Abstract
A feeding trial was performed with broilers receiving a diet of wheat-based feed (WBF), maize-based feed (MBF), or maize-based concentrates supplemented with 15% or 30% crimped kernel maize silage (CKMS-15 or CKMS-30, respectively). The aim of the study was to investigate the bacterial community compositions of the crop, gizzard, ileum, and cecum contents in relation to the feeding strategy and age (8, 15, 22, 25, 29, or 36 days). Among the four dietary treatments, bacterial diversity was analyzed for MBF and CKMS-30 by 454 pyrosequencing of the 16S rRNA gene. Since the diets had no significant influence on bacterial diversity, data were pooled for downstream analysis. With increasing age, a clear succession of bacterial communities and increased bacterial diversity were observed. Lactobacillaceae (belonging mainly to the genus Lactobacillus) represented most of the Firmicutes at all ages and in all segments of the gut except the cecum. The development of a “mature” microbiota in broilers occurred during the period from days 15 to 22. Striking increases in the relative abundances of Lactobacillus salivarius (17 to 36%) and clostridia (11 to 18%), and a concomitant decrease in the relative abundance of Lactobacillus reuteri, were found in the ileum after day 15. The concentration of deconjugated bile salts increased in association with the increased populations of L. salivarius and clostridia. Both L. salivarius and clostridia deconjugate bile acids, and increases in the abundances of these bacteria might be associated with growth reduction and gastrointestinal (GI) disorders occurring in the critical period of broiler life between days 20 and 30.
INTRODUCTION
The chicken gastrointestinal (GI) tract harbors complex communities of bacteria (1, 2). The communities are distributed throughout the GI tract, but due to differences in morphology, functionality, metabolic interactions, and microenvironment, regional heterogeneity in community composition is observed along the different GI segments (2, 3). The composition of the bacterial communities is believed to be influenced mainly by age, diet, and gut location. However, host genetics, rearing environment, stress, immune status, and interactions within bacterial communities are also important factors (1–5). The bacterial communities play a significant role in broiler growth, modulating the development of the digestive tract, influencing the production of bile acids and digestive enzymes, and consequently influencing nutrient digestion and absorption (6–9). Further, they stimulate gut immune functions (10, 11) and prevent the colonization of the GI tract with avian-pathogenic or zoonotic bacteria via competitive exclusion and the production of bacteriocins (9, 12). GI health problems related to subclinical necrotic enteritis and nonspecific small intestinal overgrowth of certain intestinal bacteria (dysbacteriosis) (13) have increased since antibiotic growth promoters (AGPs) as feed additives were banned in the European Union (14, 15) GI health problems in broilers typically occur between the ages of 20 to 30 days and result in wet litter, nonspecific enteritis, poor weight gain, and decreased nutrient digestibility and absorption (13, 16).
In order to control GI problems, different dietary interventions have been suggested; among these, fermented feed has gained increasing attention in animal nutrition. Fermented feed has a low pH (3.5 to 4.5) and contains numerous lactic acid bacteria (LAB) (108 to 109 CFU/g feed) (17, 18) that have been shown to improve chicken gut health (19, 20). The low pH of the fermented feed results in acidification of the upper digestive tract, supporting its function as a barrier against acid-sensitive pathogenic bacteria such as Escherichia coli, Salmonella spp., and Campylobacter spp. (19, 21, 22). Like other fermented products, crimped kernel maize silage (CKMS) is characterized by a low pH, high lactic and acetic acid concentrations, and high LAB numbers. CKMS is used in Danish pig production, primarily as an ingredient of fermented liquid feed, with positive results on gut health. However, the influence of CKMS on the broiler gut microbiota has not yet been investigated. A comprehensive understanding of the development of the GI bacterial community in relation to age and diet is essential in order to address the role of bacteria in the etiology of GI problems and to develop dietary interventions to resolve these problems (4). Therefore, the aim of this study was to investigate the bacterial community composition in different segments of the broiler GI tract in relation to age and diet, with a focus on the use of CKMS in maize-based diets.
MATERIALS AND METHODS
Experimental design.
The trial was performed in accordance with the guidelines of The Animal Experiments Inspectorate (Danish Veterinary and Food Administration). A total of 480 day-old male broiler chicks (Ross 308) were included. They were divided into four groups with four replicates and were distributed randomly into 16 pens (30 birds/pen). The birds were wing-tagged and were reared in pens with a floor area of 1.7 m2 (the floor was covered with a thin layer of wood shavings as bedding material), with automatic control of temperature, light, and humidity. At the age of 14 days, all birds were vaccinated (Nobilis Gumboro D78 Vet; Merck Animal Health) against infectious bursal disease (IBD) via the drinking water. Fresh feed and water were provided ad libitum throughout the experiment.
Compositions of diets.
The starter diet (day 0 to day 8) was the same for all groups and contained 22.5% crude protein (CP) and 11.4 MJ metabolizable energy (ME) per kg of feed (Table 1). In the growing period (day 9 to day 36), the birds were fed one of four grower diets. Group 1 received a wheat-based feed (WBF) containing 150 g/kg whole wheat on days 8 to 28 and 300 g/kg whole wheat on days 29 to 36. The WBF diet was included in the trial because it represents the conventional feeding strategy in Denmark. Group 2 received maize-based feed (MBF) containing 58% maize. For groups 3 and 4, some of the maize was replaced by 15% and 30% crimped kernel maize silage (CKMS), respectively, on a dry matter (DM) basis; these diets are referred to as CKMS-15 and CKMS-30, respectively. All four grower diets were formulated to be isocaloric (11.58 MJ/kg) and isonitrogenous (CP, 19.78%). Additionally, 10% whole wheat was added to all maize-based diets in order to increase the feed structure and ensure optimal gizzard function (23). Whole wheat and CKMS (after thawing overnight) were added and mixed with the pelleted compound feed on the farm prior to feeding. All four pelleted compound feeds were formulated by taking the DM content of CKMS and added whole wheat into consideration in order to obtain similar nutritional compositions after mixing. No enzymes or coccidiostats were used in the entire feeding trial. In all four ready-mixed grower diets, DM, dietary protein, ash, and fat were analyzed using the methods described by Engberg et al. (23), and short-chain fatty acids (SCFA) and nonstarch polysaccharides (NSP) were analyzed using the methods described by Jensen et al. (77) and Bach Knudsen (24), respectively. Starches and sugars were analyzed using the methods described by Bach Knudsen (24) and Jacobsen (25), respectively. The apparent metabolizable energy (AME), expressed in megajoules per kilogram, was calculated as (0.3431 × % fat) + (0.1551 × % crude protein) + (0.1669 × % starch) + (0.1301 × % total sugar) (26). The concentration of phosphorus was measured as described by Carlson and Poulsen (27). Lactic acid bacteria (LAB) were enumerated in the ready-mixed grower diets using MRS agar as described below.
TABLE 1.
Compositions of the starter and grower experimental diets
| Component | Concn in: |
|||||
|---|---|---|---|---|---|---|
| Starter feed for all groups (days 0–8) | Concentrated grower feeda |
|||||
| WBF |
MBF (days 9–36) | CKMS-15 (days 9–36) | CKMS-30 (days 9–36) | |||
| Days 9–28 | Days 29–36 | |||||
| Feed ingredients (g/kg) | ||||||
| Ready-mixed | ||||||
| Wheat | 383.8 | 588.1 | 496.4 | 52.4 | 169.4 | 268.8 |
| Maize | 200.0 | 580.0 | 430.0 | 280.0 | ||
| Rapeseed (LL), milled | 20.0 | 20.0 | 20.0 | 15.0 | 15.0 | 20.0 |
| Soybean meal (toasted) | 323.7 | 310.2 | 381.6 | 301.1 | 329.7 | 366.1 |
| Soybean oil | 22.9 | 38.8 | 50.0 | 10.0 | 10.0 | 13.7 |
| Calcium carbonate | 10.9 | 10.2 | 12.4 | 9.9 | 10.8 | 12.1 |
| Monocalcium phosphate | 17.3 | 15.4 | 18.7 | 15.5 | 17.1 | 18.9 |
| NaCl | 1.9 | 2.3 | 2.7 | 1.5 | 1.9 | 2.2 |
| Sodium-bicarbonate | 2.5 | 1.8 | 2.2 | 2.7 | 2.7 | 2.9 |
| Lysine hydrochloride (100%) | 6.6 | 3.6 | 4.3 | 3.3 | 3.8 | 4.3 |
| dl-Methionine (100%) | 4.7 | 4.1 | 5.0 | 3.7 | 4.1 | 4.7 |
| Threonine (98%) | 1.7 | 1.3 | 1.6 | 0.9 | 1.1 | 1.3 |
| Vitamin and mineral mixtureb | 4.0 | 4.2 | 5.1 | 4.0 | 4.4 | 5.0 |
| Added prior to feeding | ||||||
| Whole wheat | 150.0 | 300.0 | 100.0 | 100.0 | 100.0 | |
| Crimped kernel maize silage | 150.0 | 300.0 | ||||
| Composition of feed as determined by analysisc | ||||||
| Dry matter (g/kg) | 872.4 | 873.7 | 821.3 | 765.9 | ||
| Energy or component/kg of dry matter | ||||||
| AMEd (MJ) | 13.88 | 13.64 | 13.63 | 13.55 | ||
| Crude protein (N × 6.25) (g) | 221.3 | 220.6 | 214.4 | 217.5 | ||
| Starch (g) | 443.2 | 475.0 | 484.9 | 477.3 | ||
| Total sugar (g) | 49.3 | 42.7 | 43.1 | 42.0 | ||
| Total NSP (g) | 113 | 108 | 108 | 107 | ||
| Lignin (g) | 18.0 | 12.0 | 16.0 | 13.0 | ||
| Fat (g) | 70.4 | 50.6 | 48.1 | 48.4 | ||
| Ash (g) | 53.8 | 53.6 | 54.0 | 53.9 | ||
| Phosphorus (g) | 7.0 | 7.4 | 7.4 | 7.5 | ||
| Organic acid (mmol/kg of total feed) | ||||||
| Short-chain fatty acids | ||||||
| Acetic acid | 6.2 | 6.4 | 13.8 | 18.5 | ||
| Propionic acid | 0.0 | 0.0 | 3.2 | 6.8 | ||
| Benzoic acid | 0.0 | 0.0 | 0.4 | 1.1 | ||
| Lactic acid | 2.1 | 2.0 | 26.7 | 66.3 | ||
WBF, wheat-based feed; MBF, maize-based feed; CKMS-15, maize-based feed with 15% crimped kernel maize silage; CKMS-30, maize-based feed with 30% crimped kernel maize silage.
The vitamin and mineral mixture provided the following nutrients (with measurements expressed per kilogram of feed): retinol (retinyl acetate), 12,000 IU; cholecalciferol, 5,000 IU; vitamin E (dl-α-tocopherol acetate), 50 IU; vitamin E (synthetic), 54.9 IU; menadione, 3 mg; thiamine, 2 mg; riboflavin, 6 mg; pyridoxine, 4 mg; d-pantothenic acid, 13 mg; niacin, 55 mg; betaine hydrochloride, 260 mg; folic acid, 2 mg; biotin, 200 μg; cyanocobalamin, 16 μg; calcium d-pantothenate, 1.08 g; FeSO4·7H2O, 20 mg; ZnO, 100 mg; MnO, 120 mg; CuSO4·5H2O, 18 mg; KI, 560 μg; Na2SeO3, 300 μg; CoCO3, 500 μg.
Analysis was conducted on the complete feed after it had been mixed with CKMS and whole wheat. The composition of the starter diet was not analyzed.
AME, apparent metabolizable energy.
Fermentation of crimped kernel maize.
CKMS was obtained from a commercial pig farm in southern Denmark. The maize was harvested and crimped in November 2012 and was then ensiled for approximately 8 weeks with the addition of an organic-acid mixture containing formic, propionic, and benzoic acids and ammonium formate (Kemira AIV Pro, Helsinki, Finland). After ensiling, CKMS was stored immediately in 20-kg evacuated plastic bags in the freezer (−20°C) to maintain similar qualities of silage throughout the feeding trial. DM, pH, and microbial counts (coliform bacteria, lactose-negative bacteria, LAB, yeasts, and molds) were analyzed before and after the ensiling as described by Engberg et al. (23). In summary, DM was analyzed following the freeze-drying of samples. Coliforms and lactose-negative bacteria were enumerated on MacConkey agar incubated at 38°C for 24 h. LAB were counted on MRS agar incubated in an anaerobic cabinet at 38°C for 48 h. Yeasts and molds were enumerated on malt chloramphenicol agar (MCA) incubated aerobically at 25°C for 48 h. Predominant LAB isolated from the MRS plates were further identified by 16S rRNA gene sequencing according to a method described previously (28). The BLAST algorithm was used to determine the closest relatives of the retrieved sequences. The CKMS was tested for the presence of mycotoxins according to the methods described by Sørensen and Sondergaard (29).
Sampling.
At each of six time points (days 8, 15, 22, 25, 29, and 36), four birds were randomly selected from each pen and were killed by cervical dislocation. The birds were weighed individually, and the contents of the crop, gizzard, ileum (the intestinal segment caudal to Meckel's diverticulum), and cecum were collected. Equal amounts of intestinal contents from the four chickens (4 g/bird) of each pen were pooled by segment. The samples were mixed thoroughly and were divided into 2 subsamples for DNA-based and culture-based analysis, respectively. Samples for DNA analysis were immediately frozen in liquid nitrogen and were then stored at −80°C until analysis. The samples for bacterial culture were analyzed for pH using a glass electrode and a reference electrode before bacterial cultures were made.
Bacterial culture and short-chain fatty acid analysis.
Coliform bacteria, lactose-negative enterobacteria, LAB, Clostridium perfringens, and total anaerobes in the crop, gizzard, ileum, and cecum contents were enumerated as described by Engberg et al. (23). The concentrations of SCFA and lactic acid in the crop, gizzard, ileum, and cecum contents were measured as described by Canibe et al. (18).
DNA extraction.
In order to compare the gut bacterial compositions of animals receiving the maize-based diets expected to be most diverse, it was decided to extract DNA from the intestinal contents of birds receiving MBF and from those of birds receiving CKMS-30. DNA was extracted from the crop, gizzard, ileum, and cecum contents of broilers using QIAamp DNA stool minikits (Qiagen). Briefly, approximately 200 mg (400 mg for the ileum) of digesta was placed in a 2-ml microcentrifuge tube. After bead beating for 2 min with 700 μl stool lysis buffer (buffer ASL, provided in the kit), the preparation was centrifuged for 1 min at 7,000 × g, transferred to a 2-ml tube containing 500 μl buffer ASL, vortexed for 1 min, and heated for 5 min at 95°C. The preparation was allowed to cool before the addition of 140 μl lysozyme solution (10 mg/ml) and was then kept at 37°C for 30 min. After centrifugation at 20,000 × g for 1 min, the supernatant was transferred to a new microcentrifuge tube. From this point, the QIAamp DNA stool minikit protocol was followed with the addition of the InhibitEx tablet supplied with the kit.
Bacterial community compositions of MBF and CKMS-30 birds as determined by pyrosequencing of 16S rRNA genes.
The phylogenetic compositions of the bacterial communities were determined by pyrosequencing of the 16S rRNA genes amplified from DNA extracted from digesta. Regions V1 to V3 of the bacterial 16S rRNA gene were amplified from DNA using a two-step protocol as described previously (30). The first amplification was carried out for 20 cycles using the primer set comprising 8fAll (5′-GRGTTYGATYMTGGCTCAG-3′) and HDA2 (5′-GTATTACCGCGGCTGCTGGCAC-3′) (31). The following conditions were used to amplify the 16S rRNA sequences for pyrosequencing: 94°C for 30 s, 56°C for 30 s, 72°C for 45 s, and a final extension step of 72°C for 5 min. This product was diluted 1:5 with PCR-grade water, and 1 μl was used as the template in a 25-μl secondary PCR mixture. The secondary PCR was carried out for 25 cycles using primer 8fAll with 454 sequencing Lib-A adapter sequence A (CGTATCGCCTCCCTCGCGCCATCAGGRGTTYGATYMTGGCTCAG) and primer HDA2 with 454 sequencing Lib-A adapter sequence B plus a 10-base barcode (shown as 10 N′s) (CTATGCGCCTTGCCAGCCCGCTCAGNNNNNNNNNNGTATTACCGCGGCTGCTGGCAC) using conditions identical to those of the primary PCR except that the extension temperature of 72°C was held for 1 min instead of 45 s. Products were cleaned using a NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, Germany) and were quantified using a Qubit double-stranded DNA (dsDNA) high-sensitivity (HS) assay kit and a Qubit fluorometer (both from Invitrogen). Equivalent quantities of the PCR product from each sample were pooled to obtain a set volume of 100 ng/μl, and the pooled products were sent to Macrogen (South Korea) for unidirectional sequencing from the reverse primer using the Roche 454 genome sequencer with titanium chemistry.
Sequencing results were analyzed using Quantitative Insights into Microbial Ecology (QIIME), version 1.8.0, and UPARSE pyrosequencing pipeline packages with default settings (32, 33). Sequences were excluded from analysis if they had an average quality score of <25, contained one or more ambiguous bases, or had >2 mismatches with the sequencing primer. After splitting into bar-coded samples and initial quality filtering, the sequences were passed through the QIIME pipeline using default parameters, including chimera checking. Sequences were filtered to a fixed length of 250 bp, and reads were clustered at 97% sequence identity into operational taxonomic units (OTUs). Taxonomic data were generated for the OTUs using the RDP classifier after filtering out the cyanobacteria by checking against the Greengenes database. Furthermore, BLAST within the NCBI database was used for assigning additional taxonomy data by checking the representative sequences.
Bile acid analysis.
The specific phylogenetic composition of the ileal microbiota has been shown to influence the deconjugation of bile salts and hence lipid digestion (28, 34). Therefore, the concentrations of conjugated and deconjugated bile acids in ileal samples were quantified by reversed-phase high-performance liquid chromatography (HPLC) with pulsed amperometric detection as described previously (35, 36).
Statistical analysis.
The nonparametric statistical tests ANOSIM (analysis of similarity) and ADONIS, based on ANOVA (analysis of variance), were used in order to determine if there were differences between the two treatments (MBF and CKMS-30) based on a categorical variable found in the metadata mapping file. The compare_categories.py script of QIIME, using a distance matrix generated with the unweighted UniFrac metric, was used. A nonparametric t test was used to compare the rarefaction curves (generated by alpha diversity indices using compare_alpha_diversity.py script of QIIME) of samples collected at different ages and from different gut segments.
The effects of diet on pH, microbial counts, SCFA and lactic acid concentrations, relative abundances (expressed as percentages) of bacteria, and bile acids in the intestinal contents over a range of days were analyzed using the mixed-model procedure of SAS for Windows, version 9.3 (SAS Institute, 2012) applying the model Yij = μ + αi + βj + (αβ)ij + Up + εijp, where Yij is the observed response, μ is the overall mean, αi is the effect of treatments (WBF, MBF, CKMS-15, and CKMS-30), βj is the effect of days, (αβ)ij is the effect of the interaction between treatments and days, Up is the variance component that accounts for the correlation between measurements made in the same pen, and εijp is the residual error.
Relative abundance measures of sequencing data were used for statistical analysis, since these data were normalized by the number of sequences per sample. Comparison between the means for diet groups was performed with a t test, and pairwise comparisons were carried out after correction for multiplicity using the Bonferroni-Holm adjustment (37).
Statistical analysis of the results of body weight and bird mortality was conducted using the General Linear Model (GLM) procedure of SAS according to the general model Yi = μ + αi + εi, where Yi is the observed dependent variable, μ is the overall mean, α is the effect of treatments (MBF, CKMS-15, and CKMS-30), and εi is the random error.
Results are reported as least square means (LSMeans) with a pooled standard error (SE). Probability values of ≤0.05 were accepted as indicating significant differences between means.
RESULTS
Biochemical and microbial analyses of the maize and compound feeds.
The pH of the crimped maize decreased from 5.5 to 4.2 after ensiling. During ensiling, LAB counts increased from 7.54 to 8.23 log10 CFU/g, while the counts of coliform bacteria, lactose-negative enterobacteria, yeasts, and molds decreased below 3.00 log10 CFU/g. Three predominant LAB species, namely, Lactobacillus plantarum subsp. plantarum, Lactobacillus brevis, and Lactobacillus nantensis, were identified in CKMS by 16S rRNA gene sequencing of isolates. Among the ready-mixed grower diets, LAB counts were highest in CKMS-30 (6.89 log10 CFU/g), followed by CKMS-15 (6.54 log10 CFU/g), MBF (3.60 log10 CFU/g), and WBF (3.00 log10 CFU/g). Tests for mycotoxins in the CKMS showed low concentrations of deoxynivalenol (87 μg/kg DM) and enniatin B (38 μg/kg DM).
Production performance.
Final body weight at day 36 was higher (P = 0.02) for MBF-fed broilers (2,918 g/bird) than for CKMS-30-fed broilers (2,610 g/bird). The weight of broilers fed WBF or CKMS-15 was recorded as 2,501 g/bird or 2,752 g/bird, respectively. Broiler mortality was significantly lower (P < 0.001) in groups fed CKMS. No mortality was recorded in the CKMS-30 group, while mortality was 0.8% in the CKMS-15 group. Higher mortality was observed in broilers fed WBF (6.7%) and MBF (4.2%).
Culture-based enumerations of bacteria, pH, and short-chain fatty acid concentrations.
Bacterial counts (see Table S1 in the supplemental material) revealed only minor differences between the dietary treatments. In the gizzard, the counts of LAB (P = 0.008) and total anaerobic bacteria (P < 0.001) were lower when birds were fed maize-based diets than when they received WBF. Counts of coliforms and lactose-negative enterobacteria in the ileum and cecum tended to be lower in birds fed maize-based diets. The number of LAB in the gizzard was higher when birds received WBF (P = 0.008) than when they received maize-based diets. In the other segments, no differences were found between the dietary groups with respect to LAB. The results further showed an influence of broiler age on bacterial counts. In the crop, the numbers of coliforms (P = 0.005) and lactose-negative enterobacteria (P = 0.006) decreased with age and were significantly lower on day 29 and day 36 (see Table S1). No differences in bacterial counts were observed in the gizzard. In ileal contents, lactose-negative enterobacteria increased approximately 10-fold from day 8 to day 15 (P < 0.001) and decreased afterwards. Likewise, in cecal contents, the numbers of coliform bacteria (P < 0.001), lactose-negative enterobacteria (P < 0.001), and LAB (P < 0.001) decreased gradually as the birds grew older.
The pH of gizzard contents (see Table S2 in the supplemental material) was lowest (pH 3), followed by that of crop contents (pH 5). In birds fed CKMS, the pH of crop contents tended to be lower (P = 0.08) than those for the other groups. The cecal pH was lowest when birds were fed WBF (P < 0.001). Except for the crop, the pH of the intestinal contents varied with bird age. In the gizzard (P = 0.009) and cecum (P < 0.001), the pH increased throughout the growth period and was significantly higher at day 36 than at day 8. Ileal pH was lowest during the period from day 21 to day 25 and increased again until day 36 (P < 0.001). The major product of microbial fermentation was lactic acid in the crop, gizzard and ileum, whereas it was acetic acid in the cecum (see Table S2). The concentration of propionic acid increased from day 8 to day 36. A similar trend was observed with respect to acetic acid in ileal contents. In the cecum, lactic acid (P < 0.001) and succinic acid (P < 0.001) concentrations declined rapidly at day 15 and remained low throughout the growing period, while the butyric acid concentration increased with bird age (P < 0.001). In general, the concentration of organic acids was highest in the cecum and lowest in the gizzard. The influence of diets on organic-acid concentrations in different GI segments was minimal. Since the influence of diet on microbial counts and organic-acid concentrations appeared limited, it was decided to conduct 16S rRNA gene pyrosequencing only on samples from birds receiving either of two extreme treatments based on maize, i.e., MBF and CKMS-30.
Compositions of bacterial communities in birds fed MBF or CKMS-30 as determined by pyrosequencing of 16S rRNA gene amplicons.
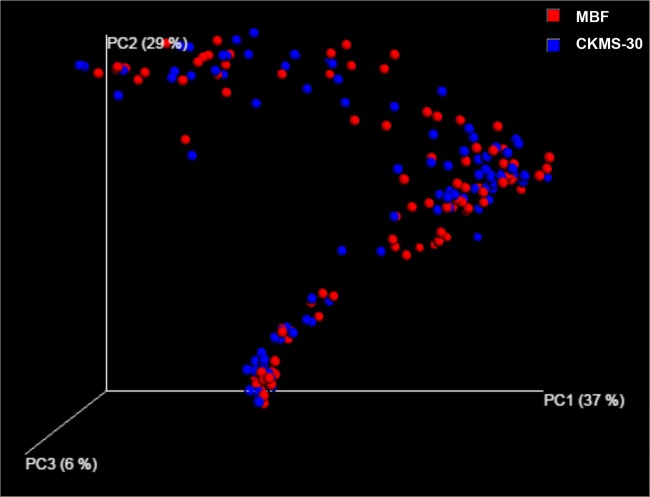
After quality filtering using UPARSE, an average of 1,490 or 1,321 sequences was recovered from each bar-coded sample, with a total of 143,081 or 126,798 sequences obtained from birds fed MBF or CKMS-30, respectively, for downstream analysis. The sequences were clustered into 292 OTUs or 308 OTUs for MBF or CKMS-30, respectively, using a 97% similarity cutoff. Alpha diversity analysis revealed comparable bacterial communities for the two dietary groups (P = 0.23). Likewise, unweighted UniFrac-based distance matrices (beta diversity) showed no difference in microbiota composition between the two diets (Fig. 1). The statistical test run by QIIME further confirmed that there were no significant differences regarding beta diversity (P, 0.07 by ANOSIM and 0.08 by ADONIS) between the MBF and CKMS-30 groups. Comparable results were obtained when individual but similar gut segments of MBF and CKMS-30 birds were compared separately, e.g., the crop (P, 0.36 by ANOSIM and 0.35 by ADONIS), gizzard (P, 0.42 by ANOSIM and 0.61 by ADONIS), ileum (P, 0.19 by ANOSIM and 0.30 by ADONIS), and cecum (P, 0.55 by ANOSIM and 0.68 by ADONIS). Since no apparent differences were observed between the MBF and CKMS-30 groups, further analyses were carried out on a pooled data set to study the succession of bacterial communities in the different GI segments.
FIG 1.
Principal coordinate analysis plot of unweighted pairwise UniFrac distances showing clustering of bacterial groups in the intestinal contents of broilers fed MBF (red spheres) or CKMS-30 (blue spheres). The plot for each dietary treatment represents the pooled data for six sampling days (days 8, 15, 22, 25, 29 and 36) and four intestinal segments (the crop, gizzard, ileum, and cecum).
Temporal variation of bacterial community compositions in the crop, gizzard, ileum, and cecum as determined by pyrosequencing of 16S rRNA gene amplicons.
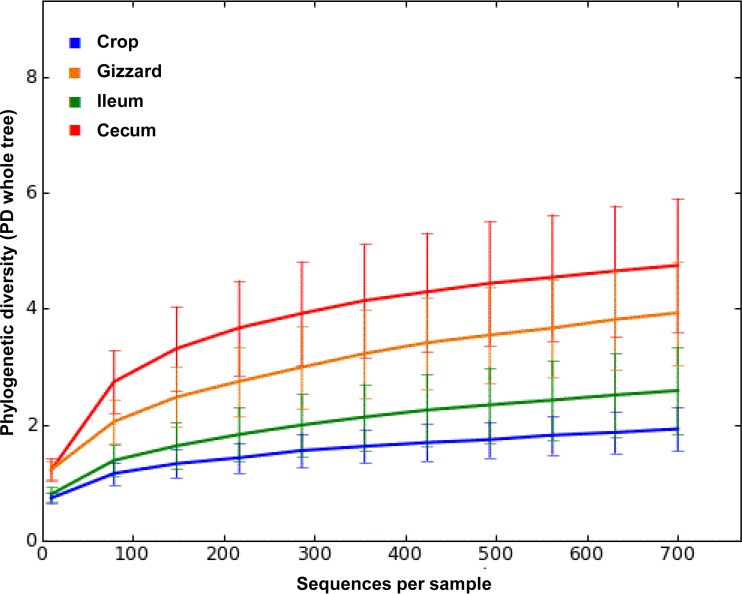
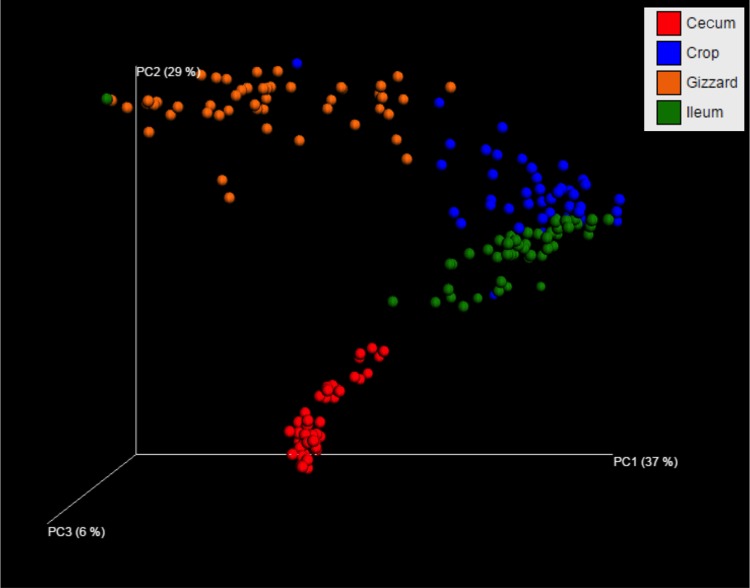
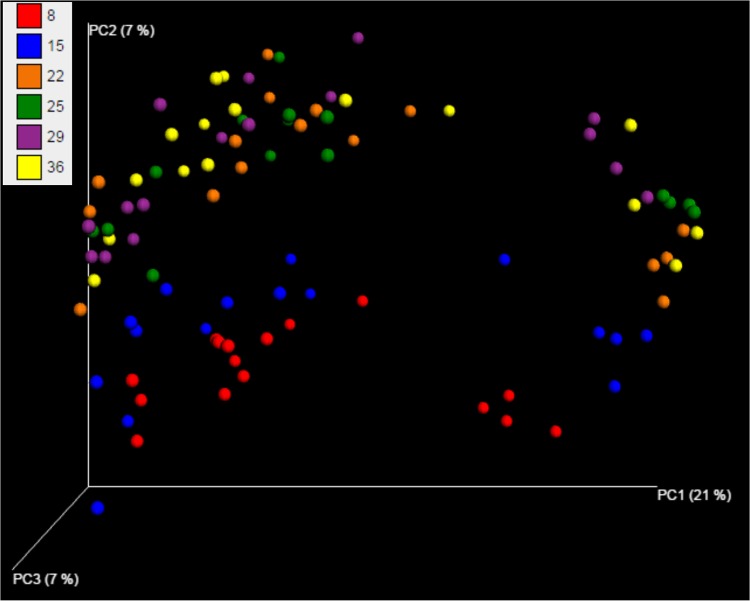
A total of 277,294 reads were obtained from the 192 samples covering an average of 1,444 reads per sample after quality filtering using UPARSE. At 97% similarity, the filtered sequences were clustered into 324 OTUs. The rarefaction curves for alpha diversity by the PD_whole_tree (Fig. 2) and Chao1, “Observed Species,” and Shannon and Simpson (not shown) diversity indices further demonstrated that the greatest diversity of the microbiota was present in the cecum, followed by the gizzard, ileum, and crop. The alpha diversities for the GI segments were significantly different from each other, as shown by PD_whole_tree (P = 0.02) and other diversity indices (P = 0.01) (data not shown). Higher alpha diversity was observed with increasing age of the birds, as demonstrated by various diversity metrics (data not shown). There was a clear increase (P < 0.001) in alpha diversity from day 22 onward as shown by different matrices, but a significant drop in diversity was observed at day 29 (see Table S3 in the supplemental material). Furthermore, beta diversity metrics clearly indicated the difference in the bacterial communities among GI segments (Fig. 3) and age groups (Fig. 4). Statistical analysis of beta diversity further demonstrated the differences between GI segments (P, 0.001 by both ANOSIM and ADONIS) and birds of different ages (P, 0.001 by both ANOSIM and ADONIS).
FIG 2.
Alpha diversity rarefaction curves measuring phylogenetic diversity (PD) with respect to sequence accumulation per sample of four intestinal segments (crop, gizzard, ileum, and cecum). The alpha diversity plot for each segment represents the pooled data for six sampling days (days 8, 15, 22, 25, 29, and 36) from both dietary treatments (MBF and CKMS-30).
FIG 3.
Principal coordinate analysis plot of unweighted pairwise UniFrac distances showing clustering of bacterial groups present in the crop (blue spheres), gizzard (orange spheres), ileum (green spheres), and cecum (red spheres) contents. The plot for each segment represents the pooled data for all sampling days (days 8, 15, 22, 25, 29, and 36) and two dietary treatments (MBF and CKMS-30).
FIG 4.
Principal coordinate analysis plot of unweighted pairwise UniFrac distances showing clustering of bacterial groups present on six sampling days (days 8, 15, 22, 25, 29, and 36). The plot represents the pooled data for four intestinal segments (crop, gizzard, ileum, and cecum) and two dietary treatments (MBF and CKMS-30).
In all age groups, the Firmicutes were the predominant phylum throughout the GI tract, representing 97 to 98% of the bacterial community in the crop (see Table S4 in the supplemental material), 87 to 98% in the gizzard (see Table S4), 94 to 98% in the ileum (see Table S5 in the supplemental material), and 58 to 96% in the cecum (see Table S6 in the supplemental material). The relative abundance of Firmicutes varied only in the gizzard (P = 0.031) and cecum (P < 0.001), where it decreased as broilers grew older. Actinobacteria were the only phylum identified in the crop with an abundance of <1% during the entire growth period (see Table S4). In the gizzard, Proteobacteria decreased with age (P = 0.01), but Actinobacteria became dominant (P < 0.001) in older broilers (see Table S4). In the ileum (day 8 and day 15) and cecum (day 8), Proteobacteria were the second most abundant phylum, but Actinobacteria dominated the ileum, whereas Bacteroidetes became dominant in the ceca of older broilers. In the cecum (see Table S6), the levels of Bacteroidetes (P < 0.001), which were detected only from day 15 (2%), were increased at day 22 (24%) and day 29 (36%).
Within the phylum Firmicutes, Lactobacillaceae was the most abundant family in the crop (94 to 98%), gizzard (59 to 86%), and ileum (61 to 73%), and abundances were stable throughout the growing period of broilers. In the gizzard, the relative abundances of Lachnospiraceae, Enterococcaceae, and Comamonadaceae were higher in the early growth period, but gradually the abundance of microbiota shifted toward Planococcaceae, Clostridiaceae, Staphylococcaceae, Brevibacteriaceae, and Corynebacteriaceae in the final growth period of broilers. The Ruminococcaceae (4 to 9%) were relatively stable in the gizzard at all ages (see Table S4 in the supplemental material). In the ileum, Enterococcaceae (P < 0.001) showed moderate abundance (25%) at day 7 but declined sharply below 1% at day 15 and remained low throughout the growing period, whereas Clostridiaceae increased from 5% at day 8 to 19% at day 36 (see Table S5 in the supplemental material). In contrast to the crop, gizzard and ileum, where Lactobacillaceae were dominant, Ruminococcaceae and Lachnospiraceae, belonging to the phylum Firmicutes, and Rikenellaceae (phylum Bacteroidetes) were most abundant in the cecum. The abundances of Lachnospiraceae, Lactobacillaceae, and Enterobacteriaceae shifted significantly in favor of Rikenellaceae and Clostridiaceae as broilers grew older (see Table S6 in the supplemental material). The Rikenellaceae sharply increased from 2% (day 15) to 24% (day 22) and then to 36% (day 29). In contrast, Lachnospiraceae and Ruminococcaceae declined by 8% and 11%, respectively, at day 22. A 10% decrease in the relative abundance of Ruminococcaceae was observed on day 29.
At the genus level, Corynebacterium and Staphylococcus represented most of the Corynebacteriaceae and Staphylococcaceae, respectively, in the crop, but their abundances remained below 1% (see Table S4 in the supplemental material). At day 8, the gizzard was dominated mainly by Roseburia (6%) and Ruminococcus (7%), but gradually their abundances decreased below 2% as the broilers grew older. In the meantime, bacterial genera started to alter at day 22, with increased abundances of Clostridium, Staphylococcus, Acinetobacter, and Corynebacterium (see Table S4). In the ileum, the relative abundance of Enterococcus decreased sharply from 25% at day 8 to below 1% at day 15 and remained low thereafter. However, increases in the relative abundances of Clostridium (1% to 18%) and Streptococcus (1% to 5%) were observed during the growth of broilers from day 8 to day 36 (see Table S5 in the supplemental material). In the cecum, Ruminococcus (16 to 23%) was the dominant genus, and its levels remained stable throughout the growing period. As the broilers grew older, the relative abundances of Roseburia (20% to 8%), Blautia (10% to 2%), and Escherichia (5% to 0.2%) decreased, whereas the relative abundances of Faecalibacterium (0.4% to 8%), Alistipes (0 to 20%), and Clostridium (0.7% to 4%) increased. A significant influence of diet on the abundances of Streptococcus in ileal (P = 0.004) and cecal (P = 0.03) contents was observed. The abundance of Streptococcus was higher in broilers receiving MBF than in those receiving CKMS-30 (data not shown).
Temporal variation of Lactobacillus species in the crop, gizzard, ileum, and cecum.
Lactobacillus represented most of the Lactobacillaceae (99 to 100%) in all GI segments. In the crop (94 to 98%) and ileum (61 to 73%), the genus Lactobacillus was dominant and remained stable during the entire broiler production period. In the gizzard, Lactobacillus abundance decreased gradually from day 15 (86%) to day 36 (58%), whereas in the cecum, it decreased from 17% to 8% at day 15 and remained low (2 to 3%) during the rest of the growth period (P < 0.001). At all ages, four Lactobacillus species (namely, Lactobacillus johnsonii, Lactobacillus reuteri, Lactobacillus crispatus, and Lactobacillus salivarius) were present in all segments (Table 2). In general, L. johnsonii was dominant through most of the growing period of broilers. Although L. plantarum and L. brevis were present in high numbers in birds fed CKMS-30 (6.89 log10 CFU/g), their relative abundances in the GI tract were very low. They were not detected in crop contents, whereas their abundances in the gizzard, ileum, and cecum were <0.3% (data not shown). The relative abundances of L. reuteri and L. crispatus were higher during the early growth period (up to day 15), whereas L. salivarius dominated the final growth phase at the expense of L. reuteri (from day 22) in all the GI segments. In cecal contents, the abundance of L. salivarius was influenced by the diet and was higher (P = 0.001) in broilers fed CKMS-30 than in those fed MBF (Table 2). Sharp increases in the relative abundances of L. salivarius were observed from day 15 to day 22 in the crop (26%), gizzard (8%), and ileum (25%), and levels remained high throughout the growing period.
TABLE 2.
Relative abundancesa of Lactobacillus species in the crops, gizzards, ilea, and ceca of broilers at different ages
| Intestinal segment and species | Relative abundance with the following diet: |
SE for abundance by diet | Relative abundance on day: |
SE for abundance by day |
P value for effectb of: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MBF | CKMS-30 | 8 | 15 | 22 | 25 | 29 | 36 | Day | Diet | |||
| Crop | ||||||||||||
| L. johnsonii | 50.0 | 47.6 | 4.3 | 47.7 | 56.4 | 51.6 | 46.3 | 50.5 | 40.4 | 10.1 | 0.719 | 0.598 |
| L. reuteri | 14.5 | 15.0 | 3.4 | 19.8 AB | 23.8 A | 8.7 B | 17.0 AB | 8.3 B | 10.9 AB | 4.4 | 0.005 | 0.868 |
| L. crispatus | 5.3 | 6.0 | 4.2 | 23.3 A | 4.2 B | 1.8 B | 0.5 B | 1.5 B | 2.8 B | 7.7 | 0.047 | 0.901 |
| L. salivarius | 24.8 | 23.9 | 5.5 | 2.2 B | 7.0 B | 32.6 A | 32.7 A | 33.6 A | 37.9 A | 7.6 | <0.001 | 0.869 |
| Gizzard | ||||||||||||
| L. johnsonii | 37.3 | 36.3 | 5.8 | 36.0 AB | 55.4 A | 33.1 AB | 31.1 B | 32.4 AB | 32.9 AB | 8.3 | 0.049 | 0.871 |
| L. reuteri | 5.6 | 7.2 | 1.8 | 7. 0AB | 11.8 A | 4.1 AB | 6.2 AB | 3.2 B | 6.0 AB | 2.7 | 0.053 | 0.412 |
| L. crispatus | 2.9 | 2.9 | 1.2 | 6.5 | 3.6 | 1.7 | 0.9 | 1.6 | 3.2 | 2.5 | 0.261 | 0.987 |
| L. salivarius | 8.0 | 8.1 | 1.5 | 0.3 B | 3.7 AB | 12.3 A | 14.5 A | 10.7 AB | 6.8 AB | 3.8 | 0.006 | 0.935 |
| Ileum | ||||||||||||
| L. johnsonii | 38.5 | 37.9 | 5.3 | 34.8 | 45.1 | 39.6 | 48.8 | 34.6 | 26.5 | 8.5 | 0.139 | 0.908 |
| L. reuteri | 4.6 B | 6.4 A | 0.4 | 10. 0 A | 10.0 A | 3.6 B | 5.0 AB | 3.2 B | 1.6 B | 1.3 | 0.003 | <0.001 |
| L. crispatus | 5.7 | 5.6 | 2.3 | 15.6 | 7.2 | 3.0 | 2.2 | 2.2 | 3.7 | 5.2 | 0.100 | 0.988 |
| L. salivarius | 16.7 | 19.5 | 3.7 | 0.9 B | 0.9 B | 26.5 A | 17.1 AB | 27.3 A | 35.8 A | 6.6 | <0.001 | 0.496 |
| Cecum | ||||||||||||
| L. johnsonii | 3.0 | 2.4 | 0.4 | 7.0 A | 4.0 B | 1.4 BC | 1.4 BC | 0.8 C | 1.1 C | 0.9 | <0.001 | 0.230 |
| L. reuteri | 1.2 | 2.2 | 0.5 | 6.7 A | 2.6 B | 0.2 B | 0.1 B | 0.2 B | 0.3 B | 1.1 | <0.001 | 0.074 |
| L. crispatus | 0.7 | 0.8 | 0.5 | 3.6 A | 0.8 B | 0.1 B | 0.01 B | 0.0 | 0.1 B | 0.8 | <0.001 | 0.839 |
| L. salivarius | 0.6 B | 1.4 A | 0.1 | 0.1 B | 0.3 B | 1.2 B | 1.5 A | 1.3 A | 1.8 A | 0.4 | <0.001 | 0.001 |
Expressed as LSMeans of percentages. Only Lactobacillus species with a relative abundance of >1% on one of the six sampling days are presented here. Values followed by different capital letters in the same row differ significantly (P ≤ 0.05).
No significant interaction between day and diet was observed.
Ileal bile acid concentrations.
The composition of the microbiota of the ileum is known to be associated with the deconjugation of bile salts in that region (28, 34, 38). We observed that the proportion of deconjugated bile acids was higher in the ilea of birds receiving CKMS-30 (P = 0.018) than in those of the other groups (Table 3). An age-related difference was found with regard to the total bile acid concentration (P = 0.001): the highest concentrations were observed on day 8 and day 36. The proportion of deconjugated bile acids decreased from day 8 to day 15 and then increased again at day 22 to remain relatively stable during the rest of the growth period (P = 0.021).
TABLE 3.
Bile acid concentrations and proportions of conjugated and deconjugated bile acids in ileal contents of broilers fed wheat-based feed, maize-based feed, or maize-based feed with 15% or 30% crimped kernel maize silage at different agesa
| Bile acids | Concn or proportion with the following diet: |
SE for diet | Concn or proportion on the following day: |
SE for day |
P valueb for effect of: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WBF | MBF | CKMS-15 | CKMS-30 | 8 | 15 | 22 | 25 | 29 | 36 | Diet | Day | |||
| Total (mmol/kg) | 59.1 | 51.0 | 49.9 | 58.5 | 4.6 | 61.9 A | 42.4 C | 47.6 B | 57.3 AB | 49.5 B | 69.0 A | 4.2 | 0.143 | 0.001 |
| Total deconjugated (%) | 36.4 B | 38.8 AB | 37.8 B | 43.6 A | 2.0 | 42.6 A | 33.7 B | 40.2 A | 40.6 A | 39.5 A | 38.3 AB | 2.5 | 0.018 | 0.021 |
| Total conjugated (%) | 63.6 A | 61.2 AB | 62.2 A | 56.4 B | 2.0 | 57.4 B | 66.3 A | 59.8 B | 59.3 B | 60.5 B | 61.7 AB | 2.5 | 0.018 | 0.021 |
Results are expressed as LSMeans. Values followed by different capital letters in the same row differ significantly (P ≤ 0.05).
No significant interaction between day and diet was observed.
DISCUSSION
Fermented feed has been suggested as a promising nutritional strategy to modulate the gut microbiota, resulting in an effective acidification of the upper digestive tract and thus preventing acid-sensitive, potentially pathogenic bacteria from colonizing the intestine (19, 21, 22, 39, 40). However, as determined by culture-dependent and culture-independent methods, the influence of CKMS on the gut bacterial community was quite limited. Considering the lower mortality of birds receiving CKMS, this result may be surprising. However, the difference in mortality has been found to be closely related to lower drinking water intake, improved litter quality, and improved footpad health (41).
It should be noted that in this study, fermented maize was used as a feed supplement at a maximum inclusion level of 30%. Dietary supplementation with CKMS at that concentration contributes a Lactobacillus concentration of only ∼7 log10 CFU/g feed, which is approximately 1,000 times lower than that in fermented compound feed (9 log10 to 10 log10 CFU/g feed) as reported by Engberg et al. (19). The minimal effect of the diet used in this study was further supported by similar pH values and organic-acid concentrations in intestinal contents (see Table S2 in the supplemental material). However, due to the addition of propionic acid during the ensiling process, the concentration of propionic acid in CKMS was higher than that in unfermented maize, which was reflected by higher propionate concentrations in the crops of birds receiving CKMS. The concentration of propionic acid in the crop increased with age due to the increased feed intake of the growing broiler. The higher concentrations of acetate in the crops of birds fed CKMS can likewise be explained by the fermentation process, where acetate is a fermentation product of a variety of bacteria.
Since higher LAB counts were found in CKMS-30 (6.89 log10 CFU/g) than in MBF (3.60 log10 CFU/g), differences between LAB numbers in the GI contents of the two dietary groups were expected. However, considering the high Lactobacillus numbers naturally present in crop contents (109 CFU/g), the contribution of CKMS, providing Lactobacillus spp. at about 107 CFU/g of diet, will be approximately 1%. Likewise, differences in the abundances of different Lactobacillus species were expected, especially in crop contents, since CKMS provided L. plantarum, L. brevis, and L. nantensis. However, these species were found to have negligible abundances (0.3% in all GI tract segments) or were not identified at this location (Table 2). It would be interesting to understand the fate of these species and the possible modulatory action of probiotics containing Lactobacillus spp. on the gut microbiota. Based on our results, the modulatory action of Lactobacillus spp. provided as probiotic feed additives or fermented feed will possibly be more effective if used in higher concentrations, exceeding 109 CFU/g of feed.
The present study showed a dynamic heterogeneity of the gut microbiota in relation to the different GI segments (crop, gizzard, ileum, and cecum), along with the development of bacterial diversity in growing broilers. Differences in bacterial composition between different GI segments were observed (Fig. 3). These differences have been reported to be related to differences in gut morphology, pH, oxygen concentration, nutrient availability, and the presence of bile acids and digestive enzymes (42). As in previous reports (43–45), the highest bacterial diversity was found in the cecum (Fig. 2), a finding supported by both higher concentrations and a higher diversity of organic acids in cecal contents (see Table S2 in the supplemental material).
The dominant bacteria in the crop belonged to the genus Lactobacillus, with a relative abundance of 97 to 98% (see Table S4 in the supplemental material), which is in agreement with earlier reports (46, 47). The crop is a habitat unlike other parts of the digestive tract. Lactobacilli adhere to the surface of the nonsecretory, squamous epithelium and multiply to produce a biofilm. The production of lactic acid by the lactobacilli produces a hostile environment for other bacteria. The numerical dominance of the lactobacilli in the crop is a normal feature that has been extensively documented in the scientific literature (48–50). Lactobacilli are shed from the crop epithelium, pass to the remainder of the gut, and hence dominate the collections of bacteria found in the small intestine. They are not dominant in the cecum, as verified by our data (2 to 3% of mature microbiota), due to the predominance of obligately anaerobic bacteria.
In contrast to previous studies reporting the presence of only four bacterial genera (Enterococcus, Streptococcus, Lactobacillus, and Escherichia) in the gizzard (51–53), our results revealed a relatively complex bacterial community at this location, comprising 5 phyla and 17 genera (see Table S4 in the supplemental material). The sequencing of 16S rRNA in this study, as opposed to the limitations of the culturing techniques in the previous studies, is responsible for obtaining a more diverse bacterial population in the gizzard. Compared to that in the other segments, the bacterial composition of gizzard contents seems to vary considerably (Fig. 3). However, due to the low pH in the gizzard, low bacterial numbers (see Table S1 in the supplemental material) and decreased bacterial activity were recorded, as indicated by low lactic and acetic acid concentrations (see Table S2 in the supplemental material). Compared to the cecum, lower bacterial activity, in terms of lower organic-acid concentrations, was found in the ileum (see Table S2), which can be explained by a shorter transit time of digesta at that location (42).
As in previous reports (54–56), bacterial diversity increased with broiler age, as indicated by the presence of a more complex bacterial community structure in older broilers (see Table S3 in the supplemental material). As shown in Fig. 4, the bacterial composition was different from day 22 onward from that observed up to day 15. No difference in beta diversity was observed between day 22 and day 36, indicating the development of a “mature” microbiota after day 15. Bacteria belonging to the genus Lactobacillus are the early colonizers of the entire GI tract in the young bird (57, 58). Throughout the whole production period, the abundance of lactobacilli remained relatively stable in the crop, gizzard, and ileum, whereas in the cecum, Lactobacillus spp. were replaced by other bacteria beginning at day 15. These findings are supported by the LAB counts (see Table S1 in the supplemental material) and lactic acid concentrations (see Table S2 in the supplemental material), which remained constant in the crop, gizzard, and ileum throughout the growing period. However, in the cecum, a decline in the relative abundance of Lactobacillus spp. with increasing age of the broilers was accompanied by a decreased LAB count (see Table S1). Lactic acid concentrations in the cecum are usually low in the older bird, since lactic acid is a substrate for butyrate-producing bacteria, such as Clostridium, Faecalibacterium, and Ruminococcus spp. (59, 60).
Clostridium has been reported to be a major genus in the cecum, ranging from 20 to 47% depending on the age of the bird (55, 61, 62). In the present study, the relative abundance of Clostridium spp. in the cecum, estimated by DNA sequencing, was below 5% throughout the growing period. This is in agreement with the low cecal counts of C. perfringens (see Table S1 in the supplemental material). The genus Bacteroides was not detected in the ileum or cecum at any time point, in contrast to reports by other authors of abundances of 1 to 20% in the ileum (55, 63) and 18 to 22% in the cecum (55, 63–65). In the present study, the genus Alistipes, belonging to the same phylum (Bacteroidetes), was found to have an abundance similar to the reported Bacteroides abundances (up to 36%) in the cecum. In comparing the results of these studies, it should be noted that the rearing environment, dietary ingredients, growth rate, and age influence the gut microbiota of poultry (4, 55, 56, 66, 67). Therefore, a direct comparison of the results from this study with those from the others may be difficult.
With respect to different Lactobacillus species, a clear temporal variation was found throughout the GI tract (Table 2). The increase in the abundance of L. salivarius in the period after day 15 was associated with a shift in Lactobacillus species abundances in which L. salivarius replaced L. reuteri and L. crispatus in older broilers. The abundances of L. reuteri and L. salivarius were in accordance with the report that L. reuteri is an early colonizer, whereas L. salivarius is detected in older birds (58). This study further determined the direct influence of the Lactobacillus composition of the upper digestive tract (crop and gizzard) on the succession of Lactobacillus species in the ileum and cecum (Table 2). A similar succession of Lactobacillus species starting from the crop and continuing to the ileum has been observed previously (58). The crop can be considered a seeding organ for the lower digestive tract, which implies that a modulation of the composition of the Lactobacillus population in the upper digestive tract may be a useful tool for achieving a desired Lactobacillus population in the lower gut, especially the ileum, in order to maintain gut health.
Lactobacillus species are commonly used as probiotics in poultry nutrition due to their potential role in decreasing colonization by enteropathogens through competitive exclusion, antagonistic activity, and the production of bacteriocins (9, 68). Among others, L. salivarius has gained increasing attention as a promising probiotic species (69–71), and the bacteriocins produced by L. salivarius have been reported to decrease the colonization of broilers by enteropathogens such as Campylobacter and Salmonella spp. (70, 72). Although L. salivarius is used as a probiotic, its ability to deconjugate bile acids has been overlooked. When L. salivarius deconjugates bile acids, they lose their ability to emulsify lipids and form micelles, thus leading to poor fat digestion and depressed broiler growth (28, 73, 74). The increased relative abundance of L. salivarius in the ileum coincided with the period between days 20 and 30. The relative abundance of the genus Clostridium also increased from 8% to 18% between day 22 and day 36. The genus Clostridium includes potentially pathogenic species, such as C. perfringens, known to play a role in the development of dysbacteriosis (16, 75). As reported by Engberg et al. (76) and Guban et al. (74), significantly lower levels of both L. salivarius and C. perfringens were recorded in broilers receiving feed supplemented with antimicrobial drugs than in those receiving a nonsupplemented control. The final body weight of broilers receiving CKMS-30 was 300 g lower than that of broilers fed MBF (P = 0.02). As shown in Table 3, birds fed CKMS-30 had higher concentrations of deconjugated bile acids in the ileum than bird fed MBF. This suggests that L. salivarius (Table 2) is associated with bile acid deconjugation and probably contributed to the impaired growth of birds receiving CKMS-30.
In summary, the inclusion of CKMS in a maize-based diet had no influence on the diversity of the gut microbiota, which did not differ significantly from that with the control maize diet, but the results demonstrate a significant alteration in the abundances of specific bacterial populations in relation to the age of broilers. The significant increases in the ileal abundances of L. salivarius and clostridia occurring after day 15 could be a factor contributing to the reduced growth performance of broilers after the withdrawal of AGPs as feed additives. It is important, therefore, to test the effects of new nutritional strategies for broilers on the composition of the microbiota, particularly during the period of 20 to 30 days after hatching.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Trine Poulsen, Thomas Rebsdorf, and Karin Durup for skillful technical assistance. Senior Scientist Søren Krogh Jensen is thanked for the analysis of bile acids in intestinal contents.
This work has been kindly funded by the Danish Ministry of Food, Agriculture and Fisheries and the Fjerkræafgiftsfond.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02549-15.
REFERENCES
- 1.Wei S, Morrison M, Yu Z. 2013. Bacterial census of poultry intestinal microbiome. Poult Sci 92:671–683. doi: 10.3382/ps.2012-02822. [DOI] [PubMed] [Google Scholar]
- 2.Yeoman CJ, Chia N, Jeraldo P, Sipos M, Goldenfeld ND, White BA. 2012. The microbiome of the chicken gastrointestinal tract. Anim Health Res Rev 13:89–99. doi: 10.1017/S1466252312000138. [DOI] [PubMed] [Google Scholar]
- 3.Choi JH, Kim GB, Cha CJ. 2014. Spatial heterogeneity and stability of bacterial community in the gastrointestinal tracts of broiler chickens. Poult Sci 93:1942–1950. doi: 10.3382/ps.2014-03974. [DOI] [PubMed] [Google Scholar]
- 4.Gabriel I, Lessire M, Mallet S, Guillot JF. 2006. Microflora of the digestive tract: critical factors and consequences for poultry. World's Poult Sci J 62:499–511. doi: 10.1017/S0043933906001115. [DOI] [Google Scholar]
- 5.Yeoman CJ, White BA. 2014. Gastrointestinal tract microbiota and probiotics in production animals. Annu Rev Anim Biosci 2:469–486. doi: 10.1146/annurev-animal-022513-114149. [DOI] [PubMed] [Google Scholar]
- 6.Choct M. 2009. Managing gut health through nutrition. Br Poult Sci 50:9–15. doi: 10.1080/00071660802538632. [DOI] [PubMed] [Google Scholar]
- 7.Yegani M, Korver DR. 2008. Factors affecting intestinal health in poultry. Poult Sci 87:2052–2063. doi: 10.3382/ps.2008-00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers JR, Gong J. 2011. The intestinal microbiota and its modulation for Salmonella control in chickens. Food Res Int 44:3149–3159. doi: 10.1016/j.foodres.2011.08.017. [DOI] [Google Scholar]
- 9.Lan Y, Verstegen MWA, Tamminga S, Williams BA. 2005. The role of the commensal gut microbial community in broiler chickens. World's Poult Sci J 61:95–104. doi: 10.1079/wps200445. [DOI] [Google Scholar]
- 10.Brisbin JT, Gong J, Sharif S. 2008. Interactions between commensal bacteria and the gut-associated immune system of the chicken. Anim Health Res Rev 9:101–110. doi: 10.1017/S146625230800145X. [DOI] [PubMed] [Google Scholar]
- 11.Lee K, Lillehoj HS, Siragusa GR. 2010. Direct-fed microbials and their impact on the intestinal microflora and immune system of chickens. J Poult Sci 47:106–114. doi: 10.2141/jpsa.009096. [DOI] [Google Scholar]
- 12.Schneitz C. 2005. Competitive exclusion in poultry—30 years of research. Food Control 16:657–667. doi: 10.1016/j.foodcont.2004.06.002. [DOI] [Google Scholar]
- 13.Teirlynck E, Gussem MDE, Dewulf J, Haesebrouck F, Ducatelle R, Van Immerseel F. 2011. Morphometric evaluation of dysbacteriosis in broilers. Avian Pathol 40:139–144. doi: 10.1080/03079457.2010.543414. [DOI] [PubMed] [Google Scholar]
- 14.Barton MD. 2000. Antibiotic use in animal feed and its impact on human health. Nutr Res Rev 13:279–299. doi: 10.1079/095442200108729106. [DOI] [PubMed] [Google Scholar]
- 15.Wegener HC. 2003. Antibiotics in animal feed and their role in resistance development. Curr Opin Microbiol 6:439–445. doi: 10.1016/j.mib.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Wilson J, Tice G, Brash ML, Hilaire SS. 2005. Manifestations of Clostridium perfringens and related bacterial enteritides in broiler chickens. World's Poult Sci J 61:435–449. doi: 10.1079/wps200566. [DOI] [Google Scholar]
- 17.Canibe N, Jensen BB. 2003. Fermented and nonfermented liquid feed to growing pigs: effect on aspects of gastrointestinal ecology and growth performance. J Anim Sci 81:2019–2031. [DOI] [PubMed] [Google Scholar]
- 18.Canibe N, Højberg O, Badsberg JH, Jensen BB. 2007. Effect of feeding fermented liquid feed and fermented grain on gastrointestinal ecology and growth performance in piglets. J Anim Sci 85:2959–2971. [DOI] [PubMed] [Google Scholar]
- 19.Engberg RM, Hammershøj M, Johansen NF, Abousekken MS, Steenfeldt S, Jensen BB. 2009. Fermented feed for laying hens: effects on egg production, egg quality, plumage condition and composition and activity of the intestinal microflora. Br Poult Sci 50:228–239. doi: 10.1080/00071660902736722. [DOI] [PubMed] [Google Scholar]
- 20.Rehman H, Vahjen W, Kohl-Parisini A, Ijaz A, Zentek J. 2009. Influence of fermentable carbohydrates on the intestinal bacteria and enteropathogens in broilers. World's Poult Sci J 65:75–89. doi: 10.1017/s0043933909000063. [DOI] [Google Scholar]
- 21.Heres L, Engel B, Van Knapen F, De Jong MC, Wagenaar JA, Urlings HA. 2003. Fermented liquid feed reduces susceptibility of broilers for Salmonella enteritidis. Poult Sci 82:603–611. doi: 10.1093/ps/82.4.603. [DOI] [PubMed] [Google Scholar]
- 22.Heres L, Engel B, Van Knapen F, Wagenaar JA, Urlings BA. 2003. Effect of fermented feed on the susceptibility for Campylobacter jejuni colonisation in broiler chickens with and without concurrent inoculation of Salmonella enteritidis. Int J Food Microbiol 87:75–86. doi: 10.1016/S0168-1605(03)00055-2. [DOI] [PubMed] [Google Scholar]
- 23.Engberg RM, Hedemann MS, Steenfeldt S, Jensen BB. 2004. Influence of whole wheat and xylanase on broiler performance and microbial composition and activity in the digestive tract. Poult Sci 83:925–938. doi: 10.1093/ps/83.6.925. [DOI] [PubMed] [Google Scholar]
- 24.Bach Knudsen KE. 1997. Carbohydrate and lignin contents of plant materials used in animal feeding. Anim Feed Sci Technol 67:319–338. doi: 10.1016/S0377-8401(97)00009-6. [DOI] [Google Scholar]
- 25.Jacobsen E. 1981. Sukker og stivelse (LHK)—ny analysemetode. Medd Biotek Institut ATV 98:39–54. [Google Scholar]
- 26.Fisher C. 2000. Advances in feed evaluation for poultry, p 243–268. In Moughan PJ, Verstegen MWA, Visser-Reyneveld MI (ed), Feed evaluation: principles and practice. Wageningen Pers, Wageningen, The Netherlands. [Google Scholar]
- 27.Carlson D, Poulsen HD. 2003. Phytate degradation in soaked and fermented liquid feed—effect of diet, time of soaking, heat treatment, phytase activity, pH and temperature. Anim Feed Sci Technol 103:141–154. doi: 10.1016/S0377-8401(02)00288-2. [DOI] [Google Scholar]
- 28.Knarreborg A, Engberg RM, Jensen SK, Jensen BB. 2002. Quantitative determination of bile salt hydrolase activity in bacteria isolated from the small intestine of chickens. Appl Environ Microbiol 68:6425–6428. doi: 10.1128/AEM.68.12.6425-6428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sørensen JL, Sondergaard TE. 2014. The effects of different yeast extracts on secondary metabolite production in Fusarium. Int J Food Microbiol 170:55–60. doi: 10.1016/j.ijfoodmicro.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 30.Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8:125. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tannock GW, Lawley B, Munro K, Pathmanathan SG, Zhou SJ, Makrides M, Gibson RA, Sullivan T, Prosser CG, Lowry D. 2013. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl Environ Microbiol 79:3040–3048. doi: 10.1128/AEM.03910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 34.Cole CB, Fuller R. 1984. Bile acid deconjugation and attachment of chicken gut bacteria: their possible role in growth depression. Br Poult Sci 25:227–231. doi: 10.1080/00071668408454861. [DOI] [PubMed] [Google Scholar]
- 35.Dekker R, Van der Meer R, Olieman C. 1991. Sensitive pulsed amperometric detection of free and conjugated bile acids in combination with gradient reversed-phase HPLC. Chromatographia 31:549–553. [Google Scholar]
- 36.Knarreborg A, Lauridsen C, Engberg RM, Jensen SK. 2004. Dietary antibiotic growth promoters enhance the bioavailability of α-tocopheryl acetate in broilers by altering lipid absorption. J Nutr 134:1487–1492. [DOI] [PubMed] [Google Scholar]
- 37.Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand J Statist 6:65–70. [Google Scholar]
- 38.Harrow SA, Ravindran V, Butler RC, Marshall JW, Tannock GW. 2007. Real-time quantitative PCR measurement of ileal Lactobacillus salivarius populations from broiler chickens to determine the influence of farming practices. Appl Environ Microbiol 73:7123–7127. doi: 10.1128/AEM.01289-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canibe N, Jensen BB. 2012. Fermented liquid feed—microbial and nutritional aspects and impact on enteric diseases in pigs. Anim Feed Sci Technol 173:17–40. doi: 10.1016/j.anifeedsci.2011.12.021. [DOI] [Google Scholar]
- 40.Heres L, Wagenaar JA, van Knapen F, Urlings BA. 2003. Passage of Salmonella through the crop and gizzard of broiler chickens fed with fermented liquid feed. Avian Pathol 32:173–181. doi: 10.1080/0307945021000071597. [DOI] [PubMed] [Google Scholar]
- 41.Ranjitkar S, Karlsson AH, Petersen MA, Bredie WL, Petersen JS, Engberg RM. 9 November 2015. The influence of feeding crimped kernel maize silage on broiler production, nutrient digestibility and meat quality. Br Poult Sci doi: 10.1080/00071668.2015.1115468. [DOI] [PubMed] [Google Scholar]
- 42.Rehman HU, Vahjen W, Awad WA, Zentek J. 2007. Indigenous bacteria and bacterial metabolic products in the gastrointestinal tract of broiler chickens. Arch Anim Nutr 61:319–335. doi: 10.1080/17450390701556817. [DOI] [PubMed] [Google Scholar]
- 43.Gong J, Forster RJ, Yu H, Chambers JR, Wheatcroft R, Sabour PM, Chen S. 2002. Molecular analysis of bacterial populations in the ileum of broiler chickens and comparison with bacteria in the cecum. FEMS Microbiol Ecol 41:171–179. doi: 10.1111/j.1574-6941.2002.tb00978.x. [DOI] [PubMed] [Google Scholar]
- 44.Sergeant MJ, Constantinidou C, Cogan TA, Bedford MR, Penn CW, Pallen MJ. 2014. Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS One 9:e91941. doi: 10.1371/journal.pone.0091941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanley D, Geier MS, Hughes RJ, Denman SE, Moore RJ. 2013. Highly variable microbiota development in the chicken gastrointestinal tract. PLoS One 8:e84290. doi: 10.1371/journal.pone.0084290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan LL, Hagen KE, Tannock GW, Korver DR, Fasenko GM, Allison GE. 2003. Detection and identification of Lactobacillus species in crops of broilers of different ages by using PCR-denaturing gradient gel electrophoresis and amplified ribosomal DNA restriction analysis. Appl Environ Microbiol 69:6750–6757. doi: 10.1128/AEM.69.11.6750-6757.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuller R. 1977. The importance of lactobacilli in maintaining normal microbial balance in the crop. Br Poult Sci 18:85–94. [DOI] [PubMed] [Google Scholar]
- 48.Brooker BE, Fuller R. 1975. Adhesion of lactobacilli to the chicken crop epithelium. J Ultrastruct Res 52:21–31. [DOI] [PubMed] [Google Scholar]
- 49.Fuller R, Brooker BE. 1974. Lactobacilli which attach to the crop epithelium of the fowl. Am J Clin Nutr 27:1305–1312. [DOI] [PubMed] [Google Scholar]
- 50.Fuller R. 1975. Nature of the determinant responsible for the adhesion of lactobacilli to chicken crop epithelial cells. J Gen Microbiol 87:245–250. [DOI] [PubMed] [Google Scholar]
- 51.Qaisrani SN, Van Krimpen MM, Kwakkel RP, Verstegen MWA, Hendriks WH. 2015. Dietary factors affecting hindgut protein fermentation in broilers: a review. World's Poult Sci J 71:139–160. doi: 10.1017/s0043933915000124. [DOI] [Google Scholar]
- 52.Janczyk P, Halle B, Souffrant WB. 2009. Microbial community composition of the crop and ceca contents of laying hens fed diets supplemented with Chlorella vulgaris. Poult Sci 88:2324–2332. doi: 10.3382/ps.2009-00250. [DOI] [PubMed] [Google Scholar]
- 53.Sekelja M, Rud I, Knutsen SH, Denstadli V, Westereng BTN, Rudi K. 2012. Abrupt temporal fluctuations in the chicken fecal microbiota are explained by its gastrointestinal origin. Appl Environ Microbiol 78:2941–2948. doi: 10.1128/AEM.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Danzeisen JL, Kim HB, Isaacson RE, Tu ZJ, Johnson TJ. 2011. Modulations of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment. PLoS One 6:e27949. doi: 10.1371/journal.pone.0027949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu J, Idris U, Harmon B, Hofacre C, Maurer JJ, Lee MD. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl Environ Microbiol 69:6816–6824. doi: 10.1128/AEM.69.11.6816-6824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knarreborg A, Simon MA, Engberg RM, Jensen BB, Tannock GW. 2002. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl Environ Microbiol 68:5918–5924. doi: 10.1128/AEM.68.12.5918-5924.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barnes EM. 1979. The intestinal microflora of poultry and game birds during life and after storage. J Appl Bacteriol 46:407–419. [DOI] [PubMed] [Google Scholar]
- 58.Tannock GW. 2004. A special fondness for lactobacilli. Appl Environ Microbiol 70:3189–3194. doi: 10.1128/AEM.70.6.3189-3194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duncan SH, Hold GL, Harmsen HJ, Stewart CS, Flint HJ. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol 52:2141–2146. doi: 10.1099/00207713-52-6-2141. [DOI] [PubMed] [Google Scholar]
- 60.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 61.Stanley D, Denman SE, Hughes RJ, Geier MS, Crowley TM, Chen H, Haring VR, Moore RJ. 2012. Intestinal microbiota associated with differential feed conversion efficiency in chickens. Appl Microbiol Biotechnol 96:1361–1369. doi: 10.1007/s00253-011-3847-5. [DOI] [PubMed] [Google Scholar]
- 62.Zhu XY, Zhong T, Pandya Y, Joerger RD. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl Environ Microbiol 68:124–137. doi: 10.1128/AEM.68.1.124-137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mohd Shaufi MA, Sieo CC, Chong CW, Gan HM, Ho YW, Sunway B. 2015. Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses. Gut Pathog 7:4. doi: 10.1186/s13099-015-0051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gong J, Si W, Forster RJ, Huang R, Yu H, Yin Y, Yang C, Han Y. 2007. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol Ecol 59:147–157. doi: 10.1111/j.1574-6941.2006.00193.x. [DOI] [PubMed] [Google Scholar]
- 65.Torok VA, Allison GE, Percy NJ, Ophel-Keller K, Hughes RJ. 2011. Influence of antimicrobial feed additives on broiler commensal posthatch gut microbiota development and performance. Appl Environ Microbiol 77:3380–3390. doi: 10.1128/AEM.02300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh KM, Shah T, Deshpande S, Jakhesara SJ, Koringa PG, Rank DN, Joshi CG. 2012. High through put 16S rRNA gene-based pyrosequencing analysis of the fecal microbiota of high FCR and low FCR broiler growers. Mol Biol Rep 39:10595–10602. doi: 10.1007/s11033-012-1947-7. [DOI] [PubMed] [Google Scholar]
- 67.Engberg RM, Hedemann MS, Jensen BB. 2002. The influence of grinding and pelleting of feed on the microbial composition and activity in the digestive tract of broiler chickens. Br Poult Sci 43:569–579. doi: 10.1080/0007166022000004480. [DOI] [PubMed] [Google Scholar]
- 68.Jin LZ, Ho YW, Abdullah N, Jalaludin S. 1998. Growth performance, intestinal microbial populations, and serum cholesterol of broilers fed diets containing Lactobacillus cultures. Poult Sci 77:1259–1265. [DOI] [PubMed] [Google Scholar]
- 69.Messaoudi S, Madi A, Prevost H, Feuilloley M, Manai M, Dousset X, Connil N. 2012. In vitro evaluation of the probiotic potential of Lactobacillus salivarius SMXD51. Anaerobe 18:584–589. doi: 10.1016/j.anaerobe.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 70.Messaoudi S, Manai M, Kergourlay G, Prévost H, Connil N, Chobert JM, Dousset X. 2013. Lactobacillus salivarius: bacteriocin and probiotic activity. Food Microbiol 36:296–304. doi: 10.1016/j.fm.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 71.Neville BA, O'Toole PW. 2010. Probiotic properties of Lactobacillus salivarius and closely related Lactobacillus species. Future Microbiol 5:759–774. doi: 10.2217/fmb.10.35. [DOI] [PubMed] [Google Scholar]
- 72.Stern NJ, Svetoch EA, Eruslanov BV, Perelygin VV, Mitsevich EV, Mitsevich IP, Pokhilenko VD, Levchuk VP, Svetoch OE, Seal BS. 2006. Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob Agents Chemother 50:3111–3116. doi: 10.1128/AAC.00259-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramasamy K, Abdullah N, Wong MC, Karuthan C, Ho YW. 2010. Bile salt deconjugation and cholesterol removal from media by Lactobacillus strains used as probiotics in chickens. J Sci Food Agric 90:65–69. doi: 10.1002/jsfa.3780. [DOI] [PubMed] [Google Scholar]
- 74.Guban J, Korver DR, Allison GE, Tannock GW. 2006. Relationship of dietary antimicrobial drug administration with broiler performance, decreased population levels of Lactobacillus salivarius, and reduced bile salt deconjugation in the ileum of broiler chickens. Poult Sci 85:2186–2194. doi: 10.1093/ps/85.12.2186. [DOI] [PubMed] [Google Scholar]
- 75.Hafez HM. 2011. Enteric diseases of poultry with special attention to Clostridium perfringens. Pak Vet J 31:175–184. [Google Scholar]
- 76.Engberg RM, Hedemann MS, Leser TD, Jensen BB. 2000. Effect of zinc bacitracin and salinomycin on intestinal microflora and performance of broilers. Poult Sci 79:1311–1319. doi: 10.1093/ps/79.9.1311. [DOI] [PubMed] [Google Scholar]
- 77.Jensen MT, Cox RP, Jensen BB. 1995. Microbial production of skatole in the hind gut of pigs given different diets and its relation to skatole deposition in backfat. Anim Sci 61:293–304. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.