Abstract
Sacbrood virus (SBV) is one of the most destructive viruses in the Asian honeybee Apis cerana but is much less destructive in Apis mellifera. In previous studies, SBV isolates infecting A. cerana (AcSBV) and SBV isolates infecting A. mellifera (AmSBV) were identified as different serotypes, suggesting a species barrier in SBV infection. In order to investigate this species isolation, we examined the presence of SBV infection in 318 A. mellifera colonies and 64 A. cerana colonies, and we identified the genotypes of SBV isolates. We also performed artificial infection experiments under both laboratory and field conditions. The results showed that 38 A. mellifera colonies and 37 A. cerana colonies were positive for SBV infection. Phylogenetic analysis based on RNA-dependent RNA polymerase (RdRp) gene sequences indicated that A. cerana isolates and most A. mellifera isolates formed two distinct clades but two strains isolated from A. mellifera were clustered with the A. cerana isolates. In the artificial-infection experiments, AcSBV negative-strand RNA could be detected in both adult bees and larvae of A. mellifera, although there were no obvious signs of the disease, demonstrating the replication of AcSBV in A. mellifera. Our results suggest that AcSBV is able to infect A. mellifera colonies with low prevalence (0.63% in this study) and pathogenicity. This work will help explain the different susceptibilities of A. cerana and A. mellifera to sacbrood disease and is potentially useful for guiding beekeeping practices.
INTRODUCTION
Honeybees play vital roles in agriculture by providing pollination services to food crops and producing honey and other hive products (1). European honeybees (Apis mellifera) and Asian honeybees (Apis cerana) are two truly domesticated bee species in global apiculture (2, 3), but they are commonly threatened by a myriad of parasites and pathogens (4). Sacbrood virus (SBV), the first virus identified in honeybees (5), is a single-stranded, positive-sense RNA virus belonging to the genus Iflavirus in the family Iflaviridae (6). The virus primarily affects the brood stage of honeybees, causing failure of pupation and larval death (7), but also affects adult bees without obvious signs of the disease (8–10). Sacbrood is characterized by the diseased-infected larva being encased in a tough, fluid-filled sac formed from the larval cuticle. The transmission of SBV can occur horizontally, vertically, or both (11). The prevalence of this virus has been investigated in various countries (4, 12–20). SBV infection appears to be less detrimental to A. mellifera colonies than to A. cerana colonies (21). In addition, it is unusual for SBV infection in A. mellifera colonies to cause colony death (22). However, SBV is deadly to Asian honeybees, and serious mortality rates for A. cerana due to SBV infection have been reported in Vietnam, Thailand, India, China, and South Korea (7, 23–26). Large-scale A. cerana colony losses caused by SBV infection were first reported in Guangdong Province, China, in 1972 (27). Infection also resulted in serious A. cerana colony losses in South Korea (26, 28).
Previous studies reported that SBV isolates infecting A. cerana (AcSBV) and SBV isolates infecting A. mellifera (AmSBV) represent two different serotypes (29) and are chemically distinct (30), suggesting that AcSBV and AmSBV confer unique pathogenicities to their respective hosts. To understand the susceptibilities of A. cerana and A. mellifera bees to SBV isolated from different honeybee species, studies of cross-species infections with AcSBV and AmSBV were performed. In previous cross-infection studies conducted using AcSBV and AmSBV to challenge A. mellifera larvae and A. cerana larvae, AmSBV and AcSBV were pathogenic only in A. mellifera larvae and A. cerana larvae, respectively (31–33).
An A. mellifera isolate, AmSBV-Kor19 (GenBank accession number JQ390592), which is similar to A. cerana isolates across most of its genome, was reported recently (26). Roberts and Anderson (29) pointed out that cross-species SBV infection of A. cerana and A. mellifera may occur, because phylogenetic analysis of SBV revealed that AmSBV-Kor19 is grouped with other A. cerana isolates. Therefore, there have been nonidentical views on cross-species infection. The objectives of this study were (i) to survey for SBV infections in apparently healthy or weak A. mellifera and A. cerana colonies, (ii) to identify genotypes of SBV isolates from A. mellifera and A. cerana colonies, and (iii) to evaluate the infectivity of AcSBV in A. mellifera, both larvae and adults, under laboratory and field conditions.
MATERIALS AND METHODS
Sample collection for SBV detection.
To survey SBV infections in field colonies, we sampled 279 A. mellifera colonies from 20 apiaries and 38 A. cerana colonies from 7 apiaries in Zhejiang Province. These colonies were apparently healthy and strong, and 100 adult worker bees were randomly sampled from each of them. To increase the chances of positive detection of SBV, 39 weak A. mellifera colonies and 26 A. cerana colonies showing symptoms of honeybee diseases were sampled. The symptoms of honeybee diseases were defined as dead broods being observed in cells and/or significant numbers of dead adult worker bees being observed in the hives or near the entrances. From each of these colonies, 10 larvae were collected. Samples were stored at −80°C until used.
RNA extraction, RT-PCR, and sequencing.
RNA extraction and cDNA synthesis (except the cDNA synthesis for the detection of negative-strand RNA) were performed using TRIzol reagent (Adilab, Beijing, China) and ReverTra Ace qPCR RT (quantitative PCR-reverse transcription) [Note that I restored qPCR RT since that is likely how the ReverTra Ace product is worded.] master mix (catalog number FSQ201; Toyobo, Shanghai, China), respectively, by following the manufacturers' instructions. The PCR volume of 50 μl was prepared using 2× Taq PCR StarMix (GenStar, Beijing, China), according to the manufacturer's protocol, and the PCR amplification was conducted with an initial denaturation at 94°C for 2 min, 35 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s, and finally 5 min at 72°C, with the primers SB14f and SB15r (SB14f, 5′-AATGGTGCGGTGGACTATGG-3′; SB15r, 5′-TGATACAGAGCGGCTCGACA-3′) (14), amplifying 579 bp of the RNA-dependent RNA polymerase (RdRp) region. Each PCR amplification included a negative control, in which the same volume of H2O was used instead of template cDNA. The PCR products were analyzed by 2.5% agarose gel electrophoresis, and the positive products were directly sequenced using an ABI 3730XL automatic sequencing system (Ruidi, Shanghai, China). Sequence specificity analysis was performed using BLAST (NCBI).
Phylogenetic analysis.
MEGA5 was used to perform the phylogenetic analysis (34). The phylogenetic analysis was conducted with the RdRp gene sequences of SBV-positive isolates obtained in this study and other representative SBV homologous sequences, which were retrieved from the GenBank database. Phylogenetic trees were constructed with the maximum likelihood (ML) method. The bootstrap values were the results of 1,000 replicates, and the values below 50% were omitted.
Inoculation experiments. (i) AcSBV preparation and quantitation.
Larvae with overt symptoms of SBV infection were sampled from an A. cerana colony; the colony had been confirmed to be infected with SBV but not with the other six common honeybee viruses, namely, Acute bee paralysis virus (ABPV), Black queen cell virus (BQCV), Chronic bee paralysis virus (CBPV), Deformed wing virus (DWV), Israeli acute paralysis virus (IAPV), and Kashmir bee virus (KBV), using RT-PCR with the method described previously (35). The SBV isolate was identified as typical SBV infecting A. cerana, namely, AcSBV, based on the alignment of 579-bp RdRp gene sequences. The procedure to obtain concentrated AcSBV was conducted as described by Bailey (9) but with dilution with phosphate-buffered saline (PBS) (pH 7.4 ± 0.2). Ten larvae were homogenized in 10 ml PBS with a mortar and pestle and then centrifuged for 40 min at 10,000 rpm in a centrifuge (Hitachi, Japan). The supernatant was collected and used for inoculation; 180 μl of the solution was used for RNA extraction, and RT-PCR was performed again, to confirm the presence of SBV and the absence of the other common viruses described above. Sequencing of the PCR products was conducted to confirm the identity of the virus. The SBV concentration was determined by an absolute quantification assay according to the method described previously (36), with the following modifications. The primers were used for qPCR assays as reported previously (37), and the primers were used to construct recombinant plasmids as reported previously (38). The qRT-PCRs were performed in triplicate in an Eppendorf Mastercycler ep realplex system using SYBR green (Thunderbird SYBR qPCR mix; Toyobo, Shanghai, China). The AcSBV concentration for the subsequent experiments was approximately 4.0 ×105 copies/μl.
(ii) Screening for viruses to identify healthy colonies.
One hundred adult worker bees collected from A. mellifera or A. cerana colonies were checked, as a pooled sample, for the presence of SBV and the other six common honeybee viruses mentioned above. The colonies with no detectable viruses were used as the source of honeybee workers and larvae for inoculation.
(iii) Laboratory inoculation of larvae.
To determine the effects of AcSBV on the mortality and infection rates of third-instar larvae, which represent the most sensitive stage for SBV infection (39), second-instar larvae of A. cerana and A. mellifera were carefully transferred to 48-well culture plates containing 150 μl artificial diet (40) and then were placed in an incubator set at 34 ± 0.5°C, with 70% ± 5% relative humidity (Stik, Shanghai, China). The second-instar larvae were obtained as described previously (33). After 24 h, the third-instar larvae of A. cerana and A. mellifera were transferred individually to other 48-well culture plates containing 130 μl artificial diet, as described above. Fifteen microliters of AcSBV solution was added to each well for the experimental groups, and 15 μl PBS was added for the control groups. After another 24 h, the bee larvae were washed 3 times with ultrapure water and kept at −80°C for later use. Every treatment was tested with 3 replicates, and each replicate contained 20 larvae. The deaths of larvae were confirmed by identifying the color of their bodies changing from white to yellow or even dark gray and then noting no response when the larvae were touched with a soft tip. RT-PCR was performed to confirm whether the larvae were infected with AcSBV (15). In total, 15 positive samples of A. mellifera were randomly chosen to verify the identity of strains by sequencing. RNA from another 5 positive samples from each replicate was pooled and used to detect the negative-strand AcSBV RNA.
(iv) Laboratory inoculation of adult bees.
Newly emerged bees (A. mellifera and A. cerana) were individually inoculated with 3 μl AcSBV preparation (experimental groups) or PBS containing 10% honey (control groups), using a Hamilton syringe to feed the bees. Fifty honeybees per cage were kept at 34 ± 0.5°C and 70% ± 5% relative humidity in an incubator (Stik, Shanghai, China) and were fed pollen that had been treated with UV light for 24 h and 50% (vol/vol) sucrose solution. Four treatments, each with 3 replicates. were established. Dead bees were recorded and removed daily until day 10 after inoculation. Four bees were collected from each cage at days 0, 2, 6, and 10 and were used to quantify the AcSBV. RNA from another 6 positive adult workers from days 2, 6, and 10 was pooled and used to detect the negative-strand AcSBV RNA.
(v) Quantification of AcSBV in adult bees by real-time qPCR.
The titers of AcSBV in adult bees (A. mellifera and A. cerana) were quantified by a two-step SYBR green real-time qPCR assay. RNA extraction, cDNA synthesis, and a qPCR assay with a final total volume of 20 μl were carried out as described above. The cDNA was generated from 1 μg of total RNA and diluted 1:5. The primer pairs for AcSBV (22) and the reference gene (β-actin) (41) were reported previously. The PCR program was as follows: 1 min at 95°C and 40 cycles of 95°C for 15 s and 60°C for 1 min, followed by a melt curve analysis. PCR amplification for each sample was performed in triplicate, to address the variability of the analysis process. The relative titers of AcSBV were calculated according to the 2−ΔΔCT method (42).
(vi) Colony inoculation.
In order to investigate the ability of AcSBV to infect A. mellifera in the field, three A. mellifera colonies that were free of SBV infection, as confirmed by RT-PCR, were selected for the inoculation experiment. One liter of sugar syrup mixed with 2 ml AcSBV solution was fed to each colony. At day 5 after feeding, the clinical symptoms of SBV were observed in the colonies, and 20 larvae that were ready to pupate and 20 adult worker bees were randomly collected from each experimental colony. Each larva or bee was checked by RT-PCR for the presence of AcSBV, according to the procedure introduced above (15). PCR products of 5 positive samples were randomly selected to confirm the identity of SBV by sequencing. RNA of 10 positive adult workers and 10 positive larvae from each colony was pooled and used to detect the negative-strand AcSBV RNA.
Tagged RT-PCR for detection of negative-strand AcSBV RNA.
Negative-strand (replicative intermediate) RNA has been regarded as an indicator of ongoing replication of positive-strand RNA viruses (43). Therefore, detection of negative-strand RNA is indicative of viral replication. Tagged RT-PCR has been developed to improve the specific detection of negative-strand RNA (44) and has already been used to confirm the replication of honeybee viruses (45–47). The reverse transcription (RT) reaction was performed using the ReverTra Ace RT-qPCR kit (catalog number FSQ101; Toyobo, Shanghai, China), by following the standard protocol with modifications. The reaction was performed at 55°C for 30 min in the presence of the primer Tag-SB7f (5′-agcctgcgcaccgtggGGAGATGTTAGAAATACCAACCGATTCC-3′ [lowercase letters indicate the Tag primer]); the reaction volume was 20 μl, with 4 μg RNA. The RT product was purified with the MinElute reaction cleanup kit (Qiagen, Germany), according to the manufacturer's instructions, which can avoid false-positive results (48), and then was used to conduct PCR assays using the primer pair Tag (forward primer, 5′-AGCCTGCGCACCGTGG-3′) (45) and SB8r (reverse primer, 5′-CCATTAAACAAATCGGTATAAGAGTCCACT-3′). The reaction mixtures were first denatured at 94°C for 30 s and then subjected to 35 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 30 s, with a final extension step at 72°C for 5 min. The RT reaction without the primer Tag-SB7f served as the negative control. At the end of the PCR, 5 μl product per reaction was used for analysis on a 2.5% agarose gel.
Data analysis.
The normality of data sets was tested using the Shapiro-Wilk test. Data sets with normal distribution were compared with the parametric Student's t test. Data sets with nonnormal distribution were analyzed using the lower-power nonparametric Mann-Whitney U test. A chi-square test was performed to compare SBV infection rates in A. mellifera and A. cerana colonies. Kaplan-Meier survival curves were used to plot and to interpret the survival data. Survival curves were compared using log rank tests. Data are shown as averages ± standard deviations (SDs). P values of <0.05 were considered statistically significant.
Nucleotide sequence accession numbers.
The genomic sequence of SBV reported in this paper has been submitted to the GenBank database under accession numbers KT205289 to KT205304 and KT734782 to KT734791.
RESULTS
Rates of SBV infection in A. mellifera and A. cerana colonies.
Of the 279 A. mellifera colonies and 38 A. cerana colonies examined with adult bees, SBV was identified in 29 (10.39%) and 11 (28.95%), respectively, showing a significant difference between species (χ2 = 8.83; P = 0.003). In the A. mellifera colonies showing disease symptoms (see “Sample collection for SBV detection”above for the definition), the SBV infection rate was 23.08%; in the diseased A. cerana colonies, the infection rate was found to be 100% (χ2 = 37.14; P < 0.001).
Phylogenetic analysis.
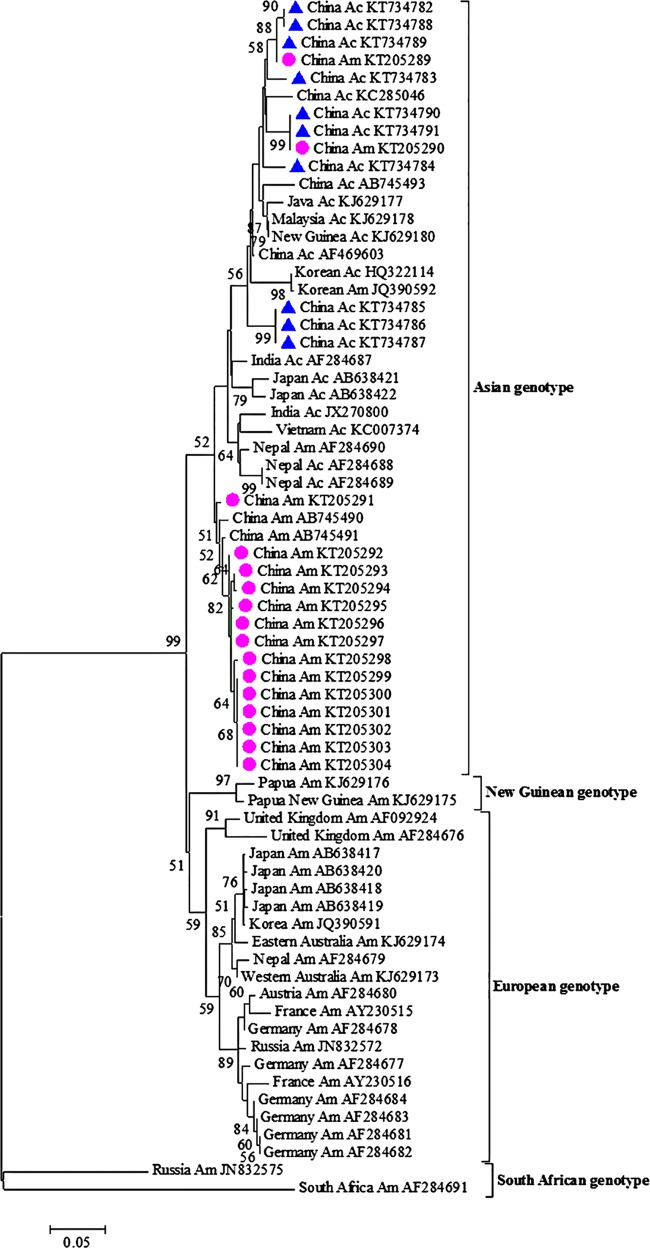
The evolutionary tree of SBV isolates is presented in Fig. 1, with representative homologous sequences retrieved from the NCBI database and obtained in the survey. As reported by Roberts and Anderson (29) and Grabensteiner et al. (14), the phylogenetic tree of SBV based on RdRp gene sequences had four clades, corresponding to four genotypes of SBV, namely, Asian genotype, New Guinean genotype, European genotype, and South African genotype. A. mellifera SBV isolates were clustered with the two previously reported SBV isolates isolated from A. mellifera in China (49), forming a separate clade. A. cerana SBV strains isolated in this study were clustered with other SBV strains isolated from A. cerana in Asian countries. However, it must be noted that two A. mellifera SBV isolates were clustered with A. cerana SBV isolates and two previously reported A. mellifera isolates (GenBank accession numbers JQ390592 and AF284690), but in this case the occurrence rate was only 0.63%.
FIG 1.
Maximum likelihood consensus tree of SBV isolates. A 386-bp region of the RNA-dependent RNA polymerase gene was used. The tree was constructed with a Tamura 3-parameter distance model. The statistical significance of a particular tree topology is evaluated by bootstrap resampling of the sequences 1,000 times, and bootstrap values of <50% are omitted. Numbers on the nodes indicate clade credibility values. New isolates of A. mellifera (circles) and A. cerana (triangles) described in this study are indicated. Strains are annotated with respect to country of isolation, virus host, and GenBank accession number. Am, A. mellifera; Ac, A. cerana.
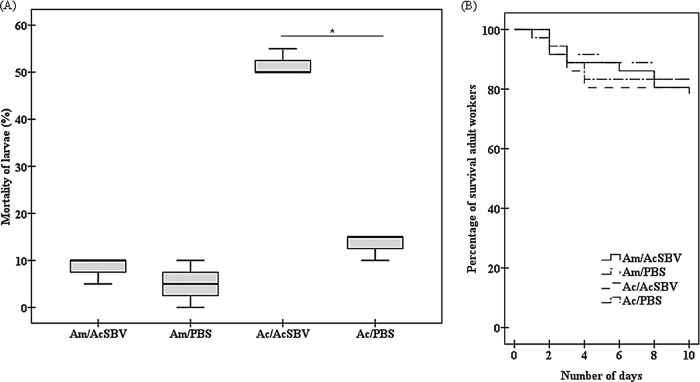
Mortality and infection rates of larvae after laboratory AcSBV challenge.
Infection was identified in 100% of A. mellifera and A. cerana larvae challenged with AcSBV. No clinical symptoms were observed in A. mellifera larvae infected with AcSBV. No significant difference in mortality rates for A. mellifera larvae was found between the AcSBV-inoculated larvae (8.3%) and the control group (5.0%) (U = 2.500; P = 0.346) (Fig. 2A). However, the mortality rate of A. cerana larvae (51.7%) challenged with AcSBV was significantly higher than that of A. cerana larvae (13.3%) without the AcSBV challenge (U = 0; P = 0.043) (Fig. 2A).
FIG 2.
Larval mortality rates at 24 h after inoculation (A) and survival rates of adult workers (B) of A. cerana (Ac) and A. mellifera (Am) inoculated with AcSBV under laboratory conditions. In panel A, the boxes represent first quartiles and third quartiles, the horizontal lines indicate the median, and the whiskers represent the minimum and maximum for all observed values. Mortality rates of larvae and survival rates of adult workers were calculated from 20 larvae and 50 adult workers per treatment group, respectively. Am/PBS, A. mellifera larvae fed PBS; Am/AcSBV, A. mellifera larvae infected with AcSBV; Ac/PBS, A. cerana larvae fed PBS; Ac/AcSBV, A. cerana larvae infected with AcSBV. The data are averages from three replicates. *, P < 0.05.
Survival of inoculated adult bees.
Log rank tests indicated that there was no significant difference in survival between A. mellifera fed AcSBV and PBS (χ2 = 0.912; P = 0.340) (Fig. 2B). In addition, AcSBV treatment did not significantly affect the survival of A. cerana, compared with the control group treated with PBS (χ2 = 1.210; P = 0.271) (Fig. 2B).
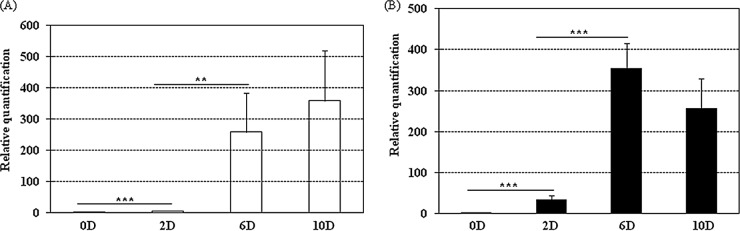
AcSBV proliferation in inoculated adult bees.
The genotypes of strains were verified to be AcSBV, and the samples of A. mellifera and A. cerana used for qRT-PCR analysis were all positive. The proliferation of AcSBV in A. mellifera and A. cerana is presented in Fig. 3A and B, respectively. In terms of AcSBV in A. mellifera, the AcSBV titers on days 2, 6, and 10 showed 6.42 ± 0.73-fold, 259.18 ± 124.34-fold, and 359.39 ± 161.76-fold changes, respectively, compared to the day 0 titer. In addition, there were significant differences in AcSBV titers for day 0 versus day 2 (U = 0; P < 0.001) and day 2 versus day 6 (U = 14.000; P = 0.004), but no significant difference was observed for day 6 versus day 10 (U = 52.000; P = 0.833). Compared with day 0, the titers of AcSBV in A. cerana on days 2, 6, and 10 showed 33.63 ± 10.54-fold, 353.25 ± 63.18-fold, and 255.96 ± 74.56-fold changes, respectively. Furthermore, significant differences were found for day 0 versus day 2 (U = 12.000; P < 0.001) and day 2 versus day 6 (U = 2.000; P < 0.001), but there was no significant difference between day 6 and day 10 (t = 0.952; P = 0.353). In addition, it should be noted that the increases in AcSBV titers in A. mellifera were less than those in A. cerana during the first 6 days after inoculation. Surprisingly, however, the increases in AcSBV titers within 10 days were more substantial in A. mellifera.
FIG 3.
Relative quantification of AcSBV in A. mellifera (A) and A. cerana (B) adult workers inoculated under laboratory conditions. Twelve bees were collected on days 0, 2, 6, and 10 and used for quantification. D, day. **, P < 0.01; ***, P < 0.001.
Infection in A. mellifera colonies fed AcSBV.
At day 5 after feeding, no clinical symptoms of SBV were observed in the colonies. However, the infection rates for larvae and adult bees in the colonies fed AcSBV solution were found to be 90.5% (54/60 larvae) and 62.2% (37/60 adult bees), respectively (U = 2.000; P = 0.268).
Presence of AcSBV negative-strand RNA.
Figure 4 shows the results of detection of AcSBV negative-strand RNA in A. mellifera adult bees and larvae inoculated under laboratory and field conditions. AcSBV negative-strand RNA could be detected in all A. mellifera samples randomly collected in the experiments. When the Tag-SB7f primers were omitted from the cDNA synthesis, no false-positive PCR signals occurred. Compared with the samples inoculated under laboratory conditions, the nucleic acid bands (Fig. 4C and D) of samples inoculated under field conditions were much weaker.
FIG 4.
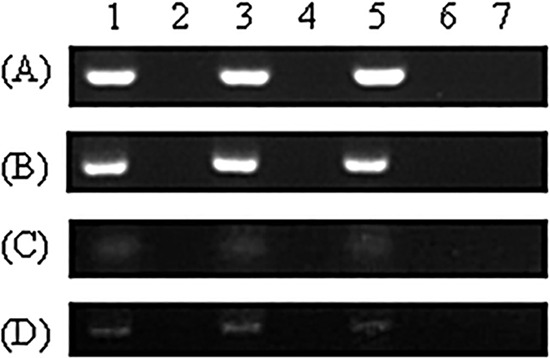
Detection of negative-strand RNA of AcSBV in A. mellifera larvae and adult workers. Lanes 1, 3, and 5, samples obtained on days 2, 6, and 10, respectively; lanes 2, 4, and 6, negative controls without Tag-SB7f primers; lane 7, negative control without the DNA template. Lanes 1, 3, and 5 in panel A represent pooled samples of 6 bees from each of 3 replicates on each day; in panels B, C, and D, results of one replicate are shown. (A) Adult worker bees challenged with AcSBV in the laboratory. (B) Larvae challenged with AcSBV in the laboratory. (C) Adult worker bees challenged with AcSBV in a colony. (D) Larvae challenged with AcSBV in a colony.
DISCUSSION
Most previous studies (31–33) concluded that there was no cross-species infection between AcSBV and AmSBV, based on the pathogenicity in larvae during the course of the experiments. However, an A. mellifera isolate, AmSBV-Kor19 (GenBank accession number JQ390592), was found to be distinct from other A. mellifera isolates in the genome (26) and clustered together with A. cerana isolates in the phylogenetic tree (29). One explanation is that this special A. mellifera isolate was the SBV that typically infects A. cerana or the SBV was recently transferred from A. cerana.
In the present study, most A. mellifera SBV isolates were clustered with the two previously reported SBV strains isolated from A. mellifera in China (49), forming a separate clade. This suggests a consequence of the climatic conditions of China and the resulting changes in host biology. However, two special AmSBV isolates were found among 38 A. mellifera isolates, which were recovered from 318 A. mellifera colonies. Similar to AmSBV-Kor19, these two AmSBV isolates were grouped together with A. cerana isolates in the phylogenetic tree. These results supported the hypothesis that cross-species infection of AcSBV may occur, but with low occurrence rates.
Our inoculation experiments with AcSBV under laboratory and field conditions confirmed the hypothesis. Unlike previous studies in which only death and pupation of larvae were observed (31–33), we employed another diagnostic method by examining the presence of AcSBV negative-strand RNA. This method allowed us to detect the replication of virus even when the pathogenicity was very low.
Although the A. mellifera larvae infected with AcSBV under either laboratory or field conditions had no obvious signs of the disease, as demonstrated in previously reported experiments (31–33), AcSBV negative-strand RNA could be detected in A. mellifera larvae (Fig. 4B and D), which suggests that the genome of AcSBV is able to replicate in A. mellifera larvae. However, additional studies must be performed to determine whether a large dose induces clinical disease.
In agreement with previous results (8–10), SBV affects adult bees without obvious signs of the disease, although the virus titers increased significantly. The titers of AcSBV in A. mellifera and A. cerana adult workers inoculated under laboratory conditions had similar growth trends, increasing significantly in the first 6 days after inoculation and reaching a plateau (Fig. 3A and B). Although the increases in AcSBV titers in A. mellifera were less than those in A. cerana during the first 6 days after inoculation, the increases in AcSBV titers in A. mellifera within 10 days were more substantial. We suspect that, when infecting A. mellifera, AcSBV may take time to adjust to the new host during the initial infection phase and then replicates efficiently by an unknown mechanism. This result indicates that AcSBV successfully replicated in A. mellifera adult workers. In addition, the AcSBV negative-strand RNA could be detected in A. mellifera adult workers inoculated in the laboratory (Fig. 4A) and under field conditions (Fig. 4C). As with the larvae, compared with the samples prepared under laboratory conditions, the nucleic acid bands (Fig. 4C and D) of samples inoculated under field conditions were much weaker. This may be due to the different amounts of virus inoculated into the bees and/or different levels of resistance of the bees to the virus under different conditions. Nevertheless, these results suggest that AcSBV is able to replicate in A. mellifera adult worker bees.
Therefore, by quantifying the titers of AcSBV in A. mellifera adult worker bees and detecting AcSBV negative-strand RNA in adult worker bees and larvae in artificial inoculation experiments, we concluded that AcSBV is able to infect A. mellifera, which is different from previous reports. This is of significance not only for the study of the pathogenicity of SBV but also for the protection of A. mellifera and A. cerana. In agreement with a previous investigation performed in China by Ai et al. (35), the infection rate of A. cerana colonies was much higher than that of A. mellifera colonies. With the finding that AcSBV can infect A. mellifera colonies, as shown in this study, A. cerana colonies serve as a large reservoir of SBV infection for A. mellifera colonies. Although the AcSBV infection rate in A. mellifera colonies was very low and symptoms were not seen, this effect should not be neglected and efforts should still be made to monitor the dynamic changes of AcSBV in A. mellifera colonies. Due to beekeeping practices, the numbers of A. mellifera colonies are much larger than those of A. cerana colonies in most beekeeping areas in Asia. Therefore, although the AcSBV infection rate in A. mellifera colonies is very low (0.63% in this study), the fact that A. mellifera colonies serve as a reservoir for SBV infection for A. cerana colonies should not be neglected, especially when the two honeybee species are located in close proximity.
Although quantification at the nucleic acid level with the qRT-PCR approach is one of the most common methods used for the quantification of virus, it should be noted that the stringency of this technique may be doubted, since nucleic acids are not necessarily part of an infectious particle (50). Our data on SBV detection in field colonies, virus RNA genome quantification, and negative-strand RNA detection have strongly supported our conclusion, while it remains unclear whether new proteins were produced and new virus particles were assembled. It would be worthwhile to quantify virus proteins or particles to determine whether new proteins are produced or new particles are assembled. In addition, because A. mellifera colonies very commonly are infected by a variety of bee viruses and rarely are infected by only SBV, purified AmSBV was not obtained for cross-species inoculation in our study. The ability of AmSBV to infect A. cerana should be studied in the future whenever possible.
ACKNOWLEDGMENTS
We thank the three anonymous reviewers for their constructive comments in improving the manuscript.
Funding Statement
This work was supported by earmarked funds from the Modern Agro-industry Technology Research System (grant CARS-45) and the Science and Technology Department of Zhejiang Province, China (grant 2012C12906-19).
REFERENCES
- 1.Pearce F. 2014. Honeybee trade is hotbed for carrying disease into wild. New Sci 221:16. doi: 10.1016/S0262-4079(14)60357-2. [DOI] [Google Scholar]
- 2.Gallai N, Salles JM, Settele J, Vaissière BE. 2009. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol Econ 68:810–821. doi: 10.1016/j.ecolecon.2008.06.014. [DOI] [Google Scholar]
- 3.Garibaldi LA, Steffan-Dewenter I, Kremen C, Morales JM, Bommarco R, Cunningham SA, Carvalheiro LG, Chacoff NP, Dudenhöffer JH, Greenleaf SS, Holzschuh A, Isaacs R, Krewenka K, Mandelik Y, Mayfield MM, Morandin LA, Potts SG, Ricketts TH, Szentgyörgyi H, Viana BF, Westphal C, Winfree R, Klein AM. 2011. Stability of pollination services decreases with isolation from natural areas despite honey bee visits. Ecol Lett 14:1062–1072. doi: 10.1111/j.1461-0248.2011.01669.x. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Zhao Y, Hammond J, Hsu HT, Evans J, Feldlaufer M. 2004. Multiple virus infections in the honey bee and genome divergence of honey bee viruses. J Invertebr Pathol 87:84–93. doi: 10.1016/j.jip.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 5.White GF. 1917. Sacbrood. US Dept Agric Bull 431:1–55. [Google Scholar]
- 6.King AMQ, Adams MJ, Lefkowitz EJ, Carstens EB (ed). 2012. Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, Philadelphia, PA. [Google Scholar]
- 7.Nguyen NTB, Le TH. 2013. Complete genome sequence of sacbrood virus strain SBM2, isolated from the honeybee Apis cerana in Vietnam. Genome Announc 1(1):e00076-12. doi: 10.1128/genomeA.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson DL, Gibbs AJ. 1989. Transpuparial transmission of Kashmir bee virus and sacbrood virus in the honey bee (Apis mellifera). Ann Appl Biol 114:1–7. doi: 10.1111/j.1744-7348.1989.tb06781.x. [DOI] [Google Scholar]
- 9.Bailey L. 1969. The multiplication and spread of sacbrood virus of bees. Ann Appl Biol 63:483–491. doi: 10.1111/j.1744-7348.1969.tb02844.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee PE, Furgala B. 1967. Viruslike particles in adult honey bees (Apis mellifera Linnaeus) following injection with sacbrood virus. Virology 32:11–17. doi: 10.1016/0042-6822(67)90247-4. [DOI] [PubMed] [Google Scholar]
- 11.Chen YP, Siede R. 2007. Honey bee viruses. Adv Virus Res 70:33–80. doi: 10.1016/S0065-3527(07)70002-7. [DOI] [PubMed] [Google Scholar]
- 12.Bailey L. 1967. The incidence of virus diseases in the honey bee. Ann Appl Biol 60:43–48. doi: 10.1111/j.1744-7348.1967.tb05920.x. [DOI] [PubMed] [Google Scholar]
- 13.Dall DJ. 1985. Inapparent infection of honey bee pupae by Kashmir and sacbrood bee viruses in Australia. Ann Appl Biol 106:461–468. doi: 10.1111/j.1744-7348.1985.tb03136.x. [DOI] [Google Scholar]
- 14.Grabensteiner E, Ritter W, Carter MJ, Davison S, Pechhacker H, Kolodziejek J, Boecking O, Derakhshifar I, Moosbeckhofer R, Licek E, Nowotny N. 2001. Sacbrood virus of the honeybee (Apis mellifera): rapid identification and phylogenetic analysis using reverse transcription-PCR. Clin Diagn Lab Immunol 8:93–104. doi: 10.1128/CDLI.8.1.93-104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tentcheva D, Gauthier L, Zappulla N, Dainat B, Cousserans F, Colin ME, Bergoin M. 2004. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl Environ Microbiol 70:7185–7191. doi: 10.1128/AEM.70.12.7185-7191.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antúnez K, D'Alessandro B, Corbella E, Ramallo G, Zunino P. 2006. Honeybee viruses in Uruguay. J Invertebr Pathol 93:67–70. doi: 10.1016/j.jip.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen S, Nicolaisen M, Kryger P. 2008. Incidence of acute bee paralysis virus, black queen cell virus, chronic bee paralysis virus, deformed wing virus, Kashmir bee virus and sacbrood virus in honey bees (Apis mellifera) in Denmark. Apidologie 39:310–314. doi: 10.1051/apido:2008007. [DOI] [Google Scholar]
- 18.Kukielka D, Sánchez-Vizcaíno JM. 2009. One-step real-time quantitative PCR assays for the detection and field study of sacbrood honeybee and acute bee paralysis viruses. J Virol Methods 161:240–246. doi: 10.1016/j.jviromet.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 19.Ma M, Li M, Yuan C, Li P, Zhang T, Su Y, Qu Z. 2010. Development of a RT-PCR method for determination of Chinese sacbrood virus. China J Biol 4:031. [Google Scholar]
- 20.Freiberg M, De Jong D, Message D, Cox-Foster D. 2012. First report of sacbrood virus in honey bee (Apis mellifera) colonies in Brazil. Genet Mol Res 11:3310–3314. doi: 10.4238/2012.September.12.14. [DOI] [PubMed] [Google Scholar]
- 21.Anderson DL. 1995. Viruses of Apis cerana and Apis mellifera, p 160–171. In Kevan P. (ed), The asiatic hive bee: apiculture, biology, and role in sustainable development in tropical and subtropical Asia. Enviroquest, Cambridge, Ontario, Canada. [Google Scholar]
- 22.Blanchard P, Guillot S, Antùnez K, Köglberger H, Kryger P, de Miranda JR, Franco S, Chauzat M-P, Thiéry R, Ribière M. 2014. Development and validation of a real-time two-step RT-qPCR TaqMan® assay for quantitation of sacbrood virus (SBV) and its application to a field survey of symptomatic honey bee colonies. J Virol Methods 197:7–13. doi: 10.1016/j.jviromet.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Zhang Y, Yan X, Han R. 2010. Prevention of Chinese sacbrood virus infection in Apis cerana using RNA interference. Curr Microbiol 61:422–428. doi: 10.1007/s00284-010-9633-2. [DOI] [PubMed] [Google Scholar]
- 24.Rana B, Garg I, Khurana SP, Verma L, Agrawal H. 1986. Thai sacbrood virus of honeybees (Apis cerana indica F) in north-west Himalayas. Indian J Virol 2:127–131. [Google Scholar]
- 25.Bailey L, Ball BV, Perry JN. 1983. Association of viruses with two protozoal pathogens of the honey bee. Ann Appl Biol 103:13–20. doi: 10.1111/j.1744-7348.1983.tb02735.x. [DOI] [Google Scholar]
- 26.Choe SE, Nguyen LTK, Noh JH, Kweon CH, Reddy KE, Koh HB, Chang KY, Kang SW. 2012. Analysis of the complete genome sequence of two Korean sacbrood viruses in the honey bee, Apis mellifera. Virology 432:155–161. doi: 10.1016/j.virol.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Zhang G, Han R. 2008. Advances on sacbrood of honeybees. China J Biol Control 24:130–137. [Google Scholar]
- 28.Kim HK, Choi YS, Lee ML, Lee MY, Lee KG, Ahn NH. 2008. Detection of sacbrood virus (SBV) from the honeybee in Korea. Korean J Apic 23:103–109. [Google Scholar]
- 29.Roberts JMK, Anderson DL. 2014. A novel strain of sacbrood virus of interest to world apiculture. J Invertebr Pathol 118:71–74. doi: 10.1016/j.jip.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Bailey L. 1982. Viruses of honeybees. Bee World 63:165–173. doi: 10.1080/0005772X.1982.11097891. [DOI] [Google Scholar]
- 31.Dong C, Liu J, Yuan Y, Zhang G, Guo Z. 1984. Experiments of serum and cross infection between Apis cerana sacbrood virus and Apis meliifera sacbrood virus. Apicult China 3:8–9. [Google Scholar]
- 32.Verma L, Rana B, Verma S. 1990. Observations on Apis cerana colonies surviving from Thai sacbrood virus infestation. Apidologie 21:169–174. doi: 10.1051/apido:19900301. [DOI] [Google Scholar]
- 33.Zhang Y, Zhang G, Huang X, Han R. 2014. Proteomic analysis of Apis cerana and Apis mellifera larvae fed with heterospecific royal jelly and by CSBV challenge. PLoS One 9:e102663. doi: 10.1371/journal.pone.0102663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ai H, Yan X, Han R. 2012. Occurrence and prevalence of seven bee viruses in Apis mellifera and Apis cerana apiaries in China. J Invertebr Pathol 109:160–164. doi: 10.1016/j.jip.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Chen Y, Zhang S, Chen S, Li W, Yan L, Shi L, Wu L, Sohr A, Su S. 2013. Viral infection affects sucrose responsiveness and homing ability of forager honey bees, Apis mellifera. PLoS One 8:e77354. doi: 10.1371/journal.pone.0077354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson RM, Evans JD, Robinson GE, Berenbaum MR. 2009. Changes in transcript abundance relating to colony collapse disorder in honey bees (Apis mellifera). Proc Natl Acad Sci U S A 106:14790–14795. doi: 10.1073/pnas.0906970106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen YP, Pettis JS, Collins A, Feldlaufer MF. 2006. Prevalence and transmission of honeybee viruses. Appl Environ Microbiol 72:606–611. doi: 10.1128/AEM.72.1.606-611.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey L, Gibbs A, Woods R. 1964. Sacbrood virus of the larval honey bee (Apis mellifera Linnaeus). Virology 23:425–429. doi: 10.1016/0042-6822(64)90266-1. [DOI] [PubMed] [Google Scholar]
- 40.Vandenberg JD, Shimanuki K. 1987. Technique for rearing worker honeybees in the laboratory. J Apic Res 26:90–97. [Google Scholar]
- 41.Simone M, Evans JD, Spivak M. 2009. Resin collection and social immunity in honey bees. Evolution 63:3016–3022. doi: 10.1111/j.1558-5646.2009.00772.x. [DOI] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Rueckert RR. 2001. Picornaviridae: the viruses and their replication, p 1679–1728. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), 2001. Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 44.Craggs JK, Ball JK, Thomson BJ, Irving WL, Grabowska AM. 2001. Development of a strand-specific RT-PCR based assay to detect the replicative form of hepatitis C virus RNA. J Virol Methods 94:111–120. doi: 10.1016/S0166-0934(01)00281-6. [DOI] [PubMed] [Google Scholar]
- 45.Yue C, Genersch E. 2005. RT-PCR analysis of deformed wing virus in honeybees (Apis mellifera) and mites (Varroa destructor). J Gen Virol 86:3419–3424. doi: 10.1099/vir.0.81401-0. [DOI] [PubMed] [Google Scholar]
- 46.Gisder S, Aumeier P, Genersch E. 2009. Deformed wing virus: replication and viral load in mites (Varroa destructor). J Gen Virol 90:463–467. doi: 10.1099/vir.0.005579-0. [DOI] [PubMed] [Google Scholar]
- 47.Li Z, Su S, Hamilton M, Yan L, Chen Y. 2014. The ability to cause infection in a pathogenic fungus uncovers a new biological feature of honey bee viruses. J Invertebr Pathol 120:18–22. doi: 10.1016/j.jip.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 48.Boncristiani HF, Di Prisco G, Pettis JS, Hamilton M, Chen YP. 2009. Molecular approaches to the analysis of deformed wing virus replication and pathogenesis in the honey bee, Apis mellifera. Virol J 6:221. doi: 10.1186/1743-422X-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang B, Peng G, Li T, Kadowaki T. 2013. Molecular and phylogenetic characterization of honey bee viruses, Nosema microsporidia, protozoan parasites, and parasitic mites in China. Ecol Evol 3:298–311. doi: 10.1002/ece3.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heider S, Metzner C. 2014. Quantitative real-time single particle analysis of virions. Virology 462:199–206. doi: 10.1016/j.virol.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]