ABSTRACT
There is a growing recognition of the roles of marine microenvironments as reservoirs of biodiversity and as sites of enhanced biological activity and in facilitating biological interactions. Here, we examine the bacterial community inhabiting free-living and particle-associated seawater microenvironments at the Pivers Island Coastal Observatory (PICO). 16S rRNA gene libraries from monthly samples (July 2013 to August 2014) were used to identify microbes in seawater in four size fractions: >63 μm (zooplankton and large particles), 63 to 5 μm (particles), 5 to 1 μm (small particles/dividing cells), and <1 μm (free-living prokaryotes). Analyses of microbial community composition highlight the importance of the microhabitat (e.g., particle-associated versus free-living lifestyle) as communities cluster by size fraction, and the microhabitat explains more of the community variability than measured environmental parameters, including pH, particle concentration, projected daily insolation, nutrients, and temperature. While temperature is statistically associated with community changes in the <1-μm and 5- to 1-μm fractions, none of the measured bulk seawater environmental variables are statistically significant in the larger-particle-associated fractions. These results, combined with high particle-associated community variability, especially in the largest size fraction (i.e., >63 μm), suggest that particle composition, including eukaryotes and their associated microbiomes, may be an important factor in selecting for specific particle-associated bacteria.
IMPORTANCE By comparing levels of particle-associated and free-living bacterial diversity at a coastal location over the course of 14 months, we show that bacteria associated with particles are generally more diverse and appear to be less responsive to commonly measured environmental variables than free-living bacteria. These diverse and highly variable particle-associated communities are likely driven by differences in particle substrates both within the water column at a single time point and due to seasonal changes over the course of the year.
INTRODUCTION
There is a growing interest in how microscale features, including living phytoplankton cells, phytoplankton exudate, and detrital particles, affect bacterial diversity and activity (1–3). Within these microenvironments, particles are relatively well studied as they can be readily physically separated from free-living bacteria using filtration. Thus, particles have been identified as an important habitat for estuarine and coastal bacteria, with up to 60% of water column bacterial cells attached to particles (4, 5), likely due to the presence of nutrients at levels up to 3 orders of magnitude higher than those found in bulk seawater (6). These high-nutrient microenvironments allow elevated bacterial production and organic matter degradation in a background of low levels of bulk nutrient (7–9). Moreover, particles with different compositions, e.g., living zooplankton versus exopolysaccharides, may represent a diverse range of microenvironments, all of which are distinct from the conditions experienced by free-living cells.
Although free-living and particle-associated bacteria are typically defined operationally based on filtration properties, these two populations have physiological, genomic, and phylogenetic differences (10–12). Particle-attached bacteria are more metabolically active, with higher levels of extracellular enzymes, adhesion proteins, and antagonistic compounds (12–15). Likely to take advantage of compositionally varied patches of organic matter, particle-associated bacteria also generally have larger genomes with a variety of metabolic and regulatory capabilities (1). In contrast, free-living bacteria usually have smaller genomes with streamlined metabolic and regulatory functions that enable efficient growth under oligotrophic conditions (16). Due to this physiological, genomic, and phylogenetic specialization, we might expect differences in particle-attached and free-living bacterial communities.
However, whether particles harbor a distinct microbial community is still controversial, as some researchers have reported a high degree of microenvironmental specialization (11, 17–20), while others observed significant overlap of the free-living and attached bacterial communities (21–23). Taxa found in both environments could be generalists but potentially exhibit fine-scale phylogenetic niche partitioning that cannot be resolved at the 16S rRNA gene level (24). Additionally, associations with specific microenvironments exist in a background of seasonally and episodically changing environmental conditions that also select for specific bacterial communities (25–27). Compared to the conditions encountered by free-living bacteria, attachment and the high-nutrient environment of particles may help to buffer the impacts of changes in bulk seawater environmental parameters such as temperature or dissolved nutrients (28, 29), potentially resulting in greater temporal uniformity in the particle-attached microbial community.
In order to examine variation in particle-attached and free-living marine bacteria over an annual cycle, we sampled microbial communities at the Pivers Island Coastal Observatory (PICO), a temperate, coastal time series with strong annual patterns in environmental parameters (30). We measured a suite of environmental variables and collected three sizes of particles as well as free-living bacteria monthly for over a year. We then assessed the microbial communities in these samples using 16S rRNA gene fragment libraries. This repeated sampling enables us to link bacterial communities and environmental variables to investigate drivers of the observed diversity patterns.
MATERIALS AND METHODS
Sample collection.
Water samples were collected as part of the Pivers Island Coastal Observatory (PICO) time series adjacent to the Beaufort Inlet, Beaufort, NC, USA (34°71.81′N, 76°67.08′W). Three years of weekly samples were collected from January 2011 to December 2013, while size-fractionated samples were collected monthly from 3 July 2013 to 6 August 2014. Time-series measurements were conducted using water collected with a Niskin bottle centered at 1 m (30, 31). Methods for determination of water temperature, pH, salinity, dissolved inorganic nutrient concentrations, and chlorophyll a concentrations as well as bacterioplankton and phytoplankton abundances were described previously (30, 32). All field measurements consisted of at least two replicate samples. The particle size distribution (1 to 30 μm) was measured using a Coulter Counter (Beckman Coulter MultiSizer III). To characterize particle-associated and free-living microbial communities, the large-particle-associated bacterial communities (>63 μm), small-particle-associated bacterial communities (63 to 5 μm), ambiguous bacterial communities (5 to 1 μm), and free-living bacterial communities (<1 μm) were separated by sequential gravity filtration using a 63-μm-pore-size plankton net followed by 5-μm-pore-size (Advantec MFS), 1-μm-pore-size (GE Water & Process Technologies), and 0.2-μm-pore-size (Pall Supor-200) filters. Large particles collected on the 63-μm-pore-size plankton net from 10 liters of seawater were resuspended in autoclaved artificial seawater and concentrated on 0.22-μm-pore-size filters. Filtered volumes were chosen such that the filtration rate did not become visibly lower during collection, and filters were rinsed with autoclaved artificial seawater to remove additional material smaller than the filter pore size (see Table S1 in the supplemental material). All filters were stored at −80°C until DNA extraction.
DNA extraction and library preparation.
We extracted DNA from the size-fractionated samples using a Puregene Yeast/Bacteria kit (Qiagen) according to the manufacturer's instructions supplemented with three rounds (60 s each) of bead beating. Microbial communities were characterized using a dual index sequencing approach (33) with the following portions of the primers targeting the V3-to-V4 region of the bacterial and archaeal 16S rRNA genes: for 16S F V3, CCTACGGGNGGCWSCAG; and for 16S R V4, GGACTACNVGGGTWTCTAAT (34). PCR mixtures contained 0.4 U of Q5 DNA polymerase (NEB) as well as a final concentration of 200 μM deoxynucleoside triphosphates (dNTPs), 2 mM MgCl2, and 0.5 μM primers. PCRs were performed using thermocycling with the following protocol: 98°C for 30 s followed by 35 cycles at 98°C for 10 s, 55°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 2 min. Triplicate reaction mixtures per sample were pooled and gel purified. Paired-end 250-bp sequencing of barcoded amplicons was performed on an Illumina MiSeq running v2 chemistry at the Duke Center for Genomic and Computational Biology.
Sequence processing.
We used USEARCH v7 for quality control of sequence reads and to merge paired-end reads and then performed operational taxonomic unit (OTU) clustering using the UPARSE algorithm (35). We first trimmed low-quality bases from the sequences using a 10-nucleotide (nt) window with a Q30 running-quality threshold. Paired-end sequences with a ≥10-nt overlap and no mismatches were then merged. We performed a final filtering step to discard low-quality merged sequences with a length of <400 bp and/or a maximum expected error of >1, which resulted in 2,358,138 reads. Merged sequences were clustered into OTUs of 98.5% pairwise identity to the centroid, resulting in OTUs in which all members were at least 97% similar. Taxonomies were assigned to the most abundant sequence in each OTU with a confidence threshold score of 0.8 using RDP Classifier (36). Rare OTUs (<5 total observations) and OTUs matching mitochondrial or plastid sequences based on RDP Classifier assignment were excluded, resulting in 1,479,742 total sequences and 12,373 OTUs. Libraries contained between 10,941 and 42,618 sequences; to control for uneven sequencing effort, we normalized the data by rarefaction to 10,941 sequences per sample.
Microbial community analysis.
For each sample, all alpha diversity calculations and error estimates were averaged over five rarefactions using the “vegan” package in R, including estimates of Shannon's diversity (37). Community compositions were compared for each pair of samples using weighted Bray-Curtis dissimilarity in MacQIIME 1.8.0 (38) and visualized using nonmetric multidimensional scaling (NMDS) ordination using the “ggplot2” R package. Analysis of similarity (ANOSIM) was used to determine the dissimilarity between the bacterial communities from each size fraction (39). Variance partitioning analysis (VPA) assessed the contributions of environmental variables (projected daily insolation, chlorophyll a, turbidity, salinity, temperature, silicate, ammonia, phosphate, and pH) and microhabitats (>63 μm, 63 to 5 μm, 5 to 1 μm, and <1 μm) to the microbial community variability (37). Canonical correspondence analysis (CCA) was then used to identify the environmental factors associated with community composition changes (40). Prior to the CCA, the “step” function in vegan (37) was used to automatically select constraints in the model using forward stepwise selection and Akaike's information criterion with 999 permutation tests at each step. We repeated the forward stepwise selection with the bacterial community from each size fraction. As no terms were statistically significant in the stepwise selection for the >63-μm and 63- to 5-μm communities, we applied the variable set for the <1-μm fraction (temperature, projected no-sky daily insolation, silicate, pH, and particle number) to all four size fractions using the “cca” function in vegan (37). To evaluate the significance of the canonical axes, we used a Monte Carlo test with 999 permutations. Statistical significance of the individual environmental variables was assessed using the marginal effect of the terms. We then used the R package “indicspecies” to determine the strength of the association between taxa and the different size fractions (41). To focus on key indicator taxa, OTUs were identified that were significantly associated with a particular size fraction (P < 0.01) and had an average relative abundance of at least 0.05% across all samples in that size fraction.
Accession numbers.
16S rRNA gene sequences were deposited in the NCBI Sequence Read Archive under accession numbers SRP068349 and PRJNA309156. Environmental metadata are available as part of the Pivers Island Coastal Observatory (PICO) database through BCO-DMO project 2281.
RESULTS AND DISCUSSION
Here, we examined particle-associated and free-living microbial communities from coastal seawater as part of the Pivers Island Coastal Observatory (PICO) time series. We sampled monthly for over 1 year (July 2013 to August 2014) in order to understand the temporal dynamics of particulate and free-living bacterial communities in relation to environmental variables. We sequentially filtered seawater to produce size fractions which corresponded roughly to zooplankton, large particles, and debris (>63 μm); to particles and phytoplankton (63 to 5 μm); to small particles and large/dividing cells (5 to 1 μm); and to free-living bacteria (<1 μm) (24). Although the 5-to-1-μm size fraction is somewhat ambiguous, it serves to clearly separate truly particle-attached (>5 μm) and free-living bacteria. To identify the microbial communities present, we sequenced the V3-to-V4 region of the 16S rRNA gene and clustered OTUs at 97% similarity using UPARSE (35) and rarefied each library to 10,941 reads using QIIME (38).
Over the 14 months of sampling, Shannon's diversity (H′) was generally highest in the two largest size fractions (>63 and 63 to 5 μm; Fig. 1). Unlike the case in smaller size fractions (5 to 1 μm and <1 μm), rarefaction curves do not show saturation in these larger size fractions, suggesting that we did not capture the full diversity of particle-attached communities (see Fig. S1 in the supplemental material). Nevertheless, the two particulate size fractions (>63 μm and 63 to 5 μm) were consistently more diverse than the free-living bacterial community (P < 0.05; ANOVA) (Fig. 1). The >63-μm (mean H′, 5.92; standard deviation [SD], 1.04) and 63- to 5-μm (mean H′, 6.23; SD, 0.35) bacterial communities were also more diverse than the whole-seawater bacterial communities from 3 years of weekly sampling at this study site (mean H′, 4.97; SD, 0.41). A large portion of the bacterial taxa observed in the >63-μm (45.5% of OTUs; 29.9% of sequences) and 63-to-5-μm (34.0% of OTUs; 7.9% of sequences) fractions were never detected in the weekly time series. In contrast, 85.3% of OTUs and 99.4% of sequences observed in the <1-μm fraction were present in the weekly samples. Thus, even long-term sampling may not capture the full diversity of relatively rare particle-associated microbes.
FIG 1.

Plot of the Shannon's diversity for 16S rRNA gene libraries from different seawater size fractions (>63 μm, 63 to 5 μm, 5 to 1 μm, and <1 μm) for 14 monthly samples from the Pivers Island Coastal Observatory (PICO) time series (July 2013 to August 2014). Points represent mean values and error bars 1 standard deviation of Shannon's diversity computed based on five replicates. Jan, January; Feb, February; Mar, March; Apr, April; Jun, June; Jul, July; Aug, August; Sep, September; Oct, October; Nov, November; Dec, December.
Comparing the bacterial communities using nonmetric multidimensional scaling (NMDS) shows that the samples clustered by size fraction (Fig. 2A) and that all size fractions were statistically distinct (see Table S2 in the supplemental material). Thus, although these size divisions are operational, they harbor distinct microbial communities. As in many coastal sites, the Alphaproteobacteria, Gammaproteobacteria, and Bacteriodetes were the major phyla in all fractions, representing at least 34.8% of total reads in each library (Fig. 2B). The free-living communities were dominated by Alphaproteobacteria (23.8% to 55.2% of libraries; Fig. 2B), including Rhodobacterales (6.9% to 17.4%) and “Candidatus Pelagibacter” (SAR11) (4.1% to 36.2%), while the proteobacteria in the >63-μm and 63- to 5-μm fractions were largely Gammaproteobacteria and Deltaproteobacteria. Thus, despite some overall similarities, marine microenvironments do contain distinct microbial communities.
FIG 2.

(A) Nonmetric multidimensional scaling (NMDS) ordination computed based on weighted Bray-Curtis dissimilarity for 16S rRNA gene libraries from seawater size fractions of >63 μm, 63 to 5 μm, 5 to 1 μm, and <1 μm from PICO monthly samples (July 2013 to August 2014). Each symbol corresponds to a distinct time point and size fraction. Samples marked with asterisks were visual outliers in March and July 2014. Ellipses represent 95% intervals around centroids for each size fraction. 2D, two dimensional. (B) Bar plot showing the average phylum-level contribution for each fraction. Phyla that on average comprised less than 0.05% of the libraries are grouped as “Other Bacteria.”
As noted previously, the temporal variability in both the diversity and composition of the largest (>63-μm) size fraction was much higher than of other microenvironments (Fig. 1 and 2) and the within-fraction Bray-Curtis dissimilarity was significantly larger than observed in other size fractions (P < 0.001 [two-tailed t test]). This increased variability might have been due to changes in particle abundance or composition (see Fig. S2 in the supplemental material) (12, 42). However, we did not find a correlation between the number of particles (1 to 30 μm in size) and the community composition, suggesting that particle composition or origin may be more important than abundance. We also note that the 10-liter volumes sampled for the >63-μm libraries were much larger than those collected for other size fractions (see Table S1); thus, the >63-μm libraries integrated over a number of discrete particles and likely different particle types. Although the other size fractions were sampled at much lower volumes, smaller particles and free-living bacteria are likely more spatially homogeneous; thus, smaller volumes may adequately capture the diversity present in these size fractions (43). Even though particles within a single sampling point likely represent a broad spectrum of habitats, their microbial communities are also more variable over time than free-living bacterial communities.
Even with high month-to-month variability, the >63-μm fraction exhibited notable outliers in May and July 2014, when the bacterial community composition diverged in the NMDS (indicated with asterisks in Fig. 2) and also decreased in Shannon's diversity (Fig. 1). These time periods did not correspond to any unusual disturbance as measured by our time series or local weather stations. In fact, a large storm in August 2014 which produced high particle loads, decreased salinity, and increased nutrient concentrations did not result in a notably distinct community (Fig. 2; see also Table S3 in the supplemental material). This lack of particulate community response to a disturbance such as a storm suggests that these divergent communities likely represent changes in particle types (including organisms) rather than the resuspension of particles from the benthos or export from land. Examining the bacterial communities of these outliers, the most abundant OTU in the May 2014 >63-μm sample belonged to the genus Staphylococcus (10% of total sequence reads). This genus is best known for containing human pathogens, but Staphylococcus species have also been found on copepods (3). In contrast, the July 2014 sample contained a dominant OTU from the order Thiohalorhabdales which comprised 23% of the total reads; this group is common on marine particles (43, 44) and has been found in association with sponges (45), suggesting a potential association with a eukaryotic host. As many families present in the >63-μm fraction, including Vibrionaceae, Alteromonadaceae, and Staphylococcaceae, were previously detected in the copepod microbiome (3), associations with specific copepods or other marine organisms, which vary in their prevalences, could explain the observed patterns. In this system, associations with specific particle types, likely eukaryotic hosts, rather than with particles with distinct geographic origins may explain some of the dramatic changes in the particle-associated bacterial community. However, as microbial community changes are often linked to environmental variables such as chlorophyll, temperature, etc., we wanted to assess the potential of these variables to influence the community composition in different size fractions.
Although bacterial communities cluster by size fraction (Fig. 2A), a microbial community within a given size fraction may respond to environmental variables such as light, temperature, and nutrients (25, 27, 46). However, across all samples, variance partitioning indicates that the microhabitat (>63 μm, 63 to 5 μm, 5 to 1 μm, and <1 μm; 50.4% of variation explained) plays a more important role than environmental factors (levels of projected daily insolation, chlorophyll a, turbidity, salinity, temperature, silicate, ammonia, phosphate, and pH; 11.9% of variation explained) in structuring size-fractionated bacterial communities. While measured seawater environmental factors do not have high explanatory power for the variability in size-fractionated bacterial communities, we postulate that free-living bacteria are likely more responsive to bulk-water properties than particle-attached bacteria, due to their exposure to and presumable reliance on these resources. Indeed, much of the microbial community variation for free-living bacteria is explained in the first two canonical correspondence analysis (CCA) axes (47.8%; Fig. 3D), but these CCA axes explain less variability as the size fraction increases (Fig. 3A to C). For the <1-μm and 5-to-1-μm fractions, temperature is the strongest environmental driver of bacterial composition (marginal effect of the terms; P < 0.01). Yet none of the measured environmental variables was statistically significant for the >63-μm and 63- to 5-μm fractions. Previous studies have shown that surface attachment and nutrients may protect bacteria from environmental variability, including temperature extremes (4, 47, 48). Thus, particle-associated communities may be protected from some environmental conditions or may respond to factors not measured here, including physical substrates, refuges from predation, higher nutrient levels, or unique particulate chemical constituents/conditions not found in the water column. Since the particle-attached bacterial community exhibits high temporal variability despite weakened sensitivity to bulk water environmental parameters, we hypothesize that community composition is most influenced by temporal changes in particle types.
FIG 3.
Canonical correspondence analysis (CCA) ordination diagram of the first two axes for size-fractionated bacterial community samples from the PICO time series. The percentage of the variation in the size-fractionated community explained by each axis is indicated in parentheses after the axis label. The constrained sets of environmental variables analyzed are indicated as vectors: temperature, projected no-sky daily insolation (Insolation), silicate, pH, and number of particles 1 to 30 μm in diameter (Particles). The size-fractionated community profiles from each time point are represented as circles; the gradient colors of the circles indicate times of year. Environmental variables marked with asterisks are statistically significant as assessed by the marginal effect of the terms (P < 0.05). (A) >63 μm. (B) 63 to 5 μm. (C) 5 to 1 μm. (D) <1 μm.
As particle-associated and free-living bacteria communities appear to differentially respond to bulk seawater changes, we wanted to better understand similarities and differences in the microbial community composition in these habitats. We observed size fraction-based OTU specialization; across our entire time series, adjacent size fractions have more OTUs in common (10.5% to 46.6%) than nonadjacent fractions (8.5% to 26.9%; Fig. 4). In addition, comparing the particle-associated and free-living bacterial fractions, the number of OTUs exclusive to a specific fraction was higher in the >63-μm fraction (24.3%) and the 63- to 5-μm fraction (18.7%) than in the <1-μm fraction (2.8%; Fig. 4A). Thus, free-living bacteria were often found in the larger size fractions but particulate OTUs were not generally found in the smallest size fraction. In a previous study, a large overlap of particle-attached and free-living bacteria was observed; 25% of the OTUs, comprising 94% of sequence reads, were found in both the particle-attached and the free-living size fractions (22). We similarly observed that a higher percentage of sequence reads than OTUs was shared across multiple size fractions, indicating that the most abundant taxa are shared across different size fractions (Fig. 4; see also Table S4 in the supplemental material). OTUs found in multiple size fractions can be explained by several mechanisms: bacterial taxa can live in several habitats or habitats that span multiple size fractions, or closely related taxa can exhibit sub-OTU-level habitat specialization (1, 24, 31). Although we carefully chose the filtered volumes to prevent clogging and rinsed the filters with autoclaved artificial seawater to remove additional material smaller than the filter pore size, we cannot rule out the possibility that the sampling process could introduce cells into other size fractions. Our data obtained here, with limited OTU overlap of fractions, suggest that particle-associated bacteria are often microenvironmental specialists but that abundant taxa are often shared between different size fractions. We also identified indicative taxa using associations between the relative abundance of an OTU and a particular size fraction (see Table S5). Consistent with other studies, many of the indicative OTUs in the free-living fraction (<1 μm) belonged to “Ca. Pelagibacter” (SAR11), a group of small, free-living bacteria. In contrast, the 63- to 5-μm fraction contains indicative OTUs belonging to the Planctomycetes, Bacteroidetes, and Verrucomicrobia, while many of the indicative phylotypes in the >63-μm size fraction belong to the anoxygenic photosynthetic Chromatiales and sulfate-reducing Desulfobacterales. The presence of these bacteria, which thrive in reduced environments, suggests the existence of anoxic microzones in larger particles and zooplankton digestive tracts (49). Thus, although there is some overlap of the taxa present in each size fraction, there are specific taxa which define each size fraction over a seasonal cycle that may offer clues to the lifestyles of organisms in these marine microenvironments.
FIG 4.
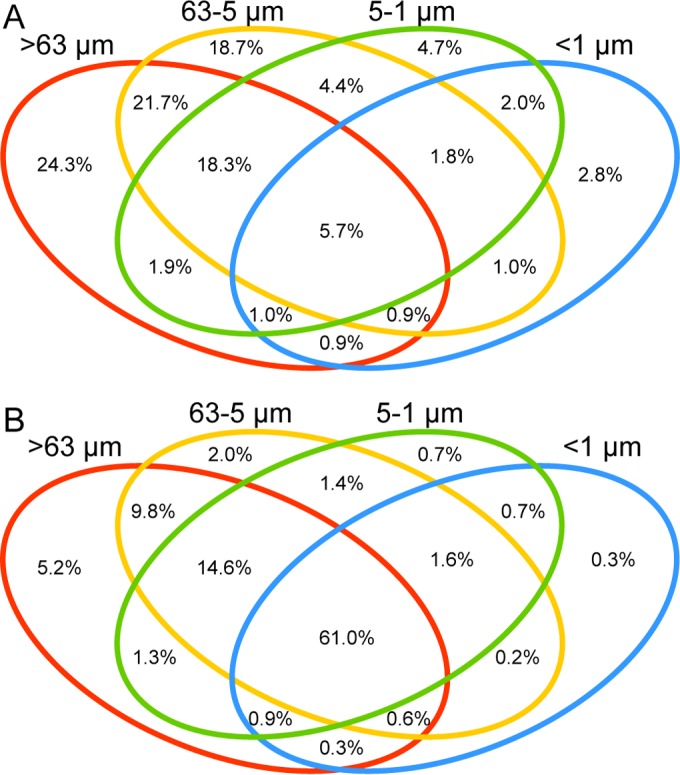
Venn diagrams showing comparisons between size fractions >63 μm, 63 to 5 μm, 5 to 1 μm, and <1 μm averaged across all time points. (A) The percentages of shared and unique operational taxonomic units (OTUs; 97% similarity) for all size fractions. (B) The percentages of total sequence present in shared and unique OTUs for all size fractions.
Conclusions.
Our sequential filtration approach reveals that over an annual cycle, particle-attached bacteria are on average both more diverse and more variable over time than free-living bacteria. However, particle-attached bacterial dynamics are not associated with commonly measured environmental variables, suggesting there are a number of unknown drivers of particle-attached bacterial diversity, potentially including associations with metazoans or other microbes and/or other differences in particle composition (50). The high bacterial community variability on large particles (>63 μm) along with taxa that are associated with anoxic conditions and/or eukaryotes suggests that the specific particulate substrate is important (42, 51). Due to the important role of particle-attached bacteria in biogeochemical cycling of carbon and other nutrients, it is critical to understand the dynamics and environmental dependence of bacteria associated with these water column microenvironments.
Supplementary Material
ACKNOWLEDGMENTS
We thank the entire PICO team for help with sampling. We further thank Patricia Tester and Rance Hardison for use of and assistance with the Coulter Counter.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00395-16.
REFERENCES
- 1.Yawata Y, Cordero OX, Menolascina F, Hehemann J-H, Polz MF, Stocker R. 2014. Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci U S A 111:5622–5627. doi: 10.1073/pnas.1318943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ, Dearth SP, Van Mooy BA, Campagna SR, Kujawinski EB, Armbrust EV, Moran MA. 2015. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci U S A 112:453–457. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerdts G, Brandt P, Kreisel K, Boersma M, Schoo K, Wichels A. 2013. The microbiome of North Sea copepods. Helgoland Mar Res 67:757–773. doi: 10.1007/s10152-013-0361-4. [DOI] [Google Scholar]
- 4.Crump BC, Baross JA, Simenstad CA. 1998. Dominance of particle-attached bacteria in the Columbia River estuary, USA. Aquat Microb Ecol 14:7–18. doi: 10.3354/ame014007. [DOI] [Google Scholar]
- 5.Garneau M-È, Vincent WF, Terrado R, Lovejoy C. 2009. Importance of particle-associated bacterial heterotrophy in a coastal Arctic ecosystem. J Mar Syst 75:185–197. doi: 10.1016/j.jmarsys.2008.09.002. [DOI] [Google Scholar]
- 6.Blackburn N, Fenchel T. 1999. Influence of bacteria, diffusion and sheer on micro-scale nutrient patches, and implications for bacterial chemotaxis. Mar Ecol Prog Ser 189:1–7. doi: 10.3354/meps189001. [DOI] [Google Scholar]
- 7.Azam F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694–696. doi: 10.1126/science.280.5364.694. [DOI] [Google Scholar]
- 8.Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF. 2008. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci U S A 105:4209–4214. doi: 10.1073/pnas.0709765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stocker R. 2012. Marine microbes see a sea of gradients. Science 338:628–633. doi: 10.1126/science.1208929. [DOI] [PubMed] [Google Scholar]
- 10.Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, DeMaere MZ, Ting L, Ertan H, Johnson J, Ferriera S, Lapidus A, Anderson I, Kyrpides N, Munk AC, Detter C, Han CS, Brown MV, Robb FT, Kjelleberg S, Cavicchioli R. 2009. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci U S A 106:15527–15533. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong EF, Franks DG, Alldredge AL. 1993. Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr 38:924–934. doi: 10.4319/lo.1993.38.5.0924. [DOI] [Google Scholar]
- 12.Karner M, Herndl GJ. 1992. Extracellular enzymatic-activity and secondary production in free-living and marine-snow-associated bacteria. Mar Biol 113:341–347. [Google Scholar]
- 13.Smith DC, Simon M, Alldredge AL, Azam F. 1992. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359:139–141. doi: 10.1038/359139a0. [DOI] [Google Scholar]
- 14.Grossart H-P, Tang KW, Kiørboe T, Ploug H. 2007. Comparison of cell-specific activity between free-living and attached bacteria using isolates and natural assemblages. FEMS Microbiol Lett 266:194–200. doi: 10.1111/j.1574-6968.2006.00520.x. [DOI] [PubMed] [Google Scholar]
- 15.Long RA, Azam F. 2001. Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol 67:4975–4983. doi: 10.1128/AEM.67.11.4975-4983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polz MF, Hunt DE, Preheim SP, Weinreich DM. 2006. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos Trans R Soc Lond B Biol Sci 361:2009–2021. doi: 10.1098/rstb.2006.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allgaier M, Brückner S, Jaspers E, Grossart HP. 2007. Intra- and inter-lake variability of free-living and particle-associated Actinobacteria communities. Environ Microbiol 9:2728–2741. doi: 10.1111/j.1462-2920.2007.01385.x. [DOI] [PubMed] [Google Scholar]
- 18.Moeseneder MM, Winter C, Herndl GJ. 2001. Horizontal and vertical complexity of attached and free-living bacteria of the eastern Mediterranean Sea, determined by 16S rDNA and 16S rRNA fingerprints. Limnol Oceanogr 46:95–107. doi: 10.4319/lo.2001.46.1.0095. [DOI] [Google Scholar]
- 19.Acinas SG, Anton J, Rodriguez-Valera F. 1999. Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol 65:514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith MW, Allen LZ, Allen AE, Herfort L, Simon HM. 30 May 2013. Contrasting genomic properties of free-living and particle-attached microbial assemblages within a coastal ecosystem. Front Microbiol doi: 10.3389/fmicb.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghiglione J, Mevel G, Pujo-Pay M, Mousseau L, Lebaron P, Goutx M. 2007. Diel and seasonal variations in abundance, activity, and community structure of particle-attached and free-living bacteria in NW Mediterranean Sea. Microb Ecol 54:217–231. doi: 10.1007/s00248-006-9189-7. [DOI] [PubMed] [Google Scholar]
- 22.Crespo BG, Pommier T, Fernández-Gómez B, Pedrós-Alió C. 2013. Taxonomic composition of the particle-attached and free-living bacterial assemblages in the Northwest Mediterranean Sea analyzed by pyrosequencing of the 16S rRNA. Microbiologyopen 2:541–552. doi: 10.1002/mbo3.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollibaugh JT, Wong PS, Murrell MC. 2000. Similarity of particle-associated and free-living bacterial communities in northern San Francisco Bay, California. Aquat Microb Ecol 21:103–114. doi: 10.3354/ame021103. [DOI] [Google Scholar]
- 24.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I. 2012. Defining seasonal marine microbial community dynamics. ISME J 6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cram JA, Chow C-ET, Sachdeva R, Needham DM, Parada AE, Steele JA, Fuhrman JA. 2015. Seasonal and interannual variability of the marine bacterioplankton community throughout the water column over ten years. ISME J 9:563–580. doi: 10.1038/ismej.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Swais H, Dunn KA, Bielawski JP, Li WK, Walsh DA. 2015. Seasonal assemblages and short-lived blooms in coastal north-west Atlantic Ocean bacterioplankton. Environ Microbiol 17:3642–3661. doi: 10.1111/1462-2920.12629. [DOI] [PubMed] [Google Scholar]
- 28.Reidl J, Klose KE. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol Rev 26:125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 29.Huq A, West P, Small E, Huq M, Colwell R. 1984. Influence of water temperature, salinity, and pH on survival and growth of toxigenic Vibrio cholerae serovar O1 associated with live copepods in laboratory microcosms. Appl Environ Microbiol 48:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson ZI, Wheeler BJ, Blinebry SK, Carlson CM, Ward CS, Hunt DE. 2013. Dramatic variability of the carbonate system at a temperate coastal ocean site (Beaufort, North Carolina, USA) is regulated by physical and biogeochemical processes on multiple timescales. PLoS One 8:e85117. doi: 10.1371/journal.pone.0085117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yung C-M, Vereen MK, Herbert A, Davis KM, Yang J, Kantorowska A, Ward CS, Wernegreen JJ, Johnson ZI, Hunt DE. 2015. Thermally adaptive tradeoffs in closely-related marine bacterial strains. Environ Microbiol 17:2421–2429. doi: 10.1111/1462-2920.12714. [DOI] [PubMed] [Google Scholar]
- 32.Hunt DE, Lin Y, Church MJ, Karl DM, Izzo LK, Tringe S, Johnson ZI. 2013. Relationship between abundance and specific activity of bacterioplankton in open ocean surface waters. Appl Environ Microbiol 79:177–184. doi: 10.1128/AEM.02155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hugoni M, Taib N, Debroas D, Domaizon I, Dufournel IJ, Bronner G, Salter I, Agogué H, Mary I, Galand PE. 2013. Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc Natl Acad Sci U S A 110:6004–6009. doi: 10.1073/pnas.1216863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q, Garrity G, Tiedje J, Cole J. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara R, Simpson GL, Solymos P, Henry M, Stevens H. 2012. Vegan: Community Ecology Package. R package version 2.1-14/r2120 https://cran.r-project.org/web/packages/vegan/index.html. [Google Scholar]
- 38.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–117. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 40.ter Braak CJ, Verdonschot PF. 1995. Canonical correspondence analysis and related multivariate methods in aquatic ecology. Aquat Sci 57:255–289. doi: 10.1007/BF00877430. [DOI] [Google Scholar]
- 41.De Cáceres M, Legendre P. 2009. Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
- 42.Bižić-Ionescu M, Zeder M, Ionescu D, Orlić S, Fuchs BM, Grossart HP, Amann R. 2015. Comparison of bacterial communities on limnic versus coastal marine particles reveals profound differences in colonization. Environ Microbiol 17:3500–3514. doi: 10.1111/1462-2920.12466. [DOI] [PubMed] [Google Scholar]
- 43.Padilla CC, Ganesh S, Gantt S, Huhman A, Parris DJ, Sarode N, Stewart FJ. 2015. Standard filtration practices may significantly distort planktonic microbial diversity estimates. Front Microbiol 6:547. doi: 10.3389/fmicb.2015.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ganesh S, Bristow LA, Larsen M, Sarode N, Thamdrup B, Stewart FJ. 2015. Size-fraction partitioning of community gene transcription and nitrogen metabolism in a marine oxygen minimum zone. ISME J 9:2682–2696. doi: 10.1038/ismej.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Voogd NJ, Cleary DF, Polónia AR, Gomes NC. 2015. Bacterial community composition and predicted functional ecology of sponges, sediment and seawater from the Thousand Islands reef complex, West Java, Indonesia. FEMS Microbiol Ecol 91:fiv019. doi: 10.1093/femsec/fiv019. [DOI] [PubMed] [Google Scholar]
- 46.Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. 2006. Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci U S A 103:13104–13109. doi: 10.1073/pnas.0602399103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amako K, Shimodori S, Imoto T, Miake S, Umeda A. 1987. Effects of chitin and its soluble derivatives on survival of Vibrio cholerae O1 at low temperature. Appl Environ Microbiol 53:603–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiebe W, Sheldon W, Pomeroy L. 1992. Bacterial growth in the cold: evidence for an enhanced substrate requirement. Appl Environ Microbiol 58:359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alldredge AL, Cohen Y. 1987. Can microscale chemical patches persist in the sea—microelectrode studies of marine snow, fecal pellets. Science 235:689–691. doi: 10.1126/science.235.4789.689. [DOI] [PubMed] [Google Scholar]
- 50.Hunt DE, Ward CS. 2015. A network-based approach to disturbance transmission through microbial interactions. Front Microbiol 6:1182. doi: 10.3389/fmicb.2015.01182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Preheim SP, Boucher Y, Wildschutte H, David LA, Veneziano D, Alm EJ, Polz MF. 2011. Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ Microbiol 13:265–275. doi: 10.1111/j.1462-2920.2010.02328.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



