Abstract
Gram-positive bacteria carry out intercellular communication using secreted peptides. Important examples of this type of communication are the enterococcal sex pheromone systems, in which the transfer of conjugative plasmids is controlled by intercellular signaling among populations of donors and recipients. This review focuses on the pheromone response system of the conjugative plasmid pCF10. The peptide pheromones regulating pCF10 transfer act by modulating the ability of the PrgX transcription factor to repress the transcription of an operon encoding conjugation functions. Many Gram-positive bacteria regulate important processes, including the production of virulence factors, biofilm formation, sporulation, and genetic exchange using peptide-mediated signaling systems. The key master regulators of these systems comprise the RRNPP (RggRap/NprR/PlcR/PrgX) family of intracellular peptide receptors; these regulators show conserved structures. While many RRNPP systems include a core module of two linked genes encoding the regulatory protein and its cognate signaling peptide, the enterococcal sex pheromone plasmids have evolved to a complex system that also recognizes a second host-encoded signaling peptide. Additional regulatory genes not found in most RRNPP systems also modulate signal production and signal import in the enterococcal pheromone plasmids. This review summarizes several structural studies that cumulatively demonstrate that the ability of three pCF10 regulatory proteins to recognize the same 7-amino-acid pheromone peptide arose by convergent evolution of unrelated proteins from different families. We also focus on the selective pressures and structure/function constraints that have driven the evolution of pCF10 from a simple, single-peptide system resembling current RRNPPs in other bacteria to the current complex inducible plasmid transfer system.
BACKGROUND AND SIGNIFICANCE
In 1965, Tomasz (1) described “a new type of regulatory mechanism in bacteria,” in which the control of competent cell genetic transformation in pneumococci was expressed in a density-dependent fashion (1). He reported that the culture medium of cells grown to the optimal density for maximum competence contained a soluble factor capable of inducing competence expression when added to low-density noncompetent cultures. Conceptually, the phenomenon of density-dependent pneumococcal competence expression mediated by intercellular signaling molecules is very similar to the autoinduction of light production in marine Vibrio species described a few years later by Nealson (2). These seminal studies initiated a paradigm shift in microbial research, changing the concept of normal bacterial behavior from single cells acting independently to coordinated behaviors of microbial populations via communication between individuals. Quorum sensing, in which a single cell type monitors its population density to coordinate activity (3), is perhaps the best studied mechanism for the modulation of multicellular behaviors by intercellular signaling, which is more broadly termed sociomicrobiology (4).
Enterococcus faecalis is a major cause of opportunistic infections of hospital patients, and E. faecalis clinical isolates are notorious for their carriage of antibiotic resistance genes (5, 6). These are frequently disseminated by conjugation. In 1978, Dunny et al. (7) reported that donor/recipient clumping and conjugative transfer of plasmids in Enterococcus (formerly Streptococcus) faecalis could be induced by low-molecular-weight signaling molecules excreted by recipient cells and sensed by plasmid-containing donor cells; it was suggested that these signals served as bacterial sex pheromones. A few years later, the Clewell et al. (8) and Suzuki et al. (9) research groups reported the identification of several different molecules that mediated signaling for various plasmids; these signals were unmodified hydrophobic peptides 7 to 8 amino acid residues in length. These studies were the first demonstrations that the prevalent extracellular signaling molecules of Gram-positive bacteria were oligopeptides, in contrast to the acyl-homoserine-lactone signals that frequently mediate quorum sensing in Gram-negative microbes (10). Both the peptide-mediated signaling mechanisms and the peptide signals themselves fall into two categories. Some signals are secreted as unmodified peptides processed from longer precursors, while others are both processed and posttranslationally modified (11–13). Likewise, sensing of peptide signals can involve either signal transduction across the membrane or signal import, followed by binding to a cytoplasmic receptor protein, which is often a transcription factor (14).
The enterococcal sex pheromone systems function by import of a signaling pheromone peptide encoded by the chromosome. For simplicity, we use “C” as an abbreviation for all conjugation/clumping-inducing peptide pheromones, where cCF10 is the peptide that specifically induces cells carrying pCF10, cAD1 induces those carrying pAD1, etc. Mature C is processed by host-encoded proteins, and all known members of the sex pheromone family are processed from the cleaved signal peptides of secreted lipoproteins (15, 16). Binding of imported C by its cytoplasmic receptor initiates the pheromone response in the donor; the presence of C in the growth medium of donor cells thus serves as a cue for the presence of recipients (Fig. 1). Peptide binding modulates the ability of the C receptor (PrgX in the case of pCF10) to regulate the transcription of an operon containing conjugation genes (17). However, the enterococcal sex pheromone systems have several additional layers of complexity, including a second plasmid-encoded peptide (inhibitor [I]) that competes directly with C for binding to the same receptor (17, 18). In addition, several layers of posttranscriptional regulation greatly amplify the direct effects of the peptides on the expression of conjugation genes (17). The remainder of this essay will focus on the tetracycline-resistant pheromone-responsive plasmid pCF10 to illustrate the salient features of many sex pheromone plasmids (19) and to explore how the current complex systems may have evolved from simpler progenitor systems similar to the peptide-regulated RRNPP signaling systems that have now been implicated in the control of virulence, developmental processes, and horizontal gene transfer in numerous Gram-positive pathogens (20, 21).
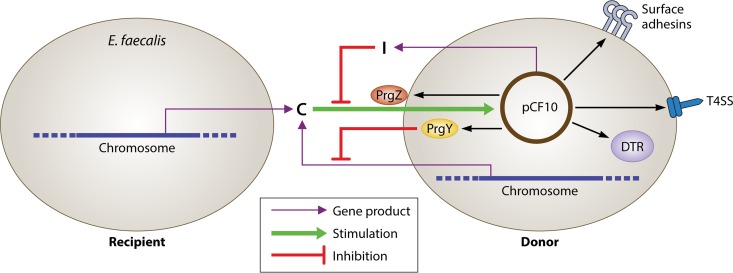
FIG 1.
Diagram of the signaling circuits in the E. faecalis pCF10 conjugation system (adapted from Annual Review of Genetics [19]). Recipient and donor have similar chromosomes, but the donor also carries pCF10. The plasmid confers a response to the chromosomally encoded peptide C, which induces conjugation. The plasmid encodes the antagonistic peptide I, which inhibits C competitively. Two constitutively expressed pCF10 gene products, PrgZ and PrgY, function in pheromone import and in reduction of the amount of active C excreted by plasmid-carrying cells, respectively, as detailed in the text. Imported C interacts with PrgX (not shown) in the cytoplasm to induce a conjugation response. Pheromone induction of donor cells results in the synthesis of conjugation-related gene products, including surface adhesin proteins, type 4 secretion proteins (T4SS), and DNA transfer proteins (DTR).
OVERVIEW OF THE PEPTIDE-MEDIATED REGULATION OF pCF10 CONJUGATION
Figure 2 depicts a simplified map of the pheromone-inducible conjugation genes of pCF10 (22). The prgQ operon confers production of >30 polypeptides and regulatory RNAs required for regulated expression of conjugation. The pheromone receptor PrgX controls the initiation of transcription of this long operon from the prgQ promoter; the interaction of I with PrgX reduces transcription, whereas the interaction of C with PrgX allows for increased transcription. It is important to note that the direct effects of the peptides on control of the prgQ promoter by PrgX are actually quite modest, but they are greatly amplified by several posttranscriptional mechanisms, which are described elsewhere (23–27). Determination of the structures of Apo-PrgX and of PrgX bound to I or C, along with extensive genetic and biochemical analyses, indicates that Apo-PrgX and PrgX-I complexes repress transcription from the prgQ promoter, while PrgX-C complexes are impaired in repression (28, 29). It was originally suggested that the replacement of I by C in PrgX-DNA complexes disrupts PrgX tetramers within repressing complexes, allowing RNA polymerase to access the prgQ promoter (28–30). Very recent data (Y. Chen, A. Bandyopadhyay, B. K. Kozlowicz, H. A. H. Haemig, A. Tai, W.-S. Hu, and G. Dunny, unpublished data) suggest that PrgX forms tetramers when complexed with either peptide, but conformational differences of the DNA-bound tetramers account for differential repression. In both models, the ultimate induction state of a donor cell is dependent on the relative intracellular levels of I and C in donor cells. Interestingly, all of the peptide-controlled transcription factors of the RRNPP family appear to have a structure that is very similar to that of PrgX (20, 21, 31), and in most cases, the gene organization of the determinants for regulatory protein and the cognate regulatory peptide is similar to that of prgX and prgQ. Below, we focus on the evolutionary processes that likely shaped the emergence of the dual-peptide-controlled pCF10 system and how it may have evolved from a simple RRNPP-like system to its present complex state.
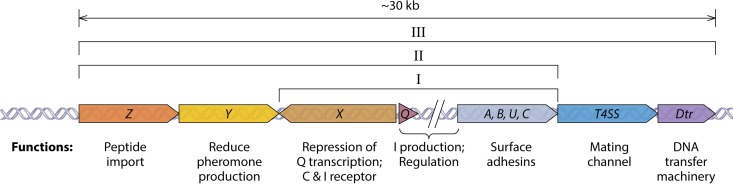
FIG 2.
Genetic organization of pheromone-inducible conjugation genes found on enterococcal plasmids (approximate size of the entire region indicated at the top). This map depicts the prg genes of pCF10 with single-letter designations, but similar gene content and organization are found on other well-studied plasmids, such as pAD1 and pPD1 (17). The left portion of the map shows conserved genes involved in pheromone sensing, and the relative locations of the genes of the pheromone-inducible prgQ operon encoding the I peptide, surface adhesin gene module (ABUC), downstream type IV secretion system (T4SS) genes, and conjugative DNA transfer genes (Dtr) are shown. The prgQ gene encodes the production of I, whereas an ∼1-kb segment between prgQ and prgA encodes two small open reading frames (ORFs) and small RNAs (sRNAs) that regulate the expression of downstream genes posttranscriptionally (65). The sizes of the individual genes are not drawn to scale. I, the putative origin of the system as a surface protein module negatively regulated by quorum sensing through the X/Q cassette; this gene pair resembles RRNPP systems recently identified in numerous Gram-positive pathogens (21, 31). II shows how the system became more complex as it acquired the ability to enable its host cell to recognize C as an indicator of close proximity of plasmid-free recipients (mate sensing). At the mechanistic level, the C peptide competes with I, which functions as a classic quorum-sensing signal of donor density (self-sensing) (64). Evolution of the ability to differentially respond to these two antagonistic peptides was accompanied by the acquisition of genes encoding an oligopeptide binding protein, PrgZ, which binds both C and I with high affinity and increases their import via the Opp ABC transporter (37, 38), and PrgY, a predicted membrane peptidase that reduces the production of endogenous C by the host cell (36). III depicts the acquisition of T4SS and Dtr genes conferring conjugative transfer ability. There is high conservation of the regions indicated by I and II among many pheromone plasmids, suggesting that they all arose from a common ancestor, but step III likely occurred multiple times to link different conjugation gene cassettes to the pheromone-inducible aggregation module.
HOW AND WHY DID THE pCF10 SYSTEM BECOME SO COMPLEX?
The key functional components of the pCF10 system, which are also found in other pheromone plasmids (17, 32), are illustrated in Fig. 2. It is likely that current pheromone-inducible conjugation systems originated from a system with a single I-regulated Q-X-like module. This module likely controlled the expression of adjacent genes for surface adhesins, as similar surface adhesion gene content and organization are conserved with other pheromone-controlled systems (17). Contemporary pheromone plasmids may have a common ancestor that includes contiguous genes corresponding to prgZ through prgQ and extending through the downstream cassette of LPXTG-anchored cell surface protein genes (prgABC) and the small regulatory prgU gene (22, 33); this gene cluster is indicated by the Roman numeral II in Fig. 2. Prior1 to the acquisition of the ability to recognize the peptide signal C, the I-autoregulated X-Q surface protein cluster may have functioned to increase the ability of the host bacterium to attach to other bacterial or metazoan host cells at low density while reducing these interactions at high bacterial density to enable escape from stagnant communities and recolonization of new niches.
The next major event in the evolution of the system was probably the ability to recognize the host-encoded C peptide as an indicator of the presence of plasmid-free enterococci in close proximity. In strict evolutionary biology parlance (34, 35), C would be classified as a cue rather than a signal, since the sensing system seems to have hijacked this molecule produced from a gene not linked to the sensing genes in physical proximity or in function. In the case of pCF10, the C peptide is produced by processing the cleaved signal peptide of a predicted secreted lipoprotein CcfA, whose function has not been demonstrated (15); likewise, all known pheromone-responsive plasmids analyzed to date encode a response to a specific peptide encoded by one of the >50 potential lipoprotein genes in the organism (16). As indicated in step II, the system acquired additional components that recognize C; PrgY prevents self-induction of donors by decreasing the amount of mature C released (36), and PrgZ binds both C and I and facilitates their import into the cell via a chromosomally encoded peptide transporter (37, 38). PrgX also needed to evolve to recognize C and I. These 3 proteins are all from different families and share only 9 to 13% sequence identity and no significant homology at the structural level. We have structural data on the interactions of PrgZ with C (37) and of PrgX with both C and I (28, 29), but to date, there are no structural data available on PrgY.
PrgZ belongs to the family of substrate-binding proteins found in ABC transporters, G protein-coupled receptors (GPCRs), and DNA binding proteins (39, 40). It is likely that PrgZ evolved from a chromosomal oligopeptide-binding protein. Previous experiments have shown that the oligopeptide-binding protein OppA of E. faecalis can facilitate the import of C, even though a higher concentration of C is required than is produced by recipients under normal physiological conditions (38). PrgZ can bind both C and I and has a typical Venus flytrap fold of a cluster C substrate-binding protein (37), with C bound within an internal cavity (Fig. 3a). C is firmly bound to PrgZ via 10 direct hydrogen bonds. These bonds are mostly formed with the peptide backbone, with one exception, an H-bond that is formed with the side chain of Thr3, giving an explanation for results from genetic screens that Thr3 of C was important for PrgZ binding (41). Further H-bonds between C and PrgZ are formed via bridging water molecules, and there is also a salt bridge that anchors the N terminus of C. Although no structure of PrgZ complexed with I is available, it is highly likely that I binds in the same way as C (37). This is expected due to the similarities of PrgZ to other oligopeptide-binding proteins (37, 40, 42, 43).
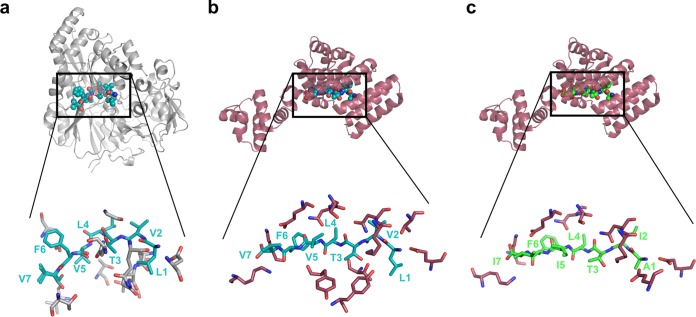
FIG 3.
Comparison of the peptide binding of PrgZ (gray) complexed with C (a) and PrgX (dark red) complexed with C (b) or I (c). Each upper subfigure shows the full protein structure in a cartoon representation, with the bound ligand in spheres, C in teal, and I in green. The lower enlarged representations at the bottom show the ligand (as sticks) with interacting protein residues. As can be seen by comparing panels b and c, the C peptide has a very different structure when bound to PrgZ compared with its structure when bound to PrgX.
As noted previously, PrgX serves as the primary cytoplasmic receptor for both peptides and acts as the master transcription regulator of the prgQ operon. PrgX has a conserved helix-turn-helix (HTH) domain responsible for its interaction with DNA (28, 29). The binding site for C and I is located in the larger dimerization domain (Fig. 3b and c), where both peptides form β-sheet-like interactions with PrgX and have a similar amount of H-bonds to PrgX. There is no conservation in the binding site between PrgX and PrgZ at either the sequence level or structurally. The two peptides favor different conformational states of PrgX, with the consequence that one alpha-helix is preferentially stabilized (I bound) or unwound (C bound) (28, 29). These different conformations either favor transcriptional repression when inhibitor is bound, or they reduce the level of repression of PrgX when C is bound. While the basis for the differential effects of the two peptides on the conformation of PrgX is not completely understood, it probably relates to the N-terminal regions of the two peptides. The bulky leucine residue at the N terminus of C likely crowds the surrounding PrgX residues in the binding pocket to a greater extent than the alanine at the N terminus of I (for a partial illustration, compare the PrgX residues surrounding the bound peptide N termini in Fig. 3b and c). These differences may indirectly affect the conformation of the C terminus of PrgX.
PrgY is required to prevent cells carrying pCF10 from being self-induced by their own endogenous pheromone (36). Its amino acid sequence, in conjunction with genetic and biochemical studies, suggest that a C-terminal subdomain anchors the protein in the membrane with the N-terminal region outside the cell; the external N-terminal subdomain confers the ability to specifically bind the mature C peptide (36, 44) and may contribute to its degradation. Initial studies of PrgY suggested that similar proteins, none with known functions, were present in organisms from all kingdoms, and that the protein phylogenies correlated with those of the host organisms (36). Recently, an important new study provided new insights into the structure/function relationships of these proteins. Zhang et.al. (45) identified Tiki as a protease family playing a critical role in cell growth and development via specific cleavage of the Wnt protein (45). PrgY is homologous to the human Tiki metalloprotease, both having a pair of GX2H motifs and a conserved glutamate residue, and it is predicted to have structural similarity to the so-called EraA/ChaN-like family of proteins (46). The structure of PrgY has not been determined, but structural modeling using Phyre2 gives a model with a 96% confidence level over most of the extracellular domain (Fig. 4). This model does not contain any structural motif that resembles the pheromone-binding site of either PrgZ or PrgX. From the homology to the Tiki metalloproteases, we can deduce which residues likely form the active site in PrgY, with some of those specific residues, like His21, having previously been verified to be important for function (36, 44). To date, only PrgY and Tiki are known to have specific interactions with polypeptide substrates.
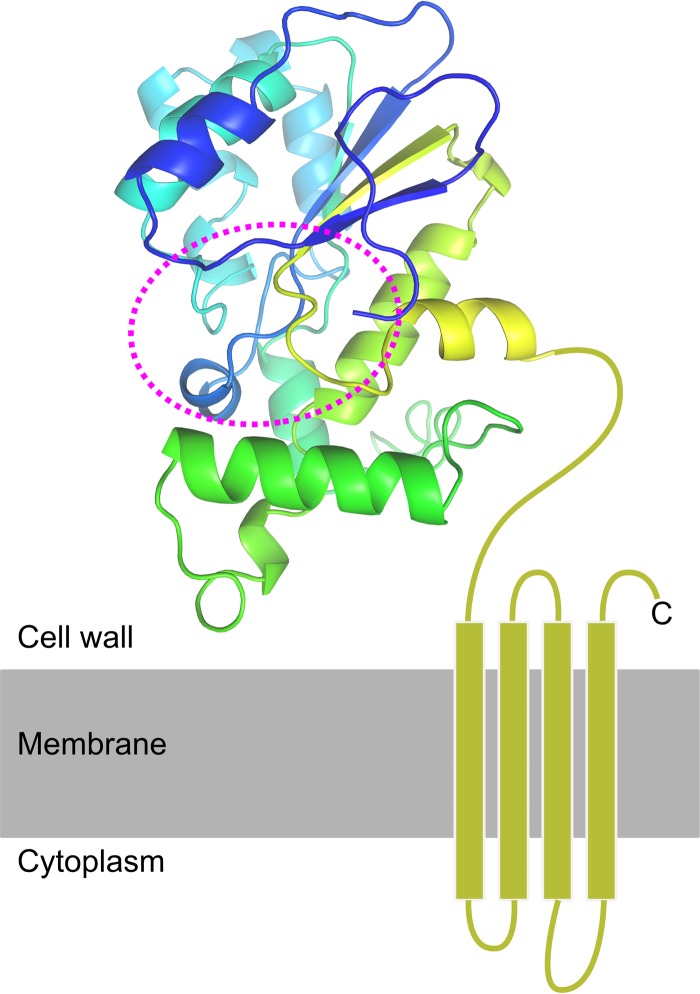
FIG 4.
Predicted structure of PrgY. The extracellular part of PrgY, here shown as a cartoon representation, was modeled using Phyre2 and colored from the N terminus (blue) toward the C-terminal end of the model (yellow). The C-terminal domain, which could not be modeled, is predicted to contain 4 transmembrane helices, shown here as rectangles in a membrane. The predicted active site, based on the homology of PrgY to the Tiki metalloproteases (46), is shown within the dashed line.
The cumulative analysis suggests that the pCF10 system did not independently evolve these 3 different components from a single protein with a peptide-binding motif. More likely, an ancestral system, i.e., the inhibitor-regulated Q-X module, at some point acquired genes that coded for the early versions of PrgY and PrgZ, and those proteins then evolved specific binding affinity to the cognate C and I peptides, as illustrated in Fig. 2. The downstream genes predicted to encode the type 4 secretion systems (T4SSs) and DNA transfer (Dtr) machinery required for conjugation are located immediately downstream from the surface protein cassettes in other pheromone plasmids, but these T4SS loci show considerable divergence (22). This suggests that pheromone-inducible aggregation cassettes became linked to the additional components required for conjugation on multiple occasions (Fig. 2, step III). Interestingly, the available data suggest that the downstream conjugation functions for all known plasmids are transcriptionally regulated by the peptide signals even though they became linked to the upstream regions in multiple events (22).
REMAINING QUESTIONS AND FUTURE DIRECTIONS
While significant questions about the molecular mechanisms of pheromone-mediated control of conjugation remain, the most compelling areas for future study may be the analysis of structure/function relationships of the key regulatory components and investigations of how these systems function in the natural environment, including their impacts on maintenance and dissemination of the plasmid itself, and on the fitness of the bacterial hosts. The significance of such studies is heightened by the fact that considerable experimental and theoretical investigations of the evolutionary aspects of the well-studied acyl-homoserine-lactone autoinducer systems in Gram-negative alphaproteobacteria have been carried out (reviewed in reference 47). For example, a recent study by Cornforth et al. (48) provided evidence that combining two separate quorum-sensing systems allowed for greater resolution of the local environment of a bacterium, both in terms of sensing social (microbial cell population density) and physical (diffusion, etc.) parameters. In contrast, much less attention has been directed toward the naturally occurring and more complex intercellular communication system represented by pCF10. While pCF10 was discovered because of its role in the transmission of antibiotic resistance (49), enterococcal pheromone-responsive plasmids frequently do not carry resistance genes (50, 51), suggesting that they may encode other traits that increase the fitness of their host cells, relative to the costs of plasmid maintenance. It is interesting to consider how the expression of the pheromone-inducible conjugation genes may impact host fitness and how this relates to the regulatory properties of the system. Published data from our group (52–57) and others (58–61) suggest that the expression of aggregation substance proteins, such as PrgB, can increase colonization and virulence by promoting biofilm formation and attachment to host tissues, and by increasing resistance to phagocytic killing. Notably, there are still no direct data on how PrgB or other inducible proteins might impact fitness in the gut. On the other hand, the overexpression of these genes likely has very high costs for the induced cell, including the energy required for synthesizing conjugation proteins, the likely inhibition of growth in cells trapped in large aggregates, and cell death and lysis due to toxic effects of overexpressed gene products on highly induced cells (62). Interestingly, clusters of genes related to the plasmid-encoded pheromone-inducible adhesins/transfer determinants have been identified within genomic islands in the chromosomes of some strains, but these chromosomal determinants are not capable of transfer unless a coresident pheromone plasmid integrates and mobilizes them via an Hfr-like mechanism (63).
Numerous studies have documented the extremely tight regulation of the pheromone system (19). The system not only avoids spurious induction but also limits the duration of induction due to the fact that the induction process itself dramatically increases inhibitor production, leading to rapid shut off of the response after a short period of induction (64). Furthermore, the inhibitor can function as a classic quorum sensor of donor density; at high donor densities, donors are poorly induced even by high concentrations of C (64). These cumulative effects of the inhibitor apparently limit the extent of induction in mixed populations of donors and recipients. This raises the question of whether the system may have evolved to maintain mixed populations of donors and recipients in shared niches in the natural environment of the bacteria, e.g., the intestinal tract. The maintenance of recipient populations by limiting their conversion to donors should result in a steady supply of C within the niche, whose inducing capacity is limited by the inhibitor. In this scenario, basal levels of expression of the inducible genes could be maintained within the mixed population, providing the previously described benefits (note that induction of a few donors can coaggregate recipients and uninduced donors in close proximity) while minimizing costs of overexpression. The pheromone system has thus evolved under strong conflicting selective pressures for an extremely sensitive detection system to induce expression while simultaneously limiting the extent and duration of induction. This may have driven convergent evolution of the three unrelated proteins with vital, but distinct, regulatory functions to recognize the same peptide. Direct experimental testing of these ideas in the mammalian gastrointestinal (GI) tract, along with further mechanistic and structural studies of regulatory components, is in progress and might yield insights into more effective approaches to reduce the spread of antibiotic resistance and to impair the ability of resistant strains to overgrow and disrupt the gut microbiota of hospital patients.
ACKNOWLEDGMENTS
The research for this review was supported by U.S. PHS grant GM49530 to G.M.D. and by the Kempe Foundations grant JCK-1524 to R.P.-A.B.
Biographies

Gary M. Dunny received his B.S. and Ph.D. from the University of Michigan and spent 11 years at Cornell University as a postdoctoral fellow and as a faculty member before moving to the University of Minnesota in 1989, where he is currently professor of microbiology. He has studied conjugation, cell signaling, and adaptation in enterococci using genetics, biochemistry, and microscopic imaging for his entire career.

Ronnie Per-Arne Berntsson studied biotechnology at Chalmers University in Gothenburg, Sweden. In 2010, he received his Ph.D. in biochemistry from the University of Groningen, the Netherlands, after working in the groups of Bert Poolman and Dirk-Jan Slotboom on studies of ABC transporters and their domains. After his Ph.D., he moved to Stockholm University, Sweden, where he received an EMBO fellowship to do postdoctoral research in the group of Pål Stenmark on botulinum neurotoxins and their receptors. In 2015, he became an assistant professor at the Department of Medical Biochemistry and Biophysics at Umeå University, Sweden. His laboratory studies the function, structure, and regulation of type 4 secretion systems in Gram-positive bacteria.
REFERENCES
- 1.Tomasz A. 1965. Control of the competent state in Pneumococcus by a hormone-like cell product: an example for a new type of regulatory mechanism in bacteria. Nature 208:155–159. doi: 10.1038/208155a0. [DOI] [PubMed] [Google Scholar]
- 2.Nealson KH. 1999. Early observations defining quorum-dependent gene expression, p 277–290. In Dunny GM, Winans SC (ed), Cell-cell signaling in bacteria. ASM Press, Washington, DC. [Google Scholar]
- 3.Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176:269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsek MR, Greenberg EP. 2005. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol 13:27–33. doi: 10.1016/j.tim.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Kristich CJ, Rice LB, Arias CA. 6 February 2014. Enterococcal infection—treatment and antibiotic resistance. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye & Ear Infirmary, Boston, MA: http://www.ncbi.nlm.nih.gov/books/NBK190420/. [PubMed] [Google Scholar]
- 6.Miller WR, Munita JM, Arias CA. 2014. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther 12:1221–1236. doi: 10.1586/14787210.2014.956092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunny GM, Brown BL, Clewell DB. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci USA 75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mori M, Sakagami Y, Narita M, Isogai A, Fujino M, Kitada C, Craig RA, Clewell DB, Suzuki A. 1984. Isolation and structure of the bacterial sex pheromone, cAD1, that induces plasmid transfer in Streptococcus faecalis. FEBS Lett 178:97–100. doi: 10.1016/0014-5793(84)81248-X. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki A, Mori M, Sakagami Y, Isogai A, Fujino M, Kitaga C, Craig RA, Clewell DB. 1984. Isolation and structure of bacterial sex pheromone, cPD1. Science 226:849–850. doi: 10.1126/science.6436978. [DOI] [PubMed] [Google Scholar]
- 10.Fuqua C, Parsek MR, Greenberg EP. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu Rev Genet 35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 11.Chandler JR, Dunny GM. 2004. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides 25:1377–1388. doi: 10.1016/j.peptides.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 12.Dunny GM, Leonard BAB. 1997. Cell-cell communication in Gram-positive bacteria. Annu Rev Microbiol 51:527–564. [DOI] [PubMed] [Google Scholar]
- 13.Dunny GM, Winans SC. 1999. Cell-cell signaling in bacteria. ASM Press, Washington, DC. [Google Scholar]
- 14.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 15.Antiporta MH, Dunny GM. 2002. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J Bacteriol 184:1155–1162. doi: 10.1128/jb.184.4.1155-1162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clewell DB, An FY, Flannagan SE, Antiporta M, Dunny GM. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol Microbiol 35:246–248. doi: 10.1046/j.1365-2958.2000.01687.x. [DOI] [PubMed] [Google Scholar]
- 17.Clewell DB, Weaver KE, Dunny GM, Coque TM, Francia MV, Hayes F. 9 February 2014. Extrachromosomal and mobile elements in enterococci: transmission, maintenance, and epidemiology. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye & Ear Infirmary, Boston, MA: http://www.ncbi.nlm.nih.gov/books/NBK190430/. [PubMed] [Google Scholar]
- 18.Nakayama J, Ruhfel RE, Dunny GM, Isogai A, Suzuki A. 1994. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J Bacteriol 176:7405–7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunny GM. 2013. Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu Rev Genet 47:457–482. doi: 10.1146/annurev-genet-111212-133449. [DOI] [PubMed] [Google Scholar]
- 20.Cook LC, Federle MJ. 2014. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Rev 38:473–492. doi: 10.1111/1574-6976.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parashar V, Aggarwal C, Federle MJ, Neiditch MB. 2015. Rgg protein structure-function and inhibition by cyclic peptide compounds. Proc Natl Acad Sci U S A 112:5177–5182. doi: 10.1073/pnas.1500357112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirt H, Manias DA, Bryan EM, Klein JR, Marklund JK, Staddon JH, Paustian ML, Kapur V, Dunny GM. 2005. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J Bacteriol 187:1044–1054. doi: 10.1128/JB.187.3.1044-1054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae T, Kozlowicz BK, Dunny GM. 2004. Characterization of cis-acting prgQ mutants: evidence for two distinct repression mechanisms by Qa RNA and PrgX protein in pheromone-inducible enterococcal plasmid pCF10. Mol Microbiol 51:271–281. [DOI] [PubMed] [Google Scholar]
- 24.Chatterjee A, Johnson CM, Shu CC, Kaznessis YN, Ramkrishna D, Dunny GM, Hu WS. 2011. Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation. Proc Natl Acad Sci U S A 108:9721–9726. doi: 10.1073/pnas.1101569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunny GM, Johnson CM. 2011. Regulatory circuits controlling enterococcal conjugation: lessons for functional genomics. Curr Opin Microbiol 14:174–180. doi: 10.1016/j.mib.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson CM, Haemig HH, Chatterjee A, Wei-Shou H, Weaver KE, Dunny GM. 2011. RNA-mediated reciprocal regulation between two bacterial operons is RNase III dependent. mBio 2(5):e00189-11. doi: 10.1128/mBio.00189-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson CM, Manias DA, Haemig HA, Shokeen S, Weaver KE, Henkin TM, Dunny GM. 2010. Direct evidence for control of the pheromone-inducible prgQ operon of Enterococcus faecalis plasmid pCF10 by a countertranscript-driven attenuation mechanism. J Bacteriol 192:1634–1642. doi: 10.1128/JB.01525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozlowicz BK, Shi K, Gu ZY, Ohlendorf DH, Earhart CA, Dunny GM. 2006. Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides. Mol Microbiol 62:958–969. doi: 10.1111/j.1365-2958.2006.05434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi K, Brown CK, Gu ZY, Kozlowicz BK, Dunny GM, Ohlendorf DH, Earhart CA. 2005. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc Natl Acad Sci U S A 102:18596–18601. doi: 10.1073/pnas.0506163102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozlowicz BK. 2005. The molecular mechanism and peptide signaling response of PrgX used to control pheromone-induced conjugative transfer of pCF10. Ph.D. dissertation. University of Minnesota, Minneapolis, MN. [Google Scholar]
- 31.Declerck N, Bouillaut L, Chaix D, Rugani N, Slamti L, Hoh F, Lereclus D, Arold ST. 2007. Structure of PlcR: insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria. Proc Natl Acad Sci U S A 104:18490–18495. doi: 10.1073/pnas.0704501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clewell DB, Dunny GM. 2002. Conjugation and genetic exchange in enterococci, p 265–300. In Gilmore MS, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB (ed), The enterococci: pathogenesis, molecular biology and antibiotic resistance. ASM Press, Washington, DC. [Google Scholar]
- 33.Kozlowicz BK, Dworkin M, Dunny GM. 2006. Pheromone-inducible conjugation in Enterococcus faecalis: a model for the evolution of biological complexity? Int J Med Microbiol 296:141–147. doi: 10.1016/j.ijmm.2006.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Platt TG, Fuqua C. 2010. What's in a name? The semantics of quorum sensing. Trends Microbiol 18:383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller L, Surette MG. 2006. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol 4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 36.Chandler JR, Flynn AR, Bryan EM, Dunny GM. 2005. Specific control of endogenous cCF10 pheromone by a conserved domain of the pCF10-encoded regulatory protein PrgY in Enterococcus faecalis. J Bacteriol 187:4830–4843. doi: 10.1128/JB.187.14.4830-4843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berntsson RP, Schuurman-Wolters GK, Dunny G, Slotboom DJ, Poolman B. 2012. Structure and mode of peptide binding of pheromone receptor PrgZ. J Biol Chem 287:37165–37170. doi: 10.1074/jbc.M112.386334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leonard BA, Podbielski A, Hedberg PJ, Dunny GM. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci U S A 93:260–264. doi: 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berntsson RP, Doeven MK, Fusetti F, Duurkens RH, Sengupta D, Marrink SJ, Thunnissen AM, Poolman B, Slotbloom DJ. 2009. The structural basis for peptide selection by the transport receptor OppA. EMBO J 28:1332–1340. doi: 10.1038/emboj.2009.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berntsson RP, Smits SH, Schmitt L, Slotbloom DJ, Poolman B. 2010. A structural classification of substrate-binding proteins. FEBS Lett 584:2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 41.Fixen KR, Chandler JR, Le T, Kozlowicz BK, Manias DA, Dunny GM. 2007. Analysis of the amino acid sequence specificity determinants of the enterococcal cCF10 sex pheromone in interactions with the pheromone-sensing machinery. J Bacteriol 189:1399–1406. doi: 10.1128/JB.01226-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levdikov VM, Blagova EV, Brannigan JA, Wright L, Vagin AA, Wilkinson AJ. 2005. The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. J Mol Biol 345:879–892. doi: 10.1016/j.jmb.2004.10.089. [DOI] [PubMed] [Google Scholar]
- 43.Tame JR, Murshudov GN, Dodson EJ, Neil TK, Dodson GG, Higgins CF, Wilkinson AJ. 1994. The structural basis of sequence-independent peptide binding by OppA protein. Science 264:1578–1581. doi: 10.1126/science.8202710. [DOI] [PubMed] [Google Scholar]
- 44.Chandler JR, Dunny GM. 2008. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J Bacteriol 190:1172–1183. doi: 10.1128/JB.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Abreu JG, Yokota C, MacDonald BT, Singh S, Coburn KL, Cheong SM, Zhang MM, Ye QZ, Hang HC, Steen H, He X. 2012. Tiki1 is required for head formation via Wnt cleavage-oxidation and inactivation. Cell 149:1565–1577. doi: 10.1016/j.cell.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bazan JF, Macdonald BT, He X. 2013. The TIKI/TraB/PrgY family: a common protease fold for cell signaling from bacteria to metazoa? Dev Cell 25:225–227. doi: 10.1016/j.devcel.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevens AM, Schuster M, Rumbaugh KP. 2012. Working together for the common good: cell-cell communication in bacteria. J Bacteriol 194:2131–2141. doi: 10.1128/JB.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cornforth DM, Popat R, McNally L, Gurney J, Scott-Phillips TC, Ivens A, Diggle SP, Brown SP. 2014. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc Natl Acad Sci U S A 111:4280–4284. doi: 10.1073/pnas.1319175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunny G, Funk C, Adsit J. 1981. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid 6:270–278. doi: 10.1016/0147-619X(81)90035-4. [DOI] [PubMed] [Google Scholar]
- 50.Clewell DB. 1993. Bacterial sex pheromone-induced plasmid transfer. Cell 73:9–12. doi: 10.1016/0092-8674(93)90153-H. [DOI] [PubMed] [Google Scholar]
- 51.Clewell DB. 1999. Sex pheromone systems in enterococci, p 47–66. In Dunny GM, Winans SC (ed), Cell-cell signaling in bacteria. ASM Press, Washington, DC. [Google Scholar]
- 52.Chandler JR, Hirt H, Dunny GM. 2005. A paracrine peptide sex pheromone also acts as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc Natl Acad Sci U S A 102:15617–15622. doi: 10.1073/pnas.0505545102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chuang ON, Schlievert PM, Wells CL, Manias DA, Tripp TJ, Dunny GM. 2009. Multiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence. Infect Immun 77:539–548. doi: 10.1128/IAI.01034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chuang-Smith ON, Wells CL, Henry-Stanley MJ, Dunny GM. 2010. Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an ex vivo model of cardiac valve colonization. PLoS One 5:e15798. doi: 10.1371/journal.pone.0015798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hirt H, Erlandsen SL, Dunny GM. 2000. Heterologous inducible expression of Enterococcus faecalis pCF10 aggregation substance Asc10 in Lactococcus lactis and Streptococcus gordonii demonstrates contribution to cell hydrophobicity and adhesion to fibrin. J Bacteriol 182:2299–2306. doi: 10.1128/JB.182.8.2299-2306.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olmsted SB, Dunny GM, Erlandsen SL, Wells CL. 1994. A plasmid-encoded surface protein on Enterococcus faecalis augments its internalization by cultured intestinal epithelial cells. J Infect Dis 170:1549–1556. [DOI] [PubMed] [Google Scholar]
- 57.Schlievert PM, Gahr PJ, Assimacopoulos AP, Dinges MM, Stoehr JA, Harmala JW, Hirt H, Dunny GM. 1998. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect Immun 66:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kreft B, Marre R, Schramm U, Wirth R. 1992. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells. Infect Immun 60:25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Süssmuth SD, Muscholl-Silberhorn A, Wirth R, Susa M, Marre R, Rozdzinski E. 2000. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect Immun 68:4900–4906. doi: 10.1128/IAI.68.9.4900-4906.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chow JW, Thal LA, Perri MB, Vazquez JA, Donabedian SM, Clewell DB, Zervos MJ. 1993. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob Agents Chemother 37:2474–2477. doi: 10.1128/AAC.37.11.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rakita RM, Vanek NN, Jacques-Palaz K, Mee M, Mariscalco MM, Dunny GM, Snuggs M, van Winkle WB, Simon SI. 1999. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect Immun 67:6067–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhatty M, Cruz MR, Frank KL, Gomez JA, Andrade F, Garsin DA, Dunny GM, Kaplan HB, Christie PJ. 2015. Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance) and PrgC contribute to plasmid transfer, biofilm formation and virulence. Mol Microbiol 95:660–677. doi: 10.1111/mmi.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manson JM, Hancock LE, Gilmore MS. 2010. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc Natl Acad Sci U S A 107:12269–12274. doi: 10.1073/pnas.1000139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chatterjee A, Cook LCC, Shu C-C, Chen Y, Manias DA, Ramkrishna D, Dunny GM, Hu W-S. 2013. Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc Natl Acad Sci U S A 110:7086–7090. doi: 10.1073/pnas.1212256110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bensing BA, Manias DA, Dunny GM. 1997. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act post-transcriptionally to activate expression of downstream conjugation functions. Mol Microbiol 24:285–294. doi: 10.1046/j.1365-2958.1997.3301710.x. [DOI] [PubMed] [Google Scholar]