ABSTRACT
Infectious salmon anemia virus (ISAV) is the etiological agent of the disease by the same name and causes major losses in the salmon industry worldwide. Epizootic ISAV outbreaks have occurred in Norway and, to a lesser degree, in Canada. In 2007, an ISAV outbreak in Chile destroyed most of the seasonal production and endangered the entire Chilean salmon industry. None of the existing prophylactic approaches have demonstrated efficacy in providing absolute protection from or even a palliative effect on ISAV proliferation. Sanitary control measures for ISAV, based on molecular epidemiology data, have proven insufficient, mainly due to high salmon culture densities and a constant presence of a nonpathogenic strain of the virus. This report describes an alternative treatment approach based on interfering peptides selected from a phage display library. The screening of a phage display heptapeptide library resulted in the selection of a novel peptide with significant in vitro antiviral activity against ISAV. This peptide specifically interacted with the viral hemagglutinin-esterase protein, thereby impairing virus binding, with plaque reduction assays showing a significant reduction in viral yields. The identified peptide acts at micromolar concentrations against at least two different pathogenic strains of the virus, without detectable cytotoxic effects on the tested fish cells. Therefore, antiviral peptides represent a novel alternative for controlling ISAV and, potentially, other fish pathogens.
IMPORTANCE Identifying novel methods for the efficient control of infectious diseases is imperative for the future of global aquaculture. The present study used a phage display heptapeptide library to identify a peptide with interfering activity against a key protein of the infectious salmon anemia virus (ISAV). A piscine orthomyxovirus, ISAV is a continuous threat to the commercial sustainability of cultured salmon production worldwide. The complex epidemiological strategy of this pathogen has made prophylactic control extremely difficult. The identified antiviral peptide efficiently impairs ISAV infection in vitro by specifically blocking hemagglutinin-esterase, a pivotal surface protein of this virus. Peptide synthesis could further modify the primary structure of the identified peptide to improve specific activity and stability. The present results form the foundation for developing a new pharmacological treatment against ISAV.
INTRODUCTION
Infectious salmon anemia (ISA) is a highly aggressive disease that primarily affects Atlantic salmon (Salmo salar), one of the most commercially relevant farmed fish. Outbreaks of ISA usually occur during the later stages of the production process, seriously impacting industry sustainability due to high mortality rates (1). The etiological agent of ISA is a relatively new virus within the Orthomyxovirus family and is the only member of the Isavirus genus (2). Termed the infectious salmon anemia virus (ISAV), this pathogen has been reported in all salmon-producing countries, most notably in Canada, Norway, Scotland, the Faroe Islands, and Chile (3–7). With specific regard to Chile, an epizootic ISA outbreak took place in 2007 that reduced salmon production by 64% (8).
This virus is horizontally transmitted from infected fish, either through water or living vectors, suggesting that transmission can occur between farms (9, 10). On the other hand, the recent detection of infective ISAV in the ovarian fluid and unfertilized eggs of systemically infected brood stocks devoid of clinical signs suggests the worrying possibility that vertical transmission may also occur (11). All epizootic outbreaks are caused by viral strains with deletions in a specific highly polymorphic region (HPR) of segment 6 in the viral genome (12). However, a nonpathogenic strain of ISAV that contains a full-length HPR (HPR0) has been consistently detected at fish farms, possibly representing the precursor strain from which the virulent types arise (13–15).
Continued growth in global demand pushes the aquaculture industry to culture fish at higher densities, which places fish under stress and, in turn, increases the risks of infection and disease. Stressful rearing conditions are particularly relevant for ISAV as the predominant HPR0 variant may undergo a deletion in the hemagglutinin-esterase (HE) gene, transitioning to an HPR-deleted type, as a result of infection and the high replication rate in immunosuppressed hosts, thus generating known or novel viral strains (15–17).
Efficient, continuous ISAV management and monitoring programs in culture centers, which use molecular procedures to characterize virus type and loads and which appropriately handle infected fish and mortalities, have helped attenuate the spread of this virus (18). As a result of these programs, 23% to 60% of Chilean fish farms have been protected, according to theoretical epidemiology and estimates based on reproduction numbers (19). Although vaccination is regularly an important component in fish health management, it has not provided the expected protection against ISAV. A reasonable explanation for this could be that confined, high-density growth conditions, among other aspects, may generate chronic stress, which could negatively impact primary and adaptive immune responses (20, 21).
Similarly to the influenza virus, the most studied of the members of the Orthomyxovirus family, ISAV attachment and fusion to the host cell are mediated by two viral membrane proteins, the joint activity of which confers infectivity potential (22). Specifically, HE is coded by genomic segment 6 and carries the receptor-binding domain (hemagglutinin) and receptor-destroying activity (esterase), much like influenza C virus (22). However, ISAV fusion activity is related to a second membrane-bound protein, which is termed the fusion protein (F) and is coded by segment 5 (23). The two proteins contain polymorphic regions that appear related to the pathogenicity and virulence potential of different strains. Specifically, insertions flanking the putative cleavage site in the F protein and/or deletions in the HPR of the HE protein are recognized molecular markers for high virulence (24, 25). Also, similarly to the influenza virus, the viral cellular receptor of ISAV is an N-acetylneuraminic acid, or sialic acid (Sia) (26).
A novel alternative to prophylactic measures, such as vaccines and enzymatic inhibitors, is the use of viral entry blockers. Sialic acid-containing polymers and lipids can block the interaction between HE and its receptor in the influenza virus (27). Furthermore, peptides derived from the suppressor of cytokine signaling-1 protein and a fibroblast growth factor have shown in vivo antiviral activity against various influenza strains from different H subtypes, inhibiting the adhesion of viral particles to target cells (28, 29). In a more straightforward approach, Sia-mimicking peptides identified from a peptide library act against H1 and H3 type influenza strains at micromolar levels (27). Considering the high conservation of the receptor-binding site on different ISAV strains, this type of peptide may be an ideal tool for developing novel antiviral strategies against this virus. This alternative offers the additional benefit of peptide synthesis plasticity, which allows the chemical introduction of modifications to improve molecule functionality for a particular application.
In this study, a phage display heptapeptide library was screened to identify molecules that specifically interact with ISAV HE or F and that could interfere with the functions of these proteins. The selected potential peptide sequences were chemically synthesized and tested in vitro for antiviral activity. Through this approach, a novel peptide molecule with remarkable in vitro antiviral activity against different ISAV strains was characterized. This peptide specifically interacted with the ISAV HE protein, impairing virus adhesion and cell infection and resulting in more than 90% viral inhibition. Given its infection-preventing nature, this novel peptide provides an attractive starting point for the further development of antiviral solutions in aquaculture, particularly for alternatives independent of the fish immune system.
MATERIALS AND METHODS
Fish cell and ISAV cultures.
Atlantic salmon kidney (ASK) cells (ATCC CRL2747) (30) were cultured at 20°C in Leibovitz's L-15 media with glutamine (Gibco) and supplemented with 200 U ml−1 penicillin, 200 μg ml−1 streptomycin, 0.5 μg ml−1 amphotericin, and 10% fetal bovine serum (Gibco). Cells were grown to 80% confluence and subcultured accordingly.
Field isolates of ISAV corresponding to the HPR3 and HPR7b types were obtained from the Laboratorio de Genetica e Inmunologia Molecular. For viral infection and propagation, an 80% confluent ASK cell monolayer was washed twice with L-15 media and covered with a viral dilution prepared in L-15 media. After 4 h, the virus was removed, and the cells were washed twice with L-15 media and cultured in 2% fetal bovine serum L-15 media with antibiotics at 17°C. After 7 days, the cell supernatant was filtered (using a 0.45-μm-pore-size filter), and the virus was collected. A plaque assay was used for virus titer determinations 12 days postinfection (dpi), as previously described (31).
Spodoptera frugiperda (Sf21) cells and baculovirus.
Spodoptera frugiperda (Sf21) cells were grown in adherent and suspension cultures using SF-900 III media (Gibco) supplemented with 1% fetal bovine serum at 28°C. Suspension cells were seeded at 0.3 × 106 cells ml−1, agitated at 150 rpm in 500-ml flasks, and subcultured upon reaching 8 × 106 to 9 × 106 cells ml−1. Adherent cells were cultured in 75-cm2 culture bottles and subcultured at 80% confluence.
Cloning and recombinant baculovirus (rBaculovirus).
The ISAV HE and F genes were amplified through PCR using a cDNA sample from an ISAV HPR7b infection in ASK cells. Three copies of the FLAG peptide sequence (3×FLAG) were added to the 3′ end of the open reading frames. The 3×FLAG sequence was amplified from the p3×FLAG cytomegalovirus (CMV) 14 vector (Sigma-Aldrich). The 3×FLAG-tagged HE and 3×FLAG-tagged F sequences were cloned into the pFastBac1 donor vector between the NotI/XhoI and EcoRI/XhoI sites, respectively. Accordingly, PCR primers were designed to add restriction sites to the constructs (Table 1). The PCRs were performed in a 20-μl reaction volume using a cDNA sample (1 μl) from infected cells or 5 ng of plasmid as the template, 1× HF Phusion buffer (NEB), 200 μM deoxynucleoside triphosphates (dNTPs), a 0.5 μM concentration of each primer, 1 M betaine, and 1 U of Phusion DNA polymerase (NEB). The thermal conditions for PCR were as follows: 3 min at 95°C, followed by 40 cycles of 30 s at 93°C, 30 s at 55°C, and 2 min at 72°C, with a final extension stage of 5 min at 72°C. RNA from ISAV-infected cells was extracted 12 dpi using TRIzol reagent (Life Technologies) according to the manufacturer's instructions. cDNA was synthesized from 5 μg of total RNA with Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen) using 500 ng of oligo(dT)12–18, a 1 mM concentration of each dNTP, and 200 U of M-MLV reverse transcriptase according to the manufacturer's instructions. The PCR products were cloned in the pCR2.1 vector using a TOPO TA cloning kit (Invitrogen). The cloned sequences were verified by sequencing (Macrogen, South Korea) using M13 primers. After digestion with restriction enzymes (Promega) and gel electrophoresis, the gene fragments and the linearized vector were purified with a GeneJET gel extraction kit (Thermo Fisher). A mixture containing 10 ng of the pFastBac1 vector, 50 ng of the HE or F gene insertion, and 10 ng of the p3×FLAG sequence was ligated using a rapid DNA ligation kit (Thermo Fisher) according to the manufacturer's instructions. Recombinant plasmids were isolated from transformed DH5α cells grown in LB media with a GeneJET plasmid DNA kit (Thermo Fisher).
TABLE 1.
Primers used to amplify the open reading frames from segments 5 and 6 of ISAV and to add restriction sites for cloninga
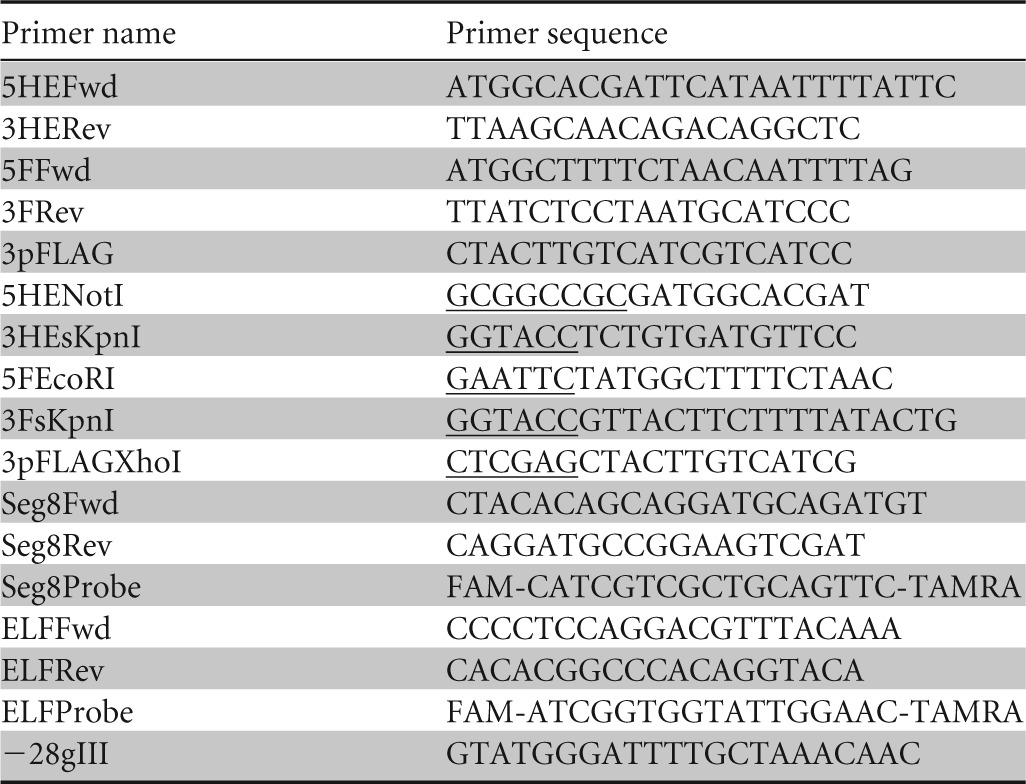
The segment 8 (Seg8) and elongation factor 1α (ELF) primers were used for gene expression analyses. The −28gIII primer was used for phage-DNA sequencing. The restriction sites for cloning are underlined. FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
Recombinant bacmids (rBacmid) were obtained through site-specific recombination of the donor plasmids pFastBac_ISAVHE and pFastBac_ISAVF with a baculovirus shuttle vector contained in DH10Bac cells (Invitrogen), using the standard procedure described by the manufacturer. Recombinant bacmids were purified from a 50-ml culture of these cells on LB media using an E.Z.N.A. BAC/PAC DNA Maxi kit (Omega).
Recombinant baculovirus (rBaculovirus) was obtained by transfecting Sf21 cells with rBacmids. After 72 h, the supernatant was collected, centrifuged at 500 × g for 5 min to remove cells and debris, and used to infect new Sf21 cells on a 6-well plate. For viral amplification, cells were infected at a multiplicity of infection (MOI) of 0.05 until a third round of infection, reaching approximately 1 × 108 PFU ml−1. Viral titers were determined with standard plaque assay methods and Neutral Red staining (Sigma-Aldrich). Clear supernatants from the third round of infection were termed rBacHE and rBacF, corresponding to HE and F rBaculoviruses, respectively. The expression levels of recombinant HE and recombinant F (rHE and rF, respectively) were determined at 24, 72, and 120 h postinfection (hpi) in Sf21 cells grown on 6-well plates at 23°C and infected at an MOI of 10. Cells from each well were washed twice with Grace's insect medium (Gibco) and suspended in 100 μl of lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100). After incubation for 2 h on ice, the samples were sonicated at 20% amplitude for five 30-s repetitions, with 30 s of cooling between repetitions. The samples were then centrifuged at 12,500 × g for 30 min; the supernatant was recovered, and total protein concentration was measured using a bicinchoninic acid (BCA) assay kit and bovine serum albumin (BSA) standard (Pierce). Medium samples from each well were obtained and clarified by centrifugation at 500 × g for 5 min, and total protein content was measured as described above. Cells and medium samples corresponding to 5 μg of total protein were loaded on a 15% SDS-PAGE gel using NuPAGE lithium dodecyl sulfate (LDS) sample buffer (Life Technologies) and, after electrophoresis, were either stained with Coomassie brilliant blue G-250 or transferred to a nitrocellulose membrane (NitroBind). Western blot analysis was performed using anti-FLAG M2 antibody (Sigma) at a 1:100 dilution or anti-ISAV HE (8D2/E9) or anti-ISAV F (8B2/A4) monoclonal antibody (GrupoBios) at a 1:250 dilution and, as a secondary antibody, horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (Thermo) at a 1:7,500 dilution. Membranes were revealed using a Westar Supernova kit (Cyanagen) according to the manufacturer's instructions. The optimal time of infection for protein expression was 120 hpi, as determined by Western blotting and cell viability analyses (CellTiter 96 AQueous One Solution cell proliferation assay; Promega).
Recombinant baculoviruses were used to infect 500 ml of suspension-grown Sf21 cells at 6 × 106 ml−1 and 23°C using an MOI of 10. After 120 h, the cells were pelleted at 500 × g, washed with Grace's insect medium, and suspended in lysis buffer, as described above. After incubation for 2 h on ice, the samples were sonicated at 20% amplitude for five 30-s repetitions, with 30 s of cooling between repetitions. The samples were then centrifuged at 12,500 × g for 30 min. The supernatant was filtered through a 0.45-μm-pore-size low-protein-binding filter (Edlab). The rHE and rF were purified using an anti-FLAG M2 magnetic beads system (Sigma-Aldrich) and a Magnetight separator stand (Novagen) according to the manufacturer's instructions. Protein concentration was measured using a BCA assay kit and a BSA standard (Pierce). Purified proteins were further analyzed using SDS-PAGE and Western blotting, as described above.
Phage heptapeptide library screening.
For the biopanning process, 50 μg of rHE or rF was used as a target. The recombinant proteins were incubated overnight in coating buffer (0.1 M NaHCO3 [pH 8.6]) in a 15-by-60-mm polystyrene petri dish. After the solution was removed, the coating buffer plus 5% BSA was used to block nonspecific binding sites through incubation for 1 h at 4°C. A 1-ml dilution containing 2 × 1011 PFU from the Ph.D.-7 phage display library (New England BioLabs) was prepared in Tris-buffered saline (TBS)–0.1% Tween 20 washing buffer and was then applied to the coated plate. After 1 h of incubation on a rocking platform, nonbinding phages were discarded, and the plate was washed 10 times with the TBS–0.1% Tween 20 washing buffer. Specific interacting phages were eluted from the plate using 1 ml of 0.2 M glycine-HCl (pH 2.2) and 1 mg ml−1 BSA. After 10 min of incubation, the solution was recovered and neutralized with 150 μl of 1 M Tris-HCl (pH 9.1). Eluted phage viruses were amplified and titers were determined according to standard methods using Escherichia coli ER2738 cells (32). To improve procedure selectivity, second and third rounds of panning were carried out using 2 × 1011 PFU of the amplified phages recovered from the previous round, newly prepared rHE- or rF-coated plates, and TBS–0.5% Tween 20 washing buffer. Titer plaques from the third round of interacting phages were used to amplify specific clones in ER2738 cells. Phage DNA was purified using previously described methods (33). Briefly, 20% polyethylene glycol (PEG)–2.5 M NaCl was used to precipitate the phages; an iodide buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 4 M sodium iodide) was used to suspend the pellet, and a 15-min incubation period at room temperature performed with absolute ethanol was used to precipitate single-stranded DNA. The peptide sequence of individual clones was determined by sequencing using the −28 gIII primer (Table 1) according to the manufacturer's instructions.
Peptide synthesis.
Peptides were synthesized with the solid-phase multiple-peptide system (34) using Fmoc amino acids (Iris Biotech) (0.65 meq/g of Rink Resin). The peptides were cleaved using trifluoroacetic acid–triisopropylsilane–1.2-ethandithiol–ultrapure water (TFA/TIS/EDT/H2O), in volume proportions of 92.5, 2.5, 2.5 and 2.5, and purified by reversed-phase high-performance liquid chromatography (RP-HPLC) with a 0% to 70% acetonitrile-water mixture gradient for 30 min at a flow rate of 1 ml min−1. The peptides were lyophilized and analyzed using electrospray ionization-mass spectrometry (ESI-MS) to confirm molecular masses. All peptides were tested for cytotoxicity in ASK cells (72 h of incubation at 1 mM) using a cell viability analysis system (CellTiter 96 AQueous One Solution cell proliferation assay; Promega) according to the manufacturer's instructions.
RT-PCR and plaque reduction assays.
For qualitative real-time PCR (qRT-PCR) assays, 2,500 PFU of ISAV was used to infect an 80% confluent monolayer well of ASK cells on a 6-well plate (∼2.5 × 105 cells), as described above (MOI of 0.01). Peptides in concentrations ranging from 0 to 500 μM were used to incubate the virus or cells in three different assays. In detail, the following four groups for ISAV infection were used, as indicated: (i) the ISAV inoculum was incubated with the peptide 30 min prior to cell infection (Pre Inc Virus); (ii) the cells were incubated with each peptide for 30 min prior to ISAV infection (Pre Inc Cells); (iii) the cells were treated for 30 min with the peptide after ISAV infection (Post Inc Cells); and (iv) as a control group, neither the ISAV inoculum or the cells were subjected to pre- or posttreatment with the peptide. The cells were washed twice with L-15 media after ISAV challenges or peptide incubations. RNA was extracted from the cells 48 hpi using TRIzol reagent (Life Technologies) and suspended in 20 μl of diethylpyrocarbonate water. An RNA sample (2 μl) was used as a template for the qRT-PCRs with Stratagene qRT-PCR III master mix according to the manufacturer's instructions. Primers were used to analyze the expression of viral genes (segment 8) (18) and of a cellular housekeeping gene (elongation factor 1α gene) (35). The fold change of viral gene expression relative to that seen with untreated controls was assessed using the threshold cycle (2−ΔΔCT) method, as previously described (36) (Table 1).
Plaque assays were used to analyze the influence of the peptides on viral infection. The titer of a 250-PFU viral inoculum was determined after 30 min of incubation with the peptides at concentrations ranging from 0 to 500 μM, as previously described (31). The plaques were counted at 12 dpi. The percentage of viral inhibition was calculated as 100(1 − N/N0), where N and N0 are the numbers of plaques in the presence and absence of peptide treatment, respectively (27). The 50% inhibitory concentration (IC50) value was obtained by interpolating the plot of log f/(1 − f) versus log[peptide], where f is the fraction of infection activity relative to the number of plaques obtained without the peptide (f = 1) (37).
Hemagglutination inhibition assays.
ASK cells grown in a 75-cm3 culture bottle were infected with ISAV HPR3 or HPR7b at an MOI of 0.01 as described above. After 7 days, the culture medium was removed, and cells were washed twice with L-15 media. The cells were suspended in 500 μl of phosphate-buffered saline (PBS) using a cell scraper. After three rounds of freezing-thawing, cell debris were removed through centrifugation at 12,000 × g for 10 min. The HE-containing supernatant was collected and stored at −80°C until use.
Prior to blood collection, 10 μl of an anticoagulant solution containing 10 mM sodium citrate was added to syringes and collection tubes. Then, blood samples (500 μl) were obtained from presmolt Salmo salar (40 g) after appropriate sedation in 0.001% benzocaine (Centrovet) for 10 min. Red blood cells (RBCs) were obtained via sedimentation after centrifugation at 110 × g for 10 min. Cells were washed three times with PBS and resuspended in the same buffer to obtain a 0.5% RBC suspension.
For the hemagglutination assay, a round-bottomed 96-well plate was loaded with 50 μl of PBS in each well. Serial 1:2 dilutions were prepared by pipetting 50 μl of the previously detailed HE-containing extract and 50 μl of PBS on each well, recovering 50 μl, and mixing again in the next well. The last well was used as a negative control (PBS only). Each well received an additional 50 μl of the 0.5% RBC suspension and was incubated for 1 h. The RBCs appeared as a diffuse suspension in hemagglutination-positive wells, while the cells formed a notable layer of sediment at the bottom of the well in hemagglutination-negative wells. The hemagglutination titer was calculated as the reciprocal dilution of the last well with a positive result, expressed as HE units per milliliter. For the hemagglutination inhibition assay, a 50-μl aliquot of the HE extract, at 160 HE U ml−1, was incubated with 0, 0.5, 5, 50, or 500 μM peptide for 30 min, followed by serial dilutions and RBC incubation as described above.
Statistical analysis.
The results are presented as the means ± standard deviations of the results of triplicate determinations. The statistical significance of the data was determined by using Student's t test. P values of <0.01, indicated by an asterisk (*), were considered significant.
RESULTS
Efficient recovery and expression of recombinant ISAV rHE and rF proteins.
Recombinant baculovirus-infected Sf21 cells were analyzed through Western blotting (Fig. 1). Viral rHE and rF proteins were clearly detected at 72 and 120 hpi, displaying the corresponding expected masses of 38.7 and 46.9 kDa, respectively (Fig. 1A). Since cell viability (above 13 × 106 cells ml−1) decreased soon after this, 120 hpi was identified as the harvesting time for protein expression, with no recombinant proteins detected in the culture media (data not shown). The purity of immunopurified rHE and rF proteins was confirmed by SDS-PAGE (Fig. 1B). Western blot analysis of the purified proteins, using ISAV HE- and F-specific antibodies, confirmed the protein identities (Fig. 1C).
FIG 1.
Recombinant protein expression evaluated using the baculovirus system. (A) Western blot analyses of rBaculovirus-infected Sf21 cells, using the anti-FLAG antibody and different sampling times postinfection. (B) Purified rHE and rF (100 ng) were analyzed on a stained SDS-PAGE gel. (C) Purified proteins were analyzed by Western blotting using anti-ISAV HE- and F-specific antibodies.
Identification of rHE- or rF-interacting phage-displayed peptides.
Purified rHE or rF was used to screen a phage display heptapeptide library. Following the third round of panning, 78 phage clones interacting with each protein were sequenced. After redundant sequences related to nonspecific interactions with the target in the rHE- and rF-interacting phages were removed, seven different peptide sequences were found for rHE and six for rF. Each sequence had a different number of reads from the total selected, and the difference was expressed as a representation percentage. The 13 peptide sequences were chemically synthesized and labeled GIMxxxx (Table 2). The de novo synthesized peptides did not induce any cytotoxicity in ASK cells in concentrations of up to 1 mM (2-fold the obtained IC50; data not shown).
TABLE 2.
Synthesized peptide sequences, targets, molecular weights, and percent representation within the total number of peptides for each target
| Peptide | Target | Sequence | Mol wt | Representation (%) |
|---|---|---|---|---|
| GIM2083 | HE | VVPWAGL | 739.92 | 12.5 |
| GIM2084 | HE | WTVHTLS | 841.97 | 25 |
| GIM2085 | HE | HMFPWRQ | 1,000.2 | 12.5 |
| GIM2086 | HE | QSWLPSL | 828.97 | 8.4 |
| GIM2087 | HE | GYKDFSA | 785.86 | 16.6 |
| GIM2088 | HE | RILITIP | 824.08 | 12.5 |
| GIM2089 | HE | WASKVAR | 815.97 | 8.4 |
| GIM2090 | F | TPARHIY | 856 | 16.6 |
| GIM2091 | F | TLLRVDN | 828.97 | 11.1 |
| GIM2092 | F | GSGNAFM | 681.77 | 27.7 |
| GIM2093 | F | MPLGFKA | 761.99 | 16.6 |
| GIM2094 | F | NERALTL | 814.94 | 16.6 |
| GIM2095 | F | VQPIPAT | 723.87 | 11.1 |
Screening for antiviral activity of the selected peptides.
All 13 peptides were initially analyzed for antiviral activity against the highly virulent HPR3 ISAV strain by using qRT-PCR to assess viral gene expression. The virus was incubated with a 50 or 500 μM concentration of each peptide for 30 min prior to infection of ASK cells. Then, the qRT-PCR inhibitory profiles of different peptides at 48 hpi were compared to data from an untreated control (Fig. 2).
FIG 2.
Peptide inhibition of ISAV infection with an HPR3 strain in ASK cells at 48 h postinfection. The antiviral activity of all peptides was tested using qRT-PCR to analyze the expression of viral segment 8 and the elongation factor 1α (ELF) housekeeping gene in cells. CT values for segment 8 and the ELF gene were used to calculate fold changes in expression of ISAV relative to the untreated-control levels (No Pep). Two peptide concentrations were used to analyze the effect of the dose. The horizontal dashed line indicates the expression level for the untreated control. *, P < 0.01.
Of the chemically synthesized peptides, only the HE-specific GIM2084 peptide showed significant activity against ISAV infection, inhibiting the expression of genomic segment 8 in a dose-dependent manner. Viral gene expression decreased to 32% and 21% at GIM2084 concentrations of 50 and 500 μM, respectively, relative to the untreated control results. In contrast, the GIM2083, GIM2085, GIM2086, GIM2087, GIM2088, GIM 2092, GIM2094, and GIM2095 peptides significantly and positively affected the expression of viral genes, with up to a 4-fold change relative to the control (GIM2088 at 50 μM). Finally, the GIM2089, GIM2090, GIM2091, and GIM2093 peptides had no significant effects on the expression of viral genes (Fig. 2).
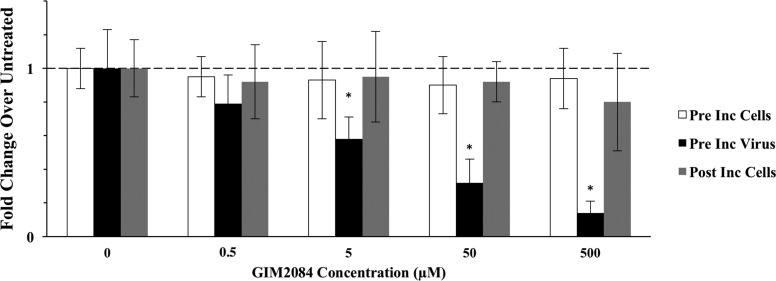
To further define the antiviral activity displayed by GIM2084, either the viral inoculum (HPR3 strain) or the target ASK cells were exposed to different concentrations of the peptide, either before or after infection. The inhibitory effect displayed by GIM2084 (i.e., decreased viral gene expression) was detected only when the virus was exposed to the peptide prior to cell infection (Fig. 3).
FIG 3.
Antiviral activity of the GIM2084 peptide prior or posterior to ISAV HPR3 infection. Different concentrations of GIM2084 were used to treat cells before infection (white bars), to treat the virus before infection (black bars), or to treat cells after infection (gray bars). The expression levels of segment 8 and ELF were used to calculate fold changes, using the untreated virus as a control (concentration = 0). The horizontal dashed line indicates the expression level for the untreated control. *, P < 0.01.
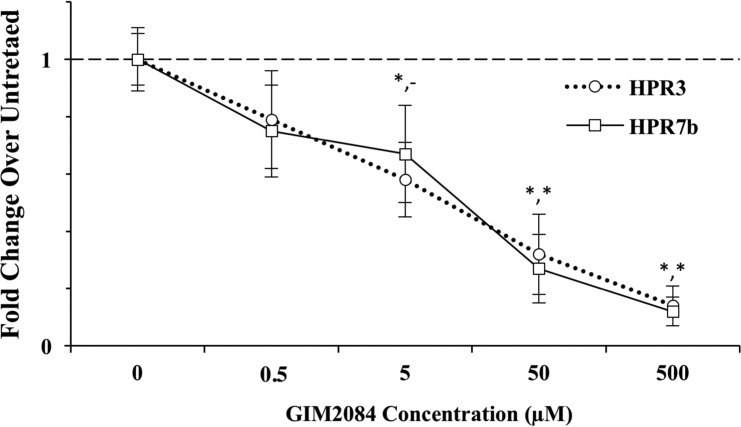
To evaluate the antiviral activity of GIM2084 against other ISAV strains, ASK cells were infected with peptide-pretreated ISAV HPR7b. The expression levels of ISAV in ASK cells infected with peptide-pretreated HPR3 and HPR7b strains were compared to the levels seen with an untreated control. Both viral strains were inhibited in a dose-dependent manner (Fig. 4).
FIG 4.
GIM2084 peptide activity against ISAV strains HPR3 and HPR7b. Virus samples were pretreated with different concentrations of GIM2084. The expression levels of segment 8 and ELF were used to calculate fold changes, using the untreated virus as a control. The horizontal dashed line indicates the expression level for the untreated controls. *, P < 0.01 (with *,- indicating significance for only HPR3 and *,* indicating significance for both HPR3 and HPR7b).
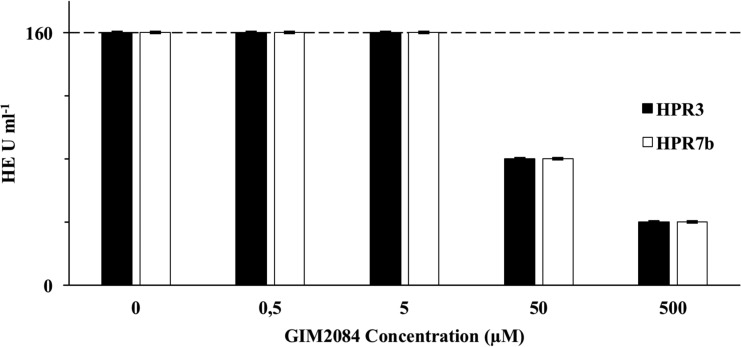
A hemagglutination inhibition assay confirmed the interaction of GIM2084 with viral HE. The peptide impaired viral protein interaction with its receptor on the surface of salmon RBCs (Fig. 5). Identical dose-dependent responses were found for the HPR3 and HPR7b viral strains, with a 4-fold reduction on hemagglutination units at 500 μM for both strains.
FIG 5.
Hemagglutination inhibition by GIM2084. A protein extract from ISAV-infected ASK cells, containing 160 HE U ml−1, was incubated with different concentrations of GIM2084 prior to a hemagglutination assay. The inhibitory effect was tested in HE-containing extracts obtained from cells infected with the HPR3 or HPR7b ISAV strain. Three replicates of the assay were performed, with exactly the same results. The horizontal dashed line indicates the hemagglutination unit concentrations for the untreated control.
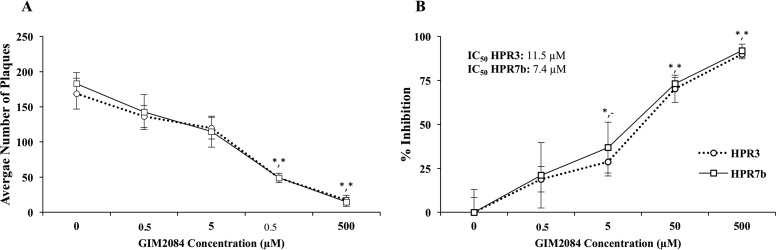
To functionally evaluate the antiviral activity of GIM2084, plaque reduction assays were performed with peptide-pretreated HPR3 and HPR7b ISAV strains. The inhibitory profiles for both strains showed a dose-dependent reduction in the number of PFU, where the IC50s for HPR3 and HPR7b were 11.5 and 7.4 μM, respectively (Fig. 6).
FIG 6.
GIM2084 peptide plaque reduction activity against two strains of ISAV. The HPR3 (○) and HPR7b (□) viral strains were incubated with different concentrations of GIM2084 prior to cell infection. (A) A higher peptide concentration was related to a lower number of plaques for both strains. (B) The proportions of inhibition reached 89% and 91% for HPR3 and HPR7b, respectively, at the highest concentration of peptide (500 μM). IC50s are indicated for both viral strains. *, P < 0.01 (with *,- indicating significance for only HPR3 and *,* indicating significance for both HPR3 and HPR7b).
DISCUSSION
The present study identified a novel peptide inhibitor of ISAV (GIM2084) that displayed significant activity against two different strains of this virus at micromolar concentrations. Specifically, the identified peptide targeted the surface HE protein, known to be pivotal for viral infectivity. This novel molecule was selected after screening a phage-displayed heptapeptide library, in a procedure specifically directed against viral surface proteins expressed in a heterologous system. Once the primary sequence of the selected peptide was determined, mass production was achieved through chemical synthesis, with synthetic peptides fully conserving the desired specific interaction.
A key element for biopanning is the use of appropriate mimics with structural properties as similar as possible to those of the original target molecule. For the influenza virus, high quantities of purified particles are used as targets in biopanning (27, 38, 39). Considering that the growth of ISAV is not comparable with that of the influenza virus, a feasible alternative is to use heterologous expression levels and to purify the selected proteins for use as targets in biopanning. Heterologous expression of ISAV HE was previously reported using the baculovirus system, with high yields of a functional protein in terms of receptor binding and destroying activities (40). This system was used in the present study and was expanded to include the expression of the F protein from ISAV, another viral membrane protein widely recognized as a virulence marker. The F protein participates in the virus-cell fusion process and is activated by cellular proteases and a low pH inside the endosome (23). This key role makes this protein an interesting target for developing inhibitory molecules.
Both HE and F were efficiently expressed in the baculovirus system, with no apparent secretion of the recombinant proteins into the culture media (data not shown). Approximately 50 μg of each protein was obtained from 300 ml of suspension-grown Sf21 cells (data not shown). SDS-PAGE and Western blot analyses using monoclonal antibodies against ISAV HE and F, as well as the determined theoretical molecular weights, validated the identities of both recombinant proteins (Fig. 1B and C).
Specific clones from a phage display heptapeptide library interacted with rHE and rF. Although 20 different peptide sequences were obtained after three consecutive biopanning rounds, only seven and six phage clones specifically interacted with rHE and rF, respectively. On the other hand, seven peptide sequences were found in the phages of both proteins and were attributed to nonspecific interactions. Each specific and nonspecific sequence corresponded to a different representation percentage of the total number, which may have been due to different affinities for the target proteins (27) (Table 2). No common sequence or pattern was found among the peptides, probably due to exclusive interaction sites for each peptide on the respective target protein. All peptides were successfully synthesized using the solid-phase multiple-peptide system (34).
Initially, qRT-PCR was used to analyze the potential antiviral activity of all chemically synthesized peptides, evaluating viral gene expression in ASK cells after infection with peptide-treated or -untreated samples. This assay showed the differentiated impact of each peptide on ISAV gene expression in vitro. Treatment with 8 of the 13 selected peptides resulted in enhanced expression of viral segment 8, with up to a 4-fold increase relative to the untreated control (GIM2088) (Fig. 2). Since the peptide library screening process was not specifically designed to obtain only HE- or F-blocking peptides but simply to obtain interacting peptides, there was no initial discrimination to prevent the selection of infection-enhancing molecules. Interestingly, most of the synthesized peptides do have an impact on ISAV gene expression, reinforcing the idea that there is some level of interaction with the virus, thus validating the present experimental approach.
Although unexpected, the observed positive effect of some peptides on ISAV infection merits a few words. In particular, this effect could be related to the conformational dynamics that both ISAV membrane proteins need to acquire to promote cell receptor recognition and trigger infection. During cell surface adhesion and activation of the fusion mechanism, the ISAV HE and F proteins become physically separated from each other on the viral surface. This is a key step for viral entry into the cell (41). In relation to the presently observed positive effect, it is reasonable to hypothesize that a peptide interacting with either HE or F prior to viral adhesion and cell infection might affect the dynamics between these two proteins, enhancing protein dissociation and selectively modifying the fusion and viral entry processes. Therefore, although the discovery of these virus-enhancing peptides was not the aim of this study, this knowledge contributes to our understanding of viral infection, which could be useful in future research.
One peptide, GIM2084, had a significant inhibitory effect on viral gene expression (Fig. 2). This peptide was highly represented among the initially selected clones (25% of total reads; Table 2). This characteristic may constitute an additional feature for further selecting interfering peptides (27). This observed high representation may be related to a high affinity for the target protein (HE), as demonstrated by a dose-dependent inhibitory effect and the low IC50 (11.5 μM) for the HPR3 strain (Fig. 2 and 6). There was a specific dependence of the inhibitory effect on the time of peptide exposure to the virus (i.e., pre- versus postinfection). Clearly, GIM2084 inhibited infection only when it was preincubated with the virus (Fig. 3), suggesting a direct interaction with the viral particle. Indeed, plaque reduction analysis sustained the significant effect of GIM2084 on the number of PFU, supporting the interpretation that the action mechanism is not related to the inhibition of viral gene expression but rather to the inactivation of infective viral particles. Plaque reduction analysis further demonstrated the antiviral activity of the peptide, with plaque formation inhibition reaching up to 89% and 91% for the aggressive HPR3 and HPR7b strains, respectively, at a peptide concentration of 500 μM (Fig. 6). The peptide showed similar dose-dependent effects for the two viral strains, with IC50s of 11.5 and 7.4 μM for HPR3 and HPR7b, respectively.
Although no experimental structural information has been reported for ISAV HE, bioinformatics analysis has identified some key elements on the primary structure of this protein, with a putative receptor-binding site being highly conserved among different strains of ISAV (22). Interestingly, GIM2084 presented similar activities in regard to gene expression and plaque reduction against the two tested strains, suggesting that the interaction site with the target protein (HE) is conserved (Fig. 4 and 6). Antiviral peptides have already been developed for influenza viruses (28, 29, 39), the action mechanisms of which block the Sia-binding site on HE (27). Given that the antiviral activity of GIM2084 is dependent on the time of application (i.e., before viral adhesion) and that the site of interaction on HE seems to be conserved among different strains, blockage of the Sia-binding site on ISAV HE may also be the action mechanism of GIM2084 (18). Hemagglutination inhibition assays supported the idea that GIM2084 has a dose-dependent, negative influence on the HE-receptor interaction in salmon RBCs, reducing the HE U ml−1 from 160 to 40 at 500 μM. These effects were identical for the two viral strains (Fig. 5), reinforcing the idea of an interaction site conserved among different HEs and suggesting that this may correspond to the highly conserved Sia-binding site.
In summary, peptide library screening for rHE and rF from ISAV was successfully used to find a molecule that blocked the initial events of viral infection in vitro. As determined by hemagglutination inhibition and plaque reduction assays, a chemically synthetized peptide specifically interacted with HE and blocked viral protein interaction with cell receptors. The impairment of viral entry is an interesting approach for viral treatments, especially considering that this technique could prevent the spread of disease and subsequent viral evolution, particularly for a waterborne virus such as ISAV.
Putative interaction with the pivotal HE protein constitutes an attractive strategy that should be considered in the development of peptide-driven prophylactic measures against viral pathogens in aquaculture. Considering that peptide-coated surfaces with antibacterial activity have shown promising results (42, 43), the capture of ISAV on peptide-loaded surfaces (e.g., nets) and the eventual inactivation of the virus in the water are possible options for the effective use of these molecules in fish farms (44). Given its action mechanism (i.e., interaction with viral surface proteins, blocking the receptor-binding site), a modified version of GIM2084 could be an ideal candidate for these types of applications. Moreover, the chemical synthesis of peptides certainly offers the possibility of designing and modifying molecules according to specific needs. Mutated versions of the original peptide could be developed to further explore the action mechanisms of GIM2084 and the importance of specific peptide residues in interactions with HE (e.g., alanine scanning). Based on these results, future research will focus on improving the antiviral activity of GIM2084 and/or on developing appropriate delivery systems for in vivo application.
ACKNOWLEDGMENTS
We gratefully acknowledge Cristian Muñoz and Omar Luna for their excellent technical assistance.
REFERENCES
- 1.Vike S, Duesund H, Andersen L, Nylund A. 2014. Release and survival of infectious salmon anaemia (ISA) virus during decomposition of Atlantic salmon (Salmo salar L.). Aquaculture 420–421:119–125. doi: 10.1016/j.aquaculture.2013.09.043. [DOI] [Google Scholar]
- 2.Krossøy B, Hordvik I, Nilsen F, Nylund A, Endresen C. 1999. The putative polymerase sequence of infectious salmon anemia virus suggests a new genus within the Orthomyxoviridae. J Virol 73:2136–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullins JE, Groman DB, Wadowska D. 1998. Infectious salmon anaemia in salt water Atlantic salmon (Salmo salar L.) in New Brunswick, Canada. Bull Eur Assoc Fish Pathol 18:110–114. [Google Scholar]
- 4.Thorud K, Djupvik HO. 1988. Infectious anaemia in Atlantic salmon (Salmo salar L.). Bull Eur Assoc Fish Pathol 8:109–111. [Google Scholar]
- 5.Rodger HD, Turnbull T, Muir F, Millar S, Richards RH. 1998. Infectious salmon anaemia (ISA) in the United Kingdom. Bull Eur Assoc Fish Pathol 18:115–116. [Google Scholar]
- 6.Anonymous. 2000. ISA hits the Faroes. Fish Farming Int 27:47. [Google Scholar]
- 7.Godoy MG, Aedo A, Kibenge MJT, Groman DB, Yason CV, Grothusen H, Lisperguer A, Calbucura M, Avendaño F, Imilán M, Jarpa M, Kibenge FSB. 2008. First detection, isolation and molecular characterization of infectious salmon anaemia virus associated with clinical disease in farmed Atlantic salmon (Salmo salar) in Chile. BMC Vet Res 4:28. doi: 10.1186/1746-6148-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibieta P, Tapia V, Venegas C, Hausdorf M, Takle H. 2011. Chilean salmon farming on the horizon of sustainability: review of the development of a highly intensive production, the ISA crisis and implemented actions to reconstruct a more sustainable aquaculture industry, p 215–246. In Sladonja B. (ed), Aquaculture and the environment—a shared destiny. InTech, Rijeka, Croatia. doi: 10.5772/2463. [DOI] [Google Scholar]
- 9.Oelckers K, Vike S, Duesund H, Gonzalez J, Wadsworth S, Nylund A. 2014. Caligus rogercresseyi as a potential vector for transmission of infectious salmon anaemia (ISA) virus in Chile. Aquaculture 420–421:126–132. doi: 10.1016/j.aquaculture.2013.10.016. [DOI] [Google Scholar]
- 10.Aldrin M, Storvik B, Frigessi A, Viljugrein H, Jansen PA. 2010. A stochastic model for the assessment of the transmission pathways of heart and skeleton muscle inflammation, pancreas disease and infectious salmon anaemia in marine fish farms in Norway. Prev Vet Med 93:51–61. doi: 10.1016/j.prevetmed.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Marshall SH, Ramírez R, Labra A, Carmona M, Muñoz C. 2014. Bona fide evidence for natural vertical transmission of infectious salmon anemia virus in freshwater brood stocks of farmed Atlantic salmon (Salmo salar) in southern Chile. J Virol 88:6012–6018. doi: 10.1128/JVI.03670-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mjaaland S, Hungnes O, Teig A, Dannevig BH, Thorud K, Rimstad E. 2002. Polymorphism in the infectious salmon anemia virus hemagglutinin gene: importance and possible implications for evolution and ecology of infectious salmon anemia disease. Virology 304:379–391. doi: 10.1006/viro.2002.1658. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham CO, Gregory A, Black J, Simpson I, Raynard RS. 2002. A novel variant of the infectious salmon anaemia virus (ISAV) haemagglutinin gene suggests mechanisms for virus diversity. Bull Eur Assoc Fish Pathol 22:366–374. [Google Scholar]
- 14.Lyngstad TM, Kristoffersen AB, Hjortaas MJ, Devold M, Aspehaug V, Larssen RB, Jansen PA. 2012. Low virulent infectious salmon anaemia virus (ISAV-HPR0) is prevalent and geographically structured in Norwegian salmon farming. Dis Aquat Org 101:197–206. doi: 10.3354/dao02520. [DOI] [PubMed] [Google Scholar]
- 15.Christiansen DH, Østergaard PS, Snow M, Dale OB, Falk K. 2011. A low-pathogenic variant of infectious salmon anemia virus (ISAV-HPR0) is highly prevalent and causes a non-clinical transient infection in farmed Atlantic salmon (Salmo salar L.) in the Faroe Islands. J Gen Virol 92:909–918. doi: 10.1099/vir.0.027094-0. [DOI] [PubMed] [Google Scholar]
- 16.Scheel I, Aldrin M, Frigessi A, Jansen PA. 2007. A stochastic model for infectious salmon anemia (ISA) in Atlantic salmon farming. J R Soc Interface 4:699–706. doi: 10.1098/rsif.2007.0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammell KL, Dohoo IR. 2005. Risk factors associated with mortalities attributed to infectious salmon anaemia virus in New Brunswick, Canada. J Fish Dis 28:651–661. doi: 10.1111/j.1365-2761.2005.00668.x. [DOI] [PubMed] [Google Scholar]
- 18.Snow M, McKay P, McBeath AJA, Black J, Doig F, Kerr R, Cunningham CO, Nylund A, Devold M. 2006. Development, application and validation of a Taqman real-time RT-PCR assay for the detection of infectious salmon anaemia virus (ISAV) in Atlantic salmon (Salmo salar). Dev Biol (Basel) 126:133–145; discussion 325–326. [PubMed] [Google Scholar]
- 19.Mardones FO, Perez A, Valdes-Donoso MP, Carpenter TE. 2011. Farm-level reproduction number during an epidemic of infectious salmon anemia virus in southern Chile in 2007–2009. Prev Vet Med 102:175–184. doi: 10.1016/j.prevetmed.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Tort L. 2011. Stress and immune modulation in fish. Dev Comp Immunol 35:1366–1375. doi: 10.1016/j.dci.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Casado E, Estepa A, Coll JM. 2011. A comparative review on European-farmed finfish RNA viruses and their vaccines. Vaccine 29:2657–2671. doi: 10.1016/j.vaccine.2011.01.097. [DOI] [PubMed] [Google Scholar]
- 22.Müller A, Markussen T, Drabløs F, Gjøen T, Jørgensen TT, Solem ST, Mjaaland S. 2010. Structural and functional analysis of the hemagglutinin-esterase of infectious salmon anaemia virus. Virus Res 151:131–141. doi: 10.1016/j.virusres.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aspehaug V, Mikalsen AB, Snow M, Biering E. 2005. Characterization of the infectious salmon anemia virus fusion protein. J Virol 79:12544–12553. doi: 10.1128/JVI.79.19.12544-12553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godoy MG, Suarez R, Lazo ES, Llegues KO, Kibenge MJT, Wang Y, Kibenge FSB. 2014. Genetic analysis and comparative virulence of infectious salmon anemia virus (ISAV) types HPR7a and HPR7b from recent field outbreaks in Chile. Virol J 11:204. doi: 10.1186/s12985-014-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devold M, Karlsen M, Nylund A. 2006. Sequence analysis of the fusion protein gene from infectious salmon anemia virus isolates: evidence of recombination and reassortment. J Gen Virol 87:2031–2040. doi: 10.1099/vir.0.81687-0. [DOI] [PubMed] [Google Scholar]
- 26.Hellebø A, Vilas U, Falk K, Vlasak R. 2004. Infectious salmon anemia virus specifically binds to and hydrolyzes 4-O-acetylated sialic acids. J Virol 78:3055–3062. doi: 10.1128/JVI.78.6.3055-3062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsubara T, Onishi A, Saito T, Shimada A, Inoue H, Taki T, Nagata K, Okahata Y, Sato T. 2010. Sialic acid-mimic peptides as hemagglutinin inhibitors for anti-influenza therapy. J Med Chem 53:4441–4449. doi: 10.1021/jm1002183. [DOI] [PubMed] [Google Scholar]
- 28.Nicol MQ, Ligertwood Y, Bacon MN, Dutia BM, Nash AA. 2012. A novel family of peptides with potent activity against influenza A viruses. J Gen Virol 93:980–986. doi: 10.1099/vir.0.038679-0. [DOI] [PubMed] [Google Scholar]
- 29.Jones JC, Turpin EA, Bultmann H, Brandt CR, Schultz-Cherry S. 2006. Inhibition of influenza virus infection by a novel antiviral peptide that targets viral attachment to cells. J Virol 80:11960–11967. doi: 10.1128/JVI.01678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rolland JB, Bouchard D, Coll J, Winton JR. 2005. Combined use of the ASK and SHK-1 cell lines to enhance the detection of infectious salmon anemia virus. J Vet Diagn Invest 17:151–157. doi: 10.1177/104063870501700209. [DOI] [PubMed] [Google Scholar]
- 31.Castillo-Cerda MT, Cottet L, Toro-Ascuy D, Spencer E, Cortez-San Martín M. 2014. Development of plaque assay for Chilean infectious salmon anaemia virus, application for virus purification and titration in salmon ASK cells. J Fish Dis 37:989–995. doi: 10.1111/jfd.12198. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 33.Wilson RK. 1993. High-throughput purification of M13 templates for DNA sequencing. Biotechniques 15:414–416, 418–420, 422. [PubMed] [Google Scholar]
- 34.Houghten RA. 1985. General method for the rapid solid-phase synthesis of large numbers of peptides: specificity of antigen-antibody interaction at the level of individual amino acids. Proc Natl Acad Sci U S A 82:5131–5135. doi: 10.1073/pnas.82.15.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore LJ, Somamoto T, Lie KK, Dijkstra JM, Hordvik I. 2005. Characterisation of salmon and trout CD8α and CD8β. Mol Immunol 42:1225–1234. doi: 10.1016/j.molimm.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Matsubara T, Sumi M, Kubota H, Taki T, Okahata Y, Sato T. 2009. Inhibition of influenza virus infections by sialylgalactose-binding peptides selected from a phage library. J Med Chem 52:4247–4256. doi: 10.1021/jm801570y. [DOI] [PubMed] [Google Scholar]
- 38.Wu D, Li G, Qin C, Ren X. 2011. Phage displayed peptides to avian H5N1 virus distinguished the virus from other viruses. PLoS One 6:e23058. doi: 10.1371/journal.pone.0023058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajik M, Omar AR, Ideris A, Hassan SS, Yusoff K. 2009. A novel peptide inhibits the influenza virus replication by preventing the viral attachment to the host cells. Int J Biol Sci 5:543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller A, Solem ST, Karlsen CR, Jørgensen T. 2008. Heterologous expression and purification of the infectious salmon anemia virus hemagglutinin esterase. Protein Expr Purif 62:206–215. doi: 10.1016/j.pep.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Fourrier M, Lester K, Thoen E, Mikalsen A, Evensen O, Falk K, Collet B, McBeath A. 2014. Deletions in the highly polymorphic region (HPR) of infectious salmon anaemia virus HPR0 haemagglutinin-esterase enhance viral fusion and influence the interaction with the fusion protein. J Gen Virol 95:1015–1024. doi: 10.1099/vir.0.061648-0. [DOI] [PubMed] [Google Scholar]
- 42.Wales ME, Mcdaniel CS, Everett AL, Rawlins JW, Blanton MD, Wild JR, Gonzalez CF. 2006. Enhancing biocides in antimicrobial coatings: peptides. Paint Coatings Ind 22:68–78. [Google Scholar]
- 43.Fulmer PA, Wynne JH. 2011. Development of broad-spectrum antimicrobial latex paint surfaces employing active amphiphilic compounds. ACS Appl Mater Interfaces 3:2878–2884. doi: 10.1021/am2005465. [DOI] [PubMed] [Google Scholar]
- 44.Vike S, Oelckers K, Duesund H, Erga SR, Gonzalez J, Hamre B, Frette O, Nylund A. 2014. Infectious salmon anemia (ISA) virus: infectivity in seawater under different physical conditions. J Aquat Anim Health 26:33–42. doi: 10.1080/08997659.2013.864720. [DOI] [PubMed] [Google Scholar]