Abstract
Fermentation pit mud, an important reservoir of diverse anaerobic microorganisms, is essential for Chinese strong-aroma liquor production. Pit mud quality, according to its sensory characteristics, can be divided into three grades: degraded, normal, and high quality. However, the relationship between pit mud microbial community and pit mud quality is poorly understood, as are microbial associations within the pit mud ecosystem. Here, microbial communities at these grades were compared using Illumina MiSeq sequencing of the variable region V4 of the 16S rRNA gene. Our results revealed that the pit mud microbial community was correlated with its quality and environmental factors. Species richness, biodiversity, and relative and/or absolute abundances of Clostridia, Clostridium kluyveri, Bacteroidia, and Methanobacteria significantly increased, with corresponding increases in levels of pH, NH4+, and available phosphorus, from degraded to high-quality pit muds, while levels of Lactobacillus, dissolved organic carbon, and lactate significantly decreased, with normal samples in between. Furthermore, 271 pairs of significant and robust correlations (cooccurrence and negative) were identified from 76 genera using network analysis. Thirteen hubs of cooccurrence patterns, mainly under the Clostridia, Bacteroidia, Methanobacteria, and Methanomicrobia, may play important roles in pit mud ecosystem stability, which may be destroyed with rapidly increased levels of lactic acid bacteria (Lactobacillus, Pediococcus, and Streptococcus). This study may help clarify the relationships among microbial community, environmental conditions, and pit mud quality, allow the improvement of pit mud quality by using bioaugmentation and controlling environmental factors, and shed more light on the ecological rules guiding community assembly in pit mud.
INTRODUCTION
Chinese strong-aroma liquor (CSAL), a traditional alcoholic beverage, accounts for >70% of both national liquor industry production (12 billion liters) and sales volume ($80 billion) (1, 2). Its production is a typical recycling process using solid-state fermentation. In brief, fermented grains obtained from the last fermentation round are mixed with crushed raw materials (sorghum, wheat, corn, rice, and sticky rice) for distillation to give CSAL (3). After cooling, the steamed mixture is supplied with a 10 to 20% (wt/wt) Daqu starter. Next, the above-mentioned mixture is placed into a fermentation vessel (underground cuboid soil pit, 2 m wide by 3 m long by 2 m deep), sealed, and anaerobically fermented for about 60 days at 28 to 32°C (2). The inside walls of the pit are covered with fermentation pit mud (FPM), which is prepared initially by incubating the mixture of clay, spent grain, bean cake powder, and fermentation bacteria (e.g., Clostridium spp.) (2). After fermentation, the fermented grains taken out of the pit are distilled to give liquor after new raw materials are supplied, and then the mixture is applied to the next round of fermentation, as described above. It is widely believed that the flavor and quality of CSAL are positively correlated with FPM quality (4, 5), which is determined mainly by FPM microbial α-diversity (the diversity within individual samples, e.g., the Shannon index) and β-diversity (the variation in community composition).
In practice, FPM quality can be divided into three grades according to its sensory characteristics (i.e., color, odor, and texture) (see Table S1 in the supplemental material) (4, 5). Normal FPMs are characterized by earthy color, ester aroma, and a moist, soft, and uniform texture (5). High-quality FPMs, which are gray-black and have a strong ester aroma, hydrogen sulfide odor, and moist, soft, and uniform texture, are widely applied to produce high-quality CSAL (6). Degraded FPMs, however, with their silvery-gray color, white lumps or aciform crystals, and weak ester aroma, can directly lead to poor CSAL quality and an economic loss for the liquor-making company (6). Previous studies have focused on microbial communities in FPMs of different ages and from different locations in a pit. For example, microbial diversity (i.e., the Shannon index) in FPM from the middle zone of a pit is higher than that in the bottom and top zones, increasing with age, and the most frequent microorganisms are Firmicutes, Euryarchaeota, Bacteroidetes, Actinobacteria, and Proteobacteria (7–9). Moreover, members of the class Clostridia are responsible for the formation of various aromatic compounds (e.g., fatty acids, alcohols, and phenols) in CSAL (7, 10–12); Clostridium kluyveri, a major caproic acid-producing bacterium, is especially involved in the synthesis of key CSAL flavors (i.e., caproic acid and ethyl caproate) (3). However, the shifts in microbial α-diversity, β-diversity, and contents of Clostridia and C. kluyveri strains across a quality gradient in FPMs have not been well understood. Such information, compared to insensitive sensory characteristics, can contribute to a better understanding of the mechanisms of CSAL fermentation and the identification of microbial indicators for predicting the FPM quality in time.
Previous studies indicated that the decline in FPM quality occurred in combination with decreased pH and nutrients but increased fatty acids (e.g., lactic acid) in FPM, and then white lumps or aciform crystals, presumed to be calcium lactate and ferrous lactate, appeared (5, 6). However, the relationships between the biogeochemical attributes of these distinct FPMs and their microbial community are still not fully understood. Recently, network analysis has widely been applied to reveal the ecological linkages among microorganisms in complex ecosystems, such as soil and activated sludge (13, 14). However, as far as we know, it has not been reported that direct and indirect interactions among microbial taxa coexist in FPMs. Accordingly, it is very necessary to figure out these relationships/linkages to better control FPM quality and shed light on the ecological rules guiding community assembly within the FPM ecosystem.
To address the above-mentioned puzzling questions for the improvement and stabilization of FPM quality, the aims of this work were to (i) characterize the patterns of microbial community diversity and contents of Clostridia and C. kluyveri of FPMs across quality gradients in FPMs, (ii) reveal the positive (cooccurrence) and negative correlations of the microbial taxa within the FPM ecosystem, (iii) unveil the relationships between the biogeochemical attributes of FPM and its microbial community, and (iv) identify the microbial indicators for evaluating the quality of FPM.
MATERIALS AND METHODS
Sampling.
Three types of FPMs with different levels of quality, namely, degraded, normal, and high quality, which were divided based on their sensory characteristics (see Table S1 in the supplemental material), were obtained from one well-known strong aromatic liquor distillery in Jiangsu Province (30°45′ to 35°20′N, 116°18′ to 121°57′E), eastern China. Four fermentation pits were randomly selected for each type, and triplicate subsamples were collected in each pit and mixed immediately. Finally, the FPM sample from each pit was transferred immediately to sterile anaerobic bags (MGC, Japan), marked, and stored at −80°C.
Analysis of FPM biogeochemistry.
The pH of fresh FPM was measured at a 1:3 (wt/vol) ratio in ultrapure water using a Mettler Toledo pH meter. Ammonium (NH4+-N) from fresh FPM was extracted in 1 M KCl at a 1:20 (wt/vol) ratio, and its concentration was measured by a SEAL AutoAnalyzer 3 (SEAL Analytical, Hampshire, United Kingdom). The moisture content of the FPM was determined by drying the samples at 105°C for 2 h. The total nitrogen (TN) content of the air-dried FPM samples was analyzed with a Kjeltec 8400 (Foss, Sweden). After air-dried FPM with 1 M KCl at a 1:30 (wt/vol) ratio was extracted, the dissolved organic carbon (DOC) concentration was determined using an organic carbon analyzer (TOC-V CPH; Shimadzu, Japan). Available phosphorus (AP) from air-dried FPM was extracted using 0.5 M NaHCO3 at a 1:20 (wt/vol) ratio and determined based on the Olsen method (15). The lactate concentration of air-dried FPM was quantified using a high-performance liquid chromatograph (HPLC) with a UV detector (see the text in the supplemental material).
DNA extraction, PCR amplification, and sequencing.
The genomic DNA of each FPM was extracted with an E.Z.N.A. soil DNA kit (catalog no. D5625-01; Omega, USA), according to the manufacturer's protocols. The V4 hypervariable region of the 16S rRNA gene was amplified using the primer set 515F and 806R (16), and a 12-bp error-correcting barcode unique to each sample was added to the reverse primer to allow multiplex sequencing. The PCR was performed according to a previously described method (16), and the PCR products from different samples were purified and quantified as described by Ren et al. (17). Subsequently, purified amplicons were pooled at equimolar concentrations for constructing a PCR amplicon library, according to the protocols of the Illumina TruSeq DNA sample preparation LT kit (San Diego, CA), and then sequenced using a MiSeq benchtop sequencer for paired-end sequencing (2 by 250 bp; Illumina, San Diego, CA).
Real-time qPCR analysis.
To detect the populations of Clostridia, C. kluyveri, and lactic acid bacteria (LAB) in an FPM sample, a quantitative PCR (qPCR) assay was used. The specific primer sets for Clostridia, C. kluyveri, and LAB were SJ-F/SJ-R (18), CloKly1F/CloKly1R (19), and Lac1/Lac2 (20), respectively. The real-time PCR calibration curves for quantifying the above-mentioned microbial taxa were generated via 10-fold serial dilutions of corresponding plasmids (see the text in the supplemental material). Each qPCR system was prepared based on the description of SsoFast EvaGreen Supermix (Bio-Rad). The amplification conditions were as follows: preheating at 98°C for 3 min, 40 cycles of 98°C for 10 s, 58°C (for Clostridia and C. kluyveri) and 55°C (for LAB) for 10 s, and increases of 0.5°C every 5 s from 65°C to 95°C for melting curve analysis (Bio-Rad CFX96; Bio-Rad Laboratories, Hercules, CA).
Sequence processing.
All the raw MiSeq-generated 16S rRNA gene sequence data were processed in the QIIME pipeline (21). Briefly, the raw sequences were sorted based on the unique barcode of each sample, and the low-quality sequences with >0 ambiguous bases, >6 homopolymers, primer mismatches, average quality scores of <30, and lengths (excluding the primer/barcode region) of <150 bp were removed. PCR chimeras were then checked and removed using the UCHIME software (22). The remaining (good-quality) sequences were clustered into operational taxonomic units (OTUs) using a 97% identity threshold with QIIME's uclust (23). Calculation of the Shannon index and Chao1 species estimator were performed according to the method described by Tao et al. (7). The taxonomic affiliation of each OTU was analyzed by Ribosomal Database Project (RDP) Classifier at a confidence level of 80%.
Statistical and network analysis.
Principal-coordinate analysis (PCoA) based on weighted UniFrac distances was performed to evaluate the similarities of different communities. To reveal the correlations between environmental variables and abundant classes (with abundance defined as >1% of total good-quality sequences), Pearson's test was performed using SPSS Statistics 22 (IBM Corp., Armonk, NY). The classified genera in each sample were selected, and the relative abundance of each genus was presented as a percentage of the total number of classified sequences that can be identified as known genera. A heatmap was analyzed using the gplots package in R to compare the top 10 classified genera in each sample. Redundancy analysis (RDA) was performed using Canoco 5.0 software to reveal the relationships between environmental factors and 81 abundant (abundance was defined as >0.1% of the total classified genera in at least one sample) genera, which accounted for 97.3 to 99.7% of all classified genera in each sample. The statistical significance of the difference between the means of samples was tested by one-way analysis of variance (ANOVA) using SPSS Statistics 22.
To explore and visualize networks, a correlation matrix was constructed by calculating all possible pairwise Spearman's rank correlations among the above-mentioned 81 genera. Only a Spearman's correlation coefficient (ρ) of >0.6 (or less than −0.6) and statistical significance (P < 0.01) were considered a valid cooccurrence (or negative) event for a robust correlation (13, 24). All statistical analyses were carried out in the R environment with the Hmisc (Vanderbilt University) package (24, 25). Networks were visualized with the interactive platform Gephi (26), and each node and edge represented one genus and a strong and significant correlation, respectively.
Microarray data accession number.
All the raw sequences were deposited in in the DDBJ database under accession no. DRA003468.
RESULTS
Biogeochemical attributes.
Among all biogeochemical attributes, levels of TN and moisture were not significantly different in distinct FPM types. However, the pHs and DOC, NH4+, AP, and lactate contents were significantly different (Table 1). Degraded FPM had the lowest pH, NH4+, and AP concentrations but the highest concentrations of DOC and lactate (P < 0.05), which is in contrast to levels in high-quality FPMs. Besides DOC and pH, there were no significant differences between normal and high-quality samples. Further regression analysis showed that FPM pH was significantly correlated with other environmental variables (except for TN), especially DOC (R2 = 0.9296) (see Fig. S2a in the supplemental material), indicating that, as with soil (27), pH can be used as an integrated parameter reflecting the overall physicochemical information of the FPM ecosystem.
TABLE 1.
Biogeochemical attributes and contents of Clostridia, C. kluyveri, and LAB in four samples each of degraded, normal, and high-quality FPMs
| Variableb | Value for indicated FPM type (mean ± SD)a |
||
|---|---|---|---|
| Degraded | Normal | High quality | |
| DOC (g/kg) | 38.26 ± 2.98 A | 18.17 ± 3.29 B | 8.23 ± 5.57 C |
| Total N (g/kg) | 9.38 ± 2.29 A | 8.67 ± 1.32 A | 9.80 ± 2.56 A |
| NH4+ (mg/kg) | 249.88 ± 0.81 A | 269.20 ± 3.75 B | 271.77 ± 3.09 B |
| AP (mg/kg) | 205.44 ± 61.30 A | 759.78 ± 69.16 B | 805.19 ± 73.81 B |
| pH | 4.57 ± 0.1 A | 6.18 ± 0.32 B | 7.61 ± 0.59 C |
| Lactate (g/kg) | 30.18 ± 4.55 A | 3.08 ± 2.02 B | 0.85 ± 0.26 B |
| Moisture (%) | 36.90 ± 0.45 A | 31.98 ± 3.70 A | 31.07 ± 4.48 A |
| N[Clostridia] | 5.95 ± 0.64 A | 8.81 ± 1.05 B | 9.09 ± 5.08 B |
| N[C. kluyveri] | 4.02 ± 0.64 A | 6.80 ± 1.52 B | 7.22 ± 1.26 B |
| N[LAB] | 8.40 ± 0.25 A | 7.38 ± 0.16 B | 7.08 ± 0.28 B |
Values with different letters (A, B, and C) in a row indicate that they are significantly different from each other (P < 0.05).
N[Clostridia], N[C. kluyveri], and N[LAB] represent the absolute abundances (log10 16S rRNA gene copies/gram of FPM) of Clostridia, C. kluyveri, and LAB in FPM samples, respectively.
α-Diversity.
After quality filtering, a total of 170,098 good-quality sequences with an average length of 219 bp from all FPM samples were obtained, and the number of reads per sample ranged from 13,613 to 14,653. At the 97% identity level, all good-quality sequences were distributed into 4,927 OTUs, and each sample contained 774 to 1,453 OTUs. Moreover, the coverage of good-quality sequences per sample was higher than 94%, and the rarefaction curves approached the saturation plateau (see Fig. S1 in the supplemental material), indicating that members in microbial communities could be well represented. More than 97% of the good-quality sequences in each sample (except sample S9, at 91%) could be classified (Table S2), suggesting that phylogenetic classification of most uncultured microorganisms in FPMs could be well explained. The microbial richness (Chao1) and diversity (Shannon) indices of microbial communities in FPMs increased significantly from degraded to high-quality samples, but these indices did not significantly differ between normal and high-quality samples (P < 0.05) (Table 2).
TABLE 2.
Microbial community richness and diversity indices for four samples each of degraded, normal, and high-quality FPMs
| Parameter | Value for indicated FPM type (mean ± SD)a |
||
|---|---|---|---|
| Degraded | Normal | High quality | |
| No. of good-quality sequences | 14,498.75 ± 110.45 A | 13,974.75 ± 315.21 B | 14,051 ± 196.83 B |
| Coverage (%) | 96.18 ± 0.15 A | 95.10 ± 0.45 B | 94.83 ± 0.41 B |
| Observed no. of OTUs | 858.13 ± 75.87 A | 1,206.70 ± 164.85 B | 1,219.15 ± 132.25 B |
| Chao1 index | 1,711.35 ± 60.98 A | 2,127.55 ± 237.72 B | 2,227.46 ± 114.96 B |
| Shannon index | 3.54 ± 0.40 A | 6.05 ± 0.61 B | 6.45 ± 0.57 B |
Values with different letters (A and B) in a row indicate that they are significantly different from each other (P < 0.05). Except for the number of good-quality sequences, the parameters were calculated based on cutoffs of 0.03 and 12,799 sequences per sample.
β-Diversity.
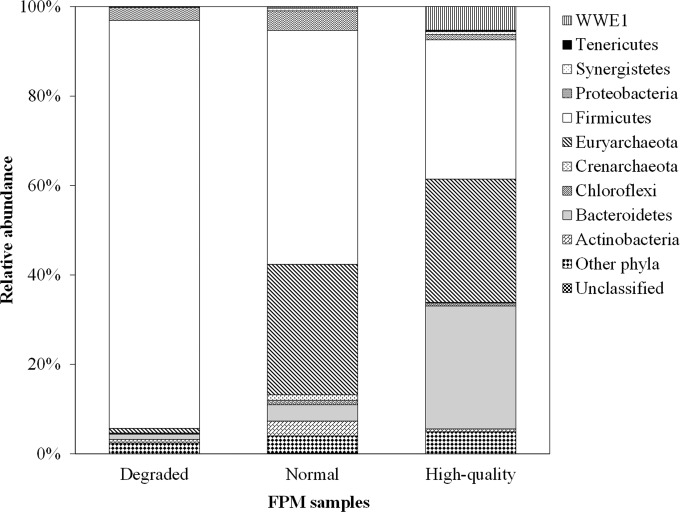
Overall, a total of 33 phyla, including 31 bacterial and 2 archaeal phyla, were detected (see Table S2 in the supplemental material), and Firmicutes (58%), Euryarchaeota (19%), and Bacteroidetes (11%) were the dominant phyla (representing >10% of total good-quality sequences). Other less abundant phyla, including the Proteobacteria (2.8%), WWE1 (1.9%), and Actinobacteria (1.6%), were found in most of the FPMs examined. Twenty of 33 phyla were shared by all FPMs, accounting for 89.21 to 98.34% in each sample. Moreover, 10 phyla represented by <0.01% of the total good-quality sequences are defined here as rare phyla, such as Fusobacteria and Deferribacteres.
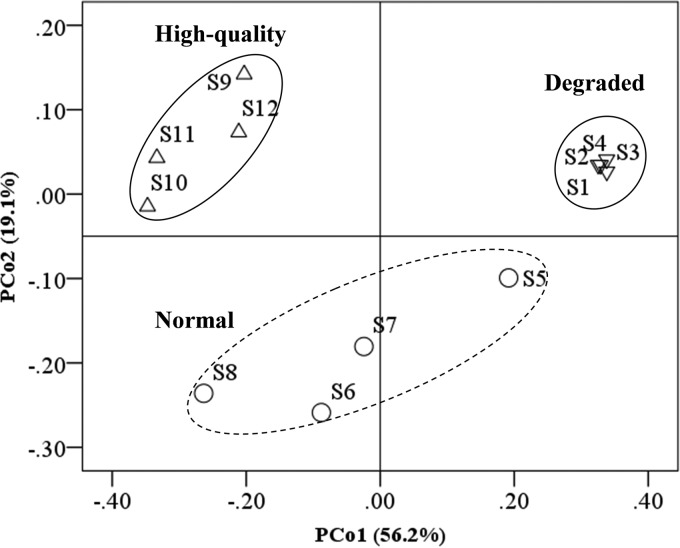
The PCoA showed that all FPM samples were expectedly clustered into degraded, normal, and high-quality clusters (Fig. 1). FPM quality was the main factor in the first principal-coordinate axis (PCo1) and contributed 56.2% of the total variation, indicating that FPM quality is frequently associated with its community composition. For instance, Firmicutes (91.29%) dominated in degraded FPMs, Firmicutes (52.31%) and Euryarchaeota (29.15%) dominated in normal FPMs, and Firmicutes (31.16%), Euryarchaeota (27.57%), and Bacteroidetes (27.51%) dominated in high-quality FPMs (Fig. 2; see Tables S2 and S5 in the supplemental material).
FIG 1.
Cluster analysis of microbial communities across a quality gradient in FPMs based on principal-coordinate analysis (PCoA) using weighted UniFrac. S1 to S12, samples 1 to 12.
FIG 2.
Abundances of different phyla in three types of FPMs. The taxa represented occurred at >1% abundance in at least one type of FPM. “Other phyla” refer to the taxa with a maximum abundance of <1% in any type of sample.
At the class level, seven of a total of 85 classes were identified as dominant (defined as >1% of the total good-quality sequences) ones, accounting for 71.54 to 95.54% in each sample, including Bacilli, Clostridia, Methanobacteria, Bacteroidia, Methanomicrobia, Cloacamonae, and Gammaproteobacteria (see Table S3 in the supplemental material), which were synchronously shared by all FPMs. However, only the relative abundance of Bacilli was significantly higher than those of other taxa in degraded FPMs (mean, 85.42%; P < 0.05), whereas Bacteroidia (27.44%) and Cloacamonae (5.27%) were significantly higher in high-quality FPMs. Two classes, Clostridia and Methanobacteria, were higher in both normal (44.84% and 25.6%, respectively) and high-quality FPMs (26.08% and 15.28%, respectively) (Table S5).
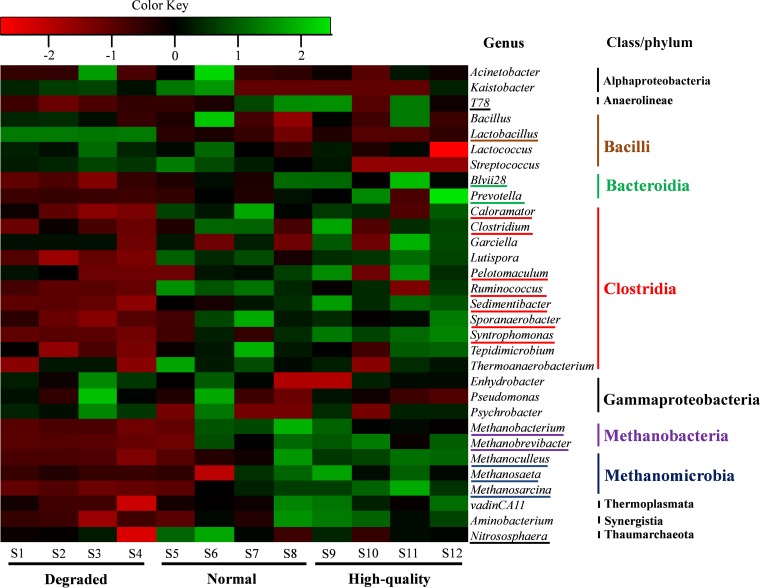
At the genus level, a total of 225 classified genera were observed in all FPM samples, and 27 genera were commonly shared by all samples, accounting for 71.17 to 98.86% of the total classified genera in each sample (see Table S4 in the supplemental material). A total of 82 unique genera were present in only one or two samples, accounting for <0.7% of the total classified sequences in each sample (except for sample S6, at 2.94%), a very minor part of microbial communities in FPMs. As shown in Fig. 3, the top 10 classified genera in each FPM sample were selected (a total of 31 genera for all the 12 samples), and their abundances were compared with those in other FPMs using heatmap analysis. Moreover, we observed 17 core genera (average abundance, >1% in at least one FPM type; the relative abundance of each genus is shown in Table S7), accounting for 89.96 to 96.06% in each FPM type, including 11 bacterial genera, namely, Lactobacillus, Ruminococcus, Caloramator, Clostridium, Sedimentibacter, Syntrophomonas, Sporanaerobacter, Pelotomaculum, T78, Prevotella, and Blvii28 group, and six archaeal genera, comprising Methanobacterium, Methanobrevibacter, Methanosaeta, Methanoculleus, Methanosarcina, and Nitrososphaera. Among them, the numbers of core genera in FPMs dramatically increased from degraded FPMs (2 genera) to high-quality FPMs (15 genera), with the number in normal FPMs (12 genera) in between (Table S7). The highest relative abundances (mean, 91.46%) of Lactobacillus spp. were found in degraded FPMs, and then abundances dramatically decreased in normal (7.05%) and high-quality (6.45%) samples, while the relative abundances of other core genera, mainly under the classes Clostridia, Bacteroidia, Methanobacteria, and Methanomicrobia, increased with FPM quality improvement.
FIG 3.
Heatmap of top 10 genera in each FPM sample. The top 10 most-abundant genera in each sample were selected (a total of 31 genera for all 12 samples) and compared with their abundances (log scale) in other samples. Underlined genera are the core genera in different FPM types.
Furthermore, the qPCR assay revealed that the absolute abundances of Clostridia and C. kluyveri were not significantly different between normal and high-quality FPMs, but they were significantly higher (∼1,000 times) than those in degraded FPMs, while the absolute abundances of LAB in degraded FPMs were significantly higher than those in normal (∼10 times) and high-quality (∼20 times) FPMs (Table 1).
Network description.
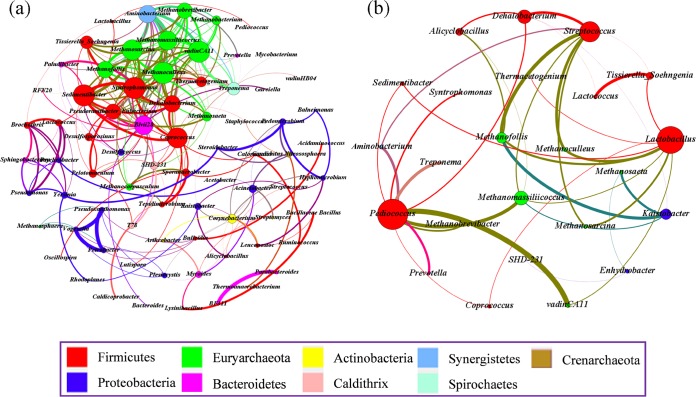
The cooccurrence patterns of FPM microorganisms were explored based on strong and significant correlations (ρ > 0.6 and P < 0.01), and a total of 75 nodes (genera) and 234 edges (pairs of significant and robust correlations) were found (Fig. 4a). There were 13 hubs (highly connected genera, ≥13 edges per node), including Sedimentibacter, Syntrophomonas, Coprococcus, Eubacterium, Methanobrevibacter, Methanosarcina, Methanoculleus, Methanomassiliicoccus, Methanofollis, vadinCA11, Aminobacterium, Blvii28, and Dehalobacter, and 10 of them were under classes Clostridia, Bacteroidia, Methanobacteria, and Methanomicrobia. Significant topological properties used in network analysis were calculated to describe the complex pattern of interrelationships among microbial genera in FPMs (28). The average network distance between all pairs of nodes (average path length) and diameter (longest distance between any two nodes) were 4.917 edges and 12 edges, respectively. Moreover, the value of modularity index was 0.488 (>0.4), suggesting that the network had a modular structure (13).
FIG 4.
Networks of cooccurring microbial genera in FPM samples based on correlation analysis. A connection indicates a statistically significant (P < 0.01) strongly positive correlation (a) (Spearman's ρ > 0.6) or a negative correlation (b) (Spearman's ρ < −0.6). For each panel, the size of each node is proportional to the number of connections, the nodes of the same color were affiliated with the same phylum, and the thickness of each connection between two nodes is proportional to the value of Spearman's correlation coefficients of >0.6 or <−0.6.
Further structural analysis also indicated that the genera from different phyla (interphylum) had a high cooccurrence incidence (64.96%, ratio of targeted edges to total edges). Among the nine different phyla, four phyla, Firmicutes, Euryarchaeota, Bacteroidetes, and Proteobacteria, with strong correlations, accounted for 42.31% (of a total 64.96%). Between any two different phyla, the cooccurrence incidence between Firmicutes and Euryarchaeota was the highest, up to 20.09%. Moreover, caproic acid-producing C. kluyveri was significantly and positively correlated with the class Methanobacteria (see Table S6 in the supplemental material). The cooccurrence incidence in the same phylum (intraphylum) was 35.04%, and the highest cooccurrence incidence (15.38%) was observed among genera under the phylum Firmicutes, followed by Euryarchaeota (10.68%). Moreover, a total of 37 pairs of significant and robust negative correlations were identified from 24 genera (Fig. 4b). Lactobacillus, Pediococcus, and Streptococcus, known as LAB (29), were hubs (≥7 edges) connected mainly with many members of the classes Clostridia, Methanobacteria, and Methanomicrobia (Fig. 4b).
Relationships between microbial communities and environmental variables.
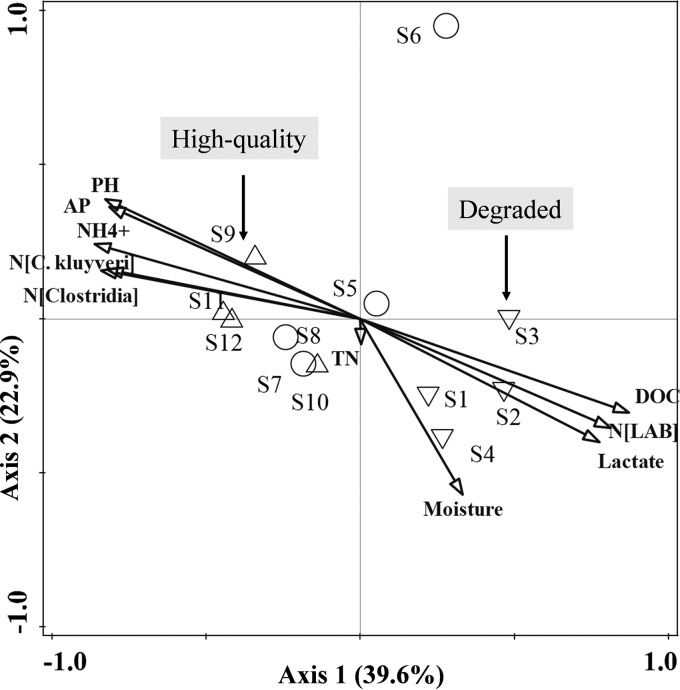
The Shannon index of diversity has widely been used in ecology (30, 31). It was most strongly correlated with FPM pH (R2 = 0.8983, P < 0.01), and peak diversity in FPMs was observed at near-neutral pHs (see Fig. S2b in the supplemental material). Redundancy analysis (RDA) was conducted using 81 abundant genera (accounting for 97.3 to 99.7%), together with environmental variables (Fig. 5; Fig. S3). Overall, the first two axes explained 62.5% of the variation in microbial compositions, and species-environment correlations for both axes were higher than 95% (pseudocanonical correlation), suggesting the remarkable correlation between microbial community structure and environmental factors. An interactive stepwise selection procedure found eight explanatory variables, namely, DOC, NH4+, pH, N[LAB] (where N[LAB] is the absolute abundance of LAB in log10 16S rRNA gene copies/gram of FPM), AP, N[Clostridia], N[C. kluyveri], and lactate, with significant contributions decreasing from 32.9% to 28.7% (P < 0.01). DOC, lactate, N[LAB], four out of five detected genera of LAB, and microbial communities in all degraded samples were positively correlated (Fig. 5; see also Fig. S3 in the supplemental material), while pH, NH4+, AP, N[Clostridia], N[C. kluyveri], about 34 genera under the classes Clostridia, Bacteroidia, Methanobacteria, and Methanomicrobia, and microbial communities in most high-quality FPM samples showed strong positive correlations. Furthermore, most normal FPMs were distributed between degraded and high-quality FPMs, indicating that their microbial communities may transform to those in degraded or high-quality FPMs under environmental stress. Pearson's test revealed that pH, AP, and NH4+ were mainly positively correlated with the relative abundances of Clostridia, Methanobacteria, Bacteroidia, and Cloacamonae but negatively correlated with Bacilli (P < 0.05). However, DOC and lactate showed contrasting correlations (Table S6).
FIG 5.
Redundancy analysis of pit mud biogeochemical attributes and microbial communities across a quality gradient in fermentation pit muds, including degraded FPMs (samples S1 to S4), normal FPMs (samples S5 to S8), and high-quality FPMs (samples S9 to S12). The arrows indicate the direction and magnitude of biogeochemical attributes associated with microbial community structures.
DISCUSSION
Microbial community and microbial indicators for evaluating the FPM quality.
Overall, FPM harbored a complex microbiota, because the high Shannon index (5.61 to 6.96) detected in high-quality FPMs (data not shown) was comparable to that (5 to 7) in some soils at a sequencing depth of 4,000 to 9,000 reads (32). Compared to about 16 phyla detected in FPMs in previous studies (7, 8), 33 phyla detected in this work might expand our understanding of the phylogenetic status of an enormous number of uncultured microbes in this ecosystem. Besides differences in numbers of samples and sequence depth, this detection may be attributed to the V4 variable region of the 16S rRNA gene used in this study, which displayed the highest number of correctly classified sequences compared to the numbers for other variable regions (33). Moreover, Firmicutes, Euryarchaeota, Bacteroidetes, Proteobacteria, and Actinobacteria have been identified as abundant phyla in FPMs (7, 8, 10), which was corroborated by our study.
The PCoA revealed that FPM quality was frequently associated with its community composition. Further, ANOVA showed that microbial richness and diversity indices and relative and/or absolute abundances of Bacilli, Clostridia, Cloacamonae, Bacteroidia, Methanobacteria, C. kluyveri, and LAB were significantly different among the three FPM types. It indicated that these distinct microbial parameters could be used as indicators for evaluating FPM quality, mainly because they largely influence the results of community-based cluster analysis (34). For instance, the Shannon index has widely been applied to evaluate the stability of an ecosystem, qualities of water and soil, and the productivity of microbial consortia (31, 35). Compared to sensory characteristics reported in previous studies (4, 5), microbial indicators can more accurately reflect FPM quality in time due to their rapid response to environmental factors (e.g., physicochemical parameters and climate) (36, 37), which can help avoid the abnormal fermentation that leads to poor CSAL production.
Core genera and their shifts across a quality gradient in FPMs.
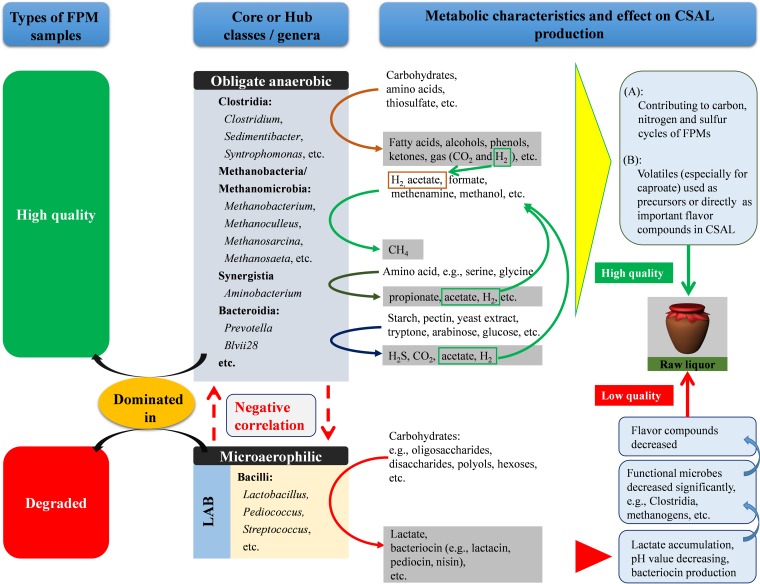
Seventeen core genera, including those representing saccharolytic, fermentation, ammonia-oxidizing, and sulfate-reducing microbes, syntrophs, and methanogens, may play important roles in not only CSAL flavor but also nutrient cycling in a fermentation system. The relative abundance of Lactobacillus and absolute abundance of LAB positively correlated with the level of lactate were significantly decreased with FPM quality improvement (Table 1 and Fig. 3), mainly because LAB (e.g., Lactobacillus) can utilize fermentable carbohydrates to produce lactate as the major end metabolite (29). Seven core bacterial genera under the class Clostridia, namely, Clostridium, Ruminococcus, Sedimentibacter, Syntrophomonas, Pelotomaculum, Sporanaerobacter, and Caloramator, have an exceptionally broad ability to ferment various substrates (e.g., carbohydrates, proteins, and thiosulfate). A wide spectrum of metabolites, such as volatile fatty acids (VFAs), alcohols, phenols, and other substances, as well as large amounts of gas (CO2 and H2), are produced (11, 38–41). Some of them, such as butyric and caproic acids, can be used as precursors to and directly as important flavor compounds of CSAL (7, 12); caproic acid-producing C. kluyveri strains in particular involve the formation of ethyl caproate, which is the key flavor compound affecting CSAL quality (12). To our knowledge, caproic acid-producing Clostridium sp. has been used as a bioaugmentation agent for improving the CSAL quality (2, 10). The relative and/or absolute abundances of the above-mentioned clostridial taxa generally increased with FPM quality improvement (Table 1; see also Table S7 in the supplemental material), which may provide a reasonable explanation for the practical experience that the production of high-quality liquor with high levels of volatile flavor substances (i.e., butyrate, caproate, and their ethyl esters) is based only on high-quality FPMs. Prevotella contributes to the degradation of proteins, xylans, starch, and peptides and produces various fatty acids (e.g., propionate, acetate, etc.) and hydrogen (42, 43). The roles of the core uncultured genera T78 and Blvii28 in FPMs were not clear. The main archaeal genera in FPMs were methanogens, including hydrogenotrophs (Methanobacterium, Methanoculleus, and Methanobrevibacter), acetotrophs (Methanosaeta), and the genus Methanosarcina, which can utilize H2, acetate, methanol, and methylamine, respectively (44, 45). As with those of core clostridial genera, the relative abundances of the above-mentioned methanogens in FPMs generally increased from degraded to high-quality FPMs, especially Methanobrevibacter (from 0.39% to 25.68%) and Methanosarcina (from 0.19% to 19.89%) (Table S7), suggesting that they are positively correlated with FPM quality. Although these methanogens are not capable of producing volatile flavor compounds (e.g., butyrate and caproate), they, combined with hydrolyzers (e.g., Bacteroidales and Ruminococcus), fermenters (e.g., Bacteroidales and Clostridiales), and acetogens (e.g., Syntrophomonas and Syntrophobacterales), contribute to carbon cycling in anaerobic fermentation systems by involving the biological anaerobic digestion process (44, 46). On the other hand, they might play important roles in the stabilities of fermentation performance and community of complex microbial consortiums in anaerobic fermentation systems by reducing VFAs (e.g., acetate, lactate, butyrate, etc.) and hydrogen stress (47, 48). Furthermore, Nitrososphaera, an ammonia-oxidizing archaeal genus, plays important roles in nitrification (49).
Interestingly, nine core genera, Lactobacillus, Clostridium, Ruminococcus (clostridial cluster IV), Sedimentibacter, Syntrophomonas, Methanobacterium, Methanobrevibacter, Methanoculleus, and Methanosarcina, also dominated in FPMs from Southwest China (Sichuan Province) (7), indicating that they might be essential for the basic functioning of FPM across China, according to the insurance hypothesis (35).
Effect of environmental variables on FPM microbial community.
Our study revealed that pH, as an integrated biogeochemical attribute, significantly increased with FPM quality improvement and was correlated with the microbial community diversity and composition. Community diversity tended to be higher and then stable at near-neutral pHs with increasing FPM pH (4.46 to 8.24) (see Fig. S2b in the supplemental material). Interestingly, this finding was consistent with the trend of changes in bacterial diversity across a soil pH gradient (pHs of ∼3 to 9) (50). Simultaneously, pH was strongly correlated with community composition (especially the phyla Firmicutes and Bacteroidetes) (7), which was corroborated by our results. Two common hypotheses may explain why microbial community diversity and composition were influenced by FPM pH. One is that FPM pH may not directly alter a microbial community but can directly or indirectly affect many physical and chemical FPM properties (e.g., nutrient availability, organic C characteristics, and cationic metal solubility) (51), which may result in changes in the microbial community. The other hypothesis is that in situ pH can directly impose a physiological stress on microorganisms (52), reducing the content of microbial groups that are difficult to grow if the FPM pH falls beyond a suitable scope. For instance, core genera under the classes Clostridia, Methanobacteria, Methanomicrobia, and Bacteroidia dominated in normal and high-quality FPMs with pHs of 5.73 to 8.24, mainly because the pH values for the growth of many members under the above-mentioned classes ranged from 4 to 9 (with an optimal near-neutral pH) (53–55), while acid-tolerant Lactobacillus, correlated with high levels of lactate, only dominated in degraded FPMs with pHs of 4.46 to 4.67. Among all FPM variables, the DOC level, followed by the lactate level, showed the strongest correlation with FPM pH, mainly because DOC may reflect a comprehensive level of various water-soluble fatty acids (e.g., acetate, propanoate, and lactate), the accumulation of which might lead to the decline of FPM pH (7). Furthermore, significant correlations between the microbial community and other physicochemical variables, including DOC, AP, and NH4+ levels, were also observed (Fig. 5; Fig. S3), indicating that these variables may directly or indirectly affect the microbial community. Such relationships might help to improve the qualities of FPM and CSAL by controlling the environmental variables.
Complex microbial associations within FPM ecosystem.
Using network analysis, 271 pairs of significant and robust correlations (positive and negative) were identified from 76 genera. The cooccurrence patterns revealed that community members may share niche spaces and have synergetic relationships in FPM (13). For instance, specific members of Clostridia, Bacteroidia, Methanobacteria, and Methanomicrobia act synergistically for anaerobic digestion, including carbohydrate degradation, production of VFAs and H2, and methanogenesis (46). Moreover, it is known that C. kluyveri, when cocultured with methanogens, can yield more caproate (56). Our results showed that these microbial taxa, as main hubs based on cooccurrence analysis, had high abundances in high-quality FPMs (Fig. 6), indicating that they may play important roles in ecosystem stability (57). However, the absolute abundance of LAB was significantly higher in degraded FPMs, and 3 genera of LAB showed strong negative relations with other microbes in FPMs, especially in Clostridia and methanogens. There were two possible explanations: one is that the accumulation of their major end product (lactate) resulted in a pH decline (∼4.5), which inhibits the growth of many Clostridia and methanogens without acid-resistant properties; the other is that their various bacteriocins, including nisin, lactacin, and others, prevent the growth of Gram-positive bacteria (e.g., Clostridia and Bacilli) (58). Accordingly, we inferred that Clostridia, Bacteroidia, and methanogens play important roles in FPM quality stability, while rapidly increased numbers of LAB may destroy this stability and lead to FPM degradation.
FIG 6.
Distribution of core genera (Fig. 3) and hubs (Fig. 4) within microbial communities in pit muds, and relationships between their potential metabolic properties and the quality of Chinese liquor and nutrient cycling in the pit mud ecosystem.
In conclusion, a very complex microbiota inhabiting the FPM ecosystem, associated with FPM quality, plays important roles in CSAL flavor and FPM nutrient cycling processes. We identified the relationships among microbial community, FPM quality, and environmental factors, as well as microbial indicators for the rapid assessment of FPM quality. For the first time, to our knowledge, microbial associations within this ecosystem were revealed. These findings represent a step forward in understanding the functioning and stability of the FPM-inhabiting microbial community, providing new insights into the microbial taxon associations within the FPM ecosystem, thus contributing to improvement and quality stabilization of FPMs and Chinese strong-aroma liquor. Such a study may provide a suitable strategy for illuminating the functioning and stability of other traditional food fermentation ecosystems with a complex microbial community.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the Jiangsu Province “Collaborative Innovation Center for Advanced Industrial Fermentation” industry development program. Special thanks also go to the “3C” plan of the Chinese liquor industry for its financial support and supply of samples.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03409-15.
REFERENCES
- 1.Song SY. 2014. 2013 annual working report by Baijiu (Liquor) Branch and Market Specialized Committee of China Alcoholic Drinks Industry Association. Liquor Mak Sci Technol 6:1–9. [Google Scholar]
- 2.Xu Y, Wang D, Fan WL, Mu XQ, Chen J. 2010. Traditional Chinese biotechnology. Adv Biochem Eng Biotechnol 122:189–233. doi: 10.1007/10_2008_36. [DOI] [PubMed] [Google Scholar]
- 3.Hu XL, Du H, Xu Y. 2015. Identification and quantification of the caproic acid-producing bacterium Clostridium kluyveri in the fermentation of pit mud used for Chinese strong-aroma type liquor production. Int J Food Microbiol 214:116–122. doi: 10.1016/j.ijfoodmicro.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 4.Huang H, Yang GR, Huang ZY, Wen CB. 2011. Analysis of causations of pit mud aging & its treatment and prevention in Sichuang Provinces. Sichuan Food Ferment 47:18–21. (In Chinese.) [Google Scholar]
- 5.Zhao CQ, Yang QH, Deng J, Wu HC. 2012. Detection of evaluated indexes for pit mud. Curr Biotechnol 3:212–216. (In Chinese.) [Google Scholar]
- 6.Zhang XY. 2010. Research on the rejuvenation of aged cellar mud. Liquor Making 37:53–54. [Google Scholar]
- 7.Tao Y, Li J, Rui J, Xu Z, Zhou Y, Hu X, Wang X, Liu M, Li D, Li X. 2014. Prokaryotic communities in pit mud from different-aged cellars used for the production of Chinese strong-flavored liquor. Appl Environ Microbiol 80:2254–2260. doi: 10.1128/AEM.04070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang CD, Chen Q, Wang Q, Li CH, Leng YY, Li SG, Zhou XW, Han WJ, Li JG, Zhang XH, Li YZ. 2014. Long-term batch brewing accumulates adaptive microbes, which comprehensively produce more flavorful Chinese liquors. Food Res Int 62:894–901. doi: 10.1016/j.foodres.2014.05.017. [DOI] [Google Scholar]
- 9.Wang M, Zhang W, Wang H, Liu C. 2012. Phylogenetic diversity analysis of archaeal in the pit mud with different cellar age. Chin J Appl Environ Biol 18:1043–1048. doi: 10.3724/SP.J.1145.2012.01043. [DOI] [Google Scholar]
- 10.Zheng J, Liang R, Zhang L, Wu C, Zhou R, Liao X. 2013. Characterization of microbial communities in strong aromatic liquor fermentation pit muds of different ages assessed by combined DGGE and PLFA analyses. Food Res Int 54:660–666. doi: 10.1016/j.foodres.2013.07.058. [DOI] [Google Scholar]
- 11.Tracy BP, Jones SW, Fast AG, Indurthi DC, Papoutsakis ET. 2012. Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr Opin Biotechnol 23:364–381. doi: 10.1016/j.copbio.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Fan W, Qian MC. 2006. Identification of aroma compounds in Chinese ‘Yanghe Daqu’ liquor by normal phase chromatography fractionation followed by gas chromatography/olfactometry. Flavour Frag J 21:333–342. doi: 10.1002/ffj.1621. [DOI] [Google Scholar]
- 13.Barberán A, Bates ST, Casamayor EO, Fierer N. 2012. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ju F, Xia Y, Guo F, Wang Z, Zhang T. 2014. Taxonomic relatedness shapes bacterial assembly in activated sludge of globally distributed wastewater treatment plants. Environ Microbiol 16:2421–2432. doi: 10.1111/1462-2920.12355. [DOI] [PubMed] [Google Scholar]
- 15.Olsen SR. 1954. Estimation of available phosphorus in soils by extraction with sodium bicarbonate. Circular no. 939. U.S. Department of Agriculture, Washington, DC. [Google Scholar]
- 16.Zhang Y, Cong J, Lu H, Yang C, Yang Y, Zhou J, Li D. 2014. An integrated study to analyze soil microbial community structure and metabolic potential in two forest types. PLoS One 9:e93773. doi: 10.1371/journal.pone.0093773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren G, Ren W, Teng Y, Li Z. 2015. Evident bacterial community changes but only slight degradation when polluted with pyrene in a red soil. Front Microbiol 6:22. doi: 10.3389/fmicb.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu XL, Wang HY, Wu Q, Xu Y. 2014. Development, validation and application of specific primers for analyzing the clostridial diversity in dark fermentation pit mud by PCR-DGGE. Bioresour Technol 163:40–47. doi: 10.1016/j.biortech.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Weimer PJ, Stevenson DM. 2012. Isolation, characterization, and quantification of Clostridium kluyveri from the bovine rumen. Appl Microbiol Biotechnol 94:461–466. doi: 10.1007/s00253-011-3751-z. [DOI] [PubMed] [Google Scholar]
- 20.Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol 67:2578–2585. doi: 10.1128/AEM.67.6.2578-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 24.Zhao D, Huang R, Zeng J, Yu Z, Liu P, Cheng S, Wu QL. 2014. Pyrosequencing analysis of bacterial community and assembly in activated sludge samples from different geographic regions in China. Appl Microbiol Biotechnol 98:9119–9128. doi: 10.1007/s00253-014-5920-3. [DOI] [PubMed] [Google Scholar]
- 25.Harrell FE., Jr 2008. Hmisc: Harrell Miscellaneous. R package version.3.5-2. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 26.Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks. ICWSM 8:361–362. [Google Scholar]
- 27.Aciego Pietri JC, Brookes PC. 2008. Relationships between soil pH and microbial properties in a UK arable soil. Soil Biol Biochem 40:1856–1861. doi: 10.1016/j.soilbio.2008.03.020. [DOI] [Google Scholar]
- 28.Newman ME. 2003. The structure and function of complex networks. SIAM Rev 45:167–256. doi: 10.1137/S003614450342480. [DOI] [Google Scholar]
- 29.Kandler O. 1983. Carbohydrate metabolism in lactic acid bacteria. Antonie Van Leeuwenhoek 49:209–224. doi: 10.1007/BF00399499. [DOI] [PubMed] [Google Scholar]
- 30.Stratford JP, Beecroft NJ, Slade RC, Gruning A, Avignone-Rossa C. 2014. Anodic microbial community diversity as a predictor of the power output of microbial fuel cells. Bioresource Technol 156:84–91. doi: 10.1016/j.biortech.2014.01.041. [DOI] [PubMed] [Google Scholar]
- 31.Spatharis S, Roelke DL, Dimitrakopoulos PG, Kokkoris GD. 2011. Analyzing the (mis)behavior of Shannon index in eutrophication studies using field and simulated phytoplankton assemblages. Ecol Indic 11:697–703. doi: 10.1016/j.ecolind.2010.09.009. [DOI] [Google Scholar]
- 32.Deng Y, Cui X, Hernández M, Dumont MG. 2014. Microbial diversity in hummock and hollow soils of three wetlands on the Qinghai-Tibetan Plateau revealed by 16S rRNA pyrosequencing. PLoS One 9:e103115. doi: 10.1371/journal.pone.0103115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Claesson MJ, O'Sullivan O, Wang Q, Nikkila J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O'Toole PW. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang T, Shao MF, Ye L. 2012. 454 pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J 6:1137–1147. doi: 10.1038/ismej.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sradnick A, Murugan R, Oltmanns M, Raupp J, Joergensen RG. 2013. Changes in functional diversity of the soil microbial community in a heterogeneous sandy soil after long-term fertilization with cattle manure and mineral fertilizer. Appl Soil Ecol 63:23–28. doi: 10.1016/j.apsoil.2012.09.011. [DOI] [Google Scholar]
- 36.de Menezes AB, Prendergast-Miller MT, Richardson AE, Toscas P, Farrell M, Macdonald LM, Baker G, Wark T, Thrall PH. 2014. Network analysis reveals that bacteria and fungi form modules that correlate independently with soil parameters. Environ Microbiol 17:2677–2689. doi: 10.1111/1462-2920.12559. [DOI] [PubMed] [Google Scholar]
- 37.Gray SB, Classen AT, Kardol P, Yermakov Z, Michael Miller R. 2011. Multiple climate change factors interact to alter soil microbial community structure in an old-field ecosystem. Soil Sci Soc Am J 75:2217–2226. doi: 10.2136/sssaj2011.0135. [DOI] [Google Scholar]
- 38.Hernández-Eugenio G, Fardeau ML, Cayol JL, Patel BK, Thomas P, Macarie H, Garcia JL, Ollivier B. 2002. Clostridium thiosulfatireducens sp. nov., a proteolytic, thiosulfate-and sulfur-reducing bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor. Int J Syst Evol Microbiol 52:1461–1468. [DOI] [PubMed] [Google Scholar]
- 39.Cavaleiro A, Sousa D, Alves M. 2010. Methane production from oleate: assessing the bioaugmentation potential of Syntrophomonas zehnderi. Water Res 44:4940–4947. doi: 10.1016/j.watres.2010.07.039. [DOI] [PubMed] [Google Scholar]
- 40.de Bok FA, Harmsen HJ, Plugge CM, de Vries MC, Akkermans AD, de Vos WM, Stams AJ. 2005. The first true obligately syntrophic propionate-oxidizing bacterium, Pelotomaculum schinkii sp. nov., co-cultured with Methanospirillum hungatei, and emended description of the genus Pelotomaculum. Int J Syst Evol Microbiol 55:1697–1703. doi: 10.1099/ijs.0.02880-0. [DOI] [PubMed] [Google Scholar]
- 41.Crespo C, Pozzo T, Karlsson EN, Alvarez MT, Mattiasson B. 2012. Caloramator boliviensis sp. nov., a thermophilic, ethanol-producing bacterium isolated from a hot spring. Int J Syst Evol Microbiol 62:1679–1686. doi: 10.1099/ijs.0.032664-0. [DOI] [PubMed] [Google Scholar]
- 42.Avgustin G, Wallace RJ, Flint HJ. 1997. Phenotypic diversity among ruminal isolates of Prevotella ruminicola: proposal of Prevotella brevis sp. nov., Prevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola. Int J Syst Bacteriol 47:284–288. doi: 10.1099/00207713-47-2-284. [DOI] [PubMed] [Google Scholar]
- 43.Huws SA, Kim EJ, Lee MR, Scott MB, Tweed JK, Pinloche E, Wallace RJ, Scollan ND. 2011. As yet uncultured bacteria phylogenetically classified as Prevotella, Lachnospiraceae incertae sedis and unclassified Bacteroidales, Clostridiales and Ruminococcaceae may play a predominant role in ruminal biohydrogenation. Environ Microbiol 13:1500–1512. doi: 10.1111/j.1462-2920.2011.02452.x. [DOI] [PubMed] [Google Scholar]
- 44.Demirel B, Scherer P. 2008. The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: a review. Rev Environ Sci Biotechnol 7:173–190. doi: 10.1007/s11157-008-9131-1. [DOI] [Google Scholar]
- 45.Balch WE, Fox G, Magrum L, Woese C, Wolfe R. 1979. Methanogens: reevaluation of a unique biological group. Microbiol Rev 43:260–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanwonterghem I, Jensen PD, Dennis PG, Hugenholtz P, Rabaey K, Tyson GW. 2014. Deterministic processes guide long-term synchronised population dynamics in replicate anaerobic digesters. ISME J 8:2015–2028. doi: 10.1038/ismej.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alsaker KV, Paredes C, Papoutsakis ET. 2010. Metabolite stress and tolerance in the production of biofuels and chemicals: gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol Bioeng 105:1131–1147. [DOI] [PubMed] [Google Scholar]
- 48.Chung KT. 1976. Inhibitory effects of H2 on growth of Clostridium cellobioparum. Appl Environ Microbiol 31:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C. 2011. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lauber CL, Hamady M, Knight R, Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brady NC, Weil RR. 1996. The nature and properties of soils. Prentice-Hall Inc, Upper Saddle River, NJ. [Google Scholar]
- 52.Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- 53.Jones WJ, Nagle DP Jr, Whitman WB. 1987. Methanogens and the diversity of archaebacteria. Microbiol Rev 51:135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiu YL, Sekiguchi Y, Hanada S, Imachi H, Tseng IC, Cheng SS, Ohashi A, Harada H, Kamagata Y. 2006. Pelotomaculum terephthalicum sp. nov. and Pelotomaculum isophthalicum sp. nov.: two anaerobic bacteria that degrade phthalate isomers in syntrophic association with hydrogenotrophic methanogens. Arch Microbiol 185:172–182. doi: 10.1007/s00203-005-0081-5. [DOI] [PubMed] [Google Scholar]
- 55.Suetin SV, Shcherbakova VA, Chuvilskaya NA, Rivkina EM, Suzina NE, Lysenko AM, Gilichinsky DA. 2009. Clostridium tagluense sp. nov., a psychrotolerant, anaerobic, spore-forming bacterium from permafrost. Int J Syst Evol Microbiol 59:1421–1426. doi: 10.1099/ijs.0.002295-0. [DOI] [PubMed] [Google Scholar]
- 56.Barker HA, Taha SM. 1942. Clostridium kluyverii, an organism concerned in the formation of caproic acid from ethyl alcohol. J Bacteriol 43:347–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peura S, Bertilsson S, Jones RI, Eiler A. 2015. Resistant microbial co-occurrence patterns inferred by network topology. Appl Environ Microbiol 81:2090–2097. doi: 10.1128/AEM.03660-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cintas L, Casaus M, Herranz C, Nes I, Hernández P. 2001. Review: bacteriocins of lactic acid bacteria. Food Sci Technol Int 7:281–305. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.