Abstract
Saccharomyces boulardii is a probiotic yeast that has been used for promoting gut health as well as preventing diarrheal diseases. This yeast not only exhibits beneficial phenotypes for gut health but also can stay longer in the gut than Saccharomyces cerevisiae. Therefore, S. boulardii is an attractive host for metabolic engineering to produce biomolecules of interest in the gut. However, the lack of auxotrophic strains with defined genetic backgrounds has hampered the use of this strain for metabolic engineering. Here, we report the development of well-defined auxotrophic mutants (leu2, ura3, his3, and trp1) through clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9-based genome editing. The resulting auxotrophic mutants can be used as a host for introducing various genetic perturbations, such as overexpression or deletion of a target gene, using existing genetic tools for S. cerevisiae. We demonstrated the overexpression of a heterologous gene (lacZ), the correct localization of a target protein (red fluorescent protein) into mitochondria by using a protein localization signal, and the introduction of a heterologous metabolic pathway (xylose-assimilating pathway) in the genome of S. boulardii. We further demonstrated that human lysozyme, which is beneficial for human gut health, could be secreted by S. boulardii. Our results suggest that more sophisticated genetic perturbations to improve S. boulardii can be performed without using a drug resistance marker, which is a prerequisite for in vivo applications using engineered S. boulardii.
INTRODUCTION
Saccharomyces boulardii (ATCC MYA-796) is a probiotic yeast that was isolated from lychee, a tropical fruit, in Indochina by Henri Boulard in 1923 (1). This yeast has been widely used in food and nutraceutical industries, because it is known to be effective for limiting diarrheal diseases. S. boulardii is acknowledged as generally regarded as safe (GRAS) by the Food and Drug Administration (FDA) (2). S. boulardii is the only yeast probiotic that has been proven effective in double-blind studies (3, 4), and it outperformed other known probiotics, such as Bifidobacterium and Lactobacillus, regarding immunomodulation (5). Additionally, S. boulardii can survive in the human gastrointestinal tract due to its resistance to high temperature and low pH (2, 4). S. boulardii can compete with diarrhea-causing pathogens for growth in the gut, making it effective for treating and preventing diarrhea (6).
Previously, most studies employing S. boulardii have focused on elucidating putative mechanisms of its beneficial properties and its applications as a probiotic (7–12). Based on the probiotic trait of S. boulardii, this yeast can be potentially used for in-gut production of therapeutic proteins. However, genetic manipulation of S. boulardii has been limited due to the lack of auxotrophic mutants as well as concerns over the use of drug resistance genetic markers (13). Recently, mutations of URA3, the auxotrophic marker gene used most often in yeast genetics, have been generated in S. boulardii by UV mutagenesis (14, 15) and the Cre-loxP system (16), paving the way for further engineering of this probiotic yeast. Nonetheless, there are several concerns over these approaches. For instance, UV mutagenesis is not limited to the URA3 gene only. Numerous unknown mutations other than ura3 may occur during UV treatment. These unknown mutations may lead to altered phenotypes related to probiotic traits, including undesirable ones. Hamedi et al. (14) reported that two uracil auxotrophic mutants obtained by UV treatment lost the acid resistance phenotype, which is a beneficial trait for a probiotic microbe. Alternatively, the Cre-loxP system generates ura3 mutants without altering other genes. However, because S. boulardii is an aneuploid containing two copies of URA3 on chromosome V (1), there would be two copies of 34-bp loxP fragments left in the target gene locus, leaving scars in the genome. This would result in difficulties in the repeated use of the Cre-loxP system, because the accumulated copies of the loxP scar can cause genome instability (17–19).
The clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 system (20, 21) is an emerging and powerful genomic engineering tool that has been widely used in bacteria (22), yeast (23), and mammalian cells (24). The CRISPR-Cas9 system is an advanced genome-editing tool in yeast genomic engineering compared to traditional methods due to its efficient, precise, and marker-free features (23). Therefore, in this study, we attempted to construct marker-free auxotrophic mutants of S. boulardii using the CRISPR-Cas9 system to enable metabolic engineering of this probiotic yeast. After obtaining auxotrophic mutants, we performed four types of genetic perturbations. First, we overexpressed a heterologous gene (lacZ) in S. boulardii using a pRS series plasmid. Second, we examined the localization of a target protein (red fluorescent protein [RFP] and green fluorescent protein [GFP]) in mitochondria (mito) using a mitochondrial localization signal of Saccharomyces cerevisiae. We then introduced a xylose-assimilating pathway consisting of three enzyme reactions into S. boulardii. Finally, using CRISPR-Cas9 technology, a beneficial enzyme, human lysozyme (25, 26), was shown to be secreted by S. boulardii by integration of the human lysozyme gene with the chicken secretion signal. We have received positive results from all these assays. Taken together, we demonstrate genetic/metabolic engineering of S. boulardii using the auxotrophic mutants generated by the CRISPR-Cas9 system.
MATERIALS AND METHODS
Strains and media.
Escherichia coli Top10 was used for the construction and propagation of plasmids. E. coli cells were grown in Luria-Bertani medium (5 g/liter yeast extract, 10 g/liter tryptone, 10 g/liter NaCl [pH 7.0]) at 37°C, and ampicillin (100 μg/ml) was added for selection when required. The S. boulardii strain used in this study is ATCC MYA-796. Yeast strains were grown on YP medium (10 g/liter yeast extract, 20 g/liter peptone) containing 20 g/liter glucose at 30°C. Yeast strains transformed with plasmids containing antibiotic markers were propagated on YPD (YP medium with 20 g/liter glucose) plates supplemented with the corresponding antibiotics. Yeast synthetic complete (YSC) medium (6.7 g/liter of yeast nitrogen base with ammonia sulfate plus 20 g/liter of glucose) minus the appropriate auxotrophic compounds was used for auxotrophic phenotype confirmation.
Plasmid and strain construction.
Plasmid p42K-pGPD-lacZ (Table 1) for β-galactosidase expression assays was constructed as follows: the pGPD-tCYC1 cassette from plasmid p426GPD (27) was doubly digested by SacI and KpnI and ligated with plasmid pRS42K digested with the same enzyme (28), forming plasmid p42K-pGPD (Table 1). lacZ was amplified from the genomic DNA of E. coli K-12 by using primer pair LacZ-forward-XmaI and LacZ-reverse-XhoI (Table 1). The PCR product was then digested with XmaI-XhoI and ligated to p42K-GPD; the resulting plasmid was designated p42K-pGPD-lacZ (Table 1).
TABLE 1.
Primers, plasmids, and strains used in this study
| Primer, plasmid, or strain | Primer sequencea | Source and/or reference(s) |
|---|---|---|
| Primers | ||
| LacZ-forward-Xma1 | CCGGAATTCcccgggATGACCATGATTACGGATTC | This study |
| LacZ-reverse-Xho1 | CCGGAATTCctcgagTTATTTTTGACACCAGACCA | This study |
| URA3donor-U | TCCATGGAGGGCACAGTTAAGCCGCTAAAGGCATTATAAGCCAAGTACAATTTTTTACTC | 29 |
| URA3donor-D | ACCAATGTCAGCAAATTTTCTGTCTTCGAAGAGTAAAAAATTGTACTTGGCTTATAATGC | 29 |
| TRP1donor-U | TCCGATGCTGACTTGCTGGGTATTATATGTGTGTAAAATAGAAAGAGAACAATTGACCCG | 29 |
| TRP1donor-D | TACAAGACTTGAAATTTTCCTTGCAATAACCGGGTCAATTGTTCTCTTTCTATTTTACAC | 29 |
| LEU2donor-U | CCAGGTGACCACGTTGGTCAAGAAATCACAGCCGAAGCCATTAAGTAACTTAAAGCTATT | 29 |
| LEU2donor-D | ATCGAACTTGACATTGGAACGAACATCAGAAATAGCTTTAAGTTACTTAATGGCTTCGGC | 29 |
| HIS3donor-U | GTAAAGCGTATTACAAATGAAACCAAGATTCAGATTGCGATCTCTTTAAAGGGTTAACCC | 29 |
| HIS3donor-D | TTCTGGGAAGATCGAGTGCTCTATCGCTAGGGGTTAACCCTTTAAAGAGATCGCAATCTG | 29 |
| gURA3-U | GCTCTAGAgcggccgcAGACATAAAAAACAAAAAAAGCACCACCGACTCGG | This study |
| gURA3-D | TCTACAGCGGCCGCgagctcTCT | This study |
| gTRP1-U | GCTCTAGAgcggccgcTCTTTGAAAAGATAATGTATGATTATGCTTTCAC | This study |
| gTRP1-D | GCTCTAGAggatccactagtAGACATAAAAAACAAAAAAAGCACCACCGACTCGG | This study |
| gCS8-U | TGATTCAATCATTCTTATTGGTTTTAGAGCTAGAAATAGCAAG | This study |
| gCS8-D | CAATAAGAATGATTGAATCAGATCATTTATCTTTCACTGCGGA | This study |
| CS8-IU | CAAAATTACCTACGGTAATTAGTGAAAGGCCAAAATCTAATGTTACAATAAATTAACCCTCACTAAAGGGA | This study |
| CS8-ID | GACCGTTCCCTTGTGTTGTACCAGTGGTAGGGTTCTTCTCGGTAGCTTCTGTAATACGACTCACTATAGGGC | This study |
| CS8-CKU | AGTGGAACATAGAAGGGG | This study |
| CS8-CKD | TAAGCAGCCCAGTGAAC | This study |
| mRuby-U | CCCGGGAGATCTggtaccATGGTGTCCAAAGGAGAGGAG | This study |
| mRuby-D | CCCGGGCTCGAGgagctcATTACCCTGTTATCCCTAGC | This study |
| Plasmids | ||
| p426GPD | pSR426-pTDH3-tCYC1 | 27 |
| pRS42K | pRS42K | 28 |
| p42K-pGPD | pRS42K-pTDH3-tCYC1 | This study |
| p42K-pGPD-lacZ | pRS42K-pTDH3-lacZ-tCYC1 | This study |
| Cas9-NAT | p414-TEF1p-Cas9-CYC1t-NAT1 | Addgene plasmid 64329, 29 |
| gRNA-ura-HYB | pRS42H carrying URA3 disruption gRNA cassette | Addgene plasmid 64330, 29 |
| gRNA-trp-HYB | pRS42H carrying TRP1 disruption gRNA cassette | Addgene plasmid 64331, 29 |
| gRNA-leu-HYB | pRS42H carrying LEU2 disruption gRNA cassette | Addgene plasmid 64332, 29 |
| gRNA-his-HYB | pRS42H carrying HIS3 disruption gRNA cassette | Addgene plasmid 64333, 29 |
| p42H-gURA3-gTRP1 | pRS42H carrying URA3 and TRP1 disruption gRNA cassette | This study |
| pSR6-X123 | pRS306 carrying pTDH3-XYL1-tTDH3, pPGK1-XYL2-tPGK1, and pTDH3-XYL3-tTDH3 | 37, 38 |
| pVT100U-mitoGFP | pVT100U-pADH-mitoGFP | Addgene plasmid 45054, 30 |
| pFA6a-mRuby2 | pFA6a-link-yomRuby2-SpHis5 | Addgene plasmid 44858, 31 |
| pVT100U-mito-mRuby | pVT100U-pADH-mito-yomRuby2 | This study |
| p42K-gCS8 | pRS42K carrying guide RNA for integration | This study |
| p426-pGPD-cHLY | pRS426-pTDH3-cHLY-tCYC1 | This study |
| Strains | ||
| S. boulardii | ATCC MYA-796 | ATCC |
| SB-X123 | ATCC MYA-796; XYL1 XYL2 XYL3 | This study |
| S.b(Lys+) | ATCC MYA-796; CS8-cHLY | This study |
Lowercase type in sequences indicates restriction sites. gRNA, guide RNA.
The Cas9-NAT plasmid confers nourseothricin (NAT) resistance, and four plasmids carrying guide RNA for the inactivation of each individual marker gene (URA3, HIS3, TRP1, and LEU2) are based on a pRS42H plasmid, which is resistant to hygromycin (29) (Table 1). As for the construction of tandem guide RNA plasmid p42H-gURA3-gTRP1, the gBlocks gene fragments of URA3 (29) were amplified by using primer pair gURA3-U and gURA3-D (Table 1). The resulting PCR product was digested by SacI-NotI and ligated to pRS42H, resulting in intermediate plasmid pRS42H-gURA3 (Table 1). The gBlocks gene fragments of TRP1 (29) were amplified by using primer pair gTRP1-U and gTRP1-D (Table 1), and the resulting PCR product was doubly digested by NotI-SpeI and then ligated to pRS42H-gURA3, forming tandem guide RNA plasmid p42H-gURA3-gTRP1 (Table 1) designed for simultaneous URA3 and TRP1 inactivation.
Plasmid pVT100U-mitoGFP, used for mitochondrion localization, was a gift from Benedikt Westermann (Addgene plasmid 45054) (30). mRuby2 was amplified from plasmid pFA6a-mRuby2 (Addgene plasmid 44858) (31) as a template by using primer pair mRuby-U and mRuby-D (Table 1). The resulting PCR product was doubly digested with KpnI and SacI and ligated to pVT100U-mitoGFP digested with the same enzyme to replace the GFP gene, and the constructed plasmid was designated pVT100U-mito-mRuby (Table 1).
In order to integrate target genes into the genome of S. boulardii for stable expression, guide plasmid p42K-CS8 was constructed (Table 1; see also the supplemental material for details). The plasmid was constructed by reverse PCR of a pRS42K plasmid containing a guide RNA sequence using primer pair gCS8-U and gCS8-D (Table 1). The 20-bp targeting sequence of guide RNA binds to the empty locus behind YPR015C in chromosome XVI. The target genes are integrated into this locus without affecting the function of other genes by homologous recombination. Plasmid p426-pGPD-cHLY was constructed for the secretion of human lysozyme in S. boulardii. Briefly, the gBlock of the yeast codon-optimized human lysozyme gene with a chicken lysozyme signal sequence (cHLY) for secretion (25, 26) was synthesized (IDT Inc.). The complete sequence of cHLY is listed in the supplemental material. A DNA fragment containing pGPD-cHLY was amplified from p426-pGPD-cHLY by using primers CS8-IU and CS8-ID (Table 1) and used as donor DNA for CRISPR-Cas9-based genomic integration. Primers CS8-CKU and CS8-CKD (Table 1) were used for diagnostic PCR for the correct integration of cHLY.
Transformation of yeast cells was carried out with the polyethylene glycol (PEG)-LiAc method (32). One microgram (1 μg) of DNA was used for Cas9 or guide RNA plasmid transformation; in the meantime, 4 μg of donor DNAs was used for homologous recombination. The confirmation of genomic integration was performed by yeast colony PCR.
β-Galactosidase activity assay.
β-Galactosidase activity was measured according to a protocol described previously by Ribeiro et al. (33). One unit of enzyme activity is defined as the amount of enzyme that catalyzes 1 μmol of substrate per min at 30°C. The protein concentration of the yeast cell extract was determined by the bicinchoninic acid (BCA) method (Pierce, Rockford, IL).
Colocalization of mito-mRuby with mitochondria by rhodamine 123 staining in S. boulardii.
Rhodamine 123 (catalogue number R8004; Sigma-Aldrich) is widely used as a mitochondrion-staining dye (34). The staining protocol was performed according to the manufacturer's protocols (Molecular Probes [see http://www.mobitec.de/probes/docs/media/pis/mp07530.pdf]). Briefly, yeast cells carrying plasmid pVT100U-mito-mRuby (Table 1) at ∼106 cells/ml were suspended in 50 mM sodium citrate buffer (pH 5) containing 20 g/liter glucose. Rhodamine 123 was then added to a final concentration of 30 to 50 μM. The mixture was incubated at room temperature for 20 min and washed three times in the same sodium citrate buffer with 20 g/liter glucose. The stained yeast cells were visualized by using an APO PH3 100×/1.40 objective on a Leica AF7000 wide-field fluorescence microscope equipped with an sCMOS pco.edge 5.5 camera (Leica/Nuhsbaum Inc., McHenry, IL).
Fermentation and metabolite analysis.
The xylose fermentation mixture was prepared by inoculating a preculture grown overnight (5 ml of YP medium containing 20 g/liter glucose) into 50 ml YPX40 medium (YP medium containing 40 g/liter of xylose) in a 250-ml Erlenmeyer flask with an initial optical density at 600 nm (OD600) of 1.0 and incubated at 30°C and 100 rpm. The OD600 was measured by using a spectrophotometer (Biomate 5; Thermo, NY), and extracellular metabolite concentrations were measured by high-performance liquid chromatography (HPLC) (Agilent Technologies 1200 series). The HPLC instrument was equipped with a Rezex ROA-Organic Acid H+ (8%) column (Phenomenex Inc., Torrance, CA) and a refractive index detector (RID), and the column was eluted with 0.005 N H2SO4 at a flow rate of 0.6 ml/min at 50°C.
Human lysozyme activity measurement.
Human lysozyme activity was measured according to the manufacturer's protocols (Sigma-Aldrich [see https://www.sigmaaldrich.com/technical-documents/protocols/biology/enzymatic-assay-of-lysozyme.html]), using Micrococcus lysodeikticus ATCC 4698 (catalogue number M3770; Sigma-Aldrich) as the substrate and lysozyme from chicken egg white (catalogue number L4919; Sigma-Aldrich) as a control. One unit of lysozyme will produce an ΔA450 of 0.001 per min at pH 6.24 at 25°C.
RESULTS
Construction of auxotrophic mutants of S. boulardii by using the CRISPR-Cas9 system.
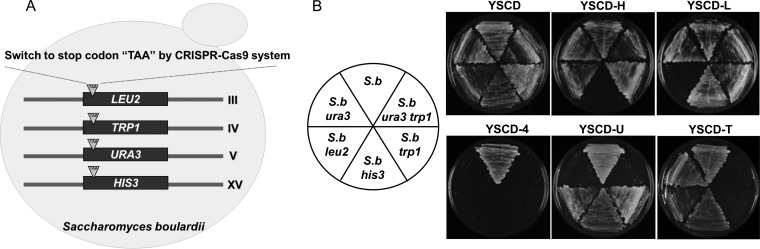
S. boulardii is a probiotic yeast that can be used as a drug delivery carrier. However, genetic manipulation of this yeast has been limited due to the lack of genome-editing tools for S. boulardii. As such, the first step of this study was to construct well-defined and scar-free auxotrophic mutants for further genetic manipulations by using existing plasmids of S. cerevisiae. Figure 1A illustrates the scheme for disrupting four auxotrophic marker genes (URA3, HIS3, TRP1, and LEU2) by the CRISPR-Cas9 system. Three nucleotides close to 5′ terminus of each open reading frame (ORF) were replaced by a stop codon, “TAA,” to minimize the change in the genome. The functions of these genes were then completely disrupted by the early termination of translation. We applied this strategy to generate mutant alleles of four commonly used auxotrophic markers in S. boulardii. Our results indicate that the CRISPR-Cas9 system developed for S. cerevisiae can work well in S. boulardii (Fig. 1B; see also Fig. S1A in the supplemental material). Notably, four auxotrophic marker genes were disrupted as designed, and the efficiency of each gene disruption was 100%, as shown in Fig. S1A in the supplemental material. In addition to the single-gene disruption, simultaneous double disruptions of URA3 and TRP1 were conducted by using a tandem guide RNA expression cassette containing both guide RNAs targeting URA3 and TRP1 (Fig. 1B; see also Fig. S1B in the supplemental material).
FIG 1.
(A) Diagram for construction of marker-free auxotrophic mutants of S. boulardii using the CRISPR-Cas9 system. (B) Confirmation of the auxotrophic phenotype of each mutant on minimal medium lacking appropriate amino acids or nucleotides. S.b, Saccharomyces boulardii; YSCD, yeast synthetic complete medium with 20 g/liter glucose as a carbon source; YSCD-H, YSCD medium minus histidine; YSCD-L, YSCD medium minus leucine; YSCD-U, YSCD medium minus uracil; YSCD-T, YSCD medium minus tryptophan; YSCD-4, YSCD medium minus histidine, leucine, uracil, and tryptophan.
To confirm the mutations in the auxotrophic strains, PCR products of URA3, HIS3, TRP1, and LEU2 in each auxotrophic strain were sequenced. Sequencing results confirmed the introduction of the TAA codon into each marker gene as designed (see Fig. S2 in the supplemental material). Also, the sequencing data were consistent with the auxotrophic phenotype of each mutant, as shown in Fig. 1B. We have also confirmed that all these auxotrophic mutants can be transformed with the pRS series plasmids (pRS423, pRS424, pRS425, and pRS426) with high efficiencies. Taken together, we not only constructed auxotrophic mutants of S. boulardii using the CRISPR-Cas9 system but also demonstrated that commonly used plasmids from S. cerevisiae can be transformed into these auxotrophic mutants.
Heterologous protein expression in S. boulardii.
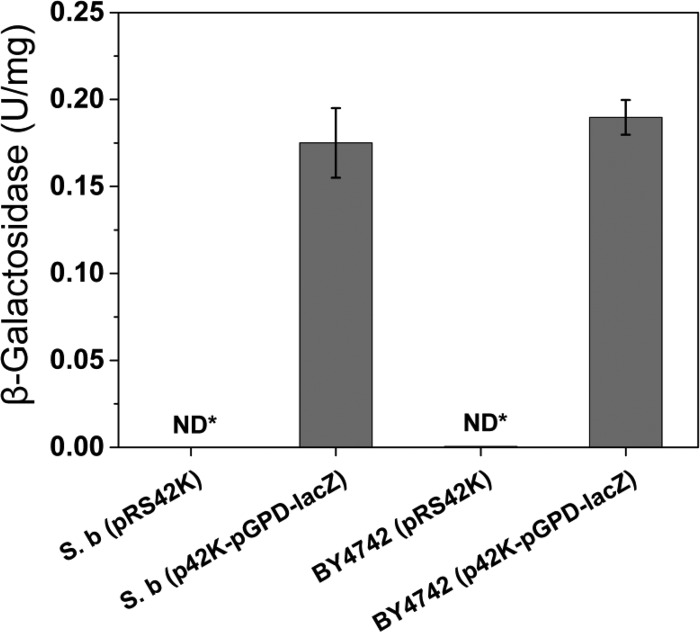
In addition to targeted gene disruption, overexpression of a heterologous gene under the control of a constitutive promoter is necessary for metabolic engineering. In order to examine the feasibility of target gene overexpression using existing genetic tools for S. cerevisiae, we constructed an overexpression cassette (p42K-pGPD-lacZ) harboring E. coli lacZ under the control of the TDH3 promoter (pGPD) in multicopy plasmid pRS42K. After transformation of the overexpression cassette, β-galactosidase activity in the crude extract of the S. boulardii transformant was measured. As shown in Fig. 2, the crude extract of S. boulardii with p42K-pGPD-lacZ showed β-galactosidase activity, while the control strain with pRS42K did not. This result indicated that a heterologous gene from E. coli could be functionally expressed in S. boulardii as it was in S. cerevisiae (Fig. 2).
FIG 2.
Expression of a heterologous protein in S. boulardii. lacZ from E. coli was overexpressed in both S. boulardii (S. b) and S. cerevisiae by using plasmid p42K-pGPD-lacZ. Transformants of the expression cassette showed β-galactosidase activities compared to control strains without the lacZ expression cassette. ND*, not detected. Results are presented as the mean values and standard deviations of data from three independent biological replicates.
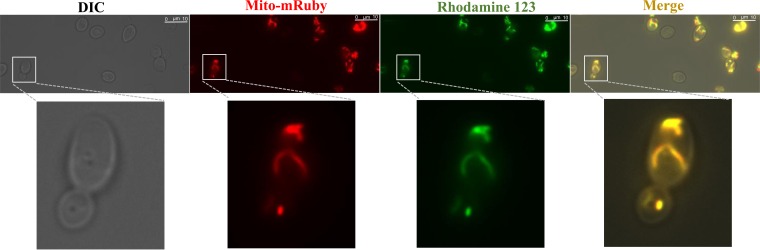
Furthermore, we examined whether a heterologous protein with a mitochondrial targeting sequence can be correctly localized in mitochondria of S. boulardii using plasmid pVT100U-mitoGFP, which is capable of mitochondrial expression of GFP in S. cerevisiae (30). To confirm the localization of the mitochondrial signal in S. boulardii using the mitochondrial staining dye rhodamine 123, which shows green signal, the GFP gene was replaced with the mRuby gene, which presents red fluorescence (31). Plasmids pVT100U-mitoGFP and pVT100U-mito-mRuby were transformed into the S. boulardii ura3 mutant. The mito-GFP and mito-RFP signals were confirmed, as shown in Fig. S3 in the supplemental material. S. boulardii cells carrying pVT100U-mito-mRuby growing in the exponential phase were collected and stained with the mitochondrial staining dye rhodamine 123 (34). The stained yeast cells were monitored under a fluorescence microscope as described in Materials and Methods. S. boulardii carrying plasmid pVT100U-mito-mRuby showed the correct localization of red fluorescence in mitochondria, as indicated by the green signal from rhodamine 123 staining (Fig. 3). These results demonstrate that existing genetic tools for S. cerevisiae can be applied to S. boulardii strains for cellular and metabolic engineering.
FIG 3.
Targeted localization of the red fluorescent protein (mRuby) in mitochondria of S. boulardii expressing mRuby with a mitochondrial targeting sequence as indicated by the mitochondrion marker rhodamine 123. The left panel shows differential interference contrast (DIC) images, the second panel from the left shows red fluorescence of mito-mRuby, the third panel from the left shows mitochondrion staining using rhodamine 123, and the right panel shows the overlay of the above-described three images to show the colocalization of mito-mRuby with mitochondria in S. boulardii.
Metabolic pathway introduction into S. boulardii.
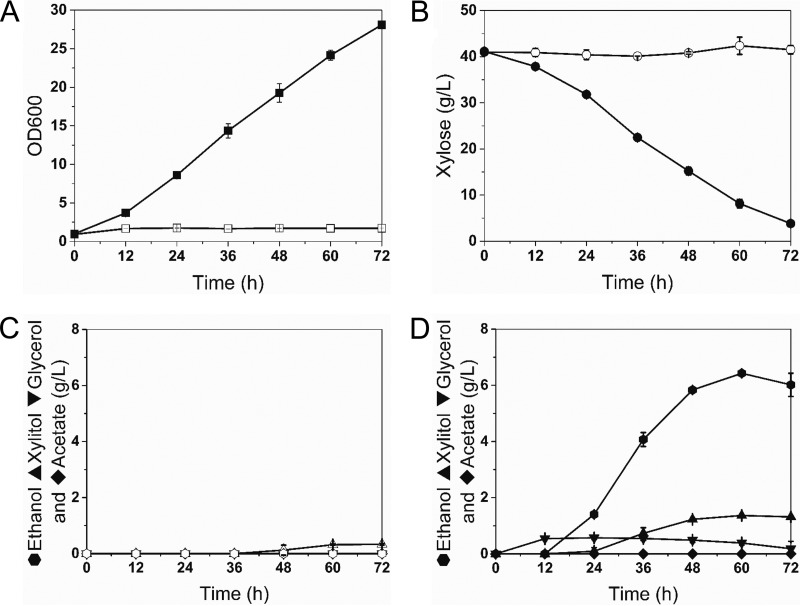
Next, we attempted to introduce a heterologous metabolic pathway into S. boulardii. Xylose is a five-carbon sugar, which is abundant in hydrolysates of the plant cell wall (35). Most yeast strains, including S. cerevisiae, cannot ferment xylose because they do not have xylose metabolic pathways consisting of xylose reductase (XR), xylitol dehydrogenase (XDH), and xylulokinase (XK). However, S. cerevisiae can be engineered to ferment xylose by introducing three genes (XYL1, XYL2, and XYL3 from Scheffersomyces stipitis) coding for XR, XDH, and XK (35–37). Similarly, we observed that S. boulardii is also incapable of naturally fermenting xylose. Thus, we introduced an integrating expression cassette (pSR6-X123) (37, 38) expressing the XYL1, XYL2, and XYL3 genes under the control of constitutive promoters of S. cerevisiae into the ura3 locus of the S. boulardii uracil auxotroph. The resulting engineered S. boulardii strain (SB-X123) showed decent xylose consumption and produced ethanol, suggesting that the xylose metabolic pathways are operational in S. boulardii (Fig. 4). Interestingly, the rate of xylose fermentation by the engineered S. boulardii strain (SB-X123) was higher than that of S. cerevisiae strains from previous studies (37, 38). Our results suggest that S. boulardii can be employed as a potential host for producing cellulosic biofuels as well as a probiotic yeast strain.
FIG 4.
Introduction of a heterologous metabolic pathway in S. boulardii. A xylose assimilation pathway from S. stipitis was introduced into S. boulardii. (A and B) Growth (A) and xylose consumption (B) of S. boulardii and SB-X123 (S. boulardii carrying XYL1, XYL2, and XYL3) in YP medium with 40 g/liter of xylose as the sole carbon source. (C and D) Metabolites for S. boulardii (C) and SB-X123 (D) during xylose fermentation. Open symbols, S. boulardii; filled symbols, SB-X123. Results are presented as the mean values and standard deviations of data from three independent biological replicates.
Secretion of human lysozyme by S. boulardii.
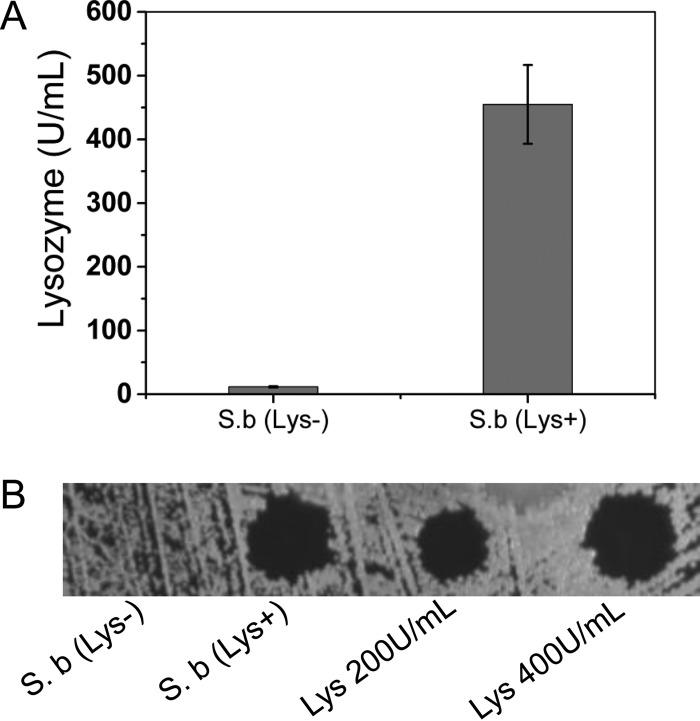
As the last step, we focused on how to engineer S. boulardii to make it a better probiotic for human beings. Human lysozyme was chosen as a target because it is widely distributed in a variety of tissues (liver, articular cartilage, and plasma) and body fluids (saliva, tears, and milk) (39). Human lysozyme will preferentially hydrolyze the −1,4 glycosidic linkages between the N-acetylmuramic acid and N-acetylglucosamine groups of the cell wall structure of Gram-positive bacteria and therefore appears to have a role in host defense (25, 26, 39). The lysozyme existing in human milk helps infants build a healthy gut environment and promotes weight gain of premature infants (40, 41). It will be more beneficial if S. boulardii gains this ability of lysozyme secretion so that it provides human lysozyme continuously in the human gut. The gene (cHLY) of human lysozyme (25) with a chicken lysozyme signal sequence (26) was synthesized (the full sequence can be found in the supplemental material) and introduced into the genome of S. boulardii by CRISPR-Cas9 technology for stable expression. As shown in Fig. 5, the S. boulardii strain carrying cHLY [strain S.b(Lys+)] gained the ability to secrete human lysozyme compared with wild-type S. boulardii [strain S.b(Lys−)]. Finally, we demonstrated that S. boulardii could be engineered to secrete a beneficial compound for human gut health.
FIG 5.
Human lysozyme is secreted by S. boulardii carrying the integrated human lysozyme gene with the chicken lysozyme signal sequence. (A) Lysozyme activity in concentrated supernatants of S.b(Lys−) and S.b(Lys+). Results are presented as the mean values and standard deviations of data from three independent biological replicates. (B) Halo assay of concentrated human lysozyme from the fermentation broth of S.b(Lys+). Totals of 200 U/ml and 400 U/ml chicken egg white lysozyme were used as controls.
DISCUSSION
In the present study, we demonstrated the feasibility of genetic/metabolic engineering of probiotic yeast S. boulardii using the newly emerging CRISPR-Cas9 technology and existing genetic tools for S. cerevisiae. Four widely used auxotrophic selection markers (URA3, HIS3, TRP1, and LEU2) were disrupted efficiently by using Cas9 nuclease and guide RNAs expressing cassettes. In addition, we report the simultaneous disruption of two genes (URA3 and TRP1) using a tandem guide RNA expression cassette. These results indicate the potential for simultaneous disruption of multiple target genes in S. boulardii to be conducted through CRISPR-Cas9 technology, which allows sophisticated and safe genetic perturbations required to engineer this yeast for food and medical applications.
After sequencing, we found that the sequences of HIS3 and URA3 are identical to those in strain S288C, which has been used for genome sequencing of S. cerevisiae. However, the sequence of TRP1 has two point mutations (T256C and C635T) compared to the sequence in S288C. Interestingly, S. boulardii LEU2 is 97% identical to that of S288C and has a 24-nucleotide insertion close to the C terminus of the region. All the sequencing data for auxotrophic markers in S. boulardii are consistent with the sequence of ATCC MYA-796 in GenBank (accession number GCA_000769245.1). None of these mutations in TRP1 and LEU2 affected the 20-bp guide RNAs that were used for Cas9 targeting.
Targeted genetic perturbation based on the CRISPR-Cas9 system has advantages over previously used methods. First, genetic perturbations of target genes can be done rapidly. As genetic perturbations of multiple genes are feasible, complex phenotypes determined by many mutations at multiple loci can be engineered. Second, the efficiencies of genetic perturbations are as high as 100%, and target genetic perturbations are made due to the precise cutting and almost no off-target effects of the CRISPR-Cas9 system in yeast. This is a key advantage compared to UV mutagenesis (14, 15), as the latter generates numerous unwanted mutations throughout the whole genome. Off-target mutation effects caused by CRISPR-Cas9 in mammalian systems have been reported previously (42, 43). Fortunately, there have been no similar reports in yeast to our knowledge. As the size of the yeast genome is ∼250 times smaller than that of the human genome, the off-target efficiency in yeast might be almost negligible. Third, the CRISPR-Cas9 system does not leave any scars or unnecessary genetic elements in the genome, whereas other existing genetic perturbation methods, such as the Cre-loxP system (16), do. Not only is the marker recycling of the Cre-loxP system mediated by the endonuclease time-consuming, but the loxP sites left on the genome also make it difficult to use the Cre-loxP system repeatedly because of lower efficiencies (44). Moreover, recombination between loxP sites can lead to unwanted chromosome rearrangements, as reported previously (17–19). While the introduction of episomal plasmids for expressing Cas9 and guide RNA is necessary, they can be easily dropped out after the desired genetic perturbations are made in the genome. Taken together, this study provides a rapid, efficient, and clean auxotrophic S. boulardii strain construction approach by using the CRISPR-Cas9 system, which allows precision engineering of S. boulardii for in vivo applications.
There have been several reports showing that S. boulardii is different from S. cerevisiae in terms of transformation procedures and limited genetic perturbation tools (2). In this study, we showed that existing plasmids and genetic tools currently used for S. cerevisiae could be readily applied to S. boulardii. The pRS series plasmids with selection markers based on auxotroph and drug resistance have been widely used for introducing genetic perturbations into S. cerevisiae (45, 46). Also, constitutive promoters, including pTDH3, pPGK1, and pTEF, have been employed for the overexpression of a target gene in conjunction with the use of the pRS series plasmids (27). While previous studies examined whether genetic tools of S. cerevisiae can be utilized for introducing genetic perturbations into S. boulardii (1, 2), the examinations were limited because four common auxotrophic strains (ura3, his3, trp1, and leu2) of S. boulardii did not exist. As we obtained isogenic single or double mutants, which can be complemented by four auxotrophic markers, we were able to perform comprehensive evaluations of S. cerevisiae genetic tools in S. boulardii. In the end, we confirmed that each auxotrophic mutant could be transformed with the corresponding pRS42X plasmids. Second, we introduced lacZ, which is commonly used as a reporter gene in E. coli, into S. boulardii. β-Galactosidase activity was detected in the S. boulardii transformants. During this assay, a pRS42K plasmid carrying the S. cerevisiae TDH3 promoter was used, indicating that the S. cerevisiae TDH3 promoter is working in S. boulardii. A previous study indicated that heterologous GFP can be expressed in S. boulardii (15). Here, mito-GFP and mito-RFP were functionally expressed and precisely localized to mitochondria, as expected, which means that even the localization of heterologous protein can be precisely achieved in S. boulardii. Next, we demonstrated that a large expression cassette containing three heterologous genes, XYL1, XYL2, and XYL3, from S. stipitis under the control of the TDH3 and PGK1 promoters could be transformed into S. boulardii and enabled xylose assimilation in S. boulardii. These results show that heterologous genes could be readily introduced into S. boulardii by using the same strategy as that used for S. cerevisiae. However, we noticed that the transformation efficiency of S. boulardii needs to be improved, as it is about 30- to 50-fold lower than that of S. cerevisiae laboratory strains by the PEG-LiAc method, which is consistent with data from a previous study (2). This efficiency may be sufficient for plasmid-based overexpression. We observed that the transformation efficiency is lower for long-fragment integration by using the CRISPR-Cas9 system, due to the aneuploidy of S. boulardii, especially the three copies of chromosome IX (1). Finally, we proved that a beneficial component, human lysozyme, could be secreted by integrating cHLY into the genome of S. boulardii by the CRISPR-Cas9 strategy. The effect of this engineered strain needs to be evaluated later.
The present study successfully showed the possibility of precisely modifying S. boulardii to achieve its function as a therapeutic delivery carrier. This is the first step, and further work shall be conducted. For instance, the introduction of metabolic pathways with beneficial final products, secretion of therapeutic proteins for the treatment of gut disorders, and gut production of vaccine in S. boulardii can be attempted, as we have an efficient and precise genetic perturbation method without the involvement of antibiotic resistance markers.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from the Energy Biosciences Institute (EBI) and the Research Initiative Program of KRIBB.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00057-16.
REFERENCES
- 1.Edwards-Ingram L, Gitsham P, Burton N, Warhurst G, Clarke I, Hoyle D, Oliver SG, Stateva L. 2007. Genotypic and physiological characterization of Saccharomyces boulardii, the probiotic strain of Saccharomyces cerevisiae. Appl Environ Microbiol 73:2458–2467. doi: 10.1128/AEM.02201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Douradinha B, Reis VC, Rogers MB, Torres FA, Evans JD, Marques ET Jr. 2014. Novel insights in genetic transformation of the probiotic yeast Saccharomyces boulardii. Bioengineered 5:21–29. doi: 10.4161/bioe.26271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotowska M, Albrecht P, Szajewska H. 2005. Saccharomyces boulardii in the prevention of antibiotic-associated diarrhoea in children: a randomized double-blind placebo-controlled trial. Aliment Pharmacol Ther 21:583–590. doi: 10.1111/j.1365-2036.2005.02356.x. [DOI] [PubMed] [Google Scholar]
- 4.Czerucka D, Piche T, Rampal P. 2007. Review article: yeast as probiotics—Saccharomyces boulardii. Aliment Pharmacol Ther 26:767–778. doi: 10.1111/j.1365-2036.2007.03442.x. [DOI] [PubMed] [Google Scholar]
- 5.Martins F, Silva A, Vieira A, Barbosa FF, Arantes RE, Teixeira M, Nicoli J. 2009. Comparative study of Bifidobacterium animalis, Escherichia coli, Lactobacillus casei and Saccharomyces boulardii probiotic properties. Arch Microbiol 191:623–630. doi: 10.1007/s00203-009-0491-x. [DOI] [PubMed] [Google Scholar]
- 6.Micklefield G. 2014. Saccharomyces boulardii in the treatment and prevention of antibiotic-associated diarrhea. MMW Fortschr Med 156(Suppl 1):S18–S22. (In German.) [PubMed] [Google Scholar]
- 7.Everard A, Matamoros S, Geurts L, Delzenne NM, Cani PD. 2014. Saccharomyces boulardii administration changes gut microbiota and reduces hepatic steatosis, low-grade inflammation, and fat mass in obese and type 2 diabetic db/db mice. mBio 5:e01011-14. doi: 10.1128/mBio.01011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Kokkotou EG, Mustafa N, Bhaskar KR, Sougioultzis S, O'Brien M, Pothoulakis C, Kelly CP. 2006. Saccharomyces boulardii inhibits ERK1/2 mitogen-activated protein kinase activation both in vitro and in vivo and protects against Clostridium difficile toxin A-induced enteritis. J Biol Chem 281:24449–24454. doi: 10.1074/jbc.M605200200. [DOI] [PubMed] [Google Scholar]
- 9.Rajput IR, Hussain A, Li YL, Zhang X, Xu X, Long MY, You DY, Li WF. 2014. Saccharomyces boulardii and Bacillus subtilis B10 modulate TLRs mediated signaling to induce immunity by chicken BMDCs. J Cell Biochem 115:189–198. doi: 10.1002/jcb.24650. [DOI] [PubMed] [Google Scholar]
- 10.Pontier-Bres R, Munro P, Boyer L, Anty R, Imbert V, Terciolo C, Andre F, Rampal P, Lemichez E, Peyron JF, Czerucka D. 2014. Saccharomyces boulardii modifies Salmonella Typhimurium traffic and host immune responses along the intestinal tract. PLoS One 9:e103069. doi: 10.1371/journal.pone.0103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villar-Garcia J, Hernandez JJ, Guerri-Fernandez R, Gonzalez A, Lerma E, Guelar A, Saenz D, Sorli L, Montero M, Horcajada JP, Knobel Freud H. 2015. Effect of probiotics (Saccharomyces boulardii) on microbial translocation and inflammation in HIV-treated patients: a double-blind, randomized, placebo-controlled trial. J Acquir Immune Defic Syndr 68:256–263. doi: 10.1097/QAI.0000000000000468. [DOI] [PubMed] [Google Scholar]
- 12.Krasowska A, Murzyn A, Dyjankiewicz A, Lukaszewicz M, Dziadkowiec D. 2009. The antagonistic effect of Saccharomyces boulardii on Candida albicans filamentation, adhesion and biofilm formation. FEMS Yeast Res 9:1312–1321. doi: 10.1111/j.1567-1364.2009.00559.x. [DOI] [PubMed] [Google Scholar]
- 13.Gay PB, Gillespie SH. 2005. Antibiotic resistance markers in genetically modified plants: a risk to human health? Lancet Infect Dis 5:637–646. doi: 10.1016/S1473-3099(05)70241-3. [DOI] [PubMed] [Google Scholar]
- 14.Hamedi H, Misaghi A, Modarressi MH, Salehi TZ, Khorasanizadeh D, Khalaj V. 2013. Generation of a uracil auxotroph strain of the probiotic yeast Saccharomyces boulardii as a host for the recombinant protein production. Avicenna J Med Biotechnol 5:29–34. [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson LE, Fasken MB, McDermott CD, McBride SM, Kuiper EG, Guiliano DB, Corbett AH, Lamb TJ. 2014. Functional heterologous protein expression by genetically engineered probiotic yeast Saccharomyces boulardii. PLoS One 9:e112660. doi: 10.1371/journal.pone.0112660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Sun H, Zhang J, Liu Q, Wang T, Chen P, Li H, Xiao Y, Wang F, Zhao X. 2015. Establishment and application of target gene disruption system in Saccharomyces boulardii. Biotechnol Bioproc E 20:26–36. doi: 10.1007/s12257-014-0197-z. [DOI] [Google Scholar]
- 17.Delneri D, Tomlin GC, Wixon JL, Hutter A, Sefton M, Louis EJ, Oliver SG. 2000. Exploring redundancy in the yeast genome: an improved strategy for use of the cre-loxP system. Gene 252:127–135. doi: 10.1016/S0378-1119(00)00217-1. [DOI] [PubMed] [Google Scholar]
- 18.Solis-Escalante D, Kuijpers NG, van der Linden FH, Pronk JT, Daran JM, Daran-Lapujade P. 2014. Efficient simultaneous excision of multiple selectable marker cassettes using I-SceI-induced double-strand DNA breaks in Saccharomyces cerevisiae. FEMS Yeast Res 14:741–754. doi: 10.1111/1567-1364.12162. [DOI] [PubMed] [Google Scholar]
- 19.Solis-Escalante D, van den Broek M, Kuijpers NG, Pronk JT, Boles E, Daran JM, Daran-Lapujade P. 2015. The genome sequence of the popular hexose-transport-deficient Saccharomyces cerevisiae strain EBY.VW4000 reveals LoxP/Cre-induced translocations and gene loss. FEMS Yeast Res 15:fou004. doi: 10.1093/femsyr/fou004. [DOI] [PubMed] [Google Scholar]
- 20.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doudna JA, Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 22.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muraki M, Jigami Y, Tanaka H, Harada N, Kishimoto F, Agui H, Ogino S, Nakasato S. 1986. Expression of synthetic human lysozyme gene in Escherichia coli. Agric Biol Chem 50:713–723. doi: 10.1271/bbb1961.50.713. [DOI] [Google Scholar]
- 26.Jigami Y, Muraki M, Harada N, Tanaka H. 1986. Expression of synthetic human-lysozyme gene in Saccharomyces cerevisiae: use of a synthetic chicken-lysozyme signal sequence for secretion and processing. Gene 43:273–279. doi: 10.1016/0378-1119(86)90216-7. [DOI] [PubMed] [Google Scholar]
- 27.Mumberg D, Muller R, Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 28.Taxis C, Knop M. 2006. System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. Biotechniques 40:73–78. doi: 10.2144/000112040. [DOI] [PubMed] [Google Scholar]
- 29.Zhang GC, Kong II, Kim H, Liu JJ, Cate JH, Jin YS. 2014. Construction of a quadruple auxotrophic mutant of an industrial polyploid Saccharomyces cerevisiae strain by using RNA-guided Cas9 nuclease. Appl Environ Microbiol 80:7694–7701. doi: 10.1128/AEM.02310-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westermann B, Neupert W. 2000. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16:1421–1427. [DOI] [PubMed] [Google Scholar]
- 31.Lee S, Lim WA, Thorn KS. 2013. Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae. PLoS One 8:e67902. doi: 10.1371/journal.pone.0067902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gietz RD, Schiestl RH, Willems AR, Woods RA. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 33.Ribeiro O, Gombert AK, Teixeira JA, Domingues L. 2007. Application of the Cre-loxP system for multiple gene disruption in the yeast Kluyveromyces marxianus. J Biotechnol 131:20–26. doi: 10.1016/j.jbiotec.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 34.Skowronek P, Krummeck G, Haferkamp O, Rodel G. 1990. Flow cytometry as a tool to discriminate respiratory-competent and respiratory-deficient yeast cells. Curr Genet 18:265–267. doi: 10.1007/BF00318391. [DOI] [PubMed] [Google Scholar]
- 35.Jeffries TW, Jin YS. 2004. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl Microbiol Biotechnol 63:495–509. doi: 10.1007/s00253-003-1450-0. [DOI] [PubMed] [Google Scholar]
- 36.Zhang GC, Liu JJ, Ding WT. 2012. Decreased xylitol formation during xylose fermentation in Saccharomyces cerevisiae due to overexpression of water-forming NADH oxidase. Appl Environ Microbiol 78:1081–1086. doi: 10.1128/AEM.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SR, Skerker JM, Kang W, Lesmana A, Wei N, Arkin AP, Jin YS. 2013. Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS One 8:e57048. doi: 10.1371/journal.pone.0057048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SR, Ha SJ, Kong II, Jin YS. 2012. High expression of XYL2 coding for xylitol dehydrogenase is necessary for efficient xylose fermentation by engineered Saccharomyces cerevisiae. Metab Eng 14:336–343. doi: 10.1016/j.ymben.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 39.Dumoulin M, Johnson RK, Bellotti V, Dobson C. 2007. Human lysozyme, p 285–308. In Uversky V, Fink A (ed), Protein misfolding, aggregation, and conformational diseases, vol 6 Springer, New York, NY. [Google Scholar]
- 40.Braun OH, Sandkuhler H. 1985. Relationships between lysozyme concentration of human milk, bacteriologic content, and weight gain of premature infants. J Pediatr Gastroenterol Nutr 4:583–586. doi: 10.1097/00005176-198508000-00015. [DOI] [PubMed] [Google Scholar]
- 41.Chandan RC, Shahani KM, Holly RG. 1964. Lysozyme content of human milk. Nature 204:76–77. doi: 10.1038/204076a0. [DOI] [PubMed] [Google Scholar]
- 42.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jakočiūnas T, Bonde I, Herrgård M, Harrison SJ, Kristensen M, Pedersen LE, Jensen MK, Keasling JD. 2015. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng 28:213–222. doi: 10.1016/j.ymben.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119–122. doi: 10.1016/0378-1119(92)90454-W. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.