Abstract
Managing ecosystems to maintain biodiversity may be one approach to ensuring their dynamic stability, productivity, and delivery of vital services. The applicability of this approach to industrial ecosystems that harness the metabolic activities of microbes has been proposed but has never been tested at relevant scales. We used a tag-sequencing approach with bacterial small subunit rRNA (16S) genes and eukaryotic internal transcribed spacer 2 (ITS2) to measuring the taxonomic composition and diversity of bacteria and eukaryotes in an open pond managed for bioenergy production by microalgae over a year. Periods of high eukaryotic diversity were associated with high and more-stable biomass productivity. In addition, bacterial diversity and eukaryotic diversity were inversely correlated over time, possibly due to their opposite responses to temperature. The results indicate that maintaining diverse communities may be essential to engineering stable and productive bioenergy ecosystems using microorganisms.
INTRODUCTION
Microalgae are one of the most productive photosynthetic organisms on the planet, using sunlight to convert CO2 and nutrients into biomass, which can be used to generate products ranging from high-value chemicals such as pigments or nutritional oils to commodities such as protein and biofuels. They can be cultivated on agricultural scales in open ponds using nonarable land and nonpotable water and as such are attractive candidates for the production of low-cost biomass (1, 2). A large limiting factor for reliable low-cost biomass production in open ponds is contamination (3–7). Managing biological contamination is costly, and while it has been achieved in open ponds for the production of high-value algal biomass (8, 9), managing algae stably in open ponds for the production of low-cost algal biomass remains challenging (10).
Agricultural pesticides or chemicals have been deployed to mitigate the challenges of contamination in algal production systems (12, 13; J. Weissman and G. Radaelli, U.S. patent application 12,321,767; R. C. McBride, C. A. Behnke, K. M. Botsch, N. A. Heaps, and C. Del Meenach, U.S. patent application 14,351,540). Approaches to managing contamination using precepts from ecology have been suggested as a viable low-cost alternative (14, 15). This perspective is informed by the idea that traits that determine fitness are not independent and often experience tradeoffs (14). For instance, Shurin et al. (16) showed that species that are good N and P competitors generally grow poorly at low light levels. Tradeoffs between other ecologically important functions have also been shown among algal taxa (17). These tradeoffs can give rise to negative associations between fitness under different conditions or abilities to perform functions such as competing for resources or resisting consumers (18–20). Tradeoffs also imply that in heterogeneous environments open to invaders, maintaining a stable monoculture will be challenging or impossible. In contrast, polycultures or ecosystems may be more stable and productive than monocultures (21). This assertion has been validated in natural and constructed algal assemblages, where increasing diversity was associated with higher productivity (22). Other experiments have indicated that assemblages of algae are more efficient at taking up nutrients and resisting invasion than monocultures (14); however, more basic research is needed to determine if consortia are a viable option for algal biomass production at industrial scales. Open ponds are very distinct from the natural environments experienced by most strains of algae, where they encounter nutrient limitation, consumers and pathogens, sinking, and fluctuating environmental conditions. Whether algae in the nutrient-replete and highly productive environments of managed open ponds follow the same patterns observed in natural communities and lab experiments is still an open question.
In this study, we monitored the bacterial and eukaryotic composition of an alga pond managed to optimize biomass productivity over the course of a year. We used 16S and internal transcribed spacer 2 (ITS2) Ion Torrent Personal Genome Machine (PGM) tag sequencing to assess the bacteria and eukaryotic taxonomic composition and diversity of the pond. We simultaneously monitored a number of aspects of ecosystem structure and function (e.g., nitrate, phosphate, dry weight [DW], and fluorescence) to examine the intra- and interrelationships of ecosystem structure with genomic composition, particularly between microbial diversity and biomass productivity. We asked whether the positive relationships among diversity, stability, and productivity observed in natural and experimental communities of algae were also seen in an engineered environment managed for bioenergy production. Our study seeks to establish the applicability of ecological principles to industrial ecosystems at scales relevant to production of biomass to generate energy or specialized products. Based on ecological theory (14, 16, 22, 23), we expect that periods of high taxonomic diversity should be associated with high and more stable biomass production.
MATERIALS AND METHODS
Pond data collection.
The algae were grown in a dirt-lined half-acre pond on the Las Cruces, NM, test site of Sapphire Energy Incorporated. The pond was filled with water in June 2011 and became colonized by green algae, and nutrients were added. The pond had a volume of 400,000 liters, and water was circulated via a pump at an average speed of 10 cm/s. The maximum depth of the pond was 30 cm. The pH of the pond was maintained at 9 via the addition of CO2, and biomass was maintained between 0.4 and 0.8g/liter by harvesting (for harvest data and biomass, see Fig. S1 in the supplemental material). The medium (i.e., initial concentrations of the pond) was made up of a salt component to simulate possible commercial-level total dissolved solids (TDS) and the salt composition of water not suitable for most agricultural practices. The composition of the medium on a per liter basis is as follows: 3.675 g NaHCO3, 4.766 g Na2SO4, 0.490 g KCl, 1.090 g NaCl, 0.518 g MgSO4·7H2O, 0.146 g NaF. The nutrient component of this medium on a per liter basis is comprised of the following: 0.3 g urea, 0.344 ml 8.5% H3PO4 (vol/vol), 0.06 ml trace [1 g sodium EDTA, 0.194 g ferric chloride, 0.072 g manganese chloride, 0.021 g zinc chloride, 0.013 g sodium molybdate, and 0.004 g cobalt(II) chloride into 1 liter deionized (DI) H2O], sterilized using a Corning 0.22-μm filter system and with 0.024 ml Fe (per liter: 336.3 g EDTA powder and 100 g Ferix-3). Ferix-3 is dry granulated ferric sulfate containing 20% Fe. Addition of nutrients such as urea, NH4, NO3, and PO4 was performed to maintain the initial state of the pond medium, an N level of 100 ppm, and a PO4 level of 40 ppm (see Fig. S2 and S3 in the supplemental material for N and PO4 addition data, together with measured urea and PO4 levels). The pond was treated on four separate occasions (days 152, 168, 177, and 190) with two commercial fungicides to address a decline in biomass that was suspected to be the result of fungal pathogen outbreak. The active ingredients in the fungicides applied were fluazinam and pyraclostrobin. A 1-ppm concentration of fluazinam was applied on days 152, 177, and 190, and 1 ppm pyraclostrobin was applied on day 168. McBride et al. (McBride et al., U.S. patent application 14,351,540) have shown the effects of fluazinam and pyraclostrobin on uncontaminated and contaminated algae at various dosage levels, including 1 ppm, by observing the culture density (optical density at 750 nm [OD750]). According to their study, fluazinam has microalga toxicity at doses greater than 7.5 ppm and pyroclostrobin shows microalga toxicity at doses greater than 15 ppm. Indeed, results in the cited document demonstrate higher optical density values at applications of 1-ppm doses of fluazinam or pyroclostrobin in contaminated algae, whereas these doses show no visible adverse effects on the optical density values in uncontaminated algae (McBride et al., U.S. patent application 14,351,540).
The pond was regularly monitored for a number of parameters, referred to as “pond ecosystem values” in this paper. Standard measurements such as temperature, pH, OD750, OD560, fluorescence at 430/685 nm, fluorescence at 363/685 nm, fluorescence at 590/650 nm, fluorescence at 450/685 nm, pond volume, variable/maximum fluorescence (Fv/Fm) via pulse-amplitude modulation (PAM), dry weight (grams per liter), alkalinity, NH4, urea, NO3, NO2, PO4, and harvest volume data were collected. OD and florescence data were collected on a SpectraMax plate reader (Molecular Devices, Sunnyvale, CA). PAM measures were collected using a PAM fluorometer (Walz, Effeltrich, Germany). Alkalinity was measured on a TitroLine (Si-Analytics, Mainz, Germany), NH4 and urea were measured using colorimetric assays (Sapphire Energy assay, similar to the assay from Seal Analytical, Mequon, WI), and NO3, NO2, and PO4 were measured using ion chromatography (IC). Dry weight was determined by standard techniques (24).
Approximately every 7 days, a biological sample was collected from the pond in a 50-ml tube. Samples were taken at a depth of around 15 cm from the same location of the pond, which was near its southwest corner. The samples were flash-frozen in liquid nitrogen within 4 h (maximum duration) of collection and stored at −80°C until processed for this evaluation. Most samples were collected within 1 h. Although the maximum duration could have skewed some prokaryotic relative abundance data, Cuthbertson et al. (25) present an acceptable window of up to 12 h without significant divergence in bacterial community, although their study suggests collection within 1 h as the optimal window, as was performed for most of our samples.
The first tag-sequencing sample used in this project corresponds to November 2011.
Sample sequencing.
The PCR-amplified products of 16S and ITS2 (see “DNA preparation” in the supplemental material) were applied for bidirectional sequencing using the Ion Torrent PGM following a modified protocol for long-amplicon (400-bp) libraries using a modified long-read Ion Xpress Plus fragment library kit (Ion Torrent Community website; Life Technologies, Carlsbad, CA). Briefly, PCR products that contained a phosphate at the 5′ end of each strand were directly ligated to a pair of Ion adaptors, P1 (universal) and A (barcoding), provided in the kit. The 34 samples derived from 16S gene PCR were ligated to Ion barcodes 1 to 34, respectively, while the 34 samples derived from ITS2 gene PCR were ligated to Ion barcodes 37 to 70, respectively, in a 96-well plate. The ligation was performed in a 25-μl reaction mixture containing 50 to 100 ng PCR sample, 2 μl ligation buffer (5×), 1 μl deoxynucleoside triphosphate (dNTP) (10 mM), 1 μl DNA ligase (5 U/μl), 2 μl nick repair polymerase, 1 μl adaptor P1, and 1 μl of barcoding adaptor A and incubated for 15 min at 15°C and then 5 min at 72°C. After cleanup and size selection using the “magnetic bead cleanup module” (Life Technologies), the ligated samples were pooled and PCR amplified in 110 μl of reaction mixture containing 100 μl HiFi Platinum Taq supermix (Life Technologies), 5 μl P1 and A primer mix, and 5 μl of pooled samples, under an initial temperature of 95°C for 5 min followed by 8 cycles of 95°C for 15 s, 58°C for 15 s, and 70°C for 1 min. After cleanup, the PCR product was quantified by quantitative PCR (qPCR). The multiple-emulsion PCRs were performed to generate template-positive Ion Sphere particles (ISPs) following the protocol of the Ion OneTouch 2 system using the Template OT2 400 kit (Life Technologies). About 25 to 30 million template-positive ISPs were loaded onto each Ion PGM 318 chip. For 16S genes, three chips were sequenced on the PGM, while for ITS2 genes, four chips were sequenced on the PGM. The FASTQ files from Torrent Server were downloaded and used for downstream data processing.
Sequence analysis.
Our ITS2 Ion Torrent PGM data contained an abundance of algal sequences. While operational taxonomic unit (OTU)-based sequence analysis approaches are widely used and provide pipelines for 16S or fungal-only ITS data in determining sample composition, they are not readily available for ITS data from other eukaryotic taxa—e.g., green algae. Grattepanche et al. (26) suggest that the cutoffs applied in OTU-based analyses are taxon dependent and that tools developed for bacterial studies (16S data analysis) are not directly applicable for all eukaryotic species (see Methods in the supplemental material for further discussion). Therefore, we mapped the 16S and ITS2 reads onto selected databases after applying certain quality controls. We also compared the results of our 16S mapping results to an OTU-based approach (see the Methods in the supplemental material) for validation purposes. We obtained Mantel test r statistics of 0.99, 0.98, 0.94, 0.94, and 0.91 with P = 0.001 for 999 repetitions for the ranks phylum, class, order, family, and genus, respectively. Diversity results from both approaches had Pearson R = 0.96 with P = 2.60 × 10−14. Therefore, we confirm that the taxonomic relative abundance and diversity results in both approaches are highly similar.
In the three chips used for 16S sequencing, we obtained 1.6, 3.8, and 4.4 million reads, while the four ITS2 chips resulted in 3.7, 4.7, 4.6, and 3.7 million reads, respectively (see Fig. S4, S5, and S6 in the supplemental material for read length distributions). In order to estimate sample compositions and associated taxonomic information, we mapped our reads to the following databases: for 16S data, we used the GreenGenes 16S sequence database (version May 2013, 1.3 million sequences) (27), and for ITS2 data, we constructed a custom database from NCBI (http://www.ncbi.nlm.nih.gov/) using the keywords “ITS2” or “internal transcribed spacer” for sequences with a length smaller than 100,000 under the “Nucleotide” database section, which resulted in 1.1 million sequences. For mapping purposes, we used the alignment software TMAP (28), optimized to deal with variable read lengths, and Ion Torrent specific error profiles (29). See Methods in the supplemental material for detailed usage of TMAP. We filtered any read having length shorter than 50 nucleotides and an error rate higher than 2.0 for 16S reads and 4.0 for ITS reads, due to their longer average size compared to 16S reads (see Fig. S6). We accepted any mapping that breached a query coverage of 70% and a percentage of identity of 95% per hit, as applied by “16S rRNA Reference Sequence Similarity Search” by NCBI (http://www.ncbi.nlm.nih.gov/genomes/16S/help.html#query). (In the supplemental material, see Results and Fig. S7 and S8 for mapping statistics.) For practical purposes, among the 26,135 and 9,631 total reference sequence hits in all chips, we picked the top 2,000 and 200, corresponding to 97.16% and 96.31% of all hit reads, for 16S and ITS2 data and used their normalized hitting read counts to represent a sample composition.
We obtained the taxonomic composition of our 16S samples using the GreenGenes taxonomy, and for ITS2 samples, we used the taxonomy database of NCBI using Biopython (30). We measured sample composition similarities across all 26 genomic samples, together with the 8 technical replicates, using Bray-Curtis dissimilarity on the top 2,000 and 200 sequences' relative abundances, for 16S and ITS2 data, respectively. We provide intrasample reproducibility assessment in Results and Fig. S9 in the supplemental material.
Here we present our results from chip 3, for both 16S and ITS2 data, due to the higher percentage of mapping reads (see Fig. S7c and S8b in the supplemental material), while simultaneously confirming reproducibility among different chips using Mantel tests (see Methods in the supplemental material), achieving r statistics in the range 0.98 to 0.99 with all other chips in both data sets.
Diversity analysis.
We used Hill numbers to measure diversity with the sensitivity parameter q = 1, which is equal to exp(Shannon entropy H) (31–34). We computed the Shannon entropy at the genus level, after using a rarefaction of 5,000 hit reads in all samples for Bacteria, Eukaryota, Viridiplantae, and Algae diversities; and 500 for fungal diversity due to the comparably small number of hits, using 100 iterations. As shown in Fig. S11 in the supplemental material, all rarefaction curves converged. We used the functions “rrarefy” and “diversity” in package “vegan” (35) in R.
Diversity estimations using the top 200/2,000 reference sequences versus all hits resulted in a Pearson R of >0.99 due to the large portion (96 to 97%) of the top sequences comprised in the community (see the rarefaction curves shown in Fig. S10 in the supplemental material).
Ecosystem variables.
We collected 15 pond ecosystem variables on a regular basis, ranging from every day for some variables to a few times a week for others, over the span of a year. We imputed the missing data points on a dry weight basis (grams per liter) using the OD750 as it had more frequent measurements and was the variable most strongly correlated with dry weight (Pearson's R = 0.85). Since dry weight is the major input in the computation of productivity, and the standard deviation (SD) in productivity was of interest, imputation was a necessary step in order to have approximately similar numbers of sample sizes (see Fig. S19 in the supplemental material) in various time windows. See Fig. S20 in the supplemental material for variance patterns in original and imputed dry weight data. Other ecosystem variables than dry weight (grams per liter) were either sufficiently sampled or did not require such preprocessing as their standard deviations were not of interest. We used the function “mice.impute.norm.predict” in the package “mice” (36) in R.
We removed the outliers (see Methods in the supplemental material) and applied linear interpolation on missing data and a 7-day central moving average smoothing. To reduce the redundancy in ecosystem variables, we identified highly positively correlated (Pearson R) groups using CAST (Cluster Affinity Search Technique) (37), with a θ = 0.5, where we measured the pairwise similarities using the Pearson correlation coefficient. After finding the highly colinear ecosystem variable clusters, we standardized (μ = 0, σ = 1) all variables and represented each such cluster using the first principal component of variables inside the cluster as a technique to represent/combine highly positively correlated variables (38, 39) and capture the maximum variance in the subject cluster. For example, the six ecosystem variables (average OD560, average OD750, DW [grams per liter], average Chloro1 fluorescence of 450/685 nm, average Green1 fluorescence of 430/685 nm, and average Cyano1 fluorescence of 383/685 nm) found in one of the ecosystem variable clusters are variables related to optical density, dry weight, and fluorescence—all sharing biological relevance to each other (see Fig. S21 in the supplemental material). Since the standardized forms of all six variables in this cluster showed similar patterns in time (i.e., have high correlation to each other), we decided to represent this cluster using their standardized first principal component, explaining 86.18% of the variance of the six ecosystem variables the cluster included.
We computed dry weight in terms of kilograms by multiplying dry weight (grams per liter) and pond volume (liter) divided by 103 and applying a 7-day central moving average smoothing. Finally, we obtained productivity (kilograms per day) by subtracting the two consecutive dry weight (kilogram) measurements in time with no harvesting in between. We also applied the same smoothing approach to our productivity variable. We chose to measure stability in terms of variability and used the standard deviation as the metric, following previous studies (40, 41). Thus, high stability is associated with low standard deviation in productivity over a window of days.
Ecosystem variables present a time series of data points, Xt. To reduce the variance in measurement, we computed statistics (mean and standard deviation) for ecosystem variables over a sliding window of length 2h + 1 days (Xt−h, …, Xt+h), centered at each time point of the sample. The choice of window size is based on a tradeoff between reduction of measurement noise versus retention of true signal, and we used a published empirical method to identify the appropriate window size (42). Specifically, we experimented with h values of 1 to 6 weeks. The noise in mean and standard deviation patterns reduced around 3 to 4 weeks and stabilized thereafter (see Fig. S22 in the supplemental material). Moreover, the difference between two distinct peaks (days 165 and 228) in the dry weight (kilogram) data (see Fig. S12 in the supplemental material) was a duration of approximately 8 weeks. Therefore, we chose windows with h = 4 weeks for our figures; however, we also report final analysis results on various window sizes (see “Relationship between algal diversity and productivity measures” in the Results section).
RESULTS
Sample dissimilarity over time.
The Bray-Curtis dissimilarities among all samples in the 16S data in Fig. 1a demonstrated two distinct time regions (days 1 to 100 and 200 to 350) in composition. Both time regions showed gradual increase in dissimilarity over time; however, samples in one of the distinct time regions were at roughly similar distances from all samples in the other. ITS2 data dissimilarities across all samples in Fig. 1b showed three main distinct regions in time (days 1 to 200, 200 to 300, and 300 to 350), with a fourth inner region (days 250 to 280). Sample compositions remained highly similar during the first 200 days (a dissimilarity of 0 to 0.2) and were highly different (dissimilarity of 0.8 to 0.9) around days 300 to 350, with an intermediary region of days 200 to 300. In both bacterial (16S) and eukaryotic (ITS2) samples, we observed that sample compositions have changed the overall compositional state and showed high/increasing dissimilarity after around day 200. This roughly corresponded to the beginning of the recovery of algal dry weight (see Fig. S12 in the supplemental material) after its sharp fall, possibly as a response to the fungicide treatment (see “Temporal bacteria and eukaryotic taxonomic profile” in the Results section for more detail).
FIG 1.
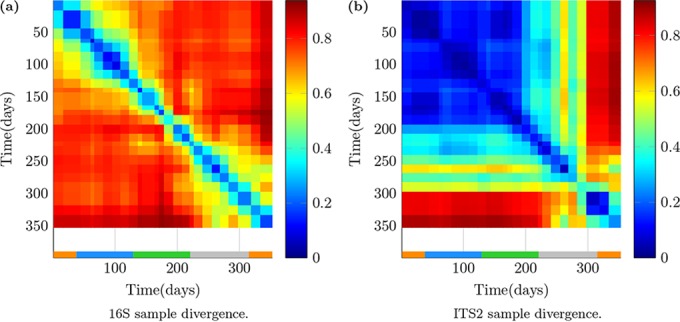
Sample dissimilarities. Panel a shows the Bray-Curtis dissimilarities among the samples between the bacterial (16S) samples, and panel b shows the dissimilarities between the eukaryotic (ITS2) samples. Seasons are denoted with a color bar atop the x axis as fall (orange), winter (blue), spring (green), and summer (silver).
Temporal bacterial and eukaryotic taxonomic profile.
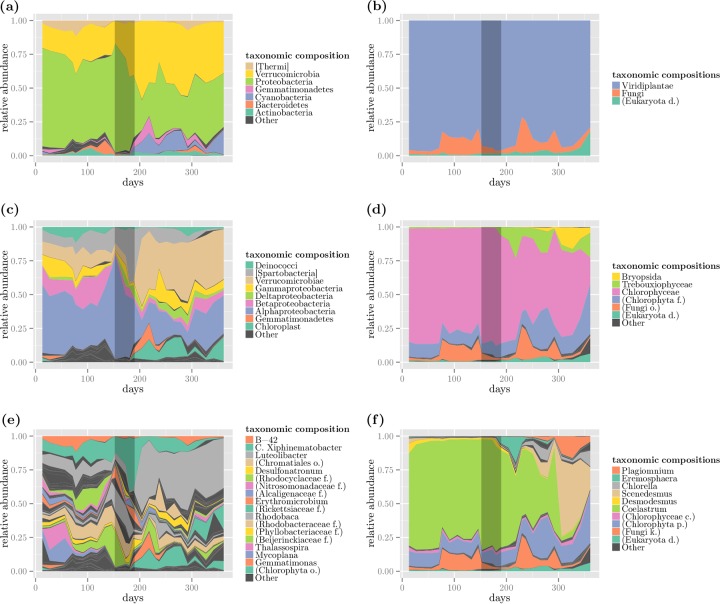
We observed the temporal taxonomic changes in the bacterial (16S) and eukaryotic (ITS2) composition of our samples using area plots for various taxonomic ranks (Fig. 2). Area plots consist of stacked relative abundances over time of taxa at different levels of taxonomic resolution. Relative abundances of less than 1% (1.3% for bacterial genera) are marked as “Other” for clarity. To examine patterns at an even finer resolution than the genus level, we also included the area plots of the top 2,000 and 200 reference sequences for 16S and ITS2 data in Fig. S13 in the supplemental material. We also placed black shading on the plots between days 152 to 190 as the duration of the four dosages of fungicide, followed by the algal dry weight recovery (day 200). We referred to the samples before day 200 as “prerecovery” and the ones after as “postrecovery.”
FIG 2.
Area plots. The plots depict the relative abundances of various taxa and are organized with increasing level of rank in their corresponding taxonomy for 16S (left-hand side) and ITS2 (right-hand side) compositions. Plots a and b represent the relative abundances at the phylum and kingdom levels, whereas plots c and d and e and f further analyze the compositions at the class and genus levels, respectively. Taxa that had no information at their respective rank are shown in parentheses using the lowest available taxonomic rank. The black shading between days 152 and 190 represents the time interval that includes the 4 time points of fungicide application.
The phylum-level 16S composition (Fig. 2a) revealed that Verrucomicrobia, Proteobacteria, and Cyanobacteria comprised the majority of the taxonomic profile. Proteobacteria decreased in the postrecovery samples, while Verrucomicrobia and Cyanobacteria increased in abundance. Analyses at the class (Fig. 2c) and genus (Fig. 2e) levels revealed that few taxa dominated the phyla present, such as the class Alphaproteobacteria in Proteobacteria and the genus Luteolibacter in Verrucomicrobia. The abundance pattern shifts in these taxa also correspond to the algal dry weight recovery, rendering Luteolibacter the most abundant genus in the postrecovery samples, which starting at day 204 reached a high of 48% and occupied 37% of the sample composition by day 350.
The eukaryotic (ITS2) taxonomy analysis at the kingdom level (Fig. 2b) shows that Viridiplantae, which mainly consist of algal species in our samples, and Fungi constitute the dominant community members across all time points. Although the class-level composition (Fig. 2d) was dominated by Chlorophyceae, the genus-level analysis (Fig. 2f) reveals striking changes in the abundance patterns of two genera: Coalestrum and Scenedesmus. The consistent dominance of Coalestrum changed in the postrecovery samples, followed by a sharp decline by day 300 to be overtaken by Scenedesmus.
The decline in the algal dry weight (kilogram) measurements (see Fig. S12 in the supplemental material) triggering the fungicide application prior to day 152 (first dosage) coincided with an increased fungal relative abundance period, whereas the time interval between days 152 and 190, where all 4 dosages have been applied, corresponds to low (lower than the overall mean) fungal relative abundances. We observed that the top five most abundant fungal sequences had high percentages of identities to Cryptomycota, Chytridiomycota, and Amoeboaphelidium sp., which were reported as algal pathogens in a previous study on a Sapphire Energy open alga pond (43). Specifically, references gi|532165669 and gi|532165968 had percentages of identity (PID) of 87%, and 89% with a Cryptomycota sp. strain. Reference gi|194354257 had 89% PID with a Chytridiomycota sp. strain (see Fig. S16 in the supplemental material for a distance tree result), whereas references gi|532165358 and gi|532166006 had 85% identity to Amoeboaphelidium sp. strain PML-2014 isolate FD01, a sequence previously reported by Letcher et al. (43) found in Sapphire Energy ponds. All hit subject sequences were the highest-scoring BLAST hits, which contained at least a phylum-level annotation, except for gi|532165358. See Fig. S15 in the supplemental material for the sequence mapping results.
Bacterial and eukaryotic diversity over time.
We measured diversity at the genus level using Hill numbers, with sensitivity parameter q = 1 after rarefaction to an equal number of subsamplings for all time samples (see Materials and Methods).
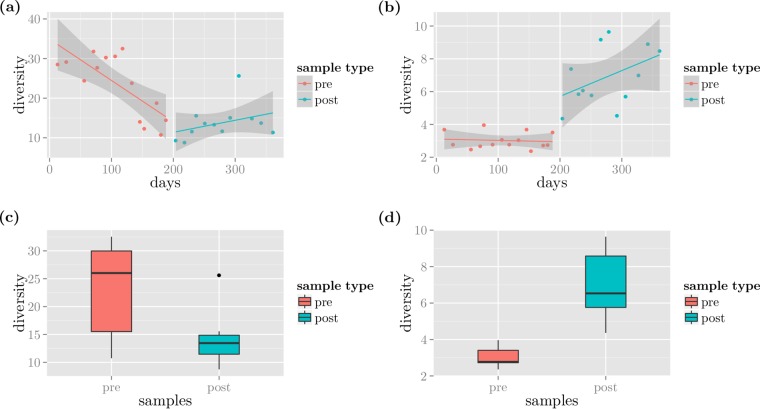
We detected a structural break in the temporal diversity trends around the algal dry weight recovery (day 200) in both data sets as shown in Fig. 3a and b. The bacterial diversity was high and decreasing in the prerecovery period, and it remained low in the postrecovery period, while the eukaryotic diversity showed the opposite trend.
FIG 3.
Diversity patterns. Panels a and b show the diversity patterns of bacterial (16S) and eukaryotic (ITS2) data, respectively, over time. Panels c (16S) and d (ITS2) show the distributions of the diversities at the two different time periods.
A Chow test revealed a significant improvement in fit was achieved by modeling the data for two subintervals rather than a regression across the entire time series available (P < 0.01 for both 16S and ITS2 data, respectively). In addition, a two-sided Wilcoxon rank sum test showed a significant difference between the median diversities in the two different periods for both bacteria and eukaryotes, where the signal was stronger (P = 3.05 × 10−3 and 2.07 × 10−7, respectively), as shown in Fig. 3c and d.
We initially observed a significant negative correlation (Pearson R = −0.56, P = < 0.01) between the bacterial and eukaryotic diversities. Controlling for temperature and fungal relative abundance (suspected algal pathogen levels and the effect of fungicide on the pathogen), however, revealed that bacterial and eukaryotic diversities had no significant explanatory value for each other (P = 0.62). We also confirmed that fungal relative abundance did not have a significant explanatory value for bacterial or eukaryotic diversity (P = 0.35 and 0.57), after controlling for temperature. We, therefore, think that the initial negative correlation between bacterial and eukaryotic diversities could be due to their different responses to temperature. See section 1.7 in the Methods in the supplemental material for controlling for confounding variables and associated model comparison.
Correlations between the pond ecosystem and taxonomic composition.
Although the pond was managed to maintain a stable environment through biomass harvesting and nutrient additions, we observed seasonal shifts in the availability of energy and nutrients. Figure S18 in the supplemental material shows seasonal patterns in temperature (an indicator of day length and light availability), the concentration of urea, and Fv/Fm (photosynthetic health). Urea availability peaked in winter (around days 100 and 400), while temperature peaked between days 200 and 300 (summer). Fv/Fm fluctuated strongly but showed apparent peaks in spring and fall (around days 150 and 350), with a decrease in summer, possibly due to the reduction in urea, similar to patterns in some natural phytoplankton communities (44). The sharp fall in Fv/Fm prior to day 200 could probably be associated with the dry weight fall (see Fig. S12 in the supplemental material).
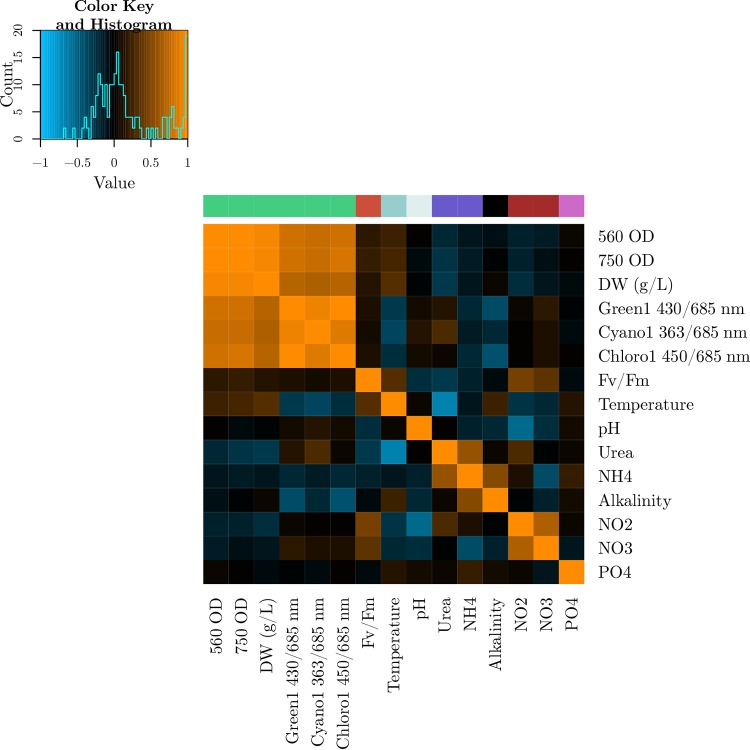
The ecosystem variables in the pond showed patterns of colinearity as well as associations with the genomic data. Figure 4 shows several variables that cluster in blocks of high correlation. We clustered the ecosystem variables using the cluster affinity search technique (CAST) (37), where pairwise similarities were measured using Pearson correlation. The 15 variables could be described by 8 independent clusters, with a θ of 0.5, which all showed the expected grouping (Table 1), including, for example, the clustering of NO2 and NO3. Figure S21 in the supplemental material displays another example of an ecosystem cluster consisting of optical density, fluorescence, and dry weight measurements, alongside their standardized first principal component. Since the first principal components of all clusters explained over 75% of their variance, as shown in Table 1, the final pond ecosystem versus taxonomic composition correlations were conducted using these first principal components.
FIG 4.
Correlation matrix of all phenotypic variables: Ecosystem variables forming a clique using the CAST algorithm are represented with a single color in the color bar, as also suggested by the orange correlation blocks in the matrix. Cell colors are based on the Pearson correlation coefficients according to the given color map.
TABLE 1.
Ecosystem clusters, associated individual ecosystem variables, and percentages of variance explained by their first principal component
| Cluster | Ecosystem variable(s) | % variance explained by 1st principal component |
|---|---|---|
| DW-fluorescence group | Avg Green1 fluorescence (430/685 nm), avg Chloro1 fluorescence (450/685 nm), avg Cyano1 fluorescence (363/685 nm), avg OD560, avg OD750, DW (g/liter) | 86.18 |
| Fv/Fm | Avg Fv/Fm | 100 |
| pH | pH probe | 100 |
| Temp | Temp probe | 100 |
| Urea-NH4 group | Urea (ppm), NH4 (ppm) | 78.64 |
| PO4 | PO4 (ppm) | 100 |
| Alkalinity | Alkalinity (ppm) | 100 |
| NO2-NO3 group | NO2 (ppm), NO3 (ppm) | 82.83 |
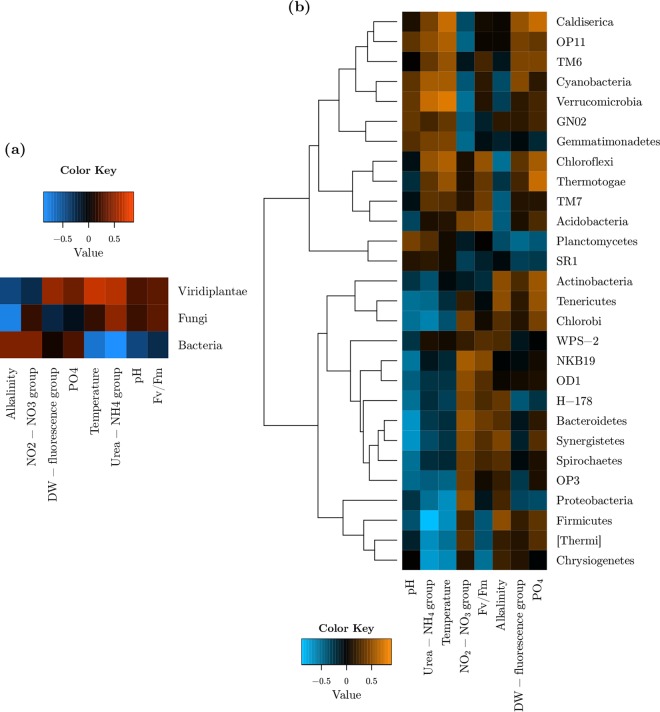
Heat maps in Fig. 5 show the Pearson correlations for kingdom diversities, and bacterial phylum relative abundances versus ecosystem clusters. Kingdom-level diversity-pond ecosystem correlation analysis (Fig. 5a) showed that Bacteria and Viridiplantae had antagonistic correlations with temperature and the urea-NH4 group. Viridiplantae, in addition, showed a positive correlation with the DW-fluorescence group as well. Diversity of Fungi, on the other hand, was positively correlated with alkalinity and urea-NH4 and negatively with DW-fluorescence.
FIG 5.
Pond ecosystem and taxonomic composition correlations. Panel a shows the correlations between ecosystem clusters and diversities at the kingdom level, whereas panel b shows bacterial phylum relative abundance correlations.
Temperature, pH, and the urea-NH4 and NO2-NO3 groups were the major ecosystem variables to show correlation with the relative abundances of bacterial phyla, as displayed in Fig. 5b. The row dendrogram also showed that there were two major clusters of bacterial relative abundance patterns at the phylum level, based on the correlations with ecosystem variables.
Relationship between algal diversity and productivity measures.
We investigated the relationship between algal diversity and the mean and standard deviation (SD) of pond productivity measurements (kilograms per day), centered at genomic sampling dates (see Materials and Methods). We removed the only non-algal genus, Plagiomnium (class Bryopsida [moss]), from the Viridiplantae composition for calculation of algal diversity. Figure 6 shows the relationship between algal diversity and pond productivity statistics. Algal diversity was positively correlated with the mean (Pearson R = 0.33, P = 1.1 × 10−1) and negatively correlated with the SD in productivity (Pearson R = −0.6, P = 1.9 × 10−3), suggesting high stability in biomass production.
FIG 6.
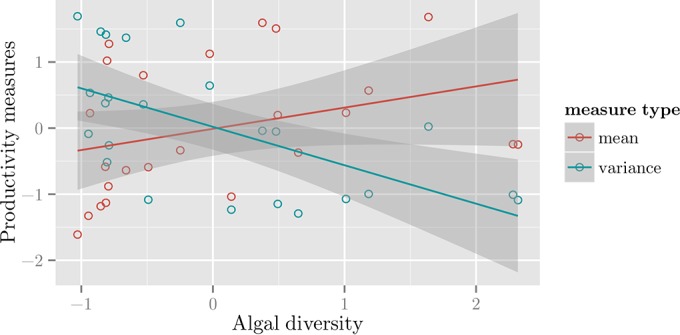
Correlations of the productivity mean and standard deviation versus algal diversity. The scatter plot shows the correlations between algal diversity versus the mean and standard deviation of productivity measurements centered around genomic sampling days for 2 h = 8 weeks (see Materials and Methods) using the regression lines. Algal diversity is positively correlated with mean productivity (Pearson R = 0.33, P = 1.1 × 10−1) and negatively correlated with standard deviation in productivity (Pearson R = −0.6, P = 1.9 × 10−3).
In order to control for temperature and fungal relative abundance (suspected algal pathogen levels and the effect of fungicide on it) as potential confounding variables, we used a model comparison using the F test to examine the explanatory value/power of algal diversity for the productivity mean and standard deviation. We conducted our analysis with various window sizes (h from 16 to 36). Our results show that algal diversity has significant explanatory value for both the productivity mean and SD (P < 0.05) for h = 22 through h = 32 (window sizes of 45 to 65 days) and the productivity SD for h = 34 and h = 36 as well, as Table 2 indicates. Temperature, however, did not have a significant explanatory value (when controlled for algal diversity and fungal relative abundance) for any of the window sizes experimented with (Table 3). Although the explanatory values of temperature for h = 24 through h = 28 had P < 0.1 for the productivity mean, they had P > 0.3 for the productivity SD in all window sizes. Since other ecosystem variables (such as urea, NH4, or PO4) were highly affected by the maintaining of the nutrient supply, unlike temperature, we refrained from adding them into a predictive model.
TABLE 2.
Explanatory values of algal diversity for algal productivity statistics, controlling for temperature and fungal relative abundancea
| h value | Productivity |
|
|---|---|---|
| Mean | SD | |
| 16 | 8.2e−02 | 9.9e−02 |
| 18 | 8.4e−02 | 7.6e−02 |
| 20 | 7.3e−02 | 5.4e−02 |
| 22 | 4.3e−02 | 4.8e−02 |
| 24 | 2.3e−02 | 4.6e−02 |
| 26 | 1.5e−02 | 4.2e−02 |
| 28 | 1.6e−02 | 1.5e−02 |
| 30 | 2.3e−02 | 1.3e−02 |
| 32 | 3.9e−02 | 1.2e−02 |
| 34 | 6.9e−02 | 9.1e−03 |
| 36 | 1e−01 | 5.4e−03 |
Results are shown for production across various h values (half window size) given as column numbers. Statistically significant explanatory values are shown in boldface.
TABLE 3.
Explanatory values of temperature for algal productivity statistics, controlling for algal diversity and fungal relative abundancea
| h value | Productivity |
|
|---|---|---|
| Mean | SD | |
| 16 | 1.1e−01 | 5.8e−01 |
| 18 | 1.3e−01 | 5.2e−01 |
| 20 | 1.4e−01 | 4.2e−01 |
| 22 | 1.2e−01 | 3.9e−01 |
| 24 | 9.3e−02 | 4.5e−01 |
| 26 | 7.8e−02 | 5.6e−01 |
| 28 | 8.5e−02 | 8.4e−01 |
| 30 | 1.3e−01 | 8.6e−01 |
| 32 | 2.1e−01 | 8.6e−01 |
| 34 | 3.4e−01 | 7.5e−01 |
| 36 | 4.6e−01 | 6.2e−01 |
Results are shown as production across various h values (half window size) given as column numbers.
DISCUSSION
Open alga ponds as an agricultural platform have the potential to revolutionize the production of low-cost biomass for food, fuel, and specialty chemicals if their productivity can be optimized and their stability maintained. Research into this effort has generated progress in terms of the scale, productivity, and stability of these ponds; however, substantive challenges remain. A novel and potentially transformative solution is to switch from the traditional agricultural paradigm of monocultures to one that deploys multiple strains (polycultures). The first step in this process is to understand if the benefits that have been ascribed to increased diversity in natural systems also occur in open ponds, which are very distinct from most natural systems as algae typically are maintained at a high density and not limited by any resource except for light. In this study, we observed the relationships between algal diversity and both algal productivity and the standard deviation of productivity in an open alga pond managed to maintain productivity but open to colonization from aerial sources of microbes. We found a positive relationship between productivity and algal diversity and a negative relationship between the standard deviation in productivity and algal diversity, suggesting that research into how to construct and manage consortia for deployment in open ponds may be an effective tool for pond management, as indicated by studies of natural and experimental systems (14, 16, 22, 23).
Our study reveals that managed open alga ponds for the production of biomass energy sustain a diversity of microbial life and a dynamic variability. The most common bacteria phyla observed in our study included the Proteobacteria, Verrucomicrobia, and Cyanobacteria, the same groups that dominate natural aquatic assemblages (45). Interestingly, the most abundant genus during the high and stable algal biomass yield period, Luteolibacter, under Verrucomicrobia, contains species that utilize algal metabolites as carbon and nutrient sources, such as Luteolibacter yonseiensis and Luteolibacter algae (46, 47). Community composition also showed seasonal shifts comparable to natural assemblages (48), even though the environment was managed to achieve relative homeostasis. Our results indicate that diversity and dynamic variability are unavoidable features of open alga ponds that should be incorporated as part of their design and management.
Kingdom-level eukaryotic taxonomic composition analysis (Fig. 2b) revealed three time intervals (days 77 to 146, days 230 to 251, and around day 292) with continuous high (higher than overall mean) fungal relative abundance, with a decrease between the first and second intervals. This decreased fungal relative abundance period (days 147 to 229) encompassed the four fungicide application time points (days 152, 168, 177, and 190). Although we observed a dry weight fall soon after the first high fungal relative abundance time interval, we did not see a similar fall in biomass during or after the other two intervals. We would like to note, however, that the algal community compositions were different across the intervals. While the first time interval coincided with low algal diversity, a more diverse algal community was observed in the other two time intervals. Indeed, Smith et al. (49) and Shurin et al. (14) discuss the possibility of crop protection against disease/predation through the use of mixed-species communities. Research also shows increased associational resistance against consumers in prey alga assemblages (50) due to various possible mechanisms (51). Although our observation supports the cited findings, control experiments would be required to deduce concrete conclusions.
Disentangling the causal association between diversity and productivity is complicated as diversity can be either a driving factor or a consequence of variation in productivity (52). A positive association between pond biomass productivity and diversity of eukaryotes may reflect several underlying processes. First, a more diverse algal community may acquire abiotic resources such as different mineral nutrients (23, 53) or wavelengths of light (54) more efficiently due to niche partitioning among species. Sampling effects of random selection of high-productivity species may occur in assembled communities. Finally, the supply of resources may determine diversity, with a loss of species during pulses of high resource supply (55). However, nutrients were supplied to our community at a constant high level throughout the course of the study, and biomass was maintained by harvesting. Alternatively diversity may not be the ultimate cause of high productivity or stability but rather may be an associated variable, for unknown reasons. However, our results agree with studies of natural systems showing positive associations between ecosystem productivity and stability and the diversity of the phytoplankton community (23, 56).
Our results showed that algal diversity had significant explanatory value for the productivity mean and standard deviation, after controlling for temperature and fungal relative abundance (and the effect of fungicide on it). We acknowledge that the effect of algal diversity on productivity and stability could be confounded by temperature and the usage of fungicide. Although controlling for temperature is simple, we believe that controlling for the possible confounding effect of fungicide is harder because it is a merely 4-time-point application. Therefore, we chose to use fungal relative abundance as an extra covariate, given the microalga toxicity values shown in the patent application by McBride et al. (7.5 and 15 ppm), which were higher than the doses used (1 ppm) (McBride et al., U.S. patent application 14,351,540).
Our observations indicate that fungal pathogens may place strong limitations on the productivity and composition of algal biofuel assemblages. These results agree well with data from other algal bioenergy studies (14, 57) and natural freshwater ecosystems (58). Fungal pathogens have been shown to be important in terminating blooms of diatoms (59, 60); however, their role in maintaining productivity is not well known. Our results indicate that fungi may impose top-down control of productivity similar in magnitude to that of mesozooplankton grazers like crustaceans and may therefore shape algal community composition.
Associations between diversity and ecosystem function varied among kingdoms. While we observed a negative correlation between temperature and bacterial diversity, eukaryotic (mostly green algae) diversity showed a positive correlation with temperature. Indeed, Stomp et al. suggest a positive association between temperature and phytoplankton richness (61). It has also been reported that many green alga genera we observed in our samples and Cyanobacteria have optima at higher temperatures, which correspond to the higher spring/summer temperatures at our research site (62). The bacterial phylum Verrucomicrobia has been shown to be positively correlated with temperature (63) and to include genera (e.g., Luteolibacter) that have potential associations with Cyanobacteria (48). Our data show increased Luteolibacter relative abundance during periods of increased Cyanobacteria relative abundance and temperature (post-day 200 in Fig. 2e), which have led to the decrease in overall bacterial diversity during higher-temperature periods particularly due to the dominance caused by the single genus Luteolibacter.
The negative correlation we observed between diversity of phytoplankton and bacteria over time provides some indications of the nature of the eukaryotic and bacteria components of the ecosystem. Producers and microbes engage in a range of pathogenic and mutualistic interactions that may drive positive or negative feedback in diversity between the two groups (64). Phytoplankton and bacterial communities show synchronous dynamics in nature, indicating that bacterial taxa are engaged in specific interactions with phytoplankton taxa (65, 66). Our data indicate that conditions favoring high phytoplankton diversity and productivity are accompanied by low bacterial diversity. The causal basis for this association is unknown; however, the correlation could be explained by an opposite response to temperature, since bacterial diversity had no explanatory power for eukaryotic diversity, after controlling for temperature. As discussed previously, the relative abundance increases in Luteolibacter and Cyanobacteria during higher temperatures, patterns also observed by Litchman et al. (62) and Woodhouse et al. (48), could have been the main reasons for diversity loss in bacteria at higher temperatures. Alongside rising temperature, continuous invasion by airborne propagules of microalgae during a high-light-availability period could be another possible reason for increased eukaryotic diversity in the post-algal dry weight recovery period (67). The data therefore give no indication of a causal association between diversity of prokaryotes and eukaryotes.
Managing consortia using traditional tools such as pesticide application could be challenging for consortium stability. The data we collected showed a dramatic impact of pesticide (fungicide) application on the fungal relative abundance and the recovery of algal dry weight. As mentioned earlier, our data do not allow us to discriminate among several possible causal relationships for this pattern. That said, the fungicide application may have reduced the fitness of the target algae and provided an opportunity for other competing green alga species to begin to enter, thus increasing diversity. Other traditional management tools for open alga ponds may similarly impact consortia in unintended ways. For example, some ponds are harvested using dissolved air flotation (DAF) technology, which is commonly used in wastewater treatment. This technology relies on the deployment of a polymer that binds to and aggregates algae based on the surface charge of that algae. The aggregates are then floated to the surface of a DAF tank and skimmed off for further concentration. Without accounting for differential selectivity of this approach on a consortium of algae, harvesting using this strategy would undoubtedly also impact the makeup and stability of a deployed consortium.
Our results indicate that ecological principles relating ecosystem productivity to community diversity are applicable to industrial ecosystems for the cultivation of photosynthetic microbes. Intensifying biomass yield and fostering resilience against the vagaries of the environment or contaminating organisms are keys to commercializing the industrial growth of microbial products (14, 15, 67, 68). Most research efforts in this area involve understanding the genetic basis for phenotypic traits related to production of specific compounds (69). Ecological engineering for productivity and stability has been proposed and discussed (22) but never demonstrated beyond the laboratory scale. Many ecological processes are highly scale and context dependent (70); therefore, principles demonstrated in tightly controlled laboratory studies must be validated on a whole-system scale under natural regimens of environmental variation in order to ascertain their applicability. Our study indicates that managing microbial polycultures for productivity and stability may form the basis of a viable industrial practice to advance the commercial potential of phytoplankton for bioenergy or other higher-value products.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kalli Lambeth for collecting samples, the Sapphire Energy, Inc., Las Cruces, production team for collecting data on ecosystem variables, and Shibu Yooseph for insightful comments.
This research was supported by NSF via grants DBI-1458557 and IIS-1318386 for Doruk Beyter and Vineet Bafna. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
V.B. is a partner in Digital Proteomics, LLC, which licenses and sells MS-related software. Digital Proteomics was not involved in any aspect of this research. D.B., R.M., J.B.S., S.M., T.C.P., F.H., Y.W.L., and V.B. were involved in designing the study, as well as providing methods and materials. D.B., R.M., T.H., P.T., S.B., F.H., and D.B. performed the research. D.B., R.M., J.B.S., and V.B. wrote the manuscript.
Three of the authors of this paper are employees of Sapphire Energy, Inc., a for-profit company, the goal of which is to commercialize the production of a renewable source of energy using algae as a production platform. V.B. holds an equity position in Digital Proteomics, LLC, a company that licenses and sells UCSD software for proteomic analysis. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03965-15.
REFERENCES
- 1.Sheehan J, Dunahay T, Benemann J, Roessler P. 1998. A look back at the US Department of Energy's aquatic species program: biodiesel from algae, vol 328 National Renewable Energy Laboratory, Golden, CO. [Google Scholar]
- 2.Waltz E. 2009. Biotech's green gold? Nat Biotechnol 27:15–18. doi: 10.1038/nbt0109-15. [DOI] [PubMed] [Google Scholar]
- 3.Chaumont D. 1993. Biotechnology of algal biomass production: a review of systems for outdoor mass culture. J Appl Phycol 5:593–604. doi: 10.1007/BF02184638. [DOI] [Google Scholar]
- 4.Richmond A. 2004. Biological principles of mass cultivation, p 125–177. In Richmond A. (ed), Handbook of microalgal culture: biotechnology and applied phycology. Blackwell Publishing, Ltd, Oxford, United Kingdom. [Google Scholar]
- 5.Tredici MR. 2004. Mass production of microalgae: photobioreactors, p 178–214. In Richmond A. (ed), Handbook of microalgal culture: biotechnology and applied phycology. Blackwell Publishing, Ltd, Oxford, United Kingdom. [Google Scholar]
- 6.Schenk PM, Thomas-Hall SR, Stephens E, Marx UC, Mussgnug JH, Posten C, Kruse O, Hankamer B. 2008. Second generation biofuels: high-efficiency microalgae for biodiesel production. Bioenergy Res 1:20–43. doi: 10.1007/s12155-008-9008-8. [DOI] [Google Scholar]
- 7.Shimamatsu H. 2004. Mass production of spirulina, an edible microalga, p 39–44. In Ang PO., Jr (ed) Asian Pacific phycology in the 21st century: prospects and challenges. Proceeding of the Second Asian Pacific Phycological Forum, Hong Kong, China, 21 to 25 June 1999 Springer, Berlin, Germany. [Google Scholar]
- 8.Lee Y-K. 2001. Microalgal mass culture systems and methods: their limitation and potential. J Appl Phycol 13:307–315. doi: 10.1023/A:1017560006941. [DOI] [Google Scholar]
- 9.Borowitzka MA, Moheimani NR. 2013. Algae for biofuels and energy, p 133–152. Springer, Berlin, Germany. [Google Scholar]
- 10.Rodolfi L, Chini Zittelli G, Bassi N, Padovani G, Biondi N, Bonini G, Tredici MR. 2009. Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol Bioeng 102:100–112. doi: 10.1002/bit.22033. [DOI] [PubMed] [Google Scholar]
- 11.Reference deleted.
- 12.Zmora O, Richmond A. 2013. Microalgae for aquaculture: microalgae production for aquaculture, p 365–379. Richmond A, Hu Q (ed) Handbook of microalgal culture: biotechnology and applied phycology, 2nd ed Blackwell Publishing, Ltd, Oxford, United Kingdom. [Google Scholar]
- 13.Lincoln E, Hall T, Koopman B. 1983. Zooplankton control in mass algal cultures. Aquaculture 32:331–337. doi: 10.1016/0044-8486(83)90230-2. [DOI] [Google Scholar]
- 14.Shurin JB, Abbott RL, Deal MS, Kwan GT, Litchman E, McBride RC, Mandal S, Smith VH. 2013. Industrial-strength ecology: trade-offs and opportunities in algal biofuel production. Ecol Lett 16:1393–1404. doi: 10.1111/ele.12176. [DOI] [PubMed] [Google Scholar]
- 15.Kazamia E, Aldridge DC, Smith AG. 2012. Synthetic ecology—a way forward for sustainable algal biofuel production? J Biotechnol 162:163–169. doi: 10.1016/j.jbiotec.2012.03.022. [DOI] [Google Scholar]
- 16.Shurin JB, Mandal S, Abbott RL. 2014. Trait diversity enhances yield in algal biofuel assemblages. J Appl Ecol 51:603–611. doi: 10.1111/1365-2664.12242. [DOI] [Google Scholar]
- 17.Litchman E, Klausmeier CA. 2008. Trait-based community ecology of phytoplankton. Annu Rev Ecol Evol Syst 39:615–639. doi: 10.1146/annurev.ecolsys.39.110707.173549. [DOI] [Google Scholar]
- 18.Chisholm SW. 1992. Phytoplankton size, p 213–237. In Falkowski PG, Woodhead AD, Vivirito K (ed), Primary productivity and biogeochemical cycles in the sea. Springer, Berlin, Germany. [Google Scholar]
- 19.Litchman E, Klausmeier CA, Schofield OM, Falkowski PG. 2007. The role of functional traits and trade-offs in structuring phytoplankton communities: scaling from cellular to ecosystem level. Ecol Lett 10:1170–1181. doi: 10.1111/j.1461-0248.2007.01117.x. [DOI] [PubMed] [Google Scholar]
- 20.Edwards KF, Klausmeier CA, Litchman E. 2011. Evidence for a three-way trade-off between nitrogen and phosphorus competitive abilities and cell size in phytoplankton. Ecology 92:2085–2095. doi: 10.1890/11-0395.1. [DOI] [PubMed] [Google Scholar]
- 21.Cardinale BJ, Srivastava DS, Duffy JE, Wright JP, Downing AL, Sankaran M, Jouseau C. 2006. Effects of biodiversity on the functioning of trophic groups and ecosystems. Nature 443:989–992. doi: 10.1038/nature05202. [DOI] [PubMed] [Google Scholar]
- 22.Stockenreiter M, Graber A-K, Haupt F, Stibor H. 2012. The effect of species diversity on lipid production by micro-algal communities. J Appl Phycol 24:45–54. doi: 10.1007/s10811-010-9644-1. [DOI] [Google Scholar]
- 23.Ptacnik R, Solimini AG, Andersen T, Tamminen T, Brettum P, Lepistö L, Willen E, Rekolainen S. 2008. Diversity predicts stability and resource use efficiency in natural phytoplankton communities. Proc Natl Acad Sci USA 105:5134–5138. doi: 10.1073/pnas.0708328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu C, Lee Y. 1997. Determination of biomass dry weight of marine microalgae. J Appl Phycol 9:189–194. doi: 10.1023/A:1007914806640. [DOI] [Google Scholar]
- 25.Cuthbertson L, Rogers GB, Walker AW, Oliver A, Hafiz T, Hoffman LR, Carroll MP, Parkhill J, Bruce KD, van der Gast CJ. 2014. Time between collection and storage significantly influences bacterial sequence composition in sputum samples from cystic fibrosis respiratory infections. J Clin Microbiol 52:3011–3016. doi: 10.1128/JCM.00764-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grattepanche J-D, Santoferrara LF, McManus GB, Katz LA. 2014. Diversity of diversity: conceptual and methodological differences in biodiversity estimates of eukaryotic microbes as compared to bacteria. Trends Microbiol 22:432–437. doi: 10.1016/j.tim.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 27.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah M, Homer N, Lyons M. 2015. Torrent Mapping Alignment Program. Thermo Fisher Scientific, Waltham, MA: https://github.com/iontorrent/TMAP Accessed 1 April 2015. [Google Scholar]
- 29.Shah M, Homer N, Lyons M. 2015. TMAP technical note. Thermo Fisher Scientific, Waltham, MA: https://github.com/iontorrent/TMAP/blob/master/doc/tmap-book.pdf Accessed 1 April 2015. [Google Scholar]
- 30.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill MO. 1973. Diversity and evenness: a unifying notation and its consequences. Ecology 54:427–432. doi: 10.2307/1934352. [DOI] [Google Scholar]
- 32.Jost L. 2006. Entropy and diversity. Oikos 113:363–375. doi: 10.1111/j.2006.0030-1299.14714.x. [DOI] [Google Scholar]
- 33.Chao A, Chiu C-H, Jost L. 2010. Phylogenetic diversity measures based on hill numbers. Philos Trans R Soc B Biol Sci 365:3599–3609. doi: 10.1098/rstb.2010.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leinster T, Cobbold CA. 2012. Measuring diversity: the importance of species similarity. Ecology 93:477–489. doi: 10.1890/10-2402.1. [DOI] [PubMed] [Google Scholar]
- 35.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2013. vegan: community ecology package. R package version 2.0-10. R Foundation for Statistical Computing, Vienna, Austria: http://CRAN.R-project.org/package=vegan. [Google Scholar]
- 36.van Buuren S, Groothuis-Oudshoorn K. 2011. mice: multivariate imputation by chained equations in R. J Stat Software 45:1–67. http://www.jstatsoft.org/v45/i03/. [Google Scholar]
- 37.Ben-Dor A, Shamir R, Yakhini Z. 1999. Clustering gene expression patterns. J Comput Biol 6:281–297. doi: 10.1089/106652799318274. [DOI] [PubMed] [Google Scholar]
- 38.Freckleton RP. 2011. Dealing with collinearity in behavioural and ecological data: model averaging and the problems of measurement error. Behav Ecol Sociobiol 65:91–101. doi: 10.1007/s00265-010-1045-6. [DOI] [Google Scholar]
- 39.Grueber C, Nakagawa S, Laws R, Jamieson I. 2011. Multimodel inference in ecology and evolution: challenges and solutions. J Evol Biol 24:699–711. doi: 10.1111/j.1420-9101.2010.02210.x. [DOI] [PubMed] [Google Scholar]
- 40.De Keersmaecker W, Lhermitte S, Honnay O, Farifteh J, Somers B, Coppin P. 2014. How to measure ecosystem stability? An evaluation of the reliability of stability metrics based on remote sensing time series across the major global ecosystems. Global Change Biol 20:2149–2161. doi: 10.1111/gcb.12495. [DOI] [PubMed] [Google Scholar]
- 41.Pimm SL. 1984. The complexity and stability of ecosystems. Nature 307:321–326. doi: 10.1038/307321a0. [DOI] [Google Scholar]
- 42.Carpenter SR, Cole JJ, Pace ML, Batt R, Brock W, Cline T, Coloso J, Hodgson JR, Kitchell JF, Seekell DA, Smith L, Weidel B. 2011. Early warnings of regime shifts: a whole-ecosystem experiment. Science 332:1079–1082. doi: 10.1126/science.1203672. [DOI] [PubMed] [Google Scholar]
- 43.Letcher PM, Lopez S, Schmieder R, Lee PA, Behnke C, Powell MJ, McBride RC. 2013. Characterization of Amoeboaphelidium protococcarum, an algal parasite new to the cryptomycota isolated from an outdoor algal pond used for the production of biofuel. PLoS One 8:e56232. doi: 10.1371/journal.pone.0056232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edwards KF, Litchman E, Klausmeier CA. 2013. Functional traits explain phytoplankton community structure and seasonal dynamics in a marine ecosystem. Ecol Lett 16:56–63. doi: 10.1111/ele.12012. [DOI] [PubMed] [Google Scholar]
- 45.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. 2011. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J, Baek GS, Woo S-G, Lee J, Yang J, Lee J. 2013. Luteolibacter yonseiensis sp. nov., isolated from activated sludge using algal metabolites. Int J Syst Evol Microbiol 63:1891–1895. doi: 10.1099/ijs.0.046664-0. [DOI] [PubMed] [Google Scholar]
- 47.Yoon J, Matsuo Y, Adachi K, Nozawa M, Matsuda S, Kasai H, Yokota A. 2008. Description of Persicirhabdus sediminis gen. nov., sp. nov., Roseibacillus ishigakijimensis gen. nov., sp. nov., Roseibacillus ponti sp. nov., Roseibacillus persicicus sp. nov., Luteolibacter pohnpeiensis gen. nov., sp. nov. and Luteolibacter algae sp. nov., six marine members of the phylum Verrucomicrobia, and emended descriptions of the class Verrucomicrobiae, the order Verrucomicrobiales and the family Verrucomicrobiaceae. Int J Syst Evol Microbiol 58:998–1007. doi: 10.1099/ijs.0.65520-0. [DOI] [PubMed] [Google Scholar]
- 48.Woodhouse JN, Kinsela AS, Collins RN, Bowling LC, Honeyman GL, Holliday JK, Neilan BA. 4 December 2015. Microbial communities reflect temporal changes in cyanobacterial composition in a shallow ephemeral freshwater lake. ISME J doi: 10.1038/ismej.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith VH, Crews T. 2014. Applying ecological principles of crop cultivation in large-scale algal biomass production. Algal Res 4:23–34. doi: 10.1016/j.algal.2013.11.005. [DOI] [Google Scholar]
- 50.Hillebrand H, Cardinale BJ. 2004. Consumer effects decline with prey diversity. Ecol Lett 7:192–201. doi: 10.1111/j.1461-0248.2004.00570.x. [DOI] [Google Scholar]
- 51.Duffy JE. 2002. Biodiversity and ecosystem function: the consumer connection. Oikos 99:201–219. doi: 10.1034/j.1600-0706.2002.990201.x. [DOI] [Google Scholar]
- 52.Cardinale BJ, Hillebrand H, Harpole W, Gross K, Ptacnik R. 2009. Separating the influence of resource availability from resource imbalance on productivity—diversity relationships. Ecol Lett 12:475–487. doi: 10.1111/j.1461-0248.2009.01317.x. [DOI] [PubMed] [Google Scholar]
- 53.Tilman D. 1981. Tests of resource competition theory using four species of Lake Michigan algae. Ecology 62:802–815. doi: 10.2307/1937747. [DOI] [Google Scholar]
- 54.Stomp M, Huisman J, de Jongh F, Veraart AJ, Gerla D, Rijkeboer M, Ibelings BW, Wollenzien UI, Stal LJ. 2004. Adaptive divergence in pigment composition promotes phytoplankton biodiversity. Nature 432:104–107. doi: 10.1038/nature03044. [DOI] [PubMed] [Google Scholar]
- 55.Irigoien X, Huisman J, Harris RP. 2004. Global biodiversity patterns of marine phytoplankton and zooplankton. Nature 429:863–867. doi: 10.1038/nature02593. [DOI] [PubMed] [Google Scholar]
- 56.Zimmerman EK, Cardinale BJ. 2014. Is the relationship between algal diversity and biomass in North American lakes consistent with biodiversity experiments? Oikos 123:267–278. doi: 10.1111/j.1600-0706.2013.00777.x. [DOI] [Google Scholar]
- 57.Carney LT, Lane TW. 2014. Parasites in algae mass culture. Front Microbiol 5:278. doi: 10.3389/fmicb.2014.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kagami M, de Bruin A, Ibelings BW, Van Donk E. 2007. Parasitic chytrids: their effects on phytoplankton communities and food-web dynamics. Hydrobiologia 578:113–129. doi: 10.1007/s10750-006-0438-z. [DOI] [Google Scholar]
- 59.Ibelings BW, De Bruin A, Kagami M, Rijkeboer M, Brehm M, Donk EV. 2004. Host parasite interactions between freshwater phytoplankton and chytrid fungi (chytridiomycota) 1. J Phycol 40:437–453. doi: 10.1111/j.1529-8817.2004.03117.x. [DOI] [Google Scholar]
- 60.Gsell AS, de Senerpont Domis LN, Naus-Wiezer SMH, Helmsing NR, Van Donk E, Ibelings BW. 2013. Spatiotemporal variation in the distribution of chytrid parasites in diatom host populations. Freshw Biol 58:523–537. doi: 10.1111/j.1365-2427.2012.02786.x. [DOI] [Google Scholar]
- 61.Stomp M, Huisman J, Mittelbach GG, Litchman E, Klausmeier CA. 2011. Large-scale biodiversity patterns in freshwater phytoplankton. Ecology 92:2096–2107. doi: 10.1890/10-1023.1. [DOI] [PubMed] [Google Scholar]
- 62.Litchman E, de Tezanos Pinto P, Klausmeier CA, Thomas MK, Yoshiyama K. 2010. Linking traits to species diversity and community structure in phytoplankton. Hydrobiologia 653:15–28. doi: 10.1007/s10750-010-0341-5. [DOI] [Google Scholar]
- 63.Lindström ES, Kamst-Van Agterveld MP, Zwart G. 2005. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl Environ Microbiol 71:8201–8206. doi: 10.1128/AEM.71.12.8201-8206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bever JD, Westover KM, Antonovics J. 1997. Incorporating the soil community into plant population dynamics: the utility of the feedback approach. J Ecol 85:561–573. doi: 10.2307/2960528. [DOI] [Google Scholar]
- 65.Rooney-Varga JN, Giewat MW, Savin MC, Sood S, LeGresley M, Martin J. 2005. Links between phytoplankton and bacterial community dynamics in a coastal marine environment. Microb Ecol 49:163–175. doi: 10.1007/s00248-003-1057-0. [DOI] [PubMed] [Google Scholar]
- 66.Kent AD, Yannarell AC, Rusak JA, Triplett EW, McMahon KD. 2007. Synchrony in aquatic microbial community dynamics. ISME J 1:38–47. doi: 10.1038/ismej.2007.6. [DOI] [PubMed] [Google Scholar]
- 67.Smith VH, Sturm BS, Denoyelles FJ, Billings SA. 2010. The ecology of algal biodiesel production. Trends Ecol Evol 25:301–309. doi: 10.1016/j.tree.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 68.Clarens AF, Resurreccion EP, White MA, Colosi LM. 2010. Environmental life cycle comparison of algae to other bioenergy feedstocks. Environ Sci Technol 44:1813–1819. doi: 10.1021/es902838n. [DOI] [PubMed] [Google Scholar]
- 69.Georgianna DR, Mayfield SP. 2012. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488:329–335. doi: 10.1038/nature11479. [DOI] [PubMed] [Google Scholar]
- 70.Carpenter SR. 1996. Microcosm experiments have limited relevance for community and ecosystem ecology. Ecology 77:677–680. doi: 10.2307/2265490. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.