Abstract
Cronobacter sakazakii is a foodborne pathogen associated with rare but often lethal infections in neonates. Powdered infant formula (PIF) represents the most frequent source of infection. Out of the identified serotypes (O1 to O7), O1, O2, and O3 are often isolated from clinical and PIF samples. Serotype-specific monoclonal antibodies (MAbs) suitable for application in enzyme immunoassays (EIAs) for the rapid detection of C. sakazakii have not yet been developed. In this study, we created specific MAbs with the ability to bind to C. sakazakii of serotypes O1, O2, and O3. Characterization by indirect EIAs, immunofluorescence, motility assays, and immunoblotting identified lipopolysaccharide (LPS) and exopolysaccharide (EPS) as the antigenic determinants of the MAbs. The established sandwich EIAs were highly sensitive and were able to detect between 2 × 103 and 9 × 106 CFU/ml. Inclusivity tests confirmed that 93% of serotype O1 strains, 100% of O2 strains, and 87% of O3 strains were detected at low cell counts. No cross-reactivity with >100 strains of Cronobacter spp. and other Enterobacteriaceae was observed, except for that with C. sakazakii serotype O3 and Cronobacter muytjensii serotype O1. Moreover, the sandwich EIAs detected C. sakazakii in PIF samples artificially contaminated with 1 to 10 bacterial cells per 10 g of sample after 15 h of preenrichment. The use of these serotype-specific MAbs not only allows the reliable detection of C. sakazakii strains but also enables simultaneous serotyping in a simple sandwich EIA method.
INTRODUCTION
Cronobacter spp. are Gram-negative opportunistic foodborne pathogens of the family Enterobacteriaceae that cause rare but severe infections in patients of all age groups. In adults, Cronobacter spp. are often associated with nosocomial infections, including pneumonia, septicemia, wound infections, and osteomyelitis, while causing invasive disease in young infants and neonates (1–4). Among the seven identified Cronobacter species, C. sakazakii, C. malonaticus, C. muytjensii, C. turicensis, C. dublinensis, C. condimenti, and C. universalis (5–8), C. sakazakii plays a prominent role due to it causing life-threatening infections in neonates (9–11). Clinically manifested infections present as necrotizing enterocolitis, sepsis, and meningitis, with a mortality rate as high as 80% (1, 12, 13). Although C. sakazakii has been isolated from a variety of different plant- and animal-based food products (14, 15), the presence in powdered infant formula (PIF) seems crucial in the infection of neonates (9, 12, 16). According to an established O-antigen serotyping scheme based on rabbit antisera and a PCR-based serotyping method (17–21), seven serotypes (O1 to O7) have been identified for C. sakazakii. Serotypes O1 and O2 seem to be most prevalent in PIF samples and in clinical cases, whereas serotype O3 has been isolated quite frequently from PIF but not as often from clinical cases (19, 22–24).
Today, the contamination of PIF by C. sakazakii is being detected using conventional microbiological methods. Optimized procedures for the isolation and identification of Cronobacter spp. (25) have been published by the International Standards Organization (ISO) and the International Dairy Federation (IDF). However, these methods are very laborious, and the isolation and identification of C. sakazakii can take up to 6 days. In addition, a rapid detection method combining real-time PCR, chromogenic agars, and biochemical tests has been published and is recommended by the U.S. FDA (Food and Drug Administration) (26). All reference detection methods are based on the identification of presumptive colonies with characteristic pigmentation. These criteria have been shown to be unreliable, since several pathogens of other genera grow as presumptive Cronobacter colonies, whereas some Cronobacter species isolates fail to grow on chromogenic agar (Enterobacter sakazakii isolation agar [ESIA] or chromogenic Cronobacter isolation agar [CCI]) or do not exhibit yellow colony pigmentation on tryptic soy agar (TSA) (27). In light of the need for a reliable and inexpensive rapid detection method, several PCR-based protocols for the identification of C. sakazakii at the genus, species, and serotype levels have been established (28). Despite their rapidity, some molecular methods lack specificity, depending on the chosen primers (22, 29). The recent reassignment of C. sakazakii serotypes O5 and O6 to C. malonaticus highlights the unreliability of PCR-based serotyping for the detection of all sequence-based variations of the O antigen of C. sakazakii (21, 22, 30). Additionally, PCR methods may be not convenient for smaller laboratories, since they require expensive equipment and highly trained employees.
Therefore, various attempts to detect Cronobacter spp. based on immunochemical methods have been made, including an indirect enzyme immunoassay (EIA) using monoclonal antibodies (MAbs) and sandwich EIAs using polyclonal rabbit or chicken antibodies. These assays allow the detection of Cronobacter spp. or C. sakazakii, regardless of the serotype (31–33).
Due to the lack of availability of high-affinity MAbs, until now, there has been no rapid detection method available for the individual serotypes of C. sakazakii (29). The objective of this study was to develop highly sensitive MAbs that are reactive with C. sakazakii serotypes O1, O2, and O3 in order to establish sandwich EIAs for the specific detection and identification of these serotypes. Lipopolysaccharide (LPS), as the most varied and abundant (70% of the outer membrane) component of the bacterial surface (34, 35), is highly immunogenic and is the best target for the development of specific antibodies. For this reason, in contrast to previous methods, not whole-cell preparations but cell-free LPS preparations were used for the immunization of mice. In order to evaluate the specificities of the produced MAbs, a large selection of Cronobacter spp. and other Enterobacteriaceae was screened in indirect EIAs. The ability of the established sandwich EIAs to specifically detect C. sakazakii was confirmed by directly analyzing artificially contaminated PIF samples after enrichment in buffered peptone water. These new assays show great promise for replacing time-consuming culturing methods and represent the first step toward the establishment of a monoclonal antibody-based C. sakazakii serotyping scheme.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. In general, all strains were cultivated in Luria-Bertani (LB) medium at 37°C, with constant agitation. For solid medium, 15 g/liter agar was added. For use in the indirect enzyme immunoassay (EIA) as a coating antigen, bacteria were harvested from 50 ml of overnight culture by centrifugation at 4,600 × g for 20 min at 4°C, washed with 15 ml of sterile phosphate-buffered saline (PBS), and resuspended in 2 ml of PBS. The preparations were stored at −20°C until further use. Strains used in the sandwich EIA to determine the limit of detection (LOD) were prepared as glycerol stocks (final concentration, 15% [vol/vol] in PBS) and stored at −80°C. To determine the number of CFU, 10-fold serial dilutions of bacterial cells were plated on LB agar plates.
TABLE 1.
Bacterial strains used in this study
| Strain | Species | Origin | Source/reference(s)a | Serotype |
|---|---|---|---|---|
| MHI 975b | Cronobacter sakazakii ATCC 29544 | Human | 6, 27 | O1 |
| MHI 996 | C. sakazakii | Baby food | LMU | O1 |
| MHI 21000b,c | C. sakazakii | Baby food | LMU | O1 |
| MHI 21001b,c,d | C. sakazakii | Baby food | LMU | O1 |
| MHI 21008b | C. sakazakii | Baby food | JLU | O1 |
| MHI 21011b | C. sakazakii | Baby food | LMU | O1 |
| MHI 21012b | C. sakazakii | Baby food | LMU | O1 |
| MHI 21028b | C. sakazakii E655 | Human | UZ | O1 |
| MHI 21030 | C. sakazakii E789 | Human | UZ | O1 |
| MHI 21038 | C. sakazakii ATCC BAA 893-1 | Milk powder | 55 | O1 |
| MHI 21039c | C. sakazakii ATCC BAA 894 | Human | 56 | O1 |
| MHI 21040 | C. sakazakii CDC 0996-77 | Human | 17 | O1 |
| MHI 21086b | C. sakazakii | Milk powder | LMU | O1 |
| MHI 21172b | C. sakazakii | Baby food | LMU | O1 |
| MHI 977b,d | C. sakazakii NCTC 8155 | Tin of dried milk | 6 | O2 |
| MHI 978 | C. sakazakii NCTC 9238 | Human | 6 | O2 |
| MHI 988b | C. sakazakii | Baby food | LMU | O2 |
| MHI 989 | C. sakazakii | Baby food | LMU | O2 |
| MHI 995b,c | C. sakazakii | Baby food | LMU | O2 |
| MHI 998 | C. sakazakii | Baby food | LMU | O2 |
| MHI 999 | C. sakazakii | Baby food | LMU | O2 |
| MHI 21003b | C. sakazakii | Baby food | JLU | O2 |
| MHI 21004b | C. sakazakii | Baby food | JLU | O2 |
| MHI 21009 | C. sakazakii | Baby food | JLU | O2 |
| MHI 21010 | C. sakazakii | Baby food | JLU | O2 |
| MHI 21027 | C. sakazakii E604 | Human | UZ | O2 |
| MHI 21029b,c | C. sakazakii E785 | Human | UZ | O2 |
| MHI 21032 | C. sakazakii E796 | Human | UZ | O2 |
| MHI 21035c | C. sakazakii E886 | Human | UZ | O2 |
| MHI 21037 | C. sakazakii E786 | Human | UZ | O2 |
| MHI 21041 | C. sakazakii CDC 1123-79 | Human | 17 | O2 |
| MHI 21042 | C. sakazakii ES5 | Human | 57 | O2 |
| MHI 21098 | C. sakazakii E767 | Milk powder | UZ | O2 |
| MHI 21122b | C. sakazakii ATCC 12868 | Unknown | 5 | O2 |
| MHI 21125 | C. sakazakii 0306-E-L-1031 | Milk | TUM | O2 |
| MHI 21126 | C. sakazakii | Animal food | LMU | O2 |
| MHI 982 | C. sakazakii | Baby food | LMU | O3 |
| MHI 990b,c,d | C. sakazakii | Baby food | LMU | O3 |
| MHI 21006b | C. sakazakii | Baby food | JLU | O3 |
| MHI 21007c | C. sakazakii | Baby food | JLU | O3 |
| MHI 21013 | C. sakazakii | Baby food | LMU | O3 |
| MHI 21014b | C. sakazakii | Baby food | LMU | O3 |
| MHI 21036 | C. sakazakii E535 | Human | UZ | O3 |
| MHI 21129 | C. sakazakii | Food | JLU | O3 |
| MHI 21130 | C. sakazakii | Food | JLU | O3 |
| MHI 21131 | C. sakazakii | Food | JLU | O3 |
| MHI 21132 | C. sakazakii | Food | JLU | O3 |
| MHI 21166c | C. sakazakii | Baby food | JLU | O3 |
| MHI 21167 | C. sakazakii | Food | JLU | O3 |
| MHI 21168 | C. sakazakii | Food | JLU | O3 |
| MHI 21169 | C. sakazakii | Baby food | JLU | O3 |
| MHI 21051 | C. sakazakii | Milk powder | LMU | O4 |
| MHI 21052 | C. sakazakii | Milk powder | LMU | O4 |
| MHI 21053 | C. sakazakii | Milk powder | LMU | O4 |
| MHI 21067 | C. sakazakii | Milk powder | LMU | O4 |
| MHI 21106 | C. sakazakii SU12_107; H1619/1 | Food | 21 | O4 |
| MHI 21107 | C. sakazakii SU12_106; H1602 | Food | 21 | O4 |
| MHI 21170 | C. sakazakii | Baby food | JLU | O4 |
| MHI 993 | C. sakazakii | Baby food | LMU | O7 |
| MHI 21066 | C. sakazakii | Milk powder | LMU | O7 |
| MHI 21109 | C. sakazakii SU12_70; H2496 | Food | 21 | O7 |
| MHI 21110 | C. sakazakii SU12_120; H1651 | Environment IFM factory | 21 | O7 |
| MHI 21111 | C. sakazakii SU12_27; A31 | Environment IFM factory | 21 | O7 |
| MHI 21133 | C. sakazakii | Food | JLU | O7 |
| MHI 21134 | C. sakazakii | Food | JLU | O7 |
| MHI 21135 | C. sakazakii | Food | JLU | O7 |
| MHI 21136 | C. sakazakii | Baby food | JLU | O7 |
| MHI 21171 | C. sakazakii | Baby food | JLU | O7 |
| MHI 21173 | C. sakazakii | Milk powder | LMU | O7 |
| MHI 21097 | C. condimenti LMG 26250T | Food | 8 | |
| MHI 979 | C. dublinensis subsp. lausannensis NCTC 9844 | Unknown | 5 | |
| MHI 980 | C. dublinensis NCTC 9846T | Unknown | 5 | |
| MHI 21093 | C. dublinensis subsp. dublinensis LMG 23823T | Milk powder | 5 | |
| MHI 21094 | C. dublinensis subsp. lausannensis LMG 23824T | Water | 5 | O2 |
| MHI 21095 | C. dublinensis subsp. lactaridi LMG 23825T | Milk powder | 5 | O1 |
| MHI 986 | C. malonaticus | Baby food | LMU | O2 |
| MHI 987 | C. malonaticus | Baby food | LMU | O2 |
| MHI 992 | C. malonaticus | Baby food | LMU | O2 |
| MHI 994 | C. malonaticus | Baby food | LMU | O2 |
| MHI 21002 | C. malonaticus | Baby food | JLU | O2 |
| MHI 21005 | C. malonaticus | Baby food | JLU | O2 |
| MHI 21091 | C. malonaticus DSM 18702 | Human | 5 | O2 |
| MHI 21031 | C. muytjensii E793 | Human | UZ | O2 |
| MHI 21096 | C. muytjensii DSM 21870 | Unknown | 5 | O2 |
| MHI 21209 | C. muytjensii E456 | Unknown | UZ | O2 |
| MHI 21212 | C. muytjensii E769 | Milk powder | UZ | O1 |
| MHI 21213 | C. muytjensii E888 | Milk powder | UZ | O1 |
| MHI 21026 | C. turicensis 3032 LMG 23827T | Neonate | 5, 58 | O1 |
| MHI 21049 | C. turicensis E625 | Baby food | UZ | O3 |
| MHI 21050 | C. turicensis E609 | Food | UZ | O3 |
| MHI 981 | C. universalis NCTC 9529T | Fresh water | 8 | O1 |
| MHI 21128 | Acinetobacter spp. | Food | LMU | |
| MHI 1004 | Aeromonas media DSM 30020 | Pasteurized milk | 59 | |
| MHI 903 | Citrobacter spp. DSM 3004 | Food | 60 | |
| MHI 701 | Escherichia coli DSM 682 | Unknown | ||
| MHI 969 | Enterobacter aerogenes | Unknown | LMU | |
| MHI 968 | Enterobacter asburiae | Unknown | LMU | |
| MHI 904 | Enterobacter cloacae DSM 30054 | Spinal fluid | ||
| MHI 21103 | Franconibacter helveticus LMG 23732T | Fruit powder | 61 | |
| MHI 21105 | Franconibacter pulveris LMG 24057T | Fruit powder | 61 | |
| MHI 910 | Hafnia alvei DSM 30097 | Unknown | 62 | |
| MHI 21024 | Klebsiella pneumoniae | Calf | LMU | |
| MHI 905 | Moellerella wisconsensis | Food | LMU | |
| MHI 991 | Morganella morganii DSM 6675 | Feces | 63 | |
| MHI 946 | Proteus vulgaris DSM 2140 | Inner ear infection | ||
| MHI 952 | Providencia stuartii DSM 6676 | Feces | ||
| MHI 1000 | Pseudomonas aeruginosa DSM 939 | Water | ||
| MHI 21046 | Salmonella enterica subsp. enterica DSM 17420 | Unknown | 64 | |
| MHI 974 | Serratia rubidaea DSM 4480 | Type strain | 65 | |
| MHI 21104 | Siccibacter turicensis LMG 23730T | Fruit powder | 61 | |
| MHI 914 | Shigella flexneri DSM 4782 | Type strain |
LMU, Chair of Hygiene and Technology of Milk, Ludwig-Maximilians-Universität München, Munich, Germany; JLU, Institute of Veterinary Food Science, Justus-Liebig-Universität Giessen, Giessen, Germany; UZ, Institute for Food Safety and Hygiene, University of Zürich, Zurich, Switzerland; TUM, Chair of Animal Hygiene, Technische Universität München, Freising, Germany.
Strains used for analysis in immunoblotting.
Strains used for analysis in motility assays.
Strains used for immunization, which were randomly chosen from a group of well-characterized food isolates.
Identification and molecular characterization of Cronobacter species.
The isolation of genomic DNA as a template for PCR was carried out according to standard protocols (36). To identify Cronobacter strains at the genus, species, and serotype levels, the proposed schema for Cronobacter spp. (29) was applied using molecular detection techniques: an alpha-glucosidase-based PCR assay was performed to confirm the bacterial genus (37), and the species of the Cronobacter strains were identified using rpoB and cgcA as the gene targets for the PCR amplification (7, 38). Only strains that were confirmed to be C. sakazakii were then subjected to multiplex PCR to identify the serotype. The PCR conditions and primers were applied as described previously by Sun et al. (20). For serotyping of the other Cronobacter species, the conditions and primers were used as described by Jarvis et al. (18).
Immunogen preparation, immunization, and hybridoma cell production.
The immunizations of mice for generating monoclonal antibodies were conducted in compliance with the German law for the protection of animals. Study permission was obtained by the Government of Upper Bavaria (permit no. 55.2-1-54-2531.6-1-08). For each of the serotypes O1, O2, and O3, one C. sakazakii strain (Table 1) was randomly chosen to prepare cell-free immunogenic extracts for the immunization of three different groups of mice. The preparation of protein and LPS extracts by treating bacterial strains with polymyxin B-sulfate and the immunization of female mice [BALB/c strain and a hybrid strain of BALB/c × (NZW × NZB)] was conducted as described before (39). The fusion of splenocytes and X63-Ag8.653 myeloma cells was performed as published by Dietrich et al. (40). The culture supernatant of the hybridoma cells was screened for serotype-specific antibodies using a standard EIA protocol (39, 41), with concentrated bacterial preparations serving as the solid phase. Positive clones were separated by limiting dilution and subsequently mass produced in miniPERM (Sarstedt, Nümbrecht, Germany) or CELLine (Integra Biosciences AG, Zizers, Switzerland) bioreactors. The immunoglobulin subtype of the MAbs was determined by using mouse MAb isotyping reagents purchased from Sigma-Aldrich (St. Louis, MO). The isolation and purification of MAbs were performed by affinity chromatography on protein A or protein G agarose (Sigma-Aldrich), as described before (41).
Monoclonal antibody characterization.
Mass-produced and purified MAbs were characterized using immunoblot analysis of LPS extracts, immunofluorescence, and motility assays.
Preparation of LPS extracts, SDS-PAGE, and immunoblot analysis.
LPS was isolated using the phenol-chloroform method described previously (39, 42, 43), with the following modifications. Before subjecting the cell suspension to the phenol-chloroform procedure, 25 μl of proteinase K (10 μg/ml) was added, and the preparation was incubated for 30 min at 55°C. The precipitated and dried extracts were dissolved in 70 μl of distilled water. For SDS-PAGE analysis, the LPS preparations were mixed with XT sample buffer and reducing agent (Bio-Rad), incubated for 10 min at 100°C, and stored at −20°C until use. Twenty microliters of each LPS extract was separated by SDS-PAGE and subsequently transferred to a polyvinylidene difluoride (PVDF) membrane. These steps and the subsequent detection of bound MAb by immunoblotting were performed as described earlier (39).
Immunofluorescence.
The reactivities of serotype-specific MAbs with membrane components of untreated live cells of all C. sakazakii serotype O1, O2, and O3 strains and cells of other bacterial species were tested using immunofluorescence microscopy, as described by Schauer et al. (39).
Motility assay.
Motility assays were performed as described previously (39). Briefly, petri dishes (6-cm diameter) with LB soft agar (0.3% agar) containing the specific MAb at a concentration of 17 μg/ml were stab-inoculated with 1 μl of an overnight culture in the centers and incubated upright at 37°C. The motility of the bacteria was determined by measuring the diameter of the motility zone after 8 h of incubation.
Inclusivity and exclusivity tests.
The inclusivity and exclusivity of the MAbs were determined by analyzing a broad range of bacterial strains in indirect EIA. The reactivities of 14 serotype O1 strains, 20 serotype O2 strains, and 15 serotype O3 strains were analyzed. In addition, a broad range of different strains, including 18 C. sakazakii strains from serotypes O4 and O7, 23 Cronobacter species strains, and 20 more distantly related bacteria, were analyzed (Table 1). In EIAs, bacterial preparations at a concentration of 1 × 107 CFU/ml served as the solid phase and were serially diluted in PBS. After incubation of EIA plates at 4°C overnight, free binding sites were blocked with 150 μl per well of 3% caseinate-PBS for 30 min. Subsequently, plates were washed (distilled water with 0.15 M NaCl and 0.02% Tween 20), and 100 μl of the MAbs was added (1 μg/ml in PBS) for 1 h. After another washing step, bound MAb was detected with 100 μl of secondary polyclonal rabbit anti-mouse immunoglobulins–horseradish peroxidase (HRP) (Dako Denmark A/S, Glostrup, Denmark) solution (1:2,000 in 1% caseinate-PBS). Following another washing step, 100 μl/well of substrate-chromogen solution (1 mmol 3,3′,5,5′-tetramethylbenzidine, 3 mmol H2O2 per liter potassium citrate buffer [pH 3.9]) was added and incubated for 20 min. The reaction was stopped by adding 100 μl of 1 M H2SO4 to each well. The absorbance was measured at 450 nm using an EIA plate reader (Ridawin, Tecan Deutschland GmbH). The absorbance data were analyzed using the Ridawin EIA calculation software (r-Biopharm GmbH, Darmstadt, Germany).
Establishment of an optimized sandwich EIA system.
In order to establish a sandwich EIA, the specific MAbs were conjugated using activated horseradish peroxidase (Roche Diagnostics, Rotkreuz, Switzerland), according to the manufacturer's instructions. To define the optimal working concentrations, the MAbs and the equivalent HRP-conjugated MAbs were applied in checkerboard titrations with and without C. sakazakii strains. To determine the limit of detection (LOD) of the assay, microtiter plates were coated with the specific MAb (10 μg/ml) in PBS at ambient temperatures overnight. Live or heat-treated (121°C for 15 min) bacterial cells were diluted in 0.1% bovine serum albumin (BSA)-PBS and subsequently detected using the HRP-conjugated specific MAbs (1C4-HRP and 2F8-HRP at 1:1,000, and 1A11-HRP at 1:2,000 in 1% caseinate-PBS solution). All other steps were conducted according to the indirect EIA method.
In order to achieve high absorbance values for bound C. sakazakii and the lowest possible absorbance values for blank samples, various compositions of the dilution buffer for the antigen were examined, including PBS, PBS containing 0.1% BSA, PBS containing different concentrations of Tween 20 (0.5% and 1%), and PBS containing both BSA and Tween 20.
Detection of C. sakazakii serotypes O1, O2, and O3 in PIF.
Powdered infant formulas (PIF), in particular, four initial milk formulas (from birth on) and three follow-on formulas (from 6th month on), were purchased from a local German drugstore. For each C. sakazakii serotype, three strains were cultivated in LB broth overnight at 37°C and plated on LB agar to determine the bacterial cell counts. To check the applicability of the developed sandwich EIA system for the sensitive detection of C. sakazakii in PIF, 10 g of sample was dissolved in 90 ml of buffered peptone water. This mixture was artificially contaminated with 1 ml of a dilution of the C. sakazakii overnight culture corresponding to 1 to 10 CFU and incubated at 37°C without shaking. After 15 h, samples were directly analyzed by sandwich EIA and, in parallel, the CFU were determined according to the ISO method (25) on chromogenic Cronobacter isolation (CCI) agar (Oxoid).
RESULTS
Identification of C. sakazakii serotypes.
Due to the recently described limitation of PCR-based methods for the determination of the different variations of genomic O-antigen gene clusters of C. sakazakii strains (22, 30), it has been proposed that C. sakazakii serotypes can reliably be determined only after the strains have been identified as members of the genus Cronobacter and species sakazakii (29). Therefore, the genus and species of a collection of 76 strains that were previously identified as C. sakazakii at our institute by classical culture methods, according to the ISO standard in reference 25, were verified using a PCR method (7, 37, 38). Thereafter, strains were serotyped using a multiplex PCR method (18, 20). Out of 76 assumed C. sakazakii strains, 70 strains were confirmed at the genus and species levels to be C. sakazakii, while six strains belonged to the species C. malonaticus. The majority of strains belonged to serotypes O2 (22 strains), O3 (15 strains), and O1 (14 strains), followed by O7 (11 strains) and O4 (7 strains) (Table 1).
Production of monoclonal antibodies.
To create serotype-specific MAbs against C. sakazakii, three different groups of five female mice each were immunized with cell-free polymyxin B extracts of the selected strains MHI 21001 (O1), MHI 977 (O2), and MHI 990 (O3). Almost all mice developed high specific antibody titers for C. sakazakii. After cell fusion, a broad range of >20 antibody-producing hybridomas that were reactive with C. sakazakii were obtained in indirect EIAs in which C. sakazakii preparations served as the solid phase. In terms of specific reactivity, MAbs were categorized into 3 groups of different reaction patterns: MAbs of the first group reacted with all serotypes of C. sakazakii and with other Enterobacteriaceae, while MAbs of the two other groups reacted in a serotype-specific manner either with single strains (group 2) or with the vast majority (group 3) of the C. sakazakii strains of a certain serotype. The titers of the MAbs were determined in indirect EIA and, finally, three high-antibody-producing hybridoma clones for which serotype-specific reactivity was observed (group 3) were selected for detailed characterization and the establishment of sandwich EIAs. These MAbs, all of the IgG class, were designated 1C4 (reactive with serotype O1), 2F8 (reactive with serotype O2), and 1A11 (reactive with serotype O3) (Table 2).
TABLE 2.
Characteristics and cross-reactivities of the produced MAbs
| MAb | IgG subtype | Serotype specificity | Target | Inclusivitya | Exclusivityb |
|---|---|---|---|---|---|
| 1C4 | IgG2a | O1 | EPS | 13/14c | 0/95 |
| 2F8 | IgG2a | O2 | LPS | 20/20 | 0/89 |
| 1A11 | IgG2b | O3 | LPS | 13/15d | 2/94e |
Number of positive indirect-EIA results for strains of the same serotype.
Number of positive results with different serotypes of C. sakazakii strains, other Cronobacter species, 17 Enterobacteriaceae strains, and three other Gram-negative strains.
No reactivity with strain MHI 21040.
No reactivity with strains MHI 982 and MHI 21129.
Reactive with MHI 21212 and MHI 21213 (both C. muytjensii serotype O1).
Inclusivity and exclusivity tests.
The specificity of the produced MAbs was determined in indirect EIAs by testing the Cronobacter species collection comprising 91 strains and additional strains of the Enterobacteriaceae family, including Franconibacter helveticus, Franconibacter pulveris, and Siccibacter turicensis, which may simultaneously occur in the same PIF sample, and other Gram-negative bacteria (Table 1). Out of 14 tested C. sakazakii O1 strains, all strains except C. sakazakii MHI 21040 (CDC0996-77) were recognized by MAb 1C4. MAb 2F8 showed binding to all 20 C. sakazakii O2 strains tested, and MAb 1A11 bound to 13 out of 15 C. sakazakii O3 strains (no reactivity with strains MHI 982 and MHI 21129). Within the C. sakazakii group, all MAbs reacted exclusively with the respective C. sakazakii serotype. The high affinity of the MAbs is documented by the fact that even after diluting the bacterial preparations 50,000 times, absorbance values of ≥1.0 were obtained in the EIA. Further, MAbs 1C4 (O1) and 2F8 (O2) showed no cross-reactivity with other Cronobacter species strains or more distantly related bacteria (Table 2). The absorbance values for these negative strains ranged between 0.008 and 0.082 at cell counts of 1 × 107 CFU/ml. MAb 1A11 (O3) additionally reacted with two C. muytjensii strains of serotype O1.
Identification of the antigenic determinant.
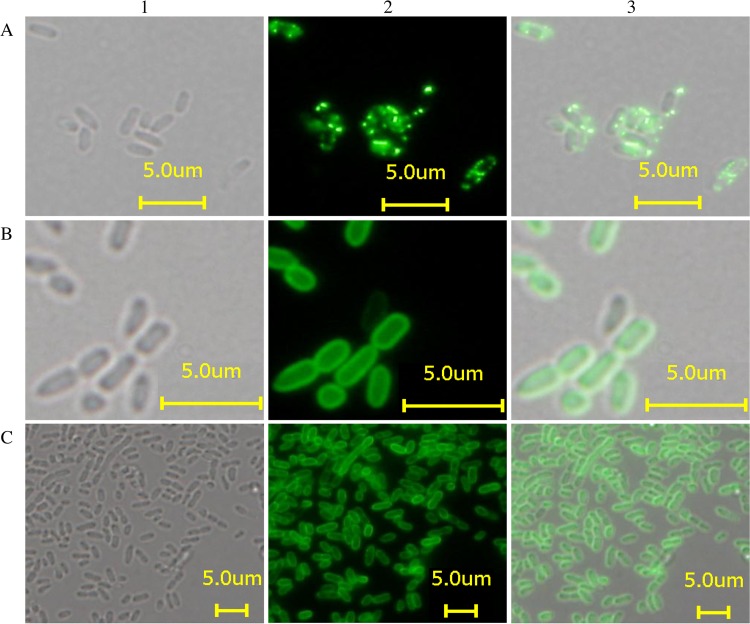
In indirect EIAs, the three developed MAbs showed a strong reactivity only with the respective C. sakazakii serotypes, suggesting that the serotype-specific antigenic determinants are represented by polysaccharides, which are components of the cell envelope of all Gram-negative bacteria and thus form an important structure for antigenic classification of the organism. This assumption was supported by immunofluorescence microscopy, in which all three antibodies showed considerable fluorescence on the surface of all 49 tested strains (Fig. 1). Two patterns were observed: evenly distributed fluorescence on the surface of C. sakazakii serotype O2 and O3 strains (Fig. 1B and C) was observed using MAbs 2F8 (O2) and 1A11 (O3). In contrast, the use of MAb 1C4 for labeling C. sakazakii serotype O1 strains resulted in a more punctuated distribution of fluorescence (Fig. 1A). Finally, immunoblotting confirmed the specificity of MAbs to a polysaccharide moiety extracted from 3 to 9 C. sakazakii strains from each serotype (O1 to O3) (Fig. 2 and Table 1). All immunoblot profiles showed a ladder pattern characteristic of polysaccharides. Furthermore, the same characteristic ladder pattern was observed after proteinase K treatment of the LPS extracts, proving that proteinaceous structures are irrelevant for antibody reactivity.
FIG 1.
Microscopic analysis of live C. sakazakii cells by phase contrast (1), immunofluorescence (2), and overlay mode (3) using serotype-specific MAbs: MHI 21172 (O1) stained with MAb 1C4 (A), MHI 21032 (O2) stained with MAb 2F8 (B), and MHI 21166 (O3) stained with MAb 1A11 (C).
FIG 2.
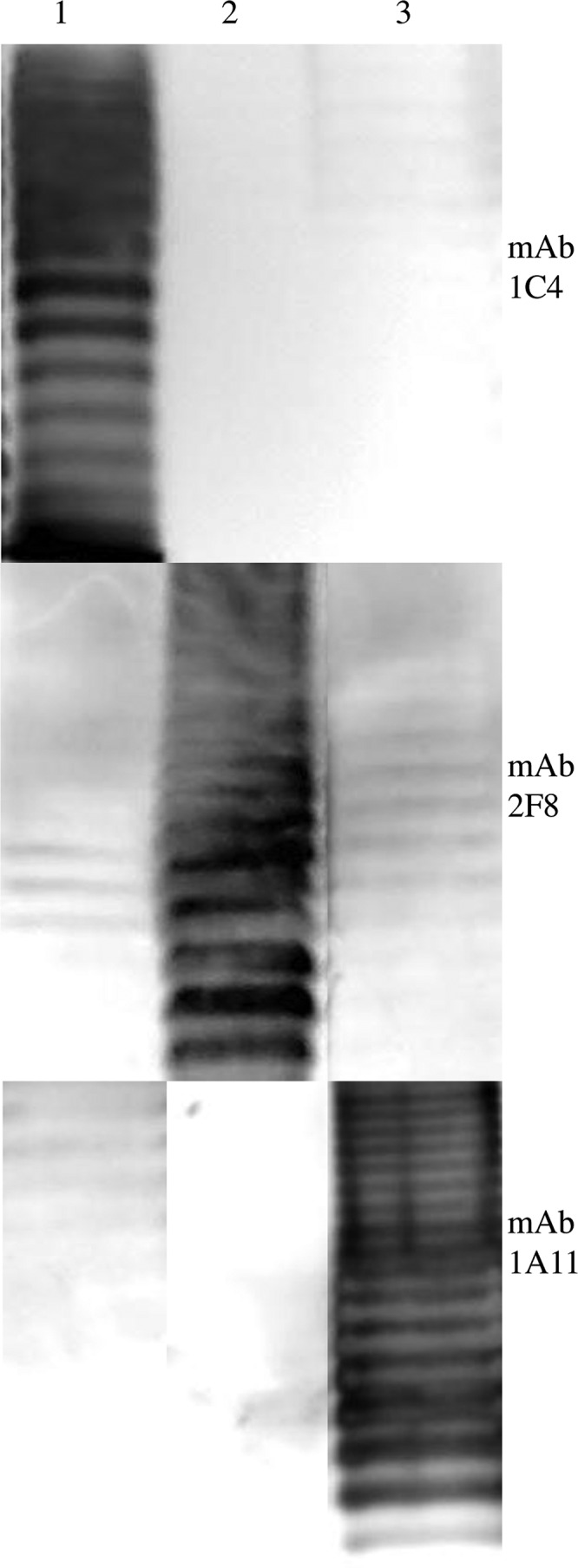
Reactivities of MAbs 1C4, 2F8, and 1A11 with (lipo)polysaccharide preparations of different serotypes of C. sakazakii strains. Lane 1, MHI 21011 (O1); lane 2, MHI 21029 (O2); lane 3, MHI 990 (O3).
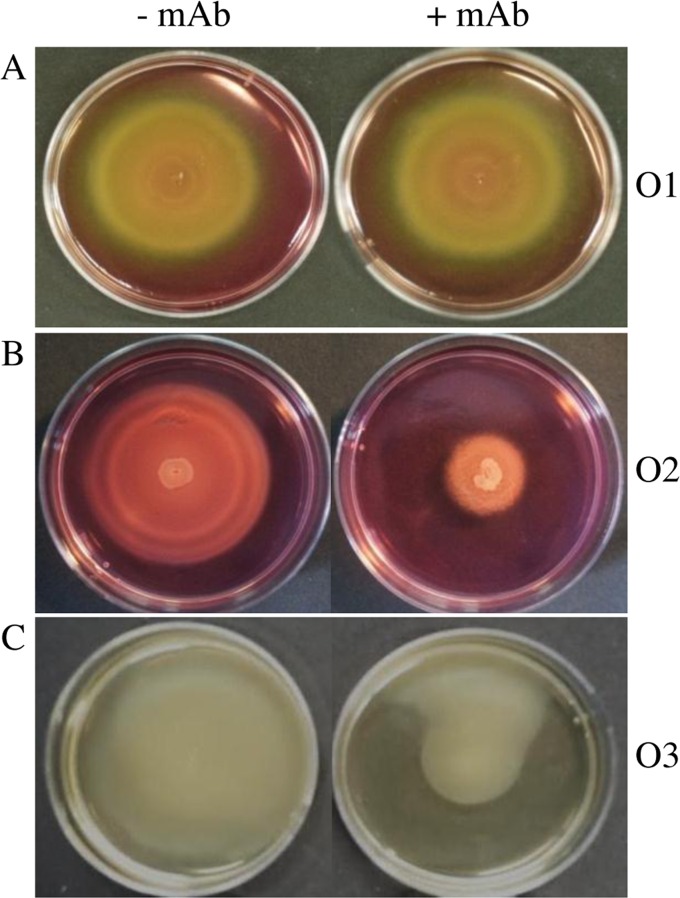
In order to further narrow down the possible antigenic determinant, and with respect to a previous publication in which a reduced flagellar motility of strains was observed after binding of LPS-specific MAb (39), the motility of Cronobacter cells on soft agar in the presence of the MAbs was analyzed for three strains of each serotype (Table 1). The addition of MAb 2F8 or 1A11 to the medium inhibited the bacterial motility of strains of the O2 and O3 serotype groups, demonstrated by a significantly reduced diameter of the diffuse growth area (Fig. 3). Overall, the motility zones were reduced by 40 to 50% compared to those of soft agar plates without added MAbs (Fig. 3B and C). In contrast, no effect on the bacterial motility of O1 serotype strains was observed by exposure to MAb 1C4 (Fig. 3A).
FIG 3.
Swimming ability of C. sakazakii. Bacterial motility was assayed in soft agar without and with specific MAbs (17 μg/ml). MAb 1C4- and MAb 2F8-containing supernatants of hybridoma cells were diluted in soft agar containing 50% Dulbecco's modified Eagle's medium (DMEM). Purified MAb 1A11 was added in soft agar without DMEM. Motility zones were measured after 8 h of incubation at 37°C. (A) MHI 21001 (O1). (B) MHI 21035 (O2). (C) MHI 990 (O3).
As MAb 1C4 did not affect bacterial motility, and LPS thus seemed unlikely to be the antigenic determinant, testing was conducted to determine whether other polysaccharides (e.g., exopolysaccharide [EPS] and capsular polysaccharide [CPS]) might be the target of MAb 1C4. Therefore, a previously published acapsular C. sakazakii strain, MHI 975 (NCTC 11467T) (44), was analyzed by immunofluorescence. The same localized fluorescence signal obtained for all other C. sakazakii strains from serotype O1 was obtained for this strain. This observation led to the assumption that for this MAb, not CPS but EPS, which is loosely attached to the bacterial surface, serves as an antigenic determinant. Altogether, these results strongly indicate that the LPS O-specific polysaccharide chain of the C. sakazakii O2 and O3 serotypes (see Fig. S1 in the supplemental material) and the EPS of the C. sakazakii O1 serotype represent the antigens specifically recognized by MAbs 1A11, 2F8, and 1C4, respectively.
Establishment of sandwich EIA.
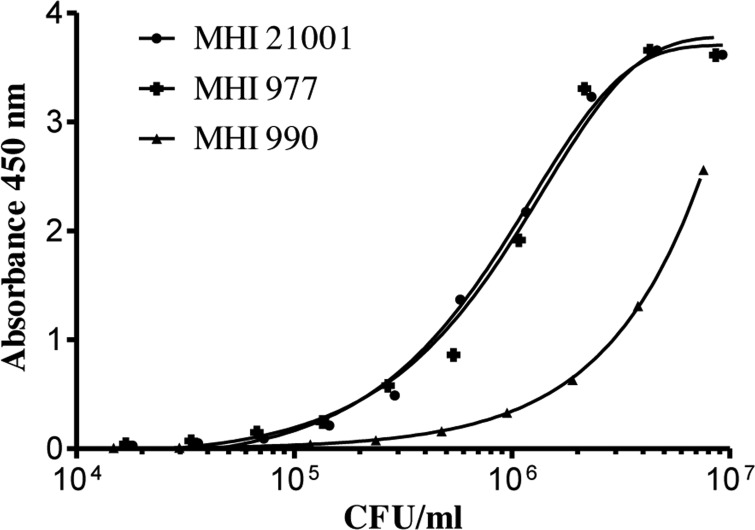
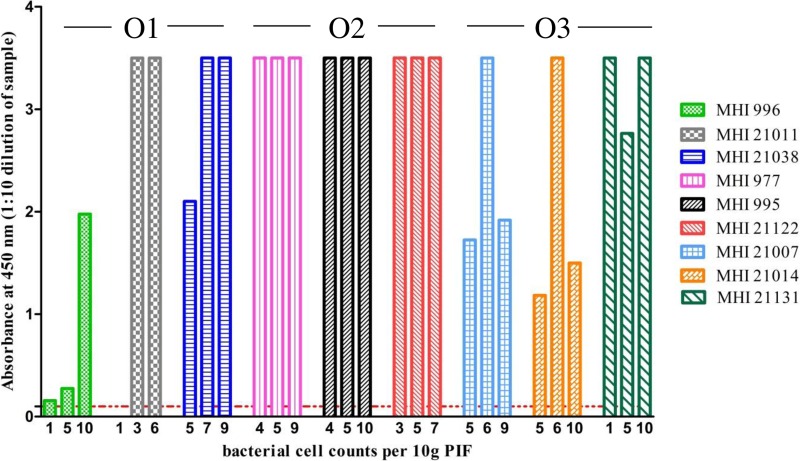
As both LPS and EPS are evenly distributed on the surface of C. sakazakii strains, it seemed reasonable to establish sandwich EIAs for the different serotypes based on each of the available specific MAbs. For this purpose, the same MAb was used both as coating and, after labeling with HRP, as detection antibody in the sandwich EIAs. The dilution of the bacterial strains in 0.1% BSA-PBS resulted in the most stable values and was subsequently used as the dilution buffer. The lowest number of detectable bacteria (LOD) was defined as twice the absorbance (at 450 nm) of the blank value (Fig. 4), which ranged from 0.06 to 0.019. For 46 out of 49 tested strains, the sensitivities of the three serotype-specific sandwich EIAs ranged from 2 × 103 CFU/ml to 9 × 106 CFU/ml, whereas 3 strains were not detectable (Table 3). More specifically, one serotype O1 strain (MHI 21011) was detectable at a cell count of 9 × 103 CFU/ml. For 6 strains of this serotype, specific detection limits of 2 × 104 to 7 × 104 CFU/ml were realized. Six strains showed slightly higher detection limits between 1 × 105 and 3 × 105 CFU/ml. Only one strain (MHI 21040) was not detectable and showed an absorbance value of 0.02 at a cell count of 1.4 × 106 CFU/ml. In comparison to the serotype O1-specific sandwich EIA, the serotype O2 assay showed even higher sensitivity: seven strains were still detectable at cell counts between 2 × 103 and 9 × 103 CFU/ml, and 11 strains had detection limits ranging from 1 × 104 to 6 × 104 CFU/ml. Only for 2 strains was a slightly higher LOD of 1 × 105 to 2 × 105 CFU/ml observed. For the majority of strains of the O3 serotype, the detection limits ranged from 1 × 104 CFU/ml to 9 × 104 CFU/ml. Two strains were just detectable at cell counts of 2 × 105 CFU/ml and 9 × 106 CFU/ml, respectively, while two further strains showed no reactivity in the assay.
FIG 4.
Sensitivity of the sandwich EIA for the detection of C. sakazakii strains MHI 21001 (O1), MHI 977 (O2), and MHI 990 (O3). MAbs 1C4, 2F8, and 1A11, respectively (10 μg/ml), served as solid phase. Bacteria were enriched overnight in LB broth at 37°C and subsequently detected in serial dilutions with 1C4-HRP, 2F8-HRP (1:1,000), and 1A11-HRP (1:2,000), respectively.
TABLE 3.
LOD of C. sakazakii strains in sandwich EIAs
| MAb (serotype) | Detection limit (CFU/ml) | Strain(s)a |
|---|---|---|
| 1C4 (O1) | 9 × 103 | MHI 21011 |
| 2 × 104 to 7 × 104 | MHI 21001,b MHI 975, MHI 21039, MHI 21028, MHI 21086, MHI 21008 | |
| 1 × 105 to 3 × 105 | MHI 21012, MHI 21038, MHI 996, MHI 21000, MHI 21172, MHI 21030 | |
| NDc | MHI 21040 | |
| 2F8 (O2) | 2 × 103 to 9 × 103 | MHI 21041, MHI 21003, MHI 998, MHI 977,b MHI 995, MHI 21004, MHI 21035 |
| 1 × 104 to 6 × 104 | MHI 21037, MHI 21010, MHI 21042, MHI 21098, MHI 999, MHI 21009, MHI 21029, MHI 21122, MHI 21027, MHI 21108, MHI 21032 | |
| 1 × 105 to 2 × 105 | MHI 988, MHI 989 | |
| 1A11 (O3) | 1 × 104 to 9 × 104 | MHI 21036, MHI 21006, MHI 21131, MHI 21014, MHI 21166, MHI 21168, MHI 21167, MHI 21007, MHI 990,b MHI 21013, MHI 21130 |
| 2 × 105 | MHI 21169 | |
| 9 × 106 | MHI 21132 | |
| NDc | MHI 982, MHI 21129 |
Listed in order of lowest to highest detection limit.
Strains used for immunization of mice.
ND, not detected.
Application of the sandwich EIA for the detection of C. sakazakii in powdered infant formula.
In order to determine the applicability of the established sandwich EIA, the effect of different PIF samples on the EIA performance was analyzed in preliminary experiments. For this purpose, 10 g of seven PIF samples, including four different initial formulas (from birth onward) and three follow-on formulas (from the 6th month on), were diluted in 90 ml of buffered peptone water and analyzed by the developed EIAs (serial 2-fold dilutions in PBS–0.1% Tween 20). While undiluted negative samples had slightly positive reactions in the EIAs, for 10-fold-diluted samples, the absorbance values ranged from 0.02 to 0.08 and were thus nearly identical to those obtained for pure-buffer solutions. Therefore, the general cutoff (background) of the assays was set at a value of 0.1.
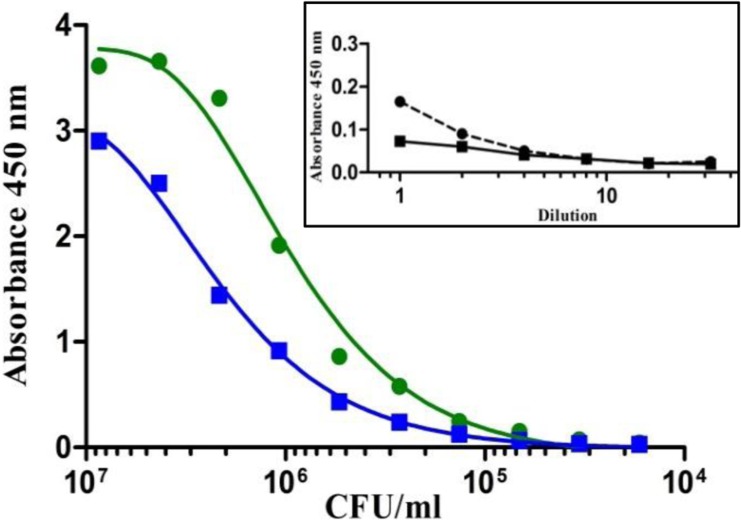
To investigate whether PIF as matrix has a negative effect on the growth of C. sakazakii and the determined sensitivity of the sandwich EIA, in a preliminary experiment, 10 g of a sample of one of the initial milk formulas was artificially contaminated with 10 CFU of C. sakazakii MHI 997 and enriched at 37°C. After 15 h, the number of CFU was determined by plating 100 μl on CCI agar, and, in parallel, the enrichment broth was analyzed in serial dilution by sandwich EIA. Under these conditions, the broth had a positive reaction in the EIA up to a dilution of 1:51,200, which corresponded to a bacterial cell count of 3 × 104 CFU/ml. These values were comparable to the previously determined LOD of 7 × 103 CFU/ml of buffer (Table 3) and underline the robustness of the developed EIA (Fig. 5).
FIG 5.
Evaluation of the established O2-specific sandwich EIA for the analysis of C. sakazakii in PIF. Shown is a comparison of the detection limit of strain MHI 977 (O2) in pure bacterial preparation in PBS (green line) and in enriched 10% PIF broth (blue line). Inset, absorbance values of buffered peptone water (black dashed line) and 10% PIF-buffered peptone water (black solid line) without Cronobacter (background). All samples were analyzed in 2-fold serial dilutions in PBS–0.1% Tween20 and probed with MAb 2F8 (10 μg/ml) and 2F8-HRP (1:1,000 in 1% caseinate–PBS).
To prove the applicability of the developed sandwich EIA for the reproducible and sensitive detection of C. sakazakii, 10 g of commercially available PIF dissolved in 90 ml of buffered peptone water was artificially contaminated each with a strain of serotypes O1, O2, and O3. The viable cell counts of the inocula used for PIF samples indicated that samples were spiked with 1 to 10 cells per 10 g of milk powder (Fig. 6) prior to enrichment, according to the ISO reference method (25), for up to 18 h at 37°C. To determine the required incubation time for detection, aliquots of the contaminated PIF samples were taken after 5 h, 8 h, 11 h, and 15 h and analyzed by sandwich EIA. In all three assays, C. sakazakii was reliably detected after 15 h and, in some cases, after only 11 h of enrichment. Longer incubation did not improve the test results. Samples could be diluted in a range of from 1:20 to >1:5,120, depending on the strain and the bacterial cell counts applied for contamination of the PIF samples (see Fig. S2 in the supplemental material). At a 10-fold dilution, all samples except for one (MHI 21011, artificially contaminated with 1 CFU per 10 g) were positive (absorbance values, >0.1), with measured signals ranging from 0.275 to 3.5 (Fig. 6).
FIG 6.
Detection of C. sakazakii serotypes O1, O2, and O3 in PIF samples. For each serotype, 3 strains were used to artificially contaminate 10 g of PIF with bacterial cell counts ranging from 1 to 10 CFU. After 15 h of enrichment, serially diluted samples were directly analyzed in the sandwich EIAs. The experiments were conducted in triplicate, as indicated by the columns. The absorbance values were dependent on the amount of C. sakazakii used for the contamination. The absorbance values for the 10-fold diluted samples ranged from 0.156 to 3.5. The cutoff value (absorbance, ≤0.1) of the EIAs is indicated by a dashed red line.
DISCUSSION
C. sakazakii is an opportunistic pathogen causing life-threatening infections in neonates. The pathogen can be isolated from clinical, environmental, and a wide range of food sources, predominantly from PIF and its associated production environments (21, 29, 45, 46).
In general, the majority of C. sakazakii isolates belong to serotypes O1 and O2 (22), followed by serotypes O3, O4, and O7 (19–22, 29, 47). The previously reported distribution of C. sakazakii serotypes agrees with the findings of this study: of the 70 C. sakazakii isolates of our strain collection, C. sakazakii O2 (22 isolates [31.43%]) was detected predominantly, followed by serotypes O3 (16 isolates [22.86%]) and O1 (14 isolates [20%]). Blažková et al. (22) reported that only C. sakazakii serotypes O1, O2, and O4 were isolated from clinical cases, while C. sakazakii serotypes O3 and O7 were not found, even though their occurrence in PIF samples has been described (22). In contrast, Yan et al. reported in two recently published studies (24, 29) that C. sakazakii serotypes O1 to O3 are the most common serotypes in clinical isolates. The distribution of C. sakazakii serotypes in clinical isolates allows the assumption that some C. sakazakii serotypes might be more pathogenic than others.
Therefore, it is important for epidemiological studies to identify not only the genus and species but also the serotype of the causative agent. Until now, serotype-specific MAbs recognizing the O-specific polysaccharide chain of LPS and reacting with untreated live cells have been described only for C. turicensis serotype O1 (39). In general, LPS and various other polysaccharides, such as CPS and EPS, have been identified as major virulence factors. They contribute to capsule and biofilm formation of Gram-negative pathogens (48, 49) and are essential for the persistence of C. sakazakii in harsh environments (30). The highly varied polysaccharide structures are strong immunogens showing a high degree of antigenic variation and are often targets for the production of antibodies. However, only limited reports are available on the production of high-affinity MAbs against LPS and their application in sensitive and robust sandwich EIAs for the detection of foodborne pathogens (50, 51). Westerman et al. (50) presented the production and characterization of specific MAbs for the LPS of Escherichia coli O157 as a valuable tool for its rapid detection. An EIA for the detection and serogroup differentiation of Salmonella spp. using O-factor-specific MAb conjugates and a broadly reactive MAb, specific for the outer core oligosaccharide of Salmonella LPS, was presented by Ng et al. (51). For C. sakazakii, a sandwich EIA based on MAbs and polyclonal antibodies (PAbs) was established previously (32), presumably being reactive with heat-stable LPS of one of the serotypes. However, the target of the antibodies and the serotypes of the tested strains ultimately were not identified.
In order to develop a MAb-based method to detect each of the C. sakazakii serotypes O1, O2, and O3, MAbs 1C4 (O1), 2F8 (O2), and 1A11 (O3) were generated and characterized. MAbs 1C4 and 2F8 reliably recognized the respective C. sakazakii serotype and showed no cross-reactivity with other Cronobacter spp. or other members of the family Enterobacteriaceae. In contrast, MAb 1A11 exhibited considerable cross-reactivity with C. muytjensii serotype O1 strains (E769 and E888). This represents an expected result, as it has been shown that this serotype shares an identical LPS biosynthetic operon with C. sakazakii serotype O3 (18, 52, 53). Accordingly, for MAb 1A11 (O3), the O-specific polysaccharide chain of LPS was identified as the antigenic determinant by immunoblotting.
Treatment of live bacteria with the LPS-specific MAbs 1A11 and 2F8 resulted in a reduced flagellum-based motility, which was also observed in a previous study for a MAb targeting C. turicensis LPS (39). In contrast, MAb 1C4 (O1) displayed a localized fluorescence signal (focus) on the bacterial surface of C. sakazakii serotype O1 and did not inhibit bacterial motility. However, a characteristic polysaccharide ladder pattern was observed in immunoblot analysis, suggesting that the EPS moiety of the cell envelope might be the antigen for MAb 1C4. MAb 1C4 reacted with all tested C. sakazakii serotype O1 strains, except MHI 21040 (Table 2). This strain is likely to possess changes in its cell surface composition, because in an as yet unpublished study, only this strain was unable to activate mitogen-activated protein (MAP) kinases, such as extracellular signal-regulated kinase (ERK), p38, and c-Jun N-terminal kinase (JNK), in human colon epithelial cells (Caco-2) (K. Schauer, unpublished data). Possibly, an altered LPS composition is also the reason for the missing reactivity of MAb 1A11 with the O3 serotype strains MHI 982 and MHI 21129. In summary, the species- and serotype-specific binding properties of the produced MAbs and their reactivities with C. sakazakii cells allow the reliable serological differentiation of C. sakazakii serotypes O1, O2, and O3.
Because of the time-consuming and laborious nature of the cultivation procedures (25) or cultivation methods followed by real-time PCR (U.S. FDA) that are presently approved as reference methods for the screening of C. sakazakii in food, there is an increased need for a complementary, easy, reliable, and rapid detection method to prevent life-threatening infections in neonates and premature infants. The application of antibodies for the identification of foodborne pathogens is an accepted approach, and enzyme immunoassays satisfy the requirements for rapid methods. The advantages of immuno-based methods have led to the production of different commercially available immunological assays for the rapid detection of pathogens, spores, and toxins (54). For the detection of Cronobacter spp., several protocols using MAbs and PAbs have been published. Park et al. (33) established a PAb-based sandwich EIA for the detection of C. muytjensii using chicken anti-C. muytjensii IgY and rabbit anti-C. muytjensii IgG, achieving a detection limit of 2.0 × 104 CFU/ml. Xu et al. (32) described two different EIA formats: an indirect EIA using MAbs specific for the genus Cronobacter, with an LOD of 105 CFU/ml, and a more sensitive (LOD, 2 × 104 CFU/ml) sandwich EIA based on PAbs specific for C. sakazakii. Hochel and Škvor (31) described the development of two PAb-based indirect competitive EIAs for the detection of C. sakazakii strain CNCTC 5739, with detection limits ranging from 0.6 × 105 CFU/ml to 3.4 × 105 CFU/ml. However, none of these methods allows a distinction between different serotypes of C. sakazakii, probably due to the fact that protein (whole-cell lysate) (33) or inactivated bacterial cells (31) were used as the immunogen. With the approach of directly using LPS preparations as the immunogen, these newly developed MAb-based sandwich EIAs enable, for the first time to our knowledge, the simultaneous detection and serotyping of C. sakazakii serotypes O1, O2, and O3. A detection limit of up to 2 × 103 CFU/ml for some strains using bacterial pure culture renders these optimized sandwich EIAs the most sensitive immunochemical detection system for C. sakazakii described so far.
The application of MAb-based sandwich EIAs to detect C. sakazakii serotypes O1, O2, and O3 in food products was investigated by spiking PIF. After enrichment for only 15 h, the LOD for the sandwich EIA was found to be as low as 1 CFU/10 g of PIF, thus considerably shortening the detection time compared to that of the classical methods. A large benefit of the established sandwich EIAs is the possibility to detect live and heat-treated cells without having to forfeit a reduction in assay sensitivity (see Fig. S3 in the supplemental material), simplifying the handling of samples at laboratories without special safety classification. Further on, samples could be directly analyzed without the need to isolate or concentrate bacterial cells from the enriched broth. These facts highlight the robustness and the simple practical application of the sandwich EIA.
In conclusion, it was possible to create three monoclonal IgG antibodies that specifically recognize and differentiate C. sakazakii serotypes O1, O2, and O3. The ability to bind to live C. sakazakii cells makes these MAbs suitable for detecting this pathogen in food, environment, and clinical samples using a developed sandwich EIA capture system. In contrast to the laborious and time-consuming traditional culture detection methods, the high specificity and sensitivity combined with the simple and rapid workflow of this method permit accurate and reliable detection of C. sakazakii serotypes O1, O2, and O3 in pure culture and powdered infant formula within 15 h. This study represents the first approach for simultaneous detecting and serotyping bacteria based on MAbs. Future work will involve the production of MAbs for C. sakazakii serotypes O4 and O7. With a complete panel of MAbs, all relevant C. sakazakii serotypes could be detected in parallel. Further on, the currently used antigenic typing system for serotyping C. sakazakii would considerably be improved.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Federal Ministry of Education and Research (BMBF) of Germany (Food Supply and Analysis [LEVERA], funding code 13N12611).
We thank Angelika Lehner for providing the C. muytjensii strains. We also thank Brunhilde Minich and Franziska Faber for excellent technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04016-15.
REFERENCES
- 1.Lai KK. 2001. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine (Baltimore) 80:113–122. [DOI] [PubMed] [Google Scholar]
- 2.Corti G, Panunzi I, Losco M, Buzzi R. 2007. Postsurgical osteomyelitis caused by Enterobacter sakazakii in a healthy young man. J Chemother 19:94–96. doi: 10.1179/joc.2007.19.1.94. [DOI] [PubMed] [Google Scholar]
- 3.Healy B, Cooney S, O'Brien S, Iversen C, Whyte P, Nally J, Callanan JJ, Fanning S. 2010. Cronobacter (Enterobacter sakazakii): an opportunistic foodborne pathogen. Foodborne Pathog Dis 7:339–350. doi: 10.1089/fpd.2009.0379. [DOI] [PubMed] [Google Scholar]
- 4.Patrick ME, Mahon BE, Greene SA, Rounds J, Cronquist A, Wymore K, Boothe E, Lathrop S, Palmer A, Bowen A. 2014. Incidence of Cronobacter spp. infections, United States, 2003–2009. Emerg Infect Dis 20:1520–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iversen C, Lehner A, Mullane N, Bidlas E, Cleenwerck I, Marugg J, Fanning S, Stephan R, Joosten H. 2007. The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov., and descriptions of Cronobacter sakazakii comb. nov., Cronobacter sakazakii subsp. sakazakii comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies 1. BMC Evol Biol 7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iversen C, Mullane N, McCardell B, Tall BD, Lehner A, Fanning S, Stephan R, Joosten H. 2008. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int J Syst Evol Microbiol 58:1442–1447. [DOI] [PubMed] [Google Scholar]
- 7.Stoop B, Lehner A, Iversen C, Fanning S, Stephan R. 2009. Development and evaluation of rpoB based PCR systems to differentiate the six proposed species within the genus Cronobacter. Int J Food Microbiol 136:165–168. doi: 10.1016/j.ijfoodmicro.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 8.Joseph S, Cetinkaya E, Drahovska H, Levican A, Figueras MJ, Forsythe SJ. 2012. Cronobacter condimenti sp. nov., isolated from spiced meat, and Cronobacter universalis sp. nov., a species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water and food ingredients. Int J Syst Evol Microbiol 62:1277–1283. doi: 10.1099/ijs.0.032292-0. [DOI] [PubMed] [Google Scholar]
- 9.Jason J. 2012. Prevention of invasive Cronobacter infections in young infants fed powdered infant formulas. Pediatrics 130:1076–1084. doi: 10.1542/peds.2012-0473. [DOI] [PubMed] [Google Scholar]
- 10.Giovannini M, Verduci E, Ghisleni D, Salvatici E, Riva E, Agostoni C. 2008. Enterobacter sakazakii: an emerging problem in paediatric nutrition. J Int Med Res 36:394–399. doi: 10.1177/147323000803600303. [DOI] [PubMed] [Google Scholar]
- 11.Caubilla-Barron J, Hurrell E, Townsend S, Cheetham P, Loc-Carrillo C, Fayet O, Prere MF, Forsythe SJ. 2007. Genotypic and phenotypic analysis of Enterobacter sakazakii strains from an outbreak resulting in fatalities in a neonatal intensive care unit in France. J Clin Microbiol 45:3979–3985. doi: 10.1128/JCM.01075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowen AB, Braden CR. 2006. Invasive Enterobacter sakazakii disease in infants. Emerg Infect Dis 12:1185–1189. doi: 10.3201/eid1208.051509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedemann M. 2009. Epidemiology of invasive neonatal Cronobacter (Enterobacter sakazakii) infections. Eur J Clin Microbiol Infect Dis 28:1297–1304. doi: 10.1007/s10096-009-0779-4. [DOI] [PubMed] [Google Scholar]
- 14.Friedemann M. 2007. Enterobacter sakazakii in food and beverages (other than infant formula and milk powder). Int J Food Microbiol 116:1–10. doi: 10.1016/j.ijfoodmicro.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 15.Baumgartner A, Grand M, Liniger M, Iversen C. 2009. Detection and frequency of Cronobacter spp. (Enterobacter sakazakii) in different categories of ready-to-eat foods other than infant formula. Int J Food Microbiol 136:189–192. doi: 10.1016/j.ijfoodmicro.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 16.van Acker J, de Smet F, Muyldermans G, Bougatef A, Naessens A, Lauwers S. 2001. Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J Clin Microbiol 39:293–297. doi: 10.1128/JCM.39.1.293-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullane N, O'Gaora P, Nally JE, Iversen C, Whyte P, Wall PG, Fanning S. 2008. Molecular analysis of the Enterobacter sakazakii O-antigen gene locus. Appl Environ Microbiol 74:3783–3794. doi: 10.1128/AEM.02302-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis KG, Grim CJ, Franco AA, Gopinath G, Sathyamoorthy V, Hu L, Sadowski JA, Lee CS, Tall BD. 2011. Molecular characterization of Cronobacter lipopolysaccharide O-antigen gene clusters and development of serotype-specific PCR assays. Appl Environ Microbiol 77:4017–4026. doi: 10.1128/AEM.00162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y, Wang M, Liu H, Wang J, He X, Zeng J, Guo X, Li K, Cao B, Wang L. 2011. Development of an O-antigen serotyping scheme for Cronobacter sakazakii. Appl Environ Microbiol 77:2209–2214. doi: 10.1128/AEM.02229-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y, Wang M, Wang Q, Cao B, He X, Li K, Feng L, Wang L. 2012. Genetic analysis of the Cronobacter sakazakii O4 to O7 O-antigen gene clusters and development of a PCR assay for identification of all C. sakazakii O serotypes. Appl Environ Microbiol 78:3966–3974. doi: 10.1128/AEM.07825-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller A, Stephan R, Fricker-Feer C, Lehner A. 2013. Genetic diversity of Cronobacter sakazakii isolates collected from a Swiss infant formula production facility. J Food Prot 76:883–887. doi: 10.4315/0362-028X.JFP-12-521. [DOI] [PubMed] [Google Scholar]
- 22.Blažková M, Javurková B, Vlach J, Göselová S, Karamonová L, Ogrodzki P, Forsythe S, Fukal L. 2015. Diversity of O antigens within the genus Cronobacter: from disorder to order. Appl Environ Microbiol 81:5574–5582. doi: 10.1128/AEM.00277-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Li C, Wu Q, Zhang J, Huang J, Yang G. 2015. Prevalence, molecular characterization, and antibiotic susceptibility of Cronobacter spp. in Chinese ready-to-eat foods. Int J Food Microbiol 204:17–23. doi: 10.1016/j.ijfoodmicro.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Yan Q, Wang J, Gangiredla J, Cao Y, Martins M, Gopinath GR, Stephan R, Lampel K, Tall BD, Fanning S. 2015. Comparative genotypic and phenotypic analysis of Cronobacter species cultured from four powdered infant formula production facilities: indication of pathoadaptation along the food chain. Appl Environ Microbiol 81:4388–4402. doi: 10.1128/AEM.00359-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ISO. 2006. ISO/TS 22964:2006 (IDF/RM 210:2006). Milk and milk products–detection of Enterobacter sakazakii. International Organization for Standardization, Geneva, Switzerland: http://www.iso.org/iso/iso_catalogue/catalogue_tc/catalogue_detail.htm?csnumber=41258. [Google Scholar]
- 26.Chen Y, Noe KE, Thompson S, Elems CA, Brown EW, Lampel KA, Hammack TS. 2012. Evaluation of a revised U.S. Food and Drug Administration method for the detection of Cronobacter in powdered infant formula: a collaborative study. J Food Prot 75:1144–1147. [DOI] [PubMed] [Google Scholar]
- 27.Iversen C, Lehner A, Mullane N, Marugg J, Fanning S, Stephan R, Joosten H. 2007. Identification of “Cronobacter” spp. (Enterobacter sakazakii). J Clin Microbiol 45:3814–3816. doi: 10.1128/JCM.01026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan Q, Fanning S. 2015. Strategies for the identification and tracking of Cronobacter species: an opportunistic pathogen of concern to neonatal health. Front Pediatr 3:38. doi: 10.3389/fped.2015.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan Q, Jarvis KG, Chase HR, Hebert K, Trach LH, Lee C, Sadowski J, Lee B, Hwang S, Sathyamoorthy V, Mullane N, Pava-Ripoll M, Iversen C, Pagotto F, Fanning S, Tall BD. 2015. A proposed harmonized LPS molecular-subtyping scheme for Cronobacter species. Food Microbiol 50:38–43. doi: 10.1016/j.fm.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Ogrodzki P, Forsythe S. 2015. Capsular profiling of the Cronobacter genus and the association of specific Cronobacter sakazakii and C. malonaticus capsule types with neonatal meningitis and necrotizing enterocolitis. BMC Genomics 16:758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hochel I, Škvor J. 2009. Characterisation of antibodies for the immunochemical detection of Enterobacter sakazakii. Czech J Food Sci 27:66–74. [Google Scholar]
- 32.Xu X, Zhang Y, Shi M, Sheng W, Du XJ, Yuan M, Wang S. 2014. Two novel analytical methods based on polyclonal and monoclonal antibodies for the rapid detection of Cronobacter spp.: development and application in powdered infant formula. LWT Food Sci Technol 56:335–340. doi: 10.1016/j.lwt.2013.11.028. [DOI] [Google Scholar]
- 33.Park S, Shukla S, Kim Y, Oh S, Hun Kim S, Kim M. 2012. Development of sandwich enzyme-linked immunosorbent assay for the detection of Cronobacter muytjensii (formerly called Enterobacter sakazakii). Microbiol Immunol 56:472–479. doi: 10.1111/j.1348-0421.2012.00466.x. [DOI] [PubMed] [Google Scholar]
- 34.Lugtenberg B, Van Alphen L. 1983. Molecular architecture and functioning of the outer membrane of Escherichia coli and other Gram-negative bacteria. Biochim Biophys Acta 737:51–115. doi: 10.1016/0304-4157(83)90014-X. [DOI] [PubMed] [Google Scholar]
- 35.Reeves PP, Wang L. 2002. Genomic organization of LPS-specific loci, p 109–135. In Hacker J, Kaper JB (ed), Pathogenicity islands and the evolution of pathogenic microbes, vol 1 Springer-Verlag, Berlin, Germany. [Google Scholar]
- 36.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 37.Lehner A, Nitzsche S, Breeuwer P, Diep B, Thelen K, Stephan R. 2006. Comparison of two chromogenic media and evaluation of two molecular based identification systems for Enterobacter sakazakii detection. BMC Microbiol 6:15. doi: 10.1186/1471-2180-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter L, Lindsey LA, Grim CJ, Sathyamoorthy V, Jarvis KG, Gopinath G, Lee C, Sadowski JA, Trach L, Pava-Ripoll M, McCardell BA, Tall BD, Hu L. 2013. Multiplex PCR assay targeting a diguanylate cyclase-encoding gene, cgcA, to differentiate species within the genus Cronobacter. Appl Environ Microbiol 79:734–737. doi: 10.1128/AEM.02898-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schauer K, Lehner A, Dietrich R, Kleinsteuber I, Canals R, Zurfluh K, Weiner K, Märtlbauer E. 2015. A Cronobacter turicensis O1 antigen-specific monoclonal antibody inhibits bacterial motility and entry into epithelial cells. Infect Immun 83:876–887. doi: 10.1128/IAI.02211-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dietrich R, Usleber E, Märtlbauer E. 1998. The potential of monoclonal antibodies against ampicillin for the preparation of a multi-immunoaffinity chromatography for penicillins. Analyst 123:2749–2754. doi: 10.1039/a805166f. [DOI] [PubMed] [Google Scholar]
- 41.Bremus A, Dietrich R, Dettmar L, Usleber E, Märtlbauer E. 2012. A broadly applicable approach to prepare monoclonal anti-cephalosporin antibodies for immunochemical residue determination in milk. Anal Bioanal Chem 403:503–515. doi: 10.1007/s00216-012-5750-z. [DOI] [PubMed] [Google Scholar]
- 42.Wang L, Hu X, Tao G, Wang X. 2012. Outer membrane defect and stronger biofilm formation caused by inactivation of a gene encoding for heptosyltransferase I in Cronobacter sakazakii ATCC BAA-894. J Appl Microbiol 112:985–997. doi: 10.1111/j.1365-2672.2012.05263.x. [DOI] [PubMed] [Google Scholar]
- 43.Kido N, Ohta M, Iida K, Hasegawa T, Ito H, Arakawa Y, Komatsu T, Kato N. 1989. Partial deletion of the cloned rfb gene of Escherichia coli O9 results in synthesis of a new O-antigenic lipopolysaccharide. J Bacteriol 171:3629–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hurrell E, Kucerova E, Loughlin M, Caubilla-Barron J, Forsythe SJ. 2009. Biofilm formation on enteral feeding tubes by Cronobacter sakazakii, Salmonella serovars and other Enterobacteriaceae. Int J Food Microbiol 136:227–231. doi: 10.1016/j.ijfoodmicro.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Mozrová V, Břeňová N, Mrázek J, Lukešová D, Marounek M. 2014. Surveillance and characterisation of Cronobacter spp. in Czech retail food and environmental samples. Folia Microbiol (Praha) 59:63–68. doi: 10.1007/s12223-013-0266-2. [DOI] [PubMed] [Google Scholar]
- 46.Pan Z, Cui JH, Lyu GP, Du XL, Qin LY, Guo YM, Xu BH, Li W, Cui ZG, Zhao C. 2014. Isolation and molecular typing of Cronobacter spp. in commercial powdered infant formula and follow-up formula. Foodborne Pathog Dis 11:456–461. doi: 10.1089/fpd.2013.1691. [DOI] [PubMed] [Google Scholar]
- 47.Xu X, Wu Q, Zhang J, Ye Y, Yang X, Dong X. 2014. Occurrence and characterization of Cronobacter spp. in powdered formula from Chinese retail markets. Foodborne Pathog Dis 11:307–312. doi: 10.1089/fpd.2013.1657. [DOI] [PubMed] [Google Scholar]
- 48.Willis LM, Whitfield C. 2013. Structure, biosynthesis, and function of bacterial capsular polysaccharides synthesized by ABC transporter-dependent pathways. Carbohydr Res 378:35–44. doi: 10.1016/j.carres.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 49.Jung JH, Choi NY, Lee SY. 2013. Biofilm formation and exopolysaccharide (EPS) production by Cronobacter sakazakii depending on environmental conditions. Food Microbiol 34:70–80. doi: 10.1016/j.fm.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 50.Westerman RB, He Y, Keen JE, Littledike ET, Kwang J. 1997. Production and characterization of monoclonal antibodies specific for the lipopolysaccharide of Escherichia coli O157. J Clin Microbiol 35:679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng SP, Tsui CO, Roberts D, Chau PY, Ng MH. 1996. Detection and serogroup differentiation of Salmonella spp. in food within 30 hours by enrichment-immunoassay with a T6 monoclonal antibody capture enzyme-linked immunosorbent assay. Appl Environ Microbiol 62:2294–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arbatsky NP, Sun Y, Shashkov AS, Wang M, Liu B, Daeva ED, Wang L, Knirel YA. 2012. Structure and genetics of the O-antigen of Cronobacter sakazakii G2726 (serotype O3) closely related to the O-antigen of C. muytjensii 3270. Carbohydr Res 355:50–55. [DOI] [PubMed] [Google Scholar]
- 53.MacLean LL, Vinogradov E, Pagotto F, Farber JM, Perry MB. 2009. Characterization of the O-antigen in the lipopolysaccharide of Cronobacter (Enterobacter) malonaticus 3267. Biochem Cell Biol 87:927–932. doi: 10.1139/O09-059. [DOI] [PubMed] [Google Scholar]
- 54.Law JWF, Ab Mutalib NS, Chan KG, Lee LH. 2015. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol 5:770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mullane NR, Ryan M, Iversen C, Murphy M, O'Gaora P, Quinn T, Whyte P, Wall PG, Fanning S. 2008. Development of multiple-locus variable-number tandem-repeat analysis for the molecular subtyping of Enterobacter sakazakii. Appl Environ Microbiol 74:1223–1231. doi: 10.1128/AEM.01726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Himelright I, Harris E, Lorch V, Anderson M, Jones T, Craig A, Kuehnert MT, Forster MA, Jensen B, Jernigan D. 2002. Enterobacter sakazakii infections associated with the use of powdered infant formula–Tennessee, 2001. MMWR Morb Mortal Wkly Rep 51:297–300. [PubMed] [Google Scholar]
- 57.Hartmann I, Carranza P, Lehner A, Stephan R, Eberl L, Riedel K. 2010. Genes involved in Cronobacter sakazakii biofilm formation. Appl Environ Microbiol 76:2251–2261. doi: 10.1128/AEM.00930-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stephan R, Lehner A, Tischler P, Rattei T. 2011. Complete genome sequence of Cronobacter turicensis LMG 23827, a food-borne pathogen causing deaths in neonates. J Bacteriol 193:309–310. doi: 10.1128/JB.01162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schubert RHW. 1968. The taxonomy and nomenclature of the genus Aeromonas Kluyver and van Niel 1936. Int J Syst Evol Microbiol 18:1–7. [Google Scholar]
- 60.Griffiths MS, Bosley JA. 1993. Assessment of macroporous polystyrene-based polymers for the immobilization of Citrobacter freundii. Enzyme Microb Technol 15:109–113. doi: 10.1016/0141-0229(93)90033-X. [DOI] [PubMed] [Google Scholar]
- 61.Stephan R, Grim CJ, Gopinath GR, Mammel MK, Sathyamoorthy V, Trach LH, Chase HR, Fanning S, Tall BD. 2014. Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris and Enterobacter turicensis as members of the genus Cronobacter and their reclassification in the genera Franconibacter gen. nov. and Siccibacter gen. nov. as Franconibacter helveticus comb. nov., Franconibacter pulveris comb. nov. and Siccibacter turicensis comb. nov., respectively. Int J Syst Evol Microbiol 64:3402–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tracey RP, Britz TJ, Van Vuuren HJJ, Lategan PM. 1986. Gas chromatographic determination of volatile metabolites formed by Obesumbacterium proteus and Hafnia species. J Inst Brew 92:446–451. [Google Scholar]
- 63.Lessel EF. 1971. Status of the name Proteus morganii and designation of the neotype strain. Int J Syst Evol Microbiol 21:55–57. [Google Scholar]
- 64.Skerman VBD, McGowan V, Sneath PHA. 1989. Approved lists of bacterial names. Int J Syst Bacteriol 30:225–420. [PubMed] [Google Scholar]
- 65.Spröer C, Mendrock U, Swiderski J, Lang E, Stackebrandt E. 1999. The phylogenetic position of Serratia, Buttiauxella and some other genera of the family Enterobacteriaceae. Int J Syst Evol Microbiol 49:1433–1438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.