ABSTRACT
As annual influenza epidemics continue to cause significant morbidity and economic burden, an understanding of viral persistence and transmission is critical for public health officials and health care workers to better protect patients and their family members from infection. The infectivity and persistence of two influenza A (H1N1) virus strains (A/New Caledonia/20/1999 and A/Brisbane/59/2007) on stainless steel (SS) surfaces were evaluated using three different surface matrices (2% fetal bovine serum, 5 mg/ml mucin, and viral medium) under various absolute humidity conditions (4.1 × 105 mPa, 6.5 × 105 mPa, 7.1 × 105 mPa, 11.4 × 105 mPa, 11.2 × 105 mPa, and 17.9 × 105 mPa) for up to 7 days. Influenza A virus was deposited onto SS coupons (7.07 cm2) and recovered by agitation and sonication in viral medium. Viral persistence was quantified using a tissue culture-based enzyme-linked immunosorbent assay (ELISA) to determine the median (50%) tissue culture infective dose (TCID50) of infectious virus per coupon. Overall, both strains of influenza A virus remained infectious on SS coupons, with an approximate 2 log10 loss over 7 days. Factors that influenced viral persistence included absolute humidity, strain-absolute humidity interaction, and time (P ≤ 0.01). Further studies on the transfer of influenza A virus from fomites by hand and the impact of inanimate surface contamination on transmission should be performed, as this study demonstrates prolonged persistence on nonporous surfaces.
IMPORTANCE This study tested the ability of two influenza A (H1N1) virus strains to persist and remain infectious on stainless steel surfaces under various environmental conditions. It demonstrated that influenza A (H1N1) viruses can persist and remain infectious on stainless steel surfaces for 7 days. Additional studies should be conducted to assess the role played by contaminated surfaces in the transmission of influenza A virus.
INTRODUCTION
Influenza virus continues to be a pathogen of significant interest, as the World Health Organization estimates that annual influenza epidemics cause approximately 5 million cases of serious illness and over 250,000 deaths per year (1). In the United States alone, it has been estimated that there are approximately 226,000 hospitalizations and 3,000 to 48,000 deaths during an influenza virus season, leading to a total economic burden that can reach $87.1 billion (2–4). Influenza A virus is an enveloped RNA virus which has been shown to be transmissible primarily by droplet, with less evidence of contact and airborne transmission being available (5, 6). Previous research has shown that fomites and surface contamination caused by large respiratory droplets may play a significant role in transmission (6–8).
Current Centers for Disease Control and Prevention health care infection control guidelines for seasonal influenza recommend performing hand hygiene before and after patient contact, after contact with contaminated surfaces or materials, and before personal protective equipment is put on and removed. Additional strategies include disinfection of potentially contaminated noncritical surfaces, including frequently touched surfaces that are in close proximity to the patient (e.g., hospital bed rails, overbed tables), with U.S. Environmental Protection Agency-registered hospital disinfectants (9, 10). However, lapses in infection control, most notably, lapses in hand hygiene implementation (11–13), may facilitate the transmission of influenza virus. Boone and Gerba (14) have shown that 50% of fomites found in homes and day care centers had influenza virus present during an active influenza season. Influenza A virus has also been shown to be transferred from stainless steel (SS) countertops to hands for up to 24 h after surface contamination (15); however, indirect transmission through contact with these surfaces has not been demonstrated.
Regardless of the precise role of fomites and surface contamination in transmission, it is important to have a clear understanding of what factors influence the persistence of pathogens on surfaces and how this persistence impacts transmission from contaminated environmental surfaces. Previous studies of persistence on environmental surfaces have found that various strains of influenza A (H1N1) virus remained infectious on SS surfaces for approximately 24 h but no longer than 72 h (15–19). In this study, we evaluated the persistence and infectivity of two influenza A (H1N1) virus strains, A/New Caledonia/20/1999 (A/NC-H1N1) and A/Brisbane/59/2007 (A/Br-H1N1), in three different substrate matrices (viral medium, 2% fetal bovine serum [FBS], and 5 mg/ml mucin) on SS surfaces stored for various times up to 7 days under a range of absolute humidity conditions.
MATERIALS AND METHODS
Cell culture and viral propagation.
Madin-Darby canine kidney (MDCK) epithelial cells (CCL-3; ATCC, Manassas, VA) that had been passaged fewer than 90 times were maintained on a medium containing Dulbecco's modified Eagle medium (DMEM; Gibco, Grand Island, NY), 2% (vol/vol) FBS (Atlanta Biologicals, Lawrenceville, GA), and 2% (vol/vol) penicillin-streptomycin (stock concentration, 10,000 units/ml penicillin G sodium and 10,000 μg/ml streptomycin sulfate; Life Technologies, Carlsbad, CA). Cells were incubated at 37°C in 5% CO2 for 24 to 48 h until they were 90% confluent. MDCK cells were rinsed with 1× phosphate-buffered saline (PBS), and the virus was added to obtain a multiplicity of infection of 0.01. The two influenza A (H1N1) virus strains used in this experiment, A/Brisbane/59/2007 (A/Br-H1N1) and A/New Caledonia/20/1999 (A/NC-H1N1), were obtained from the Lovelace Respiratory Research Institute (Albuquerque, NM). These strains were chosen because they were seasonal circulating strains and were included in the WHO yearly influenza vaccine recommendations (20). The cells were incubated at 37°C in 5% CO2 until an approximately 75% cytopathic effect (CPE) was observed. The virus was harvested, cell debris was removed by centrifugation, and aliquots of the supernatant were stored at −80°C. The starting concentration for A/NC-H1N1 was approximately 1.2 × 104 50% tissue culture infective doses (TCID50)/ml, and that for A/Br-H1N1 was 6.95 × 105 TCID50/ml.
Inoculation of stainless steel coupons and environmental conditions.
This study evaluated the persistence of the influenza A (H1N1) virus in various substrates: viral medium (DMEM, 7.5% bovine serum albumin [BSA; Life Technologies, Carlsbad, CA], 2% penicillin-streptomycin, HEPES buffer [Gibco, Grand Island, NY], and tosylsulfonyl phenylalanyl chloromethyl ketone [TPCK]-treated trypsin [Thermo Scientific, Rockford, IL]), 2% FBS, and 5 mg/ml mucin (MP Biomedicals, Solon, OH). SS sheets (24 gauge; T-304; Stewart Stainless Supply, Suwanee, GA) were punched into 7.07-cm2 coupons. SS was used as a surrogate for nonporous surfaces because it is a material commonly found in hospitals (e.g., safety grip bars, poles holding intravenous solution) and representative of household surfaces (e.g., doorknobs). Prior to inoculation, the SS coupons were washed with a low-residue cleaner, rinsed thoroughly in reverse osmosis water, soaked in 70% ethanol for 1 h, and then left to air dry. The SS coupons were placed in glass petri dishes and autoclaved at 121°C for 20 min and then an additional 20 min for drying. The three sterile SS coupons were transferred to 6-well plates that had been UV sterilized in a biosafety cabinet (BSC). Equal volumes of virus suspension were combined with the undiluted sample matrix to achieve the desired 2% FBS and 5-mg/ml mucin concentrations. Virus mixture aliquots of 100 μl were added to each SS coupon and spread with a cell spreader, making the concentration for A/NC-H1N1 equal to 1.2 × 103 TCID50/coupon and that for A/Br-H1N1 equal to 6.9 × 104 TCID50/coupon. The SS coupons were allowed to air dry for 1 h at room temperature in the BSC (sash closed, no blower). The 6-well plates were transferred to a temperature- and humidity-controlled environmental chamber (Caron, Marietta, OH) under the following conditions for up to 7 days: 4.1 × 105 mPa (18°C and 20% relative humidity [RH]), 6.5 × 105 mPa (25°C and 20% RH), 7.1 × 105 mPa (18°C and 35% RH), 11.4 × 105 mPa (25°C and 35% RH), 11.2 × 105 mPa (18°C and 55% RH), and 17.9 × 105 mPa (25°C and 55% RH). Each condition was tested once, with each time point tested in triplicate.
Sample processing.
Inoculated SS coupons were processed at 0 h (T0), 24 h (T24), 48 h (T48), 96 h (T96; 4 days), and 168 h (T168; 7 days). For sample processing, 1.9 ml of viral medium was added to the 6-well plate and, as described by Donlan et al., alternately agitated by sonication and vortexing three times for 30 s each time (21). The medium was collected and dispensed into two separate tubes. The samples were stored at −80°C until analysis.
ELISA.
The tissue cultured-based enzyme-linked immunosorbent assay (ELISA) method, which measures the ability of the virus particles to infect and replicate, has been fully described by Coulliette et al. (22). Briefly, viral samples were placed in a 96-well plate and diluted in series 1:3 in 100 μl of viral medium. MDCK cells were removed from the flask by trypsin-EDTA (Gibco, Grand Island, NY), centrifuged, and resuspended in viral medium, and the suspension was added to each well. The plate was incubated at 37°C in 5% CO2 for 17 to 19 h. The plates were rinsed with 1× PBS, fixed with 80% acetone–1× PBS, and allowed to air dry in the biosafety cabinet. The primary antibody, a mouse anti-influenza A virus monoclonal antibody (Millipore, Temecula, CA) which binds to the nucleoprotein epitope, was diluted 1:1,000 in 1× PBS–Tween 20–1% BSA, the diluted antibody was added to each well, and the plates was incubated for 1 h and washed with 1× PBS–Tween 20 (PBST). The secondary antibody, peroxidase-labeled affinity-purified goat anti-mouse IgG antibody (KPL, Gaithersburg, MD), was diluted 1:1,000 in 1× PBS–Tween 20–1% BSA, the diluted antibody was added to each well, and the plate was incubated for 1 h and washed with 1× PBST. To develop the substrate, a solution containing phosphate-citrate buffer with sodium perborate (Sigma-Aldrich, St. Louis, MO), o-phenylenediamine dihydrochloride tablets (10 mg; Sigma-Aldrich, St. Louis, MO), and hydrogen peroxide (Sigma-Aldrich, St. Louis, MO) was added to each well, the plate was incubated at room temperature, and then the reaction was completed with sulfuric acid (Sigma-Aldrich, St. Louis, MO). The 96-well plates were read at an absorbance λ of 490 nm in a Synergy II plate reader (BioTek Instruments, Winooski, VT) using the Gen5 program (versions 1.11 and 2.00).
Data analysis and statistics.
The absorbance data were exported to Microsoft Excel software (version 14; Redmond, WA). The number of TCID50 per coupon was determined using the standard Reed and Muench method (23). The limit of detection for the ELISA was 1.44 × 101 to 3.40 × 105 TCID50/ml. The number of TCID50 per coupon recovery results for each time point were log10 transformed, and the mean log10 recovery and change in log10 recovery relative to the number of TCID50 per coupon at T0 were calculated. SPSS software (version 19; Somers, NY) was used to generate box plots and descriptive statistics. SAS software (version 9.2; Cary, NC) was used to conduct linear modeling to assess the independent importance and best parameterization of each factor (i.e., sample matrix, absolute humidity [AH], and time). A generalized estimating equations (GEE) approach based on the best linear model was used to account for the correlation of the mean log10 differences due to the clustering of replicates over time, with the intercept representing the mean log10 change for the referent strata (24). The significance level for all tests performed was a P value of 0.01. It is important to note that the data for several groups could be combined for a more robust statistical analysis, as group maximum likelihood estimation (MLE) analyses showed that there was no significant difference (P > 0.01) between the levels at times of 24 and 48 h (identified as 24 to 48 h) and among the following AH groups: 6.5 × 105 mPa, 7.1 × 105 mPa, 11.4 × 105 mPa, and 11.2 × 105 mPa (identified as 11.4 × 105 mPa).
RESULTS
The mean number of TCID50 of A/NC-H1N1 recovered per coupon was 1.68 × 102 (range, 1.22 × 101 to 6.87 × 102) across all AHs and times, which is equivalent to a 13.9% recovery relative to the mean number of TCID50 per coupon in the inoculum at T0. The mean number of TCID50 of A/Br-H1N1 recovered per coupon was 1.41 × 103 (range, 2.77 × 102 to 4.89 × 103), which is equivalent to a 2.04% recovery relative to the mean number of TCID50 per coupon in the inoculum at T0. The recovery of both influenza A virus strains was compared using an MLE (not shown), which determined that the mean log10 change in recovery across all AHs and times was −0.44 (standard deviation [SD], 0.85) for A/NC-H1N1 and −1.30 (SD, 0.87) for A/Br-H1N1. Due to the significant (P < 0.001) difference in recovery between the two influenza A virus strains, where a greater change in log10 recovery was experienced for A/Br-H1N1 than for A/NC-H1N1, the remainder of the data and statistical analyses were performed by consideration of the two strains independently.
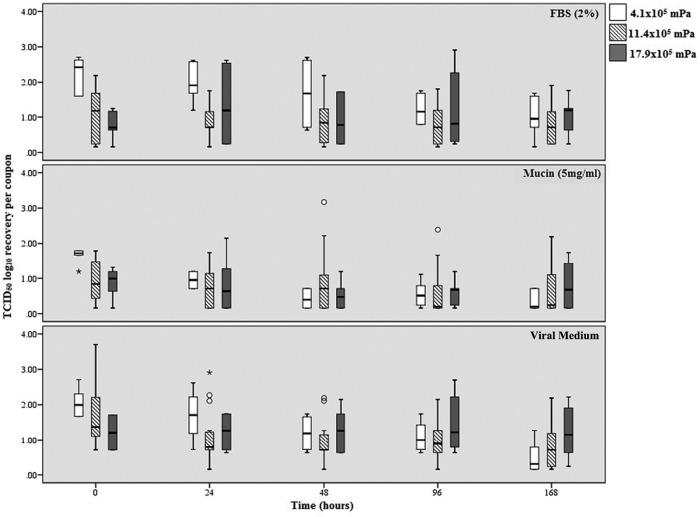
The median log10 recovery of infectious A/NC-H1N1 is presented in Fig. 1 for each matrix (FBS [2%], mucin [5 mg/ml], viral medium), time point (T0, T24, T48, T96, T168), and AH (4.1 × 105 mPa, 11.4 × 105 mPa, 17.9 × 105 mPa). The median log10 range of recovery for virus held at an AH of 4.1 × 105 mPa was 1.01 to 2.23 for virus held in FBS, 0.36 to 1.65 for virus held in mucin, and 0.50 to 2.06 for virus held in viral medium. The median log10 range of recovery for virus held at an AH of 11.4 × 105 mPa was 0.79 to 1.08 for virus held in FBS, 0.62 to 0.95 for virus held in mucin, and 0.90 to 1.69 for virus held in viral medium. The median log10 range of recovery for virus held at an AH of 17.9 × 105 mPa was 0.77 to 1.22 for virus held in FBS, 0.81 to 1.21 for virus held in mucin, and 0.88 to 1.04 for virus held in viral medium. The mean log10 change in recovery for infectious A/NC-H1N1 compared to the mean number of TCID50 of virus in the inoculum at T0 for each of the parameters is shown in Table 1. The median log10 recovery of influenza A virus at T0 was higher when it was held at an AH of 4.1 × 105 mPa than when it was held at AHs of 11.4 × 105 mPa and 17.9 × 105 mPa for all three substrates. Virus held at an AH of 17.9 × 105 mPa was the most stable across all matrices and times, whereas virus held at an AH of 4.1 × 105 mPa experienced the greatest loss in log10 recovery across all three matrices.
FIG 1.
The log10 TCID50 recovery of the A/NC-H1N1 strain per coupon over time for the three different matrices and absolute humidities. The middle bar within each box represents the median; the top and bottom of the bar represent the 75th and 25th quartiles, respectively; and the error bars represent the 95% confidence intervals. Open circles represent outliers, and asterisks represent extreme outliers (three times the range of variation).
TABLE 1.
Change in log10 TCID50 recovery of A/NC-H1N1 per SS coupon over time for each matrix and absolute humidity
| A/NC-H1N1 sample matrix | Time | Mean change in log10 TCID50a (SD) at an absolute humidity of: |
||
|---|---|---|---|---|
| 4.1 × 105 mPab | 11.4 × 105 mPac | 17.9 × 105 mPab | ||
| FBS | T24 | −0.25 (0.7) | −0.23 (0.6) | 0.57 (1.2) |
| T48 | −0.56 (0.7) | −0.18 (0.8) | 0.14 (0.8) | |
| T96 | −1.01 (0.4) | −0.31 (0.8) | 0.46 (1.3) | |
| T168 | −1.22 (0.5) | −0.32 (0.9) | 0.28 (0.7) | |
| Mucin | T24 | −0.70 (0.2) | −0.23 (0.7) | −0.04 (0.5) |
| T-48 | −1.23 (0.5) | −0.16 (0.9) | −0.35 (0.3) | |
| T96 | −1.10 (0.5) | −0.40 (0.8) | −0.27 (0.4) | |
| T168 | −1.29 (0.3) | −0.36 (0.8) | −0.07 (0.4) | |
| Viral medium | T24 | −0.36 (0.3) | −0.66 (0.7) | 0.01 (0.1) |
| T48 | −0.87 (0.3) | −0.75 (0.8) | 0.07 (0.2) | |
| T96 | −0.97 (0.1) | −0.77 (1.2) | 0.25 (0.5) | |
| T168 | −1.56 (0.5) | −1.01 (1.1) | 0.01 (0.4) | |
Data are relative to the number of TCID50 in the inoculum at T0.
n = 6.
n = 24.
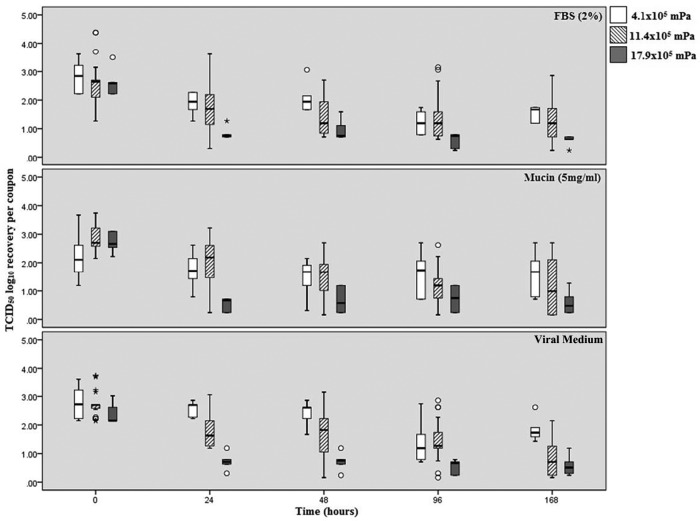
The median log10 recovery of A/Br-H1N1 is presented in Fig. 2 for each matrix (FBS [2%], mucin [5 mg/ml], viral medium), time point (T0, T24, T48, T96, T168), and AH (4.1 × 105 mPa, 11.4 × 105 mPa, 17.9 × 105 mPa). The median log10 range of recovery for virus held at 4.1 × 105 mPa was 1.53 to 2.83 for virus held in FBS, 1.60 to 2.23 for virus held in mucin, and 1.84 to 2.73 for virus held in viral medium. The median log10 range of recovery for virus held at 11.4 × 105 mPa was 2.29 to 2.61 for virus held in FBS, 1.19 to 2.84 for virus held in mucin, and 0.78 to 2.75 for virus held in viral medium. The median log10 range of recovery for virus held at 17.9 × 105 mPa was 0.59 to 2.71 for virus held in FBS, 0.58 to 2.37 for virus held in mucin, and 0.58 to 2.62 for virus held in viral medium. The mean log10 change in recovery of A/Br-H1N1 compared to the mean number of TCID50 of virus in the inoculum at T0 for each of the parameters is shown in Table 2. As for A/NC-H1N1, log10 recovery was the most stable at 17.9 × 105 mPa. In addition, influenza A virus held at an AH of 11.4 × 105 mPa experienced the greatest change in log10 recovery, with only samples in viral medium demonstrating the worst recovery, though samples in 2% FBS and 5 mg/ml mucin also demonstrated similarly low recoveries over time.
FIG 2.
The log10 TCID50 recovery of the A/Br-H1N1 strain per coupon over time for the three different matrices and absolute humidities. The middle bar within each box represents the median; the top and bottom of the bar represent the 75th and 25th quartiles, respectively; and the error bars represent the 95% confidence intervals. Open circles represent outliers, and asterisks represent extreme outliers (three times the range of variation).
TABLE 2.
Change in log10 TCID50 recovery of A/Br-H1N1 per SS coupon over time for each matrix and absolute humidity
| A/Br-H1N1 sample matrix | Time | Mean change in log10 TCID50a (SD) at an absolute humidity of: |
||
|---|---|---|---|---|
| 4.1 × 105 mPab | 11.4 × 105 mPac | 17.9 × 105 mPab | ||
| FBS | T24 | −0.93 (0.3) | −0.80 (0.5) | −1.79 (0.5) |
| T48 | −0.76 (0.5) | −1.16 (0.8) | −1.68 (0.6) | |
| T96 | −1.61 (0.2) | −1.25 (0.7) | −2.02 (0.5) | |
| T168 | −1.30 (0.7) | −1.32 (1.2) | −2.03 (0.6) | |
| Mucin | T24 | −0.49 (0.5) | −0.84 (0.8) | −2.17 (0.4) |
| T48 | −0.74 (1.3) | −1.33 (0.9) | −2.05 (0.2) | |
| T96 | −0.62 (1.4) | −1.69 (0.7) | −1.99 (0.3) | |
| T168 | −0.63 (1.4) | −1.65 (1.1) | −2.13 (0.4) | |
| Viral medium | T24 | −0.20 (0.6) | −0.94 (0.5) | −1.65 (0.3) |
| T-48 | −0.34 (0.7) | −0.99 (0.9) | −1.65 (0.5) | |
| T96 | −1.39 (0.7) | −1.27 (0.8) | −1.82 (0.4) | |
| T168 | −0.94 (0.6) | −1.95 (0.6) | −1.80 (0.3) | |
Data are relative to the number of TCID500 in the inoculum at T0.
n = 6.
n = 24.
A GEE multivariate analysis was created using the cumulative log10 change in recovery for each parameter when modeled concurrently, and the results are shown in Table 3. The model showed that AH, the AH-strain interaction, and time were factors that significantly impacted the overall change in log10 recovery (P < 0.0001) and that the virus strain did not impact persistence (P = 0.45). An MLE (not shown) showed that the sample matrix did not have an impact on influenza A virus persistence and therefore was not included in the GEE analysis. When the AH was increased from 4.1 × 105 mPa to 17.9 × 105 mPa, there was a significant (P < 0.0001) cumulative decrease in log10 recovery (−1.74 log10). Additionally, when the increase in AH occurred in combination with the A/NC-H1N1 strain, there was a significant (P < 0.0001) cumulative log10 increase (1.41 log10) compared to that for the A/Br-H1N1 strain. When time was modeled, samples had to be held for 7 days (T168) before a significant (P = 0.0013) cumulative log10 decrease in recovery (−1.05 log10) was observed.
TABLE 3.
GEE analysis of the recovery of influenza A (H1N1) virus on SS couponsa
| Parameter | Cumulative log10 change | Estimate (SE) | Confidence limit | P value (α)c |
|---|---|---|---|---|
| Intercept | −0.67 (0.11) | −0.88 to −0.46 | <0.0001 | |
| AH | ||||
| 4.1 × 105 mPab | ||||
| 11.4 × 105 mPa | −1.15 | −0.44 (0.13) | −0.69 to 0.18 | <0.0001 |
| 17.9 × 105 mPa | −1.74 | −1.07 (0.12) | −1.29 to −0.84 | <0.0001 |
| Strain | ||||
| A/NC-H1N1 | −0.77 | −0.10 (0.13) | −0.35 to 0.15 | 0.45 |
| A/Br-H1N1b | ||||
| Interactions | ||||
| 4.1–A/NC-H1N1b | ||||
| 4.1–A/Br-H1N1b | ||||
| 11.4–A/NC-H1N1 | 0.24 | 0.92 (0.18) | 0.57 to 1.27 | <0.0001 |
| 11.4–A/Br-H1N1b | ||||
| 17.9–A/NC-H1N1 | 1.41 | 2.08 (0.17) | 1.76 to 2.41 | <0.0001 |
| 17.9–A/Br-H1N1b | ||||
| Time point | ||||
| 24–48 hb | ||||
| 96 h | −0.93 | −0.26 (0.10) | −0.46 to −0.06 | 0.01 |
| 168 h | −1.05 | −0.38 (0.12) | −0.62 to −0.15 | 0.0013 |
The GEE can be used to calculate the cumulative change in the log10 TCID50 by adding the estimate for an individual parameter to the intercept value. A further model estimation can be obtained by combining the intercept (−0.67) with parameters in a given scenario; for example, at 11.4 ×105 mPa, the combination of the AH estimate (−0.44) with the A/NC-H1N1 estimate (−0.10) for 168 h (−0.38) results in a −1.59 cumulative log10 change in the TCID50 of influenza A (H1N1) virus recovered from stainless steel.
This group is the referent group and is reflective of the intercept.
The significance limit was a P value of <0.01.
DISCUSSION
Our study demonstrates that influenza A (H1N1) virus strains A/NC-H1N1 and A/Br-H1N1 persisted and remained infectious on SS coupons for 7 days (T168) under the various AH conditions. Overall, A/NC-H1N1 experienced a 1.5-log10 decrease in recovery (Table 1) across the 7 days (T168), while A/Br-H1N1 experienced a 2-log10 decrease in recovery (Table 2) across the 7 days (T168). The GEE model (Table 3) revealed that AH, the interaction between AH and the virus strain, and time were the factors that significantly affected the ability of influenza A virus to persist on SS. The sample matrix did not influence viral persistence.
The results from this study contradict those from previously published research. For example, it has previously been demonstrated that there is an increased persistence of influenza A virus in the presence of mucus (25, 26). However, in our study, the sample matrix (e.g., viral medium, FBS, mucin) did not influence influenza A virus persistence, which was also demonstrated by Coulliette et al. (22). It is important to note that the source of mucus for all three studies was different and data are not available to explain how this may impact virus persistence. Additional differences in persistence between our study and previous studies, which are summarized in Table 4, may be attributed to differences in the influenza A virus strains, viral surface recovery methods, quantification methods, sample matrices, and environmental conditions used.
TABLE 4.
Summary of published research and our study on the persistence of influenza A virus on SS surfaces
| Influenza A virus strain | AH (mPa) | Temp (°C) | RH (%) | Detection methodology | Log10 loss of recovery | Time to loss (h) | Sample size | Authors (reference) |
|---|---|---|---|---|---|---|---|---|
| A/Brazil/11/78-like | 10.8 × 105 | 27.8–28.3 | 35–50 | CPEa | 3.5 | 48 | NSb | Bean et al. (15) |
| A/PuertoRico/8/34 | 13.2 × 105 | 25.2 | 55 | CPE | 3 | 24 | 2 | Sakaguchi et al. (19) |
| A/PuertoRico/8/34 and A/Cambridge/AH04/2009 | 3.8 × 105 | 17–21 | 23–24 | Plaque assay, PCR | >4.2 | 24 | NS | Greatorex et al. (16) |
| Novel H1N1 strain (untyped) | NS | NS | NS | Antigen detection assay | >5 | 0.5 | 2 | Mukherjee et al. (17) |
| A/California/7/2009 | NS | NS | NS | HAc | >5.5 | 48 | 4 | Oxford et al. (18) |
| A/New Caledonia/20/1999 and A/Brisbane/59/2007 | 4.1 × 105, 11.4 × 105, and 17.9 × 105e | 18–25 | 20–55 | ELISAd | 2 | 168 | 1 | Perry et al. (this study) |
CPE, cytopathic effect assay.
NS, not stated.
HA, hemagglutination assay.
ELISA, enzyme-linked immunosorbent assay.
See Materials and Methods for defined temperature and relative humidity ranges.
The aforementioned previous studies (Table 4) used different strains of influenza A viruses that represent virus strains from various regions of the world that were collected at different times over a period spanning decades. Influenza virus is known for rapidly mutating, whether it is due to natural mutation or host immune selective pressure (27). This mutation could contribute to the variability between virus strains, rendering it difficult to make a general conclusion regarding persistence. In our own study, a maximum likelihood estimate (not shown) that compared the cumulative change in log10 recovery determined that a significant difference in persistence between the A/NC-H1N1 and A/Br-H1N1 strains (P ≤ 0.0001) existed. However, when the recoveries of both virus strains were simultaneously compared against all parameters in the GEE model (Table 3), there was no significant difference in recovery between the strains (P = 0.45). The A/NC-H1N1 strain had a greater decrease in log10 recovery when it was held at 4.1 × 105 mPa (−1.56 log10 [SD, 0.5 log10]), while A/Br-H1N1 had a greater decrease in log10 recovery when it was held at 17.9 × 105 mPa (−2.17 log10 [SD, 0.4 log10]). The differences in persistence between virus strains that were seen may be attributed to slight variations within the fatty acid composition in the outer phospholipid membrane of the influenza A viruses. The lipid content of influenza viruses can range from 20 to 24% (28), and it has been observed that viruses with greater lipid contents are more stable at lower RHs (29). Studies that compared the lipid contents of influenza A and B virus strains, as well as those of two influenza A virus strains, where the lipid content of one was made incomplete, have demonstrated that these slight differences do exist (30, 31). Additionally, influenza A virus strain A/Puerto Rico/8/34 has been shown to resist desiccation on surfaces for a week or longer at ambient temperature (32). This may be in part due to the protective nature of the envelope and an ability to form clumps on surfaces (29).
Methods previously used to recover influenza virus from environmental surfaces included a premoistened swab (15–17), surface rinsing (18), or a combination of residual liquid and a premoistened swab (19). To date, no recommended, standardized, or validated guidelines that describe the best methodology for recovery of viruses from any type of environmental surface are available. Therefore, it is difficult to say if a method used in a study to recover influenza A virus from environmental surfaces is the most efficient. In addition, cell culture-based methods, such as the CPE assay (15, 19), the plaque assay (16), and the hemagglutination (HA) assay (18), as well as molecular methods, like PCR (7, 14, 16), were used in previous research to determine the persistence of influenza A virus. While CPE and plaque assays can extrapolate infectivity and viability, HA assays and PCR cannot, since surface proteins and RNA can be present even after the virus is no longer viable. Our study used a tissue culture-based ELISA, which helped improve the sensitivity and removed subjectivity from interpretation of the results, as it is a direct measure of viral presence and infectivity by immunofluorescence. An analysis and/or side-by-side evaluation of methods for the recovery and determination of infectivity of influenza virus was not the focus of this study, although such an analysis would be a significant contribution to this field.
It is important to note that our study evaluated the viral persistence of different influenza virus strains in various sample matrices and environmental conditions on a nonporous surface. These combined parameters have not been previously investigated using a tissue culture-based ELISA. Although the two strains chosen for use in this study no longer circulate in humans, they act as a surrogate for the pandemic strain, which has been shown to have survival characteristics similar to those of the strains used in this study (22). Previous studies of virus persistence on environmental surfaces have found that various strains of influenza A (H1N1) virus remain infectious on stainless steel surfaces for approximately 24 h but no longer than 72 h (15–19). Inactivation studies found a minimal decrease in infectious influenza A (H1N1) virus particles on stainless steel surfaces at room temperature and 50 to 60% RH (33) but that the virus was rapidly inactivated under conditions of high heat (temperature range, 55 to 65°C) and humidity (RH range, 25 to 75%) (34). Initially, it was indicated that temperature and RH are key factors that influence viral persistence and transmission (35, 36) and that influenza virus has higher levels of persistence and increased rates of transmission at lower temperatures and RHs (29, 34–37). Recent studies have found that AH, which is an actual measure of the water vapor content of air, is better suited for determining the influence of environmental factors on viral persistence (34, 38, 39).
Overall, our study showed that two influenza A (H1N1) virus strains tested under a range of AH conditions that represent the different conditions of indoor and outdoor settings remained infectious on SS for at least 7 days, with less than a 2-log10 decrease in recovery, as measured by infectivity, being found. Further research is needed to determine the role of contaminated environmental surfaces on the transmission of influenza A virus so that improved infection control guidelines can be devised and implemented.
ACKNOWLEDGMENTS
This study was made possible by the CDC Influenza Coordination Unit. We thank Leslie Rios, Ryan Fitzgerald for laboratory assistance with this project, Jorn Winter from the CDC Influenza Division for providing protocols, and Krystle Agans from the Lovelace Respiratory Research Institute for providing strains and guidance.
The findings and conclusions in this paper are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
REFERENCES
- 1.WHO. 2003. Influenza fact sheet no. 211. WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/2003/fs211/en/. [Google Scholar]
- 2.Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. 2004. Influenza-associated hospitalizations in the United States. JAMA 292:1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- 3.CDC. 2010. Estimates of deaths associated with seasonal influenza—United States, 1976-2007. MMWR Morb Mortal Wkly Rep 59:1057–1062. http://www.cdc.gov/MMWR/preview/mmwrhtml/mm5933a1.htm. [PubMed] [Google Scholar]
- 4.Molinari N-AM, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB. 2007. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25:5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 5.Brankston G, Gitterman L, Hirji Z, Lemieux C, Gardam M. 2007. Transmission of influenza A in human beings. Lancet Infect Dis 7:257–265. doi: 10.1016/S1473-3099(07)70029-4. [DOI] [PubMed] [Google Scholar]
- 6.Weber TP, Stilianakis NI. 2008. Inactivation of influenza A viruses in the environment and modes of transmission: a critical review. J Infect 57:361–373. doi: 10.1016/j.jinf.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macias AE, de la Torre A, Moreno-Espinosa S, Leal PE, Bourlon MT, Ruiz-Palacios GM. 2009. Controlling the novel A (H1N1) influenza virus: don't touch your face! J Hosp Infect 73:280–281. doi: 10.1016/j.jhin.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 8.Boone SA, Gerba CP. 2007. Significance of fomites in the spread of respiratory and enteric viral disease. Appl Environ Microbiol 73:1687–1696. doi: 10.1128/AEM.02051-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CDC. 2003. Guidelines for environmental infection control in health-care facilities: recommendations of CDC and the Healthcare Infection Control Practices Advisory Committee (HICPAC). CDC, Atlanta, GA: http://www.cdc.gov/MMWR/preview/mmwrhtml/rr5210a1.htm. [PubMed] [Google Scholar]
- 10.CDC. 2013. Prevention strategies for seasonal influenza in healthcare settings. CDC, Atlanta, GA: http://www.cdc.gov/flu/professionals/infectioncontrol/healthcaresettings.htm. [Google Scholar]
- 11.Beckman S, Materna B, Goldmacher S, Zipprich J, D'Alessandro M, Novak D, Harrison R. 2013. Evaluation of respiratory protection programs and practices in California hospitals during the 2009-2010 H1N1 influenza pandemic. Am J Infect Control 41:1024–1031. doi: 10.1016/j.ajic.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell R, Roth V, Gravel D, Astrakianakis G, Bryce E, Forgie S, Johnston L, Taylor G, Vearncombe M, Canadian Nosocomial Infection Surveillance Program . 2013. Are health care workers protected? An observational study of selection and removal of personal protective equipment in Canadian acute care hospitals. Am J Infect Control 41:240–244. doi: 10.1016/j.ajic.2012.04.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raboud J, Shigayeva A, McGeer A, Bontovics E, Chapman M, Gravel D, Henry B, Lapinsky S, Loeb M, McDonald LC, Ofner M, Paton S, Reynolds D, Scales D, Shen S, Simor A, Stewart T, Vearncombe M, Zoutman D, Green K. 2010. Risk factors for SARS transmission from patients requiring intubation: a multicentre investigation in Toronto, Canada. PLoS One 5:e10717. doi: 10.1371/journal.pone.0010717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boone SA, Gerba CP. 2005. The occurrence of influenza A virus on household and day care center fomites. J Infect 51:103–109. doi: 10.1016/j.jinf.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Bean B, Moore BM, Sterner B, Peterson LR, Gerding DN, Balfour HH. 1982. Survival of influenza viruses on environmental surfaces. J Infect Dis 146:47–51. doi: 10.1093/infdis/146.1.47. [DOI] [PubMed] [Google Scholar]
- 16.Greatorex JS, Digard P, Curran MD, Moynihan R, Wensley H, Wreghitt T, Varsani H, Garcia F, Enstone J, Nguyen-Van-Tam JS. 2011. Survival of influenza A (H1N1) on materials found in households: implications for infection control. PLoS One 6:e27932. doi: 10.1371/journal.pone.0027932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee DV, Cohen B, Bovino ME, Desai S, Whittier S, Larson EL. 2012. Survival of influenza virus on hands and fomites in community and laboratory settings. Am J Infect Control 40:590–594. doi: 10.1016/j.ajic.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Oxford J, Berezin EN, Courvalin P, Dwyer DE, Exner M, Jana LA, Kaku M, Lee C, Letlape K, Low DE, Madani TA, Rubino JR, Saini N, Schoub BD, Signorelli C, Tierno PM, Zhong X. 2014. The survival of influenza A (H1N1) pdm09 virus on 4 household surfaces. Am J Infect Control 42:423–425. doi: 10.1016/j.ajic.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Sakaguchi H, Wada K, Kajioka J, Watanabe M, Nakano R, Hirose T, Ohta H, Aizawa Y. 2010. Maintenance of influenza virus infectivity on the surfaces of personal protective equipment and clothing used in healthcare settings. Environ Health Prev Med 15:344–349. doi: 10.1007/s12199-010-0149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Influenza Research Database. 2016. World Health Organization recommendations for composition of influenza vaccines. Influenza Research Database, Falls Church, VA: http://www.fludb.org/brc/vaccineRecommend.spg?decorator=influenza. [Google Scholar]
- 21.Donlan RM, Piede JA, Heyes CD, Sanii L, Murga R, Edmonds P, El-Sayed I, El-Sayed MA. 2004. Model system for growing and quantifying Streptococcus pneumoniae biofilms in situ and in real time. Appl Environ Microbiol 70:4980–4988. doi: 10.1128/AEM.70.8.4980-4988.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coulliette AD, Perry KA, Edwards JR, Noble-Wang JA. 2013. Persistence of the 2009 pandemic influenza A (H1N1) virus on N95 respirators. Appl Environ Microbiol 79:2148–2155. doi: 10.1128/AEM.03850-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 24.Diggle PJ, Heagerty P, Liang KY, Zeger SL. 2002. Analysis of longitudinal data. Oxford Statistical Science Series. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 25.Parker ER, Dunham WB, Mac Neal WJ. 1944. Resistance of the Melbourne strain of influenza virus to desiccation. J Lab Clin Med 29:37–42. [Google Scholar]
- 26.Thomas Y, Vogel G, Wunderli W, Suter P, Witschi M, Koch D, Tapparel C, Kaiser L. 2008. Survival of influenza virus on banknotes. Appl Environ Microbiol 74:3002–3007. doi: 10.1128/AEM.00076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiffany JM, Blough HA. 1970. Models of structure of the envelope of influenza virus. Proc Natl Acad Sci U S A 65:1105–1112. doi: 10.1073/pnas.65.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Assar SK, Block SS. 2001. Survival of microorganisms in the environment, p 1221–1242. In Block SS. (ed), Disinfection, sterilization, and preservation, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 30.Blough HA, Merlie JP, Tiffany JM. 1969. The fatty acid composition of incomplete influenza virus. Biochem Biophys Res Commun 34:831–834. doi: 10.1016/0006-291X(69)90255-1. [DOI] [PubMed] [Google Scholar]
- 31.Blough HA. 1971. Fatty acid composition of individual phospholipids of influenza virus. J Gen Virol 12:317–320. doi: 10.1099/0022-1317-12-3-317. [DOI] [PubMed] [Google Scholar]
- 32.Edward DF. 1941. Resistance of influenza virus to drying and its demonstration on dust. Lancet 238:664–666. doi: 10.1016/S0140-6736(00)72189-1. [DOI] [Google Scholar]
- 33.Noyce JO, Michels H, Keevil CW. 2007. Inactivation of influenza A virus on copper versus stainless steel surfaces. Appl Environ Microbiol 73:2748–2750. doi: 10.1128/AEM.01139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDevitt J, Rudnick S, First M, Spengler J. 2010. Role of absolute humidity in the inactivation of influenza viruses on stainless steel surfaces at elevated temperatures. Appl Environ Microbiol 76:3943–3947. doi: 10.1128/AEM.02674-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemmes JH, Winkler KC, Kool SM. 1960. Virus survival as a seasonal factor in influenza and poliomyelitis. Nature 188:430–431. doi: 10.1038/188430a0. [DOI] [PubMed] [Google Scholar]
- 36.Buckland FE, Tyrrell DAJ. 1962. Loss of infectivity on drying various viruses. Nature 195:1063–1064. doi: 10.1038/1951063a0. [DOI] [PubMed] [Google Scholar]
- 37.Lowen AC, Mubareka S, Steel J, Palese P. 2007. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog 3:e151. doi: 10.1371/journal.ppat.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaman J, Kohn M. 2009. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc Natl Acad Sci U S A 106:3243–3248. doi: 10.1073/pnas.0806852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaman J, Pitzer VE, Viboud C, Grenfell BT, Lipsitch M. 2010. Absolute humidity and the seasonal onset of influenza in the continental United States. PLoS Biol 8:e1000316. doi: 10.1371/journal.pbio.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]