ABSTRACT
Bacterial cell division is an essential and highly coordinated process. It requires the polymerization of the tubulin homologue FtsZ to form a dynamic ring (Z-ring) at midcell. Z-ring formation relies on a group of FtsZ-associated proteins (Zap) for stability throughout the process of division. In Escherichia coli, there are currently five Zap proteins (ZapA through ZapE), of which four (ZapA, ZapB, ZapC, and ZapD) are small soluble proteins that act to bind and bundle FtsZ filaments. In particular, ZapD forms a functional dimer and interacts with the C-terminal tail of FtsZ, but little is known about its structure and mechanism of action. Here, we present the crystal structure of Escherichia coli ZapD and show it forms a symmetrical dimer with centrally located α-helices flanked by β-sheet domains. Based on the structure of ZapD and its chemical cross-linking to FtsZ, we targeted nine charged ZapD residues for modification by site-directed mutagenesis. Using in vitro FtsZ sedimentation assays, we show that residues R56, R221, and R225 are important for bundling FtsZ filaments, while transmission electron microscopy revealed that altering these residues results in different FtsZ bundle morphology compared to those of filaments bundled with wild-type ZapD. ZapD residue R116 also showed altered FtsZ bundle morphology but levels of FtsZ bundling similar to that of wild-type ZapD. Together, these results reveal that ZapD residues R116, R221, and R225 likely participate in forming a positively charged binding pocket that is critical for bundling FtsZ filaments.
IMPORTANCE Z-ring assembly underpins the formation of the essential cell division complex known as the divisome and is required for recruitment of downstream cell division proteins. ZapD is one of several proteins in E. coli that associates with the Z-ring to promote FtsZ bundling and aids in the overall fitness of the division process. In the present study, we describe the dimeric structure of E. coli ZapD and identify residues that are critical for FtsZ bundling. Together, these results advance our understanding about the formation and dynamics of the Z-ring prior to bacterial cell division.
INTRODUCTION
Bacterial cell division is an essential and complex process that requires the coordinated assembly of a multiprotein molecular machine termed the divisome. The divisome is responsible for constriction of the inner and outer membranes, synthesis of septal peptidoglycan, and subsequent septum formation. In Escherichia coli, divisome proteins are recruited in a hierarchical manner and can be divided into three main groups based on their order of assembly: (i) the proto-ring, (ii) early divisome proteins, and (iii) late divisome proteins (1, 2). The successful assembly of the divisome depends on the initial formation of the Z-ring, which is comprised of the 40-kDa bacterial tubulin homologue FtsZ. FtsZ assembles into filaments in a GTP-dependent manner and a head-to-tail fashion (3–6). The filaments are then tethered to the membrane, forming the Z-ring. They act as the scaffold for divisome assembly and provide the driving force for cell constriction (7–10). Although the localization of FtsZ filaments and the requirement for recruiting other divisome proteins is well established, the organization and dynamics of the Z-ring have yet to be fully described.
FtsZ filaments are highly dynamic in vivo and in vitro, constantly undergoing polymerization, GTP hydrolysis, and depolymerization (11–16). As such, modulating the stability of the Z-ring is important for the successful formation and disassembly of the divisome. In E. coli, this process is mediated by FtsZ-associated proteins (Zap proteins). The first Zap proteins recruited during divisome assembly are ZapA, ZapB, ZapC, and ZapD and are predicted to have overlapping functions (17–21). These Zap proteins stabilize FtsZ filaments prior to cell division by slowing depolymerization, thereby inhibiting GTPase activity (17, 19, 21, 22). It has recently been shown that a fifth Zap protein, ZapE, also functions during division; however, it is thought to have a function opposing that of ZapA, ZapB, ZapC, and ZapD, as it binds to and causes the dissociation of FtsZ filaments (23). Although individually Zap proteins are not essential for divisome assembly, it is thought that the collective role they play in stabilizing FtsZ filaments prior to cell division is critical. In vitro ZapA, ZapC, and ZapD bundle FtsZ filaments by increasing the lateral interactions between individual filaments (17–19, 21). Interactions between these Zap proteins and FtsZ also result in much longer filaments because they inhibit the GTPase activity of FtsZ in vitro (17, 18, 24). ZapB does not interact directly with FtsZ but rather interacts with ZapA to cross-link FtsZ-bound ZapA, providing another level of filament bundling (22, 25, 26).
Studies of ZapA from E. coli (EcZapA) revealed that its X-ray crystallographic structure is significantly different from the previously solved Pseudomonas aeruginosa ZapA (PaZapA) structure. The orientation of the protomers within the tetrameric ZapA structure was different between EcZapA and PaZapA, which revealed a possible difference in function between the two proteins (27). ZapA specifically bundles FtsZ filaments by binding the C-terminal tail of FtsZ (25). This binding inhibits the GTPase activity of FtsZ through a charged α-helix located on each of the four protomers within the functional tetramer (25, 27). ZapD is thought to bundle FtsZ filaments in a similar manner by binding to the C-terminal tail of FtsZ and inhibiting GTPase activity. However, ZapD is observed to form dimers in solution, in contrast to tetrameric ZapA (18). Identification of specific sites of interaction between FtsZ and ZapD will provide important information about the function of ZapD and its possible overlapping role with ZapA.
Here, we present the crystal structure of ZapD from E. coli (EcZapD). The structure of EcZapD reveals a series of charged, surface-exposed residues that are candidates for involvement in FtsZ-ZapD interactions. Chemical cross-linking of FtsZ and ZapD followed by liquid chromatography-mass spectrometry (LC-MS) revealed that these two proteins associate via a series of ZapD arginine residues. Based on the EcZapD structure and the cross-linking results, nine site-directed ZapD variants were generated. Comparison of FtsZ filament sedimentation and bundling with these ZapD variants revealed specific residues that influence FtsZ bundling in vitro compared to FtsZ bundled with wild-type (WT) ZapD. Based on these results, we suggest that ZapD contains an FtsZ binding pocket that is comprised of several charged residues at the interface between the α-helical bundle and β-sandwich domains.
MATERIALS AND METHODS
Cloning, site-directed mutagenesis, and construction of the zapD deletion strain.
The zapD gene (corresponding to NCBI accession number BAB96671) was amplified from E. coli W3110 genomic DNA using the PCR primers 311F (TTCATAGAGAATTCATGCATCATCATCATCATCATAGCATCGAAGGTCGAAGTGGTATGCAGACCCAGGTCCTTTTTG) and 311R (TTACCTTCGTGAAGCTTTTAGCAACAGGCCAGTTCGAAATCCAGACGTTCCGGTACCT); this DNA fragment was then digested using EcoRI and HindIII (cut sites are underlined) and ligated into the corresponding cut sites of pBAD24 to generate pEJR031, which expresses ZapD with an N-terminal His6 tag. Site-directed mutagenesis was performed with the QuikChange lightning site-directed mutagenesis kit (Stratagene) using the primers listed in Table 1. The ΔzapD strain was made using the λ red deletion system as described previously (28, 29). The zapD gene was replaced in its entirety by the cat gene from pKD3. Gene replacement was mediated by gene products from pSIM6 when expressed in a temperature-dependent manner in E. coli W3110. Construction of the ftsZ-containing plasmid was described previously (27).
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description/genome/sequence (5′-3′) | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 hsdR17(rK− mK+) λ− | Invitrogen |
| BL21 | F− ompT gal dcm lon hsdSB(rB− mB−) λ (DE3)pLysS (Cmr)a | New England Biolabs |
| W3110 | rph-1 IN(rrnD-rrnE) | Coli Genetic Stock Centre |
| EJR02 | W3110 ΔzapD | This work |
| Plasmids | ||
| pEJR005b | pET28A with WT ftsZ from E. coli | 27 |
| pEJR031 | pBAD24 with WT His6-zapD from E. coli | This work |
| pEJR031_R16A | pBAD24 with R16A His6-zapD from E. coli | This work |
| pEJR031_R20A | pBAD24 with R20A His6-zapD from E. coli | This work |
| pEJR031_R56A | pBAD24 with R56A His6-zapD from E. coli | This work |
| pEJR031_E114A | pBAD24 with E114A His6-zapD from E. coli | This work |
| pEJR031_R116A | pBAD24 with R116A His6-zapD from E. coli | This work |
| pEJR031_R124A | pBAD24 with R124A His6-zapD from E. coli | This work |
| pEJR031_R176A | pBAD24 with R176A His6-zapD from E. coli | This work |
| pEJR031_R221A | pBAD24 with R221A His6-zapD from E. coli | This work |
| pEJR031_R225A | pBAD24 with R225A His6-zapD from E. coli | This work |
| Primers for mutagenesis | ||
| R16A_F | CAATGCGCAGCCATGTAGCCATTTTTTCATTTAGTGGATGTTCA | This work |
| R16A_R | TGAACATCCACTAAATGAAAAAATGGCTACATGGCTGCGCATTG | This work |
| R20A_F | TGCTGAATCAAAAACTCAATGGCCAGCCATGTACGCATTTTTTC | This work |
| R20A_R | GAAAAAATGCGTACATGGCTGGCCATTGAGTTTTTGATTCAGCA | This work |
| R56A_F | GTGCGGACTTCGCCGGCCTCGAAAACATCCAG | This work |
| R56A_R | CTGGATGTTTTCGAGGCCGGCGAAGTCCGCAC | This work |
| E114A_F | GAGCAATCAAACGATCTGCACGCAGAAATTGCCCG | This work |
| E114A_R | CGGGCAATTTCTGCGTGCAGATCGTTTGATTGCTC | This work |
| R116A_F | ACGCACCAGAGCAATCAAAGCATCTTCACGCAGAAATTGC | This work |
| R116A_R | GCAATTTCTGCGTGAAGATGCTTTGATTGCTCTGGTGCGT | This work |
| R124A_F | CTGGGATGCTCAGTGCCTGACGCACCAGAG | This work |
| R124A_R | CTCTGGTGCGTCAGGCACTGAGCATCCCAG | This work |
| R176A_F | GGGCCGACTGGGCAATTAAATCCAGCACCATGGT | This work |
| R176A_R | ACCATGGTGCTGGATTTAATTGCCCAGTCGGCCC | This work |
| R221A_F | GGCATAAAACGAATGGCAAAAGCGCTCTTATGTCCGGAAATTTG | This work |
| R221A_R | CAAATTTCCGGACATAAGAGCGCTTTTGCCATTCGTTTTATGCC | This work |
| R225A_F | GTGTCCAGCGGCATAAAAGCAATGGCAAAACGGCTCTT | This work |
| R225A_R | AAGAGCCGTTTTGCCATTGCTTTTATGCCGCTGGACAC | This work |
Cmr, chloramphenicol resistance.
Protein expression and purification.
FtsZ was expressed and purified as described previously (27). Purified, concentrated FtsZ was stored at 4°C for no longer than 4 days in PEM-KOH buffer [50 mM piperazine-N,N′-bis(ethanesulfonic acid)–KOH, 1 mM EDTA, 5 mM MgCl2, pH 6.5]. His6-ZapD was expressed in E. coli BL21(DE3)pLysS cells carrying the pEJR031 plasmid (Table 1). An overnight culture was diluted 1/100 into lysogeny broth (LB) medium supplemented with 150 μg/ml ampicillin and 0.2% (wt/vol) glucose. Growth continued for 3 h, and then cells were pelleted and resuspended in LB medium containing 150 μg/ml ampicillin and 0.2% (wt/vol) l-arabinose (Sigma) for induction. Expression proceeded for 1 h, after which cells were collected by centrifugation at 5,000 × g for 8 min (JA 10.1 rotor). Cell pellets were resuspended in A2 buffer (20 mM Tris-HCl, 100 mM KCl, 20 mM imidazole, 1 mM EDTA, 10% glycerol, pH 8.0) and lysed by French press. Cell debris was removed by low-speed centrifugation at 8,000 × g for 15 min (JA 10.1 rotor), and membranes were removed by high-speed centrifugation at 100,000 × g for 1 h (Ti70 rotor). His6-ZapD was purified by immobilized metal affinity chromatography as described previously for His6-ZapA (27). His6-ZapD was stable for 8 weeks at 4°C in A2 buffer.
Crystallization, data collection, structure determination, and analysis of ZapD.
ZapD crystals were grown by sitting-drop vapor diffusion. ZapD (1 μl of a 15 mg/ml solution) was mixed with an equal volume of a solution of 1.2 M ammonium sulfate, 100 mM HEPES, pH 7.5, and 10% polyethylene glycol (PEG) 600 and then was equilibrated against the same solution. Crystals grew as hexagonal prisms up to 300 μm in length. Crystals were frozen in liquid nitrogen after surface water was removed by immersion in Paratone-N oil. Data were collected at the Canadian Light Source, beamline 08ID. Crystals were highly variable in quality, with the best of the dozen tested diffracting to 2.4 Å; even the best crystals showed signs of lattice disorder. Data were processed in XDS and scaled in XSCALE (30). Crystals were of the hexagonal space group P6422. The structure was determined by molecular replacement using PHASER in PHENIX (31, 32), with the structural genomics-derived Vibrio parahaemolyticus ZapD (VpZapD) structure (Protein Data Bank [PDB] entry 2OEZ) serving as a model. The structure was rebuilt in Coot (33) and refined in PHENIX. Structure figures were prepared in PyMol.
In vitro protein cross-linking, in-gel digestion, and mass spectrometry analysis.
Reaction mixtures were prepared containing FtsZ (4.8 μM), ZapD (4.8 μM), and the chemical cross-linker glutaraldehyde (0.05% [vol/vol]) and incubated at room temperature for 5, 10, and 15 min. At each time interval, cross-linking was stopped by mixing with 5× SDS sample buffer (250 mM Tris-HCl, pH 6.8, 10% [wt/vol] SDS, 30% [vol/vol] glycerol, 5% [vol/vol] β-mercaptoethanol, 0.02% [wt/vol] bromophenol blue) and stored at −20°C for analysis. Conditions were optimized to use the lowest percentage of glutaraldehyde and the shortest incubation time that showed significant numbers of cross-linked proteins by Coomassie staining (SimplyBlue SafeStain; Invitrogen). Control reactions in which FtsZ was cross-linked without ZapD and ZapD was cross-linked without FtsZ were also performed. Samples from optimized reactions were then run on 4 to 20% precast gels (Bio-Rad) and stained with Invitrogen SimplyBlue SafeStain. Gel fragments containing apparent cross-linked FtsZ-ZapD, as detected by anti-His Western blotting (described previously [27]), were excised using a clean razor blade, and in-gel digestion was performed. Briefly, the gel slice was destained with 50 mM ammonium bicarbonate in 50% (vol/vol) acetonitrile, and cysteine residues were then reduced with 100 mM dithiothreitol (DTT) and alkylated with 55 mM iodoacetamide. After vacuum centrifugation, the gel slice was subjected to protease digestion using 20 μl of a sequencing-grade modified trypsin solution (20 μg/ml in 100 mM ammonium bicarbonate; Promega) at 37°C for 12 h. The resulting tryptic peptides were extracted from the gel slices by one aqueous wash (50 μl) followed by 2 washes (75 μl) with 5% (vol/vol) formic acid in 50% (vol/vol) acetonitrile, and the combined solution was concentrated to 10 μl using vacuum centrifugation (34).
LC-MS analyses were performed on an Agilent 1260 high-performance liquid chromatography (HPLC) system interfaced with an Agilent UHD 6530 quadruple time-of-flight (Q-TOF) mass spectrometer at the Mass Spectrometry Facility of the Advanced Analysis Centre, University of Guelph. A C18 column (100 mm by 2.1 mm by 2.7 μm; Agilent AdvanceBio Peptide Map) was used for chromatographic separation with the following solvents: water with 0.1% (vol/vol) formic acid for A and acetonitrile with 0.1% (vol/vol) formic acid for B. The following mobile-phase gradient procedure was used: initial conditions, 2% B increasing to 45% B in 40 min and then to 55% B in 10 more minutes, followed by a column wash at 95% B and 10 min of reequilibration. The first 2 and last 5 min of the gradient process were sent to waste and not the spectrometer. The flow rate was maintained at 0.2 ml/min. The mass spectrometer electrospray capillary voltage was maintained at 4.0 kV and the drying gas temperature at 350°C with a flow rate of 13 liters/min. The nebulizer pressure was 40 lb/in2, and the fragmentor was set to 150 V. Nitrogen was used both as the nebulizing and drying gas and as the collision-induced gas. The mass-to-charge ratio was scanned across the m/z range of 300 to 2,000 m/z in 4-GHz (extended dynamic range) positive-ion auto MS/MS mode. Three precursor ions per cycle were selected for fragmentation. The instrument was externally calibrated with the ESI TuneMix (Agilent). The sample injection volume was 100 μl (35).
FtsZ sedimentation assays.
FtsZ sedimentation assays were performed as described previously for FtsZ and ZapA (27). Briefly, FtsZ and ZapD were mixed together in PEM-KOH buffer and equilibrated at 30°C for 5 min, followed by GTP addition to a final concentration of 1 mM. Reaction mixtures were incubated for 5 min at 30°C to allow for bundling, and the soluble and pelleted fractions were then separated, as described previously (27). For the sedimentation assays, soluble protein fractions were collected from the supernatant of sedimented reactions, while pellet fractions were the result of resuspension of sedimented material in an equal volume of PEM-KOH buffer. All fractions were mixed with 5× SDS sample buffer for analysis by SDS-PAGE and Coomassie staining (SimplyBlue SafeStain; Invitrogen). Densitometric analysis was performed using ImageLab Software (Bio-Rad) to determine the amount of FtsZ sedimented relative to the total FtsZ from each reaction. Negative controls were run with each sedimentation assay containing FtsZ and the ZapD variant without GTP. The amount of sedimented FtsZ in this reaction was then subtracted from all other pellet fractions to account for FtsZ coming out of solution without bundling.
In vivo characterization and complementation.
For the in vivo characterization of ZapD variants, electrocompetent E. coli W3110 cells were transformed with each pBAD24 vector containing a zapD variant. In each experiment a single colony was inoculated into liquid LB medium containing 150 μg/ml ampicillin and 0.2% (wt/vol) glucose and grown overnight at 37°C. Cells were then washed in 1× phosphate-buffered saline (PBS) and resuspended at a 1/100 dilution in LB medium supplemented with ampicillin, 0.1% (vol/vol) glucose, and 0.2% (vol/vol) arabinose. After 2 h of growth, samples were taken both for analysis by SDS-PAGE/Western immunoblotting and for cell measurements. For SDS-PAGE and immunoblotting, cells were mixed with 5× SDS sample buffer and gels were loaded normalized to optical density at 600 nm (OD600). Gels were transferred to nitrocellulose membranes and blocked with 2% (wt/vol) bovine serum albumin (BSA). Membranes were incubated with a mouse anti-His6 antibody (Clontech) per the manufacturer's guidelines. Membranes were then incubated with a goat anti-mouse IgG antibody conjugated to alkaline phosphatase (Sigma) diluted 1:20,000. Washes were performed using 1× Tris-buffered saline containing 0.05% (vol/vol) Tween 20 (TBST) between all steps. Cells also were imaged using a Leica DM2000 LED light microscope equipped with a ProgRes CT3 camera (Jenoptik AG). Cell length measurements were determined using ImageJ (36), measuring >100 cells per sample.
TEM.
For imaging FtsZ filaments and FtsZ-ZapD bundles, samples were adhered to plasma-cleaned 200-mesh copper grids for 45 s. After wicking away excess sample, grids were washed once in PEM-KOH to remove excess unbound protein, washed in double-distilled water to remove reactive salts, and then stained for 30 s with 2% (wt/vol) uranyl acetate. Grids were imaged using an FEI Tecnai G2 F20 microscope operated at 120 kV. Transmission electron microscopy (TEM) micrographs were analyzed, and the mean numbers of filaments per bundle (obtained from >10 fields of view per sample; n = 50 bundles per ZapD variant) were plotted for each ZapD variant.
Gel filtration chromatography.
Gel filtration was performed using the Biologic Duoflow chromatography system (Bio-Rad) equipped with a Superdex 200 column (GE Healthcare). Purified His6-ZapD variants (500 μl of protein at 2.0 to 6.0 mg/ml) were loaded onto the column and washed with 2 column volumes of A2 buffer. Fractions containing dimeric ZapD were collected and saved for analysis by SDS-PAGE and Western immunoblotting.
Protein structure accession number.
The EcZapD structure has been deposited at the RCSB Protein Data Bank (PDB) under reference number 5DKO.
RESULTS
Structure of Escherichia coli ZapD.
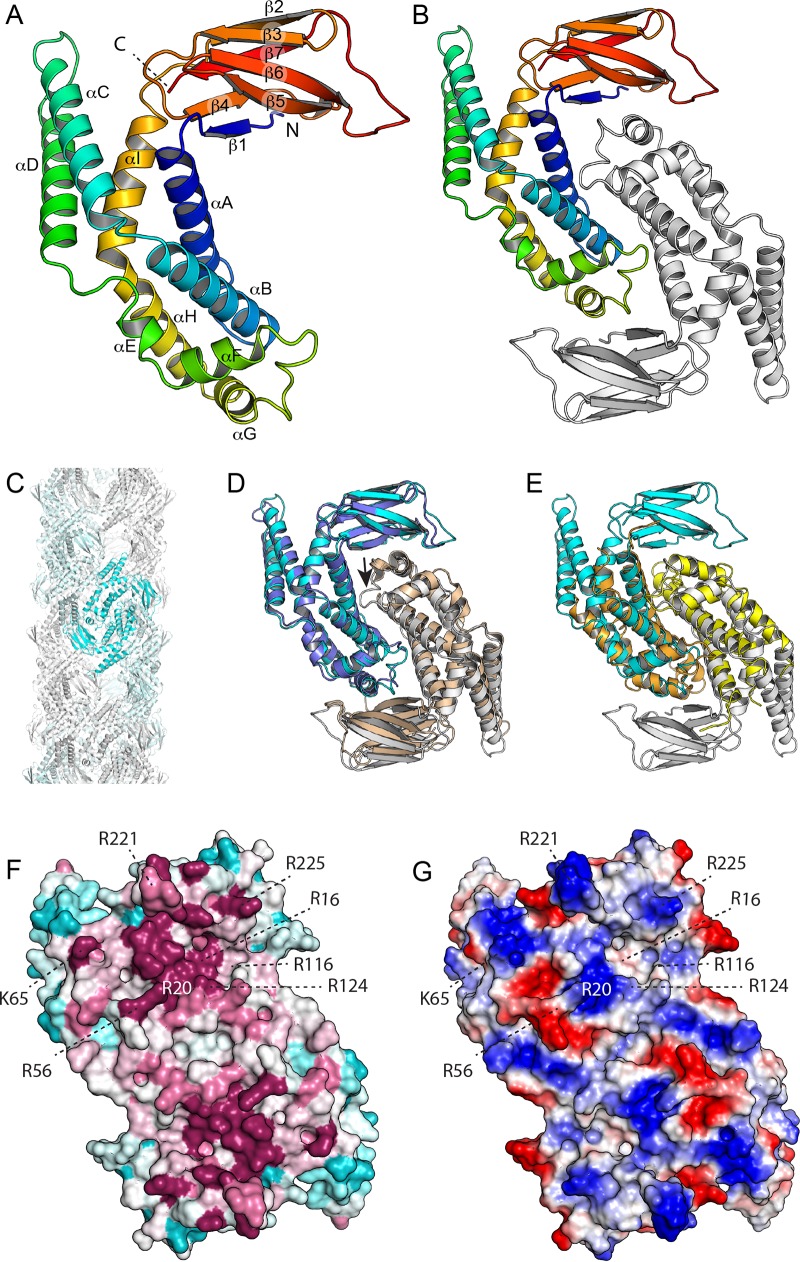
The structure of EcZapD was determined by molecular replacement at 2.4-Å resolution and refined to an Rfree of 0.288 (Table 2). The structure has a single molecule in the asymmetric unit, and despite high overall atomic displacement parameters, all residues are ordered except residues 1 and 2. The EcZapD protomer has a central α-helical domain that can be loosely described as a pair of three helical bundles with a distinct kink between them (Fig. 1A). The C terminus forms a seven-stranded jellyroll-like β-sandwich domain, with one β-strand contributed from the N terminus. This sandwich has topology 1, 4, 7, 2 on one sheet and 3, 6, 5 on the other. EcZapD forms a tight dimer in the crystal, with the α-helical domains interacting primarily through helices 1 and 2, and strands of the β-sandwich domain packing on helix 7 (Fig. 1B). Analysis by PISA indicates that this interface buries 1,871 Å2 per protomer with a dissociation free energy of −28.1 kCal/mol, indicating a stable biological interface. EcZapD dimers are further organized into large fiber-like arrangements in the crystal, with a diameter of 110 Å (Fig. 1C). However, the contacts between dimers are not strong enough to be unambiguously biologically relevant, the interfaces are poorly conserved, and we have not observed ZapD alone to form filaments by centrifugation or electron microscopy. It would therefore seem that this phenomenon is simply a crystal packing artifact.
TABLE 2.
Data collection, model refinement, and final structure statistics
| Parameter | Value(s) |
|---|---|
| Crystallographic data collection statistics | |
| Space group | P6422 |
| Cell dimensions | |
| a = b | 108.9 |
| c | 106.7 |
| Wavelength (Å) | 0.97888 |
| Resolution (Å) | 2.4 |
| Unique reflections (no.) | 26,464 |
| Redundancy | 7.9 |
| Completeness (last shell)a | 0.952 (0.961) |
| <I/σ(I)> (last shell)a | 13.7 (2.3) |
| Rsym (last shell)a | 0.078 (0.76) |
| X-ray structure refinement statistics | |
| Rcryst | 0.247 |
| Rfree | 0.288 |
| Asymmetric unit contents (no.) | |
| Protein chains | 1 |
| Water molecules | 10 |
| Other molecules | 3 SO4 |
| Avg ADPsb (Å2) | |
| Protein | 74.3 |
| Water | 61.1 |
| RMSD bond lengths (Å) | 0.003 |
| RMSD bond angles (°) | 0.0665 |
| Ramachandran favored (%) | 95.5 |
| Ramachandran outliers (%) | 0.8 |
The last shell includes all reflections between 2.46 and 2.40 Å.
ADPs, atomic displacement parameters.
FIG 1.
Structure of E. coli ZapD. (A) The structure of the ZapD protomer is colored by secondary structure progression from the N (blue) to the C terminus (red). The ZapD structure is organized as a β-sandwich domain attached to an α-helical domain. Note that αC and αD protrude from the rest of the structure. (B) Structure of the ZapD dimer. (C) ZapD packing in the crystal results in large hollow fibers 110 Å in diameter. However, we find no evidence that this organization contributes to ZapD's function in vivo. (D) Superposition of E. coli ZapD (cyan and white) and ZapD from V. parahaemolyticus (blue and white; PDB entry 2OEZ). A single protomer (blue/cyan) was superimposed by DALI, with the second protomer positioned as it maps to the dimer interface. Note that in the V. parahaemolyticus ZapD structure, the αF-αG loop is largely disordered (small arrow). (E) Superposition of VpC_cass2 (PDB entry 3JRT; yellow/orange) dimer on the E. coli ZapD structure (cyan/white). Note that the VpC_cass2 structure superposes well on the core of the ZapD helical domain. (F) E. coli ZapD colored by sequence conservation score. A multiple-sequence alignment of ZapD homologs was mapped onto the ZapD structure using ConSurf. Highly conserved residues are shown in magenta, poorly conserved residues are in cyan, and residues with intermediate conservation are shown in white. (G) Electrostatic potentials are mapped onto the EcZapD surface. Blue indicates electropositive regions, and red indicates electronegative regions.
Searching the RCSB PDB with DALI (37) reveals PDB entry 2OEZ as the only protein with strong similarity to EcZapD, with a Z score of 27.1 and root mean square deviation (RMSD) of 1.7 Å over 243 residues (Fig. 1D). This protein, the Vibrio parahaemolyticus ZapD (VpZapD) ortholog, is 40% identical to EcZapD. Its structure is labeled as a protein of unknown function (DUF1342) and is unpublished. The EcZapD β-stranded domain resembles little else in the PDB; the closest structural homologs are domains of viral shell proteins, such as the adenovirus hexon protein (PDB entry 3ZIF; Z score of 4.2; RMSD of 2.8 Å over 60 residues). The helical subdomain does, however, have some partial resemblance to various α-helical bundle proteins, by far the closest of which is an integron cassette protein, VpC_cass2 (PDB entry 3JRT; Z score of 8.5; RMSD of 3.3 Å over 119 residues), with αA, αB, αE, αF, and αG having corresponding counterparts (38) (Fig. 1E). The Z score, however, likely underestimates the relatedness, as this protein also dimerizes using a geometry very similar to that observed in EcZapD, doubling the number of corresponding elements; this broader resemblance suggests an intriguing evolutionary relationship between the two proteins. In the absence of a functional understanding of Vpc_cass2, it is unclear whether there are any functional similarities between the two proteins.
Mapping sequence conservation onto the surface of EcZapD using ConSurf (39) reveals that the face on which αE and αF are exposed is clearly more conserved than the opposite one (Fig. 1F). This face is also noticeably more basic than the opposite, less conserved face (Fig. 1G), with several key conserved arginine residues. This face has two distinct clusters of conserved basic residues, with each site spanning both protomers.
Chemical cross-linking of purified ZapD and FtsZ reveals targets for site-directed mutagenesis.
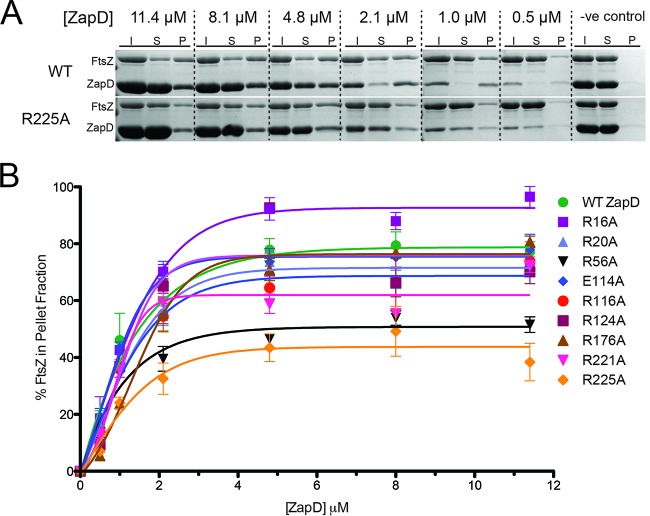
ZapD and FtsZ were overexpressed, purified, and then chemically cross-linked using glutaraldehyde at a concentration of 0.05% (vol/vol) for 5, 10, and 15 min. Control reaction mixtures containing only ZapD and glutaraldehyde incubated for 15 min (Fig. 2A, lane 4) showed monomeric ZapD, while control reactions with only FtsZ plus glutaraldehyde showed several higher-molecular-mass bands (Fig. 2A, lane 5). When both ZapD and FtsZ were incubated with glutaraldehyde, several additional high-molecular-mass bands were observed that were not present in either control reaction. The higher-molecular-mass bands from these reactions (Fig. 2A, lane 3, numbered 1 to 5) were cut out, and an in-gel protein digestion was performed prior to analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS) Q-TOF to detect and identify peptides and cross-link sites. ZapD was found cross-linked to itself and to FtsZ in these samples (Fig. 2 and Table 3). Residues R116 and R221 on ZapD, which cross-linked close to the C terminus of FtsZ, were attractive targets for investigation because the C-terminal tail of FtsZ was previously shown to be important for the ZapD-FtsZ interaction (18). Residue R225, which cross-linked to FtsZ in more than one sample, and surface-exposed charged residues within the crystal structure of ZapD were also chosen for further investigation (Table 3). In order to study the involvement of these residues in FtsZ bundling, they were individually changed to alanine by site-directed mutagenesis and the following ZapD variants were created: ZapD R16A, R20A, R56A, E114A, R116A, R124A, R176A, R221A, and R225A. ZapD variants were analyzed by gel filtration and compared to WT ZapD to confirm that each amino acid substitution did not disrupt the ZapD dimerization interface. All ZapD variants eluted in a peak corresponding to 44 kDa, except ZapD R225A, which eluted slightly earlier (see Fig. S1 in the supplemental material). This change in elution profile shifts the apparent molecular mass to ∼46 kDa. These results are consistent with previous reports showing that WT ZapD elutes as a dimer when analyzed by gel filtration (18).
FIG 2.
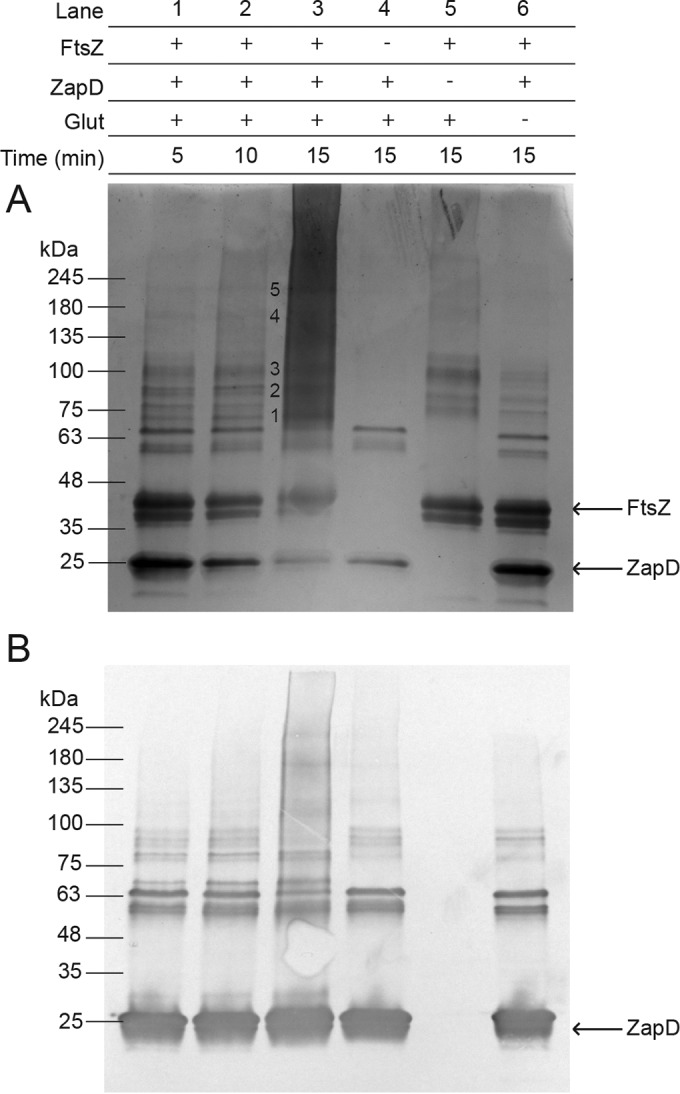
Chemical cross-linking of purified WT ZapD and FtsZ and gel filtration of WT ZapD and ZapD variants. Reaction mixtures were prepared in the presence (+) or absence (−) of purified FtsZ, ZapD, and/or the chemical cross-linker glutaraldehyde (Glut) at a concentration of 0.05% (vol/vol). Experimental reaction mixtures containing all three components were incubated for 5, 10, and 15 min, and control reaction mixtures were incubated for 15 min. The legend at the top of the figure indicates the added components for each reaction mixture, and representative images are shown below for a Coomassie-stained SDS-PAGE gel (A) and an anti-His6 Western immunoblot detecting His6-ZapD (B). Bands designated with a number were cut out for processing and analysis.
TABLE 3.
Chemical cross-linking of purified ZapD and FtsZ
| Site on: |
Scorea | Sample no.b | |
|---|---|---|---|
| ZapD | FtsZ | ||
| R116 | K380 | 44 | 1 |
| R20 | K14 | 61 | 3 |
| R70 | K155 | 60 | 3 |
| R221 | R319 | 55 | 4 |
| R225 | K51 | 42 | 4 |
| R225 | K73 | 42 | 4 |
For the score from decoy analysis, >50 equals a <5% false detection rate.
Sample number refers to the bands as labeled in Fig. 2A.
Individual ZapD residue mutations affect FtsZ sedimentation.
The ability of Zap proteins to bundle FtsZ filaments in vitro has been previously measured using FtsZ sedimentation assays (17, 19, 24, 27, 40, 41). The extent of FtsZ bundling can be determined by incubating a Zap protein with FtsZ and GTP for defined durations and concentrations. The soluble and pellet fractions then can be separated by centrifugation. The relative protein proportions of each component in the soluble and pellet fractions are determined by SDS-PAGE and Coomassie staining and compared to the total protein input for each reaction. In the presence of GTP, WT ZapD bundles FtsZ filaments to an increasing extent until saturation is reached, as seen in representative Coomassie-stained gels (Fig. 3A, top). Sedimentation appears to reach maximal levels between ZapD concentrations of 2.1 and 4.8 μM. If ZapD variants have an altered capacity to bundle FtsZ filaments, we would expect to see the effect in the sedimentation assay. Representative Coomassie-stained gels for the ZapD R225A FtsZ sedimentation assay showed a marked decrease in FtsZ sedimentation at 2.1 and 4.8 μM compared to that of WT ZapD (Fig. 3A, bottom). Sedimentation assays were performed for all ZapD variants in triplicate and analyzed by densitometry. A graph summarizing the results from the densitometric analysis provides information about the overall sedimentation profile and maximum FtsZ sedimentation (Fig. 3B). In addition, 50% effective concentration (EC50) values, which correspond to the concentration of ZapD needed to sediment 50% of FtsZ filaments in each reaction, were calculated (Table 4). For WT ZapD, bundling occurs even when ZapD is present at a low concentration relative to FtsZ (<4.8 μM, subequimolar) (Fig. 3B). When present at equimolar concentrations (4.8 μM), FtsZ bundling by WT ZapD is at its maximum. Increasing the ZapD concentration beyond equimolar does not increase the amount of FtsZ bundling, indicating the association has reached saturation. All ZapD variants showed sedimentation profiles similar to that of WT ZapD (Fig. 3B). The R20A, E114A, R116A, R124A, and R176A variants had maximum FtsZ sedimentation values and EC50s similar to those of the WT. Interestingly, the R16A variant showed an increased maximum sedimentation and a decreased EC50 compared to those of WT ZapD. In contrast, three ZapD variants, the R56A, R221A, and R225A variants, showed decreased maximum FtsZ sedimentation and increased EC50s compared to those of the WT (Table 4).
FIG 3.
ZapD-mediated FtsZ sedimentation assays. (A) Representative Coomassie-stained gels showing the sedimentation profiles of FtsZ with WT ZapD and FtsZ with ZapD R225A. All reactions were prepared with 4.8 μM FtsZ, and ZapD was added at the concentrations indicated. Fractions analyzed included the following: I, the initial protein input to each reaction; S, the soluble portion after centrifugation; and P, the pellet portion. The lane designated -ve control represents bundling assay with 4.8 μM FtsZ and ZapD in the absence of GTP. (B) Sedimentation profiles were assessed by densitometry for WT ZapD and all ZapD variants in triplicate (±standard errors). The densitometric data were plotted as the percentage of FtsZ sedimented, relative to the initial input for each reaction, versus the concentration of ZapD.
TABLE 4.
Summary of results from FtsZ bundling and sedimentation by ZapD variants
| ZapD variant | EC50a (μM) | Max sed.b (%) | No. of filaments/bundlec |
|---|---|---|---|
| WT | 1.86 | 79.5 | 12 |
| R16A variant | 1.26 | 96.5 | 11 |
| R20A variant | 2.49 | 73.6 | 12 |
| R56A variant | 5.92 | 53.9 | 7** |
| E114A variant | 1.74 | 77.9 | 16* |
| R116A variant | 2.77 | 73.8 | NA |
| R124A variant | 1.93 | 92.6 | 11 |
| R176A variant | 2.64 | 80.6 | 11 |
| R221A variant | 3.19 | 72.1 | NA |
| R225A variant | 9.08 | 49.2 | NA |
EC50 corresponds to the concentration of ZapD required to sediment 50% of FtsZ in solution.
Max sed. is the maximum observed sedimentation of FtsZ in the triplicate FtsZ sedimentation assays.
The number of filaments per bundle was counted for each sample from TEM images (n = 50). **, P value of <0.0001 compared to WT results; *, P value of <0.05 compared to WT results. NA, not applicable.
ZapD-FtsZ bundle morphology.
Previous studies have used transmission electron microscopy to show that Zap protein variants with an altered ability to bind and bundle FtsZ filaments change the bundle's appearance in vitro (17, 27, 42). It has been reported that altering even one residue involved in the protein-protein interaction can significantly affect FtsZ bundling (17, 27). Qualitatively, several ZapD variants showed altered FtsZ-bundle morphology compared to FtsZ bundled with WT ZapD (Fig. 4). ZapD R116A and R225A displayed FtsZ bundling that was dramatically different from that of WT ZapD. The negatively stained micrographs showed little organized structure and curved bundles, few close FtsZ filament associations, and no characteristic striation pattern typically associated with the bundles (Fig. 4). ZapD R56A produces noticeably thinner bundles. ZapD R221A produced uncharacteristically elongated FtsZ bundles (see Fig. S2 in the supplemental material) that were highly variable compared to WT ZapD-bundled filaments, with large areas of disorder (Fig. 4). The remaining ZapD variants tested produced FtsZ bundles that were qualitatively comparable to WT ZapD-bundled FtsZ filaments.
FIG 4.

Qualitative assessment of FtsZ bundles in the presence of WT ZapD and ZapD variants. Transmission electron micrographs of FtsZ (4.8 μM) alone (top) and bundled in the presence of WT ZapD and ZapD variants (2.1 μM). Bar, 100 nm.
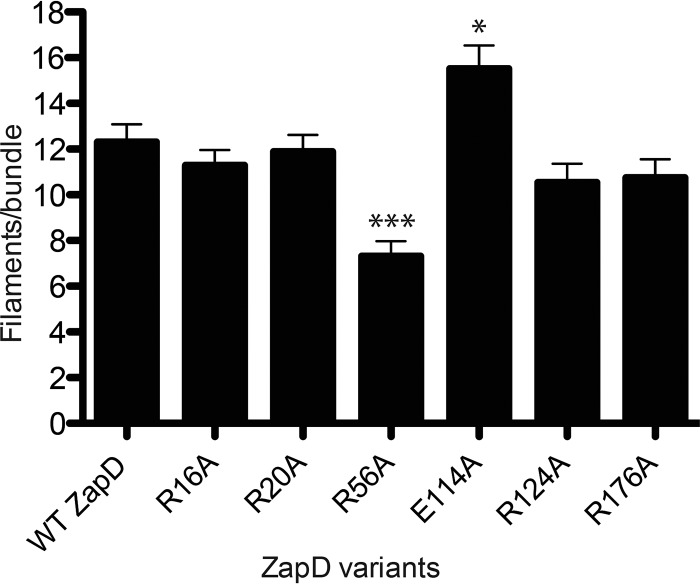
Bundling interactions between Zap proteins and FtsZ filaments have also been previously characterized by reporting the number of FtsZ filaments per bundle (27). Quantitatively, WT ZapD-bundled FtsZ has an average of 12 filaments per bundle, as measured from transmission electron micrographs (Fig. 5 and Table 4). The thin bundles produced by ZapD R56A were estimated to contain an average of 7 FtsZ filaments per bundle, whereas FtsZ bundled by ZapD E114A averaged 16 filaments. Bundles formed in the presence of ZapD R116A, R221A, and R225A variants could not be quantified due to lack of close associations. WT ZapD and ZapD R16A, R20A, R124A, and R176A variants contained similar numbers of filaments per bundle (Fig. 5 and Table 4).
FIG 5.
Quantitative assessment of FtsZ bundles in the presence of WT ZapD and ZapD variants. Shown is a histogram summarizing the mean number of FtsZ filaments per bundle. TEM images were analyzed, and the mean numbers of filaments per bundle ± standard errors were plotted for each ZapD variant. More than 10 fields of view per sample (n = 50 bundles per ZapD variant) were used to calculate means. ***, P < 0.0001; *, P < 0.05 compared to WT ZapD. The mean number of FtsZ filaments per bundle was not enumerated for ZapD R116A, R221A, and R225A, as organized bundles were not observed.
In vivo complementation of ZapD and its dominant-negative phenotype.
Zap gene deletions in E. coli typically produce very subtle phenotypes when observed by light microscopy. Deletion of the zapA gene in E. coli results in a subtle cell elongation phenotype (26, 27), whereas ΔzapD cells do not show an elongation phenotype relative to WT W3110 E. coli cells (see Fig. S3 in the supplemental material), consistent with previous reports (18). Upon complementation of the ΔzapD mutant with a plasmid encoding WT zapD, cells were elongated compared to W3110 cells and the ΔzapD and ΔzapD plus empty pBAD24 mutants (see Fig. S3A in the supplemental material). Expression of all ZapD variants in the ΔzapD background strain, with the exception of R225A, caused significant changes in cell length compared to the ΔzapD mutant expressing WT ZapD (P < 0.05). To assess for a dominant-negative phenotype, ZapD variants were expressed at low levels in E. coli W3110 cells containing a functional chromosomal copy of zapD (see Fig. S3B in the supplemental material). Here, WT ZapD-expressing cells were significantly longer than W3110 and W3110 plus empty pBAD24. Only one ZapD variant (the E114A variant) showed significant elongation (P = 0.0021 compared to the WT), while five variants (the R16A, R56A, R124A, R176A, and R221A variants) showed a significant decrease in cell length compared to cells expressing endogenous WT ZapD (P < 0.05). However, the significance of these results is unclear, since in both experiments the mean cell length of the WT ZapD-overexpressing cells was significantly different from that of control samples (P < 0.001). Expression of ZapD variants from both WT W3110 and ΔzapD mutant E. coli cells showed similar levels of protein expression (see Fig. S3C in the supplemental material).
DISCUSSION
Zap proteins are proposed to stabilize FtsZ filaments, aiding in Z-ring formation and positioning prior to bacterial cell division (24–27, 40–42). Although ZapD, which is widely conserved across proteobacteria, is not essential for cell division in E. coli, it is hypothesized that collectively ZapA, ZapB, ZapC, and ZapD are required for FtsZ filament organization and dynamics (17–21, 27, 43). E. coli ZapD requires FtsZ to localize to midcell, where it promotes Z-ring assembly via molecular mechanisms that overlap ZapA (18). Here, we solved the X-ray crystallographic structure of ZapD from E. coli in order to better understand its role in FtsZ bundling. The EcZapD structure presented here (Fig. 1) is similar to the ZapD structure from Vibrio parahaemolyticus that was previously determined as part of a structural genomics project (PDB entry 2OEZ). However, neither the structure itself nor any analyses of the implications for VpZapD function have been described in the literature. After structure resolution and data analysis using PISA software, we showed that EcZapD is a stable dimer consisting of a central α-helical dimer interface domain and two β-sandwich domains (Fig. 1). The dimeric state of our ZapD variants was confirmed by gel filtration (see Fig. S1 in the supplemental material) for WT ZapD and ZapD variants, suggesting that subtle differences observed in elution profiles between the WT and variants were not due to disrupting the dimerization interface. ZapD R225A showed a slight shift in apparent molecular mass when analyzed by gel filtration. This shift in size was not large enough to account for a change in oligomeric state and is likely due to a conformational change within the β-sheet domain upon mutation of R225 (Fig. 1). Recently, Son and Lee described the protein expression, purification, and crystallization of ZapD from E. coli (44); however, the structure was not reported. In addition, our crystallization conditions differ, as our crystals diffracted to a higher resolution (2.40 Å versus 2.95 Å) and were in the space group P6422 rather than P64 (44).
Further analysis of the EcZapD structure and ZapD-FtsZ cross-linking suggested potential interacting residues. Residues R116 and R221 on ZapD were identified to cross-link near the C terminus of FtsZ. This is consistent with previous results from work by Durand-Heredia et al., where several FtsZ truncations were created to test for interaction with full-length ZapD via a protein interaction platform in Saccharomyces cerevisiae. Their results suggest that ZapD interacts with the C-terminal tail of FtsZ (18). Of the nine ZapD variants generated in this study, five showed FtsZ bundle morphologies different from that of FtsZ bundled with WT ZapD. While the differences in the number of filaments per bundle observed for ZapD R56A and R114A variants were significant compared to that of the WT (Fig. 5), these bundling phenotypes were less severe than those of ZapD R116A, R221A, and R225A variants (Fig. 4). We therefore suggest that residues R116, R221, and R225 play a crucial role in FtsZ interactions and are likely involved in FtsZ filament bundling in vivo.
The results from the FtsZ sedimentation, bundle characterization, and cross-linking experiments are summarized in Fig. 6, where residues are labeled according to their proposed involvement in the FtsZ-ZapD interaction. Residues R116 and R225 are located close to one another and therefore are proposed to be part of a single binding pocket for FtsZ. In addition, residue R221, which is located adjacent to the putative binding pocket, seems to also be involved in FtsZ bundling. The sedimentation profile for ZapD R221A exhibited only a small increase in EC50 compared to that of WT ZapD (Table 4). However, electron micrographs of FtsZ bundled by ZapD R221A showed drastic defects in bundle morphology compared to that of WT ZapD (Fig. 4). Consequently, this ZapD variant appears to still bind FtsZ filaments efficiently but is unable to bundle filaments once bound. The FtsZ-ZapD R221A bundles also demonstrated uncharacteristically long FtsZ filaments compared to that of WT ZapD (see Fig. S2 in the supplemental material). Since ZapD is known to inhibit the GTPase activity of FtsZ (18), we suggest that this ability is retained or even enhanced in ZapD R221A. FtsZ filaments are associated when bundled by ZapD R221A, albeit loosely, and this is likely due to multiresidue interactions between these two proteins.
FIG 6.
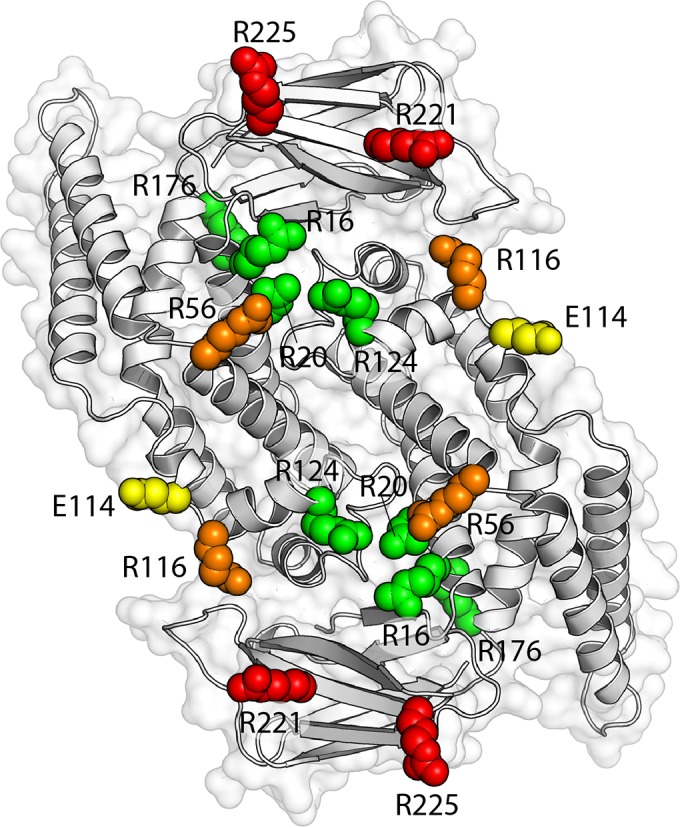
Mutant phenotypes mapped onto the EcZapD dimer structure. Orientation of the ZapD dimer as described for Fig. 1B, but here colored according to the effect of point mutations at each residue. Residues are grouped based on the results of sedimentation assays, bundle morphology, and cross-linking to FtsZ. Residues R16, R20, R124, and R176 are labeled in green. Mutation of these residues presented no significant deviations in EC50 or FtsZ bundle morphology compared to those of WT ZapD, and none of the residues form identifiable cross-links with FtsZ. Residue E114 is labeled in yellow. Mutation of E114 caused altered bundle morphology and a statistically significant increase in the number of FtsZ filaments per bundle compared to WT ZapD (16 versus 12). However, mutation of this residue did not affect the EC50, which was similar to that of WT ZapD (1.74 μM versus 1.86 μM), and E114 did not form identifiable cross-links with FtsZ. Residues R56 and R116 are labeled in orange. Mutation of R56 produced a statistically significant increase in EC50 (5.92 μM) and a statistically significant decrease in the number of FtsZ filaments per bundle compared to WT ZapD (7 versus 12), but R56 did not form identifiable cross-links with FtsZ. Mutation of R116 also demonstrated an increase in EC50 (2.77 μM) but demonstrated ineffective bundling of FtsZ. R116 also formed identifiable cross-links with FtsZ. Mutations in R221 and R225 are colored in red. Mutation of these residues produced a considerable increase in EC50s compared to that of WT ZapD (3.19 μM and 9.08 μM, respectively) and ineffective bundling of FtsZ filaments. R221 and R225 formed multiple identifiable cross-links with FtsZ.
FtsZ bundles formed through interactions with ZapD R56A appear thinner when imaged by TEM and contain significantly fewer FtsZ filaments per bundle than FtsZ-ZapD WT bundles (Fig. 4 and 5). However, the overall organization of these bundles resembles that of WT ZapD-bundled FtsZ. Residue R56 also is not located close to the proposed FtsZ binding pocket on ZapD (Fig. 6). For these reasons, we conclude that while R56 is involved in interactions with FtsZ, it plays a less integral role in FtsZ filament bundling. We hypothesize that R56 is involved in recruiting smaller-order bundles of FtsZ-ZapD. This would account for the WT arrangement and appearance of the bundles and the decreased number of FtsZ filaments per bundle. Mutation of R16 to alanine results in significantly more FtsZ in the pellet fraction than in that of WT ZapD (Fig. 3B). This result indicates that the R16A variant exhibits a gain-of-function phenotype. However, there is no apparent increase in the number of filaments per bundle, and morphologically the R16A ZapD-bundled filaments resemble those formed in the presence of WT ZapD.
Previous work by Durand-Heredia et al. indicates the cellular levels of ZapD are relatively low, with approximately 500 to 800 molecules of ZapD per cell, and represent approximately 1/5 of the known FtsZ cellular concentration (18). Using expression conditions comparable to those of Durand-Heredia et al., our results show that when WT ZapD is expressed for complementation or for overexpression at the lowest levels attainable, cells elongate. It is possible that the expression of ZapD is under tight temporal control during normal cell division, occurring only at the onset of Z-ring assembly. This agrees with the model for Z-ring formation by helical filament condensation, whereby FtsZ forms a loose helical filament at midcell covering the circumference of the cell several times (18, 25, 45, 46). When division begins, the helical filament condenses via an as-yet unknown mechanism to form a tight ring structure. In this model, ZapD would contribute to helical filament condensation. Upon elevated expression during complementation, we suggest that excess ZapD sequesters FtsZ away from midcell and neither the helical filament nor the Z-ring can form. This sequestering of FtsZ might also interfere with Z-ring formation by inhibiting binding of other essential division proteins within the C-terminal region of FtsZ. Without formation of the Z-ring, cells are unable to divide and would lead to the cell elongation phenotype seen when the ΔzapD mutant is complemented with WT ZapD. Overall, our results suggest that ZapD binds FtsZ through several charged residues located at the interface between the α-helical domain of one ZapD protomer and the β-sandwich domain of the other protomer. Taken together, these results advance our understanding of the formation and stabilization of the Z-ring prior to bacterial cell division.
Supplementary Material
ACKNOWLEDGMENTS
We thank Deborah Stewart Khursigara for critical reading of the manuscript and editorial assistance. X-ray data for EcZapD were collected at CLS by Shaun Labiuk.
Funding Statement
This work was funded by Discovery Grants from the Natural Sciences and Engineering Research Council of Canada to M.S.K. (number 327280) and C.M.K. (number 371639). A.M.B. is the recipient of a Natural Sciences and Engineering Research Council of Canada CGS Doctoral Fellowship.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00969-15.
REFERENCES
- 1.Vicente M, Rico AI. 2006. The order of the ring: assembly of Escherichia coli cell division components. Mol Microbiol 61:5–8. doi: 10.1111/j.1365-2958.2006.05233.x. [DOI] [PubMed] [Google Scholar]
- 2.Aarsman MEG, Piette A, Fraipont C, Vinkenvleugel TMF, Nguyen-Distèche M, den Blaauwen T. 2005. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol 55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- 3.Begg KJ, Hatfull GF, Donachie WD. 1980. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: cell division gene ftsQ. J Bacteriol 144:435–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.RayChaudhuri D, Park JT. 1992. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 5.Oliva MA, Cordell SC, Löwe J. 2004. Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol 11:1243–1250. doi: 10.1038/nsmb855. [DOI] [PubMed] [Google Scholar]
- 6.de Boer P, Crossley R, Rothfield L. 1992. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Trimble MJ, Brun YV, Jensen GJ. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J 26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pichoff S, Lutkenhaus J. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol 55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 9.Erickson HP. 2009. Modeling the physics of FtsZ assembly and force generation. Proc Natl Acad Sci U S A 106:9238–9243. doi: 10.1073/pnas.0902258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berezuk AM, Goodyear M, Khursigara CM. 2014. Site-directed fluorescence labeling reveals a revised N-terminal membrane topology and functional periplasmic residues in the Escherichia coli cell division protein FtsK. J Biol Chem 289:23287–23301. doi: 10.1074/jbc.M114.569624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biteen JS, Thompson MA, Tselentis NK, Bowman GR, Shapiro L, Moerner WE. 2008. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat Methods 5:947–949. doi: 10.1038/nmeth.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stricker J, Maddox P, Salmon ED, Erickson HP. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci U S A 99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Erickson HP. 2005. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J Biol Chem 280:22549–22554. doi: 10.1074/jbc.M500895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niu L, Yu J. 2008. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys J 95:2009–2016. doi: 10.1529/biophysj.108.128751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsin J, Fu R, Huang KC. 2013. Dimer dynamics and filament organization of the bacterial cell division protein FtsA. J Mol Biol 425:4415–4426. doi: 10.1016/j.jmb.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson DE, Gueiros-Filho FJ, Erickson HP. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol 186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale CA, Shiomi D, Liu B, Bernhardt TG, Margolin W, Niki H, de Boer PAJ. 2011. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J Bacteriol 193:1393–13404. doi: 10.1128/JB.01245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A. 2012. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J Bacteriol 194:3189–3198. doi: 10.1128/JB.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durand-Heredia JM, Yu HH, De Carlo S, Lesser CF, Janakiraman A. 2011. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J Bacteriol 193:1405–1413. doi: 10.1128/JB.01258-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebersbach G, Galli E, Møller-Jensen J, Löwe J, Gerdes K. 2008. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol Microbiol 68:720–735. doi: 10.1111/j.1365-2958.2008.06190.x. [DOI] [PubMed] [Google Scholar]
- 21.Gueiros-Filho FJ, Losick R. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev 16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galli E, Gerdes K. 2010. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol Microbiol 76:1514–1526. doi: 10.1111/j.1365-2958.2010.07183.x. [DOI] [PubMed] [Google Scholar]
- 23.Marteyn BS, Karimova G, Fenton AK, Gazi AD, West N, Touqui L, Prevost M-C, Betton J-M, Poyraz O, Ladant D, Gerdes K, Sansonetti PJ, Tang CM. 2014. ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. mBio 5:e00022–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohammadi T, Ploeger GEJ, Verheul J, Comvalius AD, Martos A, Alfonso C, van Marle J, Rivas G, den Blaauwen T. 2009. The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry 48:11056–11066. doi: 10.1021/bi901461p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galli E, Gerdes K. 2012. FtsZ-ZapA-ZapB interactome of Escherichia coli. J Bacteriol 194:292–302. doi: 10.1128/JB.05821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buss J, Coltharp C, Huang T, Pohlmeyer C, Wang SC, Hatem C, Xiao J. 2013. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol 89:1099–1120. doi: 10.1111/mmi.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roach EJ, Kimber MS, Khursigara CM. 2014. Crystal structure and site-directed mutational analysis reveals key residues involved in Escherichia coli ZapA function. J Biol Chem 289:23276–23286. doi: 10.1074/jbc.M114.561928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Datta S, Costantino N, Court DL. 2006. A set of recombineering plasmids for gram-negative bacteria. Gene 379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W. 2010. XDS. Acta Crystallogr Sect D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr Sect D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 35.Ma B, Zhang K, Hendrie C, Liang C, Li M, Doherty-Kirby A, Lajoie G. 2003. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom 17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 36.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sureshan V, Deshpande CN, Boucher Y, Koenig JE, Stokes HW, Harrop SJ, Curmi PMG, Mabbutt BC. 2013. Integron gene cassettes: a repository of novel protein folds with distinct interaction sites. PLoS One 8:e52934. doi: 10.1371/journal.pone.0052934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. 2005. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Small E, Marrington R, Rodger A, Scott DJ, Sloan K, Roper D, Dafforn TR, Addinall SG. 2007. FtsZ polymer-bundling by the Escherichia coli ZapA orthologue, YgfE, involves a conformational change in bound GTP. J Mol Biol 369:210–221. doi: 10.1016/j.jmb.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 41.Low HH, Moncrieffe MC, Löwe J. 2004. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J Mol Biol 341:839–852. doi: 10.1016/j.jmb.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 42.Pacheco-Gómez R, Cheng X, Hicks MR, Smith CJI, Roper DI, Addinall S, Rodger A, Dafforn TR. 2013. Tetramerization of ZapA is required for FtsZ bundling. Biochem J 449:795–802. doi: 10.1042/BJ20120140. [DOI] [PubMed] [Google Scholar]
- 43.Rico AI, Krupka M, Vicente M. 2013. In the beginning, Escherichia coli assembled the proto-ring: An initial phase of division. J Biol Chem 288:20830–20836. doi: 10.1074/jbc.R113.479519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Son SH, Lee HH. 2015. Crystallization and preliminary X-ray crystallographic analysis of Z-ring-associated protein (ZapD) from Escherichia coli. Acta Crystallogr Sect F Struct Biol Commun 71:194–198. doi: 10.1107/S2053230X15000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szwedziak P, Wang Q, Bharat TAM, Tsim M, Löwe J. 2014. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife 3:e04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lan G, Dajkovic A, Wirtz D, Sun SX. 2008. Polymerization and bundling kinetics of FtsZ filaments. Biophys J 95:4045–4056. doi: 10.1529/biophysj.108.132837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.