Abstract
FACT (facilitates chromatin transcription), an evolutionarily conserved histone chaperone involved in transcription and other DNA transactions, is upregulated in cancers, and its downregulation is associated with cellular death. However, it is not clearly understood how FACT is fine-tuned for normal cellular functions. Here, we show that the FACT subunit Spt16 is ubiquitylated by San1 (an E3 ubiquitin ligase) and degraded by the 26S proteasome. Enhanced abundance of Spt16 in the absence of San1 impairs transcriptional elongation. Likewise, decreased abundance of Spt16 also reduces transcription. Thus, an optimal level of Spt16 is required for efficient transcriptional elongation, which is maintained by San1 via ubiquitylation and proteasomal degradation. Consistently, San1 associates with the coding sequences of active genes to regulate Spt16's abundance. Further, we found that enhanced abundance of Spt16 in the absence of San1 impairs chromatin reassembly at the coding sequence, similarly to the results seen following inactivation of Spt16. Efficient chromatin reassembly enhances the fidelity of transcriptional elongation. Taken together, our results demonstrate for the first time a fine-tuning of FACT by a ubiquitin proteasome system in promoting chromatin reassembly in the wake of elongating RNA polymerase II and transcriptional elongation, thus revealing novel regulatory mechanisms of gene expression.
INTRODUCTION
In eukaryotes, DNA is packaged into nucleosomes to form chromatin. Each nucleosome within chromatin consists of ∼147 bp of DNA wrapped around a histone octamer containing one histone H3-H4 tetramer and two histone H2A-H2B dimers (1). Thus, chromatin structure plays crucial functions in regulation of DNA transactions such as transcription, replication, and DNA repair (2–4). A variety of factors are involved in altering chromatin structure and, hence, DNA transactions. These factors are generally classified as ATP-independent histone modifying enzymes, ATP-dependent chromatin remodelers, and histone chaperones. Histone modifying enzymes function through addition or removal of specific chemical groups (e.g., acetyl, methyl, ubiquitin, phospho, and SUMO), while ATP-dependent chromatin remodelers alter DNA-histone interactions or the composition of the nucleosome in an ATP-dependent manner. On the other hand, histone chaperones function by binding with nucleosomes or histones to facilitate assembly and/or disassembly of nucleosomes in an ATP-independent fashion. The histone chaperone that was first found to alter chromatin structure during transcription is FACT (facilitates chromatin transcription), which is evolutionarily conserved among eukaryotes (5, 6). In budding yeast (Saccharomyces cerevisiae), FACT is composed of Spt16 (suppressor of Ty) and Pob3 and physically interacts with nucleosomes with the assistance of the HMG (high mobility group) protein Nhp6. Likewise, FACT is also a heterodimer of Spt16 and SSRP1 (structure-specific recognition protein 1) in humans. SSRP1 contains HMG domain, while HMG domain is present in Nhp6 but not Pob3 in yeast. Thus, Pob3 is a truncated homologue of human SSRP1. Both Spt16 and Pob3 carry the pleckstrin homology (PH) domain and other protein interaction domains.
FACT facilitates nucleosomal disassembly at the promoter to stimulate the formation of preinitiation complex and, hence, transcriptional initiation (5–8). FACT also promotes nucleosomal reassembly at the coding sequence in the wake of RNA polymerase II for optimal transcriptional elongation (9–11). Therefore, FACT plays crucial roles in both transcriptional initiation and elongation, and hence, functional impairment of FACT would alter transcription and cellular growth. FACT is found to be upregulated in cancers, especially aggressive and poorly differentiated tumors, and its downregulation leads to tumor cell death (12–14). Thus, FACT has been suggested to be an attractive therapeutic target. However, it is not clearly understood how FACT is upregulated in aggressive tumor cells. Such knowledge would enrich our understanding of the molecular basis of tumorigenesis via FACT and, hence, enable the development of better or specific therapies. It is quite likely that FACT is regulated by the ubiquitin proteasome system (UPS); UPS defects lead to increased levels of FACT and, thus, to oncogenesis. To test this, we carried out experiments in yeast (Saccharomyces cerevisiae) and found that FACT is controlled by UPS, thus hinting at the UPS regulation of FACT in humans. Specifically, we found that Spt16, the FACT subunit, is ubiquitylated by San1 (which is involved in nuclear protein quality control; 15, 16) and degraded by the 26S proteasome. Enhanced abundance of Spt16 in the absence of San1 impairs transcriptional elongation, while decreased abundance of Spt16 also reduces transcription. Consistently, San1 associates with the coding sequences of the active genes to regulate Spt16's abundance for optimal transcriptional elongation. Further, we found that enhanced abundance of Spt16 in the absence of San1 impairs chromatin reassembly at the coding sequence in the wake of RNA polymerase II. Likewise, inactivation of Spt16 also reduces chromatin reassembly. Chromatin reassembly at the coding sequence in the wake of RNA polymerase II promotes the fidelity of transcriptional elongation (9–11). Collectively, our results demonstrate that a fine-tuning of the Spt16 component of FACT by UPS is essential for proper chromatin reassembly and efficient transcriptional elongation, thus revealing a novel UPS regulation of FACT with physiological relevance in gene expression, as presented below.
MATERIALS AND METHODS
Plasmids and strains.
The plasmids and yeast strains used in this study are described in the supplemental material.
Growth media.
The yeast strains were grown in YPD (yeast extract-peptone plus 2% dextrose) to an optical density at 600 nm (OD600) of 1.0 at 30°C prior to formaldehyde-based in vivo cross-linking or harvesting for RNA analysis for the studies at the ADH1, PGK1, PYK1, and PMA1 genes. For ubiquitylation analysis, yeast cells were grown in synthetic complete medium (yeast nitrogen base and complete amino acid mixture plus 2% dextrose) at 30°C to an OD600 of 0.7 and were then treated with 0.1 mM CuSO4 for 6 h. The wild-type and temperature-sensitive (ts) mutant strains of Rpt4 and Rpb1 were grown in YPD at 23°C to an OD600 of 0.85 and were then switched to 37°C for 1 h prior to harvesting. For studies at GAL genes, yeast cells were grown in YPR (yeast extract and peptone plus 2% raffinose) to an OD600 of 0.9 at 30°C and were then switched to YPG (yeast extract and peptone plus 2% galactose) for 90 min. For Spt16 overexpression, a yeast strain expressing Spt16 under the control of the GAL1 promoter was grown in YPG at 30°C. For Spt16 underexpression, a yeast strain expressing Spt16 under the control of the GAL1 promoter was initially grown in YPG at 30°C to an OD600 of 0.6 and was then transferred to YPD for 2 and 4 h. For growth curve analysis in YPD, yeast cells were grown in YPD at 30°C to an OD600 of 0.1, and then the OD600 was measured at different time points. For studies of chromatin reassembly at the GAL genes, yeast cells were grown in YPG to an OD600 of 0.9 at 30°C and were then switched to YPD for 2 or 5 min. For analysis of chromatin disassembly at the GAL genes, yeast cells were grown in YPR to an OD600 of 0.9 at 30°C and were then switched to YPG for 20, 30, and 60 min.
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed as done previously (17–21) and is briefly described in the supplemental material. The primer pairs used for PCR analysis of immunoprecipitated DNA samples are also described in the supplemental material.
WCE preparation and Western blot analysis.
To analyze the global levels of the FACT components, yeast strains expressing Myc epitope-tagged Spt16 and Pob3 components of FACT were grown in 25 ml YPD to an OD600 of 1.0. Yeast cells were then harvested, lysed, and sonicated to prepare whole-cell extract (WCE) with solubilized chromatin following the protocol as described for the ChIP assay. The WCE was run on an SDS-polyacrylamide gel and was then analyzed by Western blotting assay. An anti-Myc (9E10; Santa Cruz Biotechnology, Inc.) or antiactin (A2066; Sigma) antibody was used for Western blot analysis.
Total RNA preparation.
Total RNA was prepared from yeast cell culture as done previously (22); the method is briefly described in the supplemental material.
RT-PCR analysis.
Reverse transcription-PCR (RT-PCR) analysis was performed as done previously (22–24) and as briefly described in the supplemental material. The primer pairs used for PCR analysis of cDNAs are also described in the supplemental material.
Growth analysis in the presence of 6-AU, HU, and MMS.
Growth analysis of the yeast cells in solid media in the presence and absence of 6-azauracil (6-AU), hydroxy urea (HU), and methyl methanesulfonate (MMS) was performed as described in the supplemental material.
Ubiquitylation assay.
The ubiquitylation assay was performed as done previously (22, 25) and is briefly described in the supplemental material.
Formaldehyde-based in vivo cross-linking and co-IP assay.
The coimmunoprecipitation (co-IP) assay was performed as done previously (26, 27) and is briefly described in the supplemental material.
Immunopurification of Spt16 and Pob3.
Immunopurification was performed as done previously (26), and the method is briefly described in the supplemental material.
Protein interaction assay in vitro.
To analyze the interaction of Spt16 with San1, the yeast strain expressing Myc-tagged San1 was grown in 100 ml YPD to an OD600 of 1.0, and 800 μl WCE was subsequently prepared. WCE (400 μl) was used for immunoprecipitation as described for the ChIP assay using 10 μl anti-Myc antibody and 100 μl protein A/G plus agarose beads. Immobilized San1 on beads was thoroughly washed under high-stringency washing conditions as described for the ChIP assay, and then the washed beads with immobilized San1 were incubated with immunopurified hemagglutinin (HA)-tagged Spt16 in buffer E (50 mM Tris-HCl, 250 mM NaCl, 1% NP-40, 1 mM EDTA; pH 7.5) containing HA peptide and aprotinin for 15 min at 25°C. Subsequently, the beads were washed using buffer W (50 mM Tris-HCl, 2 mg/ml bovine serum albumin [BSA], 250 mM NaCl, 1% NP-40, 1 mM EDTA; pH 7.3) containing aprotinin four times (1 ml each time). The last wash was performed using buffer E containing aprotinin. The washed beads were then boiled in SDS-PAGE loading buffer, and the supernatant was subsequently analyzed by SDS-PAGE and Western blotting to determine the interaction between Spt16 and San1. An anti-HA-peroxidase antibody (H6533-1VL; Sigma) was used in the Western blot analysis to detect HA-tagged Spt16. Likewise, the interaction of Pob3 with San1 was analyzed using Myc-tagged San1 and immunopurified HA-tagged Pob3.
RESULTS AND DISCUSSION
San1 associates with the coding sequences of active genes to promote transcriptional elongation.
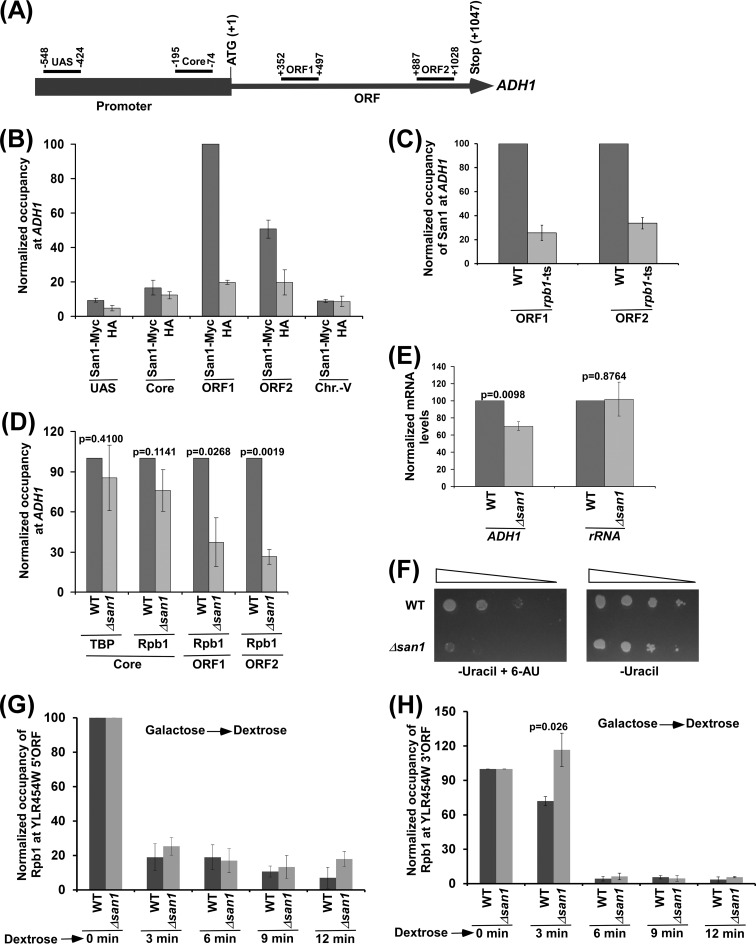
San1 has disordered N- and C-terminal regions with interspersed substrate-binding sites to recognize misfolded or unfolded proteins and a central RING (Really Interesting New Gene) domain for ubiquitylation of substrate proteins (15, 16). San1 has been well characterized and has been shown to recognize hydrophobic patches of abnormally folded or unfolded proteins for ubiquitylation and subsequent proteasomal degradation to maintain nuclear protein quality (15, 16). Since San1 is involved in nuclear protein quality control, it may regulate transcription or the nuclear phase of gene expression either directly or indirectly via the maintenance of protein quality in the nucleus. However, such a possibility has not been clearly elucidated. To determine the role of San1 in transcriptional regulation, we first analyzed the association of San1 with a constitutively active gene, ADH1. To that end, we tagged San1 by the use of Myc epitope at its chromosomal locus and then performed the ChIP assay at different regions of ADH1 (Fig. 1A), using an anti-Myc antibody against Myc-tagged San1 (Myc epitope tagging did not alter cellular growth [see Fig. S1 in the supplemental material]). An anti-HA antibody was used as a nonspecific antibody. We found that San1 is specifically associated with the coding sequence (ORF1 and ORF2 in Fig. 1A) of ADH1 (Fig. 1B; see also Fig. S2A). For a nonspecific DNA control, we also analyzed the association of San1 with a silent region within chromosome V (Chr.-V) as done in our previous studies (28) and found that San1 was not associated with an inactive region within chromosome V (Fig. 1B; see also Fig. S2A). Moreover, an anti-Myc ChIP signal was not observed at the ADH1 coding sequence, using the yeast strain that did not have Myc-tagged protein (see Fig. S2B). Further, we found that the ts inactivation of the largest subunit (Rpb1 [essential for transcription]) of RNA polymerase II impaired the association of San1 with the ADH1 coding sequence (Fig. 1C). Thus, San1 associates with the coding sequence but not the promoter in a transcription-dependent manner.
FIG 1.
San1 associates with the coding sequence of ADH1 to promote the engagement of elongating RNA polymerase II into transcription. (A) Schematic diagram showing the locations of different primer pairs at ADH1 for the ChIP analysis. The numbers are presented with respect to the position of the first nucleotide of the initiation codon (+1). UAS, upstream activation sequence. (B) ChIP analysis of San1 association with ADH1. Yeast cells expressing Myc-tagged San1 were grown in YPD at 30°C to an OD600 of 1.0 prior to formaldehyde-based in vivo cross-linking. Immunoprecipitated DNA was analyzed by PCR using the primer pairs targeted to different locations of ADH1 and an inactive region within chromosome V (Chr.-V). The ratio of immunoprecipitate to input in the autoradiogram was measured. The maximum ratio was set to 100, and other ratios were normalized to 100. The normalized ratio (represented as normalized or relative occupancy or ChIP signal) is plotted in the form of a histogram. (C) San1 association with the ADH1 coding sequence is impaired in the rpb1-ts mutant following 1 h ts inactivation at 37°C. (D) ChIP analysis of the association of TBP and Rpb1 with ADH1 in the Δsan1 and wild-type strains, using an anti-TBP antibody against TBP and 8WG16 antibody (Covance) against the carboxy-terminal domain of Rpb1. (E) RT-PCR analysis of ADH1 mRNA levels in the Δsan1 and wild-type strains (with 18S rRNA as control). (F) Growth analysis of the Δsan1 and wild-type strains in the solid SC-uracil medium containing 2% dextrose with or without 6-AU (100 μg/ml) at 30°C. (G and H) ChIP analysis of Rpb1 association with 5′ (G) and 3′ (H) ends of the ∼8-kb-long YLR454W coding sequence (under the control of the GAL1 promoter) upon switching the carbon source in the growth media from galactose to dextrose. ORF, open reading frame.
Since San1 associates with the ADH1 coding sequence, it may enhance the engagement of RNA polymerase II to promote transcriptional elongation. To test this, we analyzed the level of RNA polymerase II at the coding sequence of ADH1 in the Δsan1 and wild-type strains, using an antibody (8WG16; Covance) against the carboxy-terminal domain of Rpb1 of RNA polymerase II. Rpb1p is essential to maintain the structural and functional integrities of RNA polymerase II and thus serves as a representative component for the ChIP analysis of RNA polymerase II association with active gene. Our ChIP analysis revealed that the association of RNA polymerase II with the ADH1 coding sequence was significantly decreased in the absence of San1 (Fig. 1D; see also Fig. S2C in the supplemental material). Such a reduction in the association of RNA polymerase II with the ADH1 coding sequence is not due to decreased stability of Rpb1 in the Δsan1 strain, as revealed by Western blot analysis (see Fig. S2D; the actin level was monitored as a loading control). However, the reduction in the association of RNA polymerase II with the ADH1 coding sequence in the Δsan1 strain could have indirectly resulted from impaired preinitiation complex (PIC) formation at the promoter in the absence of San1. To test this possibility, we analyzed the recruitment of TBP (a PIC component that nucleates the formation of PIC by a set of general transcription factors at the core promoter; 29, 30) to the ADH1 promoter in the Δsan1 and wild-type strains. We found that the recruitment of TBP to the ADH1 promoter was not significantly altered in the absence of San1 (Fig. 1D; see also Fig. S2E). Likewise, the recruitment of RNA polymerase II (a PIC component that joins PIC toward the end [30]) to the ADH1 promoter was not significantly altered in the Δsan1 strain in comparison to the wild-type equivalent (Fig. 1D; see also Fig. S2C). Thus, San1 does not regulate the PIC formation at the ADH1 promoter but rather plays a direct role in facilitating the association of RNA polymerase II with the coding sequence. Consistently, San1 travelled with elongating RNA polymerase II at ADH1 but was not recruited to the promoter (Fig. 1B; see also Fig. S2A and B).
Since the association of RNA polymerase II with the ADH1 coding sequence was decreased in the absence of San1, transcription of ADH1 is likely to be impaired in the Δsan1 strain. To test this, we performed RT-PCR analysis and determined the levels of ADH1 mRNAs in the Δsan1 and wild-type strains. We found that the level of ADH1 mRNA was significantly decreased in the Δsan1 strain in comparison to the wild-type equivalent (Fig. 1E; see also Fig. S2F in the supplemental material). As a control, we analyzed the levels of 18S rRNAs in the wild-type and Δsan1 strains and found that the 18S rRNA level was not altered in the absence of San1 (Fig. 1E; see also Fig. S2F). Further, we observed that the growth of the Δsan1 cells was impaired in solid synthetic complete (SC)-uracil medium containing 6-AU (Fig. 1F). 6-AU decreases nucleotide pools for transcriptional elongation by RNA polymerase II, which results in a slow (or impaired) growth phenotype upon deletion or mutation of the factor involved in transcriptional elongation (31). Hence, impaired growth of the Δsan1 strain in the presence of 6-AU supports the idea of a role of San1 in stimulation of transcriptional elongation. This is further substantiated by the slow movement of the last wave of RNA polymerase II on the ∼8-kb-long YLR454W coding sequence (that is under the control of galactose-inducible GAL1 promoter; 32) upon switching off transcription (by transferring cells from galactose- to dextrose-containing growth media) in the Δsan1 strain (Fig. 1G and H).
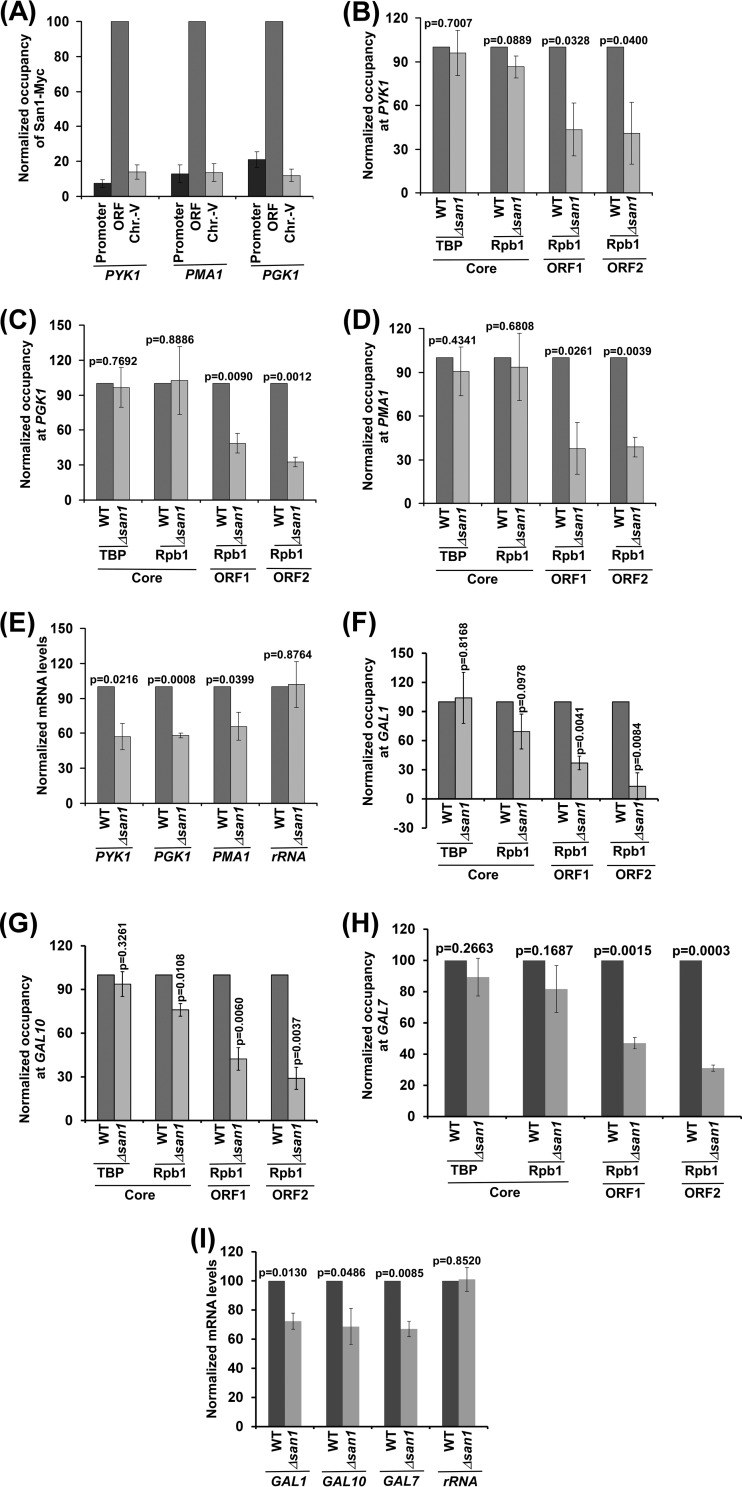
The results presented above support the hypothesis that San1 associates with the coding sequence of a constitutively active ADH1 gene and enhances the engagement of elongating RNA polymerase II to promote transcription. To determine whether San1 also associates with the coding sequences of other active genes to promote their transcriptional elongation, we performed ChIP analysis of San1 at several active genes such as PYK1, PGK1, and PMA1. We found that San1 specifically associated with the coding sequences but not promoters of these genes (Fig. 2A). Further, the association of RNA polymerase II with the coding sequences of these genes was enhanced by the presence of San1 (Fig. 2B to D). However, the recruitment of TBP and RNA polymerase II to the promoters of these genes (and hence PIC formation) was not significantly altered in the Δsan1 strain in comparison to the wild-type equivalent (Fig. 2B to D). Thus, similarly to the ADH1 results, we found that San1 specifically associated with the coding sequences of the PYK1, PGK1, and PMA1 genes to promote the engagement of elongating RNA polymerase II but not of initiating RNA polymerase II (or PIC formation). Consistently, transcription of these genes was significantly impaired in the Δsan1 strain (Fig. 2E), and the Δsan1 strain grew slowly in comparison to the wild-type equivalent in liquid growth curve analysis (see Fig. S2G in the supplemental material). Likewise, the engagement of elongating RNA polymerase II with the inducible GAL genes such as GAL1, GAL7, and GAL10 (and hence transcription) was also facilitated by San1 (Fig. 2F to I). Collectively, our results demonstrate an important function of a nucleus-localized E3 ubiquitin ligase, San1, in enhancing the association of RNA polymerase II with the coding sequence to promote transcriptional elongation.
FIG 2.
San1 associates with the coding sequences but not promoters of the PGK1, PMA1, and PYK1 genes to facilitate the association of elongating RNA polymerase II and, hence, transcription. (A) ChIP analysis of San1 association with PGK1, PMA1, and PYK1. (B to D) ChIP analysis of the association of TBP and Rpb1p with PYK1, PGK1, and PMA1 in the Δsan1 and wild-type strains. ORF1 and ORF2 represent two different locations in the coding sequence toward the 5′ and 3′ ends, respectively. (E) RT-PCR analysis of PGK1, PMA1, and PYK1 mRNA levels in the Δsan1 and wild-type strains. (F to H) ChIP analysis of the association of TBP and Rpb1 with GAL1, GAL10, and GAL7 in the Δsan1 and wild-type strains following 90 min of transcriptional induction in galactose. (I) RT-PCR analysis of GAL1, GAL10, and GAL7 mRNA levels in the Δsan1 and wild-type strains following 90 min of transcriptional induction in galactose.
San1 is involved in ubiquitylation and proteasomal degradation of the Spt16 component of FACT.
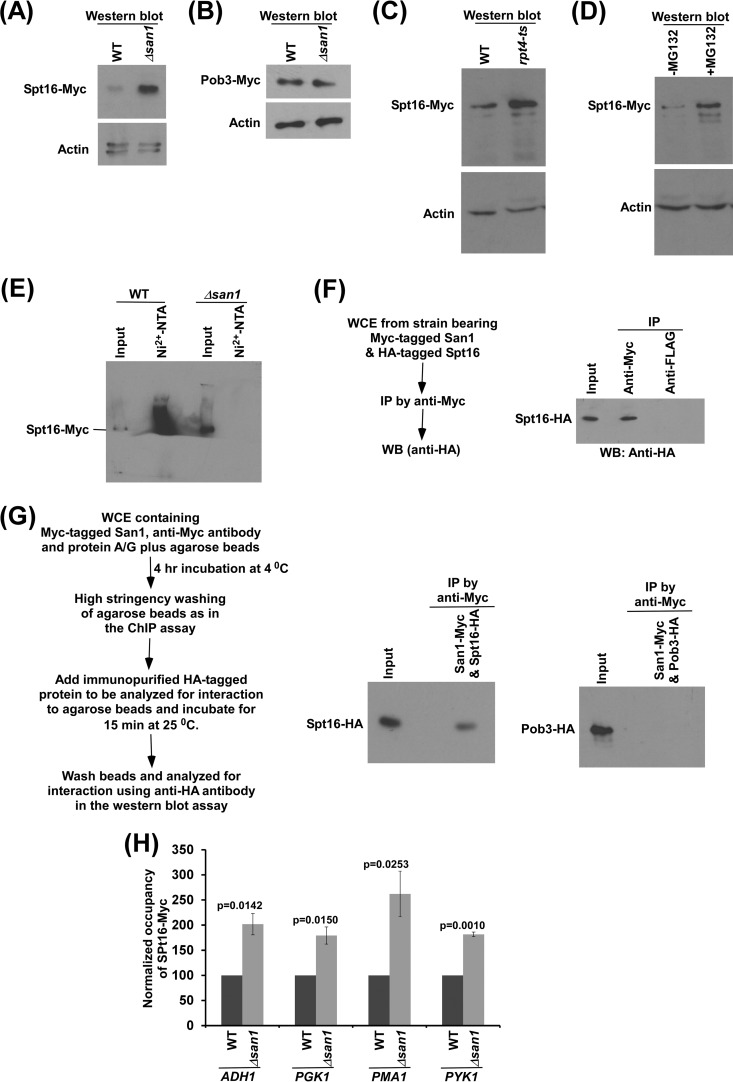
The protein interaction database (16, 33–36) revealed the interaction of San1 with the Spt16 component of FACT that is involved in promoting transcriptional elongation (5, 6). On the basis of this information, we hypothesize that San1 regulates transcriptional elongation, possibly by controlling the stability or abundance of Spt16 via ubiquitylation and proteasomal degradation. To test this, we analyzed the global levels of both components (i.e., Spt16 and Pob3) of FACT in the presence and absence of San1. For this purpose, we tagged both Spt16 and Pob3 by the use of Myc epitope in the Δsan1 and wild-type strains (such epitope tagging did not alter cellular growth [see Fig. S1 in the supplemental material]) and then performed Western blot analysis, using an anti-Myc antibody against Myc-tagged Spt16 and Pob3. We found that the global level of Spt16 was significantly increased in the absence of San1, while the stability or abundance of Pob3 was not altered (Fig. 3A and B). The level of actin was monitored as a loading control, and the level did not change in the absence of San1 (Fig. 3A and B). Thus, our results demonstrate that San1 regulates the stability or abundance of the Spt16 component of FACT.
FIG 3.
San1 interacts with Spt16 for ubiquitylation and proteasomal degradation. (A and B) Western blot analysis of the global levels of Spt16 (A) and Pob3 (B) in the Δsan1 and wild-type strains (with actin as a loading control). (C) Western blot analysis of Spt16's abundance in the rpt4-ts and wild-type strains. Both the wild-type and ts mutant strains expressing Myc-tagged Spt16 were grown in YPD at 23°C to an OD600 of 0.85 and were then switched to 39°C for 1 h before harvesting for Western blot analysis. (D) Western blot analysis of Spt16's abundance in the presence and absence of MG132. Yeast cells with a null mutation of PDR5 were grown in YPD at 30°C to an OD600 of 0.7 and were then treated with MG132 (75 μM) for 2 h prior to harvesting. (E) Ubiquitylation analysis of Spt16. Yeast strains expressing Myc-tagged Spt16 and hexahistidine-tagged ubiquitin were grown in synthetic complete medium at 30°C to an OD600 of 0.7 and were then treated with CuSO4 at a final concentration of 0.1 mM for 6 h. Precipitation was carried out by the use of Ni2+-NTA–agarose beads, and Western blot analysis was performed using an anti-Myc antibody against Myc-tagged Spt16. (F) Formaldehyde-based in vivo cross-linking and co-IP assay. WB, Western blot. (G) Analysis of interaction of San1 with immunopurified Spt16 or Pob3 (right panel) and schematic outlines of the experimental strategy to analyze protein-protein interactions in vitro (left panel). (H) ChIP analysis of the association of Spt16 with the coding sequences of ADH1, PGK1, PMA1 and PYK1 (ORF2) in the wild-type and Δsan1 strains.
San1 might be regulating Spt16's stability via ubiquitylation and proteasomal degradation. To test this, we analyzed whether Spt16 is degraded by the 26S proteasome. For this purpose, we tagged Spt16 by the use of a Myc epitope at its C terminus in the chromosomal locus in the wild-type and ts mutant strains of Rpt4, a component of the 26S proteasome that is essential for its proteolytic activity (29, 37, 38), and determined Spt16 levels in these strains at the nonpermissive temperature. We found that the global level of Spt16 was dramatically enhanced in the rpt4-ts mutant strain (Fig. 3C). However, the level of actin was not changed in the rpt4-ts mutant strain in comparison to the wild-type equivalent (Fig. 3C). Thus, Spt16 has much higher turnover than actin, and such turnover is impaired in the rpt4-ts mutant strain. Therefore, our results support the idea that the 26S proteasome is involved in the degradation of Spt16. To complement this further, we also analyzed the stability of Spt16 in the presence and absence of MG132, which inhibits the proteolytic function of the proteasome. For this purpose, we deleted a multidrug resistance gene, PDR5, in the yeast strain expressing Myc-tagged Spt16 and then analyzed the level of Spt16 with or without MG132 treatment, as done previously (28). We found that the stability of Spt16 was significantly increased following MG132 treatment (Fig. 3D). However, the actin level was not changed in the presence of MG132 (Fig. 3D). Thus, taken together, our results support the idea that Spt16's stability or abundance is regulated by the 26S proteasome.
Since the 26S proteasome is involved in targeted degradation of the ubiquitylated proteins, Spt16 is thus likely to be ubiquitylated for its proteasomal degradation in the presence of San1. To test this, we analyzed the ubiquitylation status of Spt16 in the wild-type strain. To this end, we introduced a plasmid expressing hexahistidine-tagged ubiquitin under the control of the CUP1 promoter (pUB221) in the wild-type strain expressing Myc-tagged Spt16. Using this strain, we performed the ubiquitylation assay as described previously (22, 25). Ubiquitin and ubiquitylated proteins were precipitated from the WCE using nickel-nitrilotriacetic acid (Ni2+-NTA)–agarose beads, which bound to the hexahistidine tag attached to ubiquitin. The precipitate was analyzed by Western blotting assay for the presence of Spt16 using an anti-Myc antibody against Myc-tagged Spt16. We observed Spt16 in the precipitate (lane 2 in Fig. 3E), thus supporting the idea of ubiquitylation of Spt16 in the presence of San1. To determine whether San1 is involved in ubiquitylation of Spt16, we next analyzed the ubiquitylation status of Spt16 in the absence of San1. We found that ubiquitylation of Spt16 was impaired in the Δsan1 strain (lane 4 in Fig. 3E). The Δsan1 strain contained a significantly high level of Spt16 in comparison to the wild-type equivalent (Fig. 3E; compare lane 1 with lane 3), as the stability of Spt16 was increased in the absence of San1 (Fig. 3A). Even in the presence of a high abundance of Spt16 in the Δsan1 strain, Spt16 was not found in the precipitate (lane 4 in Fig. 3E). Together, these results support the idea that San1 is involved in ubiquitylation of Spt16.
Since San1 is involved in ubiquitylation of Spt16, it is likely to interact with Spt16. To test this, we performed an in vivo formaldehyde cross-linking-based co-IP assay as done previously (26). For this purpose, we tagged San1 and Spt16 by the use of Myc and HA epitopes, respectively, in the wild-type strain and then performed the co-IP assay. Our co-IP analysis revealed that San1 interacts with Spt16 in vivo (Fig. 3F). To determine whether San1 interacts with Spt16 in vitro, we immunopurified HA-tagged Spt16 and Myc-tagged San1 and then analyzed their interactions, as done previously (26). We found that San1 interacts with Spt16 but not with the Pob3 component of FACT (Fig. 3G). Taken together, our results support the idea that San1 interacts with the Spt16 component of FACT for ubiquitylation and proteasomal degradation.
Increased stability or abundance of Spt16 is associated with a decreased level of RNA polymerase II at the coding sequences of active genes and, hence, with transcriptional elongation.
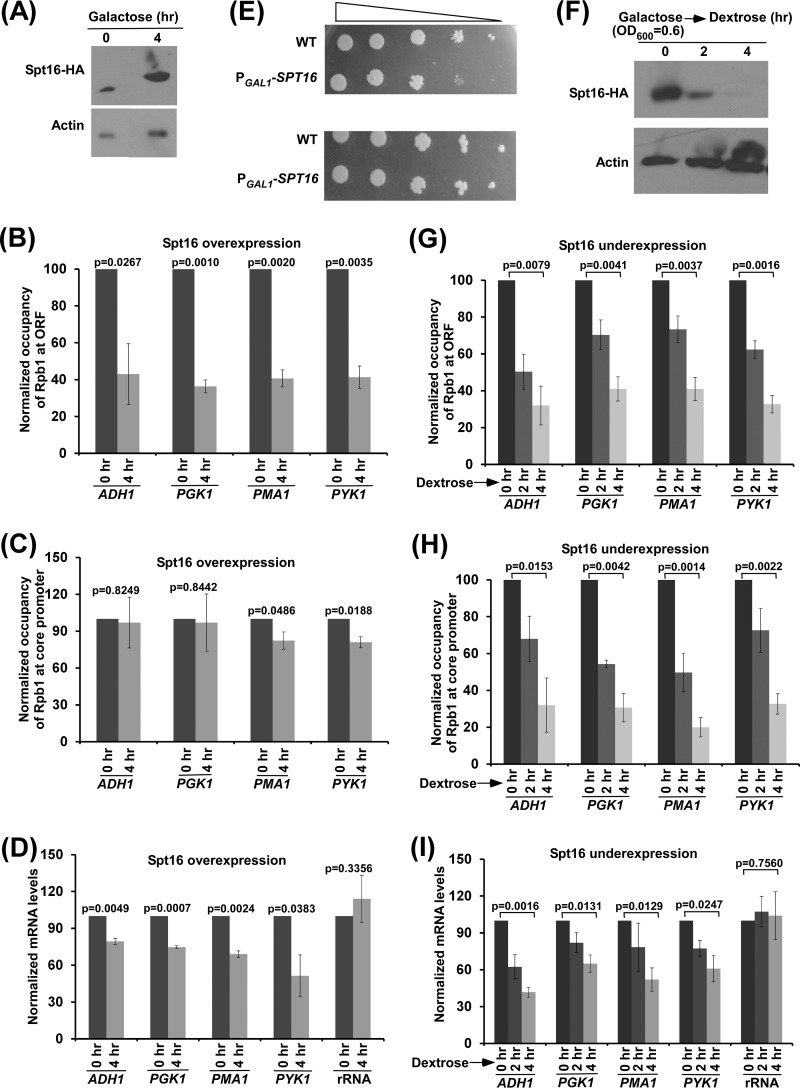
Since San1 interacts with Spt16 for ubiquitylation and proteasomal degradation (Fig. 3A to G) and associates with active coding sequence (Fig. 1B and 2A), the level of Spt16 at the active coding sequence is likely to be increased in the absence of San1. To test this, we analyzed the association of Myc-tagged Spt16 with the coding sequences of ADH1, PGK1, PMA1, and PYK1 genes in the wild-type and Δsan1 strains and found a significantly increased level of Spt16 at the coding sequences of these genes in the Δsan1 strain (Fig. 3H). Since the association of RNA polymerase II with the coding sequences of these genes (and with transcription) was decreased in the absence of San1 (Fig. 1D and E and 2B to E), it is likely that increased abundance of Spt16 in the Δsan1 strain impairs RNA polymerase II association with the coding sequence (and hence transcription). To test this, we analyzed the association of RNA polymerase II with the coding sequences of ADH1, PGK1, PYK1, and PMA1 following overexpression of Spt16. To this end, we replaced the endogenous promoter of SPT16 with GAL1 and overexpressed Spt16 in galactose-containing growth medium (Fig. 4A). Subsequently, we analyzed the level of RNA polymerase II at the promoters and coding sequences of ADH1, PGK1, PYK1, and PMA1. Our ChIP analysis revealed that the association of RNA polymerase II with the coding sequence was significantly decreased in the presence of a high abundance of Spt16 (Fig. 4B and C; see also Fig. S3A and B in the supplemental material). Consistently, transcription of these genes was also impaired following overexpression of Spt16 (Fig. 4D; see also Fig. S3C). Thus, a high abundance of Spt16 is associated with decreased levels of RNA polymerase II at the coding sequence (and hence of transcription), similarly to the results determined for the Δsan1 strain that has a high abundance of Spt16. Consistently, we found that yeast cells grew slowly following overexpression of Spt16 in solid growth medium containing 6-AU (Fig. 4E), thus supporting the idea of impairment of transcriptional elongation in the presence of a high abundance of Spt16.
FIG 4.
Effect of high or low abundance of Spt16 on the engagement of elongating RNA polymerase II in transcription. (A) Spt16 expression analysis under the control of GAL1 promoter (PGAL1-SPT16) in galactose-containing growth medium (or YPG) by Western blotting assay. Yeast cells were initially grown at 30°C to an OD600 of 0.6 and were then collected immediately (0 h) and after 4 h. (B and C) ChIP analysis of Rpb1 association with ADH1, PGK1, PMA1, and PYK1 (ORF2 [B] and core promoter [C]) following overexpression of Spt16 in YPG. (D) RT-PCR analysis of ADH1, PGK1, PMA1, and PYK1 mRNAs following overexpression of Spt16 in YPG. (E) Growth analysis of the yeast strains expressing SPT16 under the control of the GAL1 promoter (PGAL1-SPT16) or of its own endogenous promoter (WT) in the solid SC-uracil medium containing 2% galactose with (top panel) or without (bottom panel) 6-AU (100 μg/ml) at 30°C. (F) Analysis of Spt16 expression under the control of GAL1 promoter in dextrose-containing growth medium (or YPD) by Western blotting assay. (G and H) ChIP analysis of Rpb1 association with the coding sequences (ORF2 [G]) and promoters (H) of the ADH1, PGK1, PMA1, and PYK1 genes following underexpression of Spt16 in YPD. (I) RT-PCR analysis of ADH1, PGK1, PMA1, and PYK1 mRNAs following underexpression of Spt16 in YPD.
Decreased abundance or stability of Spt16 is associated with reduced levels of RNA polymerase II at the active genes and, hence, transcription.
Similarly to the results associated with increased abundance, a decreased level of Spt16 is likely to impair the association of RNA polymerase II with the active genes and transcription, since Spt16 is required for transcription (5–11). To test this, we analyzed the association of RNA polymerase II with the coding sequences of ADH1, PGK1, PYK1, and PMA1 following decreased expression of Spt16 (which is under the control of the GAL1 promoter) in dextrose-containing growth medium (Fig. 4F). We found that the association of RNA polymerase II with the coding sequences of these genes was decreased following a reduction of the abundance of Spt16 (Fig. 4G; see also Fig. S3D in the supplemental material). Similarly, decreased association of RNA polymerase II with the promoters of these genes was also observed in the absence of Spt16 (Fig. 4H; see also Fig. S3E), in agreement with the fact that Spt16 is required for transcription initiation (5–8). Thus, a decreased level or decreased abundance of Spt16 is associated with a reduction of the association of RNA polymerase II with the active genes. Consistently, transcription of these genes was also reduced following decreased expression of Spt16 (Fig. 4I; see also Fig. S3F).
Taken together, our results support the idea that increased or decreased abundance of Spt16 is associated with a reduced level of RNA polymerase II at the active genes and, hence, transcription. Thus, an optimal level of Spt16 is required for efficient transcription. Our results support the idea that San1-mediated ubiquitylation and proteasomal degradation or UPS regulation maintains an appropriate cellular level of Spt16 for optimal transcription.
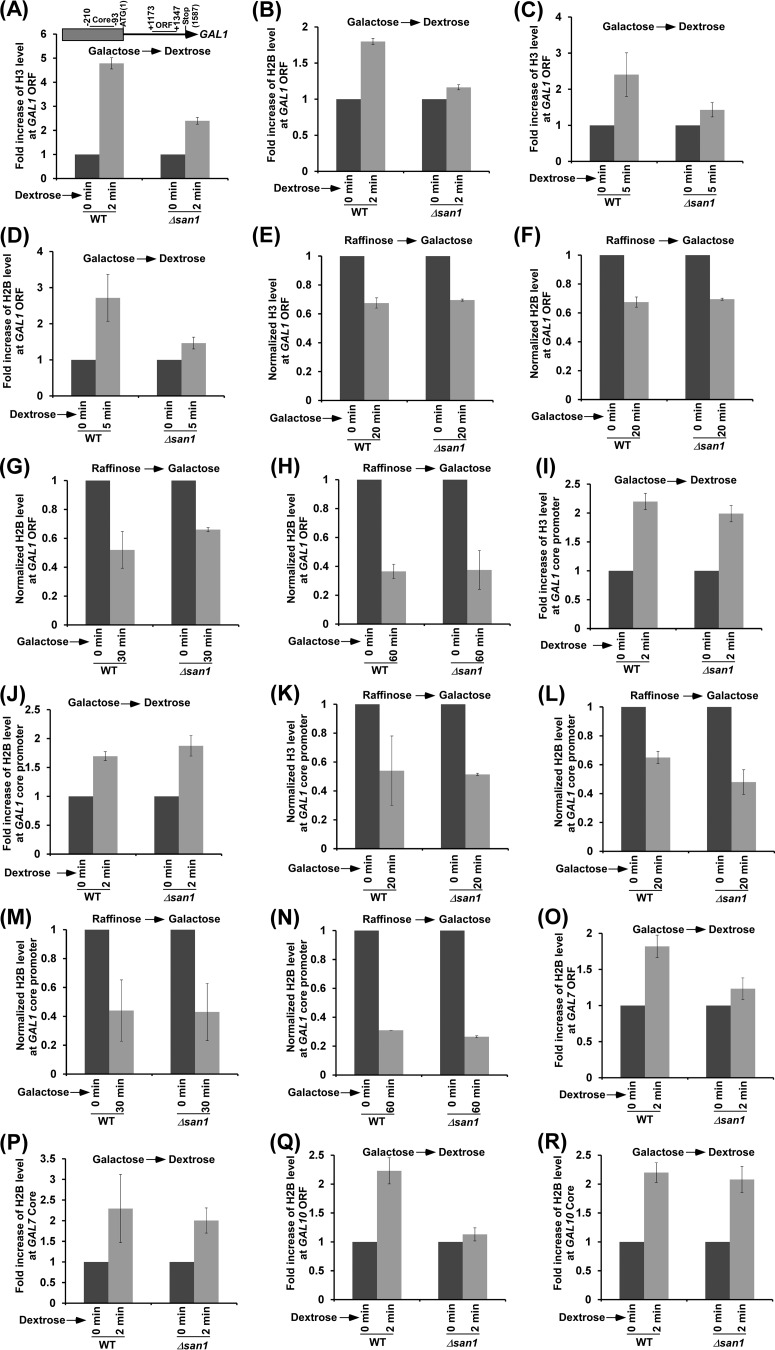
Chromatin reassembly in the wake of elongating RNA polymerase II is impaired in the absence of San1 (or in the presence of a high abundance of Spt16).
Spt16 has been previously implicated in nucleosomal disassembly and reassembly at the active gene (5–11). Spt16 is involved in chromatin disassembly at the promoter to facilitate transcriptional initiation (5–8). During transcriptional elongation, Spt16 promotes chromatin reassembly in the wake of RNA polymerase II to maintain the fidelity of transcriptional elongation (9–11). Likewise, histone H2B ubiquitylation is also involved in transcriptional elongation via enhancement of chromatin reassembly in the wake of elongating RNA polymerase II (10, 39–41). Moreover, histone H2B ubiquitylation (or Rad6/Bre1) and Spt16 interact functionally to promote chromatin reassembly and transcriptional elongation (10). On the basis of these results, we hypothesize that a high abundance of Spt16 in the absence of San1 would impair chromatin reassembly in the wake of elongating RNA polymerase II and would reduce transcriptional elongation. To test this, we analyzed the deposition of histone H2A-H2B dimer and H3-H4 tetramer at the GAL1 coding sequence (Fig. 5A) in the Δsan1 and wild-type strains immediately after switching off GAL1 transcription in dextrose-containing growth medium (for 2 min). The levels of histones H2B and H3 were monitored as representative components of the histone H2A-H2B dimer and histone H3-H4 tetramer, respectively. We found that the deposition of histone H3-H4 tetramer as well as histone H2A-H2B dimer to the GAL1 coding sequence was significantly impaired in the Δsan1 strain immediately after switching off the transcription of GAL1 (Fig. 5A and B; see also Fig. S4A in the supplemental material). Similar results were also observed at 5 min after switching off GAL1 transcription (Fig. 5C and D). These results support the idea of a role of San1 in chromatin reassembly at the GAL1 coding sequence. Next, we analyzed the eviction of histone H2A-H2B dimer and histone H3-H4 tetramer from the GAL1 coding sequence in the Δsan1 and wild-type strains following GAL1 transcriptional induction in galactose-containing growth medium. We found that the eviction of histone H3-H4 tetramer and histone H2A-H2B dimer from the GAL1 coding sequence following 20 min of transcriptional induction was not impaired in the absence of San1 (Fig. 5E and F; see also Fig. S4B). Similar results (Fig. 5G and H) were also observed at later transcriptional induction time points (e.g., 30 and 60 min). Taken together, our results support the idea that San1 promotes chromatin reassembly at the coding sequence.
FIG 5.
San1 promotes chromatin reassembly at the coding sequence. (A to D) ChIP analysis of histone H3 (A and C) and H2B (B and D) levels at the GAL1 coding sequence (toward the 3′ end or ORF2) in the Δsan1 and wild-type strains after switching off transcription in dextrose-containing growth medium (2 [A and B] and 5 [C and D] min). (E to H) ChIP analysis of histone H3 (E [20 min]) and H2B (F [20 min], G [30 min], and H [60 min]) levels at the GAL1 coding sequence in the Δsan1 and wild-type strains after switching on transcription in galactose-containing growth medium for the indicated time periods. (I and J) ChIP analysis of histone H3 (I) and H2B (J) levels at the GAL1 core promoter in the Δsan1 and wild-type strains after switching off transcription in dextrose-containing growth medium (2 min). (K to N) ChIP analysis of histone H3 (K [20 min]) and H2B (L [20 min], M [30 min], and N [60 min]) levels at the GAL1 core promoter in the Δsan1 and wild-type strains after switching on transcription in galactose-containing growth medium for the indicated time periods. (O to R) ChIP analysis of histone H3 and H2B levels at the coding sequences (toward the 3′ end or ORF2 [O and Q]) and core promoters (P and R) of GAL7 (O and P) and GAL10 (Q and R) in the Δsan1 and wild-type strains after switching off transcription in dextrose-containing growth medium (2 min).
Since San1 does not regulate PIC formation at the core promoter (Fig. 1D and 2B, C, D, and F to H), it is not likely to regulate chromatin disassembly or reassembly at the core promoter. To test this, we analyzed the role of San1 in chromatin disassembly and reassembly at the GAL1 core promoter. We found that the deposition of histone H2A-H2B dimer and histone H3-H4 tetramer to the GAL1 core promoter was not impaired in the absence of San1 after switching off GAL1 transcription in dextrose-containing growth medium (Fig. 5I and J; see also Fig. S4C in the supplemental material). Likewise, San1 does not regulate chromatin disassembly at the GAL1 core promoter following transcriptional induction in galactose-containing growth medium (Fig. 5K to N; see also Fig. S4D). Similar results were observed at other GAL genes such as GAL7 and GAL10 (Fig. 5O to R). Thus, San1 does not regulate chromatin disassembly at the core promoter and coding sequence, while it facilitates chromatin reassembly at the coding sequence but not at the promoter.
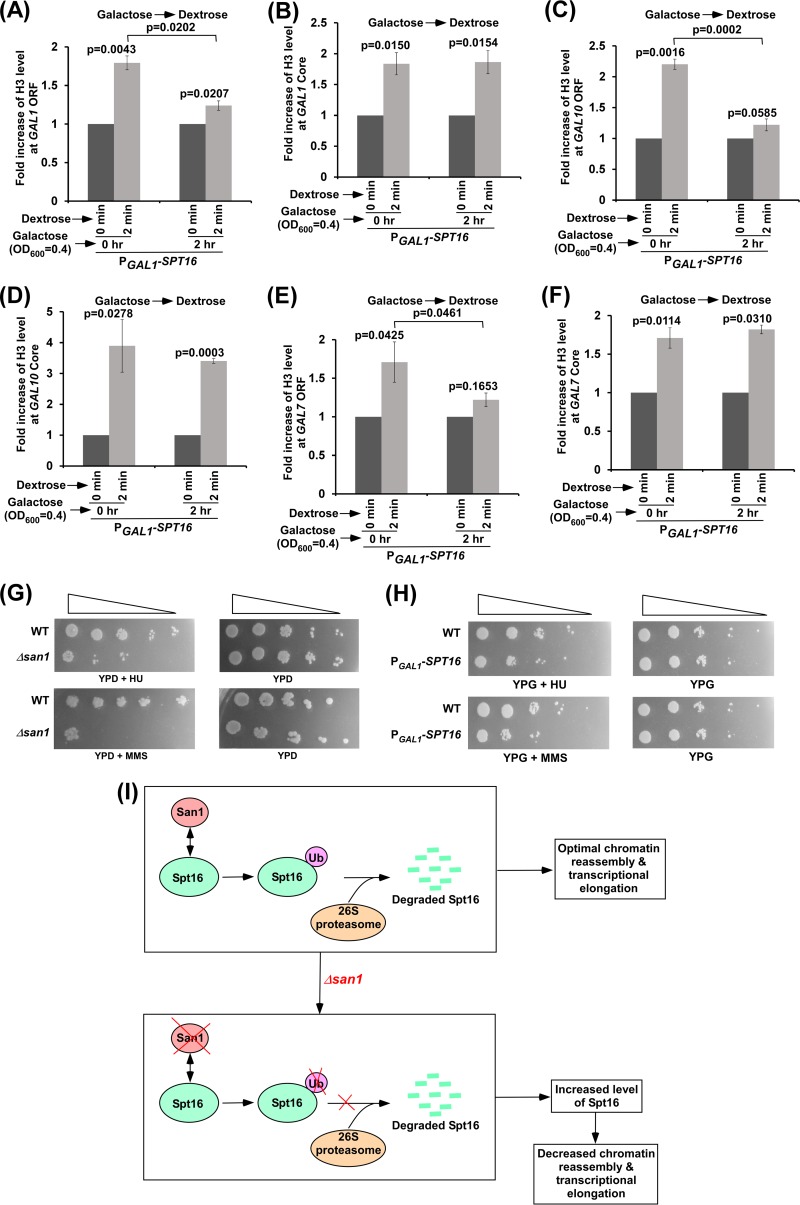
Since the loss of San1 impairs chromatin reassembly in the wake of elongating RNA polymerase II and increases Spt16's stability, a high abundance of Spt16 is likely to impair chromatin reassembly at the coding sequence. To test this, we analyzed chromatin reassembly at the GAL1 core promoter and coding sequence in the presence of a high abundance of Spt16. For this purpose, we overexpressed Spt16 under the control of the GAL1 promoter in galactose-containing growth medium and then switched off GAL1 transcription to immediately (2 min) analyze the deposition of histone H3-H4 tetramer to the GAL1 core promoter and coding sequence. Similarly to the results determined with the Δsan1 strain, we found that chromatin reassembly at the GAL1 coding sequence but not at the core promoter was impaired in the presence of a high abundance of Spt16 (Fig. 6A and B). We found similar results at other GAL genes such as GAL7 and GAL10 (Fig. 6C to F). Therefore, a high abundance of Spt16 or loss of San1 impairs chromatin reassembly at the coding sequence in the wake of RNA polymerase II.
FIG 6.
High abundance of Spt16 impairs chromatin reassembly at the coding sequence. (A and B) Chromatin reassembly analysis at the GAL1 coding sequence (A) and core promoter (B) following overexpression of Spt16. (C and D) Chromatin reassembly analysis at the GAL10 coding sequence (C) and core promoter (D) following overexpression of Spt16. (E and F) Chromatin reassembly analysis at the GAL7 coding sequence (E) and core promoter (F) following overexpression of Spt16. (G) Growth analysis of the Δsan1 and wild-type strains in the solid YPD medium with or without 0.026% MMS and HU (100 mM) at 30°C. (H) Growth analysis of the yeast strains expressing SPT16 under the control of the GAL1 promoter (PGAL1-SPT16) or of its own endogenous promoter (WT) in the solid YPG medium with or without 0.026% MMS and HU (100 mM) at 30°C. (I) Schematic diagram showing the UPS regulation of Spt16 in controlling chromatin reassembly at the coding sequence and transcriptional elongation.
Growth of the Δsan1 cells (or yeast cells overexpressing Spt16) is impaired following replication stress or DNA damage.
Since Spt16 or FACT is involved in DNA replication and repair (5, 6, 42–47), increased abundance of Spt16 in the Δsan1 strain may alter DNA replication as well as repair. To test this, we analyzed the growth of the Δsan1 strain in comparison to that of the wild-type equivalent following replication stress in the presence of HU (hydroxy urea) or DNA damage in the presence of MMS (methyl methanesulfonate). We found that the growth of the Δsan1 strain was impaired following replication stress or DNA damage (Fig. 6G), thus indicating the role of San1 in DNA replication and repair. Since the stability of Spt16 is increased in the Δsan1 strain (Fig. 3A), a high abundance of Spt16 is likely to impair cellular growth following replication stress and DNA damage. Indeed, our results reveal that a high abundance of Spt16 reduced the growth of yeast cells following HU-induced replication stress or DNA damage caused by MMS (Fig. 6H). Therefore, our results support the idea that a high abundance of Spt16 or the absence of San1 impairs DNA replication and repair. Such inhibition of DNA replication and repair in the presence of a high abundance of Spt16 or in the absence of San1 could be mediated via impairment of transcription of the genes involved in DNA replication and repair. Alternatively, increased abundance of Spt16 or loss of San1 might have been impairing chromatin assembly or disassembly during DNA replication and repair. These possibilities remain to be further elucidated.
In summary, we show here that San1 associates with the coding sequences of active genes and fine-tunes the level of Spt16 of FACT via ubiquitylation and proteasomal degradation for optimal nucleosomal reassembly at the coding sequence and transcriptional elongation (Fig. 6I). In the absence of San1, the abundance of Spt16 is increased at the coding sequence, which reduces chromatin reassembly in the wake of elongating RNA polymerase II and transcriptional elongation (Fig. 6I). However, it is not clear how enhanced abundance of Spt16 or loss of San1 impairs chromatin reassembly at the coding sequence and transcriptional elongation. It is possible that excess Spt16 in the absence of San1 alters FACT's composition or binding to partner proteins, which needs to be further investigated. Nonetheless, our results demonstrate for the first time a fine-tuning of Spt16 by the UPS in maintaining optimal chromatin reassembly and transcription elongation, thus providing novel gene regulatory mechanisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael R. Green for an anti-TBP antibody, Kevin Struhl for the yeast strain containing YLR454W under the control of the GAL1 promoter, Mary Ann Osley for a yeast strain (YKH045) expressing Flag-tagged histone H2B and HA-tagged ubiquitin, and Daniel Finley for the pUB221 plasmid expressing hexahistidine-tagged ubiquitin.
Funding Statement
The work in the Bhaumik laboratory was supported by grants from the National Institutes of Health (grants 1R15GM088798-01 and 2R15GM088798-02) and the American Heart Association (grant 15GRNT25700298).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01112-15.
REFERENCES
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Bhaumik SR, Smith E, Shilatifard A. 2007. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol 14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 3.Shukla A, Chaurasia P, Bhaumik SR. 2009. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell Mol Life Sci 66:1419–1433. doi: 10.1007/s00018-008-8605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malik S, Bhaumik SR. 2010. Mixed lineage leukemia: histone H3 lysine 4 methyltransferases from yeast to human. FEBS J 277:1805–1821. doi: 10.1111/j.1742-4658.2010.07607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler DD, Luger K. 2011. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J Biol Chem 286:18369–18374. doi: 10.1074/jbc.R110.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Formosa T. 2013. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta 1819:247–255. [PubMed] [Google Scholar]
- 7.Reinberg D, Sims RJ III. 2006. de FACTo nucleosome dynamics. J Biol Chem 281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- 8.Ransom M, Williams SK, Dechassa ML, Das C, Linger J, Adkins M, Liu C, Bartholomew B, Tyler JK. 2009. FACT and the proteasome promote promoter chromatin disassembly and transcriptional initiation. J Biol Chem 284:23461–23471. doi: 10.1074/jbc.M109.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwabish MA, Struhl K. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol 24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. 2008. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 11.Mason PB, Struhl K. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol 23:8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia H, Miecznikowski JC, Safina A, Commane M, Ruusulehto A, Kilpinen S, Leach RW, Attwood K, Li Y, Degan S, Omilian AR, Guryanova O, Papantonopoulou O, Wang J, Buck M, Liu S, Morrison C, Gurova KV. 2013. Facilitates chromatin transcription complex is an “accelerator” of tumor transformation and potential marker and target of aggressive cancers. Cell Rep 4:159–173. doi: 10.1016/j.celrep.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia H, Fleyshman D, Kolesnikova K, Safina A, Commane M, Paszkiewicz G, Omelian A, Morrison C, Gurova K. 2011. Expression of FACT in mammalian tissues suggests its role in maintaining of undifferentiated state of cells. Oncotarget 2:783–796. doi: 10.18632/oncotarget.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh FK, Kulaeva OI, Orlovsky IV, Studitsky VM. 2011. FACT in cell differentiation and carcinogenesis. Oncotarget 2:830–832. doi: 10.18632/oncotarget.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenbaum JC, Gardner RG. 2011. How a disordered ubiquitin ligase maintains order in nuclear protein homeostasis. Nucleus 2:264–270. doi: 10.4161/nucl.2.4.16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, Nelson ZW, Hetrick ED, Milac TI, Gottschling DE, Gardner RG. 2011. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell 41:93–106. doi: 10.1016/j.molcel.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR. 2006. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol Cell Biol 26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhaumik SR, Green MR. 2003. Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol 370:445–454. doi: 10.1016/S0076-6879(03)70038-X. [DOI] [PubMed] [Google Scholar]
- 19.Durairaj G, Chaurasia P, Lahudkar S, Malik S, Shukla A, Bhaumik SR. 2010. Regulation of chromatin assembly/disassembly by Rtt109p, a histone H3 Lys56-specific acetyltransferase, in vivo. J Biol Chem 285:30472–30479. doi: 10.1074/jbc.M110.113225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen R, Malik S, Frankland-Searby S, Uprety B, Lahudkar S, Bhaumik SR. 2014. Rrd1p, an RNA polymerase II-specific prolyl isomerase and activator of phosphoprotein phosphatase, promotes transcription independently of rapamycin response. Nucleic Acids Res 42:9892–9907. doi: 10.1093/nar/gku703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uprety B, Sen R, Bhaumik SR. 2015. Eaf1p is required for recruitment of NuA4 in targeting TFIID to the promoters of the ribosomal protein genes for transcriptional initiation in vivo. Mol Cell Biol 35:2947–2964. doi: 10.1128/MCB.01524-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durairaj G, Lahudkar S, Bhaumik SR. 2014. A new regulatory pathway of mRNA export by an F-box protein, Mdm30. RNA 20:133–142. doi: 10.1261/rna.042325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malik S, Durairaj G, Bhaumik SR. 2013. Mechanisms of antisense transcription initiation from the 3′ end of the GAL10 coding sequence in vivo. Mol Cell Biol 33:3549–3567. doi: 10.1128/MCB.01715-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uprety B, Kaja A, Ferdoush J, Sen R, Bhaumik SR. 2016. Regulation of antisense transcription by NuA4 histone acetyltransferase and other chromatin regulatory factors. Mol Cell Biol 36:992–1006. doi: 10.1128/MCB.00808-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muratani M, Kung C, Shokat KM, Tansey WP. 2005. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell 120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 26.Lahudkar S, Shukla A, Bajwa P, Durairaj G, Stanojevic N, Bhaumik SR. 2011. The mRNA cap-binding complex stimulates the formation of pre-initiation complex at the promoter via its interaction with Mot1p in vivo. Nucleic Acids Res 39:2188–2209. doi: 10.1093/nar/gkq1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malik S, Chaurasia P, Lahudkar S, Durairaj G, Shukla A, Bhaumik SR. 2010. Rad26p, a transcription-coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II-dependent manner in vivo. Nucleic Acids Res 38:1461–1477. doi: 10.1093/nar/gkp1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uprety B, Lahudkar S, Malik S, Bhaumik SR. 2012. The 19S proteasome subcomplex promotes the targeting of NuA4 HAT to the promoters of ribosomal protein genes to facilitate the recruitment of TFIID for transcriptional initiation in vivo. Nucleic Acids Res 40:1969–1983. doi: 10.1093/nar/gkr977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhaumik SR, Malik S. 2008. Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit Rev Biochem Mol Biol 43:419–433. doi: 10.1080/10409230802605914. [DOI] [PubMed] [Google Scholar]
- 30.Bhaumik SR. 2011. Distinct regulatory mechanisms of eukaryotic transcriptional activation by SAGA and TFIID. Biochim Biophys Acta 1809:97–108. doi: 10.1016/j.bbagrm.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL. 2007. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGArelated complex. Mol Cell 27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 32.Mason PB, Struhl K. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 33.Gardner RG, Nelson ZW, Gottschling DE. 2005. Degradation-mediated protein quality control in the nucleus. Cell 120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Xu Q, Johnston GC, Singer RA. 1993. The Saccharomyces cerevisiae Cdc68 transcription activator is antagonized by San1, a protein implicated in transcriptional silencing. Mol Cell Biol 13:7553–7565. doi: 10.1128/MCB.13.12.7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans DR, Brewster NK, Xu Q, Rowley A, Altheim BA, Johnston GC, Singer RA. 1998. The yeast protein complex containing cdc68 and pob3 mediates core-promoter repression through the cdc68 N-terminal domain. Genetics 150:1393–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fredrickson EK, Candadai SV, Tam CH, Gardner RG. 2013. Means of self-preservation: how an intrinsically disordered ubiquitin-protein ligase averts self-destruction. Mol Biol Cell 24:1041–1052. doi: 10.1091/mbc.E12-11-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. 1998. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J 17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell SJ, Gonzalez F, Joshua-Tor L, Johnston SA. 2001. Selective chemical inactivation of AAA proteins reveals distinct functions of proteasomal ATPases. Chem Biol 8:941–950. doi: 10.1016/S1074-5521(01)00060-6. [DOI] [PubMed] [Google Scholar]
- 39.Shukla A, Bhaumik SR. 2007. H2B-K123 ubiquitination stimulates RNAPII elongation independent of H3-K4 methylation. Biochem Biophys Res Commun 359:214–220. doi: 10.1016/j.bbrc.2007.05.105. [DOI] [PubMed] [Google Scholar]
- 40.Sen R, Lahudkar S, Durairaj G, Bhaumik SR. 2013. Functional analysis of Bre1p, an E3 ligase for histone H2B ubiquitylation, in regulation of RNA polymerase II association with active genes and transcription in vivo. J Biol Chem 288:9619–9633. doi: 10.1074/jbc.M113.450403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sen R, Bhaumik SR. 2013. Transcriptional stimulatory and repressive functions of histone H2B ubiquitin ligase. Transcription 4:221–226. doi: 10.4161/trns.26623. [DOI] [PubMed] [Google Scholar]
- 42.Dinant C, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, Grosbart M, Theil AF, van Cappellen WA, Kimura H, Bartek J, Fousteri M, Houtsmuller AB, Vermeulen W, Marteijn JA. 2013. Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell 51:469–479. doi: 10.1016/j.molcel.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Sand-Dejmek J, Adelmant G, Sobhian B, Calkins AS, Marto J, Iglehart DJ, Lazaro JB. 2011. Concordant and opposite roles of DNA-PK and the “facilitator of chromatin transcription” (FACT) in DNA repair, apoptosis and necrosis after cisplatin. Mol Cancer 10:74. doi: 10.1186/1476-4598-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abe T, Sugimura K, Hosono Y, Takami Y, Akita M, Yoshimura A, Tada S, Nakayama T, Murofushi H, Okumura K, Takeda S, Horikoshi M, Seki M, Enomoto T. 2011. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem 286:30504–30512. doi: 10.1074/jbc.M111.264721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, Zhang X, Feng J, Leng H, Li S, Xiao J, Liu S, Xu Z, Xu J, Li D, Wang Z, Wang J, Li Q. 2016. The histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly. Cell Rep 14:1128–1141. doi: 10.1016/j.celrep.2015.12.096. [DOI] [PubMed] [Google Scholar]
- 46.VanDemark AP, Blanksma M, Ferris E, Heroux A, Hill CP, Formosa T. 2006. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell 22:363–374. doi: 10.1016/j.molcel.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 47.Ransom M, Dennehey BK, Tyler JK. 2010. Chaperoning histones during DNA replication and repair. Cell 140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.