agency for Toxic Substances and Disease Registry, Centers for Disease Prevention and Control, Atlanta, GA: http://www.atsdr.cdc.gov/toxprofiles/tp69.pdf. [PubMed] [Google Scholar]
- 2.Haritash AK, Kaushik CP. 2009. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater 169:1–15. doi: 10.1016/j.jhazmat.2009.03.137. [DOI] [PubMed] [Google Scholar]
- 3.Kanaly RA, Harayama S. 2010. Advances in the field of high-molecular-weight polycyclic aromatic hydrocarbon biodegradation by bacteria. Microb Biotechnol 3:136–164. doi: 10.1111/j.1751-7915.2009.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball A, Truskewycz A. 2013. Polyaromatic hydrocarbon exposure: an ecological impact ambiguity. Environ Sci Pollut Res Int 20:4311–4326. doi: 10.1007/s11356-013-1620-2. [DOI] [PubMed] [Google Scholar]
- 5.Cerniglia CE. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351–368. doi: 10.1007/BF00129093. [DOI] [Google Scholar]
- 6.Jarvis IW, Dreij K, Mattsson A, Jernström B, Stenius U. 2014. Interactions between polycyclic aromatic hydrocarbons in complex mixtures and implications for cancer risk assessment. Toxicology 321:27–39. doi: 10.1016/j.tox.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Lundstedt S, White PA, Lemieux CL, Lynes KD, Lambert IB, Oberg L, Haglund P, Tysklind M. 2007. Sources, fate, and toxic hazards of oxygenated polycyclic aromatic hydrocarbons (PAHs) at PAH-contaminated sites. Ambio 36:475–485. doi: 10.1579/0044-7447(2007)36[475:SFATHO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 8.Bogan BW, Lahner LM, Sullivan WR, Paterek JR. 2003. Degradation of straight-chain aliphatic and high-molecular-weight polycyclic aromatic hydrocarbons by a strain of Mycobacterium austroafricanum. J Appl Microbiol 94:230–239. doi: 10.1046/j.1365-2672.2003.01824.x. [DOI] [PubMed] [Google Scholar]
- 9.Heitkamp MA, Franklin W, Cerniglia CE. 1988. Microbial metabolism of polycyclic aromatic hydrocarbons: isolation and characterization of a pyrene-degrading bacterium. Appl Environ Microbiol 54:2549–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hennessee CT, Seo JS, Alvarez AM, Li QX. 2009. Polycyclic aromatic hydrocarbon-degrading species isolated from Hawaiian soils: Mycobacterium crocinum sp. nov., Mycobacterium pallens sp. nov., Mycobacterium rutilum sp. nov., Mycobacterium rufum sp. nov. and Mycobacterium aromaticivorans sp. nov. Int J Syst Evol Microbiol 59:378–387. doi: 10.1099/ijs.0.65827-0. [DOI] [PubMed] [Google Scholar]
- 11.Krivobok S, Kuony S, Meyer C, Louwagie M, Willison JC, Jouanneau Y. 2003. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: evidence for two ring-hydroxylating dioxygenases. J Bacteriol 185:3828–3841. doi: 10.1128/JB.185.13.3828-3841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molina M, Araujo R, Hodson RE. 1999. Cross-induction of pyrene and phenanthrene in a Mycobacterium sp. isolated from polycyclic aromatic hydrocarbon contaminated river sediments. Can J Microbiol 45:520–529. [PubMed] [Google Scholar]
- 13.Moody JD, Freeman JP, Fu PP, Cerniglia CE. 2004. Degradation of benzo[a]pyrene by Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol 70:340–345. doi: 10.1128/AEM.70.1.340-345.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rehmann K, Noll HP, Steinberg CE, Kettrup AA. 1998. Pyrene degradation by Mycobacterium sp. strain KR2. Chemosphere 36:2977–2992. doi: 10.1016/S0045-6535(97)10240-5. [DOI] [PubMed] [Google Scholar]
- 15.Schneider J, Grosser R, Jayasimhulu K, Xue W, Warshawsky D. 1996. Degradation of pyrene, benz[a]anthracene, and benzo[a]pyrene by Mycobacterium sp. strain RJGII-135, isolated from a former coal gasification site. Appl Environ Microbiol 62:13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez PJ, Vogel TM. 1991. Substrate interactions of benzene, toluene, and para-xylene during microbial degradation by pure cultures and mixed culture aquifer slurries. Appl Environ Microbiol 57:2981–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouchez M, Blanchet D, Vandecasteele JP. 1995. Degradation of polycyclic aromatic hydrocarbons by pure strains and by defined strain associations: inhibition phenomena and cometabolism. Appl Microbiol Biotechnol 43:156–164. doi: 10.1007/BF00170638. [DOI] [PubMed] [Google Scholar]
- 18.Dean-Ross D, Moody JD, Cerniglia CE. 2002. Utilization of mixtures of polycyclic aromatic hydrocarbons by bacteria isolated from contaminated sediment. FEMS Microbiol Ecol 41:1–7. doi: 10.1111/j.1574-6941.2002.tb00960.x. [DOI] [PubMed] [Google Scholar]
- 19.Juhasz AL, Stanley GA, Britz ML. 2002. Metabolite repression inhibits degradation of benzo[a]pyrene and dibenz[a,h]anthracene by Stenotrophomonas maltophilia VUN 10,003. J Ind Microbiol Biotechnol 28:88–96. doi: 10.1038/sj.jim.7000216. [DOI] [PubMed] [Google Scholar]
- 20.McLellan SL, Warshawsky D, Shann JR. 2002. The effect of polycyclic aromatic hydrocarbons on the degradation of benzo[a]pyrene by Mycobacterium sp. strain RJGII-135. Environ Toxicol Chem 21:253–259. doi: 10.1002/etc.5620210205. [DOI] [PubMed] [Google Scholar]
- 21.McNally DL, Mihelcic JR, Lueking DR. 1999. Biodegradation of mixtures of polycyclic aromatic hydrocarbons under aerobic and nitrate-reducing conditions. Chemosphere 38:1313–1321. doi: 10.1016/S0045-6535(98)00532-3. [DOI] [Google Scholar]
- 22.Samanta SK, Chakraborti AK, Jain RK. 1999. Degradation of phenanthrene by different bacteria: evidence for novel transformation sequences involving the formation of 1-naphthol. Appl Microbiol Biotechnol 53:98–107. doi: 10.1007/s002530051621. [DOI] [PubMed] [Google Scholar]
- 23.Tiehm A, Fritzsche C. 1995. Utilization of solubilized and crystalline mixtures of polycyclic aromatic hydrocarbons by a Mycobacterium sp. Appl Microbiol Biotechnol 42:964–968. doi: 10.1007/BF00191198. [DOI] [Google Scholar]
- 24.Zeng J, Lin X, Zhang J, Li X. 2010. Isolation of polycyclic aromatic hydrocarbons (PAHs)-degrading Mycobacterium spp. and the degradation in soil. J Hazard Mater 183:718–723. doi: 10.1016/j.jhazmat.2010.07.085. [DOI] [PubMed] [Google Scholar]
- 25.Kazunga C, Aitken MD. 2000. Products from the incomplete metabolism of pyrene by polycyclic aromatic hydrocarbon-degrading bacteria. Appl Environ Microbiol 66:1917–1922. doi: 10.1128/AEM.66.5.1917-1922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lotfabad SK, Gray MR. 2002. Kinetics of biodegradation of mixtures of polycyclic aromatic hydrocarbons. Appl Microbiol Biotechnol 60:361–366. doi: 10.1007/s00253-002-1104-7. [DOI] [PubMed] [Google Scholar]
- 27.Keum YS, Seo JS, Hu Y, Li QX. 2006. Degradation pathways of phenanthrene by Sinorhizobium sp. C4. Appl Microbiol Biotechnol 71:935–941. doi: 10.1007/s00253-005-0219-z. [DOI] [PubMed] [Google Scholar]
- 28.Mallick S, Chatterjee S, Dutta TK. 2007. A novel degradation pathway in the assimilation of phenanthrene by Staphylococcus sp. strain PN/Y via meta-cleavage of 2-hydroxy-1-naphthoic acid: formation of trans-2,3-dioxo-5-(2′-hydroxyphenyl)-pent-4-enoic acid. Microbiology 153:2104–2115. doi: 10.1099/mic.0.2006/004218-0. [DOI] [PubMed] [Google Scholar]
- 29.Moody JD, Freeman JP, Doerge DR, Cerniglia CE. 2001. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl Environ Microbiol 67:1476–1483. doi: 10.1128/AEM.67.4.1476-1483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinyakong O, Habe H, Supaka N, Pinpanichkarn P, Juntongjin K, Yoshida T, Furihata K, Nojiri H, Yamane H, Omori T. 2000. Identification of novel metabolites in the degradation of phenanthrene by Sphingomonas sp. strain P2. FEMS Microbiol Lett 191:115–121. doi: 10.1111/j.1574-6968.2000.tb09327.x. [DOI] [PubMed] [Google Scholar]
- 31.Prabhu Y, Phale PS. 2003. Biodegradation of phenanthrene by Pseudomonas sp. strain PP2: novel metabolic pathway, role of biosurfactant and cell surface hydrophobicity in hydrocarbon assimilation. Appl Microbiol Biotechnol 61:342–351. doi: 10.1007/s00253-002-1218-y. [DOI] [PubMed] [Google Scholar]
- 32.Seo JS, Keum YS, Hu Y, Lee SE, Li QX. 2006. Phenanthrene degradation in Arthrobacter sp. P1-1: initial 1,2-, 3,4- and 9,10-dioxygenation, and meta- and ortho-cleavages of naphthalene-1,2-diol after its formation from naphthalene-1,2-dicarboxylic acid and hydroxyl naphthoic acids. Chemosphere 65:2388–2394. [DOI] [PubMed] [Google Scholar]
- 33.Dean-Ross D, Cerniglia CE. 1996. Degradation of pyrene by Mycobacterium flavescens. Appl Microbiol Biotechnol 46:307–312. doi: 10.1007/s002530050822. [DOI] [PubMed] [Google Scholar]
- 34.Heitkamp MA, Freeman JP, Miller DW, Cerniglia CE. 1988. Pyrene degradation by a Mycobacterium sp.: identification of ring oxidation and ring fission products. Appl Environ Microbiol 54:2556–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang Y, Gardner DR, Miller CD, Chen D, Anderson AJ, Weimer BC, Sims RC. 2006. Study of biochemical pathways and enzymes involved in pyrene degradation by Mycobacterium sp. strain KMS. Appl Environ Microbiol 72:7821–7828. doi: 10.1128/AEM.01274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vila J, López Z, Sabaté J, Minguillon C, Solanas AM, Grifoll M. 2001. Identification of a novel metabolite in the degradation of pyrene by Mycobacterium sp. strain AP1: actions of the isolate on two- and three-ring polycyclic aromatic hydrocarbons. Appl Environ Microbiol 67:5497–5505. doi: 10.1128/AEM.67.12.5497-5505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter U, Beyer M, Klein J, Rehm HJ. 1991. Degradation of pyrene by Rhodococcus sp. UW1. Appl Microbiol Biotechnol 34:671–676. doi: 10.1007/BF00167921. [DOI] [Google Scholar]
- 38.López Z, Vila J, Ortega-Calvo JJ, Grifoll M. 2008. Simultaneous biodegradation of creosote-polycyclic aromatic hydrocarbons by a pyrene-degrading Mycobacterium. Appl Microbiol Biotechnol 78:165–172. doi: 10.1007/s00253-007-1284-2. [DOI] [PubMed] [Google Scholar]
- 39.Kim YH, Freeman JP, Moody JD, Engesser KH, Cerniglia CE. 2005. Effects of pH on the degradation of phenanthrene and pyrene by Mycobacterium vanbaalenii PYR-1. Appl Microbiol Biotechnol 67:275–285. doi: 10.1007/s00253-004-1796-y. [DOI] [PubMed] [Google Scholar]
- 40.Seo JS, Keum YS, Li QX. 2012. Mycobacterium aromativorans JS19b1(T) degrades phenanthrene through C-1,2, C-3,4 and C-9,10 dioxygenation pathways. Int Biodeterior Biodegradation 70:96–103. doi: 10.1016/j.ibiod.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stingley RL, Khan AA, Cerniglia CE. 2004. Molecular characterization of a phenanthrene degradation pathway in Mycobacterium vanbaalenii PYR-1. Biochem Biophys Res Commun 322:133–146. doi: 10.1016/j.bbrc.2004.07.089. [DOI] [PubMed] [Google Scholar]
- 42.Kim SJ, Kweon O, Freeman JP, Jones RC, Adjei MD, Jhoo JW, Edmondson RD, Cerniglia CE. 2006. Molecular cloning and expression of genes encoding a novel dioxygenase involved in low- and high-molecular-weight polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol 72:1045–1054. doi: 10.1128/AEM.72.2.1045-1054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SJ, Song J, Kweon O, Holland RD, Kim DW, Kim J, Yu LR, Cerniglia CE. 2012. Functional robustness of a polycyclic aromatic hydrocarbon metabolic network examined in a nidA aromatic ring-hydroxylating oxygenase mutant of Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol 78:3715–3723. doi: 10.1128/AEM.07798-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keum YS, Seo JS, Li QX. 2005. Synthesis of bacterial metabolites of polycyclic aromatic hydrocarbons: benzochromenones, o-carboxyvinylnaphthoates, and o-substituted aryl-α-oxobutenoates. Synth Commun 35:2685–2693. [Google Scholar]
- 45.Stanier RY, Palleroni NJ, Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol 43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 46.Holt JG. (ed). 1994. Bergey's manual of determinative bacteriology. Williams and Wilkins, Baltimore, MD. [Google Scholar]
- 47.Tsukamura M, Mizuno S, Tsukamura S. 1981. Numerical analysis of rapidly growing scotochromogenic mycobacteria, including Mycobacterium obuense sp. nov., nom. rev., Mycobacterium rhodesiae sp. nov., nom. rev., Mycobacterium aichiense sp. nov., nom. rev., Mycobacterium chubuense sp. nov., nom. rev., and Mycobacterium tokaiense sp. nov., nom. rev. Int J Syst Bacteriol 31:263–275. doi: 10.1099/00207713-31-3-263. [DOI] [Google Scholar]
- 48.Kweon O, Kim SJ, Holland RD, Chen H, Kim DW, Gao Y, Yu LR, Baek S, Baek DH, Ahn H, Cerniglia CE. 2011. Polycyclic aromatic hydrocarbon metabolic network in Mycobacterium vanbaalenii PYR-1. J Bacteriol 193:4326–4337. doi: 10.1128/JB.00215-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SJ, Kweon O, Sutherland JB, Kim HL, Jones RC, Burback BL, Graves SW, Psurny E, Cerniglia CE. 2015. Dynamic response of Mycobacterium vanbaalenii PYR-1 to BP deepwater horizon crude oil. Appl Environ Microbiol 81:4263–4276. doi: 10.1128/AEM.00730-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vila J, Grifoll M. 2009. Actions of Mycobacterium sp. strain AP1 on the saturated- and aromatic-hydrocarbon fractions of fuel oil in a marine medium. Appl Environ Microbiol 75:6232–6239. doi: 10.1128/AEM.02726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding GC, Heuer H, Zühlke S, Spiteller M, Pronk GJ, Heister K, Kögel-Knabner I, Smalla K. 2010. Soil type-dependent responses to phenanthrene as revealed by determining the diversity and abundance of polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenase genes by using a novel PCR detection system. Appl Environ Microbiol 76:4765–4771. doi: 10.1128/AEM.00047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Debruyn JM, Chewning CS, Sayler GS. 2007. Comparative quantitative prevalence of mycobacteria and functionally abundant nidA, nahAc, and nagAc dioxygenase genes in coal tar contaminated sediments. Environ Sci Technol 41:5426–5432. doi: 10.1021/es070406c. [DOI] [PubMed] [Google Scholar]
- 53.O'Leary ND, O'Connor KE, Duetz W, Dobson AD. 2001. Transcriptional regulation of styrene degradation in Pseudomonas putida CA-3. Microbiology 147:973–979. doi: 10.1099/00221287-147-4-973. [DOI] [PubMed] [Google Scholar]
- 54.Vandera E, Kavakiotis K, Kallimanis A, Kyrpides NC, Drainas C, Koukkou AI. 2012. Heterologous expression and characterization of two 1-hydroxy-2-naphthoic acid dioxygenases from Arthrobacter phenanthrenivorans. Appl Environ Microbiol 78:621–627. doi: 10.1128/AEM.07137-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sho M, Hamel C, Greer CW. 2004. Two distinct gene clusters encode pyrene degradation in Mycobacterium sp. strain S65. FEMS Microbiol Ecol 48:209–220. doi: 10.1016/j.femsec.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 56.Zhang C, Anderson AJ. 2012. Polycyclic aromatic hydrocarbon degrading gene islands in five pyrene-degrading Mycobacterium isolates from different geographic locations. Can J Microbiol 58:102–111. doi: 10.1139/w11-093. [DOI] [PubMed] [Google Scholar]
- 57.Deveryshetty J, Phale PS. 2009. Biodegradation of phenanthrene by Pseudomonas sp. strain PPD: purification and characterization of 1-hydroxy-2-naphthoic acid dioxygenase. Microbiology 155:3083–3091. doi: 10.1099/mic.0.030460-0. [DOI] [PubMed] [Google Scholar]
- 58.Khan AA, Wang RF, Cao WW, Doerge DR, Wennerstrom D, Cerniglia CE. 2001. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl Environ Microbiol 67:3577–3585. doi: 10.1128/AEM.67.8.3577-3585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim SJ, Kweon O, Jones RC, Freeman JP, Edmondson RD, Cerniglia CE. 2007. Complete and integrated pyrene degradation pathway in Mycobacterium vanbaalenii PYR-1 based on systems biology. J Bacteriol 189:464–472. doi: 10.1128/JB.01310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kweon O, Kim SJ, Freeman JP, Song J, Baek S, Cerniglia CE. 2010. Substrate specificity and structural characteristics of the novel Rieske nonheme iron aromatic ring-hydroxylating oxygenases NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1. mBio 1(2):e00135-10. doi: 10.1128/mBio.00135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stingley RL, Brezna B, Khan AA, Cerniglia CE. 2004. Novel organization of genes in a phthalate degradation operon of Mycobacterium vanbaalenii PYR-1. Microbiology 150:3749–3761. doi: 10.1099/mic.0.27263-0. [DOI] [PubMed] [Google Scholar]
- 62.Seo JS, Keum YS, Li QX. 2011. Comparative protein and metabolite profiling revealed a metabolic network in response to multiple environmental contaminants in Mycobacterium aromativorans JS19b1T. J Agric Food Chem 59:2876–2882. doi: 10.1021/jf103018s. [DOI] [PubMed] [Google Scholar]
- 63.Brezna B, Khan AA, Cerniglia CE. 2003. Molecular characterization of dioxygenases from polycyclic aromatic hydrocarbon-degrading Mycobacterium spp. FEMS Microbiol Lett 223:177–183. doi: 10.1016/S0378-1097(03)00328-8. [DOI] [PubMed] [Google Scholar]
- 64.Kweon O, Kim SJ, Kim DW, Kim JM, Kim H, Ahn Y, Sutherland JB, Cerniglia CE. 2014. Pleiotropic and epistatic behavior of a ring-hydroxylating oxygenase system in the polycyclic aromatic hydrocarbon metabolic network from Mycobacterium vanbaalenii PYR-1. J Bacteriol 196:3503–3515. doi: 10.1128/JB.01945-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kazunga C, Aitken MD, Gold A, Sangaiah R. 2001. Fluoranthene-2,3- and -1,5-diones are novel products from the bacterial transformation of fluoranthene. Environ Sci Technol 35:917–922. doi: 10.1021/es001605y. [DOI] [PubMed] [Google Scholar]
- 66.Kim YH, Engesser KH, Cerniglia CE. 2003. Two polycyclic aromatic hydrocarbon o-quinone reductases from a pyrene-degrading Mycobacterium. Arch Biochem Biophys 416:209–217. doi: 10.1016/S0003-9861(03)00297-2. [DOI] [PubMed] [Google Scholar]
- 67.Kim YH, Moody JD, Freeman JP, Brezna B, Engesser KH, Cerniglia CE. 2004. Evidence for the existence of PAH-quinone reductase and catechol-O-methyltransferase in Mycobacterium vanbaalenii PYR-1. J Ind Microbiol Biotechnol 31:507–516. doi: 10.1007/s10295-004-0178-x. [DOI] [PubMed] [Google Scholar]
- 68.Vaillancourt FH, Fortin PD, Labbé G, Drouin NM, Karim Z, Agar NY, Eltis LD. 2005. Molecular basis for the substrate selectivity of bicyclic and monocyclic extradiol dioxygenases. Biochem Biophys Res Commun 338:215–222. doi: 10.1016/j.bbrc.2005.08.219. [DOI] [PubMed] [Google Scholar]