ABSTRACT
The large legume genus Mimosa is known to be associated with both alphaproteobacterial and betaproteobacterial symbionts, depending on environment and plant taxonomy, e.g., Brazilian species are preferentially nodulated by Burkholderia, whereas those in Mexico are associated with alphaproteobacterial symbionts. Little is known, however, about the symbiotic preferences of Mimosa spp. at the southern subtropical limits of the genus. In the present study, rhizobia were isolated from field-collected nodules from Mimosa species that are native to a region in southern Uruguay. Phylogenetic analyses of sequences of the 16S rRNA, recA, and gyrB core genome and the nifH and nodA symbiosis-essential loci confirmed that all the isolates belonged to the genus Cupriavidus. However, none were in the well-described symbiotic species C. taiwanensis, but instead they were closely related to other species, such as C. necator, and to species not previously known to be symbiotic (or diazotrophic), such as C. basilensis and C. pinatubonensis. Selection of these novel Cupriavidus symbionts by Uruguayan Mimosa spp. is most likely due to their geographical separation from their Brazilian cousins and to the characteristics of the soils in which they were found.
IMPORTANCE With the aim of exploring the diversity of rhizobia associated with native Mimosa species, symbionts were isolated from root nodules on five Mimosa species that are native to a region in southern Uruguay, Sierra del Abra de Zabaleta. In contrast to data obtained in the major centers of diversification of the genus Mimosa, Brazil and Mexico, where it is mainly associated with Burkholderia and Rhizobium/Ensifer, respectively, the present study has shown that all the isolated symbiotic bacteria belonged to the genus Cupriavidus. Interestingly, none of nodules contained bacteria belonging to the well-described symbiotic species C. taiwanensis, but instead they were related to other Cupriavidus species such as C. necator and C. pinatubonensis. These data suggest the existence of a higher diversity within beta-rhizobial Cupriavidus than was previously suspected, and that Mimosa spp. from Sierra del Abra de Zabaleta, may be natural reservoirs for novel rhizobia.
INTRODUCTION
Several members of the family Leguminosae (Fabaceae) are able to establish symbiotic associations with soil bacteria known as rhizobia (1–3). A distinctive characteristic of rhizobia is their ability to elicit the formation of a symbiotic organ termed a nodule on the roots (and occasionally stems) of their legume hosts, within which the bacteria convert atmospheric nitrogen into ammonium, a process catalyzed by the nitrogenase enzyme system encoded by nif genes. In return for this fixed N, the legume host supplies the bacteria with photosynthates for respiration (2, 4). The legume-rhizobial association can be highly specific, and in many cases certain legume genera are capable of establishing a symbiosis only with specific rhizobial types and vice versa (2), although there are some notable exceptions, such as the highly promiscuous Ensifer fredii strain NGR234 (5). It was long considered that all legumes were nodulated by Rhizobium and its relatives in the Alphaproteobacteria class of the Proteobacteria, such as Ensifer (Sinorhizobium) and Bradyrhizobium (6), but over the last 15 years considerable evidence has accumulated indicating that members of the genera Burkholderia and Cupriavidus in the Betaproteobacteria class are also capable of nodulating legumes, and these have been termed “beta-rhizobia” (3). Beta-rhizobia are particularly common symbionts of members of the tribe Mimoseae in the legume subfamily Mimosoideae, and this group of symbionts appears to be entirely confined to the Americas or to invasive species that have originated from it (3, 7–9). A second center of beta-rhizobial diversity has been identified in southern South Africa, where many endemic legumes in the subfamily Papilionoideae are nodulated by Burkholderia strains that have very different nodulation genes from their South American cousins, and they cannot nodulate mimosoids (10–14).
By far the largest and most well-studied legume genus associated with beta-rhizobia is Mimosa (7, 10, 15–23). Mimosa is one of the largest genera in the Leguminosae, comprising more than 500 species ranging from small herbs to trees. Most of them (496 species) are distributed in the Americas, with central Brazil, Mexico, and subtropical South America being the principal centers of diversification (24). There are also a number of invasive Mimosa species that have originated in the Americas and then been spread pantropically, where three of them (M. diplotricha, M. pigra, and M. pudica) have become serious pests. However, most Mimosa spp. are endemic or are restricted to certain biomes, and these often consist of many closely related species that associate very particularly with certain rhizobial genotypes, such as Burkholderia in Brazil (22, 23, 25) and Rhizobium/Ensifer in central Mexico (26). Interestingly, these endemics have little or no capacity to interact with Mimosa symbionts from other centers of Mimosa diversity, suggesting coevolution with their particular microsymbionts, which is likely to be a consequence of the soil microbiota pertaining to their very specialized (mainly highland) environments. In contrast to the endemic Mimosa species, the invasive ones are capable of nodulating with a wide range of symbiont types, both alpha- and beta-rhizobial (7, 9, 16, 27, 28).
In the case of Uruguay, although it is considered to be the southern limit of distribution of many legumes, including Mimosa, it is still a major center of diversity, with 40 different Mimosa species having been reported, including several endemic species (29). Little is known about what nodulates Mimosa spp. in this southernmost limit of its extent, although an endemic species, M. uragüensis, was included in the host range study of Elliott et al. (10), in which it was shown incapable of nodulating effectively with the promiscuous Burkholderia strain, B. phymatum STM815, but capable of nodulating effectively with C. taiwanensis LMG19424. This was the opposite pattern of nodulation response by most other South American (especially Brazilian) species (10, 23) and hinted that Uruguayan mimosas might have different symbiont preferences to their more northerly cousins. Such a possibility was given additional credence by the isolation in Uruguay of M. pudica-nodulating Cupriavidus strains from nodules of Parapiptadenia rigida (30), a tree that is related to Mimosa, and is included within the Piptadenia group of mimosoid legumes (8).
The principal aim of the present study was to determine the diversity of rhizobia present in Mimosa spp. native to a geographical area in Uruguay known as “Sierra del Abra de Zabaleta,” where a wild population of five different native Mimosa species has been reported (29). This area, located near to the city of Minas, has a rich history of mining activities with considerable quantities of copper, zinc, lead, and gold being extracted from its underlying strata during the 18th and 19th centuries (31). Phylogenetic analyses were conducted on the rhizobial isolates via their core genome “housekeeping” loci, 16S rRNA, recA, and gyrB to infer their evolutionary relationship vis-á-vis other bacteria, as well as via the more mobile, symbiotic-essential loci nifH and nodA, in order to determine their evolutionary relationship with symbionts of other Mimosa populations.
MATERIALS AND METHODS
Nodule collection, bacterial isolation, and growth conditions.
Nodules from naturally occurring Mimosa species were collected in “Sierra del Abra de Zabaleta,” Lavalleja Department, Uruguay (Table 1). For symbiotic bacterial isolation, root-attached nodules were placed in sealed containers and immediately transported to the laboratory, where they were stored in the fridge until isolation (usually within a day). Isolation of bacterial strains from nodules was performed using standard techniques. Briefly, washed nodules were immersed in a solution of 10 mM HgCl2 in 0.1 N HCl for 2 min, followed by seven washes with sterile-distilled water. At least three nodules per plant were individually crushed and streaked on medium 79/yeast-mannitol agar (YMA) (32) and incubated at 30°C until colonies had formed (<5 days). Single colonies were replicated, and culture purity was checked by repeated streaking onto YMA plates. Two isolates from each Mimosa species were randomly selected for further studies. Selected isolates and original host legumes are shown in Table 1. Bacteria were stored at −80°C in liquid medium 79/yeast-mannitol broth (YMB) containing 25% (vol/vol) glycerol.
TABLE 1.
Nomenclature, isolation host, identification, and nodulation abilities of strains used in this study
| Strain | Original host | Sitea | Date of collection (mo yr) | 16S rRNA best hit (refseq_rna) | Nucleotide identity (%)b | Nodulation on different Mimosa hostsc |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| M. pudica | M. polycarpa | M. magentea | M. ramulosa | M. schleidenii | ||||||
| UYMM02A | M. magentea | 2 | April 2012 | C. numazuensis NBRC 100056 (NR_113877) | 99 (1,405/1,421) | + | + | + | ND | − |
| UYMM14A | M. magentea | 1 | April 2013 | C. necator N-1 (NR_102851) | 98 (1,375/1,408) | + | + | + | ND | + |
| UYMRA05A | M. ramulosa | 1 | September 2011 | C. numazuensis NBRC 100056 (NR_113877) | 99 (1,349/1,359) | + | + | ND | + | + |
| UYMRA10A | M. ramulosa | 1 | September 2011 | C. numazuensis NBRC 100056 (NR_113877) | 99 (1,358/1,370) | + | + | + | + | ND |
| UYMS01A | M. schleidenii | 2 | April 2012 | C. necator N-1 (NR_102851) | 99 (1,364/1,380) | + | + | + | + | + |
| UYMS13B | M. schleidenii | 2 | April 2013 | C. necator N-1 (NR_102851) | 99 (1,393/1,407) | + | + | ND | + | + |
| UYMRE01A | M. reptans | 2 | April 2012 | C. numazuensis NBRC 100056 (NR_113877) | 98 (1,380/1,403) | + | + | + | ND | ND |
| UYMRE04A | M. reptans | 2 | April 2012 | C. numazuensis NBRC 100056 (NR_113877) | 98 (1,403/1,427) | + | + | + | − | + |
| UYMAM01A | M. cruenta | 3 | February 2015 | C. necator N-1 (NR_102851) | 99 (1,359/1,366) | + | + | + | + | ND |
| UYMAM01B | M. cruenta | 3 | February 2015 | C. necator N-1 (NR_102851) | 99 (1,417/1,430) | + | + | ND | + | ND |
| LMG19424 | M. pudica | C. taiwanensis | + | + | + | – | + | |||
| STM815 | Machaerium lunatum | B. phymatum | + | – | – | – | ND | |||
Collection site numbers correspond to the sites listed in Table 2.
Fractions in parentheses represent the numbers of identical nucleotides over the total of compared nucleotides.
+, nodules; −, no nodules; ND, not determined.
Microscopy and in situ immunolocalization of beta-rhizobial symbionts.
Nodules collected in the field were fixed in 2.5% (vol/vol) glutaraldehyde in 50 mM phosphate buffer (pH 7.0) and then embedded in resin for sectioning to reveal general nodule structure as described by Chen et al. (17) and Elliott et al. (10). Immunogold labeling of the sections using antibodies against B. phymatum STM815T and C. taiwanensis LMG19424T was performed as described by dos Reis et al. (23). Mimosa pudica nodules colonized by B. phymatum STM815T and M. diplotricha nodules colonized by C. taiwanensis LMG19424T were used as positive controls.
Bacterial identification and phylogenetic analyses.
An almost complete (ca. 1,400 bp) sequence of 16S rRNA was obtained after PCR amplification and sequencing using the 27F (AGAGTTTGATCMTGGCTCAG) and 1492R (TACGGYTACCTTGTTACGACTT) primer pair (33). A 350-bp intragenic fragment of the nifH gene was PCR amplified and sequenced using the PolF (TGCGAYCCSAARGCBGACTC) and PolR (TCCGGCGAGATGATGGCGAT) primer pair (34). Alternatively, the nifH-CupF1 (CCTTTTATGGTAAAGGCGGCATCGG) and nifH-CupR1 (CCATCAGCAGATCTTCCAACTGGTCC) primer pair was used for amplification and sequencing of the almost complete (792-bp) Cupriavidus sp. nifH gene. The primer pair NodAFwBt (TGCRGTGGARDCTRYGCTGGGAAA; L. Moulin, personal communication) and NodABurkR (TCAYARCTCDGGBCCGTTBCG) (27) were used for PCR amplification and sequencing of an internal 480-bp fragment of the nodA gene. The complete sequences of the gyrB and recA genes from isolates UYMMA02A and UYMS13B were obtained from the draft genome sequence of both strains (A. Iriarte et al., unpublished data).
The obtained amplicons were sequenced by Macrogen, Inc. (South Korea). Forward and reverse sequences were assembled using DNA Baser V3 Sequence Assembler. Nucleotide sequence identities were determined using the BLAST tool of the National Center for Biotechnology Information (35).
Two different pipelines were used for phylogenetic analyses. First, DNA sequences were aligned, based on amino acid sequences when available, using CLUSTALW (36). The maximum-likelihood (ML) statistical approach was used for substitution model selection with default parameters, as implemented in MEGA5 (37). Phylogenetic trees were inferred using the ML and neighbor-joining (NJ) methods, based on the Tamura three-parameter model, both of which were implemented in MEGA5 (37). The robustness of the tree branches were estimated with 100 bootstrap replicates for ML and 1,000 bootstrap replicates for NJ. As a second approach, DNA sequences were aligned, based on amino acid sequences when available, using MUSCLE (38). A Bayesian phylogenetic analysis was then performed using MRBAYES version 3.2.4 (39). Markov chain Monte Carlo (MCMC) analyses were run for at least 1 × 106 generations and up to 5 × 106 generations, sampling trees and parameters every 100 generations. Convergence statistics were checked in order to stop the analyses (average standard deviation of split frequencies < 0.015 and potential scale reduction factor 1). The first 25% of generations were discarded as burn-in, and consensus trees were calculated. A fixed rate model of nucleotide coding sequence evolution was estimated using “mixed prior,” which allows the MCMC sampler to explore all of the fixed-rate models included in MRBAYES (40). The final data set comprised 59 sequences and 1321 positions for 16S rRNA used for Fig. 2 and Fig. S1 in the supplemental material, 58 sequences and 1,339 positions for 16S rRNA used for Fig. S6, 32 sequences and 402 positions for gyrB, 29 sequences and 668 positions for recA, 51 sequences and 222 positions for nifH, and 50 sequences and 224 positions for nodA.
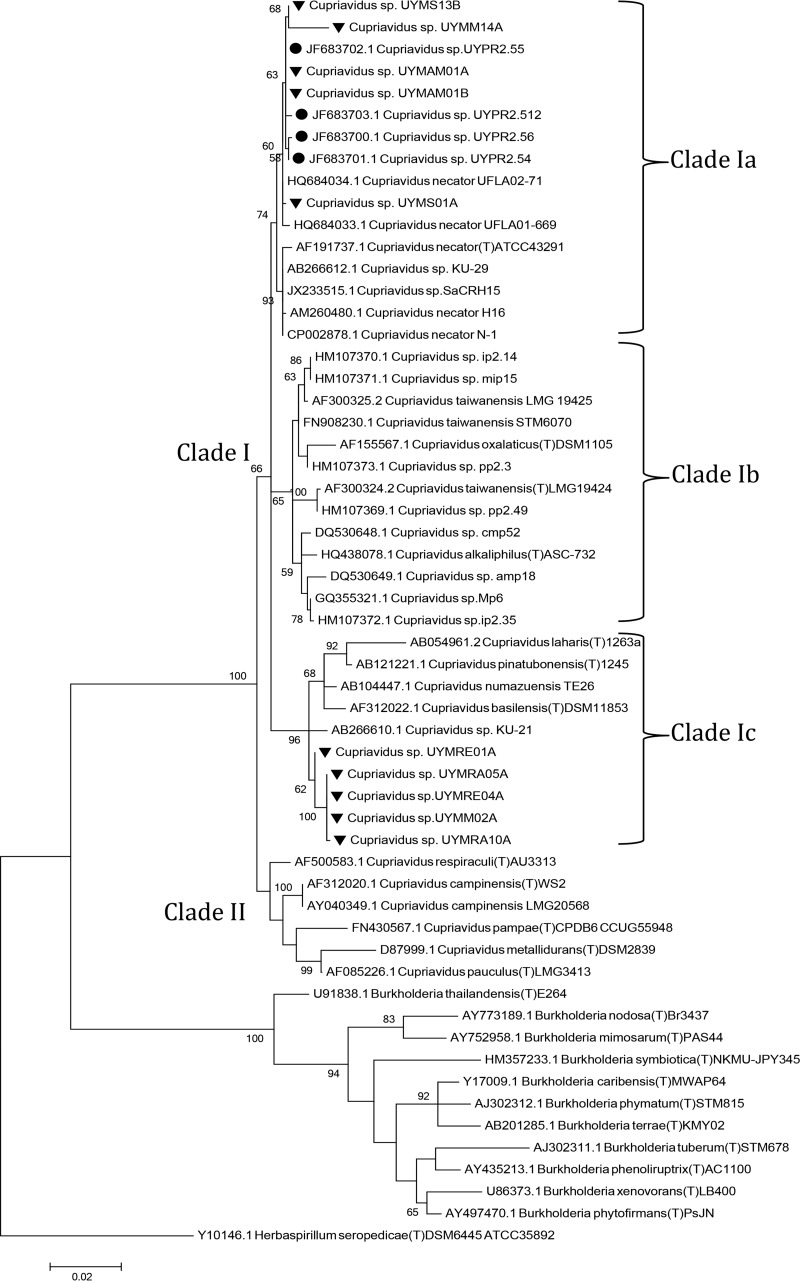
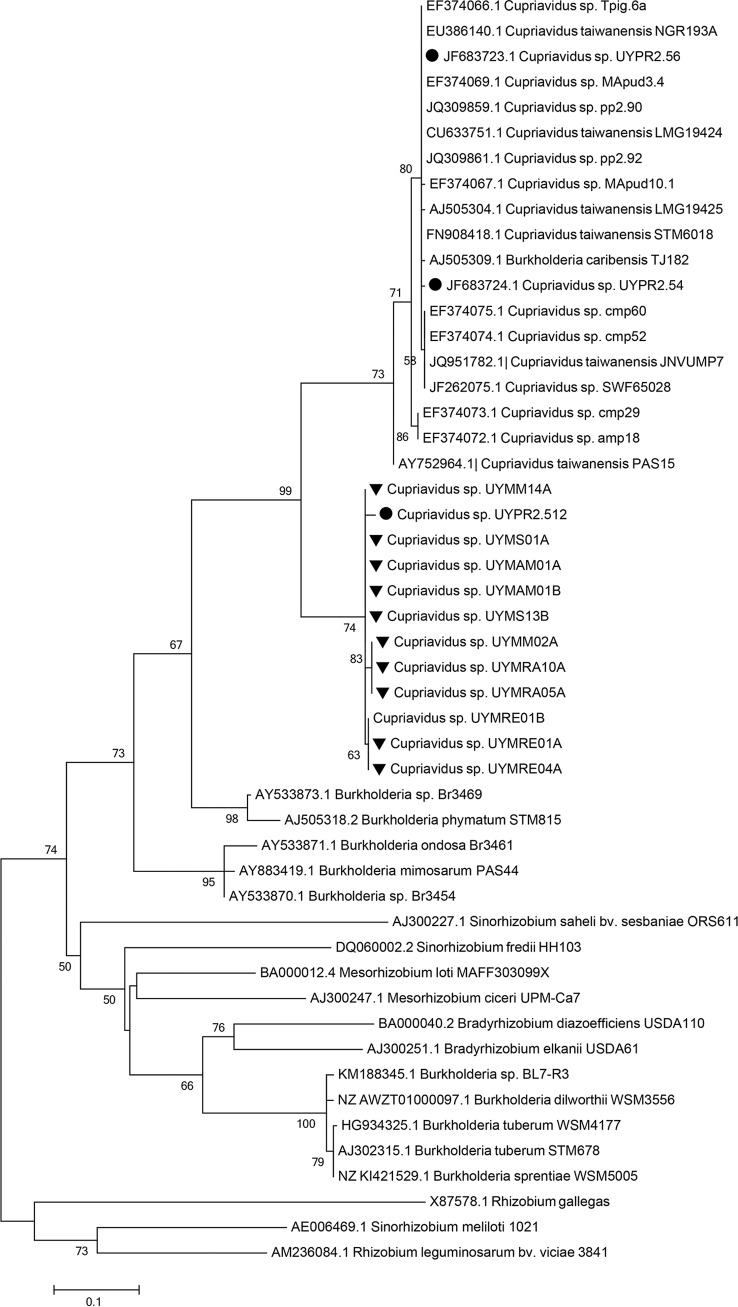
FIG 2.
16S rRNA phylogenetic tree of Mimosa symbionts isolated in this study and related betaproteobacterial species. Bootstrap support values are shown next to each node. Sequences derived from this study are marked with black triangles. Sequences derived from beta-rhizobia previously isolated in Uruguay are indicated by a black dot.
Assessment of nodulation capability.
Seeds of Mimosa spp. were scarified with concentrated sulfuric acid for 10 min and then surface sterilized with 10 mM HgCl2 in 0.1 N HCl for 2 min, followed by seven washes with sterile-distilled water. Surface-sterilized seeds were germinated on 0.8% (wt/vol) water agar, and seedlings were transferred into glass tubes containing 15 ml of Jensen's N-free medium plus 0.8% (wt/vol) agar (32). Surface-sterilized seeds were checked for the absence of contaminant bacteria by incubating two of them in TY plates at 30°C for 3 days (32). Seedlings were inoculated with 1 ml of a bacterial suspension containing ca. 107 CFU. Then, 1 ml of sterile water was added to the negative controls. Plants were grown at 26°C with a photoperiod of 16 h of light/8 h of dark. The plants were checked periodically for the presence of root nodules, and the plants were harvested 3 months after inoculation, at which time the rhizobia in the root nodules were isolated and identified by comparisons of their 16S rRNA sequences with the original strain used for their inoculation.
Rhizobial trap assays using native soil.
Equal amounts of soils from the three collection sites (Table 1) were mixed and used for nodulation of Mimosa cruenta, M. ramulosa, M. magentea, M. polycarpa, and M. pudica. Surface-sterilized seeds were germinated, transferred to 1-liter pots with soil, maintained in a greenhouse for 60 days, and watered as needed. Ten pots were used for each Mimosa species. Nodules were collected from five plants of each Mimosa species at harvest. These were surface sterilized, and rhizobial symbionts were isolated and identified by 16S rRNA sequencing as described earlier.
Physicochemical analyses of soil samples.
Soil samples were collected from the three collection sites (Table 1). Each sample was composed of five soil subsamples (1 kg each) taken from the rhizosphere of Mimosa spp. and mixed prior to physicochemical analyses in the Laboratory for Soil Analysis in INIA-La Estanzuela (http://www.inia.uy/en/products-and-services/laboratories/Laboratorio-de-Suelos-y-Agua). Soil pH levels were determined in water (41). The total organic carbon was quantified by combustion at 900°C and infrared detection of CO2 produced with LECO Truspec CN-2000 equipment (42). The ammonium content was estimated by KCl extraction and colorimetric detection (43). Nitrates were extracted with CuSO4 and measured by potentiometry with an ORION specific activity electrode (44). Phosphate content was extracted by the Bray I method (45) and detected by colorimetric measurements (46). The concentrations of water-soluble calcium, magnesium, potassium, and sodium ions in soils were determined by displacement with NH4+ and atomic emission spectrometry (47). Bioavailable metals were determined by inductively coupled plasma according to the Lindsay and Norvell diethylene triamine penta-acetic acid (DTPA) extraction method (48). Total metal content in soils was determined by using an Agilent 7500 inductively coupled plasma–optical emission spectrometer (ICP-OES; Agilent Technologies, Tokyo, Japan). Prior to the analysis, soil samples (0.8 g) were acid digested with 5 ml of concentrated HNO3 solution (70% wt/%) and 1 ml of concentrated HCl solution (34 to 37 wt%), using a MARS 6 microwave reaction system (CEM Corporation, USA). The optimized operating conditions for ICP-MS are listed in Table S2 in the supplemental material. A certified multi-element stock solution (Perkin-Elmer Pure Plus standard, product number 9300233) was used to prepare standard solutions.
Metal tolerance of isolated strains.
Tolerance of bacterial strains to metals was assessed using YMA plates containing different metal salts. Colony formation was monitored in solid medium containing 0.1, 0.5, or 2 mM CoCl2, NiCl2, or Cd(CH3COO) and 0.5, 2, or 5 mM CuSO4 or ZnSO4. Cultures were incubated 5 days at 30°C. The MIC was defined as the lowest concentration of metal salts at which no CFU was observed. Reference strains were the highly resistant bacterium Cupriavidus metallidurans CH34 (49, 50), the type strains C. taiwanensis LMG19424T (15) and B. phymatum STM815T, the P. rigida symbionts C. necator UYPR2.512 and Burkholderia sp. UY1.413 (30), and the metal-sensitive strain Escherichia coli S17-1 (51).
Nucleotide sequence accession numbers.
The sequences determined in this study were deposited in the GenBank database under the following accession numbers: KP733698 to KP733705, KP901100, KP901101, and KU248139 to KU248158 for 16S rRNA; KP733706 to KP733713, KP901096, and KP901097 for nifH; KP762489 to KP762494, KP762496, KP901098, KP901099, and KT198752 for nodA; KT198755 and KT198756 for recA; and KT198753 and KT198754 for gyrB.
RESULTS
Nodulation of native Mimosa species sampled in Lavalleja, Uruguay.
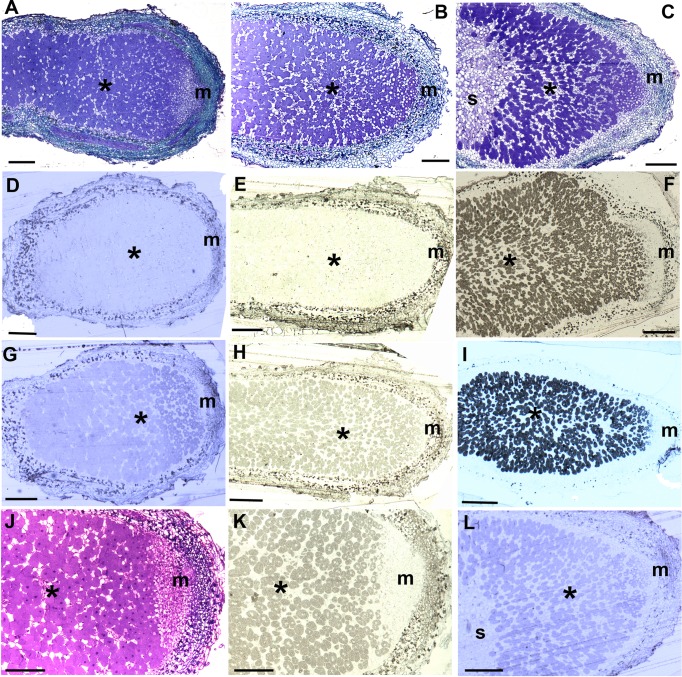
Five Mimosa species; M. cruenta (syn. M. amphigena), M. magentea, M. ramulosa, M. reptans, and M. schleidenii, naturally occurring in the hilly zone of “Sierra del Abra de Zabaleta” were examined for nodulation. Nodules were observed on most of the M. magentea, M. ramulosa, M. reptans, and M. schleidenii and on a few of the M. cruenta plants surveyed. The nodules were typically elongated and indeterminate in shape, with the characteristic pink internal coloration due to the presence of the symbiosis-essential protein leghemoglobin. Sections for light microscopy confirmed that the nodules were typical of effective Mimosa nodules (Fig. 1A, B, and C) and were similar to those reported on other species (10, 18, 19, 23, 26, 52). In order to assess the presence of Burkholderia or Cupriavidus symbionts in wild nodules, an in situ immunochemistry assay was performed with antibodies raised against the promiscuous Mimosa symbionts B. phymatum STM815T and C. taiwanensis LMG19424T (10, 19). None of the analyzed nodules reacted with the B. phymatum antibody (Fig. 1D and E), except for the positive control: M. pudica nodules containing B. phymatum (Fig. 1F). A faint positive reaction was observed for M. magentea (Fig. 1G) and M. ramulosa (Fig. 1H) treated with the C. taiwanensis LMG19424 antibody, although much weaker than the positive control: M. diplotricha nodules containing C. taiwanensis (Fig. 1I). Compared to the toluidine blue-stained serial sections, the immunogold signal could be seen to coincide with the infected tissue containing bacteroids and not with the meristem nor with the senescent tissue(s) at the nodule base (Fig. 1C, K, and L). These results suggest that the nodule symbionts were more closely related to Cupriavidus than to Burkholderia.
FIG 1.
Light microscopy sections of field-collected nodules from native Uruguayan Mimosa species that have been stained either with toluidine blue for structural analysis or have been immunogold labeled with antibodies against Burkholderia phymatum STM815T or Cupriavidus taiwanensis LMG19424T. The infected tissue with bacteroids is marked with an asterisk (*) in each section, and the meristem with an “m.” (A to C) Mimosa magentea (A), M. ramulosa (B), and M. schleidenii (C) nodules showing the structure typical of effective Mimosa nodules. (D to F) M. magentea (D), M. ramulosa (E), and M. pudica (F) nodules labeled with an antibody against B. phymatum. Note that none of the sections gave a positive reaction, except for the positive control M. pudica nodules containing B. phymatum (F). (G to I) M. magentea (G), M. ramulosa (H), and M. diplotricha (I) nodules labeled with an antibody against C. taiwanensis. Note that both native species react with this antibody but that the reaction is much weaker than that of the positive control M. diplotricha nodules containing C. taiwanensis (I). (J and K) Comparison of the meristem and infected zones of M. ramulosa nodules either stained with toluidine blue (J) or immunogold labeled with the C. taiwanensis antibody (K). Note that the immunogold signal coincides with the infected tissue containing bacteroids and not with the meristem. (L) Similarly, with M. schleidenii, the immunogold signal with this antibody coincides with the infected tissue and not with either the meristem or the senescent tissue (s) at the base, e.g., compare this to the toluidine blue-stained serial section in panel (C). All of the scale bars represent 200 μm, except for those in panels J and K, which represent 100 μm.
Symbionts of Uruguayan Mimosa species belong to Cupriavidus and are phylogenetically diverse.
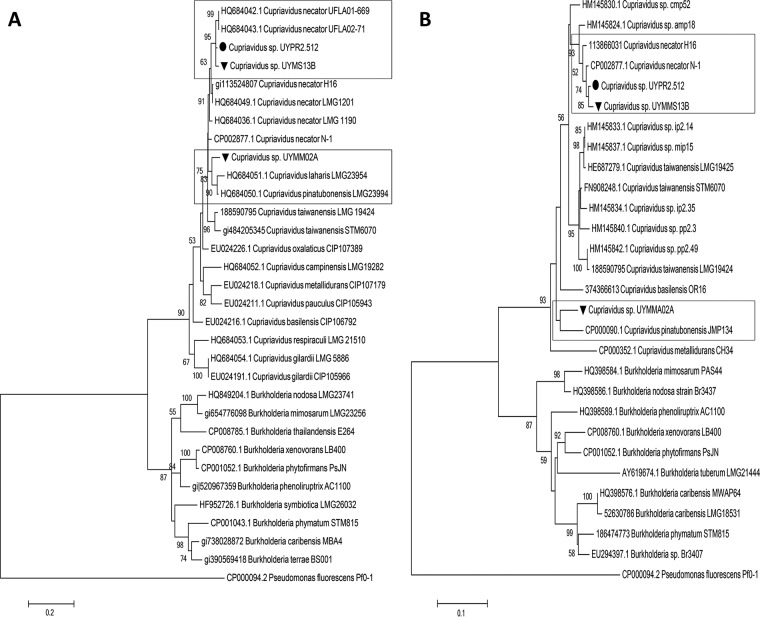
Two isolates per Mimosa species, obtained from different nodules, were randomly selected and an almost full-length 16S rRNA was sequenced from them. All the sequences displayed 98 to 99% sequence identity to Cupriavidus species (Table 1). The phylogeny of their 16S rRNA sequences was analyzed and compared to reference strains of Cupriavidus spp., including type strains, but also with various beta-rhizobia strains not yet assigned to species that were isolated from Uruguay or other countries (7, 8, 15, 18, 20–22, 28, 30, 53–57). The ML analysis shows that all the Cupriavidus were clustered in two main clades, clade I and clade II, and supported by a bootstrap value of 100 (Fig. 2). All of the symbiotic Cupriavidus strains described so far, as well as the isolates from the present study fall within clade I, in which three distinct subclades were also observed. Clade Ia comprises sequences from isolates UYMM14A, UYMS13B, UYMAM01A, UYMAM01B, and UYMS01A from the present study, two nodulating C. necator strains isolated from trap plants nodulated in pasture soils from Minas Gerais state in central Brazil (UFLA02-71 and UFLA01-669) (56), and strains (UYP2.512, UYPR2.54, and UYPR2.56) isolated from P. rigida nodules in Uruguay (30). Clade Ib comprises sequences of type strains of both the symbiotic species C. taiwanensis (15), as well as various unclassified Cupriavidus symbionts isolated from diverse Mimosa spp. (55) and the nonsymbiotic species C. oxalaticus DSM1106T (50) and C. alkaliphilus ASC-732T (isolated from the rhizosphere of nonlegume agricultural plants growing in alkaline soils in Mexico [58]). Finally, clade Ic, grouped sequences from isolates UYMRE01A, UYMRE04A, UYMRA05A, UYMRA10A, and UYMM02A from the present study, as well as sequences from other nonsymbiotic Cupriavidus species, such as C. basilensis SM11853T, isolated from fresh water sediment and able to degrade 2,6-dichlorophenol (59); C. numazuensis TE26T, a soil bacterium able to degrade trichloroethylene (60); and C. laharis 1263aT and C. pinatubonensis 1245T, both hydrogen-oxidizing bacteria isolated from mudflows on Mount Pinatubo, an active volcano in the Philippines (61). On the other hand, no symbiotic Cupriavidus were grouped in clade II, which comprised species isolated from diverse environments, including the heavy metal-resistant C. metallidurans CH34 and the C. campinensis WS2 strains isolated from industrial biotopes (59), the herbicide-degrading species C. pampae isolated from agricultural soils (62), and the C. respiraculi (63) and C. pauculus (64) type strains, which were both isolated from human clinical patients. One strain from clade Ia, UYMS13B, and one strain from clade Ic, UYMM02A, were selected for additional analyses via sequences of their gyrB and recA housekeeping genes. Maximum-likelihood (ML) phylogenetic analysis of these genes gave essentially the same distribution pattern as was observed with the 16S rRNA sequences, i.e., strain UYMS13B grouped with C. necator, whereas strain UYMM02A clustered with C. laharis and C. pinatubonensis for both genes (Fig. 3).
FIG 3.
(A and B) gyrB (A) and recA (B) phylogenetic tree of selected Mimosa symbionts and related betaproteobacterial species. Bootstrap support values are shown next to each node. Sequences derived from beta-rhizobia previously isolated in Uruguay are indicated by a black dot.
Bayesian analyses of 16S rRNA sequences failed to resolve the basal nodes of the Cupriavidus lineage with significant statistical support (posterior probability > 0.5), and are shown as a basal polytomy (see Fig. S1 in the supplemental material). Although clade Ic and Ib appear as monophyletic groups, clade II and clade Ia are distributed as basal lineages, e.g., the isolates from clade Ia reported in the present study appear to be close to some C. necator strains but also to clade Ic. However, Bayesian analysis of the recA and gyrB sequences supported the observed branching pattern obtained from the ML analysis of the housekeeping gene and 16S rRNA sequences (see Fig. S2 and S3 in the supplemental material), thereby lending strong and statistically significant support to the existence of three main groups of symbiotic strains: clades Ia, Ib, and Ic.
Phylogenetic analyses of symbiosis-related genes.
Phylogenetic analyses were conducted using partial sequences of two representative genes, nifH and nodA. The ML (Fig. 4) and Bayesian (see Fig. S4 in the supplemental material) methods both indicated that the nifH sequences from the Cupriavidus isolates clustered with P. rigida-nodulating Cupriavidus strains and with Mimosa-nodulating C. necator strains isolated in Brazil (30, 56). Consistent with their 16S rRNA sequences, the nifH sequences clustered in a monophyletic group with highly significant statistical support, as a sister branch of the clade that includes the C. taiwanensis strains LMG19424T and LMG19425, which suggests that there has been some degree of divergence in the nifH genes of these beta-rhizobial Cupriavidus strains (Fig. 4). A similar pattern was observed with the nodA genes (Fig. 5 and see Fig. S5 in the supplemental material), i.e., the nodA sequences from the Uruguayan Mimosa spp. grouped together in a cluster separate from the one formed by symbiotic C. taiwanensis strains mainly isolated from M. pigra, M. diplotricha, and M. pudica in Central America and Southeast Asia (9, 16, 28, 53, 55, 57, 65).
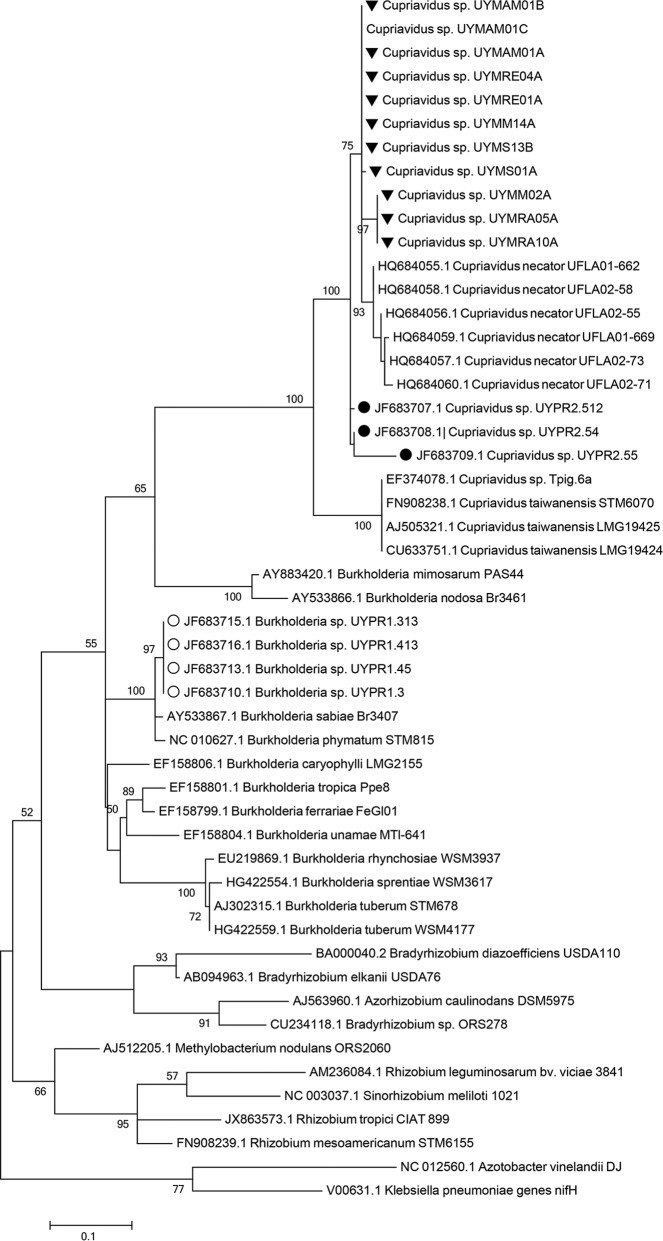
FIG 4.
nifH phylogenetic tree of Mimosa symbionts isolated in this study and related species. The numbers of interior nodes represent bootstrap support values. Sequences derived from this study are marked with black triangles. Sequences derived from Cupriavidus and Burkholderia symbionts previously isolated from Parapiptadenia rigida in Uruguay are indicated by black and white dots, respectively.
FIG 5.
nodA phylogenetic tree of Mimosa symbionts isolated in this study and related species. The numbers of interior nodes represent bootstrap support values. Sequences derived from this study are marked with black triangles. Sequences derived from beta-rhizobia previously isolated in Uruguay are indicated by a black dot.
Uruguayan Cupriavidus isolates can effectively nodulate native and non-native Mimosa species.
In order to evaluate the host specificity of the Cupriavidus isolates, the ability of a selection of them to nodulate various Mimosa spp. was tested under gnotobiotic conditions; these comprised the pantropical species M. pudica, the common South American species M. polycarpa, and the Uruguayan native species M. schleidenii, M. magentea, and M. ramulosa. After 3 months, control plants that were mock-inoculated with sterile water were not nodulated, thus demonstrating that the surface-sterilized seeds did not carry any symbiotic bacteria. Conversely, all the strains tested were able to form “pink” nodules on both M. pudica and M. polycarpa, supporting their rhizobial nature. Furthermore, M. schleidenii, M. magentea and M. ramulosa strains were all able to effectively nodulate their original hosts (Table 1). On the other hand, the M. magentea strain UYMM02A failed to nodulate M. schleidenii, but another M. magentea strain, UYMM14A, induced effective nodules on this host, while the M. reptans strain UYMRE04A nodulated M. magentea and M. schleidenii but failed to nodulate M. ramulosa, which suggests some variations in specificity even among strains from the same Mimosa species. Interestingly, of the two reference Mimosa strains, C. taiwanensis LMG19424T effectively nodulated M. polycarpa, M. magentea, and M. schleidenii, but B. phymatum STM815T failed to nodulate any of them, suggesting that in addition to strain specificity, the Mimosa species included in this study are selective in terms of their preferred symbionts (Table 1).
Mimosa spp. established in Sierra del Abra de Zabaleta soils select Cupriavidus as symbionts.
Rhizobial trap assays were performed in order to evaluate the presence of indigenous rhizobia in soils from Sierra del Abra de Zabaleta able to nodulate different Mimosa species, including both native and widespread species. After 60 days, all Mimosa roots were nodulated indicating that the soils contained active rhizobial populations. Four nodules from each Mimosa species, M. pudica, M. magentea, M. cruenta, M. polycarpa, and M. ramulosa, were randomly selected, and symbionts were isolated from them. As with nodules collected from natural populations, all of the native Mimosa nodules yielded rhizobia that were identified as Cupriavidus by mean of their 16S rRNA sequence analyses (see Table S1 in the supplemental material). Interestingly, all of the symbionts isolated from nodules on the widespread species M. polycarpa and M. pudica were also identified as Cupriavidus. Phylogenetic analyses were conducted in order to compare the sequences of the trap plant isolates with those isolated directly from nodules collected in the field (see Fig. S6 in the supplemental material). As observed for the field-collected isolates, sequences from all the trap plant isolates were grouped within the two previously observed subclades Ia and Ic (Fig. 2; see also Fig. S6 in the supplemental material). In addition, all the sequences of the trap plant isolates were separated from the group containing the symbiotic type strain C. taiwanensis LMG19424, suggesting that none of them belong to this species (see Fig. S6 in the supplemental material).
Characteristics of soils from Sierra del Abra de Zabaleta.
Soil samples were taken from the vicinity of the three collection sites where natural populations of the Mimosa spp. analyzed in the present study occur (Table 2). All three sampled soils were slightly acid (pH 5.8) and contained 2.2 to 5.0% organic matter. The measured ammonia and nitrate concentrations were in the range of 3.1 to 8.1 ppm, while the Bray method estimated that plant available phosphorous concentrations fluctuated between 3.9 to 54.6 ppm, which corresponds to the typical superficial and rocky soils of the “Sierra de las Ánimas” (http://web.renare.gub.uy/sl/coneat). The total metal ion concentrations were determined for Fe, Cu, Zn, Cd, Co, and Ni (Table 2). The concentrations of these metals were within the reported range for this area (66) and reflect the underlying strata composition defined as alkaline igneous rock (31) (http://www.mindat.org/loc-259875.html).
TABLE 2.
Physicochemical characteristics of Mimosa rhizosphere soils sampled from Sierra del Abra de Zabaleta, Uruguay
| Site | GPS localization | Altitude (m) | Soil characteristics |
Total metal ions (μg g−1) |
Bioavailable metal ions (μg g−1)d |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pHa | OMb | NO3 (μg g−1) | NH4 (μg g−1) | Pc | K (meq/100 g) | Fe | Cu | Zn | Cd | Co | Ni | Fe | Cu | Zn | Mn | |||
| S1 | S34°31′34.3″, W55°19′ 0.6″ | 142 | 5.8 | 2.2 | 3.2 | 4.4 | 3.9 | 0.32 | 23094 | 10.95 | 47.8 | 0.0294 | 10.7 | 27.7 | 113.5 | 1.00 | 0.66 | 25.8 |
| S2 | S34°30′47.2″, W55°21′16.5″ | 111 | 5.8 | 5.0 | 4 | 8.1 | 5.7 | 0.52 | 24796 | 7.2 | 42.7 | 0.0404 | 6.67 | 8.9 | 86.00 | 5.26 | 2.35 | 31.2 |
| S3 | S34°31′33.1″, W55°19′11.7″ | 163 | 5.8 | 2.6 | 3.1 | 4.9 | 54.6 | 0.44 | 136.1 | 1.46 | 1.09 | 51.4 | ||||||
pH, soil pH in water.
OM, organic matter (OM% = % total organic content × 1.72).
P, phosphates Bray-1 (μg P g−1).
Metal extracted with diethylene triamine pentaacetic acid (DPTA).
Heavy metal tolerance of Uruguayan Mimosa symbionts.
The considerable amounts of metal measured in the sampled soils prompted an analysis of the tolerance of selected bacterial isolates toward Zn2+, Cd2+, Co2+, Ni2+, and Cu2+ (Table 3). Cupriavidus metallidurans CH34 was included as a control for metal-tolerant bacterium (49), whereas E. coli S17-1 (51) was used as a control for a metal-sensitive strain. In general, symbiotic isolates from Uruguayan Mimosa spp. were consistently more tolerant to metal than E. coli S17-1 but usually more sensitive than the heavy-metal-tolerant C. metallidurans strain CH34. However, the symbiotic Cupriavidus demonstrated tolerances to Zn2+ and Cd2+ comparable to that of C. metallidurans and at least 10-fold higher than the Burkholderia strains tested (UYPR1.413 and STM815T). Tolerance to Cu2+, Co2+, and Ni2+ were variable among the tested strains and no correlation could be established between sampling site, bacterial phylogeny, and metal resistance profile.
TABLE 3.
Metal tolerance profiles of Mimosa-nodulating Cupriavidus strains from Sierra del Abra de Zabaleta, Uruguay
| Straina | MIC (mM) |
||||
|---|---|---|---|---|---|
| ZnSO4 | Cd(CH3COO)2 | CoCl2 | NiCl2 | CuSO4 | |
| UYMM02A | >5 | >2 | 0.5 | 2 | 5 |
| UYMRA10A | >5 | 2 | 0.5 | 2 | 2 |
| UYMRA05A | 5 | 2 | 0.5 | 2 | 2 |
| UYMRE04A | >5 | >2 | 2 | 0.5 | 5 |
| UYMRE01A | >5 | >2 | 2 | 0.5 | 5 |
| UYMAM01A | >5 | >2 | 0.5 | 0.5 | 2 |
| UYMAM01B | >5 | >2 | 0.5 | 0.5 | 2 |
| UYMS13B | >5 | 2 | 0.5 | 0.5 | 2 |
| UYMM14A | >5 | >2 | 0.5 | 0.5 | 5 |
| UYMS01A | >5 | >2 | 2 | 0.5 | 5 |
| UYPR2.512 | >5 | 2 | 2 | 2 | 2 |
| LMG19424T | >5 | >2 | 2 | 0.5 | 2 |
| UYPR1.413 | 0.5 | 0.1 | 0.5 | 0.5 | 5 |
| STM815 T | 0.5 | 0.1 | 0.5 | 0.5 | 5 |
| CH34 | >5 | >2 | >2 | >2 | >5 |
| S17-1 | 0.5 | 0.1 | 0.5 | 0.5 | 0.5 |
Strains isolated in the present study are highlighted in boldface. UYPR2.512, Cupriavidus necator; LMG19424T, C. taiwanensis; UYPR1.413, Burkholderia sp.; STM815 T, B. phymatum; CH34, C. metallidurans; S17-1, Escherichia coli.
DISCUSSION
Native Uruguayan Mimosa species nodulate with Cupriavidus.
The Uruguayan native species M. cruenta, M. magentea, M. reptans, M. ramulosa, and M. schleidenii were all found to be effectively nodulated in the field in the Sierra del Abra de Zabaleta, and authenticated rhizobial strains were isolated from them. This agrees with previous observations indicating that nodulation is a generic characteristic of the genus Mimosa (9, 16, 53, 55, 65, 67) and, together with the studies of dos Reis et al. (23), Gehlot et al. (28), Lammel et al. (25), and Bontemps et al. (26), has raised the total number of Mimosa species reported to nodulate to >120. An initial screening in which sections of the field-collected nodules were probed with antibodies against the Mimosa symbionts, B. phymatum STM815T and C. taiwanensis LMG19424T, was performed. This method has been used successfully in previous studies in Brazil, India, and Mexico to identify symbionts in situ and has correlated very well with data obtained from isolation and molecular identification of the symbionts (22, 23, 26, 28). Interestingly, none of the nodules from the present study reacted with the B. phymatum antibody, which is in complete contrast to studies on central Brazilian Mimosa spp., in which almost all of the 67 species examined reacted with this antibody (23), and almost all of the isolated symbionts were Burkholderia spp. (22).
The Uruguayan nodules did, however, react with the C. taiwanensis antibody, but somewhat faintly compared to positive-control M. diplotricha nodules infected with C. taiwanensis LMG19424T, which suggested that they might contain Cupriavidus but not C. taiwanensis. Indeed, this proved to be the case after molecular identification of the isolated symbionts indicated that the bacteria isolated from the nodules of the five Mimosa spp. belonged to the genus Cupriavidus, but none belonged to C. taiwanensis. The presence of rhizobia from the genus Cupriavidus has been previously reported in Uruguay where they, as well as Burkholderia, were found to be nodulating the mimosoid tree P. rigida (30). Interestingly, however, although the P. rigida-nodulating Cupriavidus strains were also capable of nodulating Mimosa effectively, it should be noted that they were sampled in northwest Uruguay, which is 400 km from sites sampled in the present study (southeast Uruguay).
Soil factors and plant taxonomy may be involved in the preference of Uruguayan Mimosa spp. for Cupriavidus.
In consideration of their thus-far-reported worldwide distribution, the apparently common occurrence of Mimosa-nodulating Cupriavidus symbionts in Uruguay is intriguing. Ralstonia taiwanensis was first isolated from nodules on invasive M. pudica and M. diplotricha in Taiwan by Chen et al. (15) and was subsequently shown to be genuinely symbiotic on these species by Chen et al. (17). Since then it, along with several other Ralstonia/Wautersia species, has been renamed, and it is now known as C. taiwanensis (50). Cupriavidus taiwanensis has been isolated from Mimosa nodules in many different parts of the world, especially from the invasive species, M. pudica and M. diplotricha, in various countries in Southeast Asia (15, 27, 28, 53, 55, 65, 68). In contrast to these invasive environments, although it has been isolated in the native range of Mimosa, i.e., Central America (20, 21), the southern United States (54), and French Guiana (7), this has generally been in low proportions compared to Burkholderia symbionts.
Cupriavidus taiwanensis is considered to have most likely originated from the Americas and to have been transported to Southeast Asia, along with its invasive hosts (55). Interestingly, however, in spite of their association with Mimosa spp. and their neotropical origin, Cupriavidus strains have not been isolated from nodules of naturally occurring Mimosa within the main centers of radiation of the genus, the Caatinga and Cerrado biomes in central Brazil (22, 23), or from Mimosa spp. sampled in southern Brazil (25); Burkholderia spp. were overwhelmingly the dominant symbionts in all these Brazilian studies. Similarly, in Mexico, which is the second largest center of radiation for the genus, no Cupriavidus strains were associated with several native and endemic Mimosa spp. (26). The apparent absence of symbiotic Cupriavidus within the centers of Mimosa radiation has been suggested to be associated with soil characteristics, e.g., in studies that have reported soil pH, C. taiwanensis is invariably found to be nodulating both native and invasive M. pudica in soils that are neutral-alkaline, whereas Burkholderia symbionts predominate in the more acidic soils (7, 23, 25, 28, 65). This might be expected, since Burkholderia is an acid-tolerating genus (11, 69), and many species of Cupriavidus are alkaline tolerant, e.g., C. alkaliphilus (58). Higher soil fertility (N-content) may also favor Cupriavidus; in competition studies for nodulation of invasive Mimosa spp., Burkholderia strains (B. mimosarum PAS44T and B. phymatum STM815T) overwhelmingly outcompeted C. taiwanensis LMG19424T until the concentration of N (ammonium or nitrate) was increased above minimal levels (16). In this context, it may therefore appear surprising that the pH of the soils from Sierra del Abra de Zabaleta in which the five Uruguayan native Mimosa spp. were found to be nodulated exclusively by Cupriavidus are slightly acid, with a pH close to 6.0. In addition, the soil organic matter was calculated to be close to 5%, which is not high, whereas the nitrogen, phosphate, and potassium contents were indicative of medium-fertility soils with nutrient concentrations below the reported ones favoring Cupriavidus nodulation of Mimosa spp. in competition assays (16).
In order to further investigate the role of these particular soils in symbiont selection, rhizobial trap assays were undertaken with seedlings of various native and widespread Mimosa spp. In agreement with the field observations, all the isolates from both the native Uruguayan and the widespread species were identified as Cupriavidus. These data, especially those concerning M. pudica, which is known to have a strong preference for Burkholderia as a symbiont in acidic soils (3, 7, 9, 16, 28, 65), suggest that the Sierra del Abra de Zabaleta soils, in spite of their slight acidity, favor the persistence and/or symbiotic competence of Cupriavidus over other rhizobial types that might be capable of nodulating Mimosa.
While the soil data suggest that pH and fertility are not the main factors involved in the selection by Uruguayan Mimosa spp. of Cupriavidus as symbionts, other factors must be involved. One possibility is soil heavy metal content, since the copper-loving genus Cupriavidus is known for its tolerance to heavy metals, such as Cu, Zn, Pb, Cd, and Ni (50), and this includes some symbiotic Cupriavidus strains (28, 79, 80). Indeed, the very high heavy metal concentration in the soils in New Caledonia (NC) has been considered one of the factors responsible for the extended nodulation of the invasive M. pudica by C. taiwanensis, with many of the isolated strains being highly tolerant to Ni, Zn, and Cr (27). The soils in the present study originated from metal-rich underlying strata and contain considerable concentrations of Zn, Cu, Co, Ni, and Fe (58; the present study). Accordingly, DPTA-extracted Zn concentrations in soils of Sierra del Abra de Zabaleta were similar to or higher than those reported for the NC soils (27). It is therefore reasonable to suggest that metal contents are also likely to play a role in the high incidence of Cupriavidus in the soils from Sierra del Abra de Zabaleta. In agreement with this hypothesis, metal resistance profiles indicated that although the tested strains were more sensitive than the model bacterium C. metallidurans CH34 toward some metals, 10-fold-higher Zn (and Cd) concentrations were needed to inhibit the growth of the Uruguayan Mimosa Cupriavidus symbionts than the symbiotic Burkholderia strains tested. As seen in the genomes of other Cupriavidus spp. (70–77), several genes encoding proteins implicated in heavy-metal resistance were also detected in the draft genomes of two Cupriavidus strains from the present study, UYMMA02A and UYMS13B (Iriarte et al., unpublished). These included homologs to three main efflux systems involved in metal detoxification, such as P-type ATPases, as well as CBA transporters and cation diffusion facilitator family transporters (78), indicating that these novel Cupriavidus strains are well adapted to deal with heavy metals. Taken together, these data suggest that metal content may have favored Cupriavidus over Burkholderia in these particular soils.
A final major factor in the selection of Cupriavidus symbionts by Uruguayan Mimosa spp. is the host taxonomy, which is known to strongly affect the selection by native and endemic Mimosa spp. of particular symbionts (22, 23, 26). This is suggested to also be of importance in the present study and by the preference of the Uruguayan species for Cupriavidus over Burkholderia strains in cross-inoculation studies (10). This then leads to the conclusion that the studied Uruguayan native Mimosa may have selected local symbiotic Cupriavidus species, resulting in highly specific associations between them, a pattern which has already been observed in other centers of radiation of the genus Mimosa, but with Burkholderia and Rhizobium/Ensifer symbionts in Brazil (22, 23, 25) and Mexico (26, 68, 81), respectively. Further experiments, such as rhizobial trap assays employing different soils and Mimosa species endemic to Uruguay, Brazil and Mexico, are needed to determine which factor(s) contribute to the selection of Cupriavidus symbionts by Mimosa.
Cupriavidus symbionts of Uruguayan Mimosa spp. do not belong to C. taiwanensis.
One of the most striking features of the present study is the fact that none of the Cupriavidus symbionts isolated, both from native Mimosa populations and trap plants assays, belonged to the well-known symbiotic species C. taiwanensis (Fig. 2 and see Fig. S6 in the supplemental material). Instead, one group was more closely related to C. necator (clade Ia, Fig. 2), and the second one to Cupriavidus species with no previously described symbiotic strains (clade Ic, Fig. 2). In the case of C. necator, symbiotic strains were isolated from nodules of Phaseolus vulgaris and Leucaena leucocephala grown as “trap plants” in pasture soils from the mining state Minas Gerais in Brazil; although the original host is unknown, these strains are capable of nodulating Mimosa spp. effectively (56), and the pastures from which they originated housed M. pudica (E. K. James and S. M. de Faria, unpublished data), so it is reasonable to suggest that these may be the original hosts. Furthermore, the present study has reported that soils from Sierra del Abra de Zabaleta contain symbiotic strains of C. necator able to nodulate both native and widespread Mimosa species (see Fig. S6 in the supplemental material), and the presence of symbiotic strains of C. necator in natural populations of various Uruguayan native Mimosa spp. has confirmed that they are indeed natural hosts for symbiotic C. necator (Table 1 and Fig. 2). These facts, taken together with the previously reported M. pudica-nodulating Cupriavidus sp. strains isolated in Uruguay from P. rigida nodules (30) that also fall into this cluster, expands the natural nodulation range of C. necator into two genera (Mimosa and Parapiptadenia) in the mimosoid tribe Mimoseae.
The identification of a third cluster of symbiotic strains related to neither C. taiwanensis nor C. necator among the symbionts isolated from M. ramulosa, M. magentea, and M. reptans is exciting, since it suggests the existence of new species of symbiotic Cupriavidus. Although further analyses are required for determination of the affiliation of these strains to species level, the phylogenetic analyses of their 16S rRNA sequences have clearly suggested that they are a compact monophyletic group close to C. basilensis, C. numazuensis, and C. pinatubonensis. Interestingly, as far as is currently known, no symbiotic representatives of these species are reported. Further support for this contention was obtained when phylogenetic analyses using the sequences of their recA and gyrB housekeeping genes were performed. While the UYMS13B sequences clustered with other “C. necator-like” strains, UYMM02A (a representative of the new symbiotic Cupriavidus species) consistently clustered in a group that included C. pinatubonensis. These results suggest the existence of a higher diversity within beta-rhizobial Cupriavidus than was previously suspected and that Mimosa spp. from Sierra del Abra de Zabaleta may be natural reservoirs for novel rhizobia.
Further confirmation of the high diversity of symbiotic Cupriavidus spp. from Uruguayan Mimosa spp. comes from the identification of the isolates originated from trap plants assays; these were all Cupriavidus spp. that were clearly divergent from C. taiwanensis (see Fig. S6 in the supplemental material). Currently, symbionts from Mimosa spp. are being isolated from diverse Uruguayan geographical localizations with the aim of validating the hypothesis that Uruguay is a center of diversity for symbiotic Cupriavidus.
With regard to the evolutionary origin of the symbiosis-related genes harbored by the new Cupriavidus strains, phylogenetic analyses showed that both the nifH and nodA phylogenies were congruent with those of the housekeeping genes and, as with the latter genes, they were clearly separate from the equivalent C. taiwanensis symbiosis-related genes. Indeed, the sequences of both the nifH and nodA genes from the Sierra del Abra de Zabaleta Mimosa strains formed a monophyletic group, regardless of their core genome and their host Mimosa species. This indicates that although vertical gene transfer has been the main mechanism for the inheritance of symbiotic traits in this group of bacteria, the various species within the group have also probably exchanged their symbiosis related genes via horizontal gene transfer (HGT). In many rhizobia, from both the alpha and beta groups, the nod and nif symbiosis-related genes are coded by operons in symbiotic plasmids (pSym), e.g., the sequence of the symbiotic plasmid pRALTA of C. taiwanensis LMG19424T revealed the most compact symbiosis-related-gene organization observed for a rhizobium with all its nod and nif genes coded on a 35-kb symbiotic island (75). This compact organization may explain the observed phylogenetic convergence between the nodA and nifH phylogenies. As previously suggested by Bontemps et al. (22) and Mishra et al. (7), these analyses indicated that both symbiosis-related genes have been possibly acquired via HGT from a common Burkholderia ancestor. In consideration of this, comparative analyses of the phylogenetic relationships of genes from symbiotic Cupriavidus spp. have led to the conclusion that pSym plasmids are monophyletic for Cupriavidus strains isolated from Mimosa spp. in the Caribbean region (57) but also (and in agreement with the present study) that Caribbean and Uruguayan symbiotic Cupriavidus strains are not monophyletic, which implies that either convergent gains or convergent losses of genes required for legume nodule symbiosis formation have occurred (57). The discovery of monophyletic symbiosis-related genes in C. necator, C. taiwanensis, and the C. pinatubonensis-related group supports the hypothesis of a common Cupriavidus ancestor with symbiotic abilities, and that subsequent speciation has led to the loss of symbiotic abilities in most species/strains over evolutionary time. Comparative genomic analyses of the isolated bacteria vis-á-vis already sequenced genomes from both symbiotic and nonsymbiotic Cupriavidus and Burkholderia strains could help to determine the origin and evolution of the symbiotic Cupriavidus family.
Supplementary Material
ACKNOWLEDGMENTS
We thank Vanesa Amarelle and Daniela Costa for invaluable help in nodule collections, Cecilia Taulé for help with nodule microscopy, Elena Peel from the Departamento de Geología, Facultad de Ciencias, Universidad de la República, for help in soil analyses, and Max Chavarría from the Universidad de Costa Rica for help in determination of the soil metal content.
The Uruguayan National Program for Development of Basic Science (PEDECIBA) and Uruguayan National Agency for Innovation and Investigation (ANII) supported this work. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04142-15.
REFERENCES
- 1.Peix A, Ramírez-Bahena MH, Velázquez E, Bedmar EJ. 2014. Bacterial associations with legumes. CRC Crit Rev Plant Sci 34:17–42. [Google Scholar]
- 2.Sprent J. 2009. Legume nodulation: a global perspective. John Wiley & Sons, Oxford, United Kingdom. [Google Scholar]
- 3.Gyaneshwar P, Hirsch AM, Moulin L, Chen W-M, Elliott GN, Bontemps C, Estrada-de Los Santos P, Gross E, Dos Reis FB, Sprent JI, Young JPW, James EK. 2011. Legume-nodulating betaproteobacteria: diversity, host range, and future prospects. Mol Plant Microbe Interact 24:1276–1288. doi: 10.1094/MPMI-06-11-0172. [DOI] [PubMed] [Google Scholar]
- 4.White J, Prell J, James EK, Poole P. 2007. Nutrient sharing between symbionts. Plant Physiol 144:604–614. doi: 10.1104/pp.107.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pueppke SG, Broughton WJ. 1999. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol Plant Microbe Interact 12:293–318. doi: 10.1094/MPMI.1999.12.4.293. [DOI] [PubMed] [Google Scholar]
- 6.Willems A. 2006. The taxonomy of rhizobia: an overview. Plant Soil 287:3–14. doi: 10.1007/s11104-006-9058-7. [DOI] [Google Scholar]
- 7.Mishra RPN, Tisseyre P, Melkonian R, Chaintreuil C, Miché L, Klonowska A, González S, Bena G, Laguerre G, Moulin L. 2012. Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol Ecol 79:487–503. doi: 10.1111/j.1574-6941.2011.01235.x. [DOI] [PubMed] [Google Scholar]
- 8.Bournaud C, de Faria SM, dos Santos JMF, Tisseyre P, Silva M, Chaintreuil C, Gross E, James EK, Prin Y, Moulin L. 2013. Burkholderia species are the most common and preferred nodulating symbionts of the Piptadenia group (tribe Mimoseae). PLoS One 8:e63478. doi: 10.1371/journal.pone.0063478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melkonian R, Moulin L, Béna G, Tisseyre P, Chaintreuil C, Heulin K, Rezkallah N, Klonowska A, González S, Simon M, Chen W-M, James EK, Laguerre G. 2014. The geographical patterns of symbiont diversity in the invasive legume Mimosa pudica can be explained by the competitiveness of its symbionts and by the host genotype. Environ Microbiol 16:2099–2111. doi: 10.1111/1462-2920.12286. [DOI] [PubMed] [Google Scholar]
- 10.Elliott GN, Chen WM, Chou JH, Wang HC, Sheu SY, Perin L, Reis VM, Moulin L, Simon MF, Bontemps C, Sutherland JM, Bessi R, De Faria SM, Trinick MJ, Prescott AR, Sprent JI, James EK. 2007. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol 173:168–180. doi: 10.1111/j.1469-8137.2006.01894.x. [DOI] [PubMed] [Google Scholar]
- 11.Garau G, Yates RJ, Deiana P, Howieson JG. 2009. Novel strains of nodulating Burkholderia have a role in nitrogen fixation with papilionoid herbaceous legumes adapted to acid, infertile soils. Soil Biol Biochem 41:125–134. doi: 10.1016/j.soilbio.2008.10.011. [DOI] [Google Scholar]
- 12.Howieson JG, De Meyer SE, Vivas-Marfisi A, Ratnayake S, Ardley JK, Yates RJ. 2013. Novel Burkholderia bacteria isolated from Lebeckia ambigua: a perennial suffrutescent legume of the fynbos. Soil Biol Biochem 60:55–64. doi: 10.1016/j.soilbio.2013.01.009. [DOI] [Google Scholar]
- 13.Lemaire B, Dlodlo O, Chimphango S, Stirton C, Schrire B, Boatwright JS, Honnay O, Smets E, Sprent J, James EK, Muasya AM. 2015. Symbiotic diversity, specificity, and distribution of rhizobia in native legumes of the Core Cape Subregion (South Africa). FEMS Microbiol Ecol 91:1–17. doi: 10.1093/femsec/fiu020, , . [DOI] [PubMed] [Google Scholar]
- 14.Liu WYY, Ridgway HJ, James TK, James EK, Chen W-M, Sprent JI, Young JPW, Andrews M. 2014. Burkholderia sp. induces functional nodules on the South African invasive legume Dipogon lignosus (Phaseoleae) in New Zealand soils. Microb Ecol 68:542–555. doi: 10.1007/s00248-014-0427-0. [DOI] [PubMed] [Google Scholar]
- 15.Chen WM, Laevens S, Lee TM, Coenye T, De Vos P, Mergeay M, Vandamme P. 2001. Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int J Syst Evol Microbiol 51:1729–1735. doi: 10.1099/00207713-51-5-1729. [DOI] [PubMed] [Google Scholar]
- 16.Elliott GN, Chou J-H, Chen W-M, Bloemberg GV, Bontemps C, Martínez-Romero E, Velázquez E, Young JPW, Sprent JI, James EK. 2009. Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ Microbiol 11:762–778. doi: 10.1111/j.1462-2920.2008.01799.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, James EK, Prescott AR, Kierans M, Sprent JI. 2003. Nodulation of Mimosa spp. by the beta-proteobacterium Ralstonia taiwanensis. Mol Plant Microbe Interact 16:1051–1061. doi: 10.1094/MPMI.2003.16.12.1051. [DOI] [PubMed] [Google Scholar]
- 18.Chen W-M, James EK, Chou J-H, Sheu S-Y, Yang S-Z, Sprent JI. 2005. Beta-rhizobia from Mimosa pigra, a newly discovered invasive plant in Taiwan. New Phytol 168:661–675. doi: 10.1111/j.1469-8137.2005.01533.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen W-M, de Faria SM, Straliotto R, Pitard RM, Simões-Araùjo JL, Chou J-H, Chou Y-J, Barrios E, Prescott AR, Elliott GN, Sprent JI, Young JPW, James EK. 2005. Proof that Burkholderia strains form effective symbioses with legumes: a study of novel Mimosa-nodulating strains from South America. Appl Environ Microbiol 71:7461–7471. doi: 10.1128/AEM.71.11.7461-7471.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett CF, Parker MA. 2005. Prevalence of Burkholderia sp. nodule symbionts on four mimosoid legumes from Barro Colorado Island, Panama. Syst Appl Microbiol 28:57–65. doi: 10.1016/j.syapm.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Barrett CF, Parker MA. 2006. Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. nodule bacteria on two Mimosa spp. in Costa Rica. Appl Environ Microbiol 72:1198–1206. doi: 10.1128/AEM.72.2.1198-1206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bontemps C, Elliott GN, Simon MF, Dos Reis FBJ, Gross E, Lawton RC, Neto NE, Loureiro MDF, De Faria SM, Sprent JI, James EK, Young PW. 2010. Burkholderia species are ancient symbionts of legumes. Mol Ecol 19:44–52. doi: 10.1111/j.1365-294X.2009.04458.x. [DOI] [PubMed] [Google Scholar]
- 23.dos Reis FB, Simon MF, Gross E, Boddey RM, Elliott GN, Neto NE, Loureiro Mde F, de Queiroz LP, Scotti MR, Chen W-M, Norén A, Rubio MC, de Faria SM, Bontemps C, Goi SR, Young JPW, Sprent JI, James EK. 2010. Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol 186:934–946. doi: 10.1111/j.1469-8137.2010.03267.x. [DOI] [PubMed] [Google Scholar]
- 24.Simon MF, Grether R, de Queiroz LP, Särkinen TE, Dutra VF, Hughes CE. 2011. The evolutionary history of Mimosa (Leguminosae): toward a phylogeny of the sensitive plants. Am J Bot 98:1201–1221. doi: 10.3732/ajb.1000520. [DOI] [PubMed] [Google Scholar]
- 25.Lammel DR, Cruz LM, Carrer H, Cardoso EJBN. 2013. Diversity and symbiotic effectiveness of beta-rhizobia isolated from subtropical legumes of a Brazilian araucaria forest. World J Microbiol Biotechnol 29:2335–2342. doi: 10.1007/s11274-013-1400-7. [DOI] [PubMed] [Google Scholar]
- 26.Bontemps C, Rogel MA, Wiechmann A, Mussabekova A, Moody S, Simon MF, Moulin L, Elliott GN, Lacercat-Didier L, Dasilva C, Grether R, Camargo-Ricalde SL, Chen W, Sprent JI, Martínez-Romero E, Young JPW, James EK. 2016. Endemic Mimosa species from Mexico prefer alphaproteobacterial rhizobial symbionts. New Phytol 209:319–333. doi: 10.1111/nph.13573. [DOI] [PubMed] [Google Scholar]
- 27.Klonowska A, Chaintreuil C, Tisseyre P, Miché L, Melkonian R, Ducousso M, Laguerre G, Brunel B, Moulin L. 2012. Biodiversity of Mimosa pudica rhizobial symbionts (Cupriavidus taiwanensis, Rhizobium mesoamericanum) in New Caledonia and their adaptation to heavy metal-rich soils. FEMS Microbiol Ecol 81:618–635. doi: 10.1111/j.1574-6941.2012.01393.x. [DOI] [PubMed] [Google Scholar]
- 28.Gehlot HS, Tak N, Kaushik M, Mitra S, Chen W-M, Poweleit N, Panwar D, Poonar N, Parihar R, Tak A, Sankhla IS, Ojha A, Rao SR, Simon MF, Dos Reis FB, Perigolo N, Tripathi AK, Sprent JI, Young JPW, James EK, Gyaneshwar P. 2013. An invasive Mimosa in India does not adopt the symbionts of its native relatives. Ann Bot 112:179–196. doi: 10.1093/aob/mct112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izaguirre P, Beyhaut R. 2003. Leguminosas en Uruguay y regiones vecinas. 2. Caesalpinioideae. Parte 3: Mimosoideae, p 1–302. Editorial Hemisferio Sur, Montevideo, Uruguay. [Google Scholar]
- 30.Taulé C, Zabaleta M, Mareque C, Platero R, Sanjurjo L, Sicardi M, Frioni L, Battistoni F, Fabiano E. 2012. New betaproteobacterial Rhizobium strains able to efficiently nodulate Parapiptadenia rigida (Benth.) Brenan. Appl Environ Microbiol 78:1692–1700. doi: 10.1128/AEM.06215-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sánchez-Bettucci L, Oyhantçabal P, Loureiro J, Ramos VA, Preciozzi F, Basei MAS. 2004. Mineralizations of the Lavalleja Group (Uruguay), a probable neoproterozoic volcano-sedimentary sequence. Gondwana Res 7:745–751. doi: 10.1016/S1342-937X(05)71060-1. [DOI] [Google Scholar]
- 32.Vincent JM. 1970. A manual for the practical study of root-nodule bacteria. IBP Handbook N15. Blackwell, Oxford, United Kingdom. [Google Scholar]
- 33.Lane DJJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY. [Google Scholar]
- 34.Poly F, Ranjard L, Nazaret S, Gourbiére F, Monrozier LJ. 2001. Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl Environ Microbiol 67:2255–2262. doi: 10.1128/AEM.67.5.2255-2262.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koichiro T, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar R. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronquist F, Huelsenbeck JP. 2003. MrBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 41.Beretta A, Bassahum D, Musselli R. 2014. ¿Medir el pH del suelo en la mezcla suelo: agua en reposo o agitando? Agrociencia Uruguay 18:90–94. [Google Scholar]
- 42.Wright AF, Bailey JS. 2001. Organic carbon, total carbon, and total nitrogen determinations in soils of variable calcium carbonate contents using a Leco CN-2000 dry combustion analyzer. Commun Soil Sci Plant Anal 32:3243–3258. doi: 10.1081/CSS-120001118. [DOI] [Google Scholar]
- 43.Rhine ED, Mulvaney RL, Pratt EJ, Sims GK. 1998. Improving the Berthelot reaction for determining ammonium in soil extracts and water. Soil Sci Soc Am J 62:473–480. doi: 10.2136/sssaj1998.03615995006200020026x. [DOI] [Google Scholar]
- 44.Gelderman RH, Beegle D. 1998. Nitrate-nitrogen, p 17–20. In Dahnke WC. (ed), Recommended chemical soil test procedures for the north central region. Missouri Agricultural Experiment Station, University of Missouri–Columbia, Columbia, MO. [Google Scholar]
- 45.Bray RH, Kurtz LT. 1945. Determination of total, organic, and available forms of phosphorus in soils. Soil Sci 59:39–46. doi: 10.1097/00010694-194501000-00006. [DOI] [Google Scholar]
- 46.Murphy J, Riley JP. 1962. A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27:31–36. doi: 10.1016/S0003-2670(00)88444-5. [DOI] [Google Scholar]
- 47.Tran TS, Simard RR. 1993. Ammonium acetate-extractable elements, p 43–50. In Carter MR. (ed), Soil sampling and methods of analysis. CRC Press, Boca Raton, FL. [Google Scholar]
- 48.Lindsay W, Norvell W. 1978. Development of a DTPA soil test for zinc, iron, manganese, and copper. Soil Sci Soc Am J 42:421–428. doi: 10.2136/sssaj1978.03615995004200030009x. [DOI] [Google Scholar]
- 49.Mergeay M, Houba C, Gerits J. 1978. Extrachromosomal inheritance controlling resistance to cadmium, cobalt, copper and zinc ions: evidence from curing in a Pseudomonas. Arch Int Physiol Biochim 86:440–442. [PubMed] [Google Scholar]
- 50.Vandamme P, Coenye T. 2004. Taxonomy of the genus Cupriavidus: a tale of lost and found. Int J Syst Evol Microbiol 54:2285–2289. doi: 10.1099/ijs.0.63247-0. [DOI] [PubMed] [Google Scholar]
- 51.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 52.Elliott GN, Chen W-M, Bontemps C, Chou J-H, Young JPW, Sprent JI, James EK. 2007. Nodulation of Cyclopia spp. (Leguminosae, Papilionoideae) by Burkholderia tuberum. Ann Bot 100:1403–1411. doi: 10.1093/aob/mcm227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen W-M, Moulin L, Bontemps C, Vandamme P, Béna G, Boivin-Masson C. 2003. Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature. J Bacteriol 185:7266–7272. doi: 10.1128/JB.185.24.7266-7272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andam CP, Mondo SJ, Parker MA. 2007. Monophyly of nodA and nifH genes across Texan and Costa Rican populations of Cupriavidus nodule symbionts. Appl Environ Microbiol 73:4686–4690. doi: 10.1128/AEM.00160-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andrus AD, Andam C, Parker MA. 2012. American origin of Cupriavidus bacteria associated with invasive Mimosa legumes in the Philippines. FEMS Microbiol Ecol 80:747–750. doi: 10.1111/j.1574-6941.2012.01342.x. [DOI] [PubMed] [Google Scholar]
- 56.Da Silva K, Florentino LA, Da Silva KB, de Brandt E, Vandamme P, de Souza Moreira FM. 2012. Cupriavidus necator isolates are able to fix nitrogen in symbiosis with different legume species. Syst Appl Microbiol 35:175–182. doi: 10.1016/j.syapm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 57.Parker MA. 2015. A single sym plasmid type predominates across diverse chromosomal lineages of Cupriavidus nodule symbionts. Syst Appl Microbiol 38:417–423. doi: 10.1016/j.syapm.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 58.Estrada-de Los Santos P, Martínez-Aguilar L, López-Lara IM, Caballero-Mellado J. 2012. Cupriavidus alkaliphilus sp. nov, a new species associated with agricultural plants that grow in alkaline soils. Syst Appl Microbiol 35:310–314. doi: 10.1016/j.syapm.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 59.Goris J, De Vos P, Coenye T, Hoste B, Jansses D, Brim H, Diels L, Mergeay M, Kersters K, Vandamme P. 2001. Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al. emend. Int J Syst Evol Microbiol 51:1773–1782. doi: 10.1099/00207713-51-5-1773. [DOI] [PubMed] [Google Scholar]
- 60.Kageyama C, Ohta T, Hiraoka K, Suzuki M, Okamoto T, Ohishi K. 2005. Chlorinated aliphatic hydrocarbon-induced degradation of trichloroethylene in Wautersia numadzuensis sp. nov. Arch Microbiol 183:56–65. doi: 10.1007/s00203-004-0746-5. [DOI] [PubMed] [Google Scholar]
- 61.Sato Y, Nishihara H, Yoshida M, Watanabe M, Rondal JD, Concepcion RN, Ohta H. 2006. Cupriavidus pinatubonensis sp. nov. and Cupriavidus laharis sp. nov., novel hydrogen-oxidizing, facultatively chemolithotrophic bacteria isolated from volcanic mudflow deposits from Mt. Pinatubo in the Philippines. Int J Syst Evol Microbiol 56:973–978. doi: 10.1099/ijs.0.63922-0. [DOI] [PubMed] [Google Scholar]
- 62.Cuadrado V, Gomila M, Merini L, Giulietti AM, Moore ERB. 2010. Cupriavidus pampae sp. nov., a novel herbicide-degrading bacterium isolated from agricultural soil. Int J Syst Evol Microbiol 60:2606–2612. doi: 10.1099/ijs.0.018341-0. [DOI] [PubMed] [Google Scholar]
- 63.Coenye T, Vandamme P, LiPuma JJ. 2003. Ralstonia respiraculi sp. nov., isolated from the respiratory tract of cystic fibrosis patients. Int J Syst Evol Microbiol 53:1339–1342. doi: 10.1099/ijs.0.02440-0. [DOI] [PubMed] [Google Scholar]
- 64.Vandamme P, Goris J, Coenye T, Hoste B, Janssens D, Kersters K, De Vos P, Falsen E. 1999. Assignment of Centers for Disease Control group IVc-2 to the genus Ralstonia as Ralstonia paucula sp. nov. Int J Syst Bacteriol 49(Pt 2):663–669. [DOI] [PubMed] [Google Scholar]
- 65.Liu X, Wei S, Wang F, James EK, Guo X, Zagar C, Xia LG, Dong X, Wang YP. 2012. Burkholderia and Cupriavidus spp. are the preferred symbionts of Mimosa spp. in Southern China. FEMS Microbiol Ecol 80:417–426. doi: 10.1111/j.1574-6941.2012.01310.x. [DOI] [PubMed] [Google Scholar]
- 66.Oyhantçabal P, Goso H, Panario D. 2004. Niveles de base de elementos potencialmente tóxicos en los suelos de la Hoja “Fuente del Puma,” Departamento de Lavalleja–Uruguay, p 1–13. In IV Congreso Uruguayo de Geología. [Google Scholar]
- 67.Wang ET, Rogel MA, García-de los Santos A, Martínez-Romero J, Cevallos MA, Martínez-Romero E. 1999. Rhizobium etli bv. mimosae, a novel biovar isolated from Mimosa affinis. Int J Syst Bacteriol 49(Pt 4):1479–1491. [DOI] [PubMed] [Google Scholar]
- 68.Verma SC, Chowdhury SP, Tripathi AK. 2004. Phylogeny based on 16S rDNA and nifH sequences of Ralstonia taiwanensis strains isolated from nitrogen-fixing nodules of Mimosa pudica, in India. Can J Microbiol 50:313–322. doi: 10.1139/w04-020. [DOI] [PubMed] [Google Scholar]
- 69.Stopnisek N, Bodenhausen N, Frey B, Fierer N, Eberl L, Weisskopf L. 2014. Genus-wide acid tolerance accounts for the biogeographical distribution of soil Burkholderia populations. Environ Microbiol 16:1503–1512. doi: 10.1111/1462-2920.12211. [DOI] [PubMed] [Google Scholar]
- 70.Poehlein A, Kusian B, Friedrich B, Daniel R, Bowien B. 2011. Complete genome sequence of the type strain Cupriavidus necator N-1. J Bacteriol 193:5017. doi: 10.1128/JB.05660-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hong KW, Thinagaran Al D, Gan HM, Yin W-F, Chan K-G. 2012. Whole-genome sequence of Cupriavidus sp. strain BIS7, a heavy-metal-resistant bacterium. J Bacteriol 194:6324. doi: 10.1128/JB.01608-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L-G, Cai L, Zhang T. 2013. Genome of Cupriavidus sp. HMR-1, a heavy metal-resistant bacterium. Genome Announc 1:e00202-12. doi: 10.1128/genomeA.00202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ray J, Waters RJ, Skerker J, Kuehl JV, Prise MN, Huang J, Chakraborty R, Arkin AP, Deutschbauer A. 2015. Complete genome sequence of Cupriavidus basilensis 4G11, isolated from the Oak Ridge field research center site. Genome Announc 3:e00322-15. doi: 10.1128/genomeA.00322-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, Benotmane MA, Leys N, Vallaeys T, Lapidus A, Monchy S, Médigue C, Taghavi S, McCorkle S, Dunn J, van der Lelie D, Mergeay M. 2010. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One 5:e10433. doi: 10.1371/journal.pone.0010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amadou C, Pascal G, Mangenot S. 2008. Genome sequence of the rhizobium Cupriavidus taiwanensis and comparative genomics of rhizobia. Genome Res 18:1472–1483. doi: 10.1101/gr.076448.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pérez-Pantoja D, De la Iglesia R, Pieper DH, González B. 2008. Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiol Rev 32:736–794. doi: 10.1111/j.1574-6976.2008.00122.x. [DOI] [PubMed] [Google Scholar]
- 77.De Meyer SE, Fabiano E, Tian R, Van Berkum P, Seshadri R, Reddy T, Markowitz V, Ivanova NN, Pati A, Woyke T, Howieson J, Kyrpides NC, Reeve W. 2015. High-quality permanent draft genome sequence of the Parapiptadenia rigida-nodulating Cupriavidus sp. strain UYPR2.512. Stand Genomic Sci 10:13. doi: 10.1186/1944-3277-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nies DH. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev 27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 79.Chen W-M, Wu C-H, James EK, Chang J-S. 2008. Metal biosorption capability of Cupriavidus taiwanensis and its effects on heavy metal removal by nodulated Mimosa pudica. J Hazard Mater 151:364–371. doi: 10.1016/j.jhazmat.2007.05.082. [DOI] [PubMed] [Google Scholar]
- 80.Ferreira PAA, Bomfeti CA, Da Silva Júnior R, Soares BL, Soares CRFS, Moreira FMDS. 2012. Eficiência simbiótica de estirpes de Cupriavidus necator tolerantes a zinco, cádmio, cobre e chumbo. Pesqui Agropecu Bras 47:85–95. doi: 10.1590/S0100-204X2012000100012. [DOI] [Google Scholar]
- 81.Rogel M, Bustos P, Santamaría RI, González V, Romero D, Cevallos MÁ, Lozano L, Castro-Mondragón J, Martínez-Romero J, Ormeño-Orrillo E, Martínez-Romero E. 2014. Genomic basis of symbiovar mimosae in Rhizobium etli. BMC Genomics 15:575–585. doi: 10.1186/1471-2164-15-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.