ABSTRACT
Tropolone, a phytotoxin produced by Burkholderia plantarii, causes rice seedling blight. To identify genes involved in tropolone synthesis, we systematically constructed mutations in the genes encoding 55 histidine kinases and 72 response regulators. From the resulting defective strains, we isolated three mutants, KE1, KE2, and KE3, in which tropolone production was repressed. The deleted genes of these mutants were named troR1, troK, and troR2, respectively. The mutant strains did not cause rice seedling blight, and complementation experiments indicated that TroR1, TroK, and TroR2 were involved in the synthesis of tropolone in B. plantarii. However, tropolone synthesis was repressed in the TroR1 D52A, TroK H253A, and TroR2 D46A site-directed mutants. These results suggest that the putative sensor kinase (TroK) and two response regulators (TroR1 and TroR2) control the production of tropolone in B. plantarii.
IMPORTANCE A two-component system is normally composed of a sensor histidine kinase (HK) and a cognate response regulator (RR) pair. In this study, HK (TroK) and two RRs (TroR1 and TroR2) were found to be involved in controlling tropolone production in B. plantarii. These three genes may be part of a bacterial signal transduction network. Such networks are thought to exist in other bacteria to regulate phytotoxin production, as well as environmental adaptation and signal transduction.
INTRODUCTION
Tropolone is a nonbenzenoid aromatic compound with a seven-member ring structure (Fig. 1) (1, 2). Tropolone derivatives include hinokitiol (β-thujaplicin), first obtained from the heartwood of Taiwanese hinoki trees by Tetsuo Nozoe in the 1930s, and stipitatic acid, which is produced by Penicillium stipitatum (3). Tropolone is produced in nature and has been detected in the filtrate of a Burkholderia species (4). These compounds show a wide range of biological activities, including antimicrobial, antifungal, antiviral, insecticidal, antiparasitic, cytotoxic, and antitumor activities (2).
FIG 1.
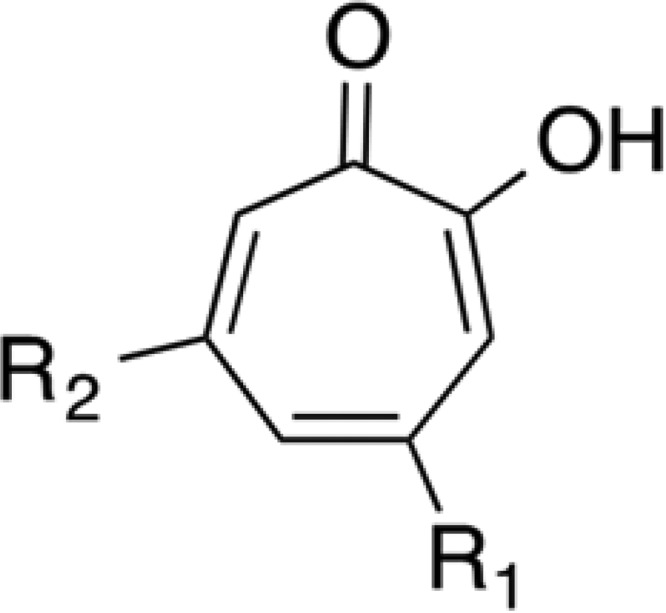
Chemical structure of tropolone and its derivatives. Tropolone, R1 H, R2 H; hinokitiol, R1 CH(CH3)2, R2 H; stipitatic acid, R1 COOH, R2 OH.
Tropolone was identified as a phytotoxin that causes bacterial seedling blight of rice (5). In addition to its antibacterial and antifungal activities, it causes chlorosis, root growth inhibition, and wilting in rice seedlings, which are symptoms caused by Burkholderia plantarii. The activities are inhibited in the presence of iron (6). When B. plantarii was cultured in iron-supplemented medium, an abundant amount of red crystals precipitated. The substance was a 3:1 complex of tropolone and Fe(III) (7), indicating that tropolone can be an iron chelator.
The synthesis of tropolone and its derivatives has since been identified in other bacteria, fungi, and plants (2). Recently, the basic genes of tropolone biosynthesis were described in Talaromyces stipitatus (P. stipitatum) (8). In marine roseobacters, the bacterial production of tropodithietic acid (TDA), a tropolone derivative, requires tdaABCDEF expression, as well as that of six additional genes (cysI, malI, paaIJK, and tdaH) (9). Tropolone and TDA production was also induced at the stationary phase (10, 11). Geng et al. (10) reported that roseobacters may use TDA as a quorum signal. Furthermore, Solis et al. (12) reported that N-acylhomoserine lactone quorum sensing and the stationary-phase RpoS, a σ factor of B. plantarii, are involved in causing rice seedling blight. These results suggested that tropolone, as well as N-acylhomoserine lactone and RpoS, can function as signaling molecules to produce tropolone via quorum-sensing pathways in B. plantarii. However, the molecular pathogenicity of B. plantarii is not well understood.
Such pathogenicity is often controlled by a two-component signal transduction system (13). To clarify the molecular pathogenicity via the complex molecular mechanism of tropolone production, we focused on a two-component system containing a sensor histidine kinase (HK) and a response regulator (RR) in B. plantarii to investigate the regulation of genes involved in tropolone synthesis. We identified three genes, those coding for a putative sensor HK (TroK) and two RRs (TroR1 and TroR2), which are involved in tropolone production.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani medium. Burkholderia plantarii MAFF301723 was precultured at 30°C (100 rpm) in PY medium (1% [wt/vol] polypeptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl [pH 7.0]) overnight and then cultured in AG medium (0.1% NH4H2PO4, 0.02% MgSO4·7H2O, 1% glucose [pH 7.0]) at 30°C (160 rpm). When necessary, selective antibiotics (25 μg/ml kanamycin; 50 μg/ml ampicillin; 50 μg/ml trimethoprim) were added to the media. Modified Hoitink agar [0.07% NH4Cl, 0.25% K2HPO4, 0.29% KH4PO4, 0.01% MgSO4, 0.6% biotin, 0.01% glucose, 0.2% Fe2(SO4)3·H2O, 1.6% agar (Nacalai Tesque, Inc.)] was used for the tropolone production assay (14).
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description or relevant genotypea | Source or reference |
|---|---|---|
| Strains | ||
| Burkholderia plantarii | ||
| MAFF301723 | Wild type | NIASb |
| KE1 | MAFF301723 troR1::pK18mobsacB | This study |
| KE2 | MAFF301723 troK::pK18mobsacB | This study |
| KE3 | MAFF301723 troR2::pK18mobsacB | This study |
| Escherichia coli DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | Thermo |
| Plasmids | ||
| pK18mobsacB | oriT oriV sacB lacZα Kmr | 20 |
| pK18-HK1-55c | 60% from ATG initiation codon of full length of 55 HK genes cloned into HindIII site of pk18mobsacB | This study |
| pK18-RR1-72c | 60% from ATG initiation codon of full length of 72 RR genes cloned into HindIII site of pk18mobsacB | This study |
| pMLBAD | pBRR1 ori araC-pBAD Tpr mob+ | 21 |
| pMLBAD-troR1+ | troR1+ cloned between NcoI and XbaI sites of pMLBAD | This study |
| pMLBAD-troK+ | troK+ cloned between NcoI and XbaI sites of pMLBAD | This study |
| pMLBAD-troR2+ | troR2+ cloned between NcoI and XbaI sites of pMLBAD | This study |
| pMLBAD-troR1+-K+ | troR1+ and troK+ cloned between NcoI and XbaI sites of pMLBAD | This study |
| pMLBAD-troK+-R2+ | troK+ and troR2+ cloned between NcoI and XbaI sites of pMLBAD | This study |
| pMLBAD-troR1+-K+-R2+ | troR1+, troK+, and troR2+ cloned between NcoI and XbaI sites of pMLBAD | This study |
| pMLBAD-troR1D47A-K+-R2+ | pMLBAD-troR1-K+-R2+ troR1D47A | This study |
| pMLBAD-troR1D52A-K+-R2+ | pMLBAD-troR1-K+-R2+ troR1D52A | This study |
| pMLBAD-troKH253A-R2+ | pMLBAD-troK-R2+ troKH253A | This study |
| pMLBAD-troKH443A-R2+ | pMLBAD-troK-R2+ troKH443A | This study |
| pMLBAD-troR2D32A | pMLBAD-troR2 troR2D32A | This study |
| pMLBAD-troR2D46A | pMLBAD-troR2 troR2D46A | This study |
Kmr, kanamycin resistant; Tpr, tropolone resistant. troR1D47A, troR1D52A, troKH253A, troKH443A, troR2D32A, and troR2D46A, troR1, troK, and troR2 genes encoding substitutions D47A, D52A, H253A, H443A, D32A, and D46A, respectively.
NIAS, National Institute of Agrobiological Science.
Fifty-five HKs and 75 RRs were identified by analysis of MAFF301723 whole-genome draft sequence data; 72 of 75 RR genes were cloned.
Construction of plasmids.
MAFF301723 genomic DNA was prepared using a Wizard genomic DNA purification kit (Promega). Genomic troR1+-troK+-troR2+, troR1+-troK+, troK+-troR2+, troR1+, troK+, and troR2+ genes were amplified from MAFF301723 genomic DNA using the Applied Biosystems 2720 thermal cycler, with primer pairs pMLBAD-troR1+-K+-R2+-F and -R, pMLBAD-troR1+-K+-F and -R, pMLBAD-troK+-R2+-F and -R, pMLBAD-troR1+-F and -R, pMLBAD-troK+-F and -R, and pMLBAD-troR2+-F and -R, respectively (data not shown), and KOD F4 Neo polymerase (Toyobo). The PCR conditions included an initial denaturation step at 94°C for 1 min, followed by 30 cycles of 98°C for 10 s, 62°C for 15 s, and 68°C for 1 min. The amplified fragments were digested with NcoI and XbaI and ligated into the corresponding sites of the digested vector pMLBAD, generating recombinant plasmids pMLBAD-troR1+-K+-R2+, pMLBAD-troR1+-K+, pMLBAD-troK+-R2+, pMLBAD-troR1+, pMLBAD-troK+, and pMLBAD-troR2+ (Table 1). Using these plasmids and specific primers, pMLBAD-troR1D47A-K+-R2+, pMLBAD-troR1D52A-K+-R2+, pMLBAD-troKH253A-R2+, pMLBAD-troKH443A-R2+, pMLBAD-troR2D32A, and pMLBAD-troR2D46A were constructed using a previously described site-directed mutagenesis method (15) by PCR, with the protocol being 94°C for 5 min and 25 cycles of 98°C for 10 s and 68°C for 6 min.
Short fragments (60% of full length) of the HK and RR genes were cloned into the HindIII sites of pK18mobsacB, generating pK18mobsacB-HK1-55 and pK18mobsacB-RR1-72, respectively (Table 1). The pK18mobsacB-HK1-55 and pK18mobsacB-RR1-72 plasmids were independently transformed by electroporation into MAFF301723 (16) and spread on Luria-Bertani agar containing kanamycin to select HK- and RR-deficient strains.
Quantitative analysis of tropolone.
Burkholderia plantarii was cultured at 30°C (100 rpm) in 10 ml of PY medium overnight. A 100-μl aliquot of the overnight culture was added to 5 ml of AG medium and incubated at 30°C (160 rpm). At stipulated time points, 50-μl aliquots of culture were collected and centrifuged (17,800 × g, 5 min, 4°C). The absorbance of the supernatant at 330 nm was measured using an ND1000 NanoDrop spectrophotometer, and tropolone was quantified using the calibration curve (Tokyo Kasei Kogyo) concentrations and their absorbance values at 330 nm. The mean and standard errors of the results from three independent experiments were statistically calculated using Excel. These values were consistent with those obtained using high-performance liquid chromatography (HPLC).
Rice seedling blight pathogenicity assays.
Unhulled rice was independently infected with MAFF301723, KE1, KE2, or KE3 in 10 ml of water and incubated at 15°C for 3 days. Following the incubation, 80 unhulled rice grains per treatment were planted on soft agar and incubated at 25°C for 1 week. Plants were then grown under lights (9 h, 0 lx; 15 h, 30,000 lx), and their disease symptoms were observed.
RESULTS AND DISCUSSION
Identification of tropolone production-deficient strains.
A total of 55 HK and 75 RR genes were identified by searching, using the BLAST algorithm, against the complete draft genome sequence of MAFF301723 (K. Nakasone, unpublished data), which is composed of chromosome I (4,144,299 bp), chromosome II (3,750,991 bp), and a plasmid (198,111 bp). HK and RR mutant strains were then constructed in B. plantarii by allelic exchange mutagenesis using pK18mobsacB-HK1-55 and pK18mobsacB-RR1-72, respectively. Among these mutants, three tropolone production-defective strains were identified on modified Hoitink agar (14). Using this method, strains producing tropolone appeared as red-brown colonies, while nonproducing strains appeared as white colonies. These tropolone production-negative strains were named KE1, KE2, and KE3, corresponding to mutations in the RR1, HK3, and RR2 genes, respectively.
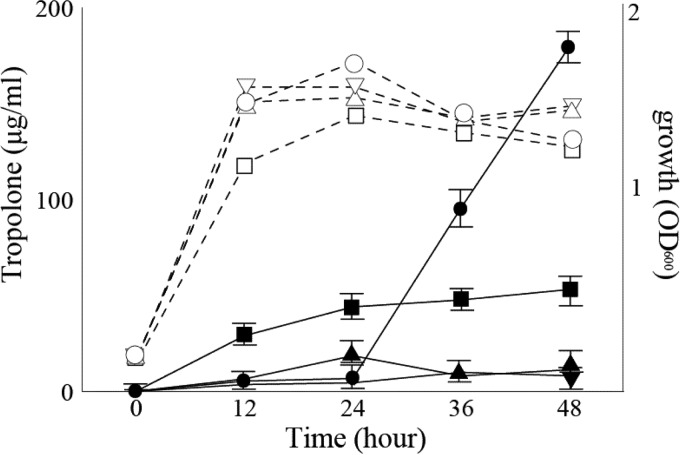
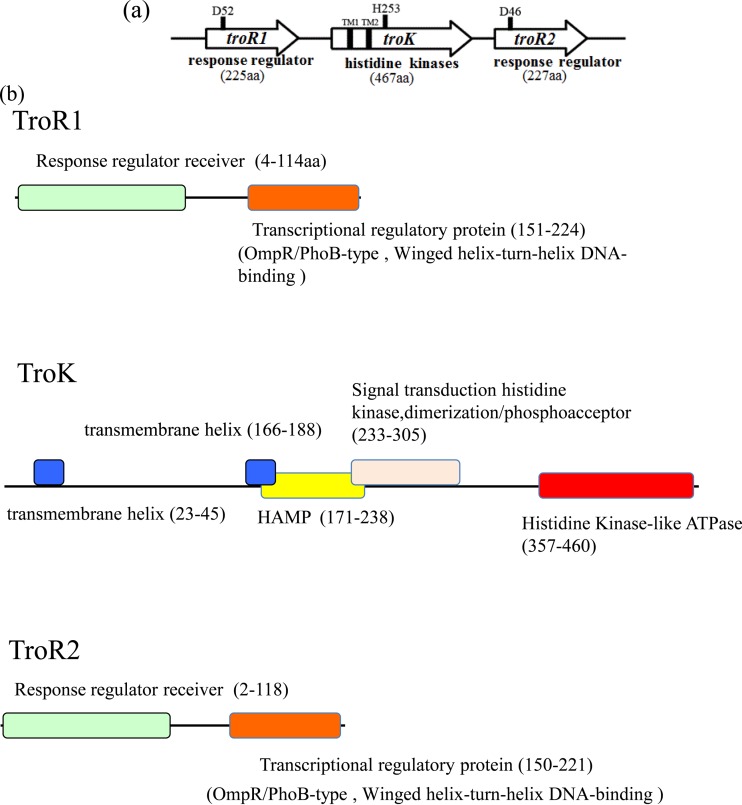
Tropolone production was evaluated in KE1, KE2, and KE3. To examine when tropolone was synthesized in B. plantarii, the wild-type strain MAFF301723 was grown in liquid medium, and tropolone concentrations excreted into the medium were measured. As shown in Fig. 2, tropolone was detected in stationary-phase cultures only. Tropolone was not detected in strains KE1 and KE3 and was detected only at a low level (53 μg/ml) at 48 h in KE2. Thus, the RR1, HK3, and RR2 genes may be involved in tropolone production during the stationary phase in B. plantarii cultured in AG medium at 30°C. Because these genes were related to tropolone production, we termed them troR1, troK, and troR2, respectively (DDBJ accession no. LC097192) (data not shown). These genes are completely consistent with the draft sequence of B. plantarii ATCC 43733 (GenBank accession no. CP007212, TroR1, bpln_1g33750; TroK, bpln_1g33740; TroR2, bpln_1g33730) (17). The domains of these proteins were analyzed using InterPro (Fig. 3). TroK is a transmembrane protein (TM1 and TM2) consisting of 467 amino acid residues. The cytoplasmic TroK possesses three domains: HAMP (amino acids [aa] 171 to 238; histidine kinases, adenylate cyclases, methyl-accepting chemotaxis proteins, and phosphatases); DHp (aa 233 to 305; dimerization-containing Hbox with phosphorylated His253); CA (aa 357 to 460; ATPases containing N, G1, and G2 boxes). TroR1 and TroR2 consist of two domains: receiver (aa 4 to 114 and 2 to 118) and transcriptional regulation (OmpR/PhoB, winged helix-turn-helix DNA binding) proteins (aa 151 to 224 and 150 to 221), respectively. These results suggest that TroK is a sensor kinase and that TroR1 and TroR2 are response regulators in a two-component system.
FIG 2.
Tropolone production is controlled by TroK, TroR1, and TroR2. Burkholderia plantarii MAFF30172 (● and ○), KE1 (▲ and △), KE2 (■ and □), and KE3 (▼ and ▽) were cultured in AG medium in the presence or absence of kanamycin (25 μg/ml) and sampled at the indicated time points for measurement of the optical density at 600 nm (OD600) (○, △, □, and ▽) and tropolone production (●, ▲, ■, and ▼). The data are presented as the means and standard errors of the results from three independent experiments.
FIG 3.
Gene organization and domain architecture. (a) troR1, troK, and troR2 are sequentially ordered. The arrows show transcriptional direction. D52, H253, and D46 are putatively phosphorylated sites which are involved in tropolone production. (b) Domain architecture of TroR1, TroK, and TroR2.
TroR1, TroK, and TroR2 are involved in tropolone production.
The troR1, troK, and troR2 genes are located adjacent to each other on the chromosome and are presumed to form an operon (data not shown). To clarify whether chromosomal deletions of upstream genes within the operon alter the expression levels of downstream genes, complementation assays were performed using six plasmids and KE1, KE2, and KE3 (Table 2). As a result, tropolone production in KE1 was recovered with only pBAD-troR1+-K+-R2+, in KE2 with two plasmids (pBAD-troR1+-K+-R2+ and pBAD-troK+-troR2+), and in KE3 with three plasmids (pBAD-troR1+-K+-troR2+, pBAD-troK+-troR2+, and pBAD-troR2+). Thus, the polar effects on downstream genes in KE1 and KE2 were confirmed, and the simultaneous expression of TroR1, TroK, and TroR2 is indispensable for tropolone production.
TABLE 2.
Complementation assaya
| Strain | Tropolone production after transformation with plasmid: |
|||||
|---|---|---|---|---|---|---|
| pMLBAD-troR1+ | pMLBAD-troK+ | pMLBAD-troR2+ | pMLBAD-troR1+-K+ | pMLBAD-troK+-R2+ | pMLBAD-troR1+-K+-R2+ | |
| KE1 | − | − | − | − | − | + |
| KE2 | − | − | − | − | + | + |
| KE3 | − | − | + | − | + | + |
Plasmids pMLBAD-troR1+, pMLBAD-troK+, pMLBAD-troR2+, pMLBAD-troR1+-K+, pMLBAD-troK+-R2+, and pMLBAD-troR1+-K+-R2+ were each independently transformed into KE1, KE2, and KE3. These transformants were cultured in PY medium overnight. They were spread on modified Hoitink agar (100 μg/ml kanamycin, 50 μg/ml tropolone, 0.02% arabinose) and incubated at 30°C. Tropolone-producing colonies appeared as red-brown (+), and tropolone-nonproducing colonies appeared as white (−). The tropolone production of the strains was also confirmed by the measurement of absorbance at 330 nm.
Effects of TroR1 D52A, TroK H253A, and TroR2 D46A on tropolone production.
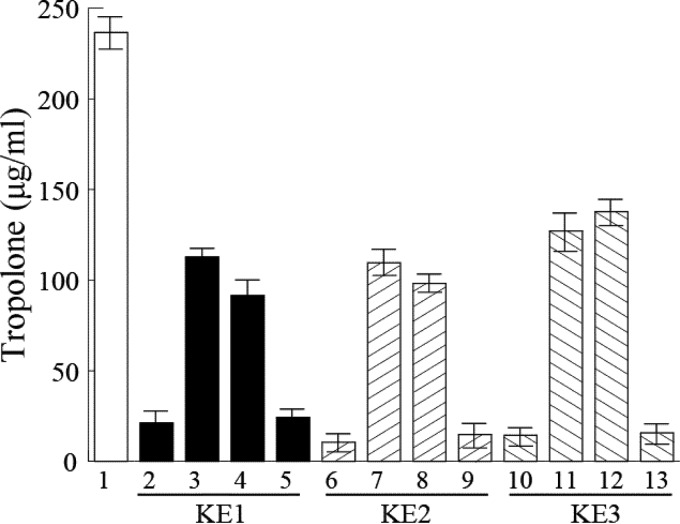
We performed a complementation assay using KE1, KE2, KE3, and plasmids encoding alanine substitution mutations at amino acid residue H253 in TroK (TroK H253; in the putative Hbox) and TroK H443 as the putative autophosphorylation sites and at TroR1 D47, TroR1 D52, TroR2 D32, and TroR2 D46 as the putative phosphorylation sites (data not shown). KE1 (lane 2), KE2 (lane 6), and KE3 (lane 10) decreased tropolone production (Fig. 4). The transformation of KE1 with pMLBAD-troR1+-K+-R2+ (lane 3) or pMLBAD-troR1D47A-K+-R2+ (lane 4) restored the production level to 50% of that of the wild type, but KE1 with pMLBAD-troR1D52A-K+-R2+ (lane 5) did not. KE2 transformed with pMLBAD-troK+-R2+ (lane 7) or pMLBAD-troKH443A-R2+ (lane 8) restored production to the same level as KE1 with pMLBAD-troR1+-K+-R2+, but KE2 with pMLBAD-troKH253A-R2 (lane 9) did not. KE3 containing pMLBAD-troR2+ (lane 11) or pMLBAD-troR2D32A (lane 12) also restored tropolone production to 50% of that of the wild type, but it was not restored with pMLBAD-troR2D46A (lane 13). As a result, TroR1 with the substitution D52A (TroR1 D52A; lane 5), TroK H253A (lane 9), and TroR2 D46A (lane 13) did not complement the tropolone production deficiencies in KE1, KE2, and KE3, respectively.
FIG 4.
Effect of TroR1 D52A, TroK H253A, and TroR2 D46A on tropolone production. MAFF301723 (lane 1), KE1 (lanes 2 to 5), KE2 (lanes 6 to 9), and KE3 (lanes 10 to 13) transformed with the indicated plasmids were cultured in AG medium at 30°C to measure tropolone production: lanes 2, 6, and 10, pMLBAD; lane 3, pMLBAD-troR1+-K+-R2+; lane 4, pMLBAD-troR1D47A-K+-R2+; lane 5, pMLBAD-troR1D52A-K+-R2+; lane 7, pMLBAD-troK+-R2+; lane 8, pMLBAD-troKH443A-R2+; lane 9, pMLBAD-troKH253A-R2+; lane 11, pMLBAD-troR2+; lane 12, pMLBAD-troR2D32A; and lane 13, pMLBAD-troR2D46A. The data are presented as the means and standard errors of the results from three independent experiments.
Pathogenesis of KE1, KE2, and KE3.
Finally, we investigated whether KE1, KE2, and KE3 could cause rice seedling blight. Unhulled rice was infected with MAFF301723, KE1, KE2, or KE3 and cultured as described in Materials and Methods. As a result, MAFF301723-infected rice showed seedling blight, while KE1-, KE2-, and KE3-infected rice seedlings did not (Fig. 5). These results confirmed that tropolone synthesis-related genes are instrumental in causing seedling blight of rice by B. plantarii, most likely through the regulatory system involving TroR1, TroK, and TroR2.
FIG 5.
Pathogenesis assay of KE1, KE2, and KE3. Unhulled rice was infected with MAFF301723, KE1, KE2, or KE3, and pathogenesis was assayed. Control, no infection.
Regulation by a two-component system containing three genes was previously reported, in CorRPS of Pseudomonas syringae (18) and CbbRRS in Rhodopseudomonas species (19). Furthermore, two-component systems containing three genes can be found using the KEGG module database (e.g., PhoRBB1, SsaA RpaAB, PixLGH, WspERF, and CcKAR). Such systems seem to be involved in bacterial signal transduction networks. In this study, we found that the regulation of tropolone production was controlled by TroR1, TroK, and TroR2. Such two-component systems, which contain three genes, are also expected to exist in other bacteria for the regulation of phytotoxin production, as well as environmental adaptation and signal transduction.
ACKNOWLEDGMENTS
This work was supported by the Research and Development Program for New BioIndustry Initiatives (2006 to 2010) of the Bio-Oriented Technology Research Advancement Institution (BRAIN), Japan, the Ministry of Education, Culture, Sports, Science and Technology-Supported Program for the Strategic Research Foundation at Private Universities, 2011 to 2015 (grant S1101035), and a grant-in-aid for science research (grant A, 20248012) from the Japan Society for the Promotion of Science. This study was also supported by a Cooperative Research Grant of the Genome Research for BioResource, NODAI Genome Research Center, Tokyo University of Agriculture.
REFERENCES
- 1.Dewar MJS. 1945. Structure of stipitatic acid. Nature 155:50–51. doi: 10.1038/155050b0. [DOI] [Google Scholar]
- 2.Bentley R. 2008. A fresh look at natural tropolonoids. Nat Prod Rep 25:118–138. doi: 10.1039/B711474E. [DOI] [PubMed] [Google Scholar]
- 3.Birkinshaw JH, Chambers AR, Raistrick H. 1942. Studies in the biochemistry of micro-organisms. Biochem J 36:242–251. doi: 10.1042/bj0360242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindberg GD, Larkin JM, Whaley HA. 1980. Production of tropolone by a Pseudomonas. J Nat Prod 43:592–594. doi: 10.1021/np50011a011. [DOI] [Google Scholar]
- 5.Azegami K, Nishiyama K, Watanabe Y, Suzuki T, Yoshida M, Nose K, Toda S. 1985. Tropolone as a root growth-inhibitor produced by a plant pathogenic Pseudomonas sp. causing seedling blight of rice. Ann Phytopath Soc Japan 51:315–317. doi: 10.3186/jjphytopath.51.315. [DOI] [Google Scholar]
- 6.Azegami K, Nishiyama K, Watanabe Y, Kadota I, Ohuchi A, Fukazawa C. 1987. Pseudomonas plantarii sp. nov., the causal agent of rice seedling blight. Int J Syst Bacteriol 37:144–152. doi: 10.1099/00207713-37-2-144. [DOI] [Google Scholar]
- 7.Azegami K, Nishiyama K, Kato H. 1988. Effect of iron limitation on “Pseudomonas plantarii” growth and tropolone and protein production. Appl Environ Microbiol 54:844–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison J, al Fahad A, Cai M, Song Z, Yehia SY, Lazarus CM, Bailey AM, Simpson TJ, Cox RJ. 2012. Genetic, molecular, and biochemical basis of fungal tropolone biosynthesis. Proc Natl Acad Sci U S A 109:7642–7647. doi: 10.1073/pnas.1201469109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geng H, Bruhn JB, Nielsen KF, Gram L, Belas R. 2008. Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl Environ Microbiol 74:1535–1545. doi: 10.1128/AEM.02339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geng H, Belas R. 2010. Expression of tropodithietic acid biosynthesis is controlled by a novel autoinducer. J Bacterial 192:4377–4387. doi: 10.1128/JB.00410-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M, Hasimoto M, Hasidoko Y. 2013. Repression of tropolone production and induction of a Burkholderia plantarii pseudo-biofilm by carot-4-en-9,10-diol, a cell-to-cell signaling disrupter produced by Trichoderma virens. PLoS One 8:e78024. doi: 10.1371/journal.pone.0078024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solis R, Bertani I, Degrassi G, Devescovi G, Venturi V. 2006. Involvement of quorum sensing and RpoS in rice seedling blight caused by Burkholderia plantarii. FEMS Microbiol Lett 259:106–112. doi: 10.1111/j.1574-6968.2006.00254.x. [DOI] [PubMed] [Google Scholar]
- 13.Gotoh Y, Eguchi Y, Watanabe T, Okamoto S, Doi A, Utsumi R. 2010. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr Opin Microbiol 13:232-239. doi: 10.1016/j.mib.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Hoitink HAJ, Sinden SL. 1970. Partial purification and properties of chlorosis inducing toxins of Pseudomonas phaseolicola and Pseudomonas glycinea. Phytopathology 60:1236–1237. doi: 10.1094/Phyto-60-1236. [DOI] [Google Scholar]
- 15.Eguchi Y, Ishii E, Yamane M, Utsumi R. 2012. The connector safA interacts with the multi-sensing domain of phoQ in Escherichia coli. Mol Microbiol 85:299–313. doi: 10.1111/j.1365-2958.2012.08114.x. [DOI] [PubMed] [Google Scholar]
- 16.Craig FF, Coote JG, Parton R, Freer JH, Gilmour NJ. 1989. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J Gen Microbiol 135:2885–2890. doi: 10.1099/00221287-135-11-2885. [DOI] [PubMed] [Google Scholar]
- 17.Seo YS, Lim JY, Park J, Kim S, Lee HH, Cheong H, Kim SM, Moon JS, Hwang I. 2015. Comparative genome analysis of rice-pathogenic Burkholderia provides insight into capacity to adapt to different environments and hosts. BMC Genomics 16:349–359. doi: 10.1186/s12864-015-1558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ullrich M, Peñaloza-Vázquez AP, Bailey AM, Bender CL. 1995. A modified two-component regulatory system is involved in temperature-dependent biosynthesis of the Pseudomonas syringae phytotoxin coronatine. J Bacteriol 177:6160–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romagnoli S, Tabita FR. 2006. A novel three-protein two-component system provides a regulatory twist on an established circuit to modulate expression of the cbb1 region of Rhodopseudomonas palustris CGA010. J Bacteriol 188:2780–2791. doi: 10.1128/JB.188.8.2780-2791.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 21.Lefebre M, Valvano MA. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl Environ Microbiol 68:5956–5964. doi: 10.1128/AEM.68.12.5956-5964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]